Abstract
All behaviors of an organism are rooted in sensory processing of signals from its environment, and natural selection shapes sensory adaptations to ensure successful detection of cues that maximize fitness. Sensory drive, or divergent selection for efficient signal transmission among heterogeneous environments, has been a useful hypothesis for describing sensory adaptations, but its current scope has primarily focused on visual and acoustic sensory modalities. Chemosensation, the most widespread sensory modality in animals that includes the senses of smell and taste, is characterized by rapid evolution and has been linked to sensory adaptations to new environments in numerous lineages. Yet, olfaction and gustation have been largely underappreciated in light of the sensory drive hypothesis. Here, we examine why chemosensory systems have been overlooked and discuss the potential of chemosensation to shed new insight on the sensory drive hypothesis and vice versa. We provide suggestions for developing a framework to better incorporate studies of chemosensory adaptation that have the potential to shape a more complete, coherent, and holistic interpretation of the sensory drive.
Keywords: chemical signaling, chemoreceptor, chemosensation, sensory drive, olfaction
Introduction
All animals must find food and reproduce while avoiding predators and pathogens that threaten their survival. Sensing, perceiving, and processing environmental cues are critical to these fitness-related behaviors (Dangles et al. 2009). However, environmental stimuli must be filtered in cluttered environments in which relevant cues may be drowned out by many extraneous signals (Endler 1992). Fine-tuned sensory systems are required to filter and maximize the detection of cues and signals that are important for survival and reproduction. In a heterogeneous world, signal and noise can be highly habitat-specific. Sensory systems can play a critical role in controlling the functional ecology of an organism as a response to new or changing environments, leading to local adaptation, sexual selection, and eventually speciation (Endler 1992; Endler and Basolo 1998; Boughman 2002).
The sensory drive hypothesis, first presented by Endler (1992), provides a framework to articulate how evolution influences signal production and signal detection in a dynamic world and predicts that selection favors mechanisms that facilitate communication depending on the environmental background. The coadaptation of highly specific signals and sensory systems with respect to background “noise” may even establish barriers to gene flow between populations in different environments and eventually lead to speciation (Boughman 2002; Fuller et al. 2005). Although there are numerous examples of visual and acoustic sensory systems that evolved via sensory drive (Kingston et al. 2001; Scott 2001; Fuller et al. 2005; Seehausen et al. 2008; Tobias et al. 2010; Wilkins et al. 2013; Price 2017), the effects of sensory drive on the evolution of other sensory modalities have been largely neglected in this context (Dangles et al. 2009; Cummings and Endler 2018). Chemosensation, although arguably the most commonly used sensory modality in animals (Hildebrand and Shepherd 1997; Yarmolinsky et al. 2009), has received little attention in the sensory drive literature, and we argue that this is largely due to a combination of methodological difficulties in assessing environmental chemical diversity and the complexity of chemical signals and perception mechanisms relative to acoustic and visual systems.
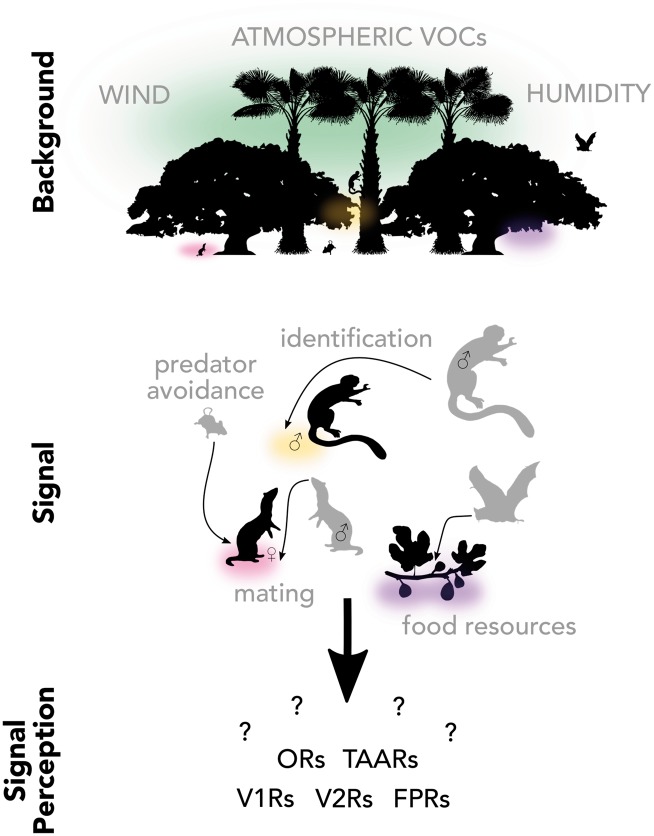
Chemosensation involves the transduction of a chemical stimulus from the environment into a neurological signal interpreted by the organism. Chemosensory systems directly interact with environmental chemical cues and regulate behaviors essential for survival and reproduction, such as finding food, avoiding predators, identifying conspecifics, caring for offspring, and attracting mates (Hart and Chao 2010; Hansson and Stensmyr 2011; Li and Liberles 2015; Meister 2015). Chemosensation is ubiquitous across the Tree of Life, from a unicellular budding yeast that initiates mating with a chemical signal (Bardwell 2004), to male orchid bees collecting environmental scents to produce their own “perfume” (Vogel 1965; Eltz et al. 1999; Roubik and Hanson 2004); from a female garter snake that chemically signals reproductive viability (Parker and Mason 2014), to felines rubbing facial pheromones to mark territory (Soini et al. 2012). This great diversity highlights the important role of chemosensation in fitness-related behaviors (Figure 1), and it is highly likely that the sensory drive hypothesis influences the evolution of the senses of smell and taste and chemical signaling in animals. Rapid rates of molecular evolution and exceptional gene turnover through duplication and loss of chemical-detecting receptor genes lay the groundwork for rapid local adaptation that may even influence sexual selection and facilitate reproductive isolation. Yet, how environmental conditions and the chemical background interfere with or facilitate signal transmission and detection are relatively unexplored. Here, we highlight the underestimated, yet pervasive role of chemosensation in sensory adaptation and how it can be tested in light of the sensory drive hypothesis.
Figure 1.
The three components of sensory drive through the perspective of olfaction and how they may relate to one another in a mammalian community. Chemical signals must be transmitted through a complex chemical background composed of a general background of volatile organic compounds (VOCs) and intra- and interspecific chemical signals, all interacting with abiotic conditions such as wind and humidity. The perceiver expresses hundreds of different genes in a number of different chemosensory receptor gene families under selection to maximize individual fitness. Sensory drive research in chemosensory systems is required to understand how changing environments influence the evolution of chemical communication. Abbreviations of mammalian chemosensory receptors: ORs: olfactory receptors; TAARs: Trace amine-associated receptors; V1Rs: vomeronasal type-1 receptors; V2Rs: vomeronasal type-2 receptors; FPRs: formyl peptide receptors.
