Abstract
Sex-ratio distortion is the most common form of non-Mendelian segregation observed in natural populations. It may occur even more frequently than direct observations suggest, because the dysgenic population consequences of a biased sex ratio are expected to result in the rapid evolution of suppressors, resulting in suppressed or “cryptic” segregation distortion. Here we report evidence for cryptic sex-ratio distortion that was discovered by introgressing segments of the genome of Drosophila mauritiana into the genome of Drosophila simulans. The autosomal suppressor of sex-ratio distortion, which is also associated with a reduction in hybrid male fertility, has been genetically localized to a region smaller than 80-kb pairs in chromosome 3.
Mendelian segregation is the foundation of almost every important principle in population genetics theory. It is also widely observed. Examples of non-Mendelian segregation—also called segregation distortion or meiotic drive—though sometimes dramatic, are rare. However, the apparent near-universality of Mendelian segregation may be misleading. Nearly 50 years ago Sandler and Novitski (1) pointed out that new mutant alleles showing segregation distortion might occur frequently within populations, but be concealed by the fixation of suppressor mutations soon after the distorters arise. In this case, the infrequency of non-Mendelian segregation in natural populations would reflect only its transience as an evolutionary phenomenon, not its rarity as a biological phenomenon.
A newly arisen segregation distorter can be fixed, lost, or become a stable polymorphism. Stable polymorphism may occur when the advantage of favored gametic transmission is balanced by lower fitness (2, 3) or suppressed by modifiers (4, 5). The evolution of modifiers of the degree of distortion depends on genetic linkage: loose linkage favors suppressors, tight linkage favors enhancers (6). Although much of our understanding of segregation distortion comes from two well studied autosomal systems, segregation distortion in Drosophila melanogaster (7) and the t-haplotype in Mus musculus (8), the multiple reports of sex-ratio distortion (9), as well as theoretical arguments (10, 11), suggest that segregation distortion of the sex chromosomes may evolve more readily than that of autosomes. Theory further suggests that the deleterious effect of a biased sex ratio results in rapid selection of Y-linked or autosomal suppressors (12, 13), thus rendering the distortion cryptic. Evidence bearing on the hypothesis that cycles of distortion and suppression may have happened multiple times in the evolutionary history of a species (14, 15) is scarce and controversial (16–22).
There is a test of the distortion-suppression hypothesis. Among genetically isolated subpopulations, each would acquire its own independent sequence of distorters and suppressors, depending on the chance order of occurrence. Because the distorters present in any one subpopulation might not be affected by suppressor alleles present in a different subpopulation, segregation distorters might be revealed in the hybrids of interspecific crosses. In tests of first-generation hybrids, this approach will fail if most suppressors are dominant. To circumvent this problem, we devised a scheme in which segments of the genome from Drosophila mauritiana are made homozygous in the genetic background of the sibling species Drosophila simulans. Here we report that the sex ratio of D. simulans is markedly distorted when a segment of the D. mauritiana third chromosome, not longer than 80 kb, is homozygous. An intriguing feature of this chromosomal segment is that it is also associated with hybrid male sterility.
Materials and Methods
Introgression Lines.
