Summary
The early midgut undergoes intensive elongation, but the underlying cellular and molecular mechanisms are unknown. The early midgut epithelium is pseudostratified and its nuclei travel between apical and basal surfaces in concert with cell cycle. Using 3D confocal imaging and 2D live imaging, we profiled behaviors of individual dividing cells. As nuclei migrate apically for mitosis, cells maintain a basal process (BP), which splits, but is inherited by only one daughter. After mitosis, some daughters directly use the inherited BP as a “conduit” to transport the nucleus basally, while >50% of daughters generate a new basal filopodium and use it as a path to return the nucleus. Post-mitotic filopodial “pathfinding” is guided by mesenchymal WNT5A. Without WNT5A, some cells fail to tether basally and undergo apoptosis, leading to a shortened midgut. Thus, these studies reveal previously unrecognized strategies for efficient post-mitotic nuclear trafficking, which is critical for early midgut elongation.
Keywords: Midgut elongation, small intestine, pseudostratified epithelium, cell division, interkinetic nuclear migration, basal process, filopodial extension, apical link, Wnt5a, apoptosis
eTOC Blurb
Wang et al. provide a detailed mechanism for early midgut elongation. By profiling behaviors of dividing cells in the epithelium, they identify two strategies, “conduit” and “pathfinding”, by which daughter nuclei return to the basal surface after apical mitosis. Loss of WNT5A perturbs the pathfinding strategy, leading to short midguts.
Introduction
The adult SI is a long tube, typically measuring over three times the body length of the individual. This remarkable length is critical for sufficient nutrition absorption. Surprisingly, over 40% of the adult SI length is attained in utero (Weaver et al., 1991). Insufficient SI growth in utero leads to a condition known as congenital short bowel syndrome (CSBS), which has high mortality due to malabsorption (Hasosah et al., 2008; van der Werf et al., 2015). Although proper fetal SI elongation is essential, little is known about the mechanisms that control this process.
The SI is derived from the midgut during development. In early mouse embryos, midgut elongation begins shortly after morphogenic movements have shaped the endodermal layer into a complete gut tube (Cervantes et al., 2009; Zorn and Wells, 2009). Midgut elongation is a continuous process, but can be divided into two distinct phases, based on the configuration of the core epithelium (Walton et al., 2016). During Phase I, the pre-villus stage (E10.5–14.5), the midgut epithelium (ME) consists of a tube with a flat lumenal surface (Kohlnhofer et al., 2016; Walton et al., 2016). At E14.5 (beginning of Phase II), dramatic changes in cell shape occur as finger-like villi emerge at the lumenal surface, convoluting and expanding the apical surface area (Freddo et al., 2016; Walton et al., 2016; Walton et al., 2012). Additionally, signaling programs that drive the proliferation and the pattern of proliferation are different in these two phases. Canonical Wnt signals are required for epithelial proliferation in Phase II, but not in Phase I (Chin et al., 2016; Korinek et al., 1998); proliferating cells are located throughout the Phase I epithelium, but as villi emerge, epithelial cells on top of the villi withdraw from the cell cycle and proliferation is progressively confined to intervillus regions (Noah et al., 2011; Walton et al., 2018). These differences emphasize the need to carefully define the mechanisms underlying midgut elongation in each phase. Phase I elongation may be particularly important since it provides the initial organ template for subsequent growth in Phase II. Defects in Phase I elongation would leave the embryo with a deficit in midgut length at the beginning of Phase II; it is unclear whether compensatory mechanisms exist in Phase II to recover from such a Phase I deficit.
For decades, the early mouse ME (during Phase I) was characterized as a stratified tube (Mathan et al., 1976; Matsumoto et al., 2002; Toyota et al., 1989). Convergence and extension-like movements were posited to drive gut elongation (Cervantes et al., 2009; Matsumoto et al., 2002; Reed et al., 2009). However, recently, two groups (Grosse et al., 2011; Yamada et al., 2013) established that the early ME is actually pseudostratified and exhibits signatures of interkinetic nuclear migration (IKNM). IKNM was initially observed and is best studied in neural epithelia. During IKNM, nuclei move in concert with the cell cycle: S-phase nuclei sit near the basal surface, then travel apically in G2, undergo mitosis near the apical surface, and finally return to basal in G1 (Guthrie et al., 1991; Sauer and Walker, 1959; Spear and Erickson, 2012). IKNM has also been documented in lung-, liver- and pancreatic-buds (Norden, 2017) but is much less studied in non-neural settings. Nevertheless, IKNM appears to be a broadly conserved feature of cell division in pseudostratified epithelia (Meyer et al., 2011).
Although it is clear that nuclei in the early ME perform IKNM (Grosse et al., 2011), the associated cell dynamics are unexplored. In particular, the tubular structure of the phase I ME, which has a limited apical surface area to accommodate mitotic figures, must utilize an efficient strategy for apical exit of the post-mitotic nuclei. To define the IKNM process at a cellular level and to determine how basal nuclear return is accomplished in the Phase I midgut epithelial tube, we acquired 3D morphologies of individual cells by confocal microscopy, and determined the sequential order of cell dynamics by 2D live imaging. These studies established that, as the nucleus moves apically for mitosis, the cell remains connected to the basal surface via a basal process (BP). During mitosis, the BP splits in two, but one BP is generally lost. The remaining BP is always inherited by one daughter cell; the other nascent daughter is born without a basal connection but with an apical link to its sister. Interestingly, during G1, daughter cell pairs return their nuclei to the basal surface in two distinct modes. In Mode I, one nucleus returns faster than the other; the daughter that inherits the BP directly uses it as a “conduit” for a quick nuclear return. In contrast, its sister takes a “pathfinding” strategy, actively projecting a filopodial process to establish a new basal path and then using that path to return its nucleus basally. Nuclear return by “pathfinding” takes twice as long as the “conduit” strategy. In Mode II, both daughters take the “pathfinding” strategy and return at a similar pace (despite the presence of an intact BP on one daughter). Overall, 64% of post-mitotic cells utilize the “pathfinding” strategy. Importantly, we found that efficient pathfinding depends on mesenchymal WNT5A. In the absence of this radial WNT5A cue, filopodial pathfinding is impaired; some cells fail to tether to the basal surface and their nuclei remain at the apical side for a prolonged period. These cells eventually undergo apoptosis, leading to a shortened midgut. Thus, WNT5A-guided basal tethering is an essential step that insures efficient apical exit of post-mitotic nuclei, which is critical for midgut epithelial tube elongation.
Results
During rapid Phase I midgut elongation, the ME is a pseudostratified and highly proliferative tube
To establish the elongation trajectory of the murine midgut during Phase I, we harvested murine gastrointestinal (GI) tracts daily between E10.5 and E14.5 (Figure 1A) and measured midgut length from the pylorus to the cecum. As shown in Figure 1B, the midgut is 1.5 millimeter (mm) in length at E10.5 and reaches 17.4 mm by E14.5. Thus, the midgut, with its epithelial core and surrounding mesenchymal investment, elongates >11-fold within this 4-day Phase I period (Figure 1B).
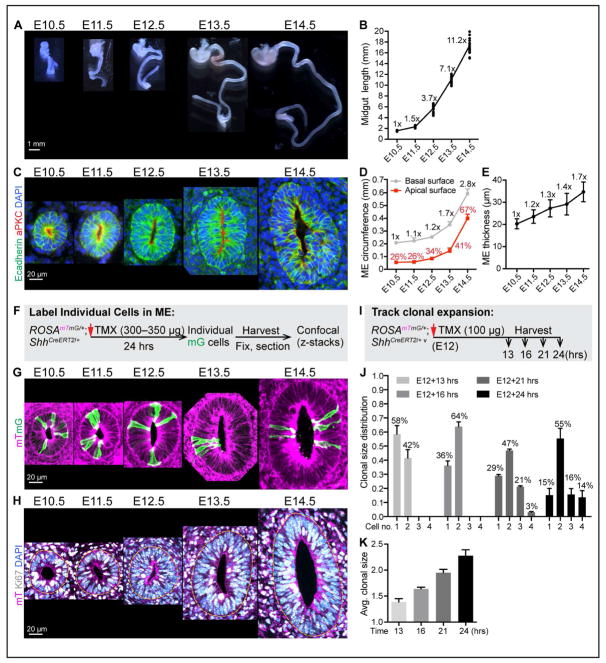
Figure 1. Midgut elongation, ME organization, cell proliferation and clonal expansion during Phase I.
(A) Isolated fetal mouse GI tracts from E10.5 to E14.5; stomach at top.
(B) Quantitation of Phase I midgut length. Length fold change over time, compared to E10.5 length, is indicated at each time point (e.g., 11.2x overall at E14.5).
(C) Structure of Phase I ME tubes. Paraffin sections of the mid-region of midguts, stained with E-cadherin (green), aPKC (red) and DAPI (nucleus, blue).
(D) Quantitation of the apical and basal circumferences of mid-region of ME tubes. Numbers on grey line indicate fold change in basal circumference over time, compared to E10.5. Numbers (%) on red line indicate the ratio of apical circumference/basal circumference at each time point. Data are represented as mean and standard deviation (SD).
(E) Epithelial thickness at the mid-region of ME tubes. Numbers indicate thickness fold change overtime, compared to E10.5 value (e.g., 1.7x at E14.5). Data are represented as mean and SD.
(F) Strategy for labeling individual cells in ROSAmTmG/+;ShhCreERT2/+ ME.
(G) 3D reconstructions from vibratome-sectioned, TMX treated ROSAmTmG/+;ShhCreERT2/+ midguts.
(H) Confocal slice of vibratome-sectioned ROSAmTmG/+ midguts labeled with mT (magenta), Ki67 (white) and DAPI (nuclei, blue). Dotted yellow line marks the basal surface of ME.
(I) Strategy for tracking clonal expansion in ROSAmTmG/+;ShhCreERT2/+ ME.
(J–K) Distribution of clonal size (number of cells within a clone) at 13, 16, 21, and 24 hrs after TMX treatment (J). Average clonal size over time (K). Data are represented as mean and SD.