Challenges in Bringing Chemosensation to Sensory Drive
Variation in signal production, signal transmission, and signal detection is shaped by natural selection in heterogeneous environments. The sensory drive hypothesis predicts that the local adaptation of signals and signal detection can evolve within populations of the same species in differing environments due to environmentally mediated divergent selection on signaling specialization. There are three predictions of the sensory drive hypothesis:
habitat-induced background “noise” influences efficient signal transmission and signal detection;
divergent selection on sensory perception in response to heterogeneous sensory environments leads to local adaptation of sensory systems; and
altered perception abilities and specializations select for signals that match changes in perception, which in turn may eventually lead to reproductive isolation between lineages.
The past and current focus on visual and acoustic systems has revealed convincing evidence for sensory drive as a mechanism in sensory-mediated local adaptation and also reproductive isolation (Boughman 2002; Dangles et al. 2009). Yet, chemosensory evolution has been virtually untested under the sensory drive hypothesis, despite its relevance in the functional ecology across animals. We argue that there are two main reasons for this: 1) chemical background, chemical signals, and chemosensory gene function and evolution are highly complex in comparison to vision and hearing, which makes the connection between environmental changes and sensory evolution more convoluted and 2) sophisticated methods to reliably determine chemical background, signals, and gene function are only recently emerging. We argue that with the development of methods to analyze and integrate environmental chemicals and chemical signals with the molecular evolution and function of chemosensory genes, we are at an exciting point in time where it is becoming increasingly feasible to test the extent of sensory drive in chemosensory evolution and thus better understand the role of sensory drive in sensory evolution in general.
Challenge #1: the complexity of the chemical background
A key component of sensory drive is the influence of the environment on the evolution of signals and signal detection. Chemical signals and cues must stand out against the chemical background, and the recipient must detect the cue in time before the signal is diffused (Figure 1). The chemical environment is a dynamic and complex system influenced by various abiotic and biotic factors. Besides spatial variation of the abundance and composition of biotic chemicals between (micro-) habitats (Guenther 1997; Kesselmeier and Staudt 1999), the chemical environment varies across time (Proffit et al. 2008; Riffell et al. 2008), and is significantly influenced by rapidly changing and highly interconnected abiotic conditions including temperature, humidity, and wind (Cetin et al. 2003). Diurnal convection dynamics of the atmosphere influence how chemicals move through the air, such that the movement of chemicals at sunrise will not be the same as at sunset (Riffell et al. 2008). Similarly, water turbulence has a critical impact on chemical concentrations in aquatic systems (Weissburg and Zimmer-Faust 1993; Zimmer-Faust et al. 1995). Furthermore, while influencing atmospheric movements and air pressure, temperature alters the chemical background on the basis of volatility (the tendency of a chemical to evaporate), which varies with molecular weight and functional groups.
In addition to the chemical background, abiotic factors also affect animal physiology and perception abilities. Humidity has been shown to be critical to olfaction in hermit crabs leading to low eletrophysiological responses of the antenna when humidity is low (Krång et al. 2012). Although this might present a special case due to a recent shift from a marine to terrestrial lifestyle, it demonstrates how the chemical environment not only changes but also is perceived differently with respect to changes in physical abiotic factors.
This complexity renders the characterization and quantification of the chemical background and the transmission efficiency of chemical signals through the background of a nontrivial task (Riffel et al. 2008). The demonstration of empirical evidence of sensory drive and the effects of the chemical background on signal transmission will require the determination of multiple complex environmental factors that are highly specific to the organism, locality, and time. Although the qualification and quantification of light and sound are highly standardized through measurements of well-known physical properties (i.e., waves, wavelengths, and amplitudes), there is presently no unbiased way of capturing the entire breadth of chemicals from the environment, presenting a significant challenge for testing the effects of sensory drive on chemosensory evolution. The detection of chemicals with current scent-trapping methods is highly dependent on the type of trap, adsorbent, and solvent used, not only complicating replication, but also biasing the outcome of the analysis (Agelopoulos and Pickett 1998). Nevertheless, these methods have been successfully implemented to study chemical ecology in a plethora of organisms (Raguso et al. 2015), and recently they have been modified for the determination of the chemical environment. Methods to capture the breadth of the volatile and nonvolatile chemical environment are emerging and becoming slowly available and applicable to macrohabitats (Barreira et al. 2017; Hellén et al. 2017). It is now possible to study the temporal changes in atmospheric volatile organic compounds across large spatial scales of the rainforest (Jokinen et al. 2015; Yáñez-Serrano et al. 2015; Alves et al. 2016) or to characterize the “volatilome” of an animal (Amann et al. 2014; Angle et al. 2016), paving the way for sensory drive research.
The relevance of the sensory drive hypothesis has become increasingly important as discoveries show that chemical cues can indeed be drowned out by environmental changes and hinder the detection of relevant odorants (Atema 1995; Riffell et al. 2008, 2014). For example, the male tobacco budworm Heliothis virescens must identify females located on host plants from long distances, but some host plant volatiles interfere with the efficient detection of pheromones emitted by females (Pregitzer et al. 2012). Conversely, the silkmoth Bombyx mori demonstrates a synergistic response with host plant cues, in which pheromone detection is enhanced by the presence of specific plant volatiles (Namiki et al. 2008). Human-produced pollutants can mask floral scents, drastically reducing signal transmission and impacting navigation in foraging moths (Riffell et al. 2014). It is becoming more obvious that the chemical background has a strong effect on signal transmission, and this is not only restricted to terrestrial habitats. In marine systems, pesticide-derived chemical pollutants disrupt olfactory signals used in navigation by salmon Oncorhynchus mykiss in British Columbia depending on concentration and pollutant mixtures (Tierney et al. 2008). Lobsters Homarus americanus generate their own currents of urine signal delivery for precise dispersal of chemical cues, creating odorant “patches” that can be more readily sampled by conspecifics in a turbid marine environment (Atema 1995).
Identification and quantification of environmental chemicals and their dynamics with respect to atmospheric and aquatic conditions over different temporal and spatial scales are essential to test the role of sensory drive in chemosensory evolution. Although quantification of the chemical background is nontrivial, substantial technological advances have been made to facilitate our understanding of chemical signaling in nature.
