Abstract
Gene flow and demographic history can play important roles in the adaptive genetic differentiation of species, which is rarely understood in the high-altitude adaptive evolution of birds. To elucidate genetic divergence of populations in the great tit complex (Parus major, P. minor and P. cinereus) at different elevations, we compared the genetic structure and gene flow in hemoglobin genes with neutral loci. Our results revealed the elevationally divergent structure of αA-globin gene, distinctive from that of the βA-globin gene and neutral loci. We further investigated gene flow patterns among the populations in the central-northern (> 1,000 m a.s.l.), south-eastern (< 1,000 m a.s.l.) regions and the Southwest Mountains (> 2,000 m a.s.l.) in China. The high-altitude (> 1,000 m a.s.l.) diverged αA-globin genetic structure coincided with higher αA-globin gene flow between highland populations, in contrast to restricted neutral gene flow concordant with the phylogeny. The higher αA-globin gene flow suggests the possibility of adaptive evolution during population divergence, contrary to the lower αA-globin gene flow homogenized by neutral loci during population expansion. In concordance with patterns of historical gene flow, genotypic and allelic profiles provide distinctive patterns of fixation in different high-altitude populations. The fixation of alleles at contrasting elevations may primarily due to highland standing variants αA49Asn/72Asn/108Ala originating from the south-western population. Our findings demonstrate a pattern of genetic divergence with gene flow in major hemoglobin genes depending on population demographic history.
Keywords: elevational divergence, gene flow, genetic structure, hemoglobin gene
Widespread species in hierarchical population structure can provide opportunities for investigating the potential influence of demographic history in shaping genetic variation arising in response to environmental change. A comprehensive example is a fitness-correlated morphological gene in the adaptive evolution of marine and freshwater three-spine stickleback Gasterosteus aculeatus responding to gene flow and demographic history (Schluter et al. 2004; Colosimo et al. 2005; Schluter and Conte 2009; Raeymaekers et al. 2014). For high-altitude adaptive evolution, genetic variation of oxygen-carrying hemoglobin (Hb) has long been used to track natural selection in native vertebrates under hypoxia stress (Perutz 1983; Monge and Leon-Velarde 1991; Scott and Milsom 2006; Storz 2007; Weber 2007). However, intraspecific genetic divergence in Hb genes may not necessarily cause adaptive changes in Hb in birds (McCracken et al. 2009a; Munoz-Fuentes et al. 2013; Cheviron et al. 2014; Galen et al. 2015; Natarajan et al. 2015). A hypothesis proposed that highland adaptation after colonization from lowlands following divergence based on the Hb gene flow from lowland to highland in Andean waterbirds Anatidae (McCracken et al. 2009b; Wilson et al. 2012). The hypothesis was subsequently supported by highland genotypes derived from ancestral lowland genotypes in functional experiments (Natarajan et al. 2015). For avian species at broad elevations, whether the genetic structure and gene flow in Hb genes can reveal intraspecific adaptive differentiation in elevations is still an open question.
To investigate the role of gene flow and demographic history in elevational genetic divergence, we performed population genetic analyses on major Hb genes and unlinked neutral loci in a widespread species complex, the great tit. The HbA tetramer, which is the major Hb isoform expressed in adult birds, comprises 2 α-chains and 2 β-chains encoded by αA- and βA-globin gene, respectively (Mairbaurl and Weber 2012; Opazo et al. 2015). It has been revealed that the adaptive genetic variants mostly presented in the βA-globin gene in high-altitude Andean birds (Projecto-Garcia et al. 2013; Galen et al. 2015; Natarajan et al. 2015). In contrast, rare causative variants were found in the αA-globin gene of cinnamon teals Anas cyanoptera and nightjars Hydropsalis ( Wilson et al. 2012; Natarajan et al. 2015; Kumar et al. 2017). The great tit complex, mainly comprising Parus major, P. minor, and P. cinereus, is known as a superspecies encircling the Qinghai–Tibet Plateau (QTP) and Central Asian desert (Kvist et al. 2003; Päckert et al. 2005; Kvist et al. 2007; Gill and Donsker 2016). This superspecies is widely distributed from sea level to above 4,000 m a.s.l. in Eurasia, with the highest habitats located in the eastern Himalaya (Gosler et al. 2017). A monophyletic clade of P. minor in eastern Himalaya (P. m. tibetanus and P. m. subtibetanus, clade B) has been further identified to compose the 5-clade mtDNA phylogeny of the great tit complex (Zhao et al. 2012). The demographic history of P. minor revealed historically stable growth in the eastern Himalaya populations before the last glacial maximum (LGM) and rapid population expansion in the East Asia during the LGM (Zhao et al. 2012; Qu et al. 2015).
In the present study, we conducted population genetic analyses for exploring the role of gene flow and demographic history in Hb genetic differentiation corresponding to changes in elevation in great tit populations. This study mainly focused on 1) comparing the difference in genetic structure of Hb genes with neutral loci; 2) examining gene flow among local populations at hierarchical elevations of P. minor; 3) identifying genotypes and profiles along elevational gradients and 4) elevational variation of allelic frequencies in Hb genes across parallel transects.
Materials and Methods
Sampling, sequencing and gametic phasing
To determine the genetic structure of all populations, we employed a total of 159 samples from all 5 clades of the 3 species (P. minor, P. cinereus and P. major) within the great tit complex (Supplementary Table S1). All the collections were carried out with permission from the State Forestry Administration of the People’s Republic of China and in accordance with the National Wildlife Conservation Law of China. To explore local population differentiation in response to changes in elevation, we analyzed a sub-dataset of P. minor populations. According to the genetic structure partitioned by elevation (Figure 1), we sampled highland (1,000–4,282 m a.s.l.) and lowland individuals (7–210 m a.s.l.) in 3 local populations of P. minor (N = 66) (Supplementary Table S1). The 3 populations were located in the Southwest Mountain region (highland Clade B, HB, N = 23), the alpine region of central China (highland Clade A, HA, N = 20), and the plain of southeast China (lowland Clade A, LA, N = 23). The HB and HA populations were adjacent to the southeast and the east of the QTP, respectively (Figure 2A).
