Abstract
The outer membranes of animal cells contain high concentrations of cholesterol, of which a small proportion is located deep within the hydrophobic core of the membrane. An automated docking procedure is described that allows the characterization of binding sites for these deep cholesterol molecules on the membrane-spanning surfaces of membrane proteins and in protein cavities or pores, driven by hydrogen bond formation. A database of this class of predicted binding site is described, covering 397 high-resolution structures. The database includes sites on the transmembrane surfaces of many G-protein coupled receptors; within the fenestrations of two-pore K+ channels and ATP-gated P2X3 channels; in the central cavities of a number of transporters, including Glut1, Glut5, and P-glycoprotein; and in deep clefts in mitochondrial complexes III and IV.
Introduction
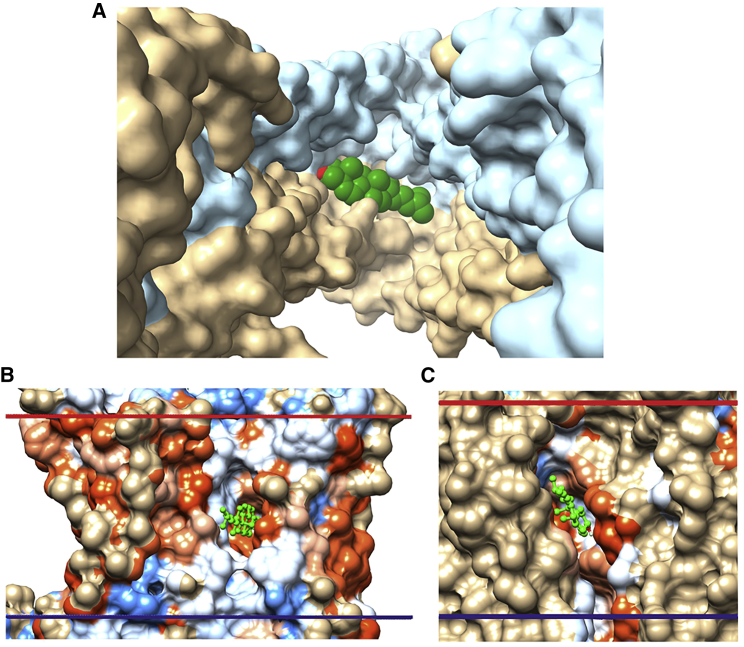
Membrane proteins play a central role in the physiology of the cell, particularly as receptors, channels, and transporters. High-resolution structural information is now available for many of these proteins in isolation, but much less is known about how they interact with the lipid bilayer component of the membrane. The membrane-spanning surfaces of membrane proteins are often pictured as bland and featureless, but in fact, the surfaces are rough, containing many crevasses and holes, and are dotted with atoms capable of forming hydrogen bonds with small polar molecules located within the hydrophobic core of the membrane (1). Some of these surface-exposed atoms will be involved already in hydrogen bonding with other atoms within the protein, and any additional, intermolecular hydrogen bonds that might form will be relatively weak (2), but others will have no intramolecular partners and could therefore form strong hydrogen bonds with a suitable partner. Fig. 1 A shows the hydrophobic region of the agonist-free β2 adrenergic receptor (3); this region contains, exposed on the surface and not involved in intramolecular hydrogen bonding, four O and nine N atoms from the backbone, three side-chain NH groups and seven side-chain OH groups, and three methionine S atoms and five side-chain SH groups.
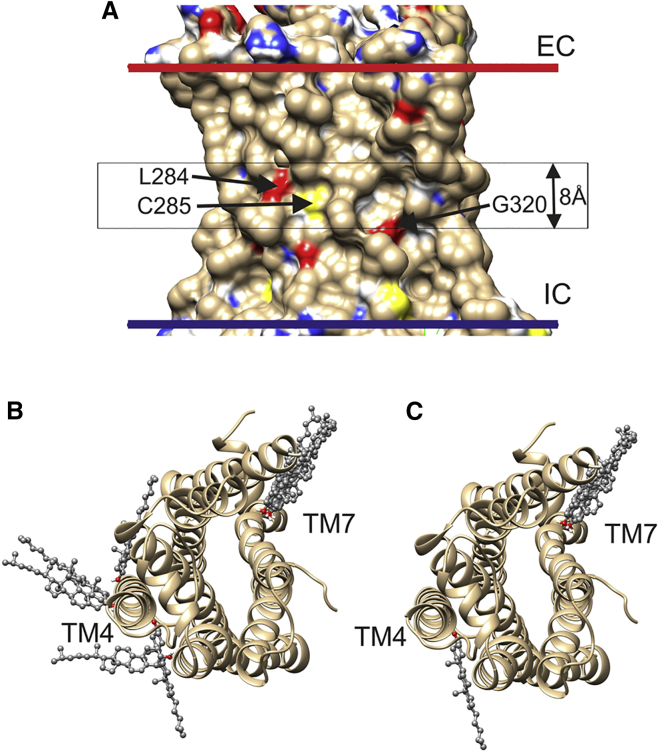
Figure 1.
Docking of cholesterol to the agonist-free β2 adrenergic receptor (PDB: 3D4S). (A) The membrane-spanning surface shows the locations of surface-exposed oxygen (red), nitrogen (blue), and sulfur (yellow) atoms not involved in intramolecular hydrogen bonding. The extracellular (EC) and intracellular (IC) sides of the hydrophobic domain of the membrane surrounding the protein, as given by the OPM database, are shown by red and blue bars, respectively. The central black box shows the position of the 8 Å slab used for docking. The three residues containing surface-exposed, non-hydrogen-bonded O and S atoms located within the box and visible in this view are labeled. (B) The 10 most energetically favorable of the 20 docking poses before selection for hydrogen bonding are shown. The view is from the EC side. (C) The six poses remaining after selection for cholesterol molecules hydrogen bonding to residues not involved in intramolecular hydrogen bonding are shown. These poses involve hydrogen bonding to Ser161 in TM4 and Gly320 in TM7. To see this figure in color, go online.