Challenge #2: the complexity of chemical signals
Visual and acoustic sensory systems process signals that can be readily quantified through well-understood inherent physical properties (e.g., electromagnetic waves and air pressure), and it is often straightforward to predict and test how these signals change in time and space. Chemosensory systems, in contrast, process signals composed of chemical molecules from a multidimensional chemical space that is virtually infinitive. Chemosensory signals can range from a single compound to complex mixtures of different compounds with diverse functional groups and varying concentrations. Pheromones, chemical signals emitted by many animal species to mediate intraspecific behaviors (Karlson and Lüscher 1959; Liberles 2014; Stowers and Kuo 2015), are often composed of dozens of compounds that may vary between individuals (Wyatt 2003). Determining which compounds are relevant with respect to sexual communication versus territoriality versus parental care versus metabolic byproduct (i.e., noise) is not trivial and often impossible. The isolation of behaviorally active compounds or compound mixtures in signals can be a tedious endeavor, due to often-unknown chemical structures of naturally occurring chemicals new to science. Although the manipulation of complex signals in visual and acoustic communication systems is relatively straightforward, the manipulation of chemical blends depends on the ability to produce or isolate the involved single compounds, which is often impossible and thus may be unfeasible. However, manipulations of signal mixtures are necessary to disentangle signal function in behavioral experiments, which may take considerable effort to conduct. For example, the functional characterization of the behavioral agents of the queen pheromone bouquet in honey bees Apis mellifera required decades of focused investigation, highlighting the difficulty in identifying the function of single components of signal mixtures. Early chemical analysis of the mandibular pheromone excreted by the queen revealed 9-oxo-(E)-2-deconoic acid as the dominant compound (Butler and Fairey 1963; Slessor et al. 2005). However, 9-oxo-(E)-2-deconoic acid alone is neither attractive to drones (male honey bees) nor does it lead to a response from worker bees. Behavioral experiments showed that for a response in workers to occur, four additional compounds are required (Slessor et al. 1988). However, it became evident that these results were not applicable to all honey bees. Although the five-ingredient mixture elicits behavioral responses in some subspecies, multiple additional subspecies-specific compounds were required to consistently elicit attraction in workers of other subspecies (Keeling et al. 2003; Slessor et al. 2005). This example further highlights how signal mixtures may vary between populations of the same species, which might be a result of local adaptation due to sensory drive. Although rarely studied, it is becoming more apparent that signals used in the same behavioral context can vary geographically within species (Ramírez et al. 2010; Pokorny et al. 2013; Duménil et al. 2014; Groot et al. 2014). In order to disentangle the evolutionary mechanisms driving this interpopulation variation in chemical signaling, future investigations must explicitly test how the environment contributes to the evolution of variation in signaling and signal perception.
Challenge #3: the complexity of the genetic basis of chemosensation
Once the chemical cue is emitted and transmitted through time and space, it must be detected and processed by the perceiver. In vertebrates and invertebrates alike, the detection of volatile and nonvolatile chemicals is based on the interaction of chemicals with proteins encoded by genes of large multi-copy chemosensory gene families expressed in sensory neurons and supporting cells of the olfactory and gustatory systems (see Kaupp 2010 for review). Although the number of genes underlying vision and acoustics is comparatively small and stable among lineages (Parker et al. 2013; Ramirez et al. 2016), the number of genetic loci involved in chemoreception can vary by orders of magnitude (Niimura 2012). Throughout animal evolution, the number of opsin paralog copies, for example, is highly conserved and rarely exceeds 10 per gene family (Ramirez et al. 2016). In contrast, tens to thousands of chemosensory genes of several fast evolving multigene families are involved in chemosensation (Nei et al. 2008; de Bruyne et al. 2010; Ota et al. 2012; Brykczynska et al. 2013; Niimura et al. 2014; Yoder et al. 2014; Derby et al. 2016). The origin of gene families involved in chemical detection occurred at different points in phylogenetic history independently in vertebrates and invertebrates (Eisthen 1992; Strausfeld and Hildebrand 1999; Grus and Zhang 2006, 2009; Sánchez-Gracia et al. 2009; Eyun et al. 2017; Brand et al. 2018). This includes several mostly G-protein-coupled receptor gene families in vertebrates (Grus and Zhang 2006; Niimura 2009, 2012) and non-G-protein-coupled receptor gene families in invertebrates (Sato et al. 2008; Cummins and Degnan 2010; Derby et al. 2016). New chemoreceptor gene families are still being discovered (Benton et al. 2009; Greer et al. 2016), emphasizing how much more there is still to learn about the molecular basis of chemosensation.
The widespread chemosensory receptor diversity is a result of rapid evolutionary rates through high gene turnover and rapid sequence diversification of homologous genes (Nei et al. 2008; Sánchez-Gracia et al. 2009; Bear et al. 2016; Brand and Ramírez 2017). The convergently evolved odorant receptor gene families in vertebrates and insects (Sato et al. 2008; Dehara et al. 2012; Niimura et al. 2014), for example, demonstrate some of the most extraordinary patterns of gene duplication and pseudogenization (i.e., gene turnover) in animals, constantly expanding and contracting over time (Nei et al. 2008). This birth–death evolution and the subsequent diversification are responsible for odorant receptors accounting for the largest gene families in animals encoding for up to 5% of the protein-coding genome in mammals, for example (Hayden et al. 2010; Niimura 2012; Niimura et al. 2014).
Although the evolutionary dynamics of chemosensory gene families in animals are well described based on an ever-increasing amount of genomic data for a diverse array of taxa (Guo and Kim 2007; Young et al. 2010; Brykczynska et al. 2013; Niimura et al. 2014, 2018; Picone et al. 2014; Yoder et al. 2014; Brand et al. 2015; Derby et al. 2016; Brand and Ramírez, 2017; Yohe et al. 2018), linking molecular patterns of sequence evolution to gene function is still a nontrivial task. One contributing factor is that the identification of ligands which individual chemosensory receptors respond to was initially limited to model organisms and experiments focused mainly on the mechanistic understanding of chemosensation (Dobritsa et al. 2003; Hallem and Carlson 2006; Touhara 2007; Wang et al. 2010; Launay et al. 2012; de Fouchier et al. 2017; Pask et al. 2017; Slone et al. 2017; Fleischer et al. 2018). Early experiments functionally characterizing the entire chemosensory receptor repertoires in mice and vinegar flies revealed that the encoding of the senses of smell and taste is more complex than the number of genes initially indicated (Malnic et al. 1999; Dobritsa et al. 2003; Hallem and Carlson 2006). The detection of a single chemical can either be dependent on a single highly specialized receptor or multiple receptors acting in a combinatorial fashion (Malnic et al. 1999; Hallem and Carlson 2006; Malnic 2007; Ullah et al. 2015). Thus, the number of compounds detected by an animal is likely vastly exceeding the number of receptor genes in the genome (Malnic et al. 1999; Hallem and Carlson 2006; Nara et al. 2011; Magklara and Lomvardas 2013; Rodriguez 2013; McClintock et al. 2014; Bushdid et al. 2016; Haverkamp et al. 2018), and the loss or gain of receptor genes might have severe effects on the sensory ecology of an organism. Indeed, the high gene turnover of chemosensory gene families among animal lineages has been linked to changes in sensory abilities including the adaptation of novel food resources (McBride 2007; McBride and Arguello 2007; Hayden et al. 2014; Goldman-Huertas et al. 2015) or specializations in the pheromone communication system (Gould et al. 2010; Ferrero et al. 2011).