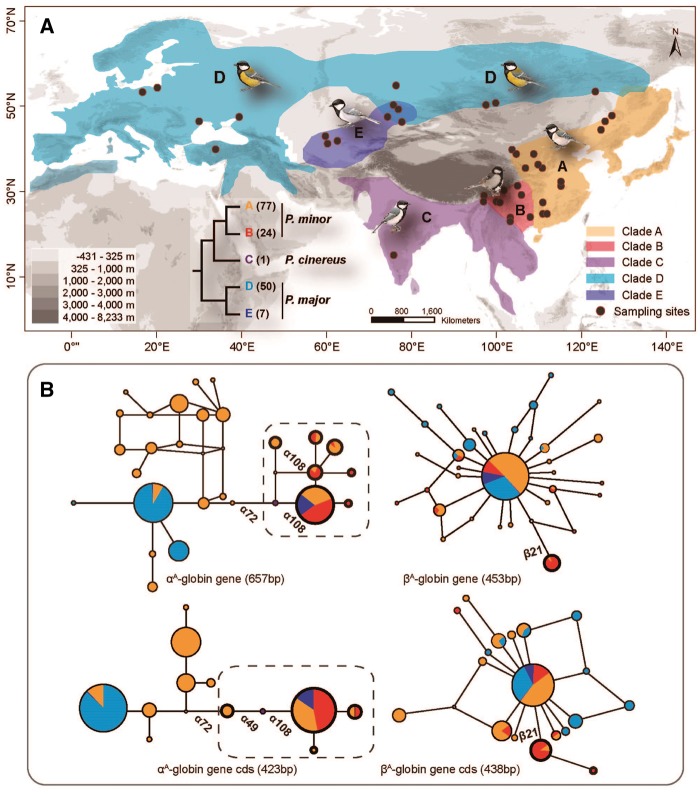
Figure 1.
Distribution of samples and unrooted haplotype networks in samples of the Great Tit complex. (A) Samples collected in species maps. Colorful shadows represent the distribution of each clade in neutral phylogeny, corresponding to the nodes in mtDNA dendrogram (Zhao et al. 2012). (B) In networks, each circle represents for a haplotype and circle size for the individual numbers with a shared haplotype. Pied colors represent for different phylogenetic clades in areas proportional to haplotype frequencies. An elevational differentiation presents in αA-globin gene in the samples above 1,000 m a.s.l of clades A, B and E (bold nodes in dash-line rectangles, Supplementary Table S1). The presence of non-synonymous substitution sites are labeled in Hb networks.
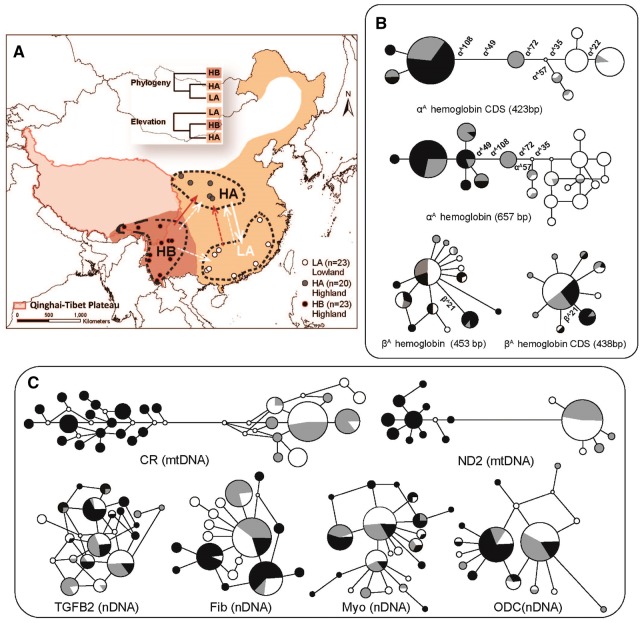
Figure 2.
Gene flow diagrams and networks in P. minor populations. (A) The collection localities of 3 local populations of clade A and B in dash circles (Figure 1). The sampling localities of HB, HA, and LA populations are in black, gray and white. The direction of gene flow between elevational differentiated populations is in red and white arrows for αA-globin gene and neutral loci, respectively. The strength of gene flow is classified into relatively strong as solid and weak as dash lines compared in αA-globin gene or neutral loci. (B-C) The genetic structures of major Hb genes and neutral reference loci (mtDNA and nucleotide introns) were illustrated in networks. Black, gray and white colors are the same individual of the 3 populations in panel A. The presence of nonsynonymous substitution sites are labeled in Hb networks (B).
We amplified and sequenced αA- and βA-globin genes and 6 unlinked reference loci (2 mtDNA and 4 autosomal introns) for all samples. The reference loci are widely used neutral markers in general phylogenetic studies, including the mitochondrial control region (CR, 539 bp), NADH dehydrogenase subunit 2 (ND2, 737 bp), beta-fibrinogen intron 5 (Fib, 281 bp), transforming growth factor beta 2 intron 5 (TGFB2, 334 bp), myoglobin intron 2 (Myo, 556 bp) and ornithine decarboxylase intron 7 (ODC, 284 bp). We designed subunit-specific primers for Hb genes (660 bp of αA-globin gene and 1,274 bp of βA-globin gene). After DNA extraction from venous blood or pectoral muscle with a DNeasy Tissue kit (QIAGEN, Hilden, Germany), we amplified the 8 genes with KD-Plus DNA polymerase (TransGen Biotech, Beijing, China). The general PCR thermal profile was 5 min at 94°C, 35 cycles of 40 s at 94 °C, 40 s at 52–64 °C, 1 min at 72 °C, and 10 min at 72 °C (with varied annealing temperatures, as shown in Supplementary Table S2). For each gene, we sequenced the PCR products on the ABI3730 sequencing system (BGI, Beijing, China).
For heterozygous autosomal genes, we inferred the gametic phases using PHASE v2.1 (Stephens et al. 2001). The PHASE algorithm was run 5 times using random seeds to obtain stable results. For the αA-globin gene (660 bp) and 4 autosomal introns mentioned above, we inferred the gametic phases of the complete sequences. For the βA-globin gene (1,274 bp), we inferred the gametic phases for the fragments spanning from exon 1 to exon 2 and partial intron 2 (453 bp), which had no recombination detected in DnaSP v5.10.1 (Librado and Rozas 2009). All substitutions that occurred in the exon 3 (126 bp) of the βA-globin gene were synonymous. The sequences with high posterior probabilities (>95%) and the consensus sequence of the cloned samples unresolved in PHASE proceeded into downstream analyses. All sequences are accessible under accession numbers MF321902-MF322504 in GenBank, NCBI, NIH (Supplementary Table S2).
