Abstract
Adenosine to inosine RNA editing is an epigenetic process that entails site-specific modifications in double-stranded RNA molecules, catalyzed by adenosine deaminases acting on RNA (ADARs). Using the multiplex microfluidic PCR and deep sequencing technique, we recently showed that exposing adolescent female rats to chronic unpredictable stress before reproduction affects editing in the prefrontal cortex and amygdala of their newborn offspring, particularly at the serotonin receptor 5-HT2c (encoded by Htr2c). Here, we used the same technique to determine whether post-stress, pre-reproductive maternal treatment with fluoxetine (5 mg/kg, 7 days) reverses the effects of stress on editing. We also examined the mRNA expression of ADAR enzymes in these regions, and asked whether social behavior in adult offspring would be altered by maternal exposure to stress and/or fluoxetine. Maternal treatment with fluoxetine altered Htr2c editing in offspring amygdala at birth, enhanced the expression of Htr2c mRNA and RNA editing enzymes in the prefrontal cortex, and reversed the effects of pre-reproductive stress on Htr2c editing in this region. Furthermore, maternal fluoxetine treatment enhanced differences in editing of glutamate receptors between offspring of control and stress-exposed rats, and led to enhanced social preference in adult offspring. Our findings indicate that pre-gestational fluoxetine treatment affects patterns of RNA editing and editing enzyme expression in neonatal offspring brain in a region-specific manner, in interaction with pre-reproductive stress. Overall, these findings imply that fluoxetine treatment affects serotonergic signaling in offspring brain even when treatment is discontinued before gestation, and its effects may depend upon prior exposure to stress.
Keywords: RNA editing, fluoxetine, stress, rat, Htr2c, intergenerational
Introduction
A-to-I RNA editing is a post-transcriptional modification mediated by adenosine deaminases acting on RNA (ADAR) enzymes [1–3]. ADARs bind to double-stranded RNA and convert adenosine (A) to inosine (I), which is read by the translational machinery as a guanosine (G). Occurring at both coding and non-coding regions, ADAR-mediated RNA editing can contribute to translational variability [4] and to the stability and self-regulation of the RNA molecule [5]. The development of high-throughput sequencing-based techniques has enabled the discovery of many novel editing sites in mammals [6].
A-to-I editing occurs in many mammalian tissues, including the brain [7, 8]. Editing changes in coding regions affect key aspects of neurotransmission [8–10]. For example, editing at ionotropic alpha-amino-propionic-acid (AMPA) and kainate (KA) glutamate receptors results in amino acid replacements that lead to significant modifications of channel gating, permeability, trafficking and maturation [11–14]. Another well-studied example of A-to-I editing is the Htr2c gene, where editing can occur at each of the five adenosines within the sequence that encodes amino acids 156–160 (sites A–E). Editing at these sites leads to altered encoding of triplet codons resulting in 32 putative isoforms of the G-protein-coupled serotonin receptor [15, 16].
A-to-I editing at mRNA encoding glutamate and serotonin receptors responds to environmental stimulation [17–25]. Using a highly accurate high-throughput targeted approach (microfluidics-based multiplex PCR and deep sequencing—mmPCR-seq [26]), we recently showed that exposing adolescent female rats to chronic unpredictable stress prior to reproduction (prereproductive stress; PRS) affects A-to-I editing in the prefrontal cortex (PFC) and amygdala of their newborn offspring. In particular, editing at the Htr2c was affected, resulting in a different pattern of Htr2c isoforms in offspring of stress-exposed versus naïve females [22]. We have previously shown that exposure of adolescent females to PRS also results in changes in behavior, stress-related plasma hormone levels, cortical gene expression and neuronal morphology in first- and second-generation offspring [27–31].
Fluoxetine (FLX) is a serotonin-specific reuptake inhibitor (SSRI) commonly prescribed for depression and related affective disorders [32]. FLX treatment affects levels of RNA editing at several sites, including the Htr2c and glutamate receptor subunits in culture [33, 34] as well as in the mouse [17, 35–37] and rat [19] brain. Moreover, FLX administration in adulthood reverses the effects of early life stress on RNA editing in the adult mouse brain [35]. Here, we used the mmPCR-seq technique to determine whether post-stress maternal treatment with FLX could reverse the effects of stress on RNA editing in the PFC and amygdala of newborn first-generation (F1) offspring. We also examined the mRNA expression of ADAR enzymes in neonatal PFC and amygdala, and asked whether social behavior in adult offspring would be altered by maternal exposure to stress and/or FLX.
Methods
Animals
Adolescent female Sprague-Dawley rats and adult males were purchased from Envigo (Jerusalem). Housing conditions (except during the stress procedure) included wood-flake bedding, ad lib food and water, 12 h artificial lighting during the day (07–19 h) and temperature maintained at 22 ± 2°C. Animals were randomly distributed across groups (see Experimental procedure below). The number of animals per group appears in the figures. Rats were handled in accordance with the NIH guidelines for the Care and Use of Laboratory Animals, 8th edition [38] and were bred and treated simultaneously to rats described in [22].
The study was approved by the University of Haifa Committee on animal experimentation (294/13, 351/14).
Experimental Procedure
The experimental procedure is depicted in Fig. 1. Briefly, adolescent [postnatal day (P)45] female rats were group housed (4–6 rats per cage) in 56 × 35 × 19 cm cages. Cages were randomly divided into control (C) and PRS groups. PRS rats underwent a 7-day chronic unpredictable stress (CUS) procedure as described previously [27–31]. Twenty-four hours later (P52), females from C and PRS groups were injected i.p. with either vehicle (VEH) or FLX (5 mg/kg, injection volume 0.5 ml), for 7 consecutive days. A week later (P66), behaviorally naïve males rat were introduced into a cage with 2 female rats and were removed 7 days later. Female rats were returned to their home cage; pregnancy was verified by weekly weighing. Each pregnant rat was moved to a 37 × 30 × 19 cm cage 7 days prior to parturition.
Figure 1:
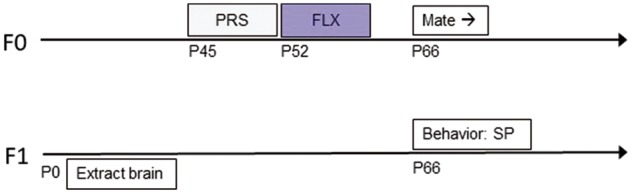
experimental design: intergenerational transmission of PRS/FLX effects
Randomly selected offspring of control and PRS rats (O-C and O-PRS, respectively) were sacrificed on the day of birth (P0) and their PFC and amygdala were extracted for RNA editing and gene expression analysis. Remaining pups were raised undisturbed until P30, then weaned and raised in same-sex, same-condition groups of 4–6. Adults (P60) male and female rats were tested for social preference (see below). Since the n’s for RNA editing analysis did not allow to examine gender effects, and no such effects were detected in the gene expression experiments, data from male and female offspring were pooled together. In the behavioral experiment, data for male and female offspring were analyzed separately.
Brain Removal and Dissection
Rats were sacrificed by decapitation and brains were removed and placed on dry ice. Brains were mounted on a cryostat and bilateral samples from PFC and amygdala from neonatal rats were removed guided by the Atlas of the Neonatal Rat Brain [39] and using 0.5 mm punches. Three punches were taken from the PFC and two punches were taken from the amygdala in each hemisphere. Punches from different rats were treated as individual samples. All samples were immediately placed on dry ice and kept at −80°C until further processing.
RNA Extraction and cDNA Preparation
RNA from brain tissue was extracted as described previously [30, 31]. Dissected brain regions were homogenized in 300 µl of TRIzol (Invitrogen, Carlsbad, CA) and 5 μl glycogen (Sigma-Aldrich, St Louis, MO), then suspended in a total of 500 µl TRIzol. After adding 100 μl chloroform to allow phase separation by centrifugation (14 000 rpm, 15′, 4°C), 250 μl ispropanol (Sigma Aldrich) was added to the aqueous phase. After 12 h in −20°C, RNA was precipitated by centrifugation (14 000 rpm, 15′). The pellet was washed in 500 μl cold 100% ethanol freshly made and stored in −20°C, recentrifuged (7600 rpm, 5′) and then rewashed in 500 μl cold 75% ethanol, recentrifuged (7600 rpm, 5′) then dried. RNA quantities were determined using a Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE) or Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA). RNA quality assessment and cDNA preparation were done as described previously [22]. The 260:280 nm absorbance ratio was measured to assess RNA quality; samples were excluded if the ratio was outside the range of 1.7–2.0, or if RNA concentration was too low. PureLink®RNA Mini Kit (Ambion) was used to further purify some of these samples. cDNA was prepared using iScript™ Advanced cDNA Synthesis Kit (Bio-Rad, Hercules, CA) or High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA) or Quanta (Bioscience, Manchester, UK). cDNA used in the mmPCR-seq experiment were purified with Agencourt® AMPure® XP beads (Beckman Coulter, Brea, CA).