With neurophysiological methods being constantly refined and adapted for use in nonmodel species (de Fouchier et al. 2017; Pask et al. 2017; Slone et al. 2017), the field is moving toward a better understanding of how selection influences chemosensory evolution on a functional level. For example, it has become evident that even single mutations in olfactory receptors can lead to adaptive shifts in olfactory tuning (Pellegrino et al. 2011; Leary et al. 2012; McBride et al. 2014). Furthermore, chemosensory genes can be highly diverse within populations of the same species, leading to variable sensitivity to chemical stimuli (Rollmann et al. 2010; Logan 2014; Mainland et al. 2014), and thus representing variation for natural selection to act on when environments and/or signals change. With an increasing understanding of the molecular dynamics and a methodological toolkit becoming available for broad application, it will be possible to understand how chemosensation is evolving with response to local environments.
Future Opportunities in Bringing Chemosensation to Sensory Drive
Through a combination of genomic, biochemical, and neurophysiological advances, the integrated study of chemosensory evolution is now becoming tractable, and testing the role of sensory drive in chemosensory system evolution represents a new frontier in evolutionary biology. We are only beginning to understand the extent of variability in the chemical environment, chemical signals, and chemosensory genes and how their interplay is shaping the evolution of chemical communication. Although challenging, we believe that studying the role of sensory drive in chemosensory systems is possible with the careful choice of study systems in combination with well-designed experiments.
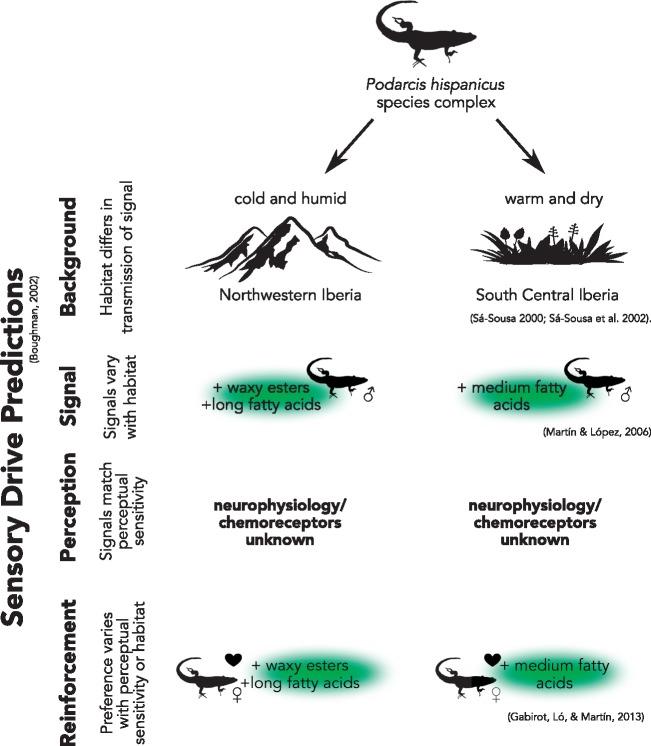
As we outlined above, there is an increasing number of examples of chemosensory-based local adaptation, chemical signal divergence, and molecular signatures of selection on chemosensory receptors. However, the majority of these studies focus on host-shift adaptation or the evolution of pheromone communication and its role in speciation, including analyses of intraspecific signaling variation among geographically (and environmentally) distinct populations (see Smadja and Butlin 2009 for summary). Although most systems partially align with the predictions of sensory drive, it is usually not explicitly tested for in the system. One such example is the Iberian wall lizard Podarcis hispanicus, a species complex that includes two populations that inhabit neighboring but differing environments (Figure 2), in which one population inhabits cold and humid environments of the highlands in northern Iberia and the other population inhabits a warmer and dryer Mediterranean climate of central and southern Iberia (Sá-Sousa 2000; Sá-Sousa et al. 2002). These divergent environmental conditions have led to differences in the chemical composition of femoral gland excretions of male lizards used for marking territory, conspecific identification, and female choice (Martín and López 2006). Males in colder, wetter climates have less volatile femoral gland excretions compared with those in drier climates, likely an environmentally driven adaptation for efficient signal deposition and signal stability (Martín and López 2006; Martín et al. 2015). Males of each respective population elicit a more aggressive response toward signals from conspecific in their own habitat (Martín and López 2006; Gabirot et al. 2012), and similarly females show preferences for signals specific to their habitat type (Gabirot et al. 2013; Martín et al. 2015). These preferences reinforce the boundaries likely initially established by more efficient signaling based on their habitat type, leading to reproductive isolation among the populations and potential cryptic speciation (Gabirot et al. 2012; Martín and López 2015; Martín et al. 2015). Accordingly, this system corroborates many of the criteria outlined by Boughman (2002) for speciation via sensory drive (Figure 2), and we encourage follow-up studies on neurophysiology and chemoreceptor evolution to test for selection driving divergent chemical perception mechanisms between habitats and to exclude genetic drift.
Figure 2.
The Iberian wall lizard Podarcis hispanicus illustrates a strong candidate for sensory drive promoting chemosensory divergence and local adaptation. Compounds of the male femoral gland excretions differ based on the environment, in which northern populations have waxier and bulkier compounds that are less volatile and enable more viable signals in the given habitat. Receptors of the perceivers are unknown, but behavioral evidence has demonstrated female preference and male–male recognition of signals based on their own environments. Silhouettes are from vecteezy and all-free-download.com.
Similar to the Iberian wall lizard, many known cases of intraspecific pheromone evolution show patterns that meet the predictions of the sensory drive theory, emphasizing the disparity between the study of chemosensory adaptations and sensory drive literature. Environmental changes are likely often a part of interpopulation divergence in chemosensory systems from latitudinal gradients (Lavagnino et al. 2008) to sympatric host shifts (Olsson et al. 2006; Tait et al. 2016) and thus have the potential to be affected by sensory drive.