The establishment of D. mauritiana lines with P[w+] inserts has been described (23); the P[w+] represents an immobile derivative of the transposable P element marked with the mini-white w+ gene. Introgression was initiated with 38 lines of D. mauritiana, each containing a copy of P[w+] inserted in the third chromosome at a different site, and a single highly inbred line of D. simulans (13w × JJ) (24). D. mauritiana males were crossed with D. simulans females, and P[w+]/+ female progeny were backcrossed to the parental D. simulans strain for five generations until 10–30% of the P[w+]/+ male progeny were fertile. These males were crossed to a D. simulans laboratory stock w; nt; III (simB), where III represents the isogenic third chromosome from 13w × JJ. By selecting for w-marked X and nt-marked second chromosomes transmitted through males, stocks were constructed that contain a single recombined third chromosome, tagged by P[w+], in a pure D. simulans genetic background. This chromosome was protected from further recombination by transmission through males. The location and extent of introgressed D. mauritiana segments on the third chromosome were characterized by genotyping 38 molecular markers (see below). Multiple independent introgressions were made with each P[w+] insert, resulting in a total of 259 lines used in the experiment. Only one of the introgression stocks used contained more than one D. mauritiana segment. The introgression lines were maintained by selecting P[w+]-tagged males each generation to cross with simB virgins. To obtain homozygous P[w+] individuals, heterozygous males and females from each introgression line were crossed. Because eye color is sensitive to the copy number and position of the P[w+] insert, heterozygous and homozygous genotypes usually are distinguishable and, in many cases, different P[w+] inserts can be distinguished easily.
Molecular Markers.
To assess the molecular extent of each introgression along the third chromosome, 38 allele-specific oligonucleotide (ASO) molecular markers across the third chromosome were developed. Genotyping was carried out essentially as in ref. 24.
Fertility Assay.
Mating tests were used to assay the segregation ratio and fertility. Each male was mated to three D. simulans w; e females (<3 days old) in a vial for 7 days. Progeny were classified by eye color and sex, and counts were made on the 13th, 16th, and 19th day after the cross was set up. Female fertility tests were conducted similarly with 8 days of mating. A female was considered fertile if she produced any progeny. Male fertility was classified into three types according to the mean progeny number per male: sterile (<5), semifertile (5–40), and fertile (>40). After mating, individuals were genotyped by ASO to determine homozygosity or heterozygosity along the entire introgressed region. All experiments were carried out at room temperature (21–24°C) with standard cornmeal–molasses medium.
Statistics.
A Wilcoxon two-sample test was used for each marker separately to test the null hypothesis of no difference in sex ratio (K, proportion of females) between two alternative genotypes. The critical value for the entire experiment was obtained by the Bonferroni method, adjusting the significance level to P′ = P/n, where P = 0.05 and n is the number of markers (25).
In the “2-P” mapping experiment described in Results, the method used for estimating the distance between two hypothetical independent sites, one controlling fertility (F) and the other sex-ratio distortion (D), was as follows. Recombinants were selected between two P[w+] inserts (P40 and P38), which are about 2,200 kb apart. Only fertile recombinants are informative, as distortion cannot be assayed on sterile flies. Numbers of recombinants are given in Fig. 4. The estimated distance between the P40 insertion and F is (13/172) × 2,200 = 166 kb, and that between F and the P38 insertion is 2,034 kb. If the order of sites is P40 insertion–D–F–P38 insertion, a crossover between D and F could be detected among the seven fertile recombinants in the 166-kb interval, whereas if the order is P40 insertion–F–D–P38 insertion, the 78 fertile recombinants in the 2,034-kb interval are informative. In the former case there are j = 7/166 = 0.042 informative recombinants per kb; in the latter case there are j = 78/2,034 = 0.038 informative recombinants per kb. Assuming a Poisson distribution, the probability of observing one or more recombinants in a subinterval of size y kb is 1 − Exp(−jy). When j = 0.038, the probability of observing no crossover between F and D is 5% when the sites are separated by a distance of 79 kb.
Figure 4.
Male fertility of introgression sublines generated by recombination in the tmy region with a 2-P selection scheme. Each circle represents the average of one subline. The diagonal line depicts a sex ratio of K = 0.5. (A) Recombinants tagged with the P40 insertion tested when heterozygous with introgression chromosome 38.9; a range of 2–20 individuals per line were tested (average, 7.2). (B) Recombinants tagged with the P38 insertion tested when heterozygous with introgression chromosome 40.6; a range of 4–12 individuals per line were tested (average, 9.0).
Results
A Cryptic Suppressor of Sex-Ratio Distortion.