Primer Preparation and mmPCR-seq
Editing sites were selected and primer preparation for mmPCR-seq was performed as described previously [22]. Briefly, we designed 48 pools of 2–3 plex multiplex PCR primers (see [22]) to amplify 146 sites. The sizes of the amplicons ranged from 150 to 350 bp. We loaded cDNA and primer pools into the 48.48 Access Array IFC (Fluidigm) and performed target amplification as described previously [26, 40]. PCR products of each sample were then subjected to a 15-cycle barcode PCR and pooled together. All pools were combined at equal volumes and purified via Agencourt® AMPure® XP beads. The library was sequenced using NextSeq 500 (Illumina, USA) with 76 bp paired-end reads. Paired-end reads were combined and mapped onto the genome (rn4) using BWA samse allowing 9 mismatches per read [41]. We aligned the sequencing reads to a combination of the reference genome and 70 bp exonic sequences surrounding known splicing junctions from available gene models (obtained from the UCSC genome browser). We quantified editing levels as described [22] by dividing the fraction of reads containing a ‘G' nucleotide by the total reads at each editing site. Only sites covered by ⩾50 mmPCR-seq reads were included. For each comparison, we excluded editing sites that had less than 3 biological replicates, and samples where >30% of editing sites were missing. Custom scripts used to process data are available upon request.
Cluster Analysis of Htr2c Isoforms from mmPCR-seq Data
A-to-I RNA editing of the Htr2c gene (Rattus norvegicus 5-hydroxytryptamine serotonin receptor 2C) occurs at 5 sites (A through E) and can result in 32 mRNA variants that translate to 24 protein isoforms. We performed editing cluster analysis as described previously [22, 42]. Briefly, we aligned the reads with samtools mpileup (v0.1.18; s [41]), to get the sequence information per genomic location, keeping the data of the original reads. Using an in-house computer program, we were able to find the editing sites in the Htr2c cluster in each read. We used only reads that included all cluster editing sites. For each sample, we summed the different combinations of actual editing locations, and found the percentage from the total number of reads that covered all the locations for each isoform. Isoforms that include the E site were not included in the calculations, since editing was not detected at this site.
Quantitative Real-Time PCR
Some of the samples used for RNA editing assessment were also assessed for mRNA expression of RNA editing enzymes and Htr2c. In some cases, additional samples were added for qRT-PCR analysis, since RNA quantities were insufficient for both RNA editing and gene expression studies. Primers (see [22]) were designed using Primer3 [43] software, and synthesized by Integrated DNA Technologies (Coralville, IA). Primer suitability was determined using standard curve analysis, melting curve analysis and linearity at threshold [44, 45]. Quantitative real-time PCR (qRT-PCR) was performed as described previously [31]. Data analysis was performed on dCt values (Ct values) normalized to the housekeeping gene hypoxanthine phosphoribosyl transferase (HPRT). ddCt was calculated relative to the control brain region (amygdala) or group (O-C/VEH). Data is represented as fold change, calculated using the ddCt method [45], with standard error of fold change values [44].
Social Preference
The social preference test is commonly used to assess rodent sociability, and has been modified and conducted as described previously [46, 47]. The arena (40lx70wx30h cm) was divided into 2 unequal compartments by a transparent perforated Plexiglas panel, allowing for intact visual and olfactory cues. The experiment rat was placed in the larger compartment and an unfamiliar con-specific partner rat of the same sex and age was placed in the smaller (40lx15wx30h cm) compartment. Rats were habituated to their respective compartments for 5 min. Twenty-four hours later, the partner rat was placed in the smaller compartment, and an unfamiliar object (plastic, 5 × 5 × 8 h, 11 × 8 × 8 h cm) was placed in the larger compartment, 10 cm diagonally from the corner of the arena. One minute later the experiment rat was placed in the arena for 5 min. Several objects and partner rats were used throughout the experiment, and were counterbalanced between groups. Time spent exploring the partner rat and the object was measured using Ethovision XT10.0 software (Noldus Information Technology, Leeburh, VA). Rats that did not complete 30 s of total exploration time (4 rats in total, 1 from each group) were excluded from the experiment.
Statistical Analyses
Data were analyzed with SPSS 23 Statistics software (IBM, Chicago, IL ) and R package version 3.2.5. A nonparametric Mann-Whitney U test with Benjamini–Hochberg multiple testing correction was used, with FDR = 0.1, was used to analyse RNA editing data [22, 42]. We used specifically constructed R package scripts (available upon request). Analysis of variance (ANOVA) and multivariate analysis of variance (MANOVA) were used to analyze gene expression, isoforms distribution and behavioral data. LSD post-hoc tests were applied when interactions were significant. Means and SE are presented in the figures. Significance level was set at P < 0.05. Results that approach significance were defined as .05 ≤ P ≤ 0.075.
Results
Editing Levels Are Lower in Neonatal PFC Compared with Amygdala
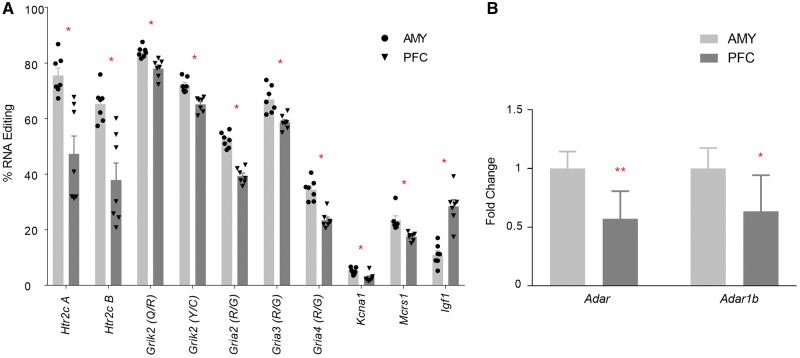
We previously showed that editing levels were generally lower in the neonatal rat PFC compared with the amygdala [22]. Here, we asked whether maternal pre-reproductive FLX treatment would affect these regional patterns. We detected editing at all 146 sites. Editing levels at 10 editing sites were different in PFC compared with the amygdala; in 9/10 sites, editing was lower in PFC (Mann–Whitney U test, FDR = 0.1; Fig. 2A; see Supplementary Table S1 for mean editing levels at each site in PFC and amygdala). mRNA expression levels of Adar and Adarb1 were also lower in PFC (one-way ANOVAs, Adar, F1,11 = 17.759, P = 0.0014; Adarb1, F1,11 = 8.788, p = 0.013; Fig. 2B). There were no regional differences in Htr2c mRNA levels (NS, not shown).
