Abstract
Hematopoietic stem cells (HSCs) maintain a quiescent state in the bone marrow to preserve their self-renewal capacity, but also undergo cell divisions as required. Organelles such as the mitochondria sustain cumulative damage during these cell divisions, and this damage may eventually compromise the cells’ self-renewal capacity. HSC divisions result in either self-renewal or differentiation, with the balance between the two directly impacting hematopoietic homeostasis; but the heterogeneity of available HSC-enriched fractions, together with the technical challenges of observing HSC behavior, has long hindered the analysis of individual HSCs, and prevented the elucidation of this process. However, recent advances in genetic models, metabolomics analyses and single-cell approaches have revealed the contributions made to HSC self-renewal by metabolic cues, mitochondrial biogenesis, and autophagy/mitophagy, which have highlighted mitochondrial quality as a key control factor in the equilibrium of HSCs. A deeper understanding of precisely how specific modes of metabolism control HSC fate at the single cell level is therefore not only of great biological interest, but will have clear clinical implications for the development of therapies for hematological disease.
Stem cells are self-renewing, and either multi- or unipotent1–5, and these unique capacities offer opportunities for stem cell-based therapies in the clinic6. Past research has implied only limited contributions by hematopoietic stem cells (HSCs) to unperturbed hematopoiesis, but HSCs are still believed essential to hematopoiesis under stress conditions such as hematopoietic recovery7–11. HSC transplantation has therefore been a key therapeutic strategy in combatting hematological disorders12–14. Like the stem cells of other tissues, HSCs basically remain quiescent to maintain their undifferentiated state, but they also undergo cell divisions as required2,3. As HSC populations are precisely controlled within certain limits in vivo, once hematopoietic recovery is complete, it is believed that HSCs return to a quiescent state, or dormancy. This suspension of the cell cycle is thought to make a critical contribution to the maintenance of stem cells’ self-renewal capacity and multipotency, as deletion of the genes involved in quiescence often leads to HSC exhaustion due to uncontrolled proliferation15–20. Indeed, the regenerative potential of HSCs may be governed by their divisional history2,3, and therefore it is believed that cell intrinsic networks involving key cell cycle regulators and the levels of Hox genes or Polycomb complex protein, along with the activity of transcriptional factors, integrate and cooperate with cumulative signals from the microenvironment to fine-tune the self-renewal capacity of HSCs and maintain whole hematopoiesis16,18,21–25. The role of cellular metabolism in regulating HSC self-renewal capacity has thus become a focus of much current stem cell research, which has yielded many new insights26–32 (Fig. 1). In this review, we will highlight the recent advances in our understanding of the intriguing relationship between cellular metabolism, mitochondrial quality control, and HSC fate decisions.
Figure 1. Overview of metabolic pathways contributing to HSC self-renewal and differentiation.
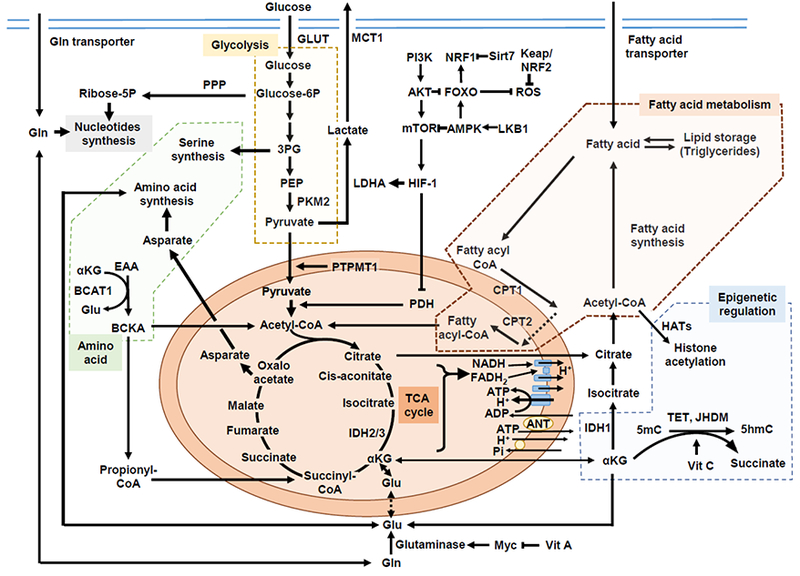
Hematopoietic stem cells (HSCs) rely on glycolysis (indicated by orange background). HIF-1α both promotes glycolysis and prevents pyruvate oxidation by suppressing the PDH complex. The PI3K-AKT pathway promotes ROS production by repressing FOXO. Fatty acid oxidation (brown background) is required for HSC selfrenewal by controlling cell fate decisions. HSCs are dependent on dietary valine and vitamin A, and Gln is converted to Glu by glutaminase, which is partly under the control of MYC. Important contributions from BCAA metabolisms regulated by BCAT1 to myeloid leukemia have been suggested (green background). The intact mitochondrial function for HSC maintenance may include metabolism-driven epigenetic changes or code. Acetyl-CoA can be a source for histone acetylation, and IDHs are a family of enzymes catalyzing the oxidative decarboxylation of isocitrate into αKG, which is a cofactor for dioxigenase enzymes, TET2 and JHDM. Vitamin C is a co-factor for the enzymatic activity of the TET family of DNA hydroxylases (blue background). Abbreviations: ΗIF-1α, hypoxia-inducible factor 1α; Glut, glucose transporter; Glucose-6P, glucose 6-phosphate; PDH, pyruvate dehydrogenase; 3PG, 3-phosphoglyceric acid; PPP, pentose phosphate pathway; PEP, phosphoenolpyruvic acid; PKM2, pyruvate kinase M2; LDHA, lactate dehydrogenase A; MCT1, monocarboxylate transporter 1; PTPMT1, PTEN-like mitochondrial phosphatase, or PTP localized to the Mitochondrion 1; TCA, tricarboxylic acid cycle; NADH, nicotinamide adenine dinucleotide; FADH, the reduced form of flavin adenine dinucleotide; ANT, adenine nucleotide translocases; Pi, inorganic phosphate; ROS, reactive oxygen species; FOXO, forkhead box Ο; PI3K, phosphoinositide 3-kinase; AKT, protein kinase B, or PKB; NRF, nuclear respiratory factor; Sirt7, sirtuin 7; LKB1, liver kinase B1; AMPK, AMP-activated protein kinase; mTOR, mammalian target of rapamycin; CoA, coenzyme A; CPT, carnitine-O-palmitoyltransferase; IDH, isocitrate dehydrogenases; Gln, glutamine; Glu, glutamate; EAA, essential amino acid (valine, leucine and isoleucine); BCAA, branched chain amino acid; BCAT1, BCAA transaminase 1; BCKA, branched chain keto acid; αKG, α-chetoglutarate; TET, ten-eleven translocation; JHDM, jmjC domain-containing histone demethylase; 5mC, 5-methylcytosine; 5hmC, 5-hydroxymethylcytosine; Vit C, vitamin C or ascorbic acid; hAT, Histone acetyltransferases;
Assessment of HSC fate
HSC cell fate decisions can be evaluated by paired daughter cell assays15,33–35. Their possible division options are: symmetric self-renewal expansion (symmetric division, SD; both daughter cells have the same function as the original cell), selfrenewal maintenance (asymmetric division, AD) and differentiation (symmetric commitment, SC; both daughter cells are differentiated from the original parent cell), and their eventual division pattern is determined by the in vivo repopulation capacity of their daughter cells. In cases where at least one daughter cell is a long-term HSC (LT-HSC), the original cell must also be an LT-HSC. However, if both daughter cells are non-LT-HSCs, interpreting the resulting data can be complex, as a cell’s original function can affect its division pattern (Fig. 2A).
