Abstract
A fundamental goal in biology is to understand how distinct cell types containing the same genetic information arise from a single stem cell throughout development. Sex determination is a key developmental process that requires a unidirectional commitment of an initially bipotential gonad towards either the male or female fate. This makes sex determination a unique model to study cell fate commitment and differentiation in vivo. We have focused this review on the accumulating evidence that epigenetic mechanisms contribute to the bipotential state of the fetal gonad and to the regulation of chromatin accessibility during and immediately downstream of the primary sex-determining switch that establishes the male fate.
Keywords: Epigenetics, Sex determination, Cell fate commitment, Plasticity, Development, Gonad
1. Introduction
Vertebrates share a common template to achieve development of two distinct sexes. Initially, male and female embryos are indistinguishable. During development, the embryonic gonad forms with the unique ability to differentiate into two alternative organs: testes (males) or ovaries (females) (Fig. 1A). Gonadal differentiation diverges based on a genetic or environmental switch that activates one pathway and represses the other. This system presents an interesting model to explore the various levels of regulation involved in fate commitment of single cells and the coordination of the entire cell community that makes up the bipotential gonad.
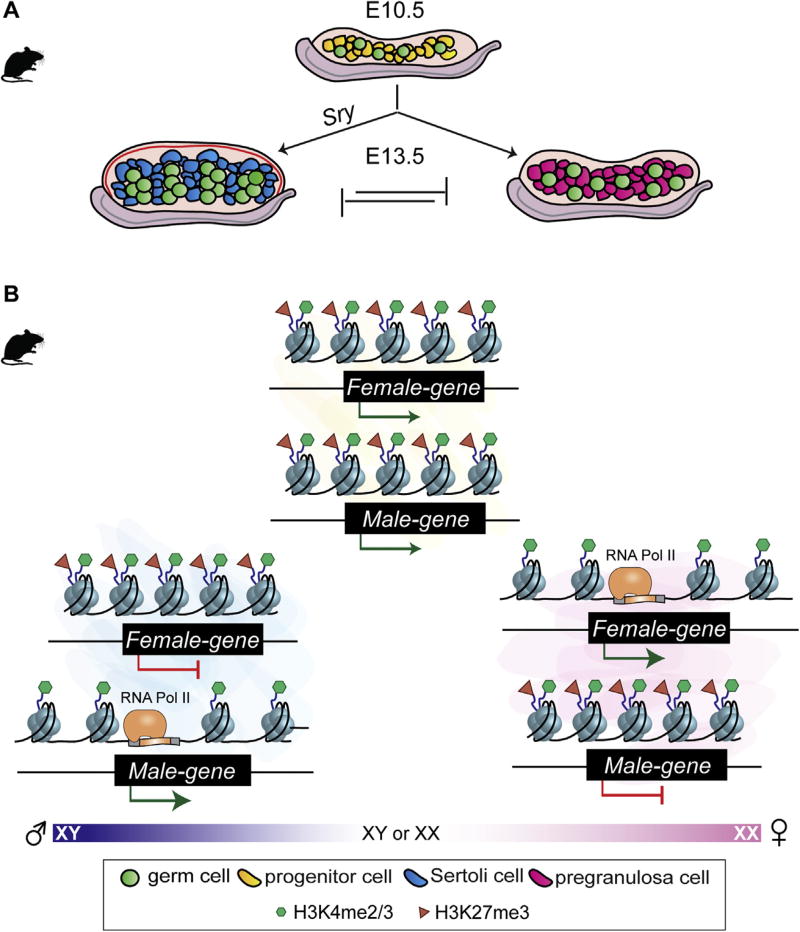
Fig. 1.
A) Overview of mammalian sex determination. In mice, gonads form at E10.5 and are indistinguishable between XY and XX individuals. The somatic progenitor cells that compose the fetal gonad (top) are initially bipotential. Following expression of the Y-encoded Sry gene at E11.5, progenitor cells differentiate into Sertoli cells and gonads develop as testes (left). In absence of Sry, progenitor cells differentiate into pregranulosa cells and gonads develop as ovaries (right). Commitment to the male or female fate requires repression of the alternate pathway. B) The chromatin landscape at the bipotential stage. Male- and female-determining genes are bivalent and co-expressed at low and similar levels in progenitor cells (top, yellow). Upregulation of male-determining genes leading to differentiation of Sertoli cells (left, blue) is accompanied by loss of the repressive H3K27me3 mark, whereas repressed female-determining genes remain bivalent. Upregulation of female-determining genes leading to differentiation of pregranulosa cells (right, pink) is accompanied by loss of the repressive H3K27me3 mark, whereas repressed male-determining genes remain bivalent.
One interesting characteristic of vertebrate sex determination is the plastic ability to switch between a male and female fate. For example, in most reptilian species, the incubation temperature of the egg controls the sex of the offspring, and switching eggs between temperatures during a critical window of development results in a switch to the opposite sex. Many fish can undergo sex reversal as adults, and although mammals do not undergo full sex reversal, the removal of certain key transcription factors can cause a switch in the identity of female cells to male cells (Ottolenghi et al., 2007; Uhlenhaut et al., 2009) or male cells to female cells (Matson et al., 2011). Importantly, gonadal cells do not switch randomly to another fate, but specifically to their developmental alternative fate.
Although multiple mechanisms are interwoven in the process of fate commitment and canalization as one sex or the other, epigenetic regulation is emerging as an important component. Epigenetics, the study of changes to gene function without changes in the DNA sequence, is capable of imposing a stable differentiation state throughout cell division. Between 1940 and 1956, Conrad Waddington proposed the concept of an epigenetic landscape to describe the process of cell fate commitment during development. He envisioned that a cell progresses towards a differentiated state through a series of fate decisions that are stabilized by changes to its epigenome. These changes maintain cell-type-specific gene expression patterns that channel the cell along certain pathways while eliminating other possible pathways the cell might take (Waddington, 1940, 1942, 1956). In this review, we will consider several lines of evidence in the field of vertebrate sex determination that suggest that epigenetic regulation is a key element in the process of commitment to and canalization of the male or female fate.
2. Are XX and XY chromatin landscapes created equal?
For many years the term “primary sex determination” has been used in vertebrates to refer to the fate decision in the bipotential gonad that leads to the commitment to testis or ovary fate. This is because (1) the first microscopic differences between male and female development occur in the gonad, and (2) once the gonad is committed to testis or ovary fate, the hormonal secretions of the testis or ovary are the dominant influence on male and female secondary sex characteristics.
However, for all heterogametic species, genetic sex (the presence of XX/XY or ZZ/ZW chromosomes) is established at fertilization long before the gonad forms, differentiates, and produces hormones. The presence of heteromorphic sex chromosomes leads to disparities in gene dosage between males and females. In flies and worms, the mechanisms that control dosage compensation are directly linked to sex determination (Cline and Meyer, 1996). In mammals, an XX/XY heterogametic species, the process of dosage compensation is not obviously linked to primary sex determination. To balance the difference in dosage of genes on the X between males and females, one X chromosome in females is inactivated in the late blastocyst stage through heterochromatin formation and spreading (Zhao et al., 2008; Pinter et al., 2012). At one time it was believed that X-inactivation silenced all genes on the inactive X. However, in recent years it has become clear that around 15% of X-linked genes in humans (Carrel and Willard, 2005) and 3.3% of X-linked genes in mice (Yang et al., 2010a) escape inactivation and are expressed at higher levels in females than in males. Conversely, males express Y-linked spermatogenesis genes that are not shared with females. Therefore, XX and XY cells harbor differences attributable to their sex chromosome complement. It is clear that if a testis is induced in an XX embryo, the embryo will develop as a phenotypic male despite the fact that the genetic sex of each cell is female (Koopman et al., 1991; Vidal et al., 2001; Qin et al., 2004; Polanco et al., 2010). This means that gonadal sex is dominant in mammalian systems. Nonetheless, the sex chromosome complement of vertebrate cells could influence their differentiation before or after steroid-producing gonads develop. In fact there is clear evidence in birds that this is the case (McQueen and Clinton, 2009).
Interestingly, a number of X escapees and Y-linked genes play crucial roles in chromatin regulation (Table 1). Chromatin is regulated at many levels by epigenetic mechanisms, such as DNA methylation and histone modifications that can alter the 3D organization of chromatin within the nucleus. Changes to chromatin structure ultimately modulate transcription factor accessibility to DNA binding sites, thereby regulating gene expression. Examples of Y-linked and X escapee genes in this category will be reviewed below.
Table 1.
Y-linked and X escapee chromatin modifiers.
| Gene | Molecular Function | References |
|---|---|---|
| Y-linked chromatin modifiers | ||
| Uty | H3K27me3-specific histone demethylase, interacts with RBBP5 and BRG1 (Shpargel et al., 2012) | Lee et al., 2007; Wang et al., 2013; Walport et al., 2014 |
| Jarid1d | H3K4-specific demethylase | Lee et al., 2007; Xu et al., 2008a |
| CDY (human) | H4-specific hyperacetylation, interacts with methylated H3K9 through its chromodomain, and recruits HDACs | Lahn et al., 2002; Kim et al., 2006a; Caron et al., 2003 |
| X escapee chromatin modifiers | ||
| Utx | H3K27me3-specific histone demethylase, interacts with RBBP5 and BRG1 | Hong et al., 2007; Xu et al., 2008b; Shpargel et al., 2012; Welstead et al., 2012; Wang et al., 2013 |
| Jarid1c | H3K4-specific demethylase | Iwase et al., 2007; Xu et al., 2008a |
| MeCP2 | Binds methylated CpGs, recruits HDACs, HP1, H3K9 methyltransferases, the DNA-demethylase MBD2, and the chromatin remodeler ATRX, and promotes CTCF binding and formation of higher-order chromatin loops | Hendrich and Bird, 1998; Nan et al., 1998; Agarwal et al., 2007; Fuks et al., 2003; Becker et al., 2013; Nan et al., 2007; Kernohan et al., 2014 |
2.1. Y-linked chromatin modifiers
The Y chromosome is the smallest chromosome and Y-encoded genes mostly function in male sex determination, testis development and fertility. Amongst the Y-encoded genes are a few chromatin modifiers: the histone demethylase Uty (ubiquitously-transcribed TPR gene on the Y chromosome), the H3K4-specific histone demethylase Jarid1d (Jumonji, AT-Rich Interactive Domain 1d), and in humans, the histone acetyltransferase CDY (Chromodomain protein Y-linked) (Dhanoa et al., 2016). Of these, Uty and Jarid1d have homologues on the X chromosome, and are reviewed in the following section.