Figure 2:
A-to-I RNA editing and ADAR gene expression in the neonatal PFC and amygdala. Significant differences in % RNA editing (A) and fold changes (means ± SE of the fold change relative to amygdala) in mRNA gene expression of editing enzymes (B) in the PFC and amygdala at P0. All samples are from offspring of control females treated with FLX prior to gestation. *P < 0.05. **P < 0.001. N’s, RNA editing; PFC 7, amygdala 7; gene expression; Adar 7, 6; Adarb1 6, 7
Maternal Treatment with FLX and Exposure to Stress Prior to Pregnancy Affect RNA Editing Enzyme and Htr2c mRNA Expression at Birth
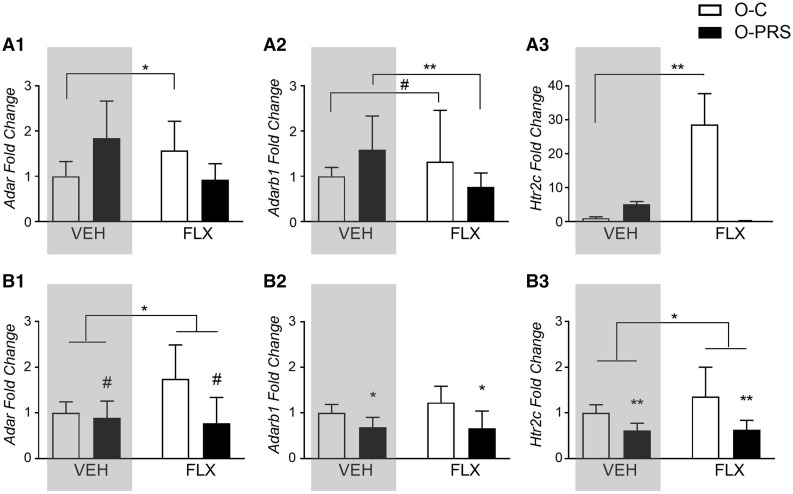
We previously showed that PRS affects mRNA expression levels of Adar and Adarb1 (which encode ADAR1 and ADAR2, respectively) differentially in the PFC and amygdala of F1 offspring at P0. The mRNA expression of Htr2c (encoding the serotonin 5HT2C receptor) in offspring is also sensitive to maternal PRS [22]. Here, we asked whether maternal post-stress FLX treatment would reverse the effects of PRS, and/or have its own impact on expression levels. The effects of PRS on expression levels were previously reported [22] and are depicted in the gray panels in Fig. 3. Since FLX-exposed offspring samples were collected at the same time as non-exposed samples, we combined the data from the previous experiment [22] with the present one and applied a 2-way ANOVA analysis for each gene. We found that in the PFC (Fig. 3A), FLX increased Adar and Adarb1 mRNA expression in offspring of control, but not PRS, dams, and reversed the PRS-induced increased in Adarb1 [Fig. 3A1: Adar, main effect of group F1,25 = 5.030, P = 0.034, group × drug interaction F1,25 = 8.528, P = 0.007, post hoc C-FLX > C-VEH (P = 0.017); Fig. 3A2: Adarb1, group × drug interaction F1,22 = 15.49, P = 0.0007, post-hoc C-FLX>C-VEH (P = 0.057), PRS-FLX< PRS-VEH (P = 0.001)]. FLX also led to an increase in Htr2c mRNA expression in O-C, but not O-PRS, rats [Fig. 3A3, main effect of drug F1,23 = 32.139, P = 0.000009, group × drug interaction F1,23 = 24.920, P = 0.0000476, post-hoc C-FLX>C-VEH P = 0.0003)]. In the amygdala, we previously observed a PRS-induced decrease in the expression of RNA-editing enzymes and Htr2c in offspring of VEH-treated rats ([22], Fig. 3Bgray panels). A similar effect of PRS was observed in offspring of FLX-treated rats (main effect of group, Fig. 3B1: Adar, F1,20 = 3.757, P = 0.067; Fig. 3B2: Adarb1, F1,21 = 12.277, P = 0.0021; Fig. 3B3: Htr2c, F1,21 = 15.93, P = 0.00066). In addition, FLX increased Adar (main effect Drug, F1,20 = 26.548, P = 0.0000484) and Htr2c expression (main effect drug, F1,21 = 7.104, P = 0.014) regardless of PRS exposure.
Figure 3:
maternal PRS- and FLX-induced changes in Adar, Adarb1 and Htr2c gene expression in the offspring brain. (A) Fold changes in mRNA gene expression of Adar (A1), Adarb1 (A2) and Htr2c (A3) in the PFC of offspring of PRS (O-PRS) and control (O-C) dams treated with VEH or FLX prior to gestation. (B) Fold changes in mRNA gene expression of Adar (B1), Adarb1 (B2) and Htr2c (B3) in the amygdala of offspring of PRS (O-PRS) and control (O-C) dams treated with VEH or FLX prior to gestation. Fold change values are presented as means ± SE of the fold change relative to O-C). #P < 0.075. *P < 0.05. **P < 0.001. Gray panels show previously published data [22]. N's, Adar, PFC: VEH, O-C 5, O-PRS 8, FLX, 6,7; amygdala: VEH, 4,9, FLX 7,5. Adarb1, PFC: VEH, O-C 5, O-PRS 8, FLX, 7,7; amygdala: VEH, 4,8, FLX 7,6. Htr2c, PFC: VEH, O-C 6, O-PRS 8, FLX, 7,8; amygdala: VEH, 4,8, FLX 6,6.
Maternal FLX Treatment Affects A-to-I RNA Editing at the Htr2c
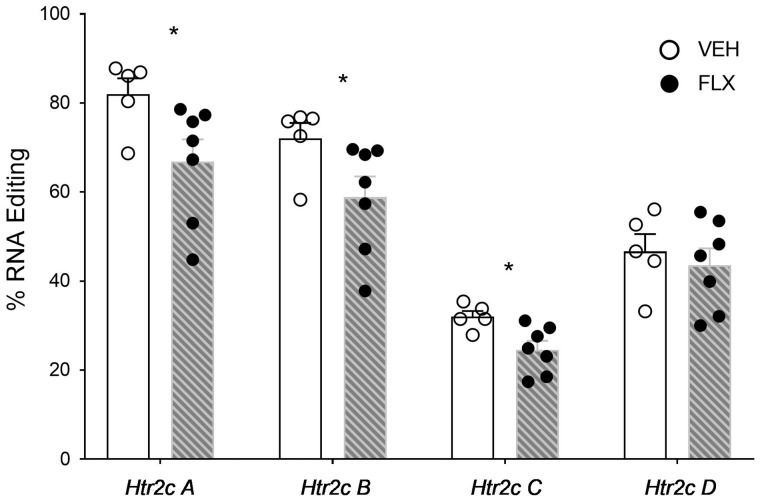
Since maternal FLX treatment affected RNA editing enzyme expression in offspring PFC and amygdala at birth, independently of PRS effects, we examined the effects of FLX on A-to-I RNA editing levels in the same regions in stress-naïve (O-C) offspring. In the PFC, editing levels at the 146 detected sites were not affected by maternal FLX (Mann–Whitney U test, FDR = 0.1; see Supplementary Table S2 for mean + SE editing levels). In the amygdala, out of 146 detected editing sites we found FLX-induced differences only at the Htr2c: maternal FLX led to a decrease in editing at the A, B and C sites (Fig. 4; Mann–Whitney U test, FDR = 0.1; mean + SE data in Supplementary Table S2).
Figure 4:
maternal FLX treatment-induced changes in A-to-I RNA editing in offspring amygdala at birth. Percent RNA editing at the Htr2c A–D sites in the amygdala of neonatal offspring of rats treated with VEH or FLX prior to reproduction. *P < 0.05. N’s, VEH 5, FLX 7
Supplementary Table S3 presents the change in % distribution of Htr2c isoforms in PFC and amygdala of O-C rats, excluding isoforms containing the E site where editing was not detected. As can be seen, there was no difference in distribution of Htr2c isoforms between VEH and FLX in PFC, but in the amygdala, the prevalence of 3 isoforms (VSVABCD, F1,8 = 8.923, P = 0.0174; VSIABC, F1,8 = 6.611, P = 0.033; VNIAB, F1,8 = 5 7.675, P = 0.024), was significantly different in offspring of FLX-treated compared with VEH-treated dams. Notably, prevalence of the highly edited isoform VSVABCD was decreased in offspring of FLX-treated dams.