In the present study, segments of the D. mauritiana third chromosome were introgressed into a homozygous w (white eye) strain of D. simulans through backcrossing and marker selection. P[w+] inserts at various positions along the D. mauritiana chromosome provided an efficient system for introgression. The limits of introgressed segments were defined by ASO markers. Each introgressed segment spanned an average of about 17% of the third chromosome, but the entire third chromosome was covered with multiple overlapping introgressions (26).
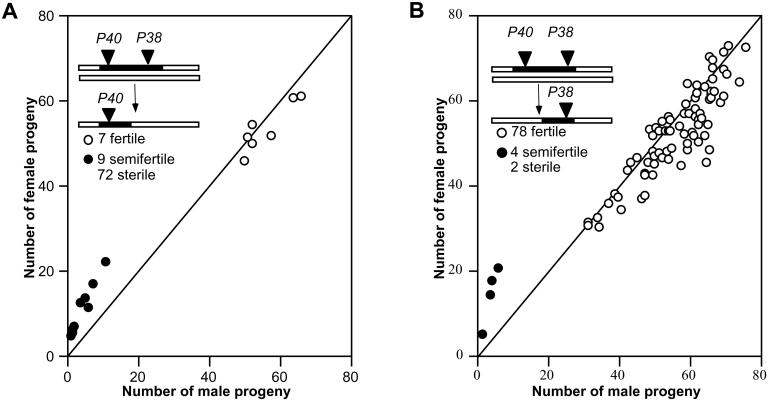
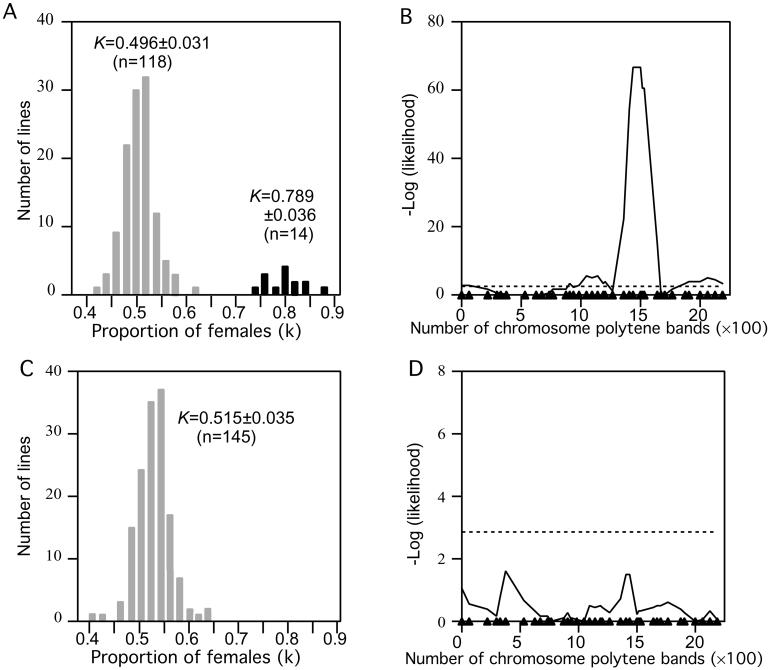
Both males and females homozygous for introgressions were tested for sex-ratio distortion by scoring the progeny from crosses with a standard D. simulans line. One group of lines showed an extreme effect on sex ratio in the progeny from homozygous males (Fig. 1A). The average sex ratio (K) among the deviant lines is ≈80% females. Fig. 1B shows the results of testing for association of the sex ratio with molecular markers across the third chromosome. The sex-ratio effect clearly maps to a region flanked by molecular markers Akt1 (89B9) and Su(Hw) (88B3). Homozygous males from all 14 nonsterile lines that cover this region gave rise to strongly female-biased progeny (K ≥ 0.70).
Figure 1.