CDY is exclusively expressed in the testes. The CDY family of proteins contains two conserved domains: a chromodomain, which interacts with the repressive H3K9me histone modification (Kim et al., 2006a), and a catalytic histone acetyltransferase domain, which has high affinity for H4 (Lahn et al., 2002). However, this catalytic domain has also been found to recruit histone deacetylases (HDACs) (Caron et al., 2003). As histone acetylation correlates with transcriptional activity, CDY may function as both a transcriptional activator and a repressor. It is speculated that CDY-mediated hyperacetylation of H4 may play an important role in facilitating the transition from histones to protamines, nuclear proteins that replace histones in sperm (Lahn et al., 2002). However, whether CDY can regulate transcription at a genome-wide level through its interaction with H3K9me and HDACs remains to be elucidated.
2.2. X escapee chromatin modifiers
Utx (ubiquitously-transcribed TPR gene on the X chromosome) is an X-linked gene that escapes X-inactivation in both mice and humans (Greenfield et al., 1998). This gene encodes a histone demethylase, which specifically removes the repressive histone modification H3K27me3 (Hong et al., 2007). Utx is critical for embryonic development, as Utx-null females die at midgestation (Shpargel et al., 2012; Welstead et al., 2012; Wang et al., 2013). Utx has a homolog on the Y chromosome, known as Uty. Although early in vitro studies showed that Uty was catalytically inactive (Lan et al., 2007), Utx-null males are viable and fertile (Shpargel et al., 2012; Welstead et al., 2012; Wang et al., 2013). Furthermore, Uty-null males phenocopy Utx-null males, suggesting that Utx and Uty are functionally redundant (Wang et al., 2013). Consistent with this interpretation, a more recent study found that human Uty is an active demethylase, but has reduced activity due to point mutations that affect substrate binding (Walport et al., 2014). Alternatively, UTX and UTY may function through H3K27me3-independent mechanisms as activators, by interacting with RBBP5, part of an H3K4me3-methyltransferase complex, and the SWI/SNF chromatin remodeler BRG1 (Shpargel et al., 2012). However, even though Utx and Uty may have similar functions, their expression levels and patterns differ in XX and XY embryos (Xu et al., 2008b), suggesting that they may regulate chromatin in a sex-specific manner.
Jarid1c (Jumonji, AT-Rich Interactive Domain 1c) is an X escapee that encodes an H3K4-specific histone demethylase that plays an important role in brain development and function. Jarid1c is more highly expressed in XX than XY mice, independent of whether an ovary or a testis is present (Xu et al., 2008a). Importantly, expression of its Y-linked homolog Jarid1d is unable to compensate for loss of Jarid1c despite its similar function, suggesting that sex-specific expression of this histone demethylase may contribute to differences in brain development and function such as neurite length and aggressive behavior (Xu et al., 2008a; Lee et al., 2007).
Another X escapee, MeCP2 (methyl CpG-binding protein 2), plays a crucial role in brain development. Although MECP2 is not itself a chromatin modifier, it has a high binding affinity to methylated CpGs and ability to interact with epigenetic machinery (Hendrich and Bird, 1998). Once bound, MECP2 can regulate chromatin structure by recruiting HDACS, heterochromatin protein 1 (HP1), H3K9 histone methyltransferases, the DNA demethylase MBP1, and the chromatin remodeler ATRX amongst others, and in one case was able to alter higher-order chromatin structure by inducing chromatin looping (Nan et al., 1998; Agarwal et al., 2007; Fuks et al., 2003; Becker et al., 2013; Nan et al., 2007; Kernohan et al., 2014).
Although there is currently no phenotypic evidence that these sex-linked chromatin modifiers directly regulate primary sex determination, sexually dimorphic expression of chromatin modifiers could directly affect the activity of many genes leading to sex-specific transcriptional programs. Whether XX and XY cells initiate development with different chromatin landscapes, and whether these differences have significant phenotypic consequences in fetal or adult life is an area of active research using models that can disentangle genetic and hormonal effects (Burgoyne et al., 1995; Arnold, 2009).
3. The chromatin landscape at the bipotential stage
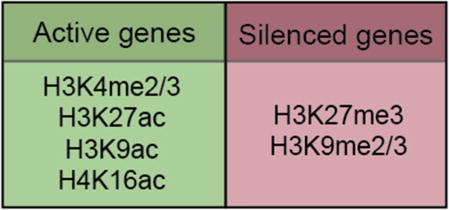
Despite differences in sex-chromosome complements, XX and XY fetal gonads are initially bipotential, and the somatic cells that compose the gonad have the full ability to differentiate into either Sertoli cells (males) or pregranulosa cells (females) (Fig. 1A) (Albrecht and Eicher, 2001). Consistent with their bipotential nature, both XX and XY progenitor cells co-express genes later associated with Sertoli- and granulosa-cell development (Nef et al., 2005; Munger et al., 2013). In other pluripotent systems, such as embryonic stem cells (ESCs), chromatin has an open configuration, which is associated with active transcription due to accessibility of transcription factor binding sites and transcription start sites. This open configuration confers pluripotent cells with a unique plasticity that enables differentiation into a wide range of lineages (Atlasi and Stunnenberg, 2017). In ESCs, promoters of developmental genes are often marked by both the active H3K4me3 and the repressive H3K27me3 histone modifications (Box 1) (Bernstein et al., 2006a; Azuara et al., 2006). Despite their antagonistic functions, this “bivalency” plays a key role in pluripotency by maintaining genes in a poised state for rapid activation or repression upon differentiation. As pluripotent cells differentiate, chromatin becomes more restricted by epigenetic deposition of DNA methylation and histone modifications that establish heritable cell-type-specific gene expression patterns (Atlasi and Stunnenberg, 2017).
Box 1.
Histone modifications mentioned in this review that mark promoters of active or silenced genes.
Based on this model, one hypothesis is that the chromatin landscape in XX and XY bipotential progenitor cells is similar, enabling equal access to promoters of sex-determining genes and regulatory elements of both sexes, and that after sex-determination, the chromatin landscape becomes more restricted to canalize either the male or female fate and repress the alternative pathway. Investigation of this hypothesis revealed that, similar to ESCs, key sex-determining genes that are co-expressed in XX and XY mouse progenitor cells are bivalent, possibly contributing to the bipotential nature of these cells (Fig. 1B) (Garcia-Moreno et al., unpublished). The upregulation of the male or female pathway is accompanied by mutually antagonistic mechanisms that repress the alternate cell fate (Kim et al., 2006b; Ottolenghi et al., 2007; Chassot et al., 2008; Uhlenhaut et al., 2009; Matson et al., 2011). At the chromatin level, this transition is reflected in a reorganization of histone marks around sex determining genes. Genes associated with the female pathway lose their repressive mark when the ovarian pathway is activated, and genes associated with the male pathway lose their repressive mark when the testis pathway is activated. However, ovary-determining genes that become transcriptionally repressed when the gonad commits to a male fate remain bivalent, similar to the fate of testis-determining genes in the ovary (Fig.1B) (Garcia-Moreno et al., unpublished). Maintaining the alternate cell fate in a state poised for activation possibly contributes to the unique capacity of supporting cells to transdifferentiation from Sertoli cells to pregranulosa cells and vice-versa, even during adulthood (Uhlenhaut et al., 2009; Matson et al., 2011). As bivalency is a conserved epigenetic mechanism (Lesch et al., 2016), it will be interesting to determine whether bivalent sex-determining genes confer plasticity to non-mammalian systems that can switch between the male and female fate in response to environmental stimuli.
Another study performed DNaseI hypersensitive site sequencing (DNaseI-seq) and chromatin immunoprecipitation followed by sequencing (ChIP-seq) for H3K27ac, a histone modification indicative of active enhancers, to generate a profile of the chromatin landscape in purified mouse Sertoli cells at embryonic day 13.5 (E13.5), soon after commitment to the male pathway (Maatouk et al., 2017). DNaseI-seq identified 28,133 nucleosome-depleted regions (NDRs) (corresponding to regulatory elements) that were unique to Sertoli cells when compared to six other tissues. As expected, these NDRs were commonly located near Sertoli-specific genes, implying that they represent regulatory elements that drive Sertoli cell differentiation. Interestingly, pregranulosa-specific genes had a similar enrichment of Sertoli-unique NDRs in their neighborhood in Sertoli cells, suggesting that the chromatin architecture between Sertoli and pregranulosa cells is similar and may reflect their shared progenitor state. However, H3K27ac-positive NDRs were significantly enriched only around Sertoli-specific genes. Together these findings indicate that as sex-determining pathways diverge, genes in Sertoli cells that are associated with the male pathway lose their repressive H3K27me3 mark and acquire H3K27ac. Simultaneously, genes associated with the female pathway remain bivalent in Sertoli cells, and retain NDRs in their enhancer/promoter regions. It is likely that these H3K27ac-negative NDRs (inactive enhancers) associated with pregranulosa-specific genes are bound by repressors that block upregulation of the female-determining pathway.
4. Epigenetic regulation of the primary switch
The balance between male and female fate in the early gonad is disrupted by master regulators of sex-determination that act as a “switch” to activate the male pathway and repress the female pathway. There is increasing evidence that these primary switches are epigenetically regulated. Below, we will review how the primary mammalian switch Sry is epigenetically regulated during sex determination, and provide evidence that the epigenetic regulation of master regulators of male sex-determination is conserved in non-mammalian systems.
4.1. The mammalian switch, Sry
In mammals, it is the presence of the Y-encoded transcription factor Sex–determining Region Y (Sry) that directs the bipotential fetal gonad towards the male fate (Sinclair et al.,1990; Gubbay et al., 1990; Koopman et al., 1991). Sry expression breaks the initial male-female balance in bipotential progenitors and directs differentiation of Sertoli cells through a downstream signaling cascade (Albrecht and Eicher, 2001). In XY mice, Sry is transiently expressed between E10.5 and E12.5 (Hacker et al., 1995). Although there are still unanswered questions about the events leading to the activation of Sry, a number of epigenetic modifiers have been identified.