To understand Phase I elongation at the cellular level, we next examined the cellular architecture of the ME during this entire period. Cross sections reveal that the Phase I ME is a flat tube (lacking villi) with a very limited apical surface area, reflected by the small ratio of apical surface vs. basal surface (Figures 1C and 1D). Over time, the wall of the ME tube gains thickness (Figure 1E) and the tube gains girth, indicated by its basal circumference (Figure 1D). Nuclear staining reveals staggered nuclei, with an apparent increase in number of nuclei staggered along the apical-basal axis, from two at E10.5–E11.5 to 3–6 at E14.5 (Figure 1C), suggesting either a stratified or pseudostratified configuration. Previous studies documented pseudostratification at some time points, but did not account for the entire 4-day window (Grosse et al., 2011; Yamada et al., 2013). To further examine this, individual cells were labeled in ROSAmTmG/+; ShhCreERT2/+ midguts. Timed pregnant dams were given a low dose of tamoxifen (TMX) 24 hours prior to harvest, to randomly induce Cre recombination and resulting membrane-targeted EGFP (mG) fluorescence in a sub-population of ME cells (Figure 1F). mG-labeled cells are easily detectable in the background of unrecombined membrane tdTomato (mT) cells (Muzumdar et al., 2007). Confocal z-stacks showed that individual mG cells touch both apical and basal surfaces at all time points (Figure 1G). Thus, throughout Phase I, the ME is pseudostratified (one cell layer thick) and the degree of pseudostratification increases over time.
Pseudostratified epithelia are generally highly proliferative (Strzyz et al., 2016). The proliferation state of the Phase I ME was examined with the proliferation marker Ki67, which labels actively cycling cells, but not G0 cells. At all time points in Phase I, the vast majority (>95%) of epithelial nuclei are positive for Ki67 (Figure 1H). Since the midgut epithelial tube gains both girth and thickness during elongation and is never stratified, it is clear that gross cell movements that involve sacrificing girth or cell layers to gain tissue length, such as convergence extension or radial intercalation, do not occur in this context. We conclude that active cell proliferation is the predominant driver for the rapid Phase I growth.
To investigate the dynamics of cell division quantitatively, we examined clonal expansion within the ME. Using the ROSAmTmG/+; ShhCreERT2/+ model, dams were given a very low dose of TMX (100 μg) at E12 and midguts were harvested 13 hrs, 16 hrs, 21 hrs and 24 hrs later (Figure 1I). This dosage of TMX activates only 10 to 30 cells per gut; mG-labeled cells sit far away from each other and are unlikely to overlap as clones expand. At 13 hrs after TMX treatment, 1-cell (58%) and 2-cell clones (42%) are observed. By 16 hrs, >60% of clones contain 2 cells. At 21 hrs, 3-cell clones are observed (21%). By 24 hrs, 14% of clones contain four cells, signifying two complete rounds of mitosis (Figure 1J). The average cell number per clone of approximately two after 21 hrs (Figure 1K) suggests that one full cell cycle (plus the effective TMX delivery time) is equal to or less than 21 hrs. The clonal expansion pattern also suggests that labeled cells do not divide synchronously.
As the nucleus migrates apically for mitosis, a thin BP maintains a basal connection; this BP splits during mitosis
The general dynamics of nuclear movement during IKNM have been well established, but the associated cellular dynamics remain obscure, especially in non-neural epithelia. Indeed, pseudostratified cells are tall, thin and densely packed, which makes it difficult to observe an entire cell in 2D images. Thus, we combined an individual cell labeling strategy with 3D reconstruction, to observe how IKNM plays out at the level of individual cells in the Phase I ME.
First, based on the clonal expansion pattern (Figure 1J), we generated 1- or 2-cell clones in ME 16 hrs after a low dose of TMX administration and acquired high-resolution 3D morphologies of individual cells in confocal z-stacks (Figure 2A). Next, to arrange the different 3D cell shapes in sequential order, their corresponding cell cycle phases were inferred. Because the nucleus occupies the majority of the cell volume, its center always sits at the widest point of the otherwise tall, thin cell. Therefore, the cell shape reflects the nuclear position and nuclear position in turn provides cell cycle phase information (S phase nuclei sit basally; M phase nuclei are apical; G1/G2 phase nuclei shuttle in between). Additionally, the mitosis marker pHH3 can refine this assignment because its nuclear staining pattern changes in different cell cycle phases. pHH3 is absent in G1 and S; it appears as a dot-like pattern in pericentromeric heterochromatin in late G2. Histone H3 phosphorylation increases during prophase and reaches maximal in metaphase; then dephosphorylation begins and pHH3 disappears by the end of M phase (Hendzel et al., 1997). Thus, with cell cycle phase information provided by both cell shape and pHH3 staining, acquired 3D cell shapes were ordered sequentially according to cell cycle progression, revealing detailed stereotypic cell behaviors in the Phase I ME.
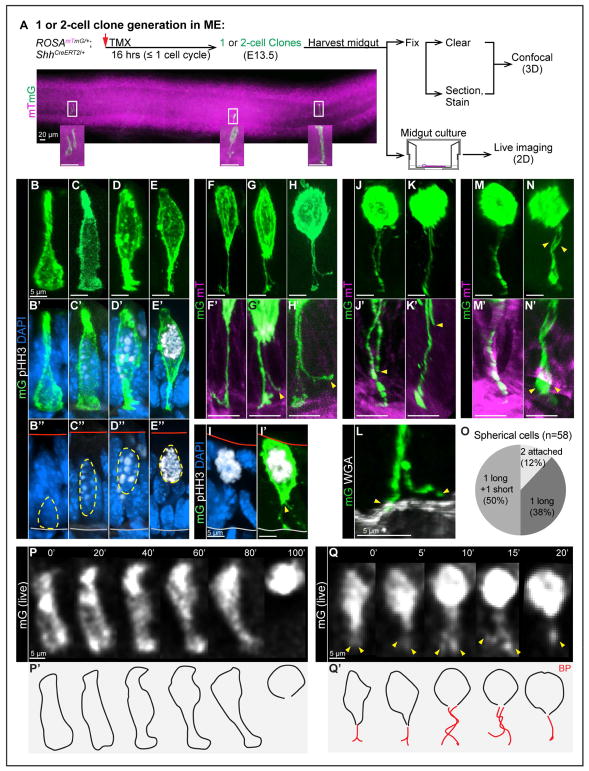
Figure 2. Individual cell behavior before and during mitosis: nuclear apical migration, BP retention, splitting and retraction.
(A) Strategy for generating one or two-cell clones in ME for confocal (3D) and live (2D) imaging. A stitched image shows a full view of one segment of midgut.
(B–E”, I–I’) 3D reconstructions of individual mG cells from vibratome-sectioned ROSAmTmG/+;ShhCreERT2/+ midguts, stained with pHH3 (white) and DAPI (nuclei, blue). Red and grey lines in B”–E” mark apical and basal surfaces, respectively.
(F–H, F’-H’, J, K, J’, K’, M, N, M’, N’) 3D reconstructions of individual mitotic mG cells from fixed, whole-mount cleared ROSAmTmG/+;ShhCreERT2/+ midguts. Images reveal BP retention (F), BP splitting (G, H), BP retraction (J, K), complete BP retraction (M), two attached BPs (N). F’–H’, J’, K’, M’, N’ are high-magnification of BP(s) in F–H, J, K, M, N, respectively.
(L) 3D reconstructions of a split BP from vibratome-sectioned ROSAmTmG/+;ShhCreERT2/+ midguts, stained with WGA (white). Only one of two end-feet (arrowheads) is attached to the basal lamina.
(O) Distribution of spherical cells with 1 long and 1 short BP, 2 attached BPs, and 1 BP.
(P, P’, Q, Q’) Successive 2D live images captured from cultured midguts. Numbers on top indicate minutes. Nuclear apical migration through the complete cell rounding; the BP becomes undetectable (P). BP splitting (arrowheads) and potential retraction at 15’. The final frame (at 20’) has one detectable BP (Q). P’–Q’ are outlines of cells in P and Q, red lines are BP(s).
In S phase, the cell exhibits a large basal surface, occupied by a pHH3 negative nucleus, and a small apical surface (Figures 2B–2B”). As the nucleus rises in late G2, indicated by a few weak pHH3 puncta, the center of the cell widens (Figures 2C–2C”). By the end of G2, when more and larger pHH3 puncta are seen, the nucleus is more apically positioned and the basal connection attenuates (Figures 2D–2D”). Before the nucleus reaches the apical surface, the cell enters prophase, indicated by highly condensed pHH3+ chromosomes (Figures 2E’ and 2E”); its basal portion shrinks into an end-foot (Figure 2E). As the nucleus continues to move apically, the cell adopts an oval shape, but remains basally connected by a thin process (100%, n=13) (Figures 2F and 2F’). This delicate process has been documented in neuronal cells and is termed the basal process (BP), while the bulbous portion of the cell that contains the nucleus is called the soma (Kosodo and Huttner, 2009).
Interestingly, before the soma adopts a rounded shape, the BP begins to split from the basal end (seen in 3 out of 6 of such cells, Figures 2G and 2G’). BP splitting continues in a basal to apical direction (10 out of 10) (Figures 2H, 2H’, and 2I’), ending before metaphase (Figures 2I and 2I’). The soma becomes spherical as mitosis proceeds. Of 58 spherical cells, 50% possess one long and one short BP (Figures 2J, 2J’, 2K, 2K’, and 2O). Staining of the basal lamina reveals that the longer BP is anchored while the other is free (Figure 2L). For 38% of spherical cells, only one BP is visible (Figures 2M, 2M’, and 2O), while in 12%, both BPs remain attached to the basal surface (Figures 2N, 2N’, and 2O). These data together suggest that 100% of cells maintain a basal connection via the BP; after BP splitting, one BP remains tethered to the basal lamina while the other one retracts or otherwise becomes severed 88% of the time.
To confirm these cell behaviors and the order of events, we turned to 2D live imaging of cultured midguts using an inverted epifluorescence microscope. ME cells were individually labeled with the strategy shown in Figure 2A. Of note, gut peristaltic movements and the transwell culture method impose a resolution limit on live imaging. Nevertheless, the resulting movies verify the temporal sequence of cell behaviors deduced from fixed midguts. That is, as nuclei move apically for mitosis (Figures 2P and 2P’), cell morphologies that were captured by confocal microscopy (Figures 2C–2E) can be observed in a step-by-step sequence. Temporally, basal-to-apical nuclear migration takes less than 100 min (Figure 2P). Frequently, the BPs are too thin to be captured by the rather low resolution afforded by live imaging. However, in some cases, BP splitting can be confirmed (Figures 2Q and 2Q’). As in 3D reconstructions (Figures 2G–2J, and 2M), the BP divides, the two processes tangle around each other, and subsequently, only one remains. The entire process of BP splitting and release takes less than 20 min (4 out of 4) (Figure 2Q).
The BP is inherited asymmetrically during cytokinesis and can serve as a conduit for quick nuclear basal return
At the end of mitosis, cytokinesis divides the cytoplasm of the parental cell into two nascent daughters, which initially remain tightly associated with one another along the cytokinetic plane (Figures 3A and 3A’). As G1 begins, the basal portions of the two daughter cells separate from each other (Figure 3B), revealing a striking phenomenon—only one of the two daughter cells inherits the BP(s) (100%, n=94) (Figure. 3B). Even when two BPs are kept after splitting (10.6%, n=94), both BPs belong to one daughter cell (Figures S1A and S1B). Live imaging confirms this asymmetric BP inheritance (Figures 3C and 3C’). Although the other daughter cell does not inherit a basal connection, it is connected to its sister via an apical link (white arrow, Figure 3B).