Comparison of genealogical structures between hb genes and neutral loci
To understand the genealogical discordance between Hb genes and neutral loci, we reconstructed unrooted networks of haplotypes for each gene in local and all samples by using the median-joining algorithm in NETWORK 4.6.1 (Bandelt et al. 1999). Networks based on the coding sequence (CDS) of the Hb genes were also reconstructed to exclude the effects of introns in Hb genes. For qualitative analysis of the genetic structures of local populations, we calculated the posterior probabilities of assigned individuals into highland and lowland populations of P. minor in STRUCTURE 2.3.4 (Pritchard et al. 2000). We performed STRUCTURE analyses on 4 different sub-datasets: all nuclear introns, the introns plus αA-globin gene, the introns plus βA-globin gene, and the introns plus αA- and βA-globin genes. The analyses were performed for 1 or 2 population models (K = 1 or 2) without prior sampling and conducted with admixture model and independent allele frequencies (α = 1, λ = 1). The algorithm was run for 100,000 iterations with a burn-in of 25,000 iterations.
We calculated polymorphism and diversity of all loci in each population and in all 3 populations of P. minor combined in DnaSP v5.10.1 (Pritchard et al. 2000). Tajima’s D (Tajima 1989) and Fu’s Fs (Fu and Li 1993; Fu 1997) statistics were computed to test deviation from neutrality. The significant deviation from neutrality is an outlier of 95% confidence interval in a null model of neutral expectation in coalescent simulation based on segregated sites and recombination rate per gene. We calculated pairwise FST values for each of 8 loci and the CDS of Hb genes in ARLEQUIN 3.5.
Genetic diversity and gene flow in local populations
We used a coalescent-based “isolation-with-migration” model implemented in IMa (Hey and Nielsen 2007) to estimate inter-population gene flow in the sampled P. minor populations. We focused on the gene flow strength in the same dataset (αA-globin or reference loci) and asymmetric gene flow ratio scaled by migration rates (m* = 2 Nm) i.e., locus-specific effective population size rather than mutation rates. We thus conducted analyses for the αA-globin gene and joint reference loci (2 mtDNA and 4 autosomal introns, Supplementary Table S3). We detected intra-locus recombination with the 4-gamete test and retained the longest non-recombining block of sequences in each locus under the IMa assumption of the absence of recombination within a locus (Hey and Nielsen 2004). According to the biasing effects of substitution models, we fitted the Hasegawa–Kishino–Yano (HKY) and Infinite Sites (IS) mutation models for mtDNA and autosomal DNA regardless of calculated models (Strasburg and Rieseberg 2010). Final parameter estimations were scaled to effective population size multiplied by mutation rate (θ = 4 Nμ) with estimated migration rates (m = m/μ) transformed to 2 Nm (M = θ × m/2) (Hey and Nielsen 2004). Given inheritance scalars of 0.25 (mtDNA) and 1.00 (autosomal DNA), we initially performed IMa runs with flat priors for each parameter. On the basis of preliminary runs, we tuned upper bounds for priors that encompassed the overall posterior distributions. We finally ran the analyses in 6 Markov-coupled chains with geometric heating (g1 = 0.8; g2 = 0.9), discarding at least 20,000 samples as “burn-in”. Each chain was continued until the effective sample size (ESS) was >100. To ensure the convergence of parameter estimates, 2 replicated runs in each analysis were conducted by using different random seeds.
Genotypic profiles of hb genes and elevational clines of hb alleles
To characterize possible genotypes differentiated between highland and lowland populations, we estimated genotypic frequencies in Hardy–Weinberg equilibrium (HWE) for haplotypes and genotypes of the 2 Hb genes in ARLEQUIN 3.5 (Excoffier and Lischer 2010). To explore the threshold of elevational genetic variation, we conducted a maximum likelihood estimation of Hb allele frequencies by assigning individuals of P. minor populations into 2 parallel elevational transects: T1 in the south and T2 in the north (Supplementary Figure S2). We used the cline shapes to acquire the profile of highland replacements and a modified Markov Chain Monte Carlo (MCMC) algorithm to determine support limits in ClineFit 2.0b (Porter et al. 1997). The 4-parameter model for the cline center (c), cline width (w), minimum allele frequency (PL) and maximum allele frequency (PR) at the ends of the cline shapes was estimated. We set a burn-in of 800 parameter tries per annealing step and 1000 tries for the second run that are combined in sampling; subsequently, 2000 replicates were saved in the sampling parameter space, with 20 replicates between each save. To assess the coincidence of the cline center and the concordance of the width, we combined the sampled parameter space of replicated runs according to the maximum 2 LnL limits and subsequently generated support plots (not shown) and sigmoidal cline graphics in Mathematica v10. The FST for each non-synonymous substitution between highland and lowland populations was calculated using locus-by-locus AMOVA in ARLEQUIN 3.5 (Excoffier and Lischer 2010).
Results
The population genetic structure along elevations
For both the great tit superspecies and P. minor, significant genetic divergence was found in Hb genes, especially for the αA-globin gene (Figures 1 and 2). The divergence of the αA-globin gene partitioned the superspecies into 2 elevational clades: the monophyletic clade of high-altitude individuals from the phylogeographic clade A, B, C, and E, and the other clade including the clade D and low-altitude individuals from the clade A (Figure 1). Similarly, the network of the αA-globin gene in P. minor populations presented the monophyletic highland clade including HA and HB, which was diverged from the lowland clade-A population (LA) (Figure 2). The βA-globin gene, however, did not exhibit any clear genetic structure. The single β21Ala/Thr substitution presented in the southwest mountains was not fixed in frequency (Figure 2 and 4). Therefore, in further analyses of genetic variance, we mainly used the local population dataset to explore a potential genetic response to the altitudinal gradient in P. minor.