Maternal FLX Treatment Can Reverse or Enhance the Effects of PRS on A-to-I Editing in the PFC
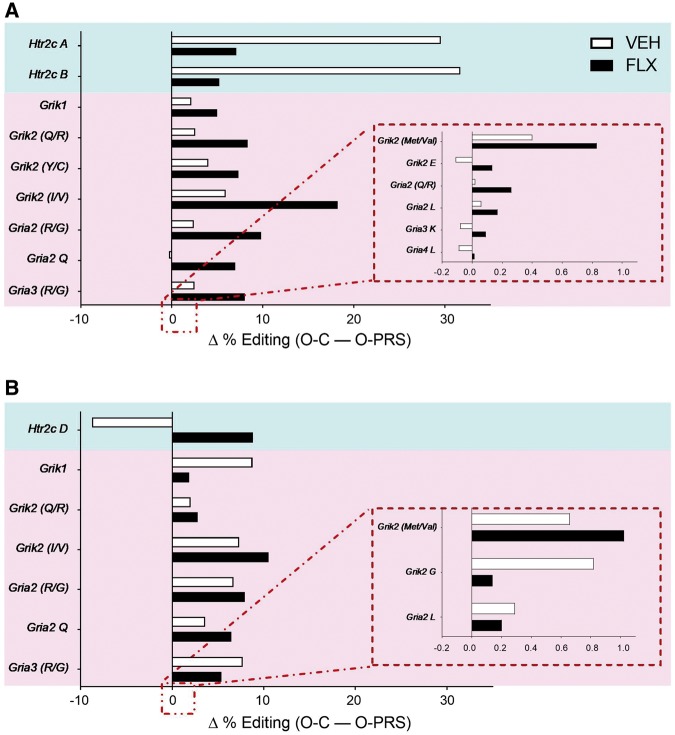
We next asked whether maternal FLX treatment would reverse some of the effects of PRS on editing in offspring PFC and amygdala. Previous analysis of multiple sites where editing affects neurotransmission and the stress response (29 sites in total; [22]), revealed significant PRS-induced changes in editing at the Htr2c in PFC [22]. Here, we assessed the difference in editing levels between O-PRS and O-C samples at each of these sites, and asked whether maternal FLX treatment would alter this difference. Figure 5 presents sites where the editing in O-PRS samples was significantly different from O-C samples (Mann–Whitney analysis, FDR = 0.1), in either VEH or FLX groups. In the PFC (Fig. 5A), PRS led to significant changes in editing at the A and B sites of the Htr2c, and FLX reversed this effect. However, FLX treatment enhanced the effect of PRS on editing at mRNA encoding AMPA and KA glutamate receptor subtypes, leading to differences between O-PRS and O-C groups that were not present in VEH samples. In the amygdala (Fig. 5B), PRS led to significant changes in editing at mRNA encoding serotonin and glutamate receptors. FLX reversed the effects of PRS at some of the sites and left differences intact at others. Unlike in the PFC, FLX did not enhance the difference between O-PRS and O-C samples at any of the sites examined (see Supplementary Table S4 for full results).
Figure 5:
maternal FLX-induced modulation of PRS effects on RNA editing levels in offspring PFC. Significant differences in RNA editing in the PFC (A) and amygdala (B) between O-C and O-PRS rats are presented as delta % editing. Dams were treated with VEH or FLX prior to reproduction. Blue panels highlight editing changes at Htr2c. Pink panels highlight editing changes in glutamate receptor subunits. Insets: Editing sites where editing differences were between −0.2 and 1. N’s, PFC: VEH, O-C 5, O-PRS 8; FLX, 7, 5; amygdala: VEH, 5, 9; FLX, 7, 6
Supplementary Table S5 presents the difference in Htr2c isoform prevalence between O-PRS and O-C samples, separately for VEH- and FLX-treated groups (differences between VEH-treated O-PRS and O-C groups were previously published [22]). As can be seen, FLX treatment affected the prevalence of isoforms in offspring PFC and amygdala in interaction with the effects of PRS. Particularly notable is the effect of FLX on the prevalence of the INI isoform (no editing at any of the Htr2c sites) in the PFC: whereas PRS had no effect on its prevalence in VEH-treated samples, it increased its prevalence by 22% when dams were treated with FLX.
Maternal FLX Treatment Leads to Enhanced Social Preference in Adult Offspring
Since FLX affected RNA editing and gene expression patterns in offspring independently of stress exposure, particularly at mRNA encoding glutamate receptors (Fig. 5), we examined the effect of pre-reproductive FLX treatment in interaction with PRS on social preference (Fig. 6A). Social preference is a task that measures the animal’s tendency to preferentially explore an unfamiliar social stimulus versus an unfamiliar inanimate object, and relies on intact NMDA receptor-mediated glutamate neurotransmission in the PFC [48, 49]. We first compared the performance of male and female rats in the task, and found that exploration duration of the social stimulus was higher in females (mean = 116.350, SE = 4.677) compared with males (mean = 94.156, SE = 4.284; main effect of sex F1,88 = 12.434, P = 0.00067). No differences in non-social stimulus exploration time were found (NS; N’s: Females; VEH, O-C 13 O-PRS 10, FLX, O-C 17 O-PRS 10).
Figure 6:
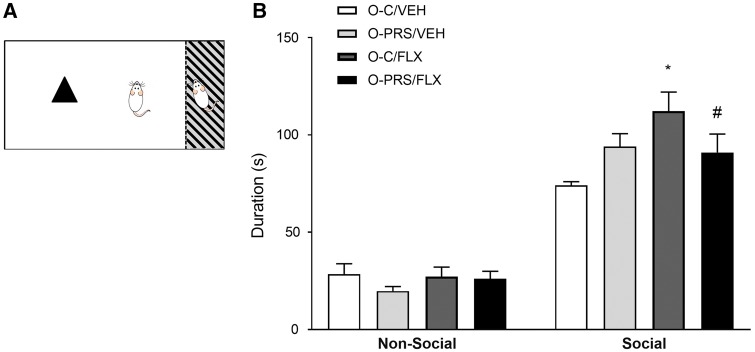
FLX maternal PRS- and FLX-induced changes in social preference in adult male offspring. (A) A representation of the social preference behavioral paradigm. Rats explore a novel inanimate (non-social) or a social stimulus for 5 min. (B) Time spent exploring social and non-social stimuli in male adult offspring of control (O-C) or stressed (O-PRS) dams treated with VEH or FLX. No differences in non-social stimulus exploration were found (NS). *P < 0.05. N’s, VEH, O-C 8, O-PRS 16; FLX, 11, 11
A separate analysis of FLX and PRS effects in males and females revealed that male, but not female, offspring were affected by pre-reproductive FLX treatment (Fig. 6, data for females not shown). A two-way ANOVA analysis of partner exploration time revealed a significant main effect of drug F1,42 = 4.488, P = 0.04, and a group × drug interaction F1,42 = 6.230, P = 0.0165. Post-hoc analysis revealed that maternal FLX treatment increased partner exploration time in O-C animals (O-C/FLX>O-C/VEH, P = 0.0043), but not in offspring of rats exposed to stress (O-PRS/FLX< O-C/FLX, P = 0.0733).
Discussion
The present study shows that treatment of female rats with the antidepressant drug FLX prior to reproduction and gestation affects RNA editing patterns, gene expression of editing enzymes and behavior in offspring. Pre-gestational FLX modulates some of the effects of prior exposure to stress, but also produces independent consequences. These findings have implications on current understanding of serotonergic signaling and its sensitivity to maternal exposure to stress and antidepressant drug treatment.
Pre-gestational treatment of adolescent female rats with FLX affects RNA editing patterns in their offspring at birth. FLX alters serotonergic transmission by blocking the serotonin transporter and increasing synaptic serotonin levels [50]. Although in our study FLX treatment was discontinued a week prior to gestation, a direct influence of the drug on the developing fetus cannot be ruled out, as previous studies have shown that FLX and particularly its active metabolite, norfluoxetine, have relatively long elimination half-lives and can remain in the plasma after drug administration is discontinued [51, 52]. FLX and other SSRIs cross the placenta, enter fetal brain tissue and are present in breast milk [53–56]. Gestational exposure to FLX was shown to induce age-specific and region-specific alterations in serotonin levels and receptor densities in offspring brain [57–59]. We showed that pre-gestational exposure to FLX increased mRNA expression of Htr2c (Fig. 3), and selectively altered editing at this receptor (Fig. 4) while leaving editing at 142 non-serotonergic sites intact. These alterations could be due to a direct interaction of FLX with developing serotonergic signaling pathways, or to indirect effects of FLX on the developing fetus which give rise to editing and gene expression changes in the neonatal brain. Another possibility is that serotonin reuptake inhibition directly affects oocytes, leading to changes in offspring gene expression patterns. Serotonin and elements of a regulatory serotonergic system are present in oocytes and may be modified by FLX treatment [60]. We previously showed that PRS alters the expression of stress-related corticotropin releasing hormone receptor type 1 (CRFR1) in oocytes and in offspring brain at birth [31]. We did not find editing changes in stress-exposed oocytes [22], but FLX-induced changes in editing, editing enzymes and molecules that regulate serotonergic activity in oocytes should be the subject of future exploration.