4.1.1. Regulation of Sry mediated by DNA methylation
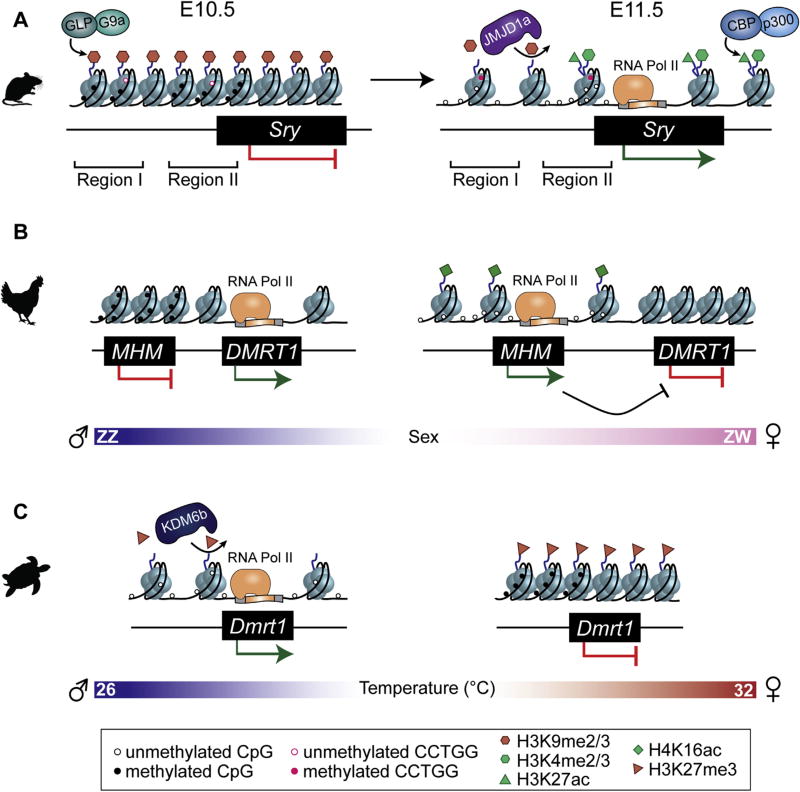
DNA methylation, which in mammals occurs primarily at cytosines located next to guanines (CpG), plays a crucial role during development by epigenetically controlling gene silencing and establishing cell-type-specific gene expression patterns during differentiation (Razin and Cedar, 1984). The methylome of the Y chromosome was first explored by Nishino et al. (2004). This group generated an in vivo methylation profile of XY mouse embryos throughout development using bisulfite sequencing, a standard technique that identifies methylated CpGs. There are 16 CpGs in the 4.5 kb region upstream of Sry that cluster into two regions. Region I contains four CpGs and overlaps the TSS of a circular transcript of Sry, which is believed to be untranslated (Dolci et al., 1997), whereas Region II contains seven CpGs and overlaps the TSS of the linear transcript of Sry, which is translated into a functional protein from E10.5–E12.5 in XY fetal gonads (Jeske et al., 1995). Bisulfite sequencing revealed that both regions were hypomethylated at the blastocyst stage and became hypermethylated in E8.5 embryos in which Sry is not yet expressed. At the time of Sry activation (E11.5), both regions lost methylation in the gonad, but not in other tissues such as the liver. Finally, at E15.5, Region II became re-methylated, whereas Region I remained hypomethylated. It is unlikely that hypomethylation at Region I is of functional significance, as this region did not induce activity in an in vitro promoter assay (Nishino et al., 2004). However, the possible three-dimensional structure of the inverted repeats around this locus may interfere with in vitro promoter assays (Gubbay et al., 1990; Capel et al., 1993). Nonetheless, this work established a clear anti-correlation between promoter CpG methylation and Sry expression in the gonad throughout sex determination (Fig. 2A).
Fig. 2.
A) Epigenetic regulation of Sry in mice. Prior to sex determination (left) Sry is repressed and enriched for the repressive GLP/G9a-mediated H3K9me2 histone modification. Region I overlaps the TSS of an untranscribed circular Sry transcript, and Region II overlaps the TSS of a functional linear Sry transcript. At E10.5, regions I and II contain methylated CpGs, and unmethylated CCTGG sites. At E11.5, Sry activation requires Jmjd1a-mediated removal of H3K9me2, CBP/p300-mediated deposition of H3K27ac, and accumulation of the active H3K4me2 at its promoter. CpGs at region I and II become demethylated, and CCTGG sites become methylated. B) Epigenetic regulation of DMRT1 in chickens. In males (ZZ, left), a region neighboring the male-determining gene DMRT1 is hypermethylated (MHM) and repressed. In females (ZW, right), MHM is hypomethylated and enriched for the active H4K16ac histone modification. The open chromatin conformation of MHM in females enables transcription of a lncRNA that inhibits expression of DMRT1. C) Epigenetic regulation of Dmrt1 in red-eared slider turtles. During embryogenesis, turtles develop as males if eggs are incubated at lower temperatures (blue, left) and as females at higher temperatures (red, right). At male-producing temperatures, Kdm6b is upregulated. KDM6b-mediated removal of the repressive H3K27me3 and demethylation of CpGs at the promoter of Dmrt1 is required for its activation and subsequent testis development. At female-producing temperatures, enrichment of H3K27me3 and CpG methylation at the promoter of Dmrt1 inhibits its expression and leads to ovary development.
To investigate the mechanistic link between methylation and Sry expression, Nishino et al. established an in vitro system by culturing primary cells isolated from E11.5 XY gonads (Nishino et al., 2004). Introduction of previously in vitro-methylated constructs containing either the circular or linear promoter regions strongly suppressed the activity of ectopically introduced Sry. These results clearly showed that Sry gene expression can be epigenetically regulated through DNA methylation-mediated silencing.
In mammals, DNA methylation occurs primarily at the CpG site. Non-CpG methylation, which occurs at CNG sites in which N can be any nucleotide, is more common in plants (Gruenbaum et al., 1981), bacteria (Yoder et al., 1997; Casadesus and Low, 2006) and yeast (Pinarbasi et al., 1996), but was observed at CC(A/T)GG motifs in various mammalian systems (Malone et al., 2001; Franchina and Kay, 2000; Agirre et al., 2003; Imamura et al., 2005). Interestingly, the promoter for both the circular and linear transcripts of Sry (regions I and II) contains a CCTGG motif (Nishino et al., 2011). In the gonad, this motif has the opposite methylation dynamic than its neighboring CpG sites, and coincides with Sry's temporal expression pattern. While CpG sites transiently lose methylation at E11.5, CCTGG sites are transiently methylated (Fig. 2A). An in vitro promoter assay further showed that CCTGG methylation can induce Sry activity, suggesting that non-CpG methylation is associated with active transcription and may play an important role in Sry activation. However, although the CCTGG site at Region II was conserved in six mouse strains, two of these strains (C57BL/6 and 129S1) did not exhibit CCTGG methylation at E11.5, suggesting that non-CpG methylation may be background-dependent and raises the possibility of strain-specific differences in epigenetic regulation of Sry (Nishino et al., 2011).
Although it is well established that DNA methylation strongly correlates with Sry's activity, the mechanisms by which its promoter CpG sites are dynamically methylated during such a precise window of development have not been described. This is partly due to the fact that in vivo disruption of DNA methyltransferases (DNMTs) leads to early embryonic lethality (Li et al., 1992; Okano et al., 1999). Warr and colleagues hypothesized that GADD45g, a protein that can induce gene expression by recruiting DNA repair factors to demethylate target genes (Barreto et al., 2007; Niehrs and Schafer, 2012), was involved in Sry's activation based on the observation that loss of Gadd45g caused complete male-to-female sex reversal in mice due to failure to activate Sry (Warr et al., 2012). However, loss of Gadd45g did not alter the normal methylation status of Sry during sex determination, suggesting that it does not activate Sry through promoter demethylation, but must function through an alternative mechanism (Gierl et al., 2012). Despite this, the same study found that demethylation at Sry's promoter begins as early as 5–6 tail somites, and is therefore one of the earliest male-specific molecular events preceding Sry expression.
Interesting data concerning the epigenetic regulation of sex determination came from cloned animals that were generated by somatic cell nuclear transfer (SCNT). Cloned animals exhibit anomalies of sex differentiation despite having a normal karyotype (Inoue et al., 2009). For example, canine SCNT results in male-to-female sex reversal in almost 23% of cases (Jeong et al., 2016). A genetic analysis of SRY + XY DSD dogs found no genetic differences that could account for sex reversal. However, the SRY locus in XY DSD dogs was hypermethylated compared to control XY dogs (Jeong et al., 2016). This hypermethylation corresponded with reduced expression of SRY (in addition to other downstream male-determining genes), and an upregulation of the ovarian developmental pathway. Although this case is specific to an artificial system, it strongly supports the idea that epigenetic regulation can control the sex-determination network.
Not much is known about the role that DNA methylation plays in regulating the human SRY gene. In one study, an epigenetic profile of the Y chromosome was generated from blood cells of two individuals from distinct populations (Singh et al., 2011). A CTCF transcription factor binding site was found in the promoter region of SRY, which overlapped an unmethylated CpG site. CTCF binds chromatin insulators and acts as a transcriptional repressor. Interestingly, this sitewas enriched for repressive histone modifications, pointing to the possibility that the human SRY gene may be silenced through recruitment of repressors and not through DNA methylation. However, caution should be taken when drawing conclusions based on analysis of a non-gonadal cell type that does not usually express SRY.