One hydrophobic molecule capable of hydrogen bonding to the transmembrane (TM) surface of a membrane protein is cholesterol, which typically makes up between 25 and 50 mol% of the lipid molecules in the plasma membranes of animal cells (4, 5, 6). Most of these cholesterol molecules are located with their −OH groups close to the glycerol backbone region of the lipid bilayer, with their hydrophobic rings in the hydrophobic core of the bilayer. Correspondingly, all membrane-protein crystal structures that include resolved cholesterol molecules show the bound cholesterol molecules with their −OH groups in what, in a lipid bilayer, would be the glycerol backbone region, as shown in Fig. S1 for the human purinergic receptor P2Y12 (7). In some cases, the −OH group of the resolved cholesterol molecule is hydrogen bonded to the protein, as in the β2 adrenergic receptor, in which the hydrogen bond is to an Arg residue, part of a suggested cholesterol consensus motif CCM; two other possible cholesterol-binding motifs, CRAC and CARC, also involve an Arg (or Lys) residue (3, 8). In other cases, the resolved cholesterol molecules do not form any hydrogen bonds with the protein, as for the P2Y12 receptor shown in Fig. S1, and it is likely that hydrogen bonds are formed to lipid or water molecules in the lipid glycerol backbone and headgroup regions.
However, not all the cholesterol molecules in a biological membrane are located with their −OH groups at the membrane-water interface; neutron diffraction studies and molecular dynamics simulations have shown a small proportion of the cholesterol molecules to be located with their −OH groups deep within the bilayer (9, 10, 11, 12, 13). It has been estimated that in a protein-containing lipid bilayer, the proportion of cholesterol molecules deep in the bilayer is approximately 3% (1). Any binding between these “deep” cholesterol molecules and the protein surface will be driven by hydrogen bonding, as there is no hydrophobic effect in the hydrophobic interior of the membrane. The standard free energy (ΔGo) for formation of an intermolecular hydrogen bond in liquid hexane is 5.3 kcals mol−1 when calculated in molar concentration units (2, 14), equal to 6.5 kcals mol−1 when calculated in mole fraction concentration units. Using the relationship ΔGo = −RTlnKd, where Kd is the dissociation constant for the hydrogen bonded complex, gives a value for Kd of 1.8 × 10−5 in mole fraction units. This means that at a mole fraction of deep cholesterol molecules of 0.009 (3% of a total cholesterol mole fraction of 30%), formation of a single hydrogen bond between a protein donor or acceptor atom and a deep cholesterol molecule would result in a >99% probability of the donor or acceptor being hydrogen bonded to a cholesterol −OH group. Cholesterol levels are generally lower in cell organelles than in the plasma membrane and could be different in different regions of a membrane if the membrane contains domains of high and low cholesterol density (13); all these factors could affect the probability of a binding site for cholesterol actually being occupied by cholesterol.
Coarse-grained molecular dynamics (CGMD) simulations of the β2 adrenergic and A2A adenosine receptors in cholesterol-containing bilayers showed that deep cholesterol molecules did indeed interact with the protein surface deep in the membrane, the interactions being between a cholesterol −OH group and some, but not all, of the potential hydrogen bond partners on the protein surface (1). No cholesterol molecules were observed to interact with the surface only via their hydrophobic moieties, and the best-localized part of a bound cholesterol molecule was observed to be its −OH group, with the ring and chain moieties adopting a range of different positions. Unfortunately, even CGMD simulations need long simulation times to ensure equilibration of the cholesterol molecules (1), making it a daunting task to extend these simulations to the full range of animal membrane proteins for which high-resolution structures are available. However, the fact that binding of deep cholesterol molecules is driven just by the cholesterol −OH group makes a molecular docking approach attractive. A comparison can be made with studies of water binding to proteins using AutoDock Vina (15) because water docking also involves just an −OH group and hydrogen bonding; in a study of a set of structures for bacterial oligopeptide-binding protein A bound to tripeptides, 97% of the crystallographically identified water molecules were correctly identified by docking, with a false positive rate of less than 1 water molecule per structure (16). It is shown here that docking of cholesterol using AutoDock Vina reproduces the results of the CGMD simulations with a speed that makes possible a complete survey of the available crystal structures.
The docking studies reported here suggest that as well as binding sites on the lipid-exposed surfaces of proteins, binding sites also occur in the fenestrations, central pores, and deep clefts present in many membrane proteins. These binding sites, deep in the membrane, will be occupied predominantly by cholesterol molecules because of the limited range of potential hydrogen bond partners to be found dissolved at high concentrations in the central core of the membrane. A number of examples of deep cholesterol binding are explored in some detail, illustrating how widespread such binding is likely to be and its potential importance for membrane protein function. Details of all the dockings are available in the Supporting Material.
Methods
Crystal structures of animal membrane proteins with resolutions of 3.5 Å or better were identified from the Membrane Proteins of Known 3D Structure (http://blanco.biomol.uci.edu/mpstruc/) and the Orientations of Proteins in Membranes (OPM; http://opm.phar.umich.edu) databases. Protein structure files were downloaded from the OPM database, as this provides structures oriented in a hydrophobic slab representing a lipid bilayer with protein coordinates centered about the middle of the hydrophobic slab (17), convenient for docking.
Docking was performed using AutoDock Vina (15) running under Chimera (18). The cholesterol ligand was prepared for docking with free rotation about the C-OH bond using AutoDock 4 (19). Ligand and solvent molecules were removed from protein structures, and proteins were prepared for docking using the routines provided in Chimera. The search box was chosen centered at x = 0, y = 0, and z =0 with a length along the z axis of 8 Å and lengths along the x and y axes sufficient to ensure free movement of the cholesterol ligand around the protein. To ensure that the use of three-dimensional grids to represent molecules in AutoDock Vina did not result in binding sites being missed close to the edge of the 8 Å search box, the docking procedure was repeated with a 12 Å search box, rejecting any dockings in which the cholesterol −OH group had an absolute z value greater than 4 Å or, allowing a little “fuzziness” at the boundary, an adjacent C atom (C2–C4) that had an absolute z value greater than 5 Å. Weighting factors for hydrogen bonds and hydrophobic effects were changed from default values of −0.59 and −0.03, respectively, to −2.0 and −0.001, respectively, as described below.
Results were analyzed using in-house Python code. Up to 20 dockings were returned by AutoDock Vina, and these were searched for dockings in which the cholesterol was hydrogen bonded to protein atoms not involved in intramolecular hydrogen bonding using the Chimera hydrogen-bond detector. For symmetric homo-oligomeric proteins, all binding has been assigned to subunit A to aid comparison between data sets. Protein cavities were identified using the CASTp server (sts.bioe.uic.edu/castp/calculation.html) (20).
A table of cholesterol dockings together with associated structure files in Protein Data Bank (PDB) format for downloading are available on the DeepCholesterol web site (https://deepcholesterol.soton.ac.uk).