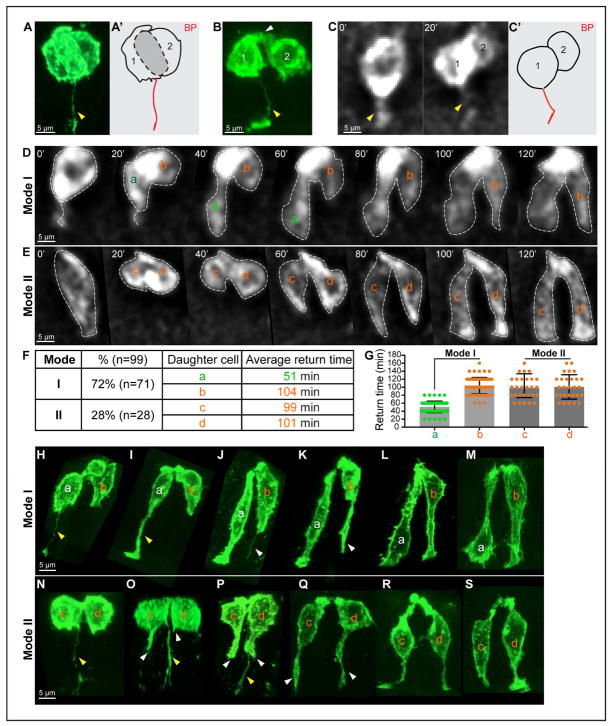
Figure 3. Daughter cell behavior during cytokinesis and G1 phase: asymmetric BP inheritance, two Modes of basal return.
(A, A’, B) 3D reconstructions of mG cells during (A) and after (B) cytokinesis from fixed, whole-mount cleared midguts. Note the asymmetric inheritance of the BP (yellow arrowhead) and an apical link (white arrowhead). A’ is a tracing of cell shapes, emphasizing the flat cytokinetic plane (grey) between the recently divided cells (1, 2) in A.
(C, C’) Successive 2D live images (20 min interval) of cytokinesis and asymmetric BP inheritance. C’ is a tracing of the cell after cytokinesis in C.
(D, E) Successive 2D live images (20 min intervals) of Mode I (D) and II (E) basal return. Daughter cells are labeled “a”, “b” (Mode I; daughter “a” has the BP) and “c”, “d” (Mode II).
(F, G) Distribution of Mode I and II, and average time for basal return of each daughter. Data are represented as mean and SD.
(H–S) 3D reconstructions of daughter pairs during G1. Mode I (H–M); Mode II (N–S). Inherited BP(s) are indicated by yellow arrowheads, de novo filopodial protrusions are indicated by white arrowheads.
Using live imaging, we were surprised to observe that basal nuclear return of the daughter cell pairs takes place in two distinct modes (Figures 3D and 3E). In Mode I, the nucleus of daughter cell (a), which maintains the BP, quickly returns to the basal surface (average 51 min, n=71), while the other nucleus (b) returns slowly (average 104 min, n=71) (Figures 3D, 3F, and 3G). In Mode II, both daughter nuclei (c) and (d) move basally simultaneously, at a pace (99–101 min, n=28) that is similar to the speed of Mode I nucleus (b) (Figures 3E–3G). Of 99 analyzed pairs of G1 daughters, Mode I is more prevalent (72%) than Mode II (28%) (Figure 3F). Of note, live imaging establishes that, in both Modes, basal return does not involve noticeable oscillation, but a directed movement to the basal side. This differs from newborn neurons in the cortex that undergo a phase of retrograde movement toward the ventricle (the apical side) before migrating to the cortical plate (the basal side) (Noctor et al., 2004).
To further investigate how daughter cell (a) in Mode I is able to return faster, we returned to the high-resolution 3D cell shapes captured by confocal microscopy. For 2-cell clones with an apical link, two morphological categories were observed. In the 1st category (Figures 3H–3M), the positions of the two sisters were staggered, with one nucleus (a) obviously occupying a more basal position than its sister (b); this corresponds to Mode I seen in live imaging (Figure 3D). In the 2nd category (Figure 3N–3S), two daughter nuclei are always located at a similar level along the apical-basal axis, corresponding to Mode II (Figure 3E). Since based on live images, daughter cells approach the basal side without oscillation in G1, 3D shapes of 2-cell clones were ordered with decreasing distances between the nucleus and the basal surface to build the return process. The results are consistent with live images and add more interesting details. In Mode I, daughter cell (a) with the BP (Figure 3H, yellow arrowhead) appears to directly use this inherited basal connection as a “conduit” to transport its nucleus basally (Figures 3I–3M). However, its sister (b) takes two steps for nuclear return. First, it grows a filopodial projection (Figures 3J and 3K, white arrowhead), potentially to explore a basal path (Figure 3L). Next, it transports its nucleus basally via this newly established bridge (Figures 3L and 3M). Of note, nucleus (b) begins to move away from the apical surface when its sister nucleus (a) is already close to the basal surface (Figure 3L). Since daughter (a)’s basal return is accomplished in ~50 min, this is consistent with the speculation that its sister (b) builds a de novo basal connection in ~50 min and then transports its nucleus to the basal side in the next 50 min. In Mode II, daughter (d), which has no BP, also generates a basal connection first (Figures 3O–3Q), similar to Mode I daughter (b). Daughter (c) inherits a BP, but does not utilize it as the returning path. Instead, it extends a filopodial protrusion from a different basal spot and finds a new path to the basal surface (Figures 3O–3Q, white arrowhead), similar to sister (d). Once filopodial protrusions from both daughters touch the basal surface, both nuclei begin to return through these newly established connections (Figures 3Q–3S).
Overall, G1-return involves only one step for Mode I daughter (a): nuclear transportation. However, Mode I daughter (b) and Mode II daughters (c) and (d) need two steps: build a new basal connection and transport the nucleus basally. These findings explain the fast and slow return in live imaging. We conclude that, during cytokinesis, only one of the two daughters (50%) inherits the BP. During G1, 36% of daughter cells directly use an inherited BP as a “conduit” to return the nucleus (“conduit” strategy); the remaining 64% of daughters build a new filopodial basal connection and use that path to deliver the nucleus to the basal surface (“pathfinding” strategy).
An apical link connects the two daughter cells as they return in G1
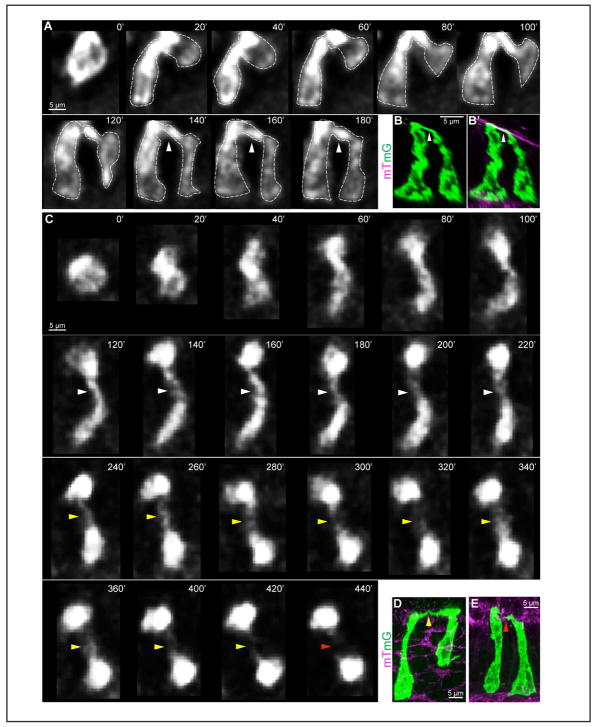
It is clear that the two sisters remain apically attached to each other during the entire time of nuclear basal return (Figures 3D, 3E, and 3H–3S). Even after both nuclei are basal, this apical membrane link persists, indicated by white arrows in live images (Figure 4A) and 3D reconstructions (Figures 4B and 4B’). As two sisters gradually separate from each other, this link is attenuated to a thin filament (yellow arrowhead, Figures 4C and 4D). Tracking this link in live imaging reveals that it can persist for 2–7 hrs after cytokinesis. When this link breaks (red arrows, Figure 4C), the two sisters are separated from each other by 1–4 cells in the ME (Figure 4E). By longer live recordings, we found that one full cell cycle length is about 16 hrs on average (14.5–17.5 hrs).
Figure 4. Retention of an apical membrane link during G1 phase.
(A) Successive 2D live images (20 min intervals) of a 2-cell clone from a lateral view with an apical link (white arrowhead) present after both nuclei are basal.
(B, B’) 3D reconstructions of a daughter pair with an apical link from whole-mount cleared midguts.
(C) Successive 2D live images (20 min intervals) of the formation (white arrowhead), attenuation (yellow arrowhead) and abscission (red arrowhead) of an apical link, as viewed from above the apical surface.
(D, E) 3D reconstructions of a daughter pair with a thinned apical link (yellow arrowhead) (D) and two separate daughter cells from whole-mount cleared midgut (E).
Mesenchymal WNT5A facilitates basal tethering of cells in the ME
During rapid Phase I elongation, nearly all cells are continuously cycling (Figure 1H) and 64% of the post-mitotic cells do not inherit or use the BP to return nuclei directly, but actively grow a filopodium towards the basal surface to build a new basal connection. This finding predicts the presence of a directional homing cue that can guide basal filopodial projection. A promising candidate is WNT5A. First, WNT5A is known to guide axon outgrowth in neuronal cells (Blakely et al., 2011; Horigane et al., 2016; Hutchins et al., 2011; Li et al., 2009). Second, Cervantes et al. found that in the Wnt5a−/− ME, some post-mitotic nuclei remain at the apical side (Cervantes et al., 2009), suggesting a problem with efficient basal tethering.