4.1.2. Regulation of Sry mediated by histone modifications
Evidence that histone modifiers are critical regulators of mammalian sex determination came from the observation that loss of the chromobox protein homologue 2 (Cbx2) leads to complete male-to-female sex reversal and development of hypoplastic gonads in mice (Katoh-Fukui et al., 1998) and humans (Biason-Lauber et al., 2009). CBX2 is part of the Polycomb-group (PcG) of proteins that typically function as transcriptional repressors and play a critical role in regulating gene silencing during development (Simon and Kingston, 2009; Margueron and Reinberg, 2011). The PcG proteins assemble into two complexes: the Polycomb Repressive Complex 1 and 2 (PRC1 and PRC2). Canonically, PRC2 first catalyzes H3K27me3 at the promoter of its target genes. PRC1 then binds H3K27me3 through its CBX2 subunit, and maintains gene repression through chromatin compaction (Bernstein et al., 2006b; Lau et al., 2017). Sex-reversal in XY Cbx2 mutants is characterized by a failure to upregulate Sry and its downstream target SRY-box 9 (Sox9) (Katoh-Fukui et al., 2012). As sex-reversal was rescued by forced expression of Sry, CBX2 was proposed to act as an early activator of Sry expression (Katoh-Fukui et al., 2012). Although some subunits of the PcG proteins have been reported to drive active transcription of certain target genes (Jacob et al., 2011; Mousavi et al., 2012; Herz et al., 2012), CBX2 has not been shown to act directly as an activator. Therefore, a role for CBX2 as an activator was unexpected. Recent evidence suggests that PRC2 may be involved in “transcriptional focusing” by spatially confining H3K27me3 + silent domains (Lu et al., 2017). It is possible that disruption of the PcG complex, such as loss of CBX2, may lead to aberrant spread of these domains resulting in silencing of underlying genes, possibly including Sry. Another possibility is that CBX2 induces Sry expression indirectly by repressing a repressor. Because the male and female pathways are mutually antagonistic, stabilization of the male fate requires active repression of female-determining genes, raising the possibility that CBX2 has a wide-spread role in the repression of the female-pathway, which may otherwise block male fate commitment.
Further evidence for the epigenetic regulation of Sry mediated through histone modifiers came from studies of the histone demethylase Jmjd1a. Jmjd1a-deficient mice exhibit male-to-female sex reversal (Kuroki et al., 2013). Jmjd1a encodes a histone demethylase that acts specifically on the repressive H3K9me2 histone modification. ChIP for H3K9me2 and the active histone modification mark H3K4me2 was performed in purified progenitor cells from gonads of XY mice at E11.5 when Sry expression is at its peak. As opposed to control XY mice, the Sry locus of Jmjd1a-deficient mice retained high enrichment of the repressive H3K9me2 mark, and had low enrichment of the active H3K4me2 mark. Consistently, Sry expression levels were reduced in mutant mice. This suggests that Jmjd1a-dependent demethylation of H3K9me2 is required for subsequent H3K4me2 accumulation at Sry's promoter, leading to an open chromatin conformation and Sry activation (Fig. 2A). Consistent with this observation, a separate study found that the Sry promoter has low levels of H3K9me3 and is enriched for the active H3K4me3 modification in E12.5 mouse testes. In contrast, adult testes have high H3K9me3 and low H3K4me3 enrichment, suggesting that the Sry locus regains H3K9me3 after it is silenced, and that this mark maintains long-term repression of Sry throughout adulthood in mice (Sinha et al., 2017). Recently, sex-reversal of Jmjd1a-deficient micewas rescued by disrupting the GLP/G9a H3K9 methyltransferase complex, pointing towards this complex as the one that catalyzes H3K9me2 at the Sry locus (Fig. 2A) (Kuroki et al., 2017). This study highlights how various epigenetic factors interact to fine-tune the expression of Sry by balancing the levels of H3K9me2 at its promoter.
In addition to accumulation of H3K4me2 at Sry's promoter, activation of Sry requires deposition of H3K27ac by the CREB-binding protein (CBP) and its paralogue p300 (Fig. 2A) (Carre et al., 2017). A recent study found that deletion of either gene in gonadal somatic cells results in partial male-to-female sex reversal, whereas loss of three of the four p300/Cbp functional alleles leads to complete male-to-female sex reversal. ChIP showed reduced levels of H3K27ac at the promoter of Sry in p300/Cbp mutants at E11.5, corresponding to reduced Sry and Sox9 expression levels. This study indicates that this histone/lysine acetyltransferase complex is a key component in the activation of Sry during testis development.
Interestingly, epigenetic profiling of human blood cells showed that the human SRY gene promoter was enriched for the active H3K9ac mark and the repressive H3K27me3 mark, but H3K9me3 was only enriched over the gene body (Singh et al., 2011). The conflicting H3K9ac and H3K9me3 modifications may be a reflection of mixed cell populations, as SRY is expressed in the B-cell lineage but not in other blood lineages. Studies of human gonadal lineages are required to understand which chromatin modifiers are involved in SRY activation during sex determination in humans. However, there is evidence that disruption of epigenetic regulation can lead to disorders of sexual development (DSDs). Consistent with the mouse studies mentioned above, one XY phenotypically normal girl with hypoplastic ovaries was found to have inherited two loss-of-function mutations in the CBX2 gene (Biason-Lauber et al., 2009). In a separate case, an SRY + XY woman with streak gonads had a hyperacetylated SRY locus relative to control XY males (Mitsuhashi et al., 2010). This hyperacetylation coincided with prolonged SRY activity, which the authors suggested might destabilize the downstream male-determining network, although further experiments are required to fully explain this case.
4.2. Evidence for epigenetic regulation of the key switch gene, Dmrt1, in other vertebrates
Several new lines of evidence suggest that epigenetic regulation of key switch genes may be a conserved element of sex determination pathways in different species. Dmrt1 (doublesex and mab3-related transcription factor 1) is a highly conserved transcription factor that plays a critical role in vertebrate sex determination (Matson and Zarkower, 2012) and has been shown to act as the master sex-determining gene in many non-mammalian systems (Masuyama et al., 2012; Smith et al., 2009; Yoshimoto et al., 2008). In mice, Dmrt1 is not required for testis-development; however, it is essential for maintaining the Sertoli cell phenotype in adult testes, as loss of Dmrt1 causes transdifferentiation of Sertoli cells into pregranulosa cells (Matson et al., 2011). Although there are currently no studies investigating the epigenetic regulation of Dmrt1 in mammals, its epigenetic regulation has been investigated in the half-smooth tongue sole, chickens, and the red-eared slider turtle.
4.2.1. Epigenetic regulation of dmrt1 in fish
dmrt1 is highly conserved in fish and plays an important role in male sex determination (Marchand et al., 2000; Guo et al., 2005). The half-smooth tongue sole Cynoglossus semilaevis is a flatfish that is a fascinating model to study the epigenetic regulation of sex determination, as it has both genetic- and temperature-dependent sex-determining mechanisms. The genetic sex of this species is determined by its Z and W chromosome composition: males carry two Z chromosomes, whereas females carry a Z and a W chromosome. In this species, dosage of the Z chromosome is the sex-determining factor. Despite the inheritance of sex chromosomes, ~14% of ZW females develop as males under normal conditions (22 °C), and the incidence of sex reversal can increase to ~73% when fish are reared at high temperatures (28 °C) (Chen et al., 2014). These sex-reversed fish are “pseudomales” that can reproduce with normal females. Importantly, the offspring of pseudomales and normal females maintain a high sex-reversal rate even under normal conditions (22 °C), suggesting that the ability to sex reverse can be inherited.
Shao et al. generated a DNA methylation profile of female, male and pseudomale fish to investigate the role of DNA methylation in the transition from genetic to environmental sex determination (Shao et al., 2014). This study identified dmrt1, which is Z-encoded in the tongue sole (Chen et al., 2014), as a critical gene that responds to temperature change through DNA methylation. Consistent with its expression pattern, dmrt1 maintains low methylation levels throughout life in male gonads, but is increasingly methylated in female gonads starting at the time of sex determination and throughout adulthood. In ZW pseudomales, the dmrt1 gene has low methylation levels similar to ZZ males, and is therefore active and able to drive testis-development. This male-specific methylation is then inherited in the ZW offspring of ZW pseudomales and ZW females, accounting for a higher rate of sex-reversal compared to ZW offspring between normal ZZ males and ZW females. This finding suggests that temperature can override genetic sex-determination by directly altering the epigenetic marks at dmrt1, and that in this species where methylation is not stripped from the genome in the early embryo, this effect is transgenerational.
4.2.2. Epigenetic regulation of DMRT1 by MHM in chickens
As in the half-smooth tongue sole, chickens have a ZZ (male) and ZW (female) sex chromosome composition. DMRT1 is also a Z-linked gene in chickens that is more highly expressed in males than in females. Overexpression of DMRT1 in chick ZW gonads induced the male pathway (Lambeth et al., 2014), whereas knockdown of DMRT1 led to feminization of male gonads (Smith et al., 2009).
A region neighboring DMRT1 is differentially methylated between males and females (Teranishi et al., 2001). In males, this region is hypermethylated (known as the male hypermethylated region or MHM) and is transcriptionally inactive, whereas in females, this region is hypomethylated and transcribed into a long non-coding RNA. In addition to being hypomethylated, the female MHM locus is enriched for the active H4K16ac mark (Bisoni et al., 2005) and has an open chromatin configuration (Itoh et al., 2011). MHM is hypothesized to inhibit DMRT1 expression in females, as the MHM transcript accumulates at the site of transcription adjacent to DMRT1 (Teranishi et al., 2001) (Fig. 2B). Consistent with this hypothesis, injection of exogenous MHM into rooster testicles led to a strong downregulation of DMRT1 and a paling of the comb color, indicative of a change in sex hormones. Importantly, transcriptional levels of other key sex-determining genes such as AMH and SOX9 were unaffected, suggesting that MHM specifically regulates DMRT1 (Yang et al., 2010b).