Concluding Remarks
Over the past 25 years, a focus on visual and acoustic systems has generated convincing evidence for the role of sensory drive as a mechanism in sensory-mediated local adaptation and potential reproductive isolation. However, the importance of sensory drive as a hypothesis that explains sensory adaptations based on environmental differences, in general, remains uncertain until the hypothesis is rigorously tested in all sensory modalities, including chemosensation. The traditional lack of studies on sensory drive in chemosensory evolution likely results from the complexity of chemosensory signals and their detection alongside the only recently emerging technology needed for the combined analysis of both phenotypic and molecular evolution of chemosensory communication. We find that precisely these technological advances have led to a deeper understanding of chemosensation and the underlying molecular basis in the recent past, paving the way for future research on chemosensory evolution with respect to the variability of natural habitats. Ultimately, we will need to link chemosensory evolution of signaling and perception to the complex chemical background of the environment to truly demonstrate how sensory drive is a critical mechanism of chemosensory evolution.
Acknowledgments
We would like to thank Becky Fuller and 2 anonymous reviewers for helpful comments on this article. We would also like to acknowledge Becky Fuller for inviting us to be a part of this special edition on “25 years of Sensory Drive,” as well as all symposia organizers and participants who celebrated the anniversary at the 2017 Evolution Meetings in Portland, Oregon, as this was a key source of inspiration.
Funding
L.R.Y. was funded in part by National Science Foundation (NSF) DEB-1701414 and NSF DEB-1442142.
References
- Agelopoulos NG, Pickett JA, 1998. Headspace analysis in chemical ecology: effects of different sampling methods on ratios of volatile compounds present in headspace samples. J Chem Ecol 24: 1161–1172. [Google Scholar]
- Alves EG, Jardine K, Tota J. Jardin A, Yãnez-Serrano AMet al. , 2016. Seasonality of isoprenoid emissions from a primary rainforest in central Amazonia. Atmos Chem Phys 16:3903–3925. [Google Scholar]
- Amann A, de Lacy Costello B, Miekisch W, Schubert J, Buszewski B. et al. , 2014. The human volatilome: volatile organic compounds, VOCs. in exhaled breath, skin emanations, urine, feces and saliva. J Breath Res 8. [DOI] [PubMed] [Google Scholar]
- Angle C, Waggoner LP, Ferrando A, Haney P, Passler T, 2016. Canine detection of the volatilome: a review of implications for pathogen and disease detection. Front Vet Sci 3:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atema J, 1995. Chemical signals in the marine environment: dispersal, detection, and temporal signal analysis. Proc Natl Acad Sci USA 92:62–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell L, 2004. A walk-through of the yeast mating pheromone response pathway. Peptides 25:1465–1476. [DOI] [PubMed] [Google Scholar]
- Barreira LMF, Duporté G, Parshintsev J. et al. , 2017. Emissions of biogenic volatile organic compounds from the boreal forest floor and understory: a study by solid-phase microextraction and portable gas chromatography-mass spectrometry. Boreal Environ Res 22:393–413. [Google Scholar]
- Bear DM, Lassance J-M, Hoekstra HE, Datta SR, 2016. The evolving neural and genetic architecture of vertebrate olfaction. Curr Biol 26:R1039–R1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB, 2009. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136:149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughman JW, 2002. How sensory drive can promote speciation. Trends Ecol Evol 17:571–577. [Google Scholar]
- Brand P, Ramírez SR, 2017. The evolutionary dynamics of the odorant receptor gene family in corbiculate bees. Genome Biol Evol 9:2023–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand P, Ramírez SR, Leese F, Quezada-Euan JJG, Tollrian R. et al. , 2015. Rapid evolution of chemosensory receptor genes in a pair of sibling species of orchid bees (Apidae: Euglossini). BMC Evol Biol 15:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand P, Robertson HM, Lin W. et al. , 2018. The origin of the odorant receptor gene family in insects. bioRxiv 10.1101/259424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruyne M, de Smart R, Zammit E, Warr CG, 2010. Functional and molecular evolution of olfactory neurons and receptors for aliphatic esters across the Drosophila genus. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 196:97–109. [DOI] [PubMed] [Google Scholar]
- Brykczynska U, Tzika AC, Rodriguez I, Milinkovitch MC, 2013. Contrasted evolution of the vomeronasal receptor repertoires in mammals and squamate reptiles. Genome Biol Evol 5:389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushdid C, Magnasco MO, Vosshall LB, Keller A, Mixture M, 2016. Humans can discriminate more than 1 trillion olfactory stimuli. Science 343:1370–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler CG, Fairey EM, 1963. The role of the queen in preventing oogenesis in worker honeybees. J Apic Res 2:14–18. [Google Scholar]
- Cetin E, Odabasi M, Seyfioglu R, 2003. Ambient volatile organic compound, VOC. concentrations around a petrochemical complex and a petroleum refinery. Sci Total Environ 312:103–112. [DOI] [PubMed] [Google Scholar]
- Cummins SF, Degnan BM, 2010. Sensory sea slugs: towards decoding the molecular toolkit required for a mollusk to smell. Commun Integr Biol 3:423–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings ME, Endler JA, 2018. 25 years of sensory drive: the evidence and its watery bias. Curr Zool 64:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangles O, Irschick DJ, Chittka L, Casas J, 2009. Variability in sensory ecology: expanding the bridge between physiology and evolutionary biology. Q Rev Biol 84:51–74. [DOI] [PubMed] [Google Scholar]
- Dehara Y, Hashiguchi Y, Matsubara K, Yanai T, Kubo M. et al. , 2012. Characterization of squamate olfactory receptor genes and their transcripts by the high-–throughput sequencing approach. Genome Biol Evol 4:602–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derby CD, Kozma MT, Senatore A, Schmidt M, 2016. Molecular mechanisms of reception and perireception in crustacean chemoreception: a comparative review. Chem Senses 41:381–398. [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Naters W, van der G, van Warr CG, Steinbrecht RA, et al. , 2003. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37:827–841. [DOI] [PubMed] [Google Scholar]