Previous studies have shown that the expression of ADAR enzymes and A-to-I RNA editing at sites encoding serotonin, glutamate and GABA receptors are altered by FLX treatment [17, 33–35, 37, 61]. Curiously, here we found that maternal treatment with FLX led to opposite effects on Htr2c editing and ADAR mRNA expression levels in offspring: whereas editing at the Htr2c A and B sites decreased, mRNA expression of editing enzymes increased. We and others [22, 23, 62–67] previously found that changes in editing levels correlated poorly with ADAR mRNA or protein expression levels. Differences between mRNA and protein expression or between expression and activity levels, compensatory or self-regulatory mechanisms could account for the non-linear relationship between editing rates and editing enzyme expression levels [68]. Furthermore, ADAR enzymes regulate additional processes, e.g. miRNA biogenesis and function [68], which could be affected by PRS and/or FLX.
Interestingly, we found that editing rates as well as ADAR mRNA expression levels in offspring of FLX-treated rats were higher in the amygdala compared with the PFC (Fig. 2), in line with our previous findings in offspring of VEH-treated rats [22] and with the maturational profile of these regions [48, 49]. This positive correlation could be due to the fact that regional differences in editing were not limited to the Htr2c, as were FLX-induced effects.
Htr2c editing affects the expression and activity levels of this G-protein-coupled receptor, so that increased editing generally results in reduced sensitivity to ligands, reduced basal activity [15, 69], decreased G-protein coupling [70] and decreased intracellular signaling [71]. Here, maternal FLX treatment decreased A and B site editing (Fig. 4) and increased Htr2c mRNA levels (Fig. 3) in the amygdala, and these effects were accompanied by decreased prevalence of the unedited INI isoform and increased prevalence of the highly edited (and presumably less functional) VSV isoform (Supplementary Table S3). Notably, the very pronounced FLX-induced increase in Htr2c mRNA expression in the PFC (>28-fold) was not accompanied by changes in receptor editing. While the present study does not enable us to determine whether changes in editing enzyme or substrate expression preceded changes in editing, one possibility is that maternal FLX treatment affected serotonergic signaling in the developing fetus and led to increased Htr2c mRNA expression in neonate offspring, which in turn impacted Htr2c editing rates and editing enzyme levels.
Pre-gestational, post-stress treatment of rats with FLX reverses some of the effects of PRS on the mRNA expression of RNA editing enzymes in offspring brain. Chronic unpredictable stress in adolescence, a vulnerable time period in neural development, is associated with the emergence of psychiatric disorders in adulthood [72]. FLX and other SSRIs are the most frequently prescribed anti-depressants, and are increasingly prescribed in adolescence and during pregnancy [73–76]. Chronic unpredictable stress is a rodent model of depression and anxiety [77], and FLX as well as other SSRIs were shown to reverse the effects of stress on depression-like behavior and HPA axis function in rodents [78]. While this is the first study to investigate the interaction between PRS and pre-gestational FLX exposure, several studies examined the consequences of perinatal FLX exposure in rodent models of depression. For example, a recent study showed that perinatal treatment (from Gestational Day 10-P21) with FLX reversed pre-gestational stress-induced abnormalities in serotonin levels and turnover in offspring PFC [79]. Other studies showed that perinatal FLX can reverse the effects of maternal stress on immobility in the forced swim test, hippocampal neurogenesis, and 5-HIAA levels in the hippocampus of juvenile or adolescent offspring [79–81].
Similarly, in the present study pre-gestational treatment with FLX reversed the consequences of PRS on editing abnormalities at the Htr2c in PFC and amygdala of offspring. Our results indicate that discontinuation of treatment prior to pregnancy does not alter FLX effects. As discussed above, this may be due to the long half-life of FLX and its metabolites and its ability to cross the placenta and enter fetal brain tissue, or to direct effects on serotonin dynamics in exposed oocytes. FLX and other SSRIs were hypothesized to exert their antidepressant actions by normalizing hypothalamic-pituitary-adrenocortical (HPA) system hyperactivity, a central clinical feature of depression. Salari et al. [81], for example, found that the effects of gestational stress on corticosterone elevations in mice were reversed by either gestational or perinatal (P10-20) FLX treatment. However, in other studies FLX treatment during gestation or in adult rodents potentiated HPA system hyperactivity on its own rather than reducing the effects of stress [81–85].
The 5-HT2C receptor is implicated in the response to chronic stress and maintains reciprocal relations with the HPA system [86, 87]. Chou-Green et al. [88] found that 5-HT2cR knockout mice are hyper-responsive to stress, and others have shown that stress alters Htr2c editing patterns. Both elevated and reduced editing levels can have deleterious effects on behavior. For example, mice expressing either the fully edited or fully unedited form of the 5-HT2c exhibit anxiogenic behavior, particularly in the BALB/c strain commonly used as a mouse model of anxiety and depression [89, 90]. In our study, FLX increased the proportion of the unedited (INI) isoform in offspring of PRS-exposed, but not control rats (Supplementary Table S5). In the context of previously published literature, our findings imply that FLX treatment leads to increased signaling at the 5-HT2c receptor particularly when the HPA axis is activated.
While reversing the effects of PRS on editing at the Htr2c, FLX treatment led to enhanced differences between C and PRS-exposed offspring at glutamate receptors, particularly in the PFC (Fig. 5). Disrupted glutamatergic neurotransmission is implicated both in the pathogenesis of affective disorders and in the outcome of pharmacological treatments [91–94]. Editing of AMPA and KA receptor subunits was shown to have a significant effect on transmission dynamics (e.g. [11, 95, 96], and is sensitive to environmental manipulations, e.g. learning and stress [23, 97]. In our hands, pre-reproductive chronic unpredictable stress did not affect editing at glutamate receptors in affected female rats or their offspring [22]. In line with previous studies that showed minor effects of FLX on glutamate receptor editing [98], in the present study we showed that pre-gestational FLX treatment on its own also had no effect on editing in newborn offspring. However, FLX treatment potentiated differences in editing of glutamate receptors between offspring of control and stress-exposed rats. This interaction between stress and drug effects seemed to be specific to glutamate receptors, was particularly pronounced in the PFC, and encompassed different editing sites at AMPA and KA receptors (Fig. 5, Supplementary Table S4). This finding may support the interaction between serotonergic and glutamatergic transmission in the modulation of stress effects, and provide a possible mechanism for plasticity in response to environmental adversity.
Deficits in social function are shared by several psychiatric disorders, including autism, depression and schizophrenia [99]. Treatment with SSRIs, including FLX, ameliorates social deficits in depressed and anxious individuals [100], and reverses aberrant social behavior in animal models of psychopathology (e.g. [101]). Here, we found that pre-gestational treatment with FLX-enhanced social preference in adult male, but not female, offspring (Fig. 6). Assuming that pre-gestationally administered FLX had a direct effect on the developing fetus, this finding can be explained by the known effects of perinatal FLX administration on the developing serotonergic system [102]. A sexually dimorphic effect of pre- or perinatal SSRI exposure on social behavior has been previously reported (see [103] for review). For example, Svirsky et al. [104] found that prenatal FLX increases aggression in male offspring and delays the onset of maternal behavior in females. Interestingly, in our study the effect of FLX on social behavior was attenuated in offspring of stress-exposed females. In agreement with this finding, exposure to prenatal stress diminished the prenatal FLX-induced increase in social (aggressive) behavior in male offspring [105]. The mechanism for this stress × drug interaction on social behavior in offspring remains to be elucidated, but may be related to opposite effects of PRS and FLX on the HPA axis. Furthermore, the interaction between PRS and FLX effects on measures of anxiety and cognitive behavior are to be explored in future studies. Finally, a relationship between FLX-induced changes in Htr2c RNA editing or expression levels at birth and FLX-induced changes in adult social behavior cannot be deduced from this study but may provide important clues for the role played by intact development of the serotonergic system in early life and social function in adulthood.
In summary, this is the first study to investigate the effects of pre-gestational treatment with FLX on offspring, as most human and animal model investigations focus on the effects of SSRI treatment during pregnancy. We found that FLX administration to female rats prior to gestation affected 5-HT2C receptor expression and editing in neonatal offspring brain, led to enhanced social preference in adult offspring, and interacted with the effects of prior exposure to stress. Chronic unpredictable stress is commonly used to model depression in humans, and FLX is often the first line of treatment for stress-related depression in adolescents and during pregnancy. Here, we demonstrate that even when discontinued prior to gestation, FLX has long-lasting effects on serotonin dynamics and on social behavior.