4.2.3. Epigenetic regulation of Dmrt1 by KDM6B in turtles
The red-eared slider turtle Trachemys scripta lacks heteromorphic sex chromosomes. Instead, the temperature of incubation acts as a sex determinant. During embryogenesis, turtles develop as males if eggs are exposed to lower temperatures (26 °C), and as females if they are exposed to higher temperatures (32 °C). Prior to sex determination, Dmrt1 expression in the embryonic gonads is sexually dimorphic, with higher levels at MPT (male-producing temperature) than at FPT (female-producing temperature) (Kettlewell et al., 2000; Czerwinski et al., 2016). Recently, Dmrt1 was shown to be both necessary and sufficient to initiate male sex determination in T. scripta (Ge et al., 2017). Moreover, Dmrt1 responds rapidly to temperature shifts, which establishes it as a master regulator of temperature-dependent sex determination. Interestingly, DNA methylation of the Dmrt1 promoter region is inversely correlated with its expression and fluctuates in response to temperature changes. For example, the methylation level of the Dmrt1 promoter greatly increased when eggs were shifted from MPT to FPT. In contrast, shifting eggs from FPT to MPT resulted in decreased promoter DNA methylation and gene activation (Ge et al., 2017) (Fig. 2C). This suggests that, similar to the tongue sole flatfish, temperature can regulate both sex-determining gene expression and promoter methylation. However, although the previous studies show an inverse correlation between CpG promoter methylation and Dmrt1 expression, whether DNA methylation is a cause or a consequence of expression changes in temperature-dependent systems has not been established.
Transcriptome sequencing of T. scripta embryonic gonads at MPT and FPT revealed that in addition to Dmrt1, Kdm6b (or Jmjd3) is also upregulated at MPT and its expression levels fluctuate in response to temperature changes (Czerwinski et al., 2016). Kdm6b is an H3K27me3-specific demethylase that can activate target genes through the removal of this repressive histone modification. Loss of Kdm6b in MPT gonads results in a complete shift from the male to the female sexual trajectory, suggesting that it also plays a key role in male sex-determination (Ge et al., in press). Knockdown of Kdm6b resulted in a significant downregulation of Dmrt1 expression in MPT gonads, pointing towards Dmrt1 as a downstream target of Kdm6b. Consistent with this hypothesis, ChIP-qPCR revealed that the Dmrt1 promoter had low levels of H3K27me3 in MPT gonads in which Kdm6b and Dmrt1 expression levels are high. In contrast, the Dmrt1 promoter was enriched for H3K27me3 in FPT gonads in which Kdm6b and Dmrt1 expression levels were low. Importantly, H3K27me3 levels at Dmrt1 increased upon loss of Kdm6b, suggesting that Kdm6b activates Dmrt1 by removing H3K27me3 at MPT leading to activation of this switch gene that induces male sexual development (Fig. 2C). This study provides the first direct mechanistic link between epigenetic and temperature regulation of sex determination.
5. Sry and other Sox genes as epigenetic regulators
In the developing testis, Sry's primary function is to upregulate Sox9 expression (Sekido and Lovell-Badge, 2008). Sox9 is part of the Sry-type HMG box (Sox) family, a group of proteins with over 60% similarity to the SRY high-mobility group (HMG) box region (Denny et al., 1992a). In mammals, several Sox genes, including Sox9, can replace Sry (Zhao and Koopman, 2012). For example, directed expression of Sox9 in XX mice induces testis development, demonstrating that Sox9 is sufficient for male sex determination (Vidal et al., 2001). Consistent with this, duplication of SOX9 in XX humans can lead to female-to-male sex reversal (Cox et al., 2011). On the other hand, Sox9-null XY gonads develop as ovaries (Barrionuevo et al., 2006), and loss-of-function mutations of the human SOX9 gene can lead to female development (Kwok et al., 1995; Stoeva et al., 2011). These studies highlight the role of Sox9 as a critical regulator of male sex determination and Sertoli cell development.
Both SRY and SOX9 are transcription factors (TFs) that recognize and bind a specific DNA sequence (Denny et al., 1992b). Although TFs typically act as direct activators when binding to DNA motifs of target genes, below we will review several studies that support a model in which SRY and SOX9 can themselves function as epigenetic regulators.
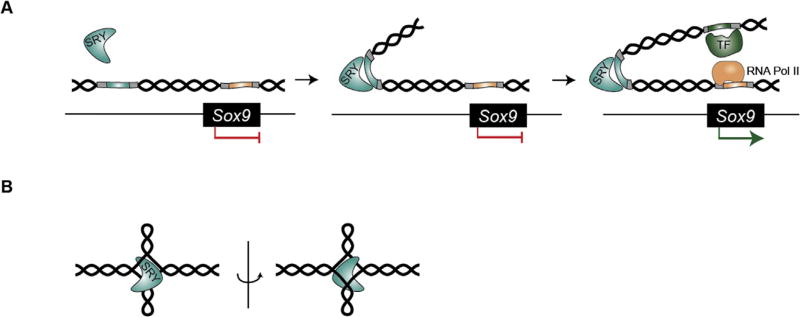
Despite Sry acting as the primary male-determining switch in mice and humans, sequence conservation between the mSry and hSRY genes is low and restricted to 79 amino acids that encode the HMG box, suggesting that this region is of high functional significance (Sinclair et al., 1990; Ferrari et al., 1992). The HMG box of SRY has an L-shaped structure formed by an extended segment and three α-helices (Read et al., 1993). Contrary to most TFs that bind the major groove of DNA, the SRY HMG box binds the minor groove of DNA inducing a 60°2013;70° bend in the double helix (Fig. 3A). In addition to DNA bending activity, HMG1 proteins can also bind to DNA cruciform structures, suggesting that SRY could also mediate attachment sites in chromatin loops (Fig. 3B) (Ferrari et al., 1992). This ability to directly modulate the 3D architecture of DNA leads to the speculation that SRY and other HMG-box containing TFs, such as the Sox genes, are core determinants of cell-type-specific chromatin landscapes. In support of this, it has been suggested that SRY might mediate effects at a distance by mechanically displacing factors associated with chromatin near the point of flexure. Alternatively, DNA bending might bring remote sites in close contact with each other to facilitate the interaction of transcription factors, or to promote the establishment of chromatin loops (Fig. 3A) (Bianchi and Beltrame, 1998). Importantly, the majority of sex-reversing point mutations in the SRY gene fall within the HMG box-encoding region (Berta et al., 1990). These mutations cause alterations to the SRY protein that reduce or ablate DNA binding affinity, suggesting that DNA binding and bending is of critical functional importance (Jager et al., 1992; Harley et al., 1992; Pontiggia et al., 1994; Murphy et al., 2001; Assumpcao et al., 2002).
Fig. 3.
Overview of SRY mechanism in mammals. A) Sry encodes a transcription factor that recognizes a specific DNA-binding motif (blue box). SRY contains an L-shaped HMG box that binds the minor groove of DNA and induces a 60–70° bend in the double helix (middle panel). It is speculated that DNA bending by SRY may mediate effects at a distance by promoting chromatin looping, which brings transcription factors in close proximity to TSS. Depicted is a putative example of SRY-mediated activation of Sox9 by binding to an upstream enhancer (right panel). B) The HMG box of SRY can bind to DNA cruciform structures regardless of DNA sequence.
In addition to directly bending DNA and altering the 3D conformation of chromatin, SRY and SOX9 can act as epigenetic regulators by interacting with chromatin-modifying complexes. In E11.5 fetal gonads, SRY interacts with Krüppel-associated box only (KRAB-O) protein and its obligatory co-repressor Krab-associating protein 1 (KAP1) (Oh et al., 2005; Peng et al., 2009). KAP1 then recruits heterochromatin protein 1 (Hp1), HDACs and the SETDB1 methyltransferase, which function as gene silencers by creating a repressive chromatin environment. Therefore, Sry may have a dual function in early sex determination by activating male-determining genes through direct binding to regulatory elements (such as binding to the testis-specific enhancer TESCO upstream of Sox9 (Sekido and Lovell-Badge, 2008)), and repressing the female pathway through recruitment of KRAB-O/KAP1 chromatin-mediated repression machinery. Interestingly, KRAB-O knockdown in Sry-expressing cultured cells resulted in reduced levels of Sox9, suggesting that KRAB-O may also mediate Sry function. However, KRAB-O knockdown mice exhibited normal testis development, possibly due to redundancy from over 100 KRAB genes that are expressed in the mouse fetal gonad (Polanco et al., 2009).
In addition to p300/CBP-mediated activation of Sry through H3K27ac deposition, SOX9 can itself epigenetically regulate target genes by interacting with this histone acetyltransferase complex (Tsuda et al., 2003). In human chondrocytes, p300/CBP significantly enhanced Sox9-mediated transcription of target genes through p300-mediated histone acetylation (Tsuda et al., 2003; Furumatsu et al., 2005). Although these studies were not performed in the developing gonad, it is possible that Sox9 functions during sex determination by acetylating SOX9 binding sites and recruiting additional activators to Sertoli-specific genes and regulatory regions. In this way, SRY and SOX9 may play critical roles during sex determination by epigenetically regulating downstream targets that further promote the divergence of the male pathway from the bipotential state.
6. Conclusions
In vertebrates, the formation of a bipotential gonad is the first and critical step for the development of sexually dimorphic organisms. The unique ability of the gonad to engage one of two possible developmental pathways during sex determination provides a valuable model to study cell fate decisions and transitions.
Accumulating evidence suggests that various levels of epigenetic regulation underlie the resolution of cell fate in the early gonad by fine-tuning the timing and expression levels of sex-determining genes. The plasticity of the bipotential gonad is reflected in (and perhaps fostered by) a poised chromatin landscape that resolves through the activation of genes associated with one fate, and the maintenance of bivalency at genes associated with the opposite fate. At present, commitment to one fate or the other does not appear to be associated with an epigenetic lock-out of the alternative pathway. Instead, evidence suggests that maintenance of cell fate depends on TFs such as SOX9, FOXL2 and DMRT1. Dependence on TFs rather than more stable repressive mechanisms may not be unusual between lineages that originate from a common precursor, especially when there is an evolutionary advantage to maintaining plasticity. In the case of the supporting cell lineage in the gonad, one possibility is that this is an evolutionary holdover from species that undergo sex reversal, but this idea has not been investigated.
The activation of genes associated with the male fate is initiated by transcription factors such as SRY and DMRT1. Although TFs initiate transcriptional changes, their ability to bind DNA is modulated by the chromatin landscape. In addition, Sry and Dmrt1 are themselves epigenetically regulated throughout sex determination, and in the case of Sry, can function as a downstream epigenetic regulator itself. The interlacing of TFs and the chromatin landscape during cell fate commitment and maintenance is an important area of future study, which will benefit from the unique characteristics of the bipotential gonad.