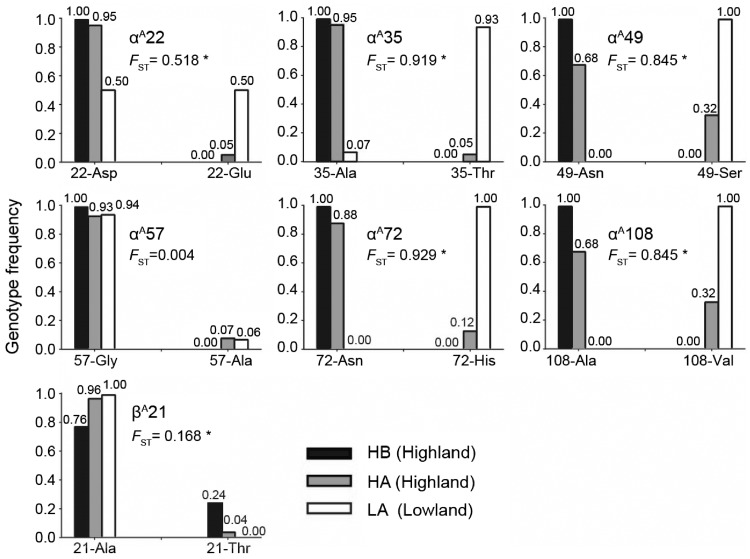
Figure 4.
Genotype frequencies in amino acid alleles of major Hb genes. FST represents for differentiation of each allele between highland and lowland populations, *P < 0.05. The striking heterozygous highland and lowland genotypes involve αA22Asp/Gly in LA population and βA21Ala/Thr in HB population, in contrast to low-level admixed in α35/57. The more homozygous αA49Asn/Ser-72Asn/His-108Ala/Val is an admixed genotype only in HA population at lower frequency while completely fixed in HB vs. LA population.
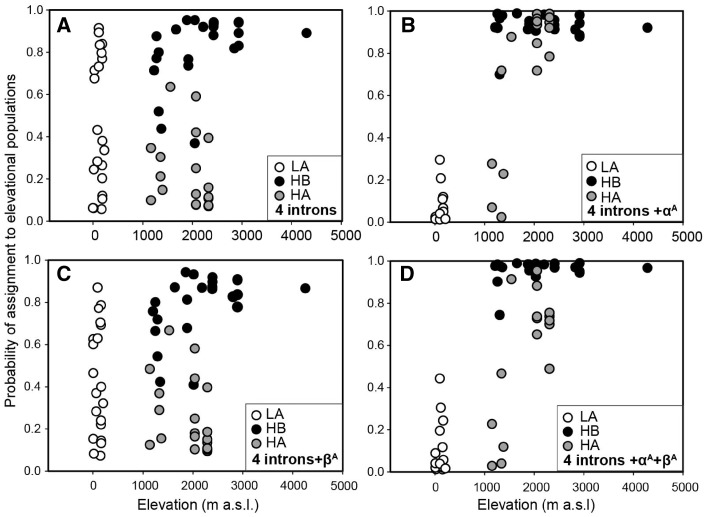
The genetic structure in differences of elevation, which could be largely due to differences in the αA-globin gene, was further confirmed by STRUCTURE analysis on local populations (Figure 3). The different assignment to lowland or highland (>1,100 m a.s.l.) populations was most dramatic in average posterior probability of the αA-globin (6.1% versus 88.2%, ±8.0% and 18.7% standard deviation [SD]) and less so of joint αA- and βA-globin genes (8.6% versus 82.6%, ±10.9% and 22.0% SD). In contrast, the assignments showed much less divergence in response to elevation either for the single βA-globin (43.6% and 55.3%) or for nuclear introns (55.7% and 44.8%).
Figure 3.
Posterior probability of individual assignment to highland and lowland populations in STRUCTURE 2.3. Black, gray, and white represent individuals from population HB, HA and LA (Fig. 2). (A) 4 introns of reference loci, (B) 4 reference introns and αA-globin gene, (C) 4 reference introns and βA-globin gene, (D) 4 reference introns and 2 hemoglobin genes. The higher divergence occurs in the dataset including αA-globin gene but lower with βA-globin gene add-in.
Gene flow among elevationally differentiated populations of P. Minor
The coalescent estimations of IMa showed unbalanced downslope gene flow (from highland to lowland) in the joint reference loci (assumed as neutral gene flow) among local P. minor populations (Table 1; Figure 2A). The highest gene flow was found in the downslope (HA to LA) neutral loci and in 16.1, 7.3 and 18.5 times higher level than the upslope (LA to HA) and the gene flow between 2 clades (HB-LA and HB-HA) (Table 1). For the αA-globin gene, in contrast, downslope gene flow between clades (HB-HA) was 23.9 times higher than upslope gene flow within the clade A (LA-HA). The lower gene flow of neutral loci is between 2 clades (HB versus HA/LA), while the lower αA-globin gene flow is between populations differed by elevations (LA versus HB/HA). Relative to the restrained neutral gene flow, the much higher gene flow in αA-globin from HB into HA is in accordance with the elevational structure of local populations.
Table 1.
Coalescent estimation of population genetic parameters among the P. minor populations using IMa
| Loci | Populations | θH | θL | mH | mL | mH* | mL* | mL*/mH* |
|---|---|---|---|---|---|---|---|---|
| (High versus lowland) | ||||||||
| All neutralloci | HA versus LA | 0.43 | 1.68 | 2.50 | 10.36 | 0.54 | 8.69 | 16.09 |
| (0.18–1.18) | (0.78–4.82) | (0.68–41.37) | (3.24–45.80) | |||||
| HB versus LA | 1.14 | 1.09 | 0.00 | 2.18 | 0.00 | 1.19 | NA | |
| (0.66–2.03) | (0.64–2.00) | (0.02–4.64) | (0.25–5.51) | |||||
| HB versus HA | 1.11 | 0.53 | 0.00 | 1.76 | 0.00 | 0.47 | NA | |
| (0.67–1.93) | (0.28–1.14) | (0.01–2.81) | (0.42–5.62) | |||||
| αA–globingene | HA versus LA | 0.43 | 0.41 | 1.03 | 0.01 | 0.22 | 0.00 | NA |
| (0.16–2.17) | (0.13–1.93) | (0.27–8.06) | (0.02–5.60) | |||||
| HB versus LA | 0.49 | 0.60 | 0.00 | 0.00 | 0.00 | 0.00 | NA | |
| (0.13–2.07) | (0.23–2.26) | (0.01–2.28) | (0.01–2.10) | |||||
| HB versus HA | 0.26 | 1.20 | 0.01 | 8.73 | 0.00 | 5.25 | NA | |
| (0.10–1.57) | (0.52–31.37) | (0.12–19.41) | (2.73–51.57) |
The neutral parameter θ = 4 Nμ, N is the effective population size of each population, μ is the mutation rate. mH, the migration rate from lowland to highland (upslope); mL, migration rate from highland to lowland (downslope). The upslope gene flow between HB and HA is from the relative lowland east QTP (HA) to the highland west QTP (HB), the opposite is downslope from HB to HA (Supplementary Figure S1 and see Appendix S3). The 95% highest posterior density (95% HPD) are shown in parentheses. mH* and mL*, were scaled m by effective population size by m × θ/2 of upslope and downslope gene flow. The mL*/mH* ratio is an index of the asymmetric gene flow. NA is an ineffective ratio of zero m values.