Results and Discussions
A rapid procedure for detecting deep cholesterol binding sites
The first step in an automated docking procedure is to define the volume around the protein to be searched. In CGMD simulations of the β2 adrenergic and A2A adenosine receptors in cholesterol-containing bilayers, it was observed that all the deep cholesterol molecules, either free or bound, were to be found in the central 8-Å-thick hydrophobic core of the bilayer (see Fig. 1 A) (1). This distinct distribution results from the anchoring of the majority of the cholesterol molecules in a membrane with their −OH groups in the glycerol backbone region of the bilayer (4). Insertion of a rigid cholesterol ring into a phospholipid bilayer reduces the mobility and increases the order of those parts of any fatty acyl chain adjacent to the ring, consequently reducing the partitioning of small molecules into that portion of the bilayer (21, 22). The average hydrophobic thickness of a eukaryotic membrane protein as estimated by the OPM database is 31 Å (17), and the hydrophobic thickness of a membrane protein generally matches the hydrophobic thickness of the surrounding lipid bilayer as defined by the distance between the glycerol backbone regions of the two sides of the bilayer (23). With a length for the cholesterol rings of 11.5 Å, the spacing between the ends of the rings, across the bilayer center, will therefore be 8 Å. This central core is also clearly visible in the crystal structure of the G-protein coupled receptor (GPCR) P2Y12, which is unique in showing two resolved cholesterol molecules, one on each side of the putative lipid bilayer around the protein (Fig. S1) (7). The two cholesterol −OH groups are located at the two hydrophobic surfaces as defined by the OPM database, and the separation across the bilayer between the ends of the two cholesterol rings is 8.5 Å. CGMD simulations also show that binding of deep cholesterol molecules to a protein surface requires a distortion of the adjacent lipid bilayer, a distortion that will be favored in the central 8 Å core of the bilayer where the groups adjacent to the protein surface will be the flexible C-terminal ends of the phospholipid fatty acyl chains and the chains of the cholesterol molecules (1). For all these reasons, the search volume was chosen to be the central 8-Å-thick section of the membrane; this also has the advantage of avoiding any cholesterol molecules that might bind at the lipid-water interface (Fig. 1 A).
Weighting values used in AutoDock Vina for the hydrophobic effect and for hydrogen bonding were derived to describe docking in an aqueous environment (15). In the center of a lipid bilayer, the hydrophobic effect will be very weak, whereas hydrogen bonding will be approximately fourfold stronger than in water (14). New weighting values were therefore developed to match the results of docking to the results of the CGMD simulations described below. It was found that matching required the absolute value for weighting for the hydrophobic effect to be below −0.0012, and that for hydrogen bonding to be between −1.9 and −2.1, compared to the default Vina values of −0.0351 and −0.587, respectively. The weighting value for the hydrophobic effect was therefore set at −0.001, and that for hydrogen bonding at −2.0. Docking energies calculated in AutoDock Vina are based on direct atom-atom interactions and take no account of whether or not a protein atom is involved in intramolecular hydrogen bonding. A Python script was therefore written to select just those dockings that involved hydrogen bonding of the cholesterol −OH group to a non-hydrogen-bonded protein donor or acceptor atom. Free rotation was allowed around the C-OH bond to allow sampling of all possible orientations of the rigid ring relative to the −OH group.
Comparison of docking and CGMD results
The validity of the weighting values used in the docking studies is shown by comparison of the docking results with those obtained previously by CGMD simulation (Tables 1 and S1). Fig. 1 B illustrates the 10 most energetically favorable docking poses from the 20 returned by AutoDock Vina for the agonist-free β2 adrenergic receptor with no selection based on hydrogen bonding, and Fig. 1 C shows the results after selecting from the 20 just those dockings that involved hydrogen bonding to protein atoms not involved in intramolecular hydrogen bonding. The selected docking poses correspond to hydrogen bonding of the cholesterol −OH proton to the backbone carbonyl oxygen of Gly320 in TM helix TM7 and to the side-chain oxygen of Ser161 in TM4 (Table 1).
Table 1.
Hydrogen-Bond Partners for Cholesterol in the β2-Adrenergic Receptor: Comparison of Docking and CGMD results
In CGMD simulations, the protein is represented by a series of beads, each bead typically corresponding to four non-hydrogen atoms (24), allowing the identification of residues close to a cholesterol −OH bead but not allowing the identification of individual atoms involved in hydrogen bonding. In the simulations for the agonist-free β2 adrenergic receptor, residues with probabilities >25% of being within 5 Å of the −OH bead of a deep cholesterol molecule fell into distinct clusters (Table 1). The first cluster consisted of Gly320 and Phe321; Gly320 contains a non-hydrogen-bonded backbone carbonyl oxygen, whereas Phe321 contains no non-hydrogen-bonded donors or acceptors, consistent with the docking results, which identified Gly320 as the hydrogen bond partner for a deep cholesterol. The second cluster consisted of three residues, of which Ser161 was the only residue with a non-hydrogen-bonded donor or acceptor, again agreeing with the docking results identifying Ser161 as a hydrogen-bond partner for a deep cholesterol (Table 1). Thus, of the eight potential surface-exposed, non-hydrogen-bonded donor and acceptor atoms located in the central 8 Å region of the membrane (Fig. 1 A), only two are identified as parts of binding sites for cholesterol by the docking protocol adopted here, and the residues containing these same two atoms are also identified as binding partners in the CGMD simulations. A common feature of many of those atoms that form sites for cholesterol binding is that they are located at the bottoms of concave surface regions or “pockets” as detected by CASTp (Figs. 2 and S2), frequently associated with binding sites (20). Calculations of ligand binding energies in AutoDock Vina are based on a statistical scoring function and so will be less reliable than those calculated using force-field methods. Nevertheless, it is comforting that the calculated binding energies in molar concentration units of 6–7 kcal mol−1 (Table 1) are comparable to the experimental value of 5.3 kcals mol−1 determined for a single hydrogen bond in a hydrophobic environment (2, 14).