For years it has been speculated that species lacking genetic cues are able to convert external signals into a phenotypic output through changes in their epigenome. Recent evidence supports this idea. The advent of whole-genome sequencing coupled with small scale techniques that assay the chromatin landscape have been instrumental in advancing our understanding of the epigenetic mechanisms that regulate primary sex-determining signals in species such as fish and reptiles. We expect this field to expand significantly in the next few years.
Although we have limited this review to studies of the initial steps in the sex-determination cascade, further studies are sure to uncover additional levels of epigenetic regulation that further promote the canalization and maintenance of the male and female pathways. In addition to advancing our understanding of the basic biological principles underlying molecular and cellular fate commitment and development, increasing our understanding of the epigenetic landscape during sex determination has implications for our ability to diagnose human disorders of sex development with unknown etiologies. Currently ~80% of patients do not receive a genetic diagnosis. In many of these undiagnosed cases, the cause may escape conventional SNP screens because it is epigenetic in nature. Epigenetic studies can be expected to define critical regulatory regions beyond the coding genome.
Acknowledgments
This work was supported by grants to BC from National Science Foundation (IOS- 1256675) and the National Institutes of Health (5R01-HD039963-13 and HD39963), and to SAGM by the National Institutes of Health (T32 NIH GM008061).
We would like to thank Isabella M. Salamone, Christopher R. Futtner, Maxwell E. Edmonds, and Corey Bunce for their comments on the manuscript. We will always be grateful to Danielle M. Maatouk, whose ideas during her brief research career left an indelible mark on our thinking.
References
- Agarwal N, Hardt T, Brero A, Nowak D, Rothbauer U, Becker A, Leonhardt H, Cardoso MC. MeCP2 interacts with HP1 and modulates its heterochromatin association during myogenic differentiation. Nucleic Acids Res. 2007;35:5402–5408. doi: 10.1093/nar/gkm599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirre X, Vizmanos JL, Calasanz MJ, Garcia-Delgado M, Larrayoz MJ, Novo FJ. Methylation of CpG dinucleotides and/or CCWGG motifs at the promoter of TP53 correlates with decreased gene expression in a subset of acute lymphoblastic leukemia patients. Oncogene. 2003;22:1070–1072. doi: 10.1038/sj.onc.1206236. [DOI] [PubMed] [Google Scholar]
- Albrecht KH, Eicher EM. Evidence that Sry is expressed in pre-Sertoli cells and Sertoli and granulosa cells have a common precursor. Dev. Biol. 2001;240:92–107. doi: 10.1006/dbio.2001.0438. [DOI] [PubMed] [Google Scholar]
- Arnold AP. Mouse models for evaluating sex chromosome effects that cause sex differences in non-gonadal tissues. J. Neuroendocrinol. 2009;21:377–386. doi: 10.1111/j.1365-2826.2009.01831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assumpcao JG, Benedetti CE, Maciel-Guerra AT, Guerra G, Jr, Baptista MT, Scolfaro MR, de Mello MP. Novel mutations affecting SRY DNA-binding activity: the HMG box N65H associated with 46,XY pure gonadal dysgenesis and the familial non-HMG box R30I associated with variable phenotypes. J. Mol. Med. (Berl.) 2002;80:782–790. doi: 10.1007/s00109-002-0376-9. [DOI] [PubMed] [Google Scholar]
- Atlasi Y, Stunnenberg HG. The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 2017 doi: 10.1038/nrg.2017.57. [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat. Cell. Biol. 2006;8(5):532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Barrionuevo F, Bagheri-Fam S, Klattig J, Kist R, Taketo MM, Englert C, Scherer G. Homozygous inactivation of Sox9 causes complete XY sex reversal in mice. Biol. Reprod. 2006;74:195–201. doi: 10.1095/biolreprod.105.045930. [DOI] [PubMed] [Google Scholar]
- Becker A, Allmann L, Hofstatter M, Casa V, Weber P, Lehmkuhl A, Herce HD, Cardoso MC. Direct homo- and hetero-interactions of MeCP2 and MBD2. PLoS One. 2013;8:e53730. doi: 10.1371/journal.pone.0053730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006a;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell. Biol. 2006b;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berta P, Hawkins JR, Sinclair AH, Taylor A, Griffiths BL, Goodfellow PN, Fellous M. Genetic evidence equating SRY and the testis-determining factor. Nature. 1990;348:448–450. doi: 10.1038/348448A0. [DOI] [PubMed] [Google Scholar]
- Bianchi ME, Beltrame M. Flexing DNA: HMG-box proteins and their partners. Am. J. Hum. Genet. 1998;63:1573–1577. doi: 10.1086/302170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biason-Lauber A, Konrad D, Meyer M, Debeaufort C, Schoenle EJ. Ovaries and female phenotype in a girl with 46,XY karyotype and mutations in the CBX2 gene. Am. J. Hum. Genet. 2009;84:658–663. doi: 10.1016/j.ajhg.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisoni L, Batlle-Morera L, Bird AP, Suzuki M, Mcqueen HA. Female-specific hyperacetylation of histone H4 in the chicken Z chromosome. Chromosome Res. 2005;13:205–214. doi: 10.1007/s10577-005-1505-4. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Thornhill AR, Boudrean SK, Darling SM, Bishop CE, Evans EP. The genetic basis of XX-XY differences present before gonadal sex differentiation in the mouse. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1995;350:260–261. doi: 10.1098/rstb.1995.0159. 253–60 discussion. [DOI] [PubMed] [Google Scholar]
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- Caron C, Pivot-Pajot C, van Grunsven LA, Col E, Lestrat C, Rousseaux S, Khochbin S. Cdyl: a new transcriptional co-repressor. EMBO Rep. 2003;4:877–882. doi: 10.1038/sj.embor.embor917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carre GA, Siggers P, Xipolita M, Brindle P, Lutz B, Wells S, Greenfield A. Loss of p300 and CBP disrupts histone acetylation at the mouse Sry promoter and causes XY gonadal sex reversal. Hum. Mol. Genet. 2017;27(1):190–198. doi: 10.1093/hmg/ddx398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Casadesus J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassot AA, Ranc F, Gregoire EP, Roepers-Gajadien HL, Taketo MM, Camerino G, de Rooij DG, Schedl A, Chaboissier MC. Activation of beta-catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum. Mol. Genet. 2008;17:1264–1277. doi: 10.1093/hmg/ddn016. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang G, Shao C, Huang Q, Liu G, Zhang P, Song W, An N, Chalopin D, Volff JN, Hong Y, Li Q, Sha Z, Zhou H, Xie M, Yu Q, Liu Y, Xiang H, Wang N, Wu K, Yang C, Zhou Q, Liao X, Yang L, Hu Q, Zhang J, Meng L, Jin L, Tian Y, Lian J, Yang J, Miao G, Liu S, Liang Z, Yan F, Li Y, Sun B, Zhang H, Zhang J, Zhu Y, Du M, Zhao Y, Schartl M, Tang Q, Wang J. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014;46:253–260. doi: 10.1038/ng.2890. [DOI] [PubMed] [Google Scholar]
- Cline TW, Meyer BJ. Vive la difference: males vs females in flies vs worms. Annu. Rev. Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- Cox JJ, Willatt L, Homfray T, Woods CG. A SOX9 duplication and familial 46,XX developmental testicular disorder. N. Engl. J. Med. 2011;364:91–93. doi: 10.1056/NEJMc1010311. [DOI] [PubMed] [Google Scholar]
- Czerwinski M, Natarajan A, Barske L, Looger LL, Capel B. A timecourse analysis of systemic and gonadal effects of temperature on sexual development of the red-eared slider turtle Trachemys scripta elegans. Dev. Biol. 2016;420:166–177. doi: 10.1016/j.ydbio.2016.09.018. [DOI] [PubMed] [Google Scholar]
- Denny P, Swift S, Brand N, Dabhade N, Barton P, Ashworth A. A conserved family of genes related to the testis determining gene, SRY. Nucleic Acids Res. 1992a;20:2887. doi: 10.1093/nar/20.11.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny P, Swift S, Connor F, Ashworth A. An SRY-related gene expressed during spermatogenesis in the mouse encodes a sequence-specific DNA-binding protein. EMBO J. 1992b;11:3705–3712. doi: 10.1002/j.1460-2075.1992.tb05455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanoa JK, Mukhopadhyay CS, Arora JS. Y-chromosomal genes affecting male fertility: a review. Vet. World. 2016;9:783–791. doi: 10.14202/vetworld.2016.783-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolci S, Grimaldi P, Geremia R, Pesce M, Rossi P. Identification of a promoter region generating Sry circular transcripts both in germ cells from male adult mice and in male mouse embryonal gonads. Biol. Reprod. 1997;57:1128–1135. doi: 10.1095/biolreprod57.5.1128. [DOI] [PubMed] [Google Scholar]
- Ferrari S, Harley VR, Pontiggia A, Goodfellow PN, Lovell-Badge R, Bianchi ME. SRY, like HMG1, recognizes sharp angles in DNA. EMBO J. 1992;11:4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchina M, Kay PH. Evidence that cytosine residues within 5'-CCTGG-3' pentanucleotides can be methylated in human DNA independently of the methylating system that modifies 5'-CG-3' dinucleotides. DNA Cell Biol. 2000;19:521–526. doi: 10.1089/104454900439755. [DOI] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Furumatsu T, Tsuda M, Yoshida K, Taniguchi N, Ito T, Hashimoto M, Ito T, Asahara H. Sox9 and p300 cooperatively regulate chromatin-mediated transcription. J. Biol. Chem. 2005;280:35203–35208. doi: 10.1074/jbc.M502409200. [DOI] [PubMed] [Google Scholar]
- Ge C, Ye J, Zhang H, Zhang Y, Sun W, Sang Y, Capel B, Qian G. Dmrt1 induces the male pathway in a turtle species with temperature-dependent sex determination. Development. 2017;144:2222–2233. doi: 10.1242/dev.152033. [DOI] [PubMed] [Google Scholar]