Genetic divergence and genotypic profiles in major Hb genes of P. Minor
We identified nucleotide polymorphisms of all genes in the P. minor populations (Supplementary Table S3). The haplotype diversity was the lowest in ND2 and βA-globin gene (Hd< 0.5) of HA and LA populations compared with other loci (average 0.726 ± 0.21 SD). The nucleotide diversity of both Hb genes and ND2 of clade A exhibited the lowest levels (π < 0.002, average 0.0052 ± 0.0050 SD). Significant negative Tajima’s D and Fu’s Fs values were present in different populations of βA-globin and reference loci (Supplementary Table S3). In αA-globin gene of all populations, insignificant negative values were predominant, indicating lower than expected neutrality. Significant negative values predominated among nuclear loci of the LA population and in βA-globin for both HA and LA populations, but for mtDNA in the HB population. In P. minor populations of clade A, significantly greater divergence was indicated by fixation indices (FST) in the αA-globin gene in contrast to other loci (FST* > 0.5 versus < 0.1, Table 2). The significant inter-clade divergence of mtDNA was consistent with genetic structure. The pairwise FST values of the αA-globin gene showed significant differentiation among populations at different elevations (Table 2).
Table 2.
Pairwise FST values for nucleotide haplotypes of the P. minor populations
| Pairwise | αA | αA-cds | βA | βA-cds | CR | ND2 | TGFB2 | Fib | Myo | ODC |
|---|---|---|---|---|---|---|---|---|---|---|
| HA versus HB | 0.15 | 0.20 | 0.07 | 0.06 | 0.81 | 0.95 | 0.15 | 0.26 | 0.02 | 0.31 |
| HA versus LA | 0.60 | 0.67 | 0.00 | 0.00 | 0.10 | 0.00 | 0.02 | 0.07 | 0.05 | 0.00 |
| HB versus LA | 0.76 | 0.87 | 0.10 | 0.13 | 0.69 | 0.95 | 0.10 | 0.15 | 0.10 | 0.29 |
Significant values (P < 0.05) are shown in bold. Sample sizes were shown in Figure 1.
All the Hb genotypes were profiled in HWE analyses (Supplementary Table S4). The genotype of each Hb gene exhibited differences in frequency between highland and lowland populations. The 6-allele genotype in the αA-globin gene was completely fixed in all individuals in the HB population (the same genotype in 46 chromosomes). Accordingly, both the observed genotypic heterozygosity of the αA-globin gene in HB was lower than that in the other 2 populations (Ho/He = 0.753, 0.924 and 0.861 in HB, HA, and LA). The βA-globin genotype of most individuals was homozygous, and a unique substitution of β21Ala/Thr occurred in HB populations at a low frequency (23.5%). From the allele-specific analysis, αA57Gly/Ala (FST = 0.004) and β21Ala/Thr (FST* = 0.168, *P < 0.05) exhibited the lowest differentiation between highland and lowland populations (Figure 4). The αA-globin genotypes of 2 distant alleles were fixed according to elevation (FST* > 0.9): α35Ala/72Asn was predominant in highland populations (HA and HB). Moreover, αA49Asn/Ser and αA108Ala/Val also exhibited high levels of elevational differentiation (FST* = 0.845).
Elevational variation in allele frequencies across parallel transects
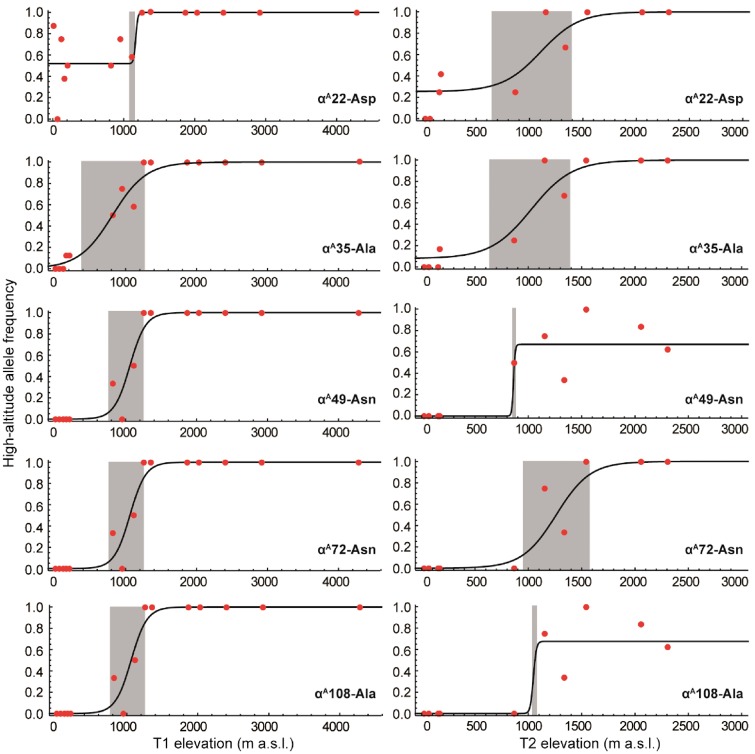
Specific αA-globin allelic profiles in local populations of P. minor were further determined by cline-fitting analyses along parallel elevational gradients (Figure 5 and Table 3). The highland alleles shifted along altitudinal gradients coincide at a threshold of 1,100 m a.s.l. (±150 m SD) that is consistent with the genetic structure of superspecies sorted by elevation. The center fittings of cline shapes were similar across parallel transects at or above 1,000 m. The average estimated cline center of T1 curves was 1028.4 (817–1,145) m a.s.l., and the cline width was 471 (59–958) m (Table 3). The fully consistent cline centers (1,060, 962–1,152 m a.s.l.) and widths (446, 14–813 m) occurred in αA49/72/108 of the T1 transects, whereas the approximately coincident centers of αA22/35/108 were in T2. The average center of T2 was slightly higher (1,131.3 m a.s.l.) and wider (707 m) than that of the T1 transect. In the more elevationally fixed alleles (α49/72/108 of T1), higher differentiation (FST = 1.00, P < 0.05) was apparent across elevational transects. Accordingly, the αA49-72-108 across T1 exhibited the highest differentiation (FST = 1.00), and αA22 across T1 had the lowest differentiation (FST = 0.36; Table 3). According to the genetic structure, the unique βA-globin replacement without an elevation pattern could not be used as the marker to reveal the elevation pattern in local populations (Figure 4 and Supplementary Figure S3).