- Duménil C, Judd GJR, Bosch D, Baldessari M, Gemeno C. et al. , 2014. Intraspecific variation in female sex pheromone of the codling moth Cydia pomonella. Insects 5:705–721., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisthen HL, 1992. Phylogeny of the vomeronasal system and of receptor cell types in the olfactory and vomeronasal epithelia of vertebrates. Microsc Res Tech 23:1–21. [DOI] [PubMed] [Google Scholar]
- Eltz T, Whitten WM, Roubik DW, Linsenmair KE, 1999. Fragrance collection, storage, and accumulation by individual male orchid bees. J Chem Ecol 25:157–176. [Google Scholar]
- Endler JA, 1992. Signals, signal conditions, and the direction of evolution. Am Nat 139:S125–S153. [Google Scholar]
- Endler JA, Basolo AL, 1998. Sensory ecology, receiver biases and sexual selection. Trends Ecol Evol 13:415–420. [DOI] [PubMed] [Google Scholar]
- Eyun S, Young Soh H, Posavi M, Munro JB, Hughes DST. et al. , 2017. Evolutionary history of chemosensory-related gene families across the arthropoda. Mol Biol Evol 34:1838–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero DM, Lemon JK, Fluegge D, Pashkovski SL, Korzan WJ. et al. , 2011. Detection and avoidance of a carnivore odor by prey. Proc Natl Acad Sci USA 108:11235–11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer J, Pregitzer P, Breer H, Krieger J, 2018. Access to the odor world: olfactory receptors and their role for signal transduction in insects. Cell Mol Life Sci 75:485–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier A, de Walker WB III, Montagné N. et al. , 2017. Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat Commun 8:15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller RC, Houle D, Travis J, 2005. Sensory bias as an explanation for the evolution of mate preferences. Am Nat 166:437–446. [DOI] [PubMed] [Google Scholar]
- Gabirot M, López P, Martín J, 2012. Differences in chemical sexual signals may promote reproductive isolation and cryptic speciation between Iberian wall lizard populations. Int J Evol Biol 2012:1–13. 10.1155/2012/698520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabirot M, López P, Martín J, 2013. Female mate choice based on pheromone content may inhibit reproductive isolation between distinct populations of Iberian wall lizards. Curr Zool 59:210–220. [Google Scholar]
- Goldman–Huertas B, Mitchell RF, Lapoint RT, Faucher CP, Hildebrand JG. et al. , 2015. Evolution of herbivory in Drosophilidae linked to loss of behaviors, antennal responses, odorant receptors, and ancestral diet. Proc Natl Acad Sci USA 112:3026–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould F, Estock M, Hillier NK, Powell B, Groot AT. et al. , 2010. Sexual isolation of male moths explained by a single pheromone response QTL containing four receptor genes. Proc Natl Acad Sci USA 107:8660–8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer PL, Bear DM, Lassance J-M, Bloom ML, Tsukahara T. et al. , 2016. A family of non-GPCR chemosensors defines an alternative logic for mammalian olfaction. Cell 165:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot AT, Schöfl G, Inglis O, Donnerhacke S, Classen A. et al. , 2014. Within-population variability in a moth sex pheromone blend: genetic basis and behavioural consequences. Proc R Soc B Biol Sci 281:20133054–20133054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grus WE, Zhang J, 2006. Origin and evolution of the vertebrate vomeronasal system viewed through system-specific genes. Bioessays 28:709–718. [DOI] [PubMed] [Google Scholar]
- Grus WE, Zhang J, 2009. Origin of the genetic components of the vomeronasal system in the common ancestor of all extant vertebrates. Mol Biol Evol 26:407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther A, 1997. Seasonal and spatial variations in natural volatile organic compound emissions. Ecol Appl 7:34–45. [Google Scholar]
- Guo S, Kim J, 2007. Molecular evolution of Drosophila odorant receptor genes. Mol Biol Evol 24:1198–1207. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR, 2006. Coding of odors by a receptor repertoire. Cell 125:143–160. [DOI] [PubMed] [Google Scholar]
- Hansson BS, Stensmyr MC, 2011. Evolution of insect olfaction. Neuron 72:698–711. [DOI] [PubMed] [Google Scholar]
- Hart AC, Chao MY, 2010. From odors to behaviors in Caenorhabditis elegans In: Menini A, editor. The Neurobiology of Olfaction. Baco Raton: CRC Press; 1–35. [PubMed] [Google Scholar]
- Haverkamp A, Hansson BS, Knaden M, 2018. Combinatorial codes and labeled lines: how insects use olfactory cues to find and judge food, mates, and oviposition sites in complex environments. Front Physiol 9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden S, Bekaert M, Crider TA, Mariani S, Murphy WJ. et al. , 2010. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res 20:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden S, Bekaert M, Goodbla A, Murphy WJ, Dávalos LM. et al. , 2014. A cluster of olfactory receptor genes linked to frugivory in bats. Mol Biol Evol 31:917–927. [DOI] [PubMed] [Google Scholar]
- Hellén H, Schallhart S, Praplan AP, Petäjä T, Hakola H, 2017. Using in situ GC-MS for analysis of C2-C7 volatile organic acids in ambient air of a boreal forest site. Atmos Meas Tech 10:281–289. [Google Scholar]
- Hildebrand JG, Shepherd GM, 1997. Mechanisms of olfactory discrimination: converging evidence for common principles across phyla. Annu Rev Neurosci 20:595–631. [DOI] [PubMed] [Google Scholar]
- Jokinen T, Berndt T, Makkonen R, Kerminen V-M, Junninen H. et al. , 2015. Production of extremely low volatile organic compounds from biogenic emissions: measured yields and atmospheric implications. Proc Natl Acad Sci USA 112:7123–7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlson P, Lüscher M, 1959. ‘ Pheromones’: a new term for a class of biologically active substances. Nature 183:55–56. [DOI] [PubMed] [Google Scholar]
- Kaupp UB, 2010. Olfactory signalling in vertebrates and insects: differences and commonalities. Nat Rev Neurosci 11:188–200. [DOI] [PubMed] [Google Scholar]
- Keeling CI, Slessor KN, Higo HA, Winston ML, 2003. New components of the honey bee (Apis mellifera L.) queen retinue pheromone. Proc Natl Acad Sci USA 100:4486–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesselmeier J, Staudt M, 1999. Biogenic volatile organic compound, VOC): an overview on emissions, physiology and ecology. J Atmos Chem 33:23–88. [Google Scholar]
- Kingston T, Lara MC, Jones G, Akbar Z, Kunz TH. et al. , 2001. Acoustic divergence in two cryptic Hipposideros species: a role for social selection? Proc R Soc B Biol Sci 268:1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krång A-S, et al. , 2012. Transition from sea to land: olfactory function and constraints in the terrestrial hermit crab Coenobita clypeatus Proc R. Soc B 279:3510–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launay G, Wade F, Pajot-Augy E, 2012. Automatic modeling of mammalian olfactory receptors and docking of odorants. Protein Eng Des Sel 25:377–386. [DOI] [PubMed] [Google Scholar]