(A and C) Sex ratio (K, proportion of females) in the progeny of introgression lines. Mean and SD of K are given (n = sample size). (B and D) Association of K with molecular markers based on Wilcoxon two-sample tests. Dashed horizontal lines represent the experiment-wide threshold for significance, and triangles on the abscissa represent positions of molecular markers. A and B, homozygous males; C and D, homozygous females.
Several biological phenomena can result in a distorted sex ratio. In the present case, simple sex-specific viability effects can be ruled out, because the distortion is observed among the progeny of males, but not among the progeny of females, homozygous for introgressions of the Akt1-Su(Hw) region (Fig. 1). In addition, there is no evidence for embryonic lethality in progeny of homozygous males. Although the homozygous males have low fertility (see below), 89.9% (n = 69) of hatched eggs survived to adulthood, compared with the control value of 80.4% (n = 919). Furthermore, explanations based on cytoplasmic factors can be eliminated because all introgression lines have the same D. simulans cytoplasm. These observations suggest that disparities in the relative numbers of functional X-bearing sperm and Y-bearing sperm is the cause of the sex-ratio distortion.
Because of the female-biased sex ratio, we designate the third-chromosomal gene associated with the distortion as too much yin (tmy). None of the 14 introgressions that yielded a biased sex ratio when homozygous also showed distortion when heterozygous (data not shown). Therefore, we infer that the tmy allele in D. simulans is dominant to the tmy allele in D. mauritiana. We denote the dominant D. simulans allele as Tmy and the recessive D. mauritiana allele as tmy. Hence, Tmy appears to be a dominant autosomal suppressor of an X-linked sex-ratio distortion in D. simulans.
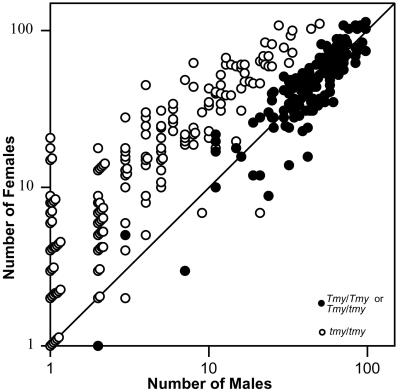
The introgressed tmy allele that causes sex-ratio distortion is also associated with male sterility. The average number of progeny per homozygous male was used to categorize each introgression line into sterile (<5), semifertile (5–40), or fertile (>40), as shown in Fig. 2. All six fertile introgressions have genotype Tmy/Tmy and show a normal sex ratio (n = 128). Twenty-two heterozygous recombinants from other lines were inferred to be Tmy/tmy males, as they show normal fertility and sex ratio like the six homozygous lines that are fertile (F[1,165] = 0.148, P = 0.701). Among the pooled group of Tmy/Tmy and Tmy/tmy males, 140 (93%) yielded progeny with a mean number ± SD of 101 ± 42 per male and a mean sex ratio of 0.50 ± 0.07. In contrast, all 14 semifertile introgressions (tmy/tmy) show a strongly female-biased sex ratio. Among 452 tested males in this category, only 217 (48%) produced progeny, yielding a mean number of 26 ± 32 progeny per fertile male (range: 1–160) and a mean sex ratio of 0.78 ± 0.19. Nearly all individual males in this group showed female-biased progeny (white circles in Fig. 3). Male fertility varies greatly, even among males from the same introgression line.