Supplementary Data
Supplementary data are available at EnvEpig online.
Funding
This work was made possible by grant support from the Israel Science Foundation (IGS, 484/10) and from the Binational Science Foundation (H.Z., Rachamimoff Travel Grant T-2014227). H.Z. was also supported by the President of Israel Scholarship for Excellence and Innovation in Science. J.B.L. was supported by NIH grant R01GM102484, and the Ellison Medical Foundation. G.R. was supported by a Stanford Graduate Fellowship.
Conflict of interest statement. None declared.
Supplementary Material
References
- 1. Gott JM, Emeson RB.. Functions and mechanisms of RNA editing. Annu Rev Genet 2000;34:499–531. [DOI] [PubMed] [Google Scholar]
- 2. Maas S, Kawahara Y, Tamburro KM, Nishikura K.. A-to-I RNA editing and human disease. RNA Biol 2006;3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bass Bl. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 2002;71:817–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pullirsch D, Jantsch MF.. Proteome diversification by adenosine to inosine RNA editing. RNA Biol 2010;7:205–12. [DOI] [PubMed] [Google Scholar]
- 5. Daniel C, Lagergren J, Ohman M.. RNA editing of non-coding RNA and its role in gene regulation. Biochimie 2015;117:22–7. [DOI] [PubMed] [Google Scholar]
- 6. Ramaswami G, Li JB.. Identification of human RNA editing sites: a historical perspective. Methods 2016;107:42–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young An, Liu Ki, Zhang R, Ramaswami G, Ariyoshi K. et al. Dynamic landscape and regulation of RNA editing in mammals. Nature 2017;550:249–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li JB, Church GM.. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat Neurosci 2013;16:1518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paz-Yaacov N, Levanon EY, Nevo E, Kinar Y, Harmelin A, Jacob-Hirsch J, Amariglio N, Eisenberg E, Rechavi G.. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc Natl Acad Sci USA 2010;107:12174–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Slotkin W, Nishikura K.. Adenosine-to-inosine RNA editing and human. Genome Med 2013;5:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Köhler M, Burnashev N, Sakmann B, Seeburg PH.. Determinants of ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: diversity by RNA editing. Neuron 1993;10:491–500. [DOI] [PubMed] [Google Scholar]
- 12. Sailer A, Swanson GT, Pérez-Otaño I, O'Leary L, Malkmus SA, Dyck RH, Dickinson-Anson H, Schiffer HH, Maron C, Yaksh TL. et al. Generation and analysis of GluR5 (Q636R) kainate receptor mutant mice. J Neurosci 1999;19:8757–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sommer B, Köhler M, Sprengel R, Seeburg PH.. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 1991;67:11–9. [DOI] [PubMed] [Google Scholar]
- 14. Vissel B, Royle GA, Christie BR, Schiffer HH, Ghetti A, Tritto T, Perez-Otano I, Radcliffe RA, Seamans J, Sejnowski T. et al. The role of RNA editing of kainate receptors in synaptic plasticity and seizures. Neuron 2001;29:217–27. [DOI] [PubMed] [Google Scholar]
- 15. Niswender CM, Copeland SC, Herrick-Davis K, Emeson RB, Sanders-Bush E.. RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J Biol Chem 1999;274:9472–8. [DOI] [PubMed] [Google Scholar]
- 16. Wang Q, O'Brien PJ, Chen CX, Cho DS, Murray JM, Nishikura K.. Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin2C receptors. J Neurochem 2000;74:1290–300. [DOI] [PubMed] [Google Scholar]
- 17. Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C.. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron 2002;34:349–56. [DOI] [PubMed] [Google Scholar]
- 18. Hackler EA, Airey DC, Shannon CC, Sodhi MS, Sanders-Bush E.. 5-HT(2C) receptor RNA editing in the amygdala of C57BL/6J, DBA/2J, and BALB/cJ mice. Neurosci Res 2006;55:96–104. [DOI] [PubMed] [Google Scholar]
- 19. Iwamoto K, Nakatani N, Bundo M, Yoshikawa T, Kato T.. Altered RNA editing of serotonin 2C receptor in a rat model of depression. Neurosci Res 2005;53:69–76. [DOI] [PubMed] [Google Scholar]
- 20. Kubota-Sakashita M, Iwamoto K, Bundo M, Kato T.. A role of ADAR2 and RNA editing of glutamate receptors in mood disorders and schizophrenia. Mol Brain 2014;7:5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh M, Singh MM, Na E, Agassandian K, Zimmerman MB, Johnson AK.. Altered ADAR 2 equilibrium and 5HT(2C) R editing in the prefrontal cortex of ADAR 2 transgenic mice. Genes Brain Behav 2011;10:637–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zaidan H, Ramaswami G, Golumbic YN, Sher N, Malik A, Barak M, Galiani D, Dekel N, Li JB, Gaisler-Salomon I. et al. A-to-I RNA editing in the rat brain is age-dependent, region-specific and sensitive to environmental stress across generations. BMC Genomics 2018;19:28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brande-Eilat N, Golumbic YN, Zaidan H, Gaisler-Salomon I.. Acquisition of conditioned fear is followed by region-specific changes in RNA editing of glutamate receptors. Stress 2015;18:309–18. [DOI] [PubMed] [Google Scholar]
- 24. Sanjana NE, Levanon EY, Hueske EA, Ambrose JM, Li JB.. Activity-dependent A-to-I RNA editing in rat cortical neurons. Genetics 2012;192:281–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Balik A, Penn AC, Nemoda Z, Greger IH.. Activity-regulated RNA editing in select neuronal subfields in hippocampus. Nucleic Acids Res 2013;41:1124–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang R, Li X, Ramaswami G, Smith KS, Turecki G, Montgomery SB, Li JB.. Quantifying RNA allelic ratios by microfluidic multiplex PCR and sequencing. Nat Methods 2014;11:51–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bock J, Poeschel J, Schindler J, Börner F, Shachar-Dadon A, Ferdman N, Gaisler-Salomon I, Leshem M, Braun K, Poeggel G. et al. Transgenerational sex-specific impact of preconception stress on the development of dendritic spines and dendritic length in the medial prefrontal cortex. Brain Struct Funct 2016;221:855–63. [DOI] [PubMed] [Google Scholar]
- 28. Leshem M, Schulkin J.. Transgenerational effects of infantile adversity and enrichment in male and female rats. Dev Psychobiol 2012;54:169–86. [DOI] [PubMed] [Google Scholar]
- 29. Shachar-Dadon A, Schulkin J, Leshem M.. Adversity before conception will affect adult progeny in rats. Dev Psychol 2009;45:9–16. [DOI] [PubMed] [Google Scholar]
- 30. Zaidan H, Gaisler-Salomon I.. Prereproductive stress in adolescent female rats affects behavior and corticosterone levels in second-generation offspring. Psychoneuroendocrinology 2015;58:120–9. [DOI] [PubMed] [Google Scholar]
- 31. Zaidan H, Leshem M, Gaisler-Salomon I.. Prereproductive stress to female rats alters corticotropin releasing factor type 1 expression in ova and behavior and brain corticotropin releasing factor type 1 expression in offspring. Biol Psychiatry 2013;74:680–7. [DOI] [PubMed] [Google Scholar]
- 32. Williams JW, Mulrow CD, Chiquette E, Noël PH, Aguilar C, Cornell J Jr.. A systematic review of newer pharmacotherapies for depression in adults: evidence report summary. Ann Intern Med 2000;132:743–56. [DOI] [PubMed] [Google Scholar]
- 33. Li B, Dong L, Wang B, Cai L, Jiang N, Peng L.. Cell type-specific gene expression and editing responses to chronic fluoxetine treatment in the in vivo mouse brain and their relevance for stress-induced anhedonia. Neurochem Res 2012;37:2480–95. [DOI] [PubMed] [Google Scholar]
- 34. Sawada J, Yamashita T, Aizawa H, Aburakawa Y, Hasebe N, Kwak S.. Effects of antidepressants on GluR2 Q/R site-RNA editing in modified HeLa cell line. Neurosci Res 2009;64:251–8. [DOI] [PubMed] [Google Scholar]