- Gierl MS, Gruhn WH, von Seggern A, Maltry N, Niehrs C. GADD45G functions in male sex determination by promoting p38 signaling and Sry expression. Dev. Cell. 2012;23:1032–1042. doi: 10.1016/j.devcel.2012.09.014. [DOI] [PubMed] [Google Scholar]
- Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, Steiner K, Tam PP, Monaco AP, Willard HF, Koopman P. The UTX gene escapes X inactivation in mice and humans. Hum. Mol. Genet. 1998;7:737–742. doi: 10.1093/hmg/7.4.737. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Naveh-Many T, Cedar H, Razin A. Sequence specificity of methylation in higher plant DNA. Nature. 1981;292:860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- Guo Y, et al. Gene structure, multiple alternative splicing, and expression in gonads of zebrafish Dmrt1. Biochem. Biophys. Res. Commun. 2005;330(3):950–957. doi: 10.1016/j.bbrc.2005.03.066. [DOI] [PubMed] [Google Scholar]
- Hacker A, Capel B, Goodfellow P, Lovell-Badge R. Expression of Sry, the mouse sex determining gene. Development. 1995;121:1603–1614. doi: 10.1242/dev.121.6.1603. [DOI] [PubMed] [Google Scholar]
- Harley VR, Jackson DI, Hextall PJ, Hawkins JR, Berkovitz GD, Sockanathan S, Lovell-Badge R, Goodfellow PN. DNA binding activity of recombinant SRY from normal males and XY females. Science. 1992;255:453–456. doi: 10.1126/science.1734522. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Mohan M, Garrett AS, Miller C, Casto D, Zhang Y, Seidel C, Haug JS, Florens L, Washburn MP, Yamaguchi M, Shiekhattar R, Shilatifard A. Polycomb repressive complex 2-dependent and -independent functions of Jarid2 in transcriptional regulation in Drosophila. Mol. Cell. Biol. 2012;32:1683–1693. doi: 10.1128/MCB.06503-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T, Kerjean A, Heams T, Kupiec JJ, Thenevin C, Paldi A. Dynamic CpG and non-CpG methylation of the Peg1/Mest gene in the mouse oocyte and preimplantation embryo. J. Biol. Chem. 2005;280:20171–20175. doi: 10.1074/jbc.M501749200. [DOI] [PubMed] [Google Scholar]
- Inoue K, Ogonuki N, Mekada K, Yoshiki A, Sado T, Ogura A. Sex-reversed somatic cell cloning in the mouse. J. Reprod. Dev. 2009;55:566–569. doi: 10.1262/jrd.09-099e. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Kampf K, Arnold AP. Possible differences in the two Z chromosomes in male chickens and evolution of MHM sequences in Galliformes. Chromosoma. 2011;120:587–598. doi: 10.1007/s00412-011-0333-x. [DOI] [PubMed] [Google Scholar]
- Iwase S, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128(6):1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Jacob E, Hod-Dvorai R, Ben-Mordechai OL, Boyko Y, Avni O. Dual function of polycomb group proteins in differentiated murine T helper (CD4þ) cells. J. Mol. Signal. 2011;6:5. doi: 10.1186/1750-2187-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager RJ, Harley VR, Pfeiffer RA, Goodfellow PN, Scherer G. A familial mutation in the testis-determining gene SRY shared by both sexes. Hum. Genet. 1992;90:350–355. doi: 10.1007/BF00220457. [DOI] [PubMed] [Google Scholar]
- Jeong YH, Lu H, Park CH, Li M, Luo H, Kim JJ, Liu S, Ko KH, Huang S, Hwang IS, Kang MN, Gong D, Park KB, Choi EJ, Park JH, Jeong YW, Moon C, Hyun SH, Kim NH, Jeung EB, Yang H, Hwang WS, Gao F. Stochastic anomaly of methylome but persistent SRY hypermethylation in disorder of sex development in canine somatic cell nuclear transfer. Sci. Rep. 2016;6:31088. doi: 10.1038/srep31088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske YW, Bowles J, Greenfield A, Koopman P. Expression of a linear Sry transcript in the mouse genital ridge. Nat. Genet. 1995;10:480–482. doi: 10.1038/ng0895-480. [DOI] [PubMed] [Google Scholar]
- Katoh-Fukui Y, Miyabayashi K, Komatsu T, Owaki A, Baba T, Shima Y, Kidokoro T, Kanai Y, Schedl A, Wilhelm D, Koopman P, Okuno Y, Morohashi K. Cbx2, a polycomb group gene, is required for Sry gene expression in mice. Endocrinology. 2012;153:913–924. doi: 10.1210/en.2011-1055. [DOI] [PubMed] [Google Scholar]
- Katoh-Fukui Y, Tsuchiya R, Shiroishi T, Nakahara Y, Hashimoto N, Noguchi K, Higashinakagawa T. Male-to-female sex reversal in M33 mutant mice. Nature. 1998;393:688–692. doi: 10.1038/31482. [DOI] [PubMed] [Google Scholar]
- Kernohan KD, Vernimmen D, Gloor GB, Berube NG. Analysis of neonatal brain lacking ATRX or MeCP2 reveals changes in nucleosome density, CTCF binding and chromatin looping. Nucleic Acids Res. 2014;42:8356–8368. doi: 10.1093/nar/gku564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettlewell JR, Raymond CS, Zarkower D. Temperature-dependent expression of turtle Dmrt1 prior to sexual differentiation. Genesis. 2000;26:174–178. [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006a;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kobayashi A, Sekido R, Dinapoli L, Brennan J, Chaboissier MC, Poulat F, Behringer RR, Lovell-Badge R, Capel B. Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol. 2006b;4:e187. doi: 10.1371/journal.pbio.0040187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- Kuroki S, Matoba S, Akiyoshi M, Matsumura Y, Miyachi H, Mise N, Abe K, Ogura A, Wilhelm D, Koopman P, Nozaki M, Kanai Y, Shinkai Y, Tachibana M. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science. 2013;341:1106–1109. doi: 10.1126/science.1239864. [DOI] [PubMed] [Google Scholar]
- Kuroki S, Okashita N, Baba S, Maeda R, Miyawaki S, Yano M, Yamaguchi M, Kitano S, Miyachi H, Itoh A, Yoshida M, Tachibana M. Rescuing the aberrant sex development of H3K9 demethylase Jmjd1a-deficient mice by modulating H3K9 methylation balance. PLoS Genet. 2017;13:e1007034. doi: 10.1371/journal.pgen.1007034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok C, Weller PA, Guioli S, Foster JW, Mansour S, Zuffardi O, Punnett HH, Dominguez-Steglich MA, Brook JD, Young ID, et al. Mutations in SOX9, the gene responsible for Campomelic dysplasia and autosomal sex reversal. Am. J. Hum. Genet. 1995;57:1028–1036. [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Tang ZL, Zhou J, Barndt RJ, Parvinen M, Allis CD, Page DC. Previously uncharacterized histone acetyltransferases implicated in mammalian spermatogenesis. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8707–8712. doi: 10.1073/pnas.082248899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeth LS, Raymond CS, Roeszler KN, Kuroiwa A, Nakata T, Zarkower D, Smith CA. Over-expression of DMRT1 induces the male pathway in embryonic chicken gonads. Dev. Biol. 2014;389:160–172. doi: 10.1016/j.ydbio.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Lau MS, Schwartz MG, Kundu S, Savol AJ, Wang PI, Marr SK, Grau DJ, Schorderet P, Sadreyev RI, Tabin CJ, Kingston RE. Mutation of a nucleosome compaction region disrupts Polycomb-mediated axial patterning. Science. 2017;355:1081–1084. doi: 10.1126/science.aah5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomblike protein. Cell. 2007;128:877–887. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Lesch BJ, Silber SJ, Mccarrey JR, Page DC. Parallel evolution of male germline epigenetic poising and somatic development in animals. Nat. Genet. 2016;48:888–894. doi: 10.1038/ng.3591. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Lu T, Heyne S, Dror E, Casas E, Leonhardt L, Boenke T, Yang C, Sagar, Arrigoni L, Dalgaard K, Teperino R, Enders L, Selvaraj M, Ruf M, Raja S, Xie H, Boenisch U, Orkin S, Lynn F, Hoffman B, Grun D, Vavouri T, Lempradl A, Pospisilik A. The Polycombdependent epigenome controls B-cell dysfunction, dedifferentiation and diabetes. bioRxiv. 2017 doi: 10.1016/j.cmet.2018.04.013. DOI:205641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatouk DM, Natarajan A, Shibata Y, Song L, Crawford GE, Ohler U, Capel B. Genome-wide identification of regulatory elements in Sertoli cells. Development. 2017;144:720–730. doi: 10.1242/dev.142554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CS, Miner MD, Doerr JR, Jackson JP, Jacobsen SE, Wall R, Teitell M. CmC(A/T)GG DNA methylation in mature B cell lymphoma gene silencing. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10404–10409. doi: 10.1073/pnas.181206898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand O, et al. DMRT1 expression during gonadal differentiation and spermatogenesis in the rainbow trout, Oncorhynchus mykiss. Biochim. Biophys. Acta. 2000;1493(1–2):180–187. doi: 10.1016/s0167-4781(00)00186-x. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama H, et al. Dmrt1 mutation causes a male-to-female sex reversal after the sex determination by Dmy in the medaka. Chromosome Res. 2012;20(1):163–176. doi: 10.1007/s10577-011-9264-x. [DOI] [PubMed] [Google Scholar]
- Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature. 2011;476:101–104. doi: 10.1038/nature10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson CK, Zarkower D. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 2012;13:163–174. doi: 10.1038/nrg3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen HA, Clinton M. Avian sex chromosomes: dosage compensation matters. Chromosome Res. 2009;17:687–697. doi: 10.1007/s10577-009-9056-8. [DOI] [PubMed] [Google Scholar]
- Mitsuhashi T, Warita K, Sugawara T, Tabuchi Y, Takasaki I, Kondo T, Hayashi F, Wang ZY, Matsumoto Y, Miki T, Takeuchi Y, Ebina Y, Yamada H, Sakuragi N, Yokoyama T, Nanmori T, Kitagawa H, Kant JA, Hoshi N. Epigenetic abnormality of SRY gene in the adult XY female with pericentric inversion of the Y chromosome. Congenital. Anom. (Kyoto) 2010;50:85–94. doi: 10.1111/j.1741-4520.2010.00274.x. [DOI] [PubMed] [Google Scholar]