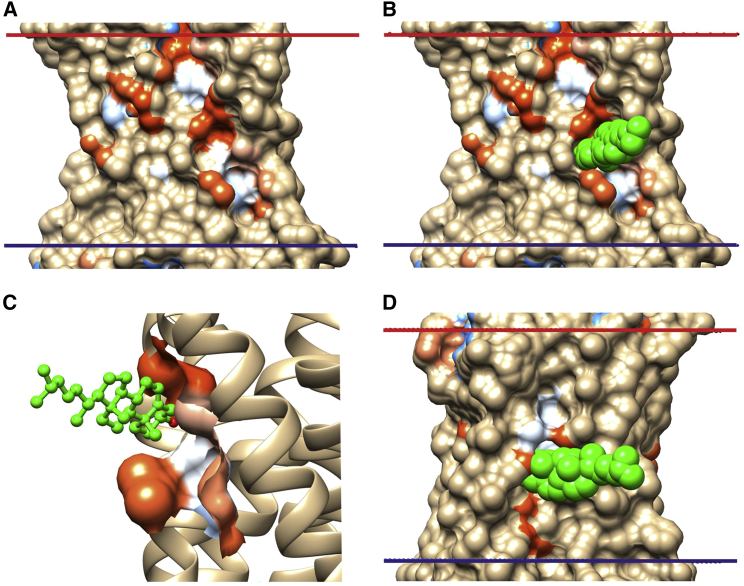
Figure 2.
The two cholesterol-binding sites identified on the agonist-free β2 adrenergic receptor. (A and B) The TM surface with surface pockets identified using CASTp (20) is shown, colored from most hydrophobic (orange) to most hydrophilic (blue), (A) without and (B) with a cholesterol molecule (green spheres) bound to Gly320. (C) An expanded view of the cholesterol binding pocket (cholesterol in ball and stick representation) is shown. (D) The TM surface with a cholesterol molecule bound to Ser161 is shown. To see this figure in color, go online.
CGMD simulations for the agonist-bound β2 adrenergic receptor show no binding to the two clusters identified for the agonist-free structure and, instead, show most favorable binding to a cluster of three residues including Glu122 (Table 1) (1). This again is consistent with the docking results, which detect hydrogen bonding to the side chain of Glu122, either alone or together with the backbone carbonyl of the adjacent Val206 (Table 1). The CGMD simulations also suggested the presence of a much weaker interaction with a second cluster consisting of Phe208 and Tyr209, both residues individually having a low probability (<25%) of being in contact with a cholesterol −OH bead (1). Neither of these residues contain surface-exposed, non-hydrogen-bonded donor or acceptor atoms and so are not detected as part of a binding site in the docking studies. It is possible that the interaction suggested by the CGMD simulation is an artifact attributable to the “stickiness” of the force fields used in the CGMD simulations (25).
The patterns of interaction detected by CGMD simulations with the A2A adenosine receptor were different from those for the β2 adrenergic receptor in that the clusters were larger and individual residues showed lower probabilities of interaction with cholesterol (1). Further, whereas cholesterol −OH groups were well localized in the clusters for the β2 adrenergic receptor, for the A2A adenosine receptor, they occupied a range of positions, sometimes with more than one molecule occupying a position in a cluster at the same time. Of the seven residues in the agonist-free receptor (PDB: 4EIY) and the six in the agonist-bound receptor (PDB: 3QAK) located in the central 8 Å region and containing surface-exposed donor or acceptor atoms not involved in intramolecular hydrogen bonding, none had a high probability of being in contact with a cholesterol −OH bead, average probabilities of contact being 6 and 7%, respectively (1); evidently, binding to the clusters detected by CGMD simulation for the A2A adenosine receptor is not driven by hydrogen bonding. Consistent with these results, docking studies failed to detect any hydrogen-bond-dependent binding of cholesterol for either the 4EIY or 3QAK structures (Table S1). Poorly localized binding of hydrophobic molecules in large hydrophobic cavities has been reported in a variety of proteins, and the interaction clusters identified in the CGMD simulations were suggested to be of this type (1); nonconventional binding sites of this type would not be detected by the docking approach adopted here.
Channels
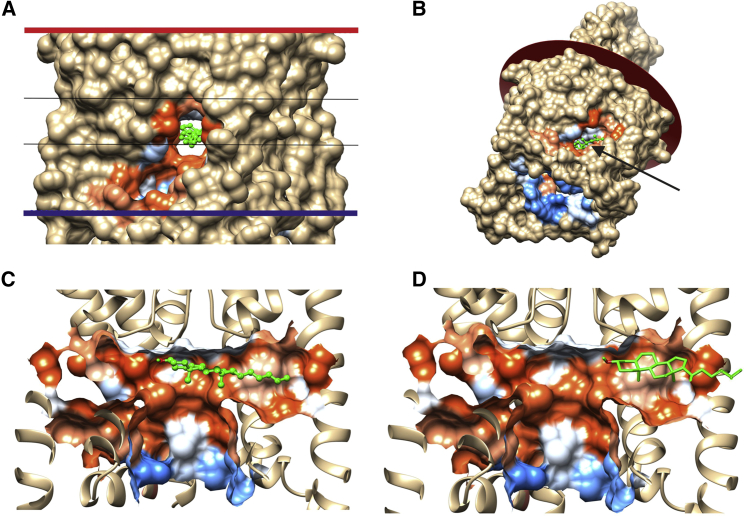
The two-pore K+ channels TWIK-1, TRAAK, TREK-1, and TREK-2 have a structure with a narrow selectivity filter on the extracellular (EC) side leading into a large central cavity open to the intracellular (IC) space, as shown in Fig. 3 for TWIK-1 (26, 27, 28, 29). The central cavity is connected to the 8 Å core of the lipid bilayer, where the deep cholesterol molecules are found, by openings at the interfaces between the two constituent subunits, just below the selectivity filter (these openings are referred to as fenestrations). The strongest docking observed for TWIK-1 is for a cholesterol molecule with its seven-carbon-long chain in the fenestration, filling its length, with the cholesterol ring and −OH group in the central pore under the selectivity filter and hydrogen bonded to either Thr225 in pore helix 2 or to Leu115 in pore helix 1, where it will block ion passage between the selectivity filter and the central cavity (Fig. 3, A and C; Table S2). The crystal structure of TWIK-1 showed electron density in the fenestration, which could be fitted to an alkyl chain (26). It has been suggested that the chain could belong to a phospholipid molecule, but although a molecular dynamics simulation showed that a phospholipid acyl chain could enter into the fenestration, the chain did not enter far enough to occlude the central pore (30); the binding site for the cholesterol alkyl chain reported here overlaps with the observed electron density. Although the only dockings involving hydrogen bonding of the cholesterol −OH group are those with the −OH group in the central pore, about a quarter of the 20 dockings returned by AutoDock Vina before selection based on hydrogen bonding showed the cholesterol molecule with its ring and −OH group, non-hydrogen bonded, within just the fenestration (Fig. 3 D). The fenestration is therefore wide enough to accommodate a cholesterol ring so that a cholesterol molecule could diffuse, −OH group first, from the central core of the lipid bilayer along the fenestration to reach the central pore. Epicholesterol (5-cholestan-3α-ol), in which the −OH group has a 3α rather than a 3β stereochemistry, has been shown to have a smaller effect than cholesterol on the function of many K+ channels (31). Docking studies show that although epicholesterol can fit into the fenestrations of TWIK-1, the −OH group fails to make any hydrogen bonds with non-hydrogen-bonded atoms in the pore (data not shown).