Figure 2. Division patterns by paired daughter cell assays.
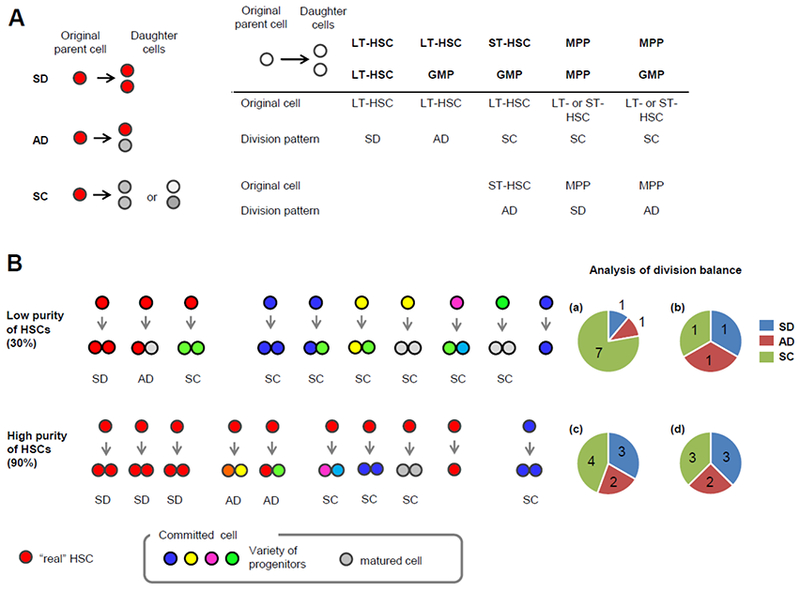
(A) Original cell function affects its division pattern. Schematic model of 3 division patterns; after SD, both daughter cells have the same function and differentiation stage as the parent cell (red), while both daughter cells appear as more committed cells (grey or pale grey) than the parent cells after SC (left). After initial division of the parent cell from the HSC-enriched fraction, the repopulation capacity and/or differentiation potential of the paired daughter cells is individually determined (e.g. by in vivo repopulation capacity, retrospectively). As the HSC-enriched fraction is a heterogeneous population, the immunophenotypically isolated single cells from this fraction can be hematopoietic progenitors or mature cells. Some examples of the combinations of the parent cells, their daughter cells and their division patterns are shown at right bottom. (B) Analysis of division patterns in homogenous and heterogeneous populations. When 10 single cells are isolated from the population with 30% purity of HSCs, 3 are generally “real” HSCs (top). In this example, each of these three HSCs undergoes SD, AD and SC, respectively (b), and 1 cell does not undergo cell division during the assay period. Because committed cells are not able to produce HSCs, the division patterns of those cells are assessed as SC. Thus, the resulting division balance of the whole compartment will be 1 SD, 1 AD and 7 SC (a), and it is difficult to extract the phenotypes of real HSCs from this low purity of HSCs. However, in the case of 90% HSC purity (bottom), the division balance of HSCs (d) can be accurately estimated from the resulting division symmetry of the isolated whole population (c). SD, symmetric division; AD, asymmetric division; SC, symmetric commitment; LT-HSC, long-term hematopoietic stem cell; ST-HSC, short-term HSC; MPP, multi-potent progenitor; GMP, granulocyte-monocyte progenitors.
Further, the homogeneity of the cell population is critical to accurate division pattern analysis. Tracking the divisions of individual cells from a heterogeneous population has proved difficult, and any contamination of non-HSCs can lead to an overestimate of the rate of SC. As an example, let us consider a 30% pure population (low purity), in which three out of ten single cells in the HSC fraction must be “real” HSCs, and a case in which one of these HSCs undergoes SD (33%), while another undergoes AD (33%) and the third undergoes SC (33%). As committed cells cannot produce HSCs upon their division, their division patterns must be regarded as SC. The resulting division balance of the entire population would therefore be SD 11%, AD 11% and SC 78% (Fig. 2B). HSCs have been identified retrospectively after single cell transplantation by clonal assays, and these assays have demonstrated the heterogeneity of currently available HSC-enriched fractions33,36–39. Unfortunately, the reported frequency of HSCs in these fractions is generally lower than 30%, and it is worth pointing out that in the case described earlier (SD: AD: SC = 1: 1: 1), an HSC purity of even ~ 40% would be regarded as low, because the overestimation of SC would lead to a significant shift in the assessed division balance (to a maximum of 44% SC in n = 27, and 41% SC in n = 50 divisions assessed, respectively. *p < 0.05 by Chi-square test). However, when we have a high purity population of real HSCs, we can more accurately determine their division pattern (Fig. 2B).
To avoid this imprecision, researchers have long sought a reliable marker for individual HSCs which is strongly associated with repopulation capacity and does not fluctuate with changes in the surrounding environment and/or cell cycle. In various attempts to detect purified HSCs, recent studies have utilized combinations of cell surface markers, the reporter Cre-recombinase, and antibody positivity; but so far, these efforts have met with only limited success8,40–45.
Division assays with markers for self-renewing HSCs
Until recently, HSC number and capacity were believed to decrease rather than increase with age, and it has proved very challenging to expand the HSC population while maintaining stem-ness. Indeed, although division patterns in hematopoietic stem and progenitor cells (HSPCs) were thought to be controlled by the balance between SC and AD15,33,34, advanced single-cell approaches have recently confirmed that HSCs are capable of symmetric self-renewing division (or SD)33,46. Analysis by the long-label retaining method with H2B-GFP (histone 2b - green fluorescent protein), for instance, has shown that HSCs can divide symmetrically at least several times throughout adult life to achieve higher density in the bone marrow47.