- Lavagnino NJ, Anholt RRH, Fanara JJ, 2008. Variation in genetic architecture of olfactory behaviour among wild-derived populations of Drosophila melanogaster. J Evol Biol 21:988–996. [DOI] [PubMed] [Google Scholar]
- Leary GP, Allen JE, Bunger PL, Luginbill JB, Linn CE. et al. , 2012. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc Natl Acad Sci USA 109:14081–14086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Liberles SD, 2015. Aversion and attraction through olfaction. Curr Biol 25:R120–R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles SD, 2014. Mammalian pheromones. Annu Rev Physiol 76:151–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DW, 2014. Do you smell what I smell? Genetic variation in olfactory perception. Biochem Soc Trans 42:861–865. [DOI] [PubMed] [Google Scholar]
- Magklara A, Lomvardas S, 2013. Stochastic gene expression in mammals: lessons from olfaction. Trends Cell Biol 23:449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainland JD, Keller A, Li YR, Zhou T, Trimmer C. et al. , 2014. The missense of smell: functional variability in the human odorant receptor repertoire. Nat Neurosci 17:114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B, 2007. Searching for the ligands of odorant receptors. Mol Neurobiol 35:175–181. [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB, 1999. Combinatorial receptor codes for odors. Cell 96:713–723. [DOI] [PubMed] [Google Scholar]
- Martín J, López P, 2006. Interpopulational differences in chemical composition and chemosensory recognition of femoral gland secretions of male lizards Podarcis hispanica: implications for sexual isolation in a species complex. Chemoecology 16:31–38. [Google Scholar]
- Martín J, López P, 2015. Condition-dependent chemosignals in reproductive behavior of lizards. Horm Behav 68:14–24. [DOI] [PubMed] [Google Scholar]
- Martín J, Ortega J, López P, 2015. Interpopulational variations in sexual chemical signals of Iberian wall lizards may allow maximizing signal efficiency under different climatic conditions. PLoS One 10:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride CS, 2007. Rapid evolution of smell and taste receptor genes during host specialization in Drosophila sechellia. Proc Natl Acad Sci USA 104:4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride CS, Arguello JR, 2007. Five Drosophila genomes reveal nonneutral evolution and the signature of host specialization in the chemoreceptor superfamily. Genetics 177:1395–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride CS, Baier F, Omondi AB, Spitzer SA, Jutomia J. et al. , 2014. Evolution of mosquito preference for humans linked to an odorant receptor. Nature 515:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock TS, Adipietro K, Titlow WB, Breheny P, Walz A. et al. , 2014. In vivo identification of eugenol-responsive and muscone-responsive mouse odorant receptors. J Neurosci 34:15669–15678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister M, 2015. On the dimensionality of odor space. Elife 4:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namiki S, Iwabuchi S, Kanzaki R, 2008. Representation of a mixture of pheromone and host plant odor by antennal lobe projection neurons of the silkmoth Bombyx mori. J Comp Physiol A Neuroethol Sensory, Neural, Behav Physiol 194:501–515. [DOI] [PubMed] [Google Scholar]
- Nara K, Saraiva LR, Ye X, Buck LB, 2011. A large-scale analysis of odor coding in the olfactory epithelium. J Neurosci 31:9179–9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M, 2008. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet 9:951–963. [DOI] [PubMed] [Google Scholar]
- Niimura Y, 2009. Evolutionary dynamics of olfactory receptor genes in chordates: interaction between environments and genomic contents. Hum Genomics 4:107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, 2012. Olfactory receptor multigene family in vertebrates: from the viewpoint of evolutionary genomics. Curr Genomics 13:103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Matsui A, Touhara K, 2014. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res 24:1485–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Matsui A, Touhara K, 2018. Acceleration of olfactory receptor gene loss in primate evolution: possible link to anatomical change in sensory systems and dietary transition. Mol Biol Evol 35:1437–1450. [DOI] [PubMed] [Google Scholar]
- Olsson SB, Linn CE, Roelofs WL, 2006. The chemosensory basis for behavioral divergence involved in sympatric host shifts. I. Characterizing olfactory receptor neuron classes responding to key host volatiles. J Comp Physiol A Neuroethol Sensory, Neural, Behav Physiol 192:279–288. [DOI] [PubMed] [Google Scholar]
- Ota T, Nikaido M, Suzuki H, Hagino-Yamagishi K, Okada N, 2012. Characterization of V1R receptor, ora. genes in Lake Victoria cichlids. Gene 499:273–279. [DOI] [PubMed] [Google Scholar]
- Parker MP, Mason RT, 2014. A novel mechanism regulating a sexual signal: the testosterone-based inhibition of female sex pheromone expression in garter snakes. Horm Behav 66:509–516. [DOI] [PubMed] [Google Scholar]
- Parker J, Tsagkogeorga G, Cotton JA. et al. , 2013. Genome-wide signatures of convergent evolution in echolocating mammals. Nature 502:228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pask GM, Slone JD, Millar JG. et al. , 2017. Specialized odorant receptors in social insects that detect cuticular hydrocarbon cues and candidate pheromones. Nat Commun 8:297.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino M, Steinbach N, Stensmyr MC, Hansson BS, Vosshall LB, 2011. A natural polymorphism alters odour and DEET sensitivity in an insect odorant receptor. Nature 478:511–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picone B, Hesse U, Panji S, Heusden P, Van Jonas M. et al. , 2014. Taste and odorant receptors of the coelacanth: a gene repertoire in transition. J Exp Zool Part B Mol Dev Evol 322:403–414. [DOI] [PubMed] [Google Scholar]
- Pokorny T, Hannibal M, Quezada-Euan JJG, Hedenström E, Sjöberg N. et al. , 2013. Acquisition of species-specific perfume blends: influence of habitat-dependent compound availability on odour choices of male orchid bees (Euglossa spp.). Oecologia 172:417–425. [DOI] [PubMed] [Google Scholar]
- Pregitzer P, Schubert M, Breer H, Hansson BS, Sachse S. et al. , 2012. Plant odorants interfere with detection of sex pheromone signals by male Heliothis virescens. Front Cell Neurosci 6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price TD, 2017. Sensory drive, color, and color vision. Am Nat 190:157–170. [DOI] [PubMed] [Google Scholar]
- Proffit M, Schatz B, Bessiere J, Chen C, Soler C. et al. , 2008. Signalling receptivity: comparison of the emission of volatile compounds by figs of Ficus hispida before, during and after the phase of receptivity to pollinators. Symbiosis 45:15–24. [Google Scholar]