Figure 3.
Cholesterol binding within fenestrations of TWIK-1 (PDB: 3UKM). (A) The TM surface showing the pocket around the central fenestration containing a bound cholesterol (green) and the 8 Å search box (black lines) is shown. (B) A tilted view shows the EC plane and the large cavity exposed on the IC side, with the bound cholesterol (green) marked by an arrow. (C) A cutaway view shows a bound cholesterol (green) in the central pore with its −OH group (red) beneath the selectivity filter and its chain in the fenestration. (D) A cutaway view shows a cholesterol molecule with its −OH group in the fenestration. To see this figure in color, go online.
In TWIK-1, the two openings to the bilayer interior, one on each side of the dimer, are symmetrical, and docking of cholesterol is observed equally in the two fenestrations. In contrast, in the two-pore domain TRAAK channel, in what is referred to as the down or nonconductive state, packing of the two monomers making up the channel is nonsymmetrical, with one of the two openings to the bilayer interior being much smaller than that in TWIK-1, whereas the other is much larger (32). In the alternative up or conductive state of the TRAAK channel, TM4 in one of the subunits moves to pack against TM2 of the other subunit, closing the larger of the two fenestrations (Fig. S3, A and B). Docking of cholesterol is only observed in the larger of the two fenestrations, and, indeed, the size of the fenestration is such that a cholesterol molecule can be docked either way round, with the cholesterol-OH hydrogen bonding to either Ile127, Leu236, Thr237, or Thr238 in the central cavity or to Leu151 on the outer surface (Fig. S3, C and D; Table S2). In all these binding modes, the cholesterol molecule is located under the selectivity filter and so could block ion movement through the channel. In the up or conductive state, no docking of cholesterol is observed in the fenestrations (Table S2). In both the up and down states of TRAAK, docking is also observed to the outer surface of the channel, to Tyr42, or to the neighboring Ser45 (Table S2).
The fenestrations in TREK-2 are similar to those in TRAAK, and, as in TRAAK, the fenestrations are closed off by movement of TM4 in the up state (28); in this state, cholesterol binds only to Tyr87 on the protein surface and does not occupy the fenestrations (Table S2). In contrast, in the down state, cholesterol binds in the fenestration, hydrogen bonding to Leu279 or Thr281 in pore helix 2, to Ile170 in pore helix 1, or to Ile194 in TM2 (Table S2). Prozac (fluoxetine) binds in the fenestrations of TREK-2 in a binding site defined by Ile194, Leu279, and Thr280 (28) so that binding of Prozac and cholesterol will be competitive. The only available structures for TREK-1 are in the up state with no large, open fenestrations, and the only binding observed is to the protein surface, to residues in TM1, TM3, or TM4 (Table S2).
In the Kv family of four subunit K+ channels, binding of cholesterol is observed to either a single site on the channel surface or to no sites (Table S2). For the homotetrameric Kir family, no binding is observed to the channel surface, but binding is seen in the central cavity of the channel, the cholesterol hydrogen bonding to either Gln141 in the pore helix or to Trp94 in TM1 (Table S2). However, unlike the two-pore K+ channels, Kir channels contain no fenestrations that would allow direct entry of cholesterol molecules from the surrounding lipid bilayer into the channel. The G-protein-gated K+ channel GIRK2 is also homotetrameric, with a structure similar to that of Kir2.2 (33, 34), and again a cholesterol molecule binds in the central pore just below the selectivity filter, hydrogen bonding to Ser181 in TM2; in the constitutionally active R201A mutant in the presence of phosphatidylinositol 4,5-bisphosphate (PIP2), four strong binding modes are observed in the central cavity, with hydrogen bonding to Tyr102 in TM1, Ser181 and Ser177 in TM2, and Glu152 in the pore helix. GIRK2, like Kir2.2, lacks any fenestrations.
In the tetrameric transient receptor potential channel family, cholesterol molecules bind to the protein surface, generally to a residue in the deep clefts between subunits such as Thr550 in TRPV1, Tyr439 and Ser503 in TRPML1, Tyr491 in TRPML3, and Thr663 and Tyr611 in PKD2, but these bound cholesterol molecules do not penetrate into the central pore (Table S2). Thr550 in TRPV1 has been shown to hydrogen bond to the ligand homovanillyl ester, whose binding site is also occupied by phosphatidylinositols (35). Similarly, in the GluA2 glutamate receptor family, cholesterol molecules bind in the clefts between subunits but do not penetrate into the central cavity to block the channel.