Our use of Tie2 positivity as a marker has allowed us to identify a purified population of HSCs, and we have demonstrated with our local transplantation protocol that single HSCs from this population exhibit high reconstitution capacity in vivo46,48. Our tracking technique allowed us to determine the function of the paired daughter cells resulting from single HSC divisions, which in turn enabled us to more accurately visualize division patterns, and distinguish self-renewal expansion from self-renewal maintenance. In these studies we found that only top hierarchical HSCs underwent SD, in which both daughter cells are HSCs and retain Tie2 positivity46.
As increasing evidence supports the essential contributions of metabolic control to HSC division patterns, determining the metabolic mode of purified HSCs is of crucial importance15,29. Single cell gene expression assays have revealed that critical roles in HSC expansion are played by fatty acid oxidation (FAO)46. The mitochondria are the primary sites of FAO, in which fatty acids are enzymatically broken down49, and as they are essential sub-cellular components in the metabolic process, their role in division patterns and the subsequent cell fates of HSCs is a question of great scientific interest (Fig. 3). Further, research has shown that during asymmetric division in mammary epithelial stem-like cells, older mitochondria are pushed into daughter cells fated to differentiation in order to maintain high-quality stem cell homeostasis50. In contrast, symmetric division requires self-clearance systems in both daughter cells, as young and old mitochondria have been found equally distributed between both29,46; however, the processes involved remain among the least understood in stem cell biology.
Figure 3. Quality control machineries in HSC division balance and hematopoietic homeostasis.

(A) In SD, mitochondria are equally segregated into two daughter cells, although their metabolic processes may differ from those of the mother cell. Upon cell division, organelles such as mitochondria are damaged, which activates mitochondrial autophagy. This activation of mitophagy promotes mitochondrial quality control, and subsequent self-renewing HSC expansion. (B) In some mammary stem-like-cell divisions, mitochondria are split unevenly between the two daughter cells, and old mitochondria are apportioned primarily to the tissue-progenitor daughter, whereas newly synthesized mitochondria are apportioned to the stem cell-like daughter. It has yet to be formally demonstrated, but asymmetric HSC division by unequal apportionment of older or damaged mitochondria could be a potential strategy for removing damaged cell components. (C) HSC activation is accompanied by mitochondria activation and a shift in metabolic activity to Oxphos (right). Healthy but active mitochondria are unselectively removed by autophagy, and these active HSCs return to replicative quiescence (left). The majority (two thirds) of HSCs from aged mice as well as some autophagy-deficient HSCs (e.g. Atg12-deficient HSCs) were not able to efficiently limit the number of active mitochondria, which drives aging phenotypes in the blood (far left). Hyperactivated mitophagy (e.g. loss of Atad3a) results in blocked hematopoietic lineage commitment at the progenitor stage and enlarged HSPC pools (far right). HSPC, hematopoietic stem and progenitor cell.
Mitochondrial autophagy, or mitophagy, is a specific form of autophagy for the selective clearance of damaged mitochondria51. In depolarized mitochondria, the degradation of PINK1 (PTEN-induced putative kinase 1) is impaired, leading to the accumulation and activation of this kinase on the mitochondrial outer membrane (MOM)52–55. PINK1 phosphorylates ubiquitin chains, which leads to the recruitment of Parkin to the mitochondria and the activation of its E3 ligase activity. Mitochondrial proteins are then poly-ubiquitinated, and these are recognized by autophagy receptors to initiate autophagosomes formation52–55. The difference in the effects observed after chronic deletion or acute knockdown of Parkin implies that adaptive mechanisms for mitophagy cannot be established after acute silencing of Parkin (and/or Pink1) genes56,57. The impact of Parkin/Pink1-knockdown has therefore been explored in the context of HSC division patterns, which have demonstrated that enhanced clearance of damaged mitochondria by FAO is a key mechanism of the self-renewing expansion of Tie2+ HSCs (Fig. 3A)46
Metabolic control in HSC homeostasis
Mitochondria are bioenergetic and biosynthetic organelles that synthesize lipids and heme, as well as iron-sulfur clusters, amino acids, and nucleotides, and play important roles in HSC homeostasis (Fig. 1)28. HSCs exhibit much lower baseline and maximal respiration than progenitor cells, even though different levels of mitochondrial content, as measured by staining from targeted fluorescent protein, have been reported due to dye flux by xenobiotic efflux pumps58–63. Enhanced respiration is nevertheless detrimental to HSC maintenance and function64–68; for example, loss of mitochondrial carrier homologue 2 (MTCH2) increases mitochondrial respiration and intracellular ROS, triggering HSC entry into the cell cycle and compromising self-renewal capacity69. In contrast, lowering mitochondrial activity by chemical mitochondrial uncoupler supports sustained repopulation capacity under culture62. The defects in cell cycle quiescence and repopulation capacity observed in HSCs with impaired HIF (hypoxia-inducible factor) - PDK (Pyruvate dehydrogenase kinase) pathways are accompanied by enhanced flux of glycolytic metabolisms in the mitochondria during the TCA (tricarboxylic acid) cycle60,70. Further, deletion of Sirtuin 7 (Sirt7) increases mitochondrial unfolded protein stress, as well as mitochondrial biogenesis and respiration, leading to impaired regenerative capacity with a loss of quiescence and a shift in metabolic process that signals cellular differentiation71,72. When HSCs differentiate, they exit from quiescence and undergo a metabolic switch to mitochondrial Oxphos. Indeed, disrupting mitochondrial Oxphos upon the loss of Ptpmt1, a mitochondrial phosphatase targeting phosphatidylinositol phosphates, blocks early HSC differentiation and results in rapid hematopoietic failure in vivo73.
The hypoxic condition has been shown to be critical to the maintenance of selfrenewal, while stress factors (e.g., infection or polyinosinic-polycytidylic acid, granulocyte-colony stimulating factor, or chronic blood loss) are now known to induce HSC cycling19,74,75. This entry into the cell cycle is associated with DNA replication, upregulated energy production via oxidative phosphorylation (Oxphos), and elevated levels of intracellular reactive oxygen species (ROS). As quiescent HSCs are generally sensitive to increased intracellular ROS, the DNA damage that accumulates with repeated cell divisions leads to reduced self-renewal capacity and, ultimately, HSC exhaustion26,27,76–82.