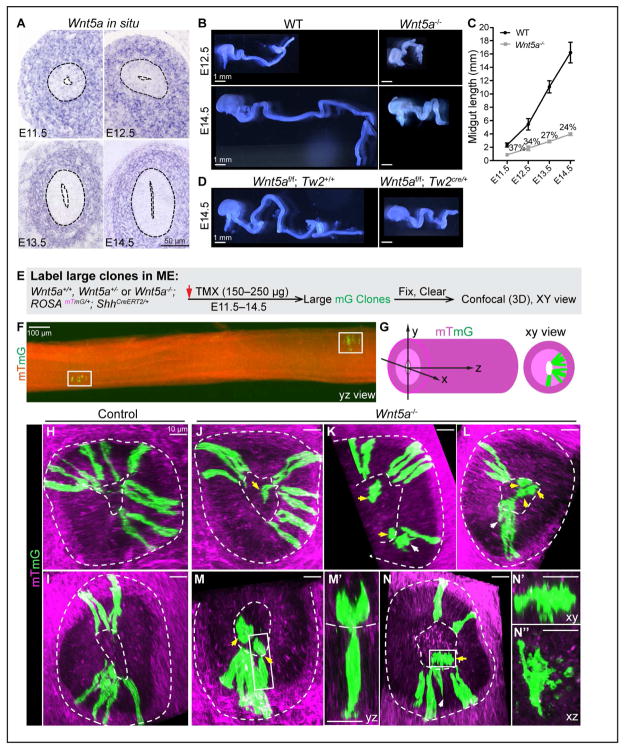
If WNT5A provides a cue for basal return, it should be expressed at the basal side of the ME or in the underlying tissue throughout the pseudostratified phase. Though previous whole-mount Wnt5a in situ hybridization hinted that this maybe the case (Cervantes et al., 2009; Lickert et al., 2001), we directly examined Wnt5a expression pattern in sectioned midguts between E11.5 and E14.5. Indeed, Wnt5a mRNA is robustly expressed within the sub-epithelial mesenchyme at all time points (Figure 5A).
Figure 5. Mesenchymal WNT5A is required for basal connection in ME.
(A) In situ hybridization of Wnt5a on midgut sections at E11.5–14.5.
(B) E12.5 and E14.5 WT and Wnt5a−/− midguts.
(C) Plot of WT and Wnt5a−/− midgut length from E11.5 to E14.5. Numbers (%) indicates the ratio of Wnt5a−/−/ WT midgut length at each time point. Data are represented as mean and SD.
(D) Loss of Wnt5a in the mesodermal compartment (Wnt5af/f;Tw2Cre/+) mirrors Wnt5a−/−.
(E) Strategy for generating large mG clones in Wnt5a+/+, Wnt5a+/− and Wnt5a−/−; ROSAmTmG/+;ShhCreERT2/+ ME.
(F) 2D lateral (yz) view of a segment of midgut containing two separate, large clones (white boxes) from a stitched image.
(G) Definition of xyz dimensions of the midgut.
(H, I) 3D confocal z-stacks (xy view) of large clones in control midguts. Apical and basal surfaces are indicated by dotted lines.
(J–M, M’, N–N”) 3D confocal z-stacks (xy view) of Wnt5a−/− large clones. White arrows mark cells in the epithelial layer without obvious basal connections. Yellow arrows indicate lumenal cells. M’, N’ and N” are higher magnification of cells indicated by the boxes in M and N, with viewing axes indicated.
It has been reported that the Wnt5a−/− midgut is short at E14.5 (Cervantes et al., 2009), but the length in early Phase I was not carefully examined. We found that the Wnt5a−/− midgut measures 37% of wild-type (WT) length at E11.5, and this deficit increases over time, progressively widening the length deficit in the mutant to 24% by E14.5 (Figures 5B and 5C). In addition, deletion of Wnt5a from mesoderm-derived tissue, using a Wnt5af/f;Twist2Cre/+ conditional model, also generates a severely shortened midgut very similar to the Wnt5a null (Figure 5D), confirming that mesenchymally secreted WNT5A ligands are required for rapid midgut elongation.
To directly test the effect of loss of WNT5A on basal tethering, we compared basal connections of individual epithelial cells in large clones from WT and Wnt5a−/− midguts. Single mG cells were labeled at E11.5 by 150–250 μg TMX gavage and midguts were harvested at E14.5 to generate large clones (Figures 5E and 5F). In sibling control (Wnt5a+/+ or Wnt5a+/−) ME, all green cells connect to the apical surface and nearly all connect to the basal surface, except for a few nascent daughters (1.3%, 4 out of 304 cells in 24 clones) (Figures 5H and 5I). In Wnt5a−/− ME, while most cells have both apical and basal contacts, some are basally unconnected (13%, 24 out of 184 cells in 19 clones) (Figures 5J–5M, 5M’, and 5N–5N”). These basally untethered cells are either within the epithelial layer (white arrows, Figures 5K, 5L, and 5N) or lumenally located (yellow arrows, Figures 5J–5N). Often, they cling atop another cell in the clone (Figure 5M’), perhaps a consequence of a maintained apical link. Some cells are surrounded by vesicles, similar to apoptotic bodies (Figures 5N––5N”). These data suggest that WNT5A may be required for proper basal tethering.
WNT5A guides “pathfinding” daughter cells to extend filopodia for basal return
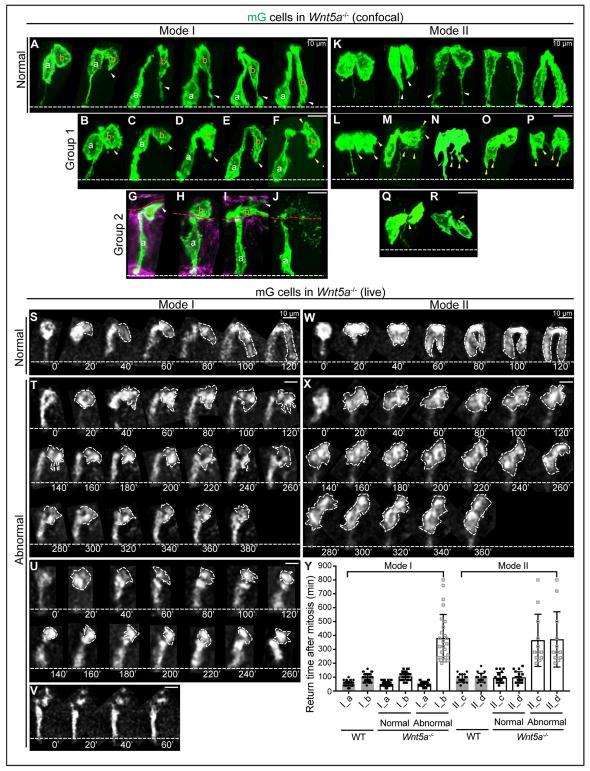
Next, we examined filopodial projections during G1 in 3D reconstructed 2-cell clones with an apical link in the Wnt5a−/− ME. Clones typical of Mode I (staggered daughter nuclei) and Mode II (two daughter nuclei at a similar level) were observed. In Mode I, some pairs appeared to be returning normally (Figure 6A). However, abnormal Mode I pairs were also seen. In some cases, daughter (a) successfully moves its nucleus basally, but daughter (b) exhibits no dominant basal filopodial protrusion (Figures 6B–6F). Another group has an even more striking phenotype, with daughter (b) entering the lumen. Some daughters (b) sit close to (Figure 6G) or atop the apical surface (Figure 6H) and extend a protrusion horizontally or randomly. Some show membrane blebbing and are surrounded by apoptotic body-like vesicles (Figures 6I and 6J).
Figure 6. WNT5A guides “pathfinding” daughter cells to extend filopodia for basal return.
(A) 3D reconstructions of normal Mode I 2-cell clones in Wnt5a−/− ME.
(B–J) Abnormal Mode I 2-cell clones in Wnt5a−/− ME. Group 1, dominant filopodia are absent (yellow arrowheads) (B–F); group 2, one daughter cell is lumenally positioned (G–J).
(K) Normal Mode II 2-cell clones in Wnt5a−/− ME.
(L–R) Abnormal Mode II 2-cell clones in Wnt5a−/− ME. Abnormal appearing filopodia are marked with yellow arrowheads. Some pairs have no basal connection at all (O, P, R).
(S) Successive 2D live tracking (20 min intervals) of cells undergoing normal-appearing Mode I basal return in Wnt5a−/− ME.
(T–V) Abnormal Mode I daughter pairs in Wnt5a−/− ME. One daughter fails to return and remains apical (T), or becomes lumenal (U), or becomes fragmented (V).
(W) Normal Mode II return in Wnt5a−/− ME.
(X) Abnormal Mode II return in Wnt5a−/− ME. Both daughters remain apical.
(Y) Quantitation of basal return time for Mode I (a,b) and Mode II (c,d) daughters in WT and Wnt5a−/− ME. Solid circles and squares reflect the return time after mitosis. Hollow squares reflect the recording time of cells that stay at the apical side. Data are represented as mean and SD.
For Mode II pairs, some appear normal (Figure 6K), while some adopt patterns that are never observed in WT. For example, daughter cells exhibit multiple radial protrusions (Figures 6L and 6M) or trifurcated protrusions toward the basal surface (Figure 6N). Some project their basal tip towards the apical surface, forming a “J” shape (Figures 6Q and 6R), while other pairs adopt odd shapes with multiple protrusions (Figures 6O and 6P). In summary, the basally-oriented filopodial outgrowth seen with high fidelity in WT “pathfinding” daughter cells in both Modes is compromised in the absence of WNT5A.
These findings were confirmed by live imaging of cultured Wnt5a−/− midguts. Most daughter pairs (70 of 108) return normally in either Mode I (n=50) (Figure 6S) or Mode II (n=20) (Figure 6W). However, in abnormal Mode I pairs (26 of 108), only “conduit” daughter (a) cells return on time (20–80 min; average 50 min), while “pathfinding” daughter (b) cells consistently send short protrusions in different directions but never make a continuous basal return during the recording (Figures 6T and 6U). Some cells sitting atop a tethered sister eventually fragment, a sign of apoptosis (Figure 6V). In abnormal Mode II pairs (12 of 108), both daughters send multiple protrusions randomly but do not return for the length of the recording. Interestingly, for the pair in Figure 6X, though the original BP is available during the entire time, it is never used for nuclear return. Combined with quantitative analysis of return times (Figure 6Y), we conclude that WNT5A depletion perturbs “pathfinding” returns, but not “conduit” returns.
Of note, loss of WNT5A does not affect apical nuclear migration (Figures S2A–S2D and S2I), BP retention (Figure S2E), BP splitting (Figures S2F, S2G, and S2J), BP retraction (Figure S2H), or asymmetric BP inheritance (Figures 6A, 6K, 6L, 6M, and 6Q). Rather, defects are specific to filopodial behavior.