- 35. Englander MT, Dulawa SC, Bhansali P, Schmauss C.. How stress and fluoxetine modulate serotonin 2C receptor pre-mRNA editing. J Neurosci 2005;25:648–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bhansali P, Dunning J, Singer SE, David L, Schmauss C.. Early life stress alters adult serotonin 2C receptor pre-mRNA editing and expression of the alpha subunit of the heterotrimeric G-protein G q. J Neurosci 2007;27:1467–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li B. Fluoxetine affects GluK2 editing, glutamate-evoked Ca(2+) influx and extracellular signal-regulated kinase phosphorylation in mouse astrocytes. J Psychiatry Neurosci 2011;36:322–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Council NR, Guide for the Care and Use of Laboratory Animals. NW Washington, DC: National Academies Press, 2010. [Google Scholar]
- 39. Ramachandra R, Subramanian T.. Neonatal Rat Brain. London: CRC Press, 2011. [Google Scholar]
- 40. Ramaswami G, Deng P, Zhang R, Anna Carbone M, Mackay TFC, Li JB.. Genetic mapping uncovers cis-regulatory landscape of RNA editing. Nat Commun 2015;6:8194.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Khermesh K, D'Erchia AM, Barak M, Annese A, Wachtel C, Levanon EY, Picardi E, Eisenberg E.. Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer's disease. RNA 2016;22:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rozen S, Skaletsky H.. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 2000;132:365–86. [DOI] [PubMed] [Google Scholar]
- 44. Bookout AL, Mangelsdorf DJ.. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl Recept Signal 2003;1:e012.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP.. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol Lett 2004;26:509–15. [DOI] [PubMed] [Google Scholar]
- 46. Lukas M, Toth I, Reber SO, Slattery DA, Veenema AH, Neumann ID.. The neuropeptide oxytocin facilitates pro-social behavior and prevents social avoidance in rats and mice. Neuropsychopharmacology 2011;36:2159–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nadler JJ, Moy SS, Dold G, Trang D, Simmons N, Perez A, Young NB, Barbaro RP, Piven J, Magnuson TR. et al. Automated apparatus for quantitation of social approach behaviors in mice. Genes Brain Behav 2004;3:303–14. [DOI] [PubMed] [Google Scholar]
- 48. Bicks LK, Koike H, Akbarian S, Morishita H.. Prefrontal cortex and social cognition in mouse and man. Front Psychol 2015;6:1805.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Finlay JM, Dunham GA, Isherwood AM, Newton CJ, Nguyen TV, Reppar PC, Snitkovski I, Paschall SA, Greene RW.. Effects of prefrontal cortex and hippocampal NMDA NR1-subunit deletion on complex cognitive and social behaviors. Brain Res 2015;1600:70–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sanchez C, Hyttel J.. Comparison of the effects of antidepressants and their metabolites on reuptake of biogenic amines and on receptor binding. Cell Mol Neurobiol 1999;19:467–89. [DOI] [PubMed] [Google Scholar]
- 51. Sawyer EK, Howell Ll.. Pharmacokinetics of fluoxetine in rhesus macaques following multiple routes of administration. Pharmacology 2011;88:44–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Raap DK, Evans S, Garcia F, Li Q, Muma NA, Wolf WA, Battaglia G, Van De Kar LD.. Daily injections of fluoxetine induce dose-dependent desensitization of hypothalamic 5-HT1A receptors: reductions in neuroendocrine responses to 8-OH-DPAT and in levels of Gz and Gi proteins. J Pharmacol Exp Ther 1999;288:98–106. [PubMed] [Google Scholar]
- 53. Pohland RC, Byrd TK, Hamilton M, Koons JR.. Placental transfer and fetal distribution of fluoxetine in the rat. Toxicol Appl Pharmacol 1989;98:198–205. [DOI] [PubMed] [Google Scholar]
- 54. Baumann P, Rochat B.. Comparative pharmacokinetics of selective serotonin reuptake inhibitors: a look behind the mirror. Int Clin Psychopharmacol 1995;10:15–21. [DOI] [PubMed] [Google Scholar]
- 55. Kristensen JH, Ilett KF, Hackett LP, Yapp P, Paech M, Begg EJ.. Distribution and excretion of fluoxetine and norfluoxetine in human milk. Br J Clin Pharmacol 1999;48:521–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Noorlander Cw, Ververs FFT, Nikkels PGJ, van Echteld CJA, Visser GHA, Smidt MP.. Modulation of serotonin transporter function during fetal development causes dilated heart cardiomyopathy and lifelong behavioral abnormalities. PLoS One 2008;3:e2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cabrera-Vera TM, Battaglia G.. Prenatal exposure to fluoxetine (Prozac) produces site-specific and age-dependent alterations in brain serotonin transporters in rat progeny: evidence from autoradiographic studies. J Pharmacol Exp Ther 1998;286:1474–81. [PubMed] [Google Scholar]
- 58. Pei S, Liu L, Zhong Z, Wang H, Lin S, Shang J.. Risk of prenatal depression and stress treatment: alteration on serotonin system of offspring through exposure to Fluoxetine. Sci Rep 2016;6:33822.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Smit-Rigter LA, Noorlander CW, von Oerthel L, Chameau P, Smidt MP, van Hooft JA.. Prenatal fluoxetine exposure induces life-long serotonin 5-HT(3) receptor-dependent cortical abnormalities and anxiety-like behaviour. Neuropharmacology 2012;62:865–70. [DOI] [PubMed] [Google Scholar]
- 60. Amireault P, Dube F.. Serotonin and its antidepressant-sensitive transport in mouse cumulus-oocyte complexes and early embryos. Biol Reprod 2005;73:358–65. [DOI] [PubMed] [Google Scholar]
- 61. Hertz L, Rothman Dl, Li B, Peng L.. Chronic SSRI stimulation of astrocytic 5-HT2B receptors change multiple gene expressions/editings and metabolism of glutamate, glucose and glycogen: a potential paradigm shift. Front Behav Neurosci 2015;9:25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gaisler-Salomon I, Kravitz E, Feiler Y, Safran M, Biegon A, Amariglio N, Rechavi G.. Hippocampus-specific deficiency in RNA editing of GluA2 in Alzheimer's disease. Neurobiol Aging 2014;35:1785–91. [DOI] [PubMed] [Google Scholar]
- 63. Behm M, Ohman M.. RNA editing: a contributor to neuronal dynamics in the mammalian brain. Trends Genet 2016;32:165–75. [DOI] [PubMed] [Google Scholar]
- 64. Behm M, Wahlstedt H, Widmark A, Eriksson M, Öhman M.. Accumulation of nuclear ADAR2 regulates adenosine-to-inosine RNA editing during neuronal development. J Cell Sci 2017;130:745–53. [DOI] [PubMed] [Google Scholar]
- 65. Deffit SN, Hundley HA.. To edit or not to edit: regulation of ADAR editing specificity and efficiency. Wiley Interdiscip Rev RNA 2016;7:113–27. [DOI] [PubMed] [Google Scholar]
- 66. Jacobs MM, Fogg RL, Emeson RB, Stanwood GD.. ADAR1 and ADAR2 expression and editing activity during forebrain development. Dev Neurosci 2009;31:223–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wahlstedt H, Daniel C, Enstero M, Ohman M.. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res 2009;19:978–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem 2010;79:321–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Price RD, Weiner DM, Chang MSS, Sanders-Bush E.. RNA editing of the human serotonin 5-HT2C receptor alters receptor-mediated activation of G13 protein. J Biol Chem 2001;276:44663–8. [DOI] [PubMed] [Google Scholar]
- 70. Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB.. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 1997;387:303–8. [DOI] [PubMed] [Google Scholar]
- 71. McGrew L, Price RD, Hackler E, Chang MSS, Sanders-Bush E.. RNA editing of the human serotonin 5-HT2C receptor disrupts transactivation of the small G-protein RhoA. Mol Pharmacol 2004;65:252–6. [DOI] [PubMed] [Google Scholar]
- 72. Lupien SJ, McEwen BS, Gunnar MR, Heim C.. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat Rev Neurosci 2009;10:434–45. [DOI] [PubMed] [Google Scholar]