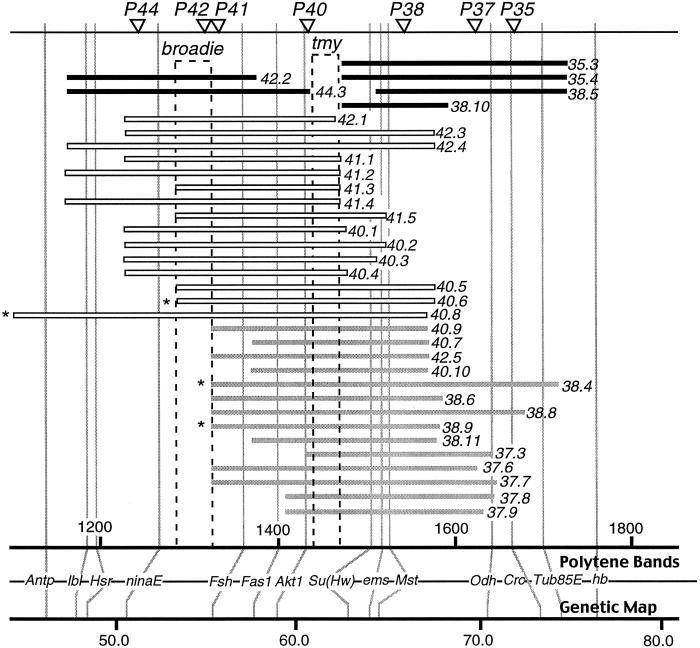
Figure 2.
Mapping tmy, the suppressor of sex-ratio distortion. Horizontal bars represent the introgressed D. mauritiana segments defined by molecular markers. P[w+] inserts are represented by triangles at the top and numbers to the right of each bar. The first two digits identify the P[w+] insert, and the number after the period identifies each independent introgression. Bar shading represents the phenotype of homozygous males. Black bars represent fertile males with no sex-ratio distortion, gray bars represent semifertile males with significant sex-ratio distortion, and white bars represent sterile males. *, lines used in the 2-P mapping scheme in Fig. 4.
Figure 3.
Phenotypes of Tmy/Tmy and Tmy/tmy males (●) differ from those of tmy/tmy males (○). The diagonal line corresponds to a sex ratio of K = 0.5. Not included are 42 homozygous tmy males that produced only female progeny and five that produced only male progeny. The symbols for individuals with the same phenotype are stacked in the graph.
The interval between Akt1 and Su(Hw), which contains tmy, is the only introgressed region of the third chromosome that, when homozygous, is associated with male sterility or semifertility. The remaining part of the third chromosome is thoroughly covered by homozygous introgressions that are fully fertile in females and males (26).
Neither the male sterility nor the sex-ratio distortion of tmy can be accounted for by a particular P-element insertion, because several different P-element inserts in this region have similar phenotypes of semisterility and sex-ratio distortion (Fig. 2). Furthermore, none of the P-element insertions show significantly female-biased progeny in the original pure stocks of D. mauritiana (data not shown).
As shown in Fig. 2, the genetic basis of the difference between the semifertile and the sterile introgressions lies in a small region in the interval between ninaE and Fsh. Evidently, this interval contains a genetic factor (which we call broadie) that causes complete male sterility in combination with tmy.
Sex-Ratio Distortion and Sterility Localize Together Genetically.
To locate tmy more precisely along chromosome 3, we mated males and females heterozygous for an introgressed chromosomal segment and its associated P[w+] insert. From this cross, 1,458 offspring homozygous for the P[w+] insert were selected for ASO genotyping and phenotypic testing. Each male offspring could be classified into one of three types: fertile with normal sex ratio, semifertile with sex-ratio distortion, or sterile. No fertile males with sex ratio distortion were observed, strongly suggesting that both phenotypes are pleiotropic effects of a single gene. These data place tmy 0.9 centimorgan (≈330 kb) distal to Akt1 and 3.5 centimorgans proximal to Su(Hw).
To further test whether tmy controls both sex-ratio distortion and sterility, additional recombinants were generated. First, an introgression line tagged with two P[w+]-inserts was constructed by recombining introgression chromosomes 40.8 and 38.4 (Fig. 2). From this 2-P line, chromosomes that had undergone recombination between the two P[w+] inserts were recovered and tested over introgression chromosomes 38.9 or 40.6, both of which contain tmy. Among 172 recombinants between the two P[w+] inserts, again only the three parental phenotypes were observed: sterile, semifertile with sex-ratio distortion (tmy), or fertile with no sex-ratio distortion (Tmy) (Fig. 4). There were no fertile males with distortion. The length of DNA between the two P[w+] inserts is estimated to be about 2,200 kb. From this length and the number of recombinants analyzed, we estimate that the probability of observing no crossover between fertility and distortion is about 5% if there are two separate sites located about 80 kb apart (see Materials and Methods). In other words, we are 95% confident that the two sites are less than 80 kb apart. This calculation further supports the notion of pleiotropy, but final proof awaits molecular cloning and analysis of tmy.