Autophagy in hematopoiesis and HSC aging
Recent studies from multiple groups have also shown that macroautophagy (hereafter called simply autophagy)83–85 has an indirect but significant effect on HSC metabolism. Self-renewing stem cells, particularly in tissues with high cellular turnover such as the blood, counterbalance an array of stresses. HSCs in particular may combat stresses to maintain life-long hematopoiesis, and therefore the repair or clearance of mitochondrial damage is supported by a range of mechanisms that are critical to their function. Autophagy is a lysosomal degradation pathway which maintains the quantity and quality of organelles and proteins by degrading them once damaged or unwanted83–85. The autophagy-related (Atg) conjugation systems, which contribute to the formation of double-membraned autophagosomes, are another crucial element in the proper regulation of autophagy to ensure mitochondrial maintenance. The Forkhead Box O 3a (FOXO3A)-driven pro-autophagy gene program is known to protect HSCs from metabolic stress86, and a small-molecule inducer of autophagy has been shown to stimulate erythropoiesis87,88. The failure of this coordinated regulation can have a profound impact, as impaired autophagy has been shown to result in HSC exhaustion, and conditional depletion of Atg7 can lead to lethal anemia89,90.
More recently, the analysis of the roles of autophagy in the hematopoietic system has extended to the context of the aging. One third of HSCs from older mice exhibit high levels of autophagy activity, and these HSCs show higher repopulation capacity. Defective autophagy by the ablation of Atg12 accelerates blood aging phenotypes, with myeloid-biased lineage distribution and elevated Oxphos. The unselective removal of “active and healthy” mitochondria by autophagy contributes to reducing oxidative metabolism, which is essential for maintaining replicative quiescence in HSCs (Fig. 3C)58.
Enhanced mitophagy in hematopoiesis
The impact of excessive mitophagy on hematopoiesis has also been explored (although not in purified HSC populations)91. Atad3a, or ATPase family AAA domain-containing protein 3a, facilitates the transportation of Pink1 from the translocase of the outer membrane (TOM) complex to the translocase of the inner membrane (TIM) complex. In healthy mitochondria, Pink1 is rapidly degraded after its import by mitochondria peptidases. Conditional deletion of Atad3a in adult hematopoietic cells leads to the accumulation of Pink1 and the enhancement of mitophagy. Atad3a conditional knockout mice exhibited blocked hematopoietic lineage commitment at the progenitor stage, and enlarged HSPC pools. Ablation of Pink1 in these mice rescued defective mitophagy, which was in turn associated with the rescue of some defective hematopoietic phenotypes found in Atad3a-deficient mice (Fig. 3C)91. Interestingly, high mitochondrial turnover capacity was found in the progenitor stages, and both defective and enhanced mitophagy led to blocked erythoid differentiation at terminal erythrocyte maturation and erythroid progenitor differentiation, respectively89,91,92. Although the contributions of autophagy at different hematopoietic stages remain to be clarified, these studies collectively demonstrate that mitophagy must be precisely controlled to ensure maintenance of HSPCs and their appropriate differentiation.
Key open questions
Beyond generating ATP for cellular energy, mitochondria are required for mtDNA maintenance and intracellular calcium homeostasis, produce key metabolites that are utilized to synthesize macromolecules (e.g., lipids and nucleotides), and function as signaling organelles (e.g. for apoptosis)93–97. They are also known to form networks, and can change shape through the combined actions of fission, fusion, and movement along cytoskeletal tracks. These dynamics likely affect cell fate choice through multiple mechanisms, but we are only beginning to understand the mitochondrial requirements for stemness. Indeed, recent studies have shown that the Prdm16 (PR domain containing 16) - Mitofusin-2 (Mfn2) axis contributes to the maintenance of HSCs with lymphoid potential by buffering calcium levels through mitochondrial tethering to the endoplasmic reticulum98,99. In addition, intact mitochondrial function for HSC maintenance may require metabolism-driven epigenetic changes or code100–104.
Autophagy (or macroautophagy) was originally characterized as a non-selective bulk degradative system; however, it has been shown that under certain conditions, autophagosomes engulf cytosolic materials selectively, and diverse autophagy pathways have been identified105. Whether selective autophagy (e.g. pexophagy, glycophagy, SQSTM1-related autophagy) or other forms of autophagy (microautophagy and chaperone-mediated autophagy) participate in HSC homeostasis remains to be determined, but it will be interesting to explore how the controlled turnover of macromolecular components and nutrients (e.g. amino acids, metals, lipids etc) by autophagy contributes to the self-renewal capacity of HSCs; it is already clear that specific autophagy activity is required at various periods of life (e.g. developmental, perinatal young and adult hematopoiesis, as well as blood aging)28,58,89. A new method of assessing the dynamic content of autophagosomes, combined with genetic approaches to elucidating the selective forms of autophagy, will enrich our understanding of the roles of autophagy in the precise control of HSC fate decisions. Another open question of high importance is how the quantitative balance between selective autophagy and other catabolic pathways is controlled, as in the case of depolarized mitochondria, which are specifically degraded by Parkin-mediated mitophagy but might also be removed by bulk nonselective autophagy. Analysis of how each pathway is quantitatively regulated and the detailed contributions of mitophagy to the physiological aging of HSCs await future investigation.
Technical challenges to study HSC division balance
Our limited knowledge of division symmetry in HSCs and progenitor cells has so far come almost exclusively from in vitro studies15,33,34,106,107; virtually nothing has been observed in vivo. Yet in vivo HSC behavior certainly differs from ex vivo, and it has been shown that cellular metabolism can be extrinsically modulated. A complete model of the bone marrow environment in vitro (i.e. oxygen levels, cell-cell interactions, cellular components of the niche, cytokines and buffer milieu) has not yet been achieved21,108, and it is known that the metabolic modes of HSCs are dramatically changed once cells are placed ex vivo: for instance, HSCs are known to adapt their mitochondrial metabolism in the hypoxic niche70,109–111. When bone marrow is harvested and maintained in a hypoxic environment, greater numbers of phenotypically-defined HSCs can be obtained than can be collected in ambient air, but this beneficial effect is lost rapidly (in as little as 30 min) after exposure to normoxia112,113. Thus the key metabolic pathways obtained from in vitro assays cannot reflect in vivo functional states. The development of new platforms to assess the division balance of single HSCs in vivo will provide a deeper understanding of both the metabolic and molecular basis of HSC fate decisions in vivo.