Figure 5.
Elevational cline fitting of αA-globin allele frequencies in parallel transects of P. minor. Highland allele frequencies at a given elevation are in red points, the support limit of cline center is in gray shade. The shift at ca 1, 100 m a.s.l. appears to be a threshold of most high-altitude Hb alleles in the P. minor populations. The coincident cline varied elevational shifts occurred in α49/72/108 in T2 transect and α22/35 in T1 transect. The genotypic frequency of α49Asn/Ser-72Asn/His-108Ala/Val in T1 transect and α72Asn/His in both transects are fixed in high/low elevations outside the cline shift regions.
Table 3.
Maximum likelihood of elevational clines of αA-globin allele frequencies across parallel transects in P. minor
| Alleles | Transect | Centre/m | Width/m | PL | PR | FST(*P < 0.05) |
|---|---|---|---|---|---|---|
| αA22 | T1 | 1145 | 59 | 0.52 | 1.00 | 0.36* |
| (1066–1262) | (0–606) | (0.41–0.63) | (0.96–1.00) | |||
| T2 | 1106 | 719 | 0.25 | 1.00 | 0.68* | |
| (440–1415) | (4–2275) | (0.00–0.44) | (0.92–1.00) | |||
| αA35 | T1 | 817 | 958 | 0.00 | 1.00 | 0.97* |
| (666–1048) | (385–1387) | (0.00–0.15) | (0.95–1.00) | |||
| T2 | 1027 | 764 | 0.08 | 1.00 | 0.86* | |
| (632–1269) | (17–1713) | (0.00–0.24) | (0.92–1.00) | |||
| αA49 | T1 | 1060 | 446 | 0.00 | 1.00 | 1.00* |
| (962–1152) | (14–813) | (0.00–0.08) | (0.96–1.00) | |||
| T2 | 861 | 31 | 0.00 | 0.67 | 0.60* | |
| (181–1203) | (0–1539) | (0.00–0.09 | (0.52–0.82) | |||
| αA72 | T1 | 1060 | 446 | 0.00 | 1.00 | 1.00* |
| (962–1152) | (14–813) | (0.00–0.08) | (0.96–1.00) | |||
| T2 | 1261 | 638 | 0.00 | 1.00 | 0.84* | |
| (1081–1449) | (329–1207) | (0.00–0.08) | (0.93–1.00) | |||
| αA108 | T1 | 1060 | 446 | 0.00 | 1.00 | 1.00* |
| (962–1152) | (14–813) | (0.00–0.08) | (0.96–1.00) | |||
| T2 | 1047 | 56 | 0.00 | 0.68 | 0.60* | |
| (846–1339) | (0.25–1272) | (0.00–0.07) | (0.53–0.83) |
Notes: The parenthesized ranges are parameter support limits up to lnLmax–lnL < 2 likelihood.
The Hb allelic variations in the great tit complex samples comprised the same 6 αA-globin substitutions and 1 βA-globin substitution as in the P. minor populations. In accordance with local populations, the allele frequencies of αA49/72/108 exhibited relatively greater fixed elevational variation and αA72 exhibited complete elevational fixation (Supplementary Figure S3). And frequencies of the same allele in highland populations exhibited different results in parallel transects that αA49/108 was flexible in T2 (Figure 5 and Supplementary Figure S3). In contrast, the αA22/35/57 lacked differentiation along the altitudinal gradient for samples from all clades (Figure 1). The βA21Thr occurred at a lower frequency from lowland to highland populations except for fixation at the peak elevation (< 36% at lower than 3,000 m a.s.l.).
Discussion
Elevational differentiation of genetic structure in αA-globin gene
We found an elevationally differentiated structure in the αA-globin gene, which is distinctive from the mtDNA phylogeny and the unstructured networks in nuclear loci. This topological mismatch between functional gene and neutral genes might be induced by incomplete lineage sorting or introgressive hybridization between sister lineages, as in sister Anatidae species (Natarajan et al. 2015). The elevationally differentiated genetic structure of the αA-globin gene is also distinctive from the unstructured βA-globin. This result differs from most Andean birds that elevational differentiated βA-globin gene is contributed to the conspecific adaptive divergence (McCracken et al. 2009b; Bulgarella et al. 2012; Galen et al. 2015; Natarajan et al. 2015; but see Wilson et al. 2012). The high- and low-altitude divergence in αA-globin gene only occurred in cinnamon teals as similar manner as βA-globin gene divergence in other waterfowl (McCracken et al. 2009b; Bulgarella et al. 2012; Wilson et al. 2012). The adaptive divergence in Hb genes of Andean waterbirds consistently contributed to the dispersal of ancestral genotypes in lowland populations by colonization into the highlands. In conclusion, the discrepancy in genetic structures between the 2 major Hb genes might be resulted from adaptive evolution in the αA-globin gene of great tits.
Gene flow patterns among local populations of P. minor
From coalescent gene flow estimation of gene flow in P. minor, we found higher than expected αA-globin gene flow between clades with restricted neutral gene flow. In contrast to background gene flow revealed by neutral loci, the inter-clade downslope gene flow of αA-globin gene is nearly absent between elevation-diverged populations within the clade. Similar patterns exhibited in the restricted βA-globin gene flow in the adaptive elevational divergence with highland-derived replacements in Andean waterbirds (McCracken et al. 2009b; Wilson et al. 2012; Natarajan et al. 2015). The αA-globin gene flow from subtropical highlands (HB) into temperate lowlands (HA) of P. minor is in a direction similar to the βA-globin gene flow from tropical highlands into temperate lowland of south-American waterfowl (McCracken et al. 2009b; Munoz-Fuentes et al. 2013). The restricted αA-globin gene flow in the same clade of P. minor may also have been homogenized by high gene flow of neutral loci contingent on population history, as in the pattern shown for major Hb genes in Andean waterfowl (McCracken et al. 2009b). Our results suggest the role of αA-globin gene flow in driving genetic divergence along elevational gradients in population history of a widespread species.