- Raguso RA, Agrawal AA, Douglas AE, Jander G, Kessler A. et al. , 2015. The raison d’être of chemical ecology. Ecology 96:617–630. [DOI] [PubMed] [Google Scholar]
- Ramírez SR, Eltz T, Fritzsch F, Pemberton R, Pringle EG. et al. , 2010. Intraspecific geographic variation of fragrances acquired by orchid bees in native and introduced populations. J Chem Ecol 36:873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez MD, Pairett AN, Pankey MS, Serb JM, Speiser DI. et al. , 2016. The last common ancestor of most bilaterian animals possessed at least nine opsins. Genome Biol Evol 8:3640–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riffell JA, Abrell L, Hildebrand JG, 2008. Physical processes and real-time chemical measurement of the insect olfactory environment. J Chem Ecol 34:837–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riffell JA, Shlizerman E, Sanders E, Abrell L, Medina B. et al. , 2014. Flower discrimination by pollinators in a dynamic chemical environment. Science 344:1515–1518. [DOI] [PubMed] [Google Scholar]
- Rodriguez I, 2013. Singular expression of olfactory receptor genes. Cell 155:274–277. [DOI] [PubMed] [Google Scholar]
- Rollmann SM, Wang P, Date P, West SA, Mackay TFC. et al. , 2010. Odorant receptor polymorphisms and natural variation in olfactory behavior in Drosophila melanogaster. Genetics 186:687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubik DW, Hanson PE, 2004. Orchid Bees of Tropical America: Biology and Field Guide. Heredia, Costa Rica: Inst Nac Biodivers, INBio. [Google Scholar]
- Sá-Sousa P, 2000. A predictive distribution model for the Iberian wall lizard, Podarcis hispanicus ( in Portugal). Herpetol J 10:1–11. [Google Scholar]
- Sá-Sousa P, Vicente L, Crespo EG, 2002. Morphological variability of Podarcis hispanica (Sauria: lacertidae) in Portugal. Amphib Reptil 23:55–69. [Google Scholar]
- Sánchez-Gracia A, Vieira FG, Rozas J, 2009. Molecular evolution of the major chemosensory gene families in insects. Heredity 103:208–216. [DOI] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB. et al. , 2008. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452:1002–1006. [DOI] [PubMed] [Google Scholar]
- Scott RJ, 2001. Sensory drive and nuptial colour loss in the three-spined stickleback. J Fish Biol 59:1520–1528. [Google Scholar]
- Seehausen O, Yohey T, Magalhaes IS, Carleton KL, Mrosso HDJ. et al. , 2008. Speciation through sensory drive in cichlid fish. Nature 455:620–627. [DOI] [PubMed] [Google Scholar]
- Slessor KN, Kaminski L-A, King GGS, Borden JH, Winston ML, 1988. Semiochemical basis of the retinue response to queen honey bees. Nature 332:354–356. [Google Scholar]
- Slessor KN, Winston ML, Le Conte Y, 2005. Pheromone communication in the honeybee (Apis mellifera L.). J Chem Ecol 31:2731–2745. [DOI] [PubMed] [Google Scholar]
- Slone JD, Pask GM, Ferguson ST, Millar JG, Berger SL. et al. , 2017. Functional characterization of odorant receptors in the ponerine ant Harpegnathos saltator Proc Natl Acad Sci USA114:8586–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smadja C, Butlin RK, 2009. On the scent of speciation: the chemosensory system and its role in premating isolation. Heredity 102:77–97. [DOI] [PubMed] [Google Scholar]
- Soini HA, Linville SU, Wiesler D, Posto AL, Williams DR. et al. , 2012. Investigation of scents on cheeks and foreheads of large felines in connection to the facial marking behavior. J Chem Ecol 38:145–156. [DOI] [PubMed] [Google Scholar]
- Stowers L, Kuo T-H, 2015. Mammalian pheromones: emerging properties and mechanisms of detection. Curr Opin Neurobiol 34:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld NJ, Hildebrand JG, 1999. Olfactory systems: common design, uncommon origins? Curr Opin Neurobiol 9:634–639. [DOI] [PubMed] [Google Scholar]
- Tait C, Batra S, Ramaswamy SS, Feder JL, Olsson SB, 2016. Sensory specificity and speciation: a potential neuronal pathway for host fruit odour discrimination in Rhagoletis pomonella. Proc R Soc B Biol Sci 283:20162101.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney KB, Sampson JL, Ross PS, Sekela MA, Kennedy CJ, 2008. Salmon olfaction is impaired by an environmentally realistic pesticide mixture. Environ Sci Technol 42:4996–5001. [DOI] [PubMed] [Google Scholar]
- Tobias JA, Aben J, Brumfield RT, Derryberry EP, Halfwerk W. et al. , 2010. Song divergence by sensory drive in Amazonian birds. Evolution 64:2820–2839. [DOI] [PubMed] [Google Scholar]
- Touhara K, 2007. Deorphanizing vertebrate olfactory receptors: recent advances in odorant–response assays. Neurochem Int 51:132–139. [DOI] [PubMed] [Google Scholar]
- Ullah I, Sjöstrand J, Andersson P, Sennblad B, Lagergren J, 2015. Integrating sequence evolution into probabilistic orthology analysis. Syst Biol 64:969–982. [DOI] [PubMed] [Google Scholar]
- Vogel S, 1965. Kesselfallen–Blumen. Umschau 65:12–17. [Google Scholar]
- Wang G, Carey AF, Carlson JR, Zwiebel LJ, 2010. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci USA 107:4418–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissburg MJ, Zimmer-Faust RK, 1993. Life and death in moving fluids : hydrodynamic effects on chemosensory-mediated predation. Ecology 74:1428–1443. [Google Scholar]
- Wilkins MR, Seddon N, Safran RJ, 2013. Evolutionary divergence in acoustic signals: causes and consequences. Trends Ecol Evol 28:156–166. [DOI] [PubMed] [Google Scholar]
- Wyatt TD, 2003. Pheromones and Animal Behaviour: Communication by Smell and Taste. In: Wyatt TD, editor. Cambridge: Cambridge University Press. [Google Scholar]
- Yáñez-Serrano AM, Nölscher AC, Williams J. et al. , 2015. Diel and seasonal changes of biogenic volatile organic compounds within and above an Amazonian rainforest. Atmos Chem Phys 15:3359–3378. [Google Scholar]
- Yarmolinsky DA, Zuker CS, Ryba NJP, 2009. Common sense about taste: from mammals to insects. Cell 139:234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder AD, Chan LM, dos Reis M, Larsen PR, Campbell CR. et al. , 2014. Molecular evolutionary characterization of a V1R subfamily unique to strepsirrhine primates. Genome Biol Evol 6:213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohe LR, Davies KTJ, Rossiter SJ, Davalos L, 2018. Expressed vomeronasal type-1 receptors, V1rs. in bats uncover conserved mechanisms of social chemical signaling. bioRxiv 10.1101/293472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Massa HF, Hsu L, Trask BJ, 2010. Extreme variability among mammalian V1R gene families. Genome Res 20:10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer-Faust RK, Finelli CM, Pentcheff ND, Wethey DS, 1995. Odor plumes and animal navigation in turbulent water flow: a field study. Biol Bull 188:111–116. [DOI] [PubMed] [Google Scholar]