Reporter systems are powerful tools for the characterization of fundamental HSC properties in vivo, with the functionality of the labeled cells validated retrospectively by clonal assays after single-cell transplantation. Theories differ regarding the contributions of HSCs to unperturbed homeostasis vs. tissue recovery conditions7,8,11, and technical considerations may influence conclusions derived from transplantation experiments; nevertheless, several studies have described murine and human HSCs as the major contributors to multi-lineage hematopoiesis both in the steady state and during cytokine response114,115. Notably, phenotypic HSCs comprise a major source of the megakaryocyte/platelet lineage in steady state conditions11,35, but these cells show “multi”-lineage differentiation capacity once they are transplanted into irradiated recipient mice11. These data imply potential differences in fate decision mechanisms between steady state and hematopoietic recovery. Perhaps most importantly in terms of our understanding of HSC metabolism, myeloablative preconditioning, such as irradiation and high-dose chemotherapy, is commonly used to create space in the niche for HSC engraftment12–14, but also severely alters the levels of ROS and other metabolic regulators, as well as the bone marrow microenvironment116. These genotoxic effects remain a substantial barrier to further clinical translation of this approach, and have raised concerns about whether transplantation results accurately reflect the true situation of the physiological metabolic mode of HSCs. A non-genotoxic method has long been sought as an alternative to current regimens, especially in the treatment of non-malignant blood diseases, and these efforts have met with some success at the pre-clinical stage116–118. New in vivo genetic tools are being developed to assess hematopoiesis with three- or even five-blood lineage resolution11,35,115, and in light of these advances, the technical challenges of exploring native HSC fate decisions will remain critical to future research.
Other recent studies have proposed an additional differentiation model in which HSCs can directly differentiate into lineage-restricted progenitors while bypassing the multipotent progenitor stage during acute conditions that demand the rapid replenishment of mature cells (e.g., respond to ablation stress)11,33,35. These findings suggest another possibility in which, as is often the case with other cell types, the first HSC divides symmetrically, and then one of its daughter cells stochastically loses its sternness (for instance, through the availability of niche positions or interaction with cytokines), which yields two daughter cells with distinct fates: one HSC and one differentiated hematopoietic cell29. The establishment of assay systems in which realtime markers are associated with HSC-specific functions (e.g., repopulation capacity) will enable researchers to accurately assess the division patterns of HSCs by prospectively tracking their division patterns. This development will be a breakthrough in identifying the key regulatory machineries of HSC fate decisions, and will significantly improve our understanding of the fundamental properties of HSCs.
Conclusion and Perspectives
HSC fate control is certain to be a central focus of ongoing research, and it is thus essential to expand our knowledge of both the mitochondrial and molecular basis of HSC fate decisions. The metabolic comparison between SD and other division modes, and the subsequent identification of specific metabolites as HSC fate-determinant, will be of particularly high interest because inducing SD may prove key to therapeutic applications for transplantation cases in which HSC expansion ex vivo is required with a limited number of donor cells. Better understanding of the molecular mechanisms and cross-links between all three division options will make possible the manipulation of HSC cell fate decisions. As the rarity of HSCs is a major hurdle for metabolic or epigenetic studies that depend on purified HSC populations, novel metabolomics and epigenomics approaches adapted to small numbers of HSCs certainly bear further exploration.
A disturbed division balance causes hematological disorders119,120, and the longterm survival rate among blood cancer patients remains stubbornly low, as most patients who have achieved remission eventually relapse. Leukemia stem cells (LSCs, also known as leukemia-initiating cells) are believed to not only drive disease initiation, progression, and drug resistance, but also contribute to relapse119–125. Elimination of every single LSC is therefore essential to a long-term cure. Upon division, LSCs can either self-renew or commit to differentiation, and shifting their division balance away from renewal and toward commitment holds great promise as a therapeutic strategy126,127. It is no surprise that the metabolic requirements of leukemogenesis and LSC function have therefore become a focus of much current research101,102,104,128–131, and the discovery of contributions to leukemogenesis by metabolism, mitochondrial biogenesis, and cytoprotective autophagy support the notion that mitochondrial quality control by autophagy may be a key determinant of division balance132. Tracking the division pattern of individual LSCs has, however, proved challenging, and the development of new techniques of single LSC assay is critical to achieving a better understanding of the molecular basis of LSC fate choice133,134. As the many metabolic pathways involved are conserved in human hematopoiesis, identifying the key metabolic cues that precisely control LSC fate and maintain stem-ness upon division could provide effective targets in strategies to enhance LSC commitment, and will therefore be of high clinical importance.
Highlights.
Specific modes of metabolism play important roles in HSC self-renewal.
Heterogeneity and technical challenges prevented the elucidation of HSC behavior.
Recent advances highlighted mitochondrial quality control as a key HSC fate factor.
A deeper understanding of HSC fate via metabolic control has clinical implications.