Given the demographic history of the great tit (Zhao et al. 2012; Qu et al. 2015), we propose a plausible route for historical population expansion. The αA-globin genetic variants may have existed in the high-altitude eastern Himalayan population and dispersed to the lower habitats in central China earlier before the inter-clade divergence in neutral loci. The strong downslope neutral gene flow, in contrast, implies the historical rapid population expansion was from the central highland into the southeast lowland population after inter-clade divergence. Thus, the restricted αA-globin gene flow within the same clade may be a result of rapid population expansion in the lowland population, offsetting the possible countervailing selection on high-altitude genetic variants after inter-clade divergence.
Elevational variation of alleles depends on demographic history
The genotypic profile of major Hb genes in HWE coincides with the genetic structure of elevational segregation in the αA-globin gene. However, the haplotypic Ho/He ratios of the αA-globin gene in the HB and LA populations deviated significantly from equilibrium. This result suggests subunit-specific allelic profiles. From elevational cline fitting, we identified the high/lowland fixed α49/72/108 genotypes that coincide with possible standing variants α49Asn/72Asn/108Ala, originating from an ancestral population in the eastern Himalaya (Tietze and Borthakur 2012; Qu et al. 2015). From the ancestral states reconstruction of Hb genes in Paridae, the highland αA-globin haplotype was inferred to be the standing genetic variant in P. minor (Zhu et al., unpublished data). In contrast, the lowland fixed β21Thr is more likely a result of newly derived mutations in highland eastern Himalaya populations and fixed after the rapid expansion. These results implied that standing genetic variation of the αA-globin gene might have been fixed in clade B before the inter-clade divergence. The heterozygous α49/72/108 genotypes in clade A, in contrast, may be a mixed result of countervailing selection on standing high-altitude variants and homogenizing neutral gene flow, as the case in Andean waterfowl (McCracken et al. 2009b; Wilson et al. 2012; Natarajan et al. 2015;). Therefore, we suggest that the αA-globin gene is likely under historical adaptive selection rather than the βA-globin gene in P. minor and the great tit complex.
It is known that adaptive divergence in a wild species might not be determined by phenotypic changes of Hb , whereas the fixation of genetic variants of functional genes eventually depends on demographic history (Olson-Manning et al. 2012; Raeymaekers et al. 2014; Ferchaud and Hansen 2016). The fixation of standing genetic variants in highland populations (HB or HA) was inferred for most alleles (8/10, except for α49/108 in T2) (Zhu et al., unpublished data) that might be homogenized by neutral gene flow. Similarly, much narrower shifts but unfixed highland α49Asn/108Ala alleles in HA and wider cline shifts (α22/35/72 of T2) may have been caused by neutral gene flow in clade A. In agreement with these results, the α22/35 were absent from altitudinal clines in the full sampling, whereas only the completely diverged α72Asn/His suggested that an ancestral standing polymorphism was fixed in all highland populations of the great tit complex. Therefore, although the same elevational differentiation was detected in the genetic structure of the superspecies, specific allele profiles of in P. minor populations were affected by local homogenizing gene flow. In a word, differed from the case of standing Hb variants originated in low-altitude Andean waterfowl (Natarajan et al. 2015), the recurrently replaced alleles in deeply diverged lineages of Eastern Himalaya and Central Asia can be interpreted as the fixation of standing high-altitude alleles in East-Himalaya.
In summary, our study highlights the necessity of integrating gene flow and demographic history into exploring potential adaptive evolution in the genetic divergence of widespread species. This preliminary investigation on genetic variants in a widespread passerine superspecies provides an ideal model to further explore the adaptive evolution of Hb genes in wild birds.
Data archiving
All DNA sequences are accessible under GenBank accession numbers (MF321902–MF322504), and sample information for all individuals is found in Table S1 of the Supplementary file.
Supplementary Material
Acknowledgments
We thank Shimiao Shao for providing comments on the early draft, Yalin Chen and Dezhi Zhang for revising the latest version of this manuscript. We also thank Per Alström for valuable comments on this work. We thank Tianlong Cai for providing the elevation maps and Ruiying Zhang for providing sample information.
Funding
This work was funded by the Strategic Priority Research Programme of the Chinese Academy of Sciences (XDB13020300), State Key Programme of Natural Science Foundation of China (NSFC 31330073), a grant from the Ministry of Science and Technology of China (2014FY210200) to F.L., and a grant from the NSFC programme (J1210002) to Y.G. and X.J.Z. G.S. is supported by the NSFC fund (31471991) and a grant (Y229YX5105) from the Key Laboratory of the Zoological Systematics and Evolution of the Chinese Academy of Sciences.
Author contributions
X.Z., Y.G., and F.L. conceived the ideas; G.S. and Y.Q. collected samples; Y.G. and Z.X. performed the experiments and analyzed the data; X.Z., Y.G., G.S., G.D., and F.L. led the writing; X.Z., Y.G., G.S., Y.Q., D.G. and F.L. discussed the results and determined the conclusions. All authors agreed on the main conclusions of the article.
Supplementary material
Supplementary material can be found at http://www.cz.oxfordjournals.org/.