Discussion
The data presented here indicate that a cryptic genetic system of sex-ratio distortion exists in D. simulans, which can be revealed by replacing a conspecific dominant autosomal suppressor (Tmy) with a nonsuppressing allele (tmy) from D. mauritiana. A possible alternative interpretation is that the recessive mauritiana allele of tmy somehow creates the distortion when it interacts with the sex chromosomes from D. simulans but not from D. mauritiana. In either case, the sex-ratio segregation distortion represents a type of hybrid incompatibility. The hypothesis of a cryptic system in D. simulans appears to be the more plausible in terms of gene action, and it is also consistent with population genetic theory, but the alternative cannot be excluded.
Several previous attempts to find segregation distortion in hybrid Drosophila populations have not been fruitful (16–18). Those studies were unable to detect dominant suppressors because at least one complement of conspecific autosomes was present in the tested males. However, several other apparent cases of cryptic sex-ratio distortion have been reported recently. (i) F1 hybrid males from crosses between different subpopulations of D. simulans yielded female-biased sex ratios, a genetic condition that appears to be suppressed within some populations by a polygenic system of factors on the Y chromosome and both major autosomes (27–29). (ii) Some recombinant inbred lines established from hybrids of D. simulans and Drosophila sechellia showed female-biased sex-ratio distortion (30). It is not clear yet whether either of these sex-ratio traits involving D. simulans share any components in common with the sex-ratio distortion reported here. (iii) The usually sterile hybrid males from crosses between subspecies of Drosophila pseudoobscura from the United States and Bogota, Columbia, can, in the presence of certain genes in the genetic background, show some slight fertility, and the progeny of these “rescued” males show a distorted sex ratio (31). (iv) Some hybrids between different strains of Drosophila subobscura show sex-ratio distortion, and male sterility as well (32). (v) In D. melanogaster, a Y-linked suppressor of the X-linked repeated sequence, Stellate, may have originated as a suppressor of Stellate-associated meiotic drive (33).
The study of simulans/mauritiana introgressions reported here provides clear evidence for a tight association between a suppressor of sex-ratio distortion and hybrid male sterility. Both traits localize to a DNA fragment of 80 kb or smaller and may be pleiotropic effects of a single gene. The simulans/sechellia study was not able to detect such an association because only fertile introgressions were produced owing to the method by which the lines were constructed (30). The report concerning the subspecies of D. pseudoobscura suggests that genes causing hybrid sex-ratio distortion may map to the same chromosomal intervals as those causing hybrid male sterility, but in that case the level of genetic resolution is somewhat uncertain (31). Similarly, although male sterility and sex-ratio distortion were observed simultaneously in crosses between different strains of D. subobscura (32), it is unclear whether they have the same genetic basis.