Acknowledgments
We are grateful to members of the Ito lab and Einstein Stem Cell Institute as well as T. Suda for their comments on HSC self-renewal. K.I. is supported by grants from the National Institutes of Health (R01DK98263, R01DK115577, and R01DK100689) and New York State Department of Health as Core Director of Einstein Single-Cell Genomics/Epigenomics (C029154). We apologize to the investigators whose work could not be cited owing to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference:
- 1.McCulloch EA & Till JE The radiation sensitivity of normal mouse bone marrow cells, determined by quantitative marrow transplantation into irradiated mice. Radiat Res 13, 115–125 (1960). [PubMed] [Google Scholar]
- 2.Weissman IL, Anderson DJ & Gage F Stem and progenitor cells: origins, phenotypes, lineage commitments, and transdifferentiations. Annu Rev Cell Dev Biol 17, 387–403, doi: 10.1146/annurev.cellbio.17.1.387 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Visvader JE & Clevers H Tissue-specific designs of stem cell hierarchies. Nat Cell Biol 18, 349–355, doi: 10.1038/ncb3332 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Morrison SJ, Shah NM & Anderson DJ Regulatory mechanisms in stem cell biology. Cell 88, 287–298 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Ramalho-Santos M & Willenbring H On the origin of the term “stem cell”. Cell Stem Cell 1, 35–38, doi: 10.1016/j.stem.2007.05.013 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Daley GQ, Goodell MA & Snyder EY Realistic prospects for stem cell therapeutics. Hematology Am Soc Hematol Educ Program, 398–418 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Sun J et al. Clonal dynamics of native haematopoiesis. Nature 514, 322–327, doi: 10.1038/nature13824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busch K et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518, 542–546, doi: 10.1038/nature14242 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Sawai CM et al. Hematopoietic Stem Cells Are the Major Source of Multilineage Hematopoiesis in Adult Animals. Immunity 45, 597–609, doi: 10.1016/j.immuni.2016.08.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito K & Frenette PS HSC Contribution in Making Steady-State Blood. Immunity 45, 464–466, doi: 10.1016/j.immuni.2016.09.002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez-Fraticelli AE et al. Clonal analysis of lineage fate in native haematopoiesis. Nature 553, 212–216, doi: 10.1038/nature25168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Appelbaum FR Hematopoietic-cell transplantation at 50. N Engl J Med 357, 1472–1475, doi: 10.1056/NEJMp078166 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Copelan EA Hematopoietic stem-cell transplantation. N Engl J Med 354, 1813–1826, doi: 10.1056/NEJMra052638 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Stranges E, Russo CA & Friedman B in Healthcare Cost and Utilization Project (HCUP) Statistical Briefs (2006). [PubMed]
- 15.Ito K et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nature medicine 18, 1350–1358, doi: 10.1038/nm.2882 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson A et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129, doi: 10.1016/j.cell.2008.10.048 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Cheng T et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 287, 1804–1808 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Pietras EM, Warr MR & Passegue E Cell cycle regulation in hematopoietic stem cells. The Journal of cell biology 195, 709–720, doi: 10.1083/jcb.201102131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trumpp A, Essers M & Wilson A Awakening dormant haematopoietic stem cells. Nature reviews. Immunology 10, 201–209, doi: 10.1038/nri2726 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Nakamura-Ishizu A, Takizawa H & Suda T The analysis, roles and regulation of quiescence in hematopoietic stem cells. Development 141, 4656–4666, doi: 10.1242/dev.106575 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Morrison SJ & Scadden DT The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334, doi: 10.1038/nature12984 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zon LI Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature 453, 306–313, doi: 10.1038/nature07038 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Frenette PS, Pinho S, Lucas D & Scheiermann C Mesenchymal stem cell: keystone of the hematopoietic stem cell niche and a stepping-stone for regenerative medicine. Annual review of immunology 31, 285–316, doi: 10.1146/annurev-immunol-032712-095919 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Scadden DT The stem-cell niche as an entity of action. Nature 441, 1075–1079, doi: 10.1038/nature04957 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Watt FM & Hogan BL Out of Eden: stem cells and their niches. Science 287, 1427–1430 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Ito K & Suda T Metabolic requirements for the maintenance of self-renewing stem cells. Nature reviews. Molecular cell biology 15, 243.-, doi: 10.1038/nrm3772 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shyh-Chang N, Daley GQ & Cantley LC Stem cell metabolism in tissue development and aging. Development 140, 2535–2547, doi: 10.1242/dev.091777 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandel NS, Jasper H, Ho TT & Passegue E Metabolic regulation of stem cell function in tissue homeostasis and organismal ageing. Nat Cell Biol 18, 823–832, doi: 10.1038/ncb3385 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Ito K & Ito K Metabolism and the Control of Cell Fate Decisions and Stem Cell Renewal. Annu Rev Cell Dev Biol 32, 399–409, doi: 10.1146/annurev-cellbio-111315-125134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilkinson AC & Yamazaki S The hematopoietic stem cell diet. Int J Hematol, doi: 10.1007/s12185-018-2451-1 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Anso E et al. The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat Cell Biol 19, 614–625, doi: 10.1038/ncb3529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cabezas-Wallscheid N et al. Vitamin A-Retinoic Acid Signaling Regulates Hematopoietic Stem Cell Dormancy. Cell 169, 807–823 e819, doi: 10.1016/j.cell.2017.04.018 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto R et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell 154, 1112–1126, doi: 10.1016/j.cell.2013.08.007 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Suda T, Suda J & Ogawa M Disparate differentiation in mouse hemopoietic colonies derived from paired progenitors. Proc Natl Acad Sci U S A 81, 2520–2524 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto R et al. Large-Scale Clonal Analysis Resolves Aging of the Mouse Hematopoietic Stem Cell Compartment. Cell Stem Cell 22, 600–607 e604, doi: 10.1016/j.stem.2018.03.013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osawa M, Hanada K, Hamada H & Nakauchi H Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 273, 242–245 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Benveniste P, Cantin C, Hyam D & Iscove NN Hematopoietic stem cells engraft in mice with absolute efficiency. Nat Immunol 4, 708–713, doi: 10.1038/ni940 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Kiel MJ et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121, doi: 10.1016/j.cell.2005.05.026 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Dykstra B et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell 1, 218–229, doi: 10.1016/j.stem.2007.05.015 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Gazit R et al. Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J Exp Med 211, 1315–1331, doi: 10.1084/jem.20130428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanjuan-Pla A et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature 502, 232–236, doi: 10.1038/nature12495 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Acar M et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526, 126–130, doi: 10.1038/nature15250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Challen GA, Boles NC, Chambers SM & Goodell MA Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell 6, 265–278, doi: 10.1016/j.stem.2010.02.002 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen JY et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature 530, 223–227, doi: 10.1038/nature16943 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carrelha J et al. Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature 554, 106–111, doi: 10.1038/nature25455 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Ito K et al. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science 354, 1156–1160, doi: 10.1126/science.aaf5530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernitz JM, Kim HS, MacArthur B, Sieburg H & Moore K Hematopoietic Stem Cells Count and Remember Self-Renewal Divisions. Cell 167, 1296–1309 e1210, doi: 10.1016/j.cell.2016.10.022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turcotte R et al. Image-guided transplantation of single cells in the bone marrow of live animals. Sci Rep 7, 3875, doi: 10.1038/s41598-017-02896-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kunau WH, Dommes V & Schulz H beta-oxidation of fatty acids in mitochondria, peroxisomes, and bacteria: a century of continued progress. Prog Lipid Res 34, 267–342 (1995). [DOI] [PubMed] [Google Scholar]