- 73. Charlton RA, Jordan S, Pierini A, Garne E, Neville AJ, Hansen AV, Gini R, Thayer D, Tingay K, Puccini A. et al. Selective serotonin reuptake inhibitor prescribing before, during and after pregnancy: a population-based study in six European regions. BJOG: Int J Obstet Gy 2015;122:1010–20. [DOI] [PubMed] [Google Scholar]
- 74. Weikum WM, Mayes LC, Grunau RE, Brain U, Oberlander TF.. The impact of prenatal serotonin reuptake inhibitor (SRI) antidepressant exposure and maternal mood on mother-infant interactions at 3 months of age. Infant Behav Dev 2013;36:485–93. [DOI] [PubMed] [Google Scholar]
- 75. Zoega H, Kieler H, Nørgaard M, Furu K, Valdimarsdottir U, Brandt L, Haglund B.. Use of SSRI and SNRI antidepressants during pregnancy: a population-based study from Denmark, Iceland, Norway and Sweden. PLoS One 2015;10:e0144474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tao R, Calley Cs, Hart J, Mayes TL, Nakonezny PA, Lu H, Kennard BD, Tamminga CA, Emslie GJ.. Brain activity in adolescent major depressive disorder before and after fluoxetine treatment. Am J Psychiatry 2012;169:381–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hollis F, Isgor C, Kabbaj M.. The consequences of adolescent chronic unpredictable stress exposure on brain and behavior. Neuroscience 2013;249:232–41. [DOI] [PubMed] [Google Scholar]
- 78. Yang Y, Hu Z, Du X, Davies H, Huo X, Fang M.. miR-16 and fluoxetine both reverse autophagic and apoptotic change in chronic unpredictable mild stress model rats. Front Neurosci 2017;11:428.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gemmel M, Kokras N, Dalla C, Pawluski JL.. Perinatal fluoxetine prevents the effect of pre-gestational maternal stress on 5-HT in the PFC, but maternal stress has enduring effects on mPFC synaptic structure in offspring. Neuropharmacology 2018;128:168–80. [DOI] [PubMed] [Google Scholar]
- 80. Rayen I, van den Hove DL, Prickaerts J, Steinbusch HW, Pawluski JL.. Fluoxetine during development reverses the effects of prenatal stress on depressive-like behavior and hippocampal neurogenesis in adolescence. PLoS One 2011;6:e24003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Salari A-A, Fatehi-Gharehlar L, Motayagheni N, Homberg JR.. Fluoxetine normalizes the effects of prenatal maternal stress on depression- and anxiety-like behaviors in mouse dams and male offspring. Behav Brain Res 2016;311:354–67. [DOI] [PubMed] [Google Scholar]
- 82. Gomez F, Garcia-Garcia L.. Anxiogenic-like effects of fluoxetine render adult male rats vulnerable to the effects of a novel stress. Pharmacol Biochem Behav 2017;153:32–44. [DOI] [PubMed] [Google Scholar]
- 83. Barden N, Reul JM, Holsboer F.. Do antidepressants stabilize mood through actions on the hypothalamic-pituitary-adrenocortical system? Trends Neurosci 1995;18:6–11. [DOI] [PubMed] [Google Scholar]
- 84. Ran Y-h, Hu X-X, Wang Y-L, Zhao N, Zhang L-M, Liu H-X, Li Y-F.. YL-0919, a dual 5-HT1A partial agonist and SSRI, produces antidepressant- and anxiolytic-like effects in rats subjected to chronic unpredictable stress. Acta Pharmacol Sin 2018;39:12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Avitsur R. Increased symptoms of illness following prenatal stress: can it be prevented by fluoxetine? Behav Brain Res 2017;317:62–70. [DOI] [PubMed] [Google Scholar]
- 86. Holmes MC, French KL, Seckl JR.. Dysregulation of diurnal rhythms of serotonin 5-HT2C and corticosteroid receptor gene expression in the hippocampus with food restriction and glucocorticoids. J Neurosci 1997;17:4056–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Heisler LK, Pronchuk N, Nonogaki K, Zhou L, Raber J, Tung L, Yeo GSH, O'Rahilly S, Colmers WF, Elmquist JK. et al. Serotonin activates the hypothalamic-pituitary-adrenal axis via serotonin 2C receptor stimulation. J Neurosci 2007;27:6956–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chou-Green JM, Holscher TD, Dallman MF, Akana SF.. Repeated stress in young and old 5-HT(2C) receptor knockout mice. Physiol Behav 2003;79:217–26. [DOI] [PubMed] [Google Scholar]
- 89. Mombereau C, Kawahara Y, Gundersen BB, Nishikura K, Blendy JA.. Functional relevance of serotonin 2C receptor mRNA editing in antidepressant- and anxiety-like behaviors. Neuropharmacology 2010;59:468–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bombail V, Qing W, Chapman KE, Holmes MC.. Prevention of 5-hydroxytryptamine2C receptor RNA editing and alternate splicing in C57BL/6 mice activates the hypothalamic-pituitary-adrenal axis and alters mood. Eur J Neurosci 2014;40:3663–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. McEwen BS. Stress and hippocampal plasticity. Annu Rev Neurosci 1999;22:105–22. [DOI] [PubMed] [Google Scholar]
- 92. Skolnick P. Antidepressants for the new millennium. Eur J Pharmacol 1999;375:31–40. [DOI] [PubMed] [Google Scholar]
- 93. Zarate CA, Quiroz J, Payne J, Manji HK.. Modulators of the glutamatergic system: implications for the development of improved therapeutics in mood disorders. Psychopharmacol Bull 2002;36:35–83. [PubMed] [Google Scholar]
- 94. Popoli M, Gennarelli M, Racagni G.. Modulation of synaptic plasticity by stress and antidepressants. Bipolar Disord 2002;4:166–82. [DOI] [PubMed] [Google Scholar]
- 95. Dingledine R, Borges K, Bowie D, Traynelis SF.. The glutamate receptor ion channels. Pharmacol Rev 1999;51:7–61. [PubMed] [Google Scholar]
- 96. Seeburg PH. A-to-I editing: new and old sites, functions and speculations. Neuron 2002;35:17–20. [DOI] [PubMed] [Google Scholar]
- 97. Bonini D, Mora C, Tornese P, Sala N, Filippini A, La Via L, Milanese M, Calza S, Bonanno G, Racagni G. et al. Acute footshock stress induces time-dependent modifications of AMPA/NMDA protein expression and AMPA phosphorylation. Neural Plast 2016;2016:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Barbon A, Popoli M, La Via L, Moraschi S, Vallini I, Tardito D, Tiraboschi E, Musazzi L, Giambelli R, Gennarelli M. et al. Regulation of editing and expression of glutamate alpha-amino-propionic-acid (AMPA)/kainate receptors by antidepressant drugs. Biol Psychiatry 2006;59:713–20. [DOI] [PubMed] [Google Scholar]
- 99. Meyer-Lindenberg A, Weinberger DR.. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci 2006;7:818–27. [DOI] [PubMed] [Google Scholar]
- 100. Vaswani M, Linda FK, Ramesh S.. Role of selective serotonin reuptake inhibitors in psychiatric disorders: a comprehensive review. Prog Neuropsychopharmacol Biol Psychiatry 2003;27:85–102. [DOI] [PubMed] [Google Scholar]
- 101. Dulawa Sc, Holick KA, Gundersen B, Hen R.. Effects of chronic fluoxetine in animal models of anxiety and depression. Neuropsychopharmacology 2004;29:1321–30. [DOI] [PubMed] [Google Scholar]
- 102. Meyer LR, Dexter B, Lo C, Kenkel E, Hirai T, Roghair RD, Haskell SE.. Perinatal SSRI exposure permanently alters cerebral serotonin receptor mRNA in mice but does not impact adult behaviors. J Matern Fetal Neonatal Med 2018;31:1393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Glover ME, Clinton SM.. Of rodents and humans: a comparative review of the neurobehavioral effects of early life SSRI exposure in preclinical and clinical research. Int J Dev Neurosci 2016;51:50–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Svirsky N, Levy S, Avitsur R.. Prenatal exposure to selective serotonin reuptake inhibitors (SSRI) increases aggression and modulates maternal behavior in offspring mice. Dev Psychobiol 2016;58:71–82. [DOI] [PubMed] [Google Scholar]
- 105. Kiryanova V, Meunier SJ, Vecchiarelli HA, Hill MN, Dyck RH.. Effects of maternal stress and perinatal fluoxetine exposure on behavioral outcomes of adult male offspring. Neuroscience 2016;320:281–96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.