Augmented apoptosis in Wnt5a−/− ME contributes to the length deficit
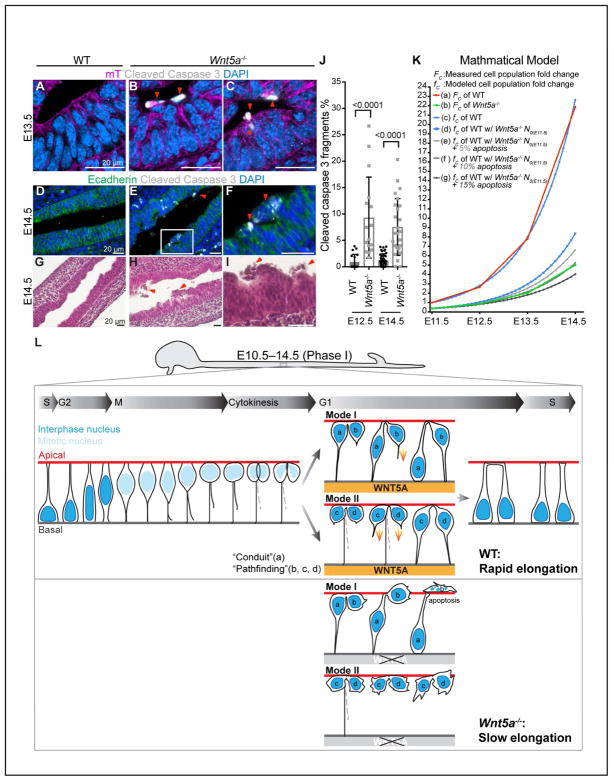
The data above indicate that impaired basal tethering can result in apoptosis in the Wnt5a−/− ME. To further confirm this, we examined cleaved-caspase 3 staining and documented a significant increase in apoptotic fragments at the ME apical side or in the lumen of Wnt5a−/− midguts (Figures 7A–7F and 7J). H&E staining confirms the apical and lumenal location of dead cells in Wnt5a−/− midguts (Figures 7H and 7I), in contrast to the clear lumen seen in WT (Figure 7G).
Figure 7. Apoptosis is increased in Wnt5a−/− midguts.
(A–C) Vibratome, cross-sectioned E13.5 Wnt5a+/+; ROSAmTmG/+ and Wnt5a−/−; ROSAmTmG/+ midguts, stained for cleaved caspase 3 (white) and DAPI (nuclei, blue). Apoptotic cell (red arrowheads) are very rare in WT ME (A) but frequently seen at the apical side of Wnt5a−/− ME (B, C).
(D–F) Paraffin, longitudinal-sectioned E14.5 WT and Wnt5a−/− midguts, stained for cleaved-caspase 3 (white), E-cadherin (green) and DAPI (nuclei, blue). F is a high-magnification of the box in E.
(G–I) H&E stained longitudinal sections of E14.5 WT (G) and Wnt5a−/− midguts (H, I). Numerous dead cells (red arrowheads) are seen in Wnt5a−/− gut lumen (H) and at the lumenal surface (I).
(J) Quantitation of cleaved-caspase 3 positive fragments on cross sections of E12.5 and E14.5 WT and Wnt5a−/− ME. Data are represented as mean and SD.
(K) Mathematical model (see STAR Methods). Measured WT (a) and Wnt5a−/− (b) ME cell population fold change; (c) fwt : modeled WT cell population fold change; (d) ft0 = wnt5a−/−: modeled WT cell population fold change but beginning with N0 of E11.5 Wnt5a−/−; (e–g) ft0 = wnt5a−/−,5–15%: modeled according to (d), but assuming 5% (e), 10% (f) or 15% (g) apoptosis.
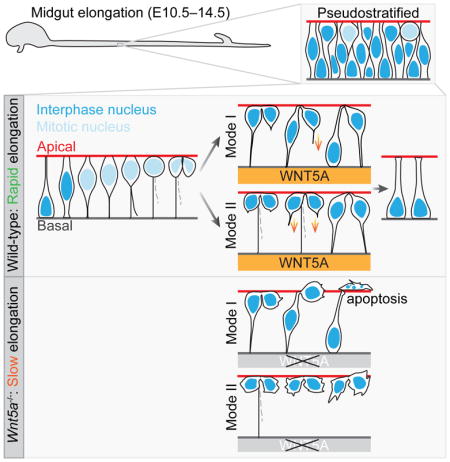
(L) Schematic summary of IKNM-associated cell division in WT and Wnt5a−/− ME. Without a WNT5A cue, some “pathfinding” daughter cells fail to return to the basal side. Apoptosis of these cells leads to the slow elongation of Wnt5a−/− midguts.
Loss of cells by apoptosis could certainly contribute to the observed Wnt5a−/− midgut length deficit (Figures 5B–5D). It is also possible that WNT5A is required for proper epithelial cell proliferation, which would also affect elongation rate. However, nearly all cells in Wnt5a−/− ME remain Ki67 positive (Figures S3A and S3B), except for a few nuclei in the most apical layer, which may be pre-apopotic (Figures S3A’–S3B”). In addition, the percentage of pHH3 positive mitotic figures relative to total nuclei in cross sections of ME (Figures S3C–S3E) and on the apical surface of opened midguts (Figures S3F–S3H) does not differ between WT and Wnt5a−/−. Moreover, no obvious delays were seen in the timing of G2 apical nuclear migration, mitosis (data not shown), or G1 return of normal pairs in Wnt5a−/− midguts (Figure 6Y). Thus, apoptosis, rather than proliferative change, appears to be the major contributor to the length deficit of Wnt5a−/− midguts.
In a highly proliferative ME, apoptosis leads to loss of single progenitors, as well as loss of all of that cell’s potential clonal offspring. To investigate whether increased apoptosis could account for the slow growth trajectory observed in Wnt5a−/− midguts, we applied a simple mathematical model for Phase I (see STAR Methods) (Figure 7K). This proliferation-based model (c) aligns with the actual WT cell population growth curve (a), supporting the conclusion that early WT midgut growth is driven by continuous cell division. Next, since Wnt5a−/− midguts show no change in proliferation, we modeled idealized growth using this WT exponential growth rate, but extrapolating from the initial measured size of the Wnt5a−/− midgut (E11.5) (d). Finally, we examined the consequences of several different apoptosis rates (e, f, g) on this idealized Wnt5a−/− growth curve (d). The model predicts that an apoptosis rate of 10% produces a growth curve (f) that closely mirrors that of the Wnt5a−/− experimental measurements (b). We conclude that the slow elongation of Wnt5a−/− midguts results primarily from increased apoptosis, secondary to improper basal tethering of post-mitotic cells.
Discussion
This work has uncovered an exquisite choreography of IKNM-associated cell division at the individual cell level in the early pseudostratified ME. Nearly all cells actively cycle, driving rapid midgut epithelial tube elongation throughout Phase I. During mitosis, 50% of daughter cells suffer temporary loss of a basal tether due to asymmetric BP inheritance. For all of these cells, as well as for a fraction of cells that inherit the BP, de novo basal connection is established by active, basally-oriented filopodial extensions. We show that mesenchymal WNT5A facilitates this process, providing an important radial cue that protects against apoptotic loss caused by failure of basal tethering (Figure 7L).
The data herein establish the behavior of the BP (retention, splitting and inheritance) in the early ME. Due to its long, thin morphology, elucidation of BP behavior during cell division has been challenging. Strong membrane fluorescence in the mTmG reporter line allowed us to visualize the BP throughout the cell cycle after high-resolution 3D reconstruction. Our studies show that, as nuclei migrate apically for mitosis, all cells retain a thin BP to maintain a basal connection. Interestingly, all mitotic cells appear to split their BP. The process of BP splitting begins when the soma adopts a spindle-like shape (pro-metaphase) and ends when the soma becomes pyriform in shape (~metaphase). This is consistent with observations in zebrafish neuroepithelial cells, in which BP splitting takes place prior to the onset of anaphase (Kosodo et al., 2008). However, in that system, only a fraction of the BPs actually split. Importantly, neuroepithelial cells undergo both proliferative divisions (responsible for expanding the proliferative population, similar to the ME) and neurogenic (fate determining) divisions; it is possible that the BP splits only during proliferative divisions.
Soon after BP splitting, one BP retracts and only one is retained in the majority of cases. Why should the cell put such effort into splitting the BP in two, if one is quickly discarded? A hint may come from the finding that, in neuroepithelial cells, the conserved cytokinesis component, anillin, which concentrates at the BP bifurcation site, migrates apically as the BP splits and then clusters at the basal-most membrane of the cell body before anaphase (Kosodo et al., 2008). Thus, BP splitting per se may serve to deliver elements of the cytokinesis machinery to the soma, making BP splitting an initiator of cytokinesis, though this remain to be functionally tested in the ME.
Following cytokinesis, the BP is inherited by one daughter. Asymmetric BP inheritance has been reported during neurogenesis and could potentially consign daughter cells to different fates (Miyata et al., 2001). However, in the Phase I midgut, the epithelium is composed of a relatively homogenous population of rapidly cycling progenitors and ownership of the BP seems unlikely to directly influence cell fate. Nevertheless, asymmetric BP inheritance allows one daughter cell to acquire its own basal connection instead of sharing the parent cell’s basal spot with its sister; this could be a prerequisite for daughter cell dispersal, which is observed in all post-mitotic clones in the Phase I ME. In addition, since clonal daughters could re-enter the cell cycle at a similar time, cell dispersal may relieve crowding generated by rounded mitotic cells in a limited apical space.
Asymmetric BP inheritance endows half of the daughter cells with path-finding flexibility but also exposes them to the risk of getting lost (apoptosis) or initiating another cell layer (stratification). Our studies have uncovered two “insurance policies” that likely minimize these potential risks. First, a long-lived apical membrane link persists between the two sister cells, even when they are 3–4 cells apart. This phenomenon has never been previously documented in the gut. Scission of this link occurs only after both sister nuclei have returned safely to the basal surface, suggesting the possible existence of a mechanism to control abscission timing. A second protection is from the basal direction: guidance by mesenchymal WNT5A. This uniquely protects “pathfinding” daughter cells, which extend filopodia to establish a new return path and thus run a greater risk of cell loss than “conduit” daughter cells. Of note, the fact that some pathfinding daughters still manage to return properly in the absence of WNT5A indicates the possible presence of additional guidance cues. Although both of these insurance policies can reduce the risks of pathfinding, it is a mystery why 28% of daughters with a BP give up the existing “conduit” and utilize the higher risk return strategy. One speculation is that, perhaps due to active trafficking of surrounding cells, the inherited BP no longer traces the best radial path for optimal return.