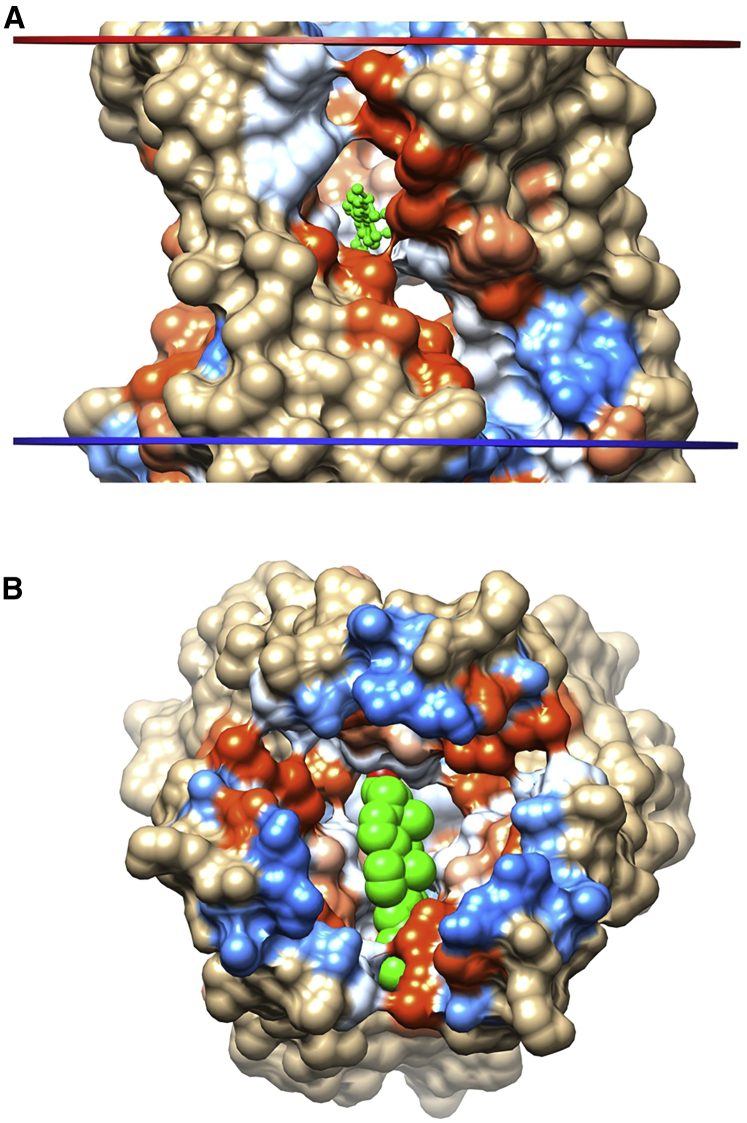
In the closed apo or antagonist-bound state of the trimeric ATP-gated P2X3 channel, binding of cholesterol is only seen to Thr336 or Ser36 on the external surface (Table S2). However, in the open channel, Thr336 becomes occluded by a neighboring subunit, and Thr330, previously occluded, now hydrogen bonds to a cholesterol molecule with its −OH group in the central cavity and its chain in the very large fenestrations or lateral portals that link the channel pore to the core of the lipid bilayer (Fig. 4); Thr330 is located at the narrowest region of the pore gate on the cytoplasmic side (36). Ions are suggested to enter the transmembrane pore via the lateral portals (36, 37); a cholesterol molecule bound to Thr330 might not completely block the portal because of the large size of the portal (Fig. 4 A). The structure of the channel in the desensitized state is more like that in the open state than that in the closed state (36), and cholesterol again binds to Thr330 and the adjacent Ser331. Very similar results are observed for the P2X4 channel, with cholesterol binding to the non-hydrogen-bonded carbonyls of Gly343 (in PDB: 4DW1) or Ala 344 (in PDB: 5WZY) at the pore constriction in the open channel, with no binding in the closed channel. In the open-channel form of the P2X4 channel of the Golf Coast tick, no binding is observed in the open channel, as the carbonyl group of the residue at the pore constriction, Val361, is hydrogen bonded within helix TM2 (38). For the P2X7 channel bound to the competitive antagonist TNP-ATP, the channel is in an expanded, incompletely activated conformation (39), and cholesterol does not bind in the pore but to −OH containing residues exposed on the protein surface (Table S2). The acid-sensing ion channel adopts a structure very similar to those of the P2X channels (40), and the closed form of the acid-sensing ion channel shows no binding of cholesterol either in the pore or to surface exposed sites.
Figure 4.
Cholesterol binding within the central pore of the open-state homotrimeric ATP-gated P2X3 channel (PDB: 5SVK). (A) The TM surface showing the large portals connecting the channel pore to the central core of the lipid bilayer is shown with a bound cholesterol (green). (B) A view of the TM domain from the EC side with the EC domains removed shows a cholesterol (spheres) bound in the channel pore. To see this figure in color, go online.
G-protein-coupled receptors
Docking was performed for 146 class A GPCR structures, representing 36 different members of the class A family, with cholesterol binding observed to 60% of them (Table S1). The frequencies with which the seven TM α-helices of the receptors provided residues acting as proton donors or acceptors to deep cholesterol molecules are all rather similar, but with TM5 having the highest probability of containing such residues (Fig. S4). In a few cases, the same residues appear in docking sites in more than two members of the family, particularly noticeable for residues at positions 5.46/5.461 and 7.47 in the Ballesteros-Weinstein numbering system (Table S5) (41).
For the 5-hydroxytrytamine 5-HT1B receptor, a cholesterol-binding site was detected involving the side chain of Thr64 in TM1 and the backbone oxygen of Ser99 in TM2, which is not hydrogen bonded in the helix because of the presence of Pro104 (Table S1); Ser99, but not Thr64, is conserved in the 5-HT1A receptor. The level of binding of 5-HT to the 5-HT1A receptor decreases on removal of cholesterol and is restored by the readdition of either cholesterol or ent-cholesterol, the mirror image of cholesterol, but not by epicholesterol (42). Docking studies with the 5-HT1B receptor and ent-cholesterol give the same results as for cholesterol, whereas docking with epicholesterol returned no binding sites (data not shown), leaving open the possibility of a functional role for binding of cholesterol to Ser99. For the 5-HT2B receptor, two cholesterol-binding sites were detected, one involving the side chain of Thr228 in TM5 and the other involving the backbone oxygen of Met63 in TM1 (Table S1). Thr228 is adjacent to the Pro-Ile-Phe motif that forms an interface between TM3, TM5, and TM6 near the base of the ligand binding pocket (43), and mutation of Thr228 to Ala resulted in a very large reduction in affinity for 5-HT (44).
Thirty structures are available for the A2A adenosine receptor with resolutions of 3.5 Å or better, and of these, 18 showed no binding sites for cholesterol in the docking analysis (Table S1). One structure (PDB: 3UZA) showed binding to the backbone oxygen of Cys185 in TM5. Although the presence of Pro189 in TM5 results in the backbone oxygen of Cys185 not forming a hydrogen bond within TM5, in all the crystal structures except for 3UZA, this backbone oxygen is hydrogen bonded to the side chain of Gln89 in TM3. The remaining 11 structures showed binding to a pair of residues, Gly56 and Val57, in TM2 with binding energies between −6.1 and −5.5 kcals mol−1 (Table S1). There are small differences in the locations of the backbone oxygens of Gly56 and Val57 and in the surface pockets reported by CASTp between structures that do and do not show binding sites for cholesterol (Fig. S5). Some of these differences in surface pockets could be due to the presence of thermostabilizing mutations in some structures, the most common of which are the StarR2 set of eight mutations, one of which, Leu54, is close to the Gly56/Val57 pair. Small differences could also arise from the variety of insertions used to aid crystallization and from the different agonists and antagonists included in the crystallization media or could be a result of the relatively low resolutions of some of the structures.