References
- Bandelt H-J, Forster P, Röhl A, 1999. Median–joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48. [DOI] [PubMed] [Google Scholar]
- Bulgarella M, Peters JL, Kopuchian C, Valqui T, Wilson RE. et al. , 2012. Multilocus coalescent analysis of haemoglobin differentiation between low– and high–altitude populations of crested ducks Lophonetta specularioides. Mol Ecol 21:350–368. [DOI] [PubMed] [Google Scholar]
- Cheviron ZA, Natarajan C, Projecto-Garcia J, Eddy DK, Jones J. et al. , 2014. Integrating evolutionary and functional tests of adaptive hypotheses: a case study of altitudinal differentiation in hemoglobin function in an Andean sparrow Zonotrichia capensis. Mol Biol Evol 31:2948–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Dickson M. et al. , 2005. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307:1928–1933. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL, 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. [DOI] [PubMed] [Google Scholar]
- Ferchaud AL, Hansen MM, 2016. The impact of selection, gene flow and demographic history on heterogeneous genomic divergence: three-spine sticklebacks in divergent environments. Mol Ecol 25:238–259. [DOI] [PubMed] [Google Scholar]
- Fu YX, 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH, 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galen SC, Natarajan C, Moriyama H, Weber RE, Fago A. et al. , 2015. Contribution of a mutational hot spot to hemoglobin adaptation in high-altitude Andean house wrens. Proc Natl Acad Sci USA 112: 13958–13963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill F, Donsker D, 2016. Ioc world bird list (v 6.4) [Cited 2016 May 21] Available from: http://www.worldbirdnames.org/
- Gosler A, Clement P, Christie DA, 2017. Great tit Parus major In: del Hoyo J, Elliott A, Sargatal J, Christie DA, Juana d, editors. Hand Book of the Birds of the World Alive. Barcelona: Lynx Edicions. [Google Scholar]
- Hey J, Nielsen R, 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. Persimilis. Genetics 167: 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R, 2007. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc Natl Acad Sci USA 104:2785–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Natarajan C, Moriyama H, Witt CC, Weber RE. et al. , 2017. Stability-mediated epistasis restricts accessible mutational pathways in the functional evolution of avian hemoglobin. Mol Biol Evol 34:1240–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist L, Arbabi T, Päckert M, Orell M, Martens J, 2007. Population differentiation in the marginal populations of the great tit (Paridae: Parus major). Biol J Linn Soc 90:201–210. [Google Scholar]
- Kvist L, Martens J, Higuchi H, Nazarenko AA, Valchuk OP. et al. , 2003. Evolution and genetic structure of the great tit Parus major complex. Proc R Soc Lond B Biol Sci 270:1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J, 2009. Dnasp v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- Mairbaurl H, Weber RE, 2012. Oxygen transport by hemoglobin. Compr Physiol 2:1463–1489. [DOI] [PubMed] [Google Scholar]
- McCracken KG, Barger CP, Bulgarella M, Johnson KP, Kuhner MK. et al. , 2009a. Signatures of high-altitude adaptation in the major hemoglobin of five species of Andean dabbling ducks. Am Nat 174:631–650. [DOI] [PubMed] [Google Scholar]
- McCracken KG, Bulgarella M, Johnson KP, Kuhner MK, Trucco J. et al. , 2009b. Gene flow in the face of countervailing selection: adaptation to high-altitude hypoxia in the βA hemoglobin subunit of yellow-billed pintails in the Andes. Mol Biol Evol 26: 815–827. [DOI] [PubMed] [Google Scholar]
- Monge C, Leon-Velarde F, 1991. Physiological adaptation to high altitude: oxygen transport in mammals and birds. Physiol Rev 71: 1135–1172. [DOI] [PubMed] [Google Scholar]
- Munoz-Fuentes V, Cortazar-Chinarro M, Lozano-Jaramillo M, McCracken KG, 2013. Stepwise colonization of the Andes by ruddy ducks and the evolution of novel beta-globin variants. Mol Ecol 22: 1231–1249. [DOI] [PubMed] [Google Scholar]
- Natarajan C, Projecto-Garcia J, Moriyama H, Weber RE, Munoz-Fuentes V. et al. , 2015. Convergent evolution of hemoglobin function in high-altitude Andean waterfowl involves limited parallelism at the molecular sequence level. PLoS Genet 11:e1005681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson-Manning CF, Wagner MR, Mitchell-Olds T, 2012. Adaptive evolution: Evaluating empirical support for theoretical predictions. Nat Rev Genet 13:867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Hoffmann FG, Natarajan C, Witt CC, Berenbrink M. et al. , 2015. Gene turnover in the avian globin gene families and evolutionary changes in hemoglobin isoform expression. Mol Biol Evol 32:871–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Päckert M, Martens J, Eck S, Nazarenko AA, Valchuk OP. et al. , 2005. The great tit Parus major: a misclassified ring species. Biol J Linn Soc 86:153–174. [Google Scholar]
- Perutz MF, 1983. Species adaptation in a protein molecule. Mol Biol Evol 1:1–28. [DOI] [PubMed] [Google Scholar]
- Porter AH, Wenger R, Geiger H, Scholl A, Shapiro AM, 1997. The Pontiac daplidice - edusa hybrid zone in northwestern Italy. Evolution 51:1561–1573. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P, 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Projecto-Garcia J, Natarajan C, Moriyama H, Weber RE, Fago A. et al. , 2013. Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc Natl Acad Sci U S A (51):20669–20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Tian S, Han N, Zhao H, Gao B. et al. , 2015. Genetic responses to seasonal variation in altitudinal stress: whole-genome resequencing of great tit in eastern Himalayas. Sci Rep 5:14256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeymaekers JAM, Konijnendijk N, Larmuseau MHD, Hellemans B, De Meester L. et al. , 2014. A gene with major phenotypic effects as a target for selection vs. homogenizing gene flow. Mol Ecol 23:162–181. [DOI] [PubMed] [Google Scholar]
- Schluter D, Clifford EA, Nemethy M, McKinnon JS, 2004. Parallel evolution and inheritance of quantitative traits. Am Nat 163:809–822. [DOI] [PubMed] [Google Scholar]
- Schluter D, Conte GL, 2009. Genetics and ecological speciation. Proc Natl Acad Sci USA 106 (1 Suppl): 9955–9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott GR, Milsom WK, 2006. Flying high: a theoretical analysis of the factors limiting exercise performance in birds at altitude. Respir Physiol Neurobiol 154:284–301. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P, 2001. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, 2007. Hemoglobin function and physiological adaptation to hypoxia in high-altitude mammals. J Mammal 88:8. [Google Scholar]
- Strasburg JL, Rieseberg LH, 2010. How robust are ‘isolation with migration’ analyses to violations of the IM model? A simulation study. Mol Biol Evol 27:297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietze DT, Borthakur U, 2012. Historical biogeography of tits (Aves: Paridae, Remizidae). Org Divers Evol (12):433–444. [Google Scholar]
- Weber RE, 2007. High-ltitude adaptations in vertebrate hemoglobins. Respir Physiol Neurobiol 158: 132–142. [DOI] [PubMed] [Google Scholar]
- Wilson RE, Peters JL, McCracken KG, 2012. Genetic and phenotypic divergence between low– and high–altitude populations of two recently diverged cinnamon teal subspecies. Evolution 67: 170–184. [DOI] [PubMed] [Google Scholar]
- Zhao N, Dai C, Wang W, Zhang R, Qu Y. et al. , 2012. Pleistocene climate changes shaped the divergence and demography of Asian populations of the great tit Parus major: evidence from phylogeographic analysis and ecological niche models. J Avian Biol 43: 297–310. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.