- 50.Katajisto P et al. Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science 348, 340–343, doi: 10.1126/science.1260384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Youle RJ & Narendra DP Mechanisms of mitophagy. Nat Rev Mol Cell Biol 12, 9–14, doi: 10.1038/nrm3028 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stolz A & Dikic I PINK1-PARKIN interplay: down to ubiquitin phosphorylation. Mol Cell 56, 341–342, doi: 10.1016/j.molcel.2014.10.022 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Sekine S & Youle RJ PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biol 16, 2, doi: 10.1186/s12915-017-0470-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lazarou M et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314, doi: 10.1038/nature14893 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okatsu K et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells 15, 887–900, doi: 10.1111/j.1365-2443.2010.01426.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams JA et al. Chronic Deletion and Acute Knockdown of Parkin Have Differential Responses to Acetaminophen-induced Mitophagy and Liver Injury in Mice. The Journal of biological chemistry 290, 10934–10946, doi: 10.1074/jbc.M114.602284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dawson TM, Ko HS & Dawson VL Genetic animal models of Parkinson’s disease. Neuron 66, 646–661, doi: 10.1016/j.neuron.2010.04.034 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ho TT et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature 543, 205–210, doi: 10.1038/nature21388 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simsek T et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7, 380–390, doi: 10.1016/j.stem.2010.07.011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takubo K et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12, 49–61, doi: 10.1016/j.stem.2012.10.011 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Almeida MJ, Luchsinger LL, Corrigan DJ, Williams LJ & Snoeck HW Dye-Independent Methods Reveal Elevated Mitochondrial Mass in Hematopoietic Stem Cells. Cell Stem Cell 21, 725–729 e724, doi: 10.1016/j.stem.2017.11.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vannini N et al. Specification of haematopoietic stem cell fate via modulation of mitochondrial activity. Nat Commun 7, 13125, doi: 10.1038/ncomms13125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Romero-Moya D et al. Cord blood-derived CD34+ hematopoietic cells with low mitochondrial mass are enriched in hematopoietic repopulating stem cell function. Haematologica 98, 1022–1029, doi: 10.3324/haematol.2012.079244 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gan B et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature 468, 701–704, doi: 10.1038/nature09595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gurumurthy S et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature 468, 659–663, doi: 10.1038/nature09572 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakada D, Saunders TL & Morrison SJ Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature 468, 653–658, doi: 10.1038/nature09571 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen C et al. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med 205, 2397–2408, doi: 10.1084/jem.20081297 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiao N et al. Hematopoietic stem cells lacking Ott1 display aspects associated with aging and are unable to maintain quiescence during proliferative stress. Blood 119, 4898–4907, doi: 10.1182/blood-2012-01-403089 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maryanovich M et al. An MTCH2 pathway repressing mitochondria metabolism regulates haematopoietic stem cell fate. Nat Commun 6, 7901, doi: 10.1038/ncomms8901 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Suda T, Takubo K & Semenza GL Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 9, 298–310, doi: 10.1016/j.stem.2011.09.010 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Mohrin M et al. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 347, 1374–1377, doi: 10.1126/science.aaa2361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mohrin M, Widjaja A, Liu Y, Luo H & Chen D The mitochondrial unfolded protein response is activated upon hematopoietic stem cell exit from quiescence. Aging Cell, e12756, doi: 10.1111/acel.12756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu WM et al. Metabolic regulation by the mitochondrial phosphatase PTPMT1 is required for hematopoietic stem cell differentiation. Cell Stem Cell 12, 62–74, doi: 10.1016/j.stem.2012.11.022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Essers MA et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 458, 904–908, doi: 10.1038/nature07815 (2009). [DOI] [PubMed] [Google Scholar]
- 75.Walter D et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature 520, 549–552, doi: 10.1038/nature14131 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Rossi DJ, Jamieson CH & Weissman IL Stems cells and the pathways to aging and cancer. Cell 132, 681–696, doi: 10.1016/j.cell.2008.01.036 (2008). [DOI] [PubMed] [Google Scholar]
- 77.Ito K et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nature medicine 12, 446–451, doi: 10.1038/nm1388 (2006). [DOI] [PubMed] [Google Scholar]
- 78.Ito K et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature 431, 997–1002, doi: 10.1038/nature02989 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Maryanovich M & Gross A A ROS rheostat for cell fate regulation. Trends in cell biology 23, 129–134, doi: 10.1016/j.tcb.2012.09.007 (2013). [DOI] [PubMed] [Google Scholar]
- 80.Miyamoto K et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell stem cell 1, 101–112, doi: 10.1016/j.stem.2007.02.001 (2007). [DOI] [PubMed] [Google Scholar]
- 81.Liang R & Ghaffari S Mitochondria and FOXO3 in stem cell homeostasis, a window into hematopoietic stem cell fate determination. J Bioenerg Biomembr 49, 343–346, doi: 10.1007/s10863-017-9719-7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Testa U, Labbaye C, Castelli G & Pelosi E Oxidative stress and hypoxia in normal and leukemic stem cells. Exp Hematol 44, 540–560, doi: 10.1016/j.exphem.2016.04.012 (2016). [DOI] [PubMed] [Google Scholar]
- 83.He C & Klionsky DJ Regulation mechanisms and signaling pathways of autophagy. Annual review of genetics 43, 67–93, doi: 10.1146/annurev-genet-102808-114910 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Galluzzi L, Pietrocola F, Levine B & Kroemer G Metabolic control of autophagy. Cell 159, 1263–1276, doi: 10.1016/j.cell.2014.11.006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ueno T & Komatsu M Autophagy in the liver: functions in health and disease. Nat Rev Gastroenterol Hepatol 14, 170–184, doi: 10.1038/nrgastro.2016.185 (2017). [DOI] [PubMed] [Google Scholar]
- 86.Warr MR et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 494, 323–327, doi: 10.1038/nature11895 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Doulatov S et al. Drug discovery for Diamond-Blackfan anemia using reprogrammed hematopoietic progenitors. Sci Transl Med 9, doi: 10.1126/scitranslmed.aah5645 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu F et al. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood 116, 4806–4814, doi: 10.1182/blood-2010-06-288589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mortensen M et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci U S A 107, 832–837, doi: 10.1073/pnas.0913170107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Riffelmacher T & Simon AK Mechanistic roles of autophagy in hematopoietic differentiation. FEBS J 284, 1008–1020, doi: 10.1111/febs.13962 (2017). [DOI] [PubMed] [Google Scholar]