One genetic theory of speciation proposes that meiotic drive underlies Haldane's rule (10, 11), which refers to the widespread finding that, when F1 hybrid sterility or inviability affects the sexes unequally, it is usually the heterogametic sex rather than the homogametic sex that is more severely affected. The hypothesis underlying the theory is that related species frequently each contain suppressed systems of meiotic drive that become reactivated in hybrids, resulting in sterility. If drive systems, such as sex-ratio distortion, are more frequent in the heterogametic sex, then so will the hybrid sterility. This theory is quite attractive because it applies equally well to species with heterogametic females as to those with heterogamatic males. This is not the case with some alternatives such as the “faster male” hypothesis (34, 35), which ascribes Haldane's rule to an accelerated rate of evolution of genes associated with male reproductive functions. The discovery of tmy provides direct evidence that links a cryptic meiotic drive system to hybrid sterility. Therefore, it lends some credence to the meiotic drive theory of Haldane's rule for sterility. One caveat is that, because the sterility and distorting effects of tmy are recessive in introgression lines, tmy may not actually contribute to F1 male sterility, although it is possible that its dominance effects are different in the distinct genetic background of F1 hybrids. In addition, the fact that only one such factor was found in the third chromosome suggests that drive/sterility associations are not numerous, although the ascertainment of such associations is difficult because it requires strong sex-ratio distortion and relatively large effects on fertility.
One limitation of the meiotic drive theory as a general explanation of Haldane's rule is that the connection between meiotic drive and inviability remains, at best, obscure. However, there is considerable evidence that sex-specific hybrid sterility and inviability have different genetic bases (36), so a common evolutionary explanation for sterility and inviability may not exist.
The meiotic drive theory of Haldane's rule for sterility can be expanded to include secondary effects. When a segregation distorter arises in a population, it may increase in frequency or even become fixed, despite possible deleterious pleiotropic effects on fertility. Suppressors of both the distortion and fertility reduction will evolve and might involve different genes. When these processes occur in allopatric populations, divergence in the genetic basis of spermatogenesis will evolve, eventually leading to hybrid incompatibilities. In this case, some of the hybrid sterility factors will be associated with distortion and others will not. Further experiments capable of detecting cryptic drive systems are needed to determine how frequently this hypothetical scenario might take place in natural populations.
Acknowledgments
We thank Lynn Stam for the development and teaching of molecular marker methods; Douda Bensasson, Cristian Castillo-Davis, Emmanouil Dermitzakis, David Haig, John Parsch, and John True for critiques, as well as Rick Fehon and his lab for logistical and spiritual support. This work was supported by National Institutes of Health Grants GM47292 and GM60035. Y.T. was a recipient of Catherine Stern Dissertation Year Fellowship of Duke University.
Abbreviation
- ASO
allele-specific oligonucleotide
References
- 1.Sandler L, Novitski E. Am Nat. 1957;41:105–110. [Google Scholar]
- 2.Edwards A W. Heredity. 1961;16:291–304. [Google Scholar]
- 3.Wu C-I. Theor Popul Biol. 1983;24:107–120. doi: 10.1016/0040-5809(83)90035-7. [DOI] [PubMed] [Google Scholar]
- 4.Prout T, Bundgaard J, Bryant S. Theor Popul Biol. 1973;4:446–465. doi: 10.1016/0040-5809(73)90020-8. [DOI] [PubMed] [Google Scholar]
- 5.Feldman M W, Otto S P. Am Nat. 1991;137:443–456. [Google Scholar]