The β2 adrenergic receptor shows cholesterol binding to both the inactive and active states but to different sites (Fig. 2; Table 1), suggesting that binding of cholesterol could result in a shift in equilibrium between the different conformational states of the receptor. The presence of cholesterol results in an increase in affinity for the partial inverse agonist timolol but not for the full agonist isoproterenol (3). Mutation of Gly320 in TM7, part of one of the binding sites in the inactive state, resulted in a halving of the affinity for isoproterenol, and the sequence NPLIY in TM7, containing the Pro residue responsible for the backbone oxygen of Gly320 not being hydrogen bonded, is conserved in the β2 adrenergic receptor family (45, 46). Mutation of Glu122, part of the binding site in the active receptor (Table 1), led to reduced affinity for both agonists and antagonists, and Glu122 has been suggested to be part of a pathway linking allosteric changes on the two sides of the receptor (47, 48). Cholesterol-binding sites are also detected on the muscarinic acetylcholine receptor family, and the presence of cholesterol has been shown to result in a large increase in the affinity of the M1 receptor for the antagonist quinuclidinylbenzylate (49).
The results for bovine rhodopsin and opsin are very different from those for most class A GPCRs in that of the 20 high resolution structures, only one (PDB: 3CAP) shows cholesterol binding (Table S1). In the 3CAP structure, both atoms of the −OH group of Thr297 are non-hydrogen bonded and exposed on the protein surface and interact with a deep cholesterol molecule, whereas in all the other structures, a simple rotation of the Thr297 side chain about the CA-CB bond results in the −OH group being hydrogen bonded either to the backbone O of Phe294 in rhodopsin or to the backbone O of Phe293 in opsin. In mammals, rhodopsin is located in disks formed by invagination of the plasma membrane so that newly formed disk membranes are rich in cholesterol, whereas the cholesterol content of older disks has fallen from the original 30 mol% to approximately 5 mol% (50). In contrast, in squid, rhodopsin is located in cholesterol-rich microvilli (51), and as shown in Table S1, strong hydrogen bonding of cholesterol to Leu85 is observed in TM2.
Mutational studies in other classes of GPCR are also consistent with an important role for some of the residues identified as being involved in cholesterol binding (Table S1). In two class B GPCRs, the corticotrophin-releasing factor receptor 1 and the glucagon receptor, interaction of cholesterol is observed with the side-chain −OH of a Thr or Ser residue at positions 2.62 or 2.63 in the numbering system of Wootten et al. (52). Mutation of Ser1892.62 in the human glucagon receptor led to a small increase in the affinity for glucagon, whereas mutation of the adjacent Val1912.64 lead to a large decrease in affinity (53). In the class C metabotropic glutamate receptor, interaction of cholesterol is observed with Ser715 and Thr719 in TM4, and mutation of either of these residues has been shown to lead to a reduced affinity for glutamate (54).
Transporters
Cholesterol binding to the Ca2+-ATPase of muscle sarcoplasmic reticulum is complex, with different patterns of binding for the different conformational states of the protein (Table S3). For a wide variety of Ca2+-bound forms, binding is observed to some or all of Ser942 (TM9), Leu797 (TM6), and Thr906/Met909 (TM8) with no binding to these residues in non-Ca2+-bound forms, reflecting changes in helix packing on binding Ca2+. Binding of thapsigargin favors the E2 conformation of the Ca2+-ATPase, and cholesterol binds to thapsigargin-bound forms predominantly at Cys268/Tyr295, again reflecting changes in helix packing. The Na+,K+-ATPase also shows a complex pattern of binding, with distinct differences between phosphorylated and nonphosphorylated forms (Table S3).
The solute carrier transporter superfamily member Glut1 in inhibitor-bound forms of the inward-open state shows binding of cholesterol to the side chain of Ser73 located at the bottom of a large hole forming part of the domain-domain interface between TM2 and TM11 (Table S3); in the inhibitor-free structure, the Ser73 side chain points into the central cavity, and no binding of cholesterol is observed. Binding to the external surface is also observed for the Glut5 fructose transporter in the open-outward form to Ser422 in cow and its equivalent Ser421 in rat, and in the open-inward form, binding is also observed to Thr452 and Thr354 (Table S3). In the mitochondrial ADP/ATP carrier, cholesterol binds to Gly224 or Arg234 in the large central cavity where the inhibitor carboxyatractyloside also binds (Table S3). Although the cholesterol content of mitochondria is generally very low, in hepatoma cells, it can increase to approximately 20 mol% in the inner mitochondrial membrane, and in reconstituted systems, high levels of cholesterol inhibit the rate of transport of ATP by the ADP/ATP carrier (55).
The P-glycoprotein, a member of the ABC transporter family, consists of two pseudosymmetric halves encoded in a single polypeptide chain. In the nucleotide-free, inward-facing state, cholesterol binds to either His60 or Tyr303, spanning the central cavity of the protein (Fig. 5 A), and to Thr765 on the membrane-exposed surface (Table S3). In the nucleotide-bound, outward-facing state, cholesterol no longer binds in the central cavity, although binding in the central cavity is observed in the nucleotide-bound, outward-facing state of the MRP-1 drug resistance protein (Table S3). The P-glycoprotein has been reported to transport cholesterol across the membrane, and the function of the P-glycoprotein is modified by cholesterol in the membrane (56, 57). The ABCB10 mitochondrial ABC transporter is a homodimer, and although cholesterol binds in the central cavity, most binding modes involve solely one or other of the two monomers, and only rarely does a cholesterol molecule span between the two halves; Ser181, one of the identified hydrogen bond partners, has been suggested to be part of a conserved binding site for amphipathic substrates (58). For the cystic fibrosis transmembrane conductance regulator, binding is observed to the outer face of the transporter and to the inner cavity, but again with no bridging between the two halves of the cavity (Table S3).
Figure 5.
Binding of cholesterol to P-glycoprotein and two mitochondrial complexes. (A) A view of the P-glycoprotein (PDB: 4Q9H) from the IC side shows a bound cholesterol (green), with the two domains colored blue and tan. (B and C) TM surfaces of cytochrome bc1 (PDB: 1BGY) and cytochrome c oxidase (PDB: 1V54), respectively, are given, showing lipid-exposed pockets with bound cholesterol molecules. To see this figure in color, go online.
Other classes of membrane protein
Results for other classes of membrane protein are listed in Table S4. Most show binding of cholesterol to the membrane-exposed surfaces of the protein. However, for the CAAX intramembrane proteases, binding is limited to the large central hydrophobic cavity. In the electron transport chain complex II, binding is to the external surface of the complex, whereas in complex III (Cytochrome bc1), binding is observed to Tyr358 in subunit C in a large cleft open to the hydrophobic core of the lipid bilayer (Fig. 5 B). In electron transport chain complex IV (cytochrome c oxidase), binding is similarly observed to Ser89 at the bottom of a deep lipid-exposed cleft (Fig. 5 C) as well as to other residues on the lipid-exposed surface of the complex.