Interestingly, several features of IKNM-associated cell division that we observed in the midgut were also seen in the branching ureteric bud (Packard et al., 2013), a simple epithelium that develops from a pseudostratified epithelium (Chi et al., 2009). In both tissues, mitotic cells retain a BP for basal connection; the BP is inherited asymmetrically, daughter cells are connected apically during their basal return, and daughter cells finally sit non-contiguously, a feature termed “mitosis-associated cell dispersal” in the ureteric bud (Packard et al., 2013). However, BP splitting was not observed in the ureteric bud and differences in post-mitotic basal nuclear return by conduit vs. pathfinding strategies were not documented. Also, in the ureteric bud, most mitotic cells actually exit the plane of the epithelium to complete mitosis in the lumen, while mitosis always occurs within the plane of the epithelium in the midgut. It is possible that these differences represent adaptions to different morphogenetic geometries (tubular elongation vs. tip branching). It will be of interest to compare cellular behaviors in additional pseudostratified epithelia to clarify which features seen here are broadly conserved and which represent tissue-specific adaptations. Of note, the early hindgut epithelium is also a pseudostratified (data not shown) and the Wnt5a−/− hindgut is short (see Figures 5B–5D), though the behavior of dividing cells and the integrity of post-mitotic pathfinding in that setting remains to be tested.
How does WNT5A provide the directional cue? Interestingly, Wnt5a+/− midguts exhibit no distinguishable phenotype (Cervantes et al., 2009) and basal tethering of epithelial cells is unaffected. In contrast, WNT5A overexpression in both epithelium and mesenchyme from E10.5 onwards, does compromise gut elongation (Bakker et al., 2012). Thus, it is possible that mesenchymal WNT5A establishes a gradient across the epithelium in a basal-to-apical direction and disruption of this gradient ruins the “homing cue”. A related question is how cells in the epithelial layer sense the WNT5A cue. ROR2 is a known WNT5A receptor and is expressed by both midgut epithelial and mesenchymal cells (Yamada et al., 2010). Both Ror2−/− and Wnt5a−/− mice exhibit a short gut, but to a different extent (63% vs. 28% of WT by E13.5), suggesting that additional receptors are involved. In this regard, RYK, a less studied WNT5A receptor, is a promising candidate (Andre et al., 2012; Macheda et al., 2012). Indeed, Ryk−/− mice also have outgrowth defects in multiple organs (Halford et al., 2000), but the gut was not examined. Further work is needed to test the involvement of RYK in gut elongation, and examine whether both ROR2 and RYK act as WNT5A sensors for “pathfinding” daughters.
Our finding that basal tethering of post-mitotic cells is critical for cell survival in the pseudostratified ME might have implications for understanding the pathogenesis of CSBS. Recently, mutations in FILAMIN A were identified in two males of a family with presumed X-linked CSBS and in another unrelated male patient with CSBS (van der Werf et al., 2013). FILAMIN A, an actin-binding protein, can interact with cytoskeletal proteins (Kim and McCulloch, 2011; Ohta et al., 1999) and acts downstream of WNT5A/ROR2 in filopodia formation (Nishita et al., 2006). Male mice lacking Filamin A die during mid-gestation from cardiac defects, but gut phenotypes were not examined (Hart et al., 2006). A next important goal is to further probe the potential involvement of a WNT5A-ROR2/RYK-FILAMIN A pathway in guiding the basal tethering of post-mitotic cells in the elongating ME.
STAR Methods
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse anti-Ecadherin | BD Biosciences | Cat# 610181; RRID: AB_397580 |
| rabbit anti-PKCζ | Santa Cruz | Cat# sc-216 RRID: AB_2300359 |
| rabbit anti-Ki67 | Thermo Fisher | Cat# RM-9106-S RRID: AB_149707 |
| mouse anti-Phospho-histone H3 | Millipore | Cat# 05-806 RRID: AB_310016 |
| rabbit Cleaved Caspase-3 (Asp175) | Cell signaling | Cat# 9661 RRID: AB_2341188 |
| rabbit anti-GFP Alexa Fluor 488 conjugated | Thermo Fisher | Cat# A-21311 RRID: AB_221477 |
| Alexa Fluor® 488 Goat Anti-Mouse IgG (H+L) | Thermo Fisher | Cat# A-11001 RRID: AB_2534069 |
| Alexa Fluor® 488 Goat Anti-Rabbit IgG (H+L) Antibody | Thermo Fisher | Cat# A-11008 RRID: AB_143165 |
| Alexa Fluor® 546 Goat Anti-Rabbit IgG (H+L) | Thermo Fisher | Cat# A-11010 RRID: AB_2534077 |
| Alexa Fluor® 555 Donkey Anti-Mouse IgG (H+L) | Thermo Fisher | Cat# A-31570 RRID: AB_2536180 |
| Alexa Fluor® 647 Goat Anti-Rabbit IgG (H+L) | Thermo Fisher | Cat# A-21244 RRID: AB_141663 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Hoechst | Life Technologies | Cat# 33258 |
| Alexa Fluor™ 647 Phalloidin | Thermo Fisher | Cat# A22287 |
| Alexa Fluor™ 647 WGA | Life Technologies | Cat# W32466 |
| ProLong™ Gold Antifade Mountant | Life Technologies | Cat# P36930 |
| Tamoxifen | Sigma | Cat# T5648 |
| Focus Clear Kit | Cedarlane Labs | Cat# F101-KIT |
| 1% Penicillin-Streptomycin-Glutamine | Gibco | Cat# 10378016 |
| Glass-bottom culture dish 35-mm | MatTek Corporation | Cat# P35G-0-20-C |
| Richard-Allan Scientific™ HistoGel™ Specimen Processing Gel | Thermo Fisher | Cat# HG-4000-012 |
| TOPO™ TA Cloning™ Kit | Thermo Fisher | Cat# K457502 |
| Blocking Reagent | Roche | Cat# 11096176001 |
| Anti-Digoxigenin-AP, Fab fragments | Roche | Cat# 11093274910 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL6/J | Charles River | Strain Code: 027 |
| Mouse: RosamTmG/+: Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | The Jackson Laboratory | RRID:IMSR_JAX:007576 |
| Mouse: ShhCreERT2/+: B6.129S6-Shhtm2(cre/ERT2)Cjt/J | The Jackson Laboratory | RRID:IMSR_JAX:005623 |
| Mouse: Wnt5a+/−: B6;129S7-Wnt5atm1Amc/J | The Jackson Laboratory | RRID:IMSR_JAX:004758 |
| Mouse: Wnt5afl/fl: B6;129S-Wnt5atm1.1Krvl/J | The Jackson Laboratory | RRID:IMSR_JAX:026626 |
| Mouse: Twist2Cre/+: B6.129X1-Twist2tm1.1(cre)Dor/J | The Jackson Laboratory | RRID:IMSR_JAX:008712 |
| Oligonucleotides | ||
| Primers: mTmG | The Jackson Laboratory | 22163; oIMR9020; oIMR9021 |
| Primers: ShhCreERT2 | The Jackson Laboratory | oIMR3798; oIMR7338; oIMR7339; oIMR8346; |
| Primers: Wnt5a | The Jackson Laboratory | 9260; 9261; oIMR7415 |
| Primers: Wnt5a flox | The Jackson Laboratory | 22492; 22493 |
| Primers: Twist2Cre | The Jackson Laboratory | 15199; 15200; oIMR9174; oIMR9176 |
| Recombinant DNA | ||
| Software and Algorithms | ||
| Imaris | Bitplane | http://www.bitplane.com/imaris |
| Fiji/ImageJ | Schindelin et al., 2012 | https://imagej.net/Fiji |
| Other | ||
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Deborah Gumucio (dgumucio@umich.edu).
Experimental Model and Subject Details
Mice and Embryo Staging
All mouse work was performed in accordance with UCUCA and University of Michigan ULAM guidelines. Mouse strains used in this study were: C57BL6/J (Charles River), RosamTmG/+ (JAX 007576) (Muzumdar et al., 2007), ShhCreERT2/+ (JAX 005623)(Harfe et al., 2004), Wnt5a+/−(JAX 004758) (Yamaguchi et al., 1999), Wnt5afl/fl (JAX 026626) (Ryu et al., 2013), Twist2Cre/+ (JAX 008712) (Sosic et al., 2003). Both males and females were analyzed. Mice were maintained in level B facility with 12-hour light/dark cycles. Stage-specific embryos were isolated from timed matings based on the observation of a copulatory plug representing E0.5.
Method Details
Genotyping
Genotyping PCRs were performed with Taq DNA Polymerase, native kit (Invitrogen 18038) and GoTaq Flexi PCR kit (Promega M8295). Primers were from Integrated DNA Technologies. PCR primer sequences and protocols were as listed on the Jackson Laboratory website.
Tamoxifen Preparation and Administration
200 mg tamoxifen (Sigma, T5648) powder was dissolved into 1 mL 100% ethanol and then into 9mL fresh corn oil (Sigma, C8267) to make the 20 mg/mL tamoxifen stock. We further diluted this stock in corn oil for a total volume of 200 μL for gavage. For example, for a 100μg tamoxifen gavage, we add 5 μL stock into 195 μL corn oil. Pregnant females were gavaged with a low dosage of tamoxifen (100–350 μg) to activate mG cells in the midgut at 12 hrs–3 days prior to sacrifice.
GI tract dissection
Embryos were removed from the decidua and placed in ice-cold Dulbecco's PBS (DPBS) with the left side facing up. An opening was made along the left abdomen to open the abdomen cavity. The liver was gently lifted and one cut was made at the esophagus. Another cut was made at the end of the large intestine where it was connected to the anus. The balled up gut was gently pulled out of the body cavity by grabbing the esophagus end. The gut was finally straightened by separating the omentum at curvature areas.
Midgut Tissue Clearing and Confocal Imaging
Midguts were dissected, straightened, and fixed in 4% paraformaldehyde (PFA) for 20 min at room temperature (RT) and rinsed with PBS. Fixed midguts were aligned on glass slides, immersed in Focus Clear solution (Cedarlane Labs) for 15–30 min to render them transparent and mounted with Mount Clear solution (Cedarlane Labs) and a coverslip (thickness #1.5). Imaging was performed using a Nikon A-1 confocal microscope. Z-stacks were 3D reconstructed using Imaris software (Bitplane).
Midgut Cultures and Time-lapse Imaging
The culture medium, 10% Fetal bovine serum, 1% Penicillin-Streptomycin-Glutamine (Gibco 10378016) in DMEM (Gibco 21063029), was prepared and equilibrated in a tissue culture incubator (5% CO2, 37°C) for 30 min. 1 mL culture medium was added in a 35 mm glass-bottomed culture dish. A transwell filter and a sterilized stainless-steel adapter ring (Costantini et al., 2011) were assembled with the culture dish. GI tracts were dissected and straightened in DPBS, and placed on the top of the transwell filter using the Wiretrol. A lid was added on the top. This entire culture unit (Figure 2A) was immediately incubated at 37°C with 5% CO 2 to allow tissue to attach on the transwell membrane. After 1–2 hrs incubation, time-lapse imaging was performed using the DeltaVision restoration microscopy system with softWoRx software. The data were processed and analyzed using Fiji software (Schindelin et al., 2012).