- 91.Jin G et al. Atad3a suppresses Pink1-dependent mitophagy to maintain homeostasis of hematopoietic progenitor cells. Nat Immunol 19, 29–40, doi: 10.1038/s41590-017-0002-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sandoval H et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature 454, 232–235, doi: 10.1038/nature07006 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vander Heiden MG, Cantley LC & Thompson CB Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033, doi: 10.1126/science.1160809 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chandel NS Evolution of Mitochondria as Signaling Organelles. Cell Metab 22, 204–206, doi: 10.1016/j.cmet.2015.05.013 (2015). [DOI] [PubMed] [Google Scholar]
- 95.Rizzuto R, De Stefani D, Raffaello A & Mammucari C Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol 13, 566–578, doi: 10.1038/nrm3412 (2012). [DOI] [PubMed] [Google Scholar]
- 96.Youle RJ & van der Bliek AM Mitochondrial fission, fusion, and stress. Science 337, 1062–1065, doi: 10.1126/science.1219855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen H & Chan DC Mitochondrial Dynamics in Regulating the Unique Phenotypes of Cancer and Stem Cells. Cell Metab 26, 39–48, doi: 10.1016/j.cmet.2017.05.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aguilo F et al. Prdm16 is a physiologic regulator of hematopoietic stem cells. Blood 117, 5057–5066, doi: 10.1182/blood-2010-08-300145 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Luchsinger LL, de Almeida MJ, Corrigan DJ, Mumau M & Snoeck HW Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature 529, 528–531, doi: 10.1038/nature16500 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Raffel S et al. BCAT1 restricts alphaKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature 551, 384–388, doi: 10.1038/nature24294 (2017). [DOI] [PubMed] [Google Scholar]
- 101.Tefferi A et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia 24, 1302–1309, doi: 10.1038/leu.2010.113 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Figueroa ME et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567, doi: 10.1016/j.ccr.2010.11.015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Agathocleous M et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549, 476–481, doi: 10.1038/nature23876 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cimmino L et al. Restoration of TET2 Function Blocks Aberrant Self-Renewal and Leukemia Progression. Cell 170, 1079–1095 e1020, doi: 10.1016/j.cell.2017.07.032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mizushima N, Levine B, Cuervo AM & Klionsky DJ Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075, doi: 10.1038/nature06639 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu M et al. Imaging hematopoietic precursor division in real time. Cell Stem Cell 1, 541–554, doi: 10.1016/j.stem.2007.08.009 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hoppe PS et al. Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature 535, 299–302, doi: 10.1038/nature18320 (2016). [DOI] [PubMed] [Google Scholar]
- 108.Passaro D, Abarrategi A, Foster K, Ariza-McNaughton L & Bonnet D Bioengineering of Humanized Bone Marrow Microenvironments in Mouse and Their Visualization by Live Imaging. J Vis Exp, doi: 10.3791/55914 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Spencer JA et al. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature 508, 269–273, doi: 10.1038/nature13034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nombela-Arrieta C et al. Quantitative imaging of haematopoietic stem and progenitor cell localization and hypoxic status in the bone marrow microenvironment. Nat Cell Biol 15, 533–543, doi: 10.1038/ncb2730 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Piccoli C et al. To breathe or not to breathe: the haematopoietic stem/progenitor cells dilemma. Br J Pharmacol 169, 1652–1671, doi: 10.1111/bph.12253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mantel CR et al. Enhancing Hematopoietic Stem Cell Transplantation Efficacy by Mitigating Oxygen Shock. Cell 161, 1553–1565, doi: 10.1016/j.cell.2015.04.054 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sjostedt S, Rooth G & Caligara F The Oxygen Tension of the Blood in the Umbilical Cord and the Intervillous Space. Arch Dis Child 35, 529–533 (1960). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sawai CM et al. Hematopoietic Stem Cells Are the Major Source of Multilineage Hematopoiesis in Adult Animals. Immunity 45, 597–609, doi: 10.1016/j.immuni.2016.08.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Upadhaya S, Reizis B & Sawai CM New genetic tools for the in vivo study of hematopoietic stem cell function. Exp Hematol 61, 26–35, doi: 10.1016/j.exphem.2018.02.004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Palchaudhuri R et al. Non-genotoxic conditioning for hematopoietic stem cell transplantation using a hematopoietic-cell-specific internalizing immunotoxin. Nat Biotechnol 34, 738–745, doi: 10.1038/nbt.3584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Waskow C et al. Hematopoietic stem cell transplantation without irradiation. Nat Methods 6, 267–269, doi: 10.1038/nmeth.1309 (2009). [DOI] [PubMed] [Google Scholar]
- 118.Taya Y et al. Depleting dietary valine permits nonmyeloablative mouse hematopoietic stem cell transplantation. Science 354, 1152–1155, doi: 10.1126/science.aag3145 (2016). [DOI] [PubMed] [Google Scholar]
- 119.Zimdahl B et al. Lis1 regulates asymmetric division in hematopoietic stem cells and in leukemia. Nat Genet 46, 245–252, doi: 10.1038/ng.2889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ito T et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 466, 765–768, doi: 10.1038/nature09171 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Corces MR, Chang HY & Majeti R Preleukemic Hematopoietic Stem Cells in Human Acute Myeloid Leukemia. Front Oncol 7, 263, doi: 10.3389/fonc.2017.00263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sarkozy C et al. Outcome of older patients with acute myeloid leukemia in first relapse. Am J Hematol 88, 758–764, doi: 10.1002/ajh.23498 (2013). [DOI] [PubMed] [Google Scholar]
- 123.Bonnet D & Dick JE Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3, 730–737 (1997). [DOI] [PubMed] [Google Scholar]
- 124.Huntly BJ & Gilliland DG Cancer biology: summing up cancer stem cells. Nature 435, 1169–1170, doi: 10.1038/4351169a (2005). [DOI] [PubMed] [Google Scholar]
- 125.Shlush LI et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature 547, 104–108, doi: 10.1038/nature22993 (2017). [DOI] [PubMed] [Google Scholar]
- 126.Morrison SJ & Kimble J Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441, 1068–1074, doi: 10.1038/nature04956 (2006). [DOI] [PubMed] [Google Scholar]
- 127.Kharas MG et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med 16, 903–908, doi: 10.1038/nm.2187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ward PS et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234, doi: 10.1016/j.ccr.2010.01.020 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Agathocleous M et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature, doi: 10.1038/nature23876 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jiang Y & Nakada D Cell intrinsic and extrinsic regulation of leukemia cell metabolism. Int J Hematol 103, 607–616, doi: 10.1007/s12185-016-1958-6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kuntz EM et al. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat Med 23, 1234–1240, doi: 10.1038/nm.4399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sumitomo Y et al. Cytoprotective autophagy maintains leukemia-initiating cells in murine myeloid leukemia. Blood 128, 1614–1624, doi: 10.1182/blood-2015-12-684696 (2016). [DOI] [PubMed] [Google Scholar]
- 133.Duarte D et al. Inhibition of Endosteal Vascular Niche Remodeling Rescues Hematopoietic Stem Cell Loss in AML. Cell Stem Cell 22, 64–77 e66, doi: 10.1016/j.stem.2017.11.006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hawkins ED et al. T-cell acute leukaemia exhibits dynamic interactions with bone marrow microenvironments. Nature 538, 518–522, doi: 10.1038/nature19801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]