- Mousavi K, Zare H, Wang AH, Sartorelli V. Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol. Cell. 2012;45:255–262. doi: 10.1016/j.molcel.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger SC, Natarajan A, Looger LL, Ohler U, Capel B. Fine time course expression analysis identifies cascades of activation and repression and maps a putative regulator of mammalian sex determination. PLoS Genet. 2013;9:e1003630. doi: 10.1371/journal.pgen.1003630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy EC, Zhurkin VB, Louis JM, Cornilescu G, Clore GM. Structural basis for SRY-dependent 46-X, Y sex reversal: modulation of DNA bending by a naturally occurring point mutation. J. Mol. Biol. 2001;312:481–499. doi: 10.1006/jmbi.2001.4977. [DOI] [PubMed] [Google Scholar]
- Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, Shu X, Kriaucionis S, Bird A. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2709–2714. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Nef S, Schaad O, Stallings NR, Cederroth CR, Pitetti JL, Schaer G, Malki S, Dubois-Dauphin M, Boizet-Bonhoure B, Descombes P, Parker KL, Vassalli JD. Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev. Biol. 2005;287:361–377. doi: 10.1016/j.ydbio.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Niehrs C, Schafer A. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol. 2012;22:220–227. doi: 10.1016/j.tcb.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Nishino K, Hattori N, Sato S, Arai Y, Tanaka S, Nagy A, Shiota K. Non-CpG methylation occurs in the regulatory region of the Sry gene. J. Reprod. Dev. 2011;57:586–593. doi: 10.1262/jrd.11-033a. [DOI] [PubMed] [Google Scholar]
- Nishino K, Hattori N, Tanaka S, Shiota K. DNA methylation-mediated control of Sry gene expression in mouse gonadal development. J. Biol. Chem. 2004;279:22306–22313. doi: 10.1074/jbc.M309513200. [DOI] [PubMed] [Google Scholar]
- Oh HJ, Li Y, Lau YF. Sry associates with the heterochromatin protein 1 complex by interacting with a KRAB domain protein. Biol. Reprod. 2005;72:407–415. doi: 10.1095/biolreprod.104.034447. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Ottolenghi C, Pelosi E, Tran J, Colombino M, Douglass E, Nedorezov T, Cao A, Forabosco A, Schlessinger D. Loss of Wnt4 and Foxl2 leads to female-to-male sex reversal extending to germ cells. Hum. Mol. Genet. 2007;16:2795–2804. doi: 10.1093/hmg/ddm235. [DOI] [PubMed] [Google Scholar]
- Peng H, Ivanov AV, Oh HJ, Lau YF, Rauscher FJ., 3rd Epigenetic gene silencing by the SRY protein is mediated by a KRAB-O protein that recruits the KAP1 co-repressor machinery. J. Biol. Chem. 2009;284:35670–35680. doi: 10.1074/jbc.M109.032086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinarbasi E, Elliott J, Hornby DP. Activation of a yeast pseudo DNA methyltransferase by deletion of a single amino acid. J. Mol. Biol. 1996;257:804–813. doi: 10.1006/jmbi.1996.0203. [DOI] [PubMed] [Google Scholar]
- Pinter SF, Sadreyev RI, Yildirim E, Jeon Y, Ohsumi TK, Borowsky M, Lee JT. Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res. 2012;22:1864–1876. doi: 10.1101/gr.133751.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanco JC, Wilhelm D, Davidson TL, Knight D, Koopman P. Sox10 gain-of-function causes XX sex reversal in mice: implications for human 22q-linked disorders of sex development. Hum. Mol. Genet. 2010;19:506–516. doi: 10.1093/hmg/ddp520. [DOI] [PubMed] [Google Scholar]
- Polanco JC, Wilhelm D, Mizusaki H, Jackson A, Browne C, Davidson T, Harley V, Sinclair A, Koopman P. Functional analysis of the SRY-KRAB interaction in mouse sex determination. Biol. Cell. 2009;101:55–67. doi: 10.1042/BC20080061. [DOI] [PubMed] [Google Scholar]
- Pontiggia A, Rimini R, Harley VR, Goodfellow PN, Lovell-Badge R, Bianchi ME. Sex-reversing mutations affect the architecture of SRY-DNA complexes. EMBO J. 1994;13:6115–6124. doi: 10.1002/j.1460-2075.1994.tb06958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Kong LK, Poirier C, Truong C, Overbeek PA, Bishop CE. Long-range activation of Sox9 in odd sex (ods) mice. Hum. Mol. Genet. 2004;13:1213–1218. doi: 10.1093/hmg/ddh141. [DOI] [PubMed] [Google Scholar]
- Razin A, Cedar H. DNA methylation in eukaryotic cells. Int. Rev. Cytol. 1984;92:159–185. doi: 10.1016/s0074-7696(08)61327-3. [DOI] [PubMed] [Google Scholar]
- Read CM, Cary PD, Crane-Robinson C, Driscoll PC, Norman DG. Solution structure of a DNA-binding domain from HMG1. Nucleic Acids Res. 1993;21:3427–3436. doi: 10.1093/nar/21.15.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- Shao C, Li Q, Chen S, Zhang P, Lian J, Hu Q, Sun B, Jin L, Liu S, Wang Z, Zhao H, Jin Z, Liang Z, Li Y, Zheng Q, Zhang Y, Wang J, Zhang G. Epigenetic modification and inheritance in sexual reversal of fish. Genome Res. 2014;24:604–615. doi: 10.1101/gr.162172.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel KB, et al. TX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 2012;8(9):e1002964. doi: 10.1371/journal.pgen.1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Singh NP, Madabhushi SR, Srivastava S, Senthilkumar R, Neeraja C, Khosla S, Mishra RK. Epigenetic profile of the euchromatic region of human Y chromosome. Nucleic Acids Res. 2011;39:3594–3606. doi: 10.1093/nar/gkq1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha P, Singh K, Sachan M. Heterogeneous pattern of DNA methylation in developmentally important genes correlates with its chromatin conformation. BMC Mol. Biol. 2017;18:1. doi: 10.1186/s12867-016-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461(7261):267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- Stoeva R, Grozdanova L, Scherer G, Krasteva M, Bausch E, Krastev T, Linev A, Stefanova M. A novel SOX9 nonsense mutation, q401x, in a case of campomelic dysplasia with XY sex reversal. Genet. Counsel. 2011;22:49–53. [PubMed] [Google Scholar]
- Teranishi M, Shimada Y, Hori T, Nakabayashi O, Kikuchi T, Macleod T, Pym R, Sheldon B, Solovei I, Macgregor H, Mizuno S. Transcripts of the MHM region on the chicken Z chromosome accumulate as non-coding RNA in the nucleus of female cells adjacent to the DMRT1 locus. Chromosome Res. 2001;9:147–165. doi: 10.1023/a:1009235120741. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Takahashi S, Takahashi Y, Asahara H. Transcriptional co-activators CREB-binding protein and p300 regulate chondrocyte-specific gene expression via association with Sox9. J. Biol. Chem. 2003;278:27224–27229. doi: 10.1074/jbc.M303471200. [DOI] [PubMed] [Google Scholar]
- Uhlenhaut NH, Jakob S, Anlag K, Eisenberger T, Sekido R, Kress J, Treier AC, Klugmann C, Klasen C, Holter NI, Riethmacher D, Schutz G, Cooney AJ, Lovell-Badge R, Treier M. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009;139:1130–1142. doi: 10.1016/j.cell.2009.11.021. [DOI] [PubMed] [Google Scholar]
- Vidal VP, Chaboissier MC, de Rooij DG, Schedl A. Sox9 induces testis development in XX transgenic mice. Nat. Genet. 2001;28:216–217. doi: 10.1038/90046. [DOI] [PubMed] [Google Scholar]
- Waddington C. Principles of Development. Allen and Unwin; London: 1956. [Google Scholar]
- Waddington CH. Organisers and Genes. Cambridge University Press; 1940. [Google Scholar]
- Waddington CH. Canalization of development and the inheritance of aquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- Walport LJ, Hopkinson RJ, Vollmar M, Madden SK, Gileadi C, Oppermann U, Schofield CJ, Johansson C. Human UTY(KDM6C) is a male-specific N-methyl lysyl demethylase. J. Biol. Chem. 2014;289:18302–18313. doi: 10.1074/jbc.M114.555052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu YC, Markoulaki S, Welstead GG, Cheng AW, Shivalila CS, Pyntikova T, Dadon DB, Voytas DF, Bogdanove AJ, Page DC, Jaenisch R. TALEN-mediated editing of the mouse Y chromosome. Nat. Biotechnol. 2013;31:530–532. doi: 10.1038/nbt.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr N, Carre GA, Siggers P, Faleato JV, Brixey R, Pope M, Bogani D, Childers M, Wells S, Scudamore CL, Tedesco M, del Barco Barrantes I, Nebreda AR, Trainor PA, Greenfield A. Gadd45gamma and Map3k4 interactions regulate mouse testis determination via p38 MAPK-mediated control of Sry expression. Dev. Cell. 2012;23:1020–1031. doi: 10.1016/j.devcel.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welstead GG, Creyghton MP, Bilodeau S, Cheng AW, Markoulaki S, Young RA, Jaenisch R. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc. Natl. Acad. Sci. U. S. A. 2012;109:13004–13009. doi: 10.1073/pnas.1210787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Deng X, Disteche CM. Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS One. 2008a;3:e2553. doi: 10.1371/journal.pone.0002553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Deng X, Watkins R, Disteche CM. Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J. Neurosci. 2008b;28:4521–4527. doi: 10.1523/JNEUROSCI.5382-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010a;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zheng J, Xu G, Qu L, Chen S, Li J, Yang N. Exogenous cMHM regulates the expression of DMRT1 and ER alpha in avian testes. Mol. Biol. Rep. 2010b;37:1841–1847. doi: 10.1007/s11033-009-9619-y. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Yoshimoto S, et al. A W-linked DMdomain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc. Natl. Acad. Sci. U. S. A. 2008;105(7):2469–2474. doi: 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Koopman P. SRY protein function in sex determination: thinking outside the box. Chromosome Res. 2012;20(1):153–162. doi: 10.1007/s10577-011-9256-x. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]