Midgut Sectioning and Immunostaining
Midguts were fixed in 4% PFA at RT for 2 hrs or at 4°C overnight. For vibratome sectioning, fixed midguts were embedded in 4–6% low-melting point agarose and sectioned at thickness of 100μm as previously described (Wang et al., 2013). Vibratome sections were permeabilized in 0.5% TritonX-100 in PBS for 20 min at RT while rocking and washed with PBS 5 min, three times. Sections were blocked with 4% goat serum, 0.1% TritonX-100 in PBS for 30 min. Primary antibody incubation was performed overnight at 4°C, followed by secondary antibody in cubation for 2 hrs at RT. Samples were mounted in Prolong Gold (Life Technologies P36930), and imaging was performed on a Nikon A-1 confocal microscope. For paraffin sectioning, fixed midguts were embedded into histogel (Thermo Scientific HG-4000-012) and dehydrated in ethanol, infused with paraffin, embedded in paraffin blocks, and sectioned at thickness of 5 μm. Sections were deparaffinized in xylene, rehydrated, washed with PBS and then boiled in 10 mM sodium citrate (pH 6) for 10 min for antigen retrieval. The staining process was same as for vibratome section staining. Imaging was performed using a Zeiss apotome microscope.
Whole-mount Staining
Isolated midguts were immediately cut into 3 segments (proximal, middle, and distal) in DPBS. Each segment was opened with a forceps and tungsten needle to expose the luminal surface. After opening, they were fixed in 4% PFA for 2 hrs at RT. Tissue were washed three times in PBS for 5 min each and then were permeabilized in 0.5% TritonX-100 in PBS for 25 min at RT while rocking. After washing in PBS, tissues were blocked with 4% goat serum, 0.1% TritonX-100 in PBS for 30 min. Primary antibody incubation was performed overnight at 4°C, followed by secondary antibody incubation for 2 hrs at RT. After washing, midguts were mounted on glass slides with the luminal surface facing up and coverslipped. Z-stacks of the luminal surfaces of opened midguts were obtained on a Zeiss apotome microscope and projected into single 2D images.
In Situ Hybridization
RNA probes were synthesized by RT-PCR, cloned into pCR4-TOPO vectors (Invitrogen) and sequenced for confirmation. Nucleotides 413–1424 of NM_009524.4 (Wnt5a, transcript variant 1) was used as probe template.
Tissue was fixed in 4% PFA and embedded in OCT for cryosections (10 μm). Sections were rehydrated, treated with 10 μg/mL proteinase K in PBS for 15 min at RT, post fixed 10 min in 4% PFA, and then treated with acetic anhydride solution. After prehybridization for 4 hrs at RT, the sections were hybridized at 70°C overnight with Wnt5a probes in 50% deionized formamide, 5 x SSC, 5x Denhardt’s solution, 250 μg/mL yeast tRNA, 0.5 mg/mL herring sperm DNA and 2% blocking reagent (Roche 11096176001). Slides were washed in 5 x SSC at 70°C , 0.2 x SSC at 70°C for 1 hr, 0.2 x SSC at RT for 5 min and Buffer 1 (100 mM Tris-HCl pH 7.5, 150 mM NaCl) at RT for 5 min. Sections were blocked for 1 hr in Buffer 1 containing 10% heat-inactivated fetal calf serum and 2% blocking reagent and incubated with alkaline phosphatase-conjugated anti-digoxigenin antibody (Roche), 1:5000 in blocking solution at 4°C overnight. After several washes in Buffer 1, the slides were equilibrated in Buffer 3 (100 mM Tris·HCl pH 9.5, 100 mM NaCl, 50 mM MgCl2) and color reaction was performed in the dark in Buffer 3 supplemented with 100 mg/mL NBT (4-nitro blue tetrazolium chloride), 50 mg/mL BCIP (5-bromo-4-chloro-3-indolyl-phosphate), 0.05% Tween-20 and 1mM levamisole for 10–20 hrs. The color reaction was stopped in PBS, and the slides were mounted with 70% glycerol/PBS.
Mathematical Modeling
We determined cell population fold change (F) as: F = FxyFZ. Fxy is the fold change in actual cell number on the xy plane (measured by counting nuclei in midgut cross sections, Figure S4), while FZ is the relative change in cell number along the z axis, as indicated by the fold change in measured midgut length Fl. The model assumes a negligible change in cell width over time. In this simplified model, midgut lengthening is driven by symmetrical divisions in a uniform population of progenitor cells. As each cell divides every 16 hrs (based on live imaging data), the cell population at the next cycle Nt + 1 will exhibit geometrical growth: Nt + 1 = kNt, where k is the net cell population fold change. Without apoptosis, 100 cells become 200 cells at the end of one cell cycle, k = 2. When 5% apoptosis (a) occurs, 5/100 cells are lost and only 95 cells divide, resulting in 190 cells after one cell cycle. Mathematically, the model with apoptosis becomes Nt + 1 = k (1 − a)Nt, where a is the apoptosis rate.
Image Analysis
Quantitation of midgut length was performed on whole-mount tissue images and analyzed using Fiji/ImageJ. Quantitation of midgut epithelial tube circumferences, epithelial thickness, apoptosis rate, mitosis rate, and number of nuclei per section were performed on images of staining on paraffin cross-sections and analyzed using Fiji/ImageJ. Clone size (the number of cells per clone) was counted on a Zeiss apotome microscope. The number of BP that the mitotic cells possess was counted on 3D reconstructed images of spherical cells, generated by Imaris. Quantitation of average time for G1 return was performed on time-lapse images and analyzed using Fiji/ImageJ. The number of cells that have a basal connection defect was counted on 3D reconstructed images of large clones generated by Imaris. Quantitation of mitosis rate on the luminal surfaces of opened midguts was performed on whole-mount pHH3 stained images and analyzed using Fiji/ImageJ.
Imaging Processing
In Figures 1A, 1C, 1G, and 1H, five separate images with different sizes were adjusted to the same scale and were combined into one image. A black rectangle was placed behind as background to group the images into one panel. In Figures 2P, 2Q, 3D, 3E, 4A, 4C, and 6S-6X, separate static images at different time points of live imaging were combined into one image with a black rectangle as the background. In Figures 3H–3S and 6A–6R, separate images were grouped into one panel with a black rectangle as the background. In Figures 5B and 5D, images of different sizes were adjusted to the same scale. One black rectangle was placed behind to align them. In Figure 2A and 5F, two separate images (same experiment) were stitched together to show a full view of one segment of midgut.
Quantification and Statistical Analysis
All statistical tests and graphs were generated in Excel or Prism 6 (GraphPad Software). T-tests were unpaired nonparametric tests (Mann-Whitney test). P < 0.05 was considered statistically significant. NS= not significant. Error bars represent the SD of a data set.
Supplementary Material
Highlights.
The early elongating midgut epithelium exhibits interkinetic nuclear migration
Mitotic cells retain a thin basal process (BP), which is inherited by one daughter
G1 nuclei return basally via a BP “conduit” or via “pathfinding” filopodia
Mesenchymal WNT5A guides filopodial pathfinding, failure of which causes apoptosis
Acknowledgments
We thank Åsa Kolterud for in situ hybridization analysis; Kate Walton, Yue Shao, Xufeng Xue for technical advice; Suzie Shoffner for help with mathematical modeling; Microscopy and Image Analysis Laboratory at University of Michigan for imaging technical assistance; and Cameron Hollingshead for discussion. This work was supported by the NIH (R01 DK089933).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Author Contributions
Conceptualization, S.W. and D.G.; Methodology, S.W., C.C. and S.S.; Validation, S.W.; Formal Analysis, S.W. and S.S.; Investigation, S.W.; Resources, D.G.; Writing – Original Draft, S.W.; Writing – Review & Editing, D.G., S.W. and S.S.; Visualization, S.W.; Supervision, D.G.; Project Administration, S.W. and D.G.; Funding Acquisition, D.G.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andre P, Wang Q, Wang N, Gao B, Schilit A, Halford MM, Stacker SA, Zhang X, Yang Y. The Wnt coreceptor Ryk regulates Wnt/planar cell polarity by modulating the degradation of the core planar cell polarity component Vangl2. The Journal of biological chemistry. 2012;287:44518–44525. doi: 10.1074/jbc.M112.414441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker ERM, Raghoebir L, Franken PF, Helvensteijn W, van Gurp L, Meijlink F, van der Valk MA, Rottier RJ, Kuipers EJ, van Veelen W, et al. Induced Wnt5a expression perturbs embryonic outgrowth and intestinal elongation, but is well-tolerated in adult mice. Dev Biol. 2012;369:91–100. doi: 10.1016/j.ydbio.2012.06.007. [DOI] [PubMed] [Google Scholar]
- Blakely BD, Bye CR, Fernando CV, Horne MK, Macheda ML, Stacker SA, Arenas E, Parish CL. Wnt5a regulates midbrain dopaminergic axon growth and guidance. PloS one. 2011;6:e18373. doi: 10.1371/journal.pone.0018373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes S, Yamaguchi TP, Hebrok M. Wnt5a is essential for intestinal elongation in mice. Dev Biol. 2009;326:285–294. doi: 10.1016/j.ydbio.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X, Michos O, Shakya R, Riccio P, Enomoto H, Licht JD, Asai N, Takahashi M, Ohgami N, Kato M, et al. Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Dev Cell. 2009;17:199–209. doi: 10.1016/j.devcel.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin AM, Tsai YH, Finkbeiner SR, Nagy MS, Walker EM, Ethen NJ, Williams BO, Battle MA, Spence JR. A Dynamic WNT/beta-CATENIN Signaling Environment Leads to WNT-Independent and WNT-Dependent Proliferation of Embryonic Intestinal Progenitor Cells. Stem cell reports. 2016;7:826–839. doi: 10.1016/j.stemcr.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F, Watanabe T, Lu B, Chi X, Srinivas S. Dissection of embryonic mouse kidney, culture in vitro, and imaging of the developing organ. Cold Spring Harbor protocols. 2011;2011 doi: 10.1101/pdb.prot5613. pdb prot5613. [DOI] [PubMed] [Google Scholar]
- Freddo AM, Shoffner SK, Shao Y, Taniguchi K, Grosse AS, Guysinger MN, Wang S, Rudraraju S, Margolis B, Garikipati K, et al. Coordination of signaling and tissue mechanics during morphogenesis of murine intestinal villi: a role for mitotic cell rounding. Integrative biology : quantitative biosciences from nano to macro. 2016 doi: 10.1039/c6ib00046k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse AS, Pressprich MF, Curley LB, Hamilton KL, Margolis B, Hildebrand JD, Gumucio DL. Cell dynamics in fetal intestinal epithelium: implications for intestinal growth and morphogenesis. Development. 2011;138:4423–4432. doi: 10.1242/dev.065789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie S, Butcher M, Lumsden A. Patterns of Cell-Division and Interkinetic Nuclear Migration in the Chick-Embryo Hindbrain. J Neurobiol. 1991;22:742–754. doi: 10.1002/neu.480220709. [DOI] [PubMed] [Google Scholar]