- 6.Hartl D L. Theor Popul Biol. 1975;7:168–174. doi: 10.1016/0040-5809(75)90012-x. [DOI] [PubMed] [Google Scholar]
- 7.Temin R G, Ganetzky B, Powers P A, Lyttle T W, Pimpinelli S, Dimitri P, Wu C-I, Hiraizumi Y. Am Nat. 1991;137:287–331. [Google Scholar]
- 8.Silver L M. Trends Genet. 1993;9:250–254. doi: 10.1016/0168-9525(93)90090-5. [DOI] [PubMed] [Google Scholar]
- 9.Jaenike J. Am Nat. 1996;148:237–254. [Google Scholar]
- 10.Hurst L D, Pomiankowski A. Genetics. 1991;128:841–858. doi: 10.1093/genetics/128.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank S. Evolution (Lawrence, Kans) 1991;45:262–267. doi: 10.1111/j.1558-5646.1991.tb04401.x. [DOI] [PubMed] [Google Scholar]
- 12.Fisher R A. The Genetical Theory of Natural Selection. New York: Dover; 1958. [Google Scholar]
- 13.Hamilton W D. Science. 1967;156:477–488. doi: 10.1126/science.156.3774.477. [DOI] [PubMed] [Google Scholar]
- 14.Crow J F. BioEssays. 1991;13:305–312. doi: 10.1002/bies.950130609. [DOI] [PubMed] [Google Scholar]
- 15.Carvalho A B, Vaz S C. Heredity. 1999;83:221–228. doi: 10.1038/sj.hdy.6886100. [DOI] [PubMed] [Google Scholar]
- 16.Coyne J A. Genetics. 1986;114:485–494. doi: 10.1093/genetics/114.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coyne J A, Orr H A. Evolution (Lawrence, Kans) 1993;47:685–687. doi: 10.1111/j.1558-5646.1993.tb02123.x. [DOI] [PubMed] [Google Scholar]
- 18.Johnson N A, Wu C-I. Genetics. 1992;130:507–511. doi: 10.1093/genetics/130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coyne J A, Charlesworth B, Orr H A. Evolution (Lawrence, Kans) 1991;45:1710–1714. doi: 10.1111/j.1558-5646.1991.tb02677.x. [DOI] [PubMed] [Google Scholar]
- 20.Frank S A. Evolution (Lawrence, Kans) 1991;45:1714–1717. doi: 10.1111/j.1558-5646.1991.tb02678.x. [DOI] [PubMed] [Google Scholar]
- 21.Charlesworth B, Coyne J A, Orr H A. Genetics. 1993;133:421–424. doi: 10.1093/genetics/133.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pomiankowski A, Hurst L D. Genetics. 1993;133:425–432. [Google Scholar]
- 23.True J R, Mercer J M, Laurie C C. Genetics. 1996;142:507–523. doi: 10.1093/genetics/142.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J J, Mercer J M, Stam L F, Gibson G, C, Laurie C C. Genetics. 1996;142:1129–1145. doi: 10.1093/genetics/142.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sokal R R, Rohlf F J. Biometry: The Principles and Practice of Statistics in Biological Research. New York: Freeman; 1995. [Google Scholar]
- 26.Tao Y. Ph.D. thesis. Durham, NC: Duke University; 2000. [Google Scholar]
- 27.Merçot H, Atlan A, Jacques M, Montchamp-Moreau C. J Evol Biol. 1995;8:283–300. [Google Scholar]
- 28.Atlan A, Merçot M, Landré C, Montchamp-Moreau C. Evolution (Lawrence, Kans) 1997;51:1884–1893. doi: 10.1111/j.1558-5646.1997.tb05111.x. [DOI] [PubMed] [Google Scholar]
- 29.Cazemajor M, Joly D, Montchamp-Moreau M. Genetics. 2000;154:229–236. doi: 10.1093/genetics/154.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dermitzakis E T, Masly J P, Waldrip H M, Clark A G. Genetics. 2000;154:687–694. doi: 10.1093/genetics/154.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orr H A, Presgraves D C. BioEssays. 2000;22:1085–1094. doi: 10.1002/1521-1878(200012)22:12<1085::AID-BIES6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 32.Hauschteck-Jungen E. Genetica. 1990;83:31–44. doi: 10.1007/BF00774686. [DOI] [PubMed] [Google Scholar]
- 33.Hurst L D. Genetics. 1996;142:641–643. doi: 10.1093/genetics/142.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu C-I, Davis A W. Am Nat. 1993;142:187–212. doi: 10.1086/285534. [DOI] [PubMed] [Google Scholar]
- 35.Wu C-I, Johnson N, Palopoli M F. Trends Ecol Evol. 1996;11:281–284. doi: 10.1016/0169-5347(96)10033-1. [DOI] [PubMed] [Google Scholar]
- 36.Turelli M, Orr H A. Genetics. 2000;154:163–1679. doi: 10.1093/genetics/154.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]