- Halford MM, Armes J, Buchert M, Meskenaite V, Grail D, Hibbs ML, Wilks AF, Farlie PG, Newgreen DF, Hovens CM, et al. Ryk-deficient mice exhibit craniofacial defects associated with perturbed Eph receptor crosstalk. Nat Genet. 2000;25:414–418. doi: 10.1038/78099. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian F, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Hart AW, Morgan JE, Schneider J, West K, McKie L, Bhattacharya S, Jackson IJ, Cross SH. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum Mol Genet. 2006;15:2457–2467. doi: 10.1093/hmg/ddl168. [DOI] [PubMed] [Google Scholar]
- Hasosah M, Lemberg DA, Skarsgard E, Schreiber R. Congenital short bowel syndrome: a case report and review of the literature. Canadian journal of gastroenterology = Journal canadien de gastroenterologie. 2008;22:71–74. doi: 10.1155/2008/590143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, VanHooser A, Ranalli T, Brinkley BR, BazettJones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Horigane S, Ageta-Ishihara N, Kamijo S, Fujii H, Okamura M, Kinoshita M, Takemoto-Kimura S, Bito H. Facilitation of axon outgrowth via a Wnt5a-CaMKK-CaMKIalpha pathway during neuronal polarization. Molecular brain. 2016;9:8. doi: 10.1186/s13041-016-0189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins BI, Li L, Kalil K. Wnt/calcium signaling mediates axon growth and guidance in the developing corpus callosum. Developmental neurobiology. 2011;71:269–283. doi: 10.1002/dneu.20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, McCulloch CA. Filamin A mediates interactions between cytoskeletal proteins that control cell adhesion. Febs Lett. 2011;585:18–22. doi: 10.1016/j.febslet.2010.11.033. [DOI] [PubMed] [Google Scholar]
- Kohlnhofer BM, Thompson CA, Walker EM, Battle MA. GATA4 regulates epithelial cell proliferation to control intestinal growth and development in mice. Cellular and molecular gastroenterology and hepatology. 2016;2:189–209. doi: 10.1016/j.jcmgh.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- Kosodo Y, Huttner WB. Basal process and cell divisions of neural progenitors in the developing brain. Development, growth & differentiation. 2009;51:251–261. doi: 10.1111/j.1440-169X.2009.01101.x. [DOI] [PubMed] [Google Scholar]
- Kosodo Y, Toida K, Dubreuil V, Alexandre P, Schenk J, Kiyokage E, Attardo A, Mora-Bermudez F, Arii T, Clarke JDW, et al. Cytokinesis of neuroepithelial cells can divide their basal process before anaphase. Embo Journal. 2008;27:3151–3163. doi: 10.1038/emboj.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Hutchins BI, Kalil K. Wnt5a Induces Simultaneous Cortical Axon Outgrowth and Repulsive Axon Guidance through Distinct Signaling Mechanisms. J Neurosci. 2009;29:5873–5883. doi: 10.1523/JNEUROSCI.0183-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Kispert A, Kutsch S, Kemler R. Expression patterns of Wnt genes in mouse gut development. Mechanisms of development. 2001;105:181–184. doi: 10.1016/s0925-4773(01)00390-2. [DOI] [PubMed] [Google Scholar]
- Macheda ML, Sun WW, Kugathasan K, Hogan BM, Bower NI, Halford MM, Zhang YF, Jacques BE, Lieschke GJ, Dabdoub A, et al. The Wnt receptor Ryk plays a role in mammalian planar cell polarity signaling. The Journal of biological chemistry. 2012;287:29312–29323. doi: 10.1074/jbc.M112.362681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathan M, Moxey PC, Trier JS. Morphogenesis of fetal rat duodenal villi. The American journal of anatomy. 1976;146:73–92. doi: 10.1002/aja.1001460104. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Hashimoto K, Yoshioka T, Otani H. Occlusion and subsequent re-canalization in early duodenal development of human embryos: integrated organogenesis and histogenesis through a possible epithelial-mesenchymal interaction. Anat Embryol. 2002;205:53–65. doi: 10.1007/s00429-001-0226-5. [DOI] [PubMed] [Google Scholar]
- Meyer EJ, Ikmi A, Gibson MC. Interkinetic Nuclear Migration Is a Broadly Conserved Feature of Cell Division in Pseudostratified Epithelia. Curr Biol. 2011;21:485–491. doi: 10.1016/j.cub.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nishita M, Yoo SK, Nomachi A, Kani S, Sougawa N, Ohta Y, Takada S, Kikuchi A, Minami Y. Filopodia formation mediated by receptor tyrosine kinase Ror2 is required for Wnt5a-induced cell migration. Journal of Cell Biology. 2006;175:555–562. doi: 10.1083/jcb.200607127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noah TK, Donahue B, Shroyer NF. Intestinal development and differentiation. Experimental cell research. 2011;317:2702–2710. doi: 10.1016/j.yexcr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, VMC, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nature neuroscience. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Norden C. Pseudostratified epithelia - cell biology, diversity and roles in organ formation at a glance. J Cell Sci. 2017;130:1859–1863. doi: 10.1242/jcs.192997. [DOI] [PubMed] [Google Scholar]
- Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP. The small GTPase RalA targets filamin to induce filopodia. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard A, Georgas K, Michos O, Riccio P, Cebrian C, Combes AN, Ju A, Ferrer-Vaquer A, Hadjantonakis AK, Zong H, et al. Luminal Mitosis Drives Epithelial Cell Dispersal within the Branching Ureteric Bud. Dev Cell. 2013;27:319–330. doi: 10.1016/j.devcel.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RA, Womble MA, Dush MK, Tull RR, Bloom SK, Morckel AR, Devlin EW, Nascone-Yoder NM. Morphogenesis of the Primitive Gut Tube Is Generated by Rho/ROCK/Myosin II-Mediated Endoderm Rearrangements. Dev Dynam. 2009;238:3111–3125. doi: 10.1002/dvdy.22157. [DOI] [PubMed] [Google Scholar]
- Ryu YK, Collins SE, Ho HYH, Zhao HQ, Kuruvilla R. An autocrine Wnt5a-Ror signaling loop mediates sympathetic target innervation. Dev Biol. 2013;377:79–89. doi: 10.1016/j.ydbio.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer ME, Walker BE. Radioautographic Study of Interkinetic Nuclear Migration in the Neural Tube. P Soc Exp Biol Med. 1959;101:557–560. doi: 10.3181/00379727-101-25014. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosic D, Richardson JA, Yu K, Ornitz DM, Olson EN. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappa B activity. Cell. 2003;112:169–180. doi: 10.1016/s0092-8674(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Spear PC, Erickson CA. Interkinetic nuclear migration: a mysterious process in search of a function. Development, growth & differentiation. 2012;54:306–316. doi: 10.1111/j.1440-169X.2012.01342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzyz PJ, Matejcic M, Norden C. Heterogeneity, Cell Biology and Tissue Mechanics of Pseudostratified Epithelia: Coordination of Cell Divisions and Growth in Tightly Packed Tissues. International review of cell and molecular biology. 2016;325:89–118. doi: 10.1016/bs.ircmb.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Toyota T, Yamamoto M, Kataoka K. Light and electron microscope study on developing intestinal mucosa in rat fetuses with special reference to the obliteration of the intestinal lumen. Archives of histology and cytology. 1989;52:51–60. doi: 10.1679/aohc.52.51. [DOI] [PubMed] [Google Scholar]
- van der Werf CS, Halim D, Verheij JB, Alves MM, Hofstra RM. Congenital Short Bowel Syndrome: from clinical and genetic diagnosis to the molecular mechanisms involved in intestinal elongation. Biochimica et biophysica acta. 2015;1852:2352–2361. doi: 10.1016/j.bbadis.2015.08.007. [DOI] [PubMed] [Google Scholar]
- van der Werf CS, Sribudiani Y, Verheij JBGM, Carroll M, O'Loughlin E, Chen CH, Brooks AS, Liszewski MK, Atkinson JP, Hofstra RMW. Congenital short bowel syndrome as the presenting symptom in male patients with FLNA mutations. Genet Med. 2013;15:310–313. doi: 10.1038/gim.2012.123. [DOI] [PubMed] [Google Scholar]
- Walton KD, Freddo AM, Wang S, Gumucio DL. Generation of intestinal surface: an absorbing tale. Development. 2016;143:2261–2272. doi: 10.1242/dev.135400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KD, Kolterud A, Czerwinski MJ, Bell MJ, Prakash A, Kushwaha J, Grosse AS, Schnell S, Gumucio DL. Hedgehog-responsive mesenchymal clusters direct patterning and emergence of intestinal villi. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:15817–15822. doi: 10.1073/pnas.1205669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KD, Mishkind D, Riddle MR, Tabin CJ, Gumucio DL. Blueprint for an intestinal villus: Species-specific assembly required. Wiley interdisciplinary reviews Developmental biology. 2018 doi: 10.1002/wdev.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Cha SW, Zorn AM, Wylie C. Par6b regulates the dynamics of apicobasal polarity during development of the stratified Xenopus epidermis. PloS one. 2013;8:e76854. doi: 10.1371/journal.pone.0076854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LT, Austin S, Cole TJ. Small intestinal length: a factor essential for gut adaptation. Gut. 1991;32:1321–1323. doi: 10.1136/gut.32.11.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Udagawa J, Hashimoto R, Matsumoto A, Hatta T, Otani H. Interkinetic nuclear migration during early development of midgut and ureteric epithelia. Anatomical science international. 2013;88:31–37. doi: 10.1007/s12565-012-0156-8. [DOI] [PubMed] [Google Scholar]
- Yamada M, Udagawa J, Matsumoto A, Hashimoto R, Hatta T, Nishita M, Minami Y, Otani H. Ror2 is Required for Midgut Elongation During Mouse Development. Dev Dynam. 2010;239:941–953. doi: 10.1002/dvdy.22212. [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Bradley A, McMahon AP, Jones S. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development. 1999;126:1211–1223. doi: 10.1242/dev.126.6.1211. [DOI] [PubMed] [Google Scholar]
- Zorn AM, Wells JM. Vertebrate endoderm development and organ formation. Annual review of cell and developmental biology. 2009;25:221–251. doi: 10.1146/annurev.cellbio.042308.113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.