Conclusions
It is shown here that Autodock Vina can be used, with modified binding parameters, to study protein binding of cholesterol molecules deep in the membrane. Errors in docking studies are generally calculated based on root mean-square deviation between a crystallographically determined bound ligand structure at some site and the structure of a ligand docked to that site. Estimating error in this way is not possible for the deep cholesterol binding sites, as there are no crystallographic data, and root mean-square deviation cannot be measured between an atomic structure (for cholesterol in docking studies) and a coarse-grained structure (cholesterol in CGMD simulations). However, the fact that the hydrogen-bond partners identified here by docking match those present at binding sites identified by CGMD simulation, with no false hits, suggest that the reliability of docking is high.
The studies reported here detect binding of deep cholesterol molecules to 60% of the 146 GPCR structures studied, including examples in which binding differs between the active and inactive states of the receptor. Binding is observed in the fenestrations of two-pore K+ channels, with the cholesterol chain in a fenestration and the cholesterol −OH and ring in the central pore, where it can block ion flow through the channel; in the TRAAK channel, binding is only observed for the nonconductive state and not for the conductive state. Conformation-specific binding is also observed in the fenestrations of the trimeric ATP-gated P2X3 channels; for a number of transporters, including Glut1, Glut5, and P-glycoprotein; and in mitochondrial complexes. These studies demonstrate the importance of the many non-hydrogen-bonded atoms exposed on the TM surfaces or in central cavities in many membrane proteins provided by the polypeptide backbone in proline-containing TM helices and by residue side chains. It is suggested that interaction between these proton donors and acceptors and molecules of cholesterol could be functionally important, the probability of a binding event being high even though only a small proportion of the cholesterol molecules in a membrane are located deep within the membrane core, because of the strength of a hydrogen bond formed in a hydrophobic environment.
Tables S1–S4 give all current predicted cholesterol binding sites. These data are also available and will be updated at the DeepCholesterol web site (https://deepcholesterol.soton.ac.uk).
Acknowledgments
The Centre for Biological Sciences, University of Southampton is thanked for computing facilities.
Editor: D. Peter Tieleman.
Footnotes
Five figures and five tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30727-6.
Supporting Citations
References (59, 60) appear in the Supporting Material.
Supporting Material
References
- 1.Genheden S., Essex J.W., Lee A.G. G protein coupled receptor interactions with cholesterol deep in the membrane. Biochim. Biophys. Acta. 2017;1859:268–281. doi: 10.1016/j.bbamem.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Feldblum E.S., Arkin I.T. Strength of a bifurcated H bond. Proc. Natl. Acad. Sci. USA. 2014;111:4085–4090. doi: 10.1073/pnas.1319827111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanson M.A., Cherezov V., Stevens R.C. A specific cholesterol binding site is established by the 2.8 A structure of the human β2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song Y., Kenworthy A.K., Sanders C.R. Cholesterol as a co-solvent and a ligand for membrane proteins. Protein Sci. 2014;23:1–22. doi: 10.1002/pro.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fantini J., Barrantes F.J. How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front. Physiol. 2013;4:31. doi: 10.3389/fphys.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hedger G., Sansom M.S.P. Lipid interaction sites on channels, transporters and receptors: recent insights from molecular dynamics simulations. Biochim. Biophys. Acta. 2016;1858:2390–2400. doi: 10.1016/j.bbamem.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang K., Zhang J., Zhao Q. Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature. 2014;509:115–118. doi: 10.1038/nature13083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gimpl G. Interaction of G protein coupled receptors and cholesterol. Chem. Phys. Lipids. 2016;199:61–73. doi: 10.1016/j.chemphyslip.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Marquardt D., Kučerka N., Katsaras J. Cholesterol’s location in lipid bilayers. Chem. Phys. Lipids. 2016;199:17–25. doi: 10.1016/j.chemphyslip.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Marquardt D., Heberle F.A., Katsaras J. Lipid bilayer thickness determines cholesterol’s location in model membranes. Soft Matter. 2016;12:9417–9428. doi: 10.1039/c6sm01777k. [DOI] [PubMed] [Google Scholar]
- 11.Marrink S.J., de Vries A.H., Wassall S.R. Cholesterol shows preference for the interior of polyunsaturated lipid membranes. J. Am. Chem. Soc. 2008;130:10–11. doi: 10.1021/ja076641c. [DOI] [PubMed] [Google Scholar]
- 12.Prasanna X., Chattopadhyay A., Sengupta D. Cholesterol modulates the dimer interface of the β2-adrenergic receptor via cholesterol occupancy sites. Biophys. J. 2014;106:1290–1300. doi: 10.1016/j.bpj.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingólfsson H.I., Carpenter T.S., Lightstone F.C. Computational lipidomics of the neuronal plasma membrane. Biophys. J. 2017;113:2271–2280. doi: 10.1016/j.bpj.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben-Tal N., Sitkoff D., Honig B. Free energy of amide hydrogen bond formation in vacuum, in water, and in liquid alkane solution. J. Phys. Chem. B. 1997;101:450–457. [Google Scholar]
- 15.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross G.A., Morris G.M., Biggin P.C. Rapid and accurate prediction and scoring of water molecules in protein binding sites. PLoS One. 2012;7:e32036. doi: 10.1371/journal.pone.0032036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lomize A.L., Pogozheva I.D., Mosberg H.I. Positioning of proteins in membranes: a computational approach. Protein Sci. 2006;15:1318–1333. doi: 10.1110/ps.062126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 19.Morris G.M., Huey R., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binkowsk T.A., Naghibzadeh S., Liang J. CASTp: computed atlas of surface topography of proteins. Nucleic Acids Res. 2003;31:3352–3355. doi: 10.1093/nar/gkg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luxnat M., Galla H.J. Partition of chlorpromazine into lipid bilayer membranes: the effect of membrane structure and composition. Biochim. Biophys. Acta. 1986;856:274–282. doi: 10.1016/0005-2736(86)90037-4. [DOI] [PubMed] [Google Scholar]
- 22.Sankaram M.B., Thompson T.E. Modulation of phospholipid acyl chain order by cholesterol. A solid-state 2H nuclear magnetic resonance study. Biochemistry. 1990;29:10676–10684. doi: 10.1021/bi00499a015. [DOI] [PubMed] [Google Scholar]
- 23.Lee A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 24.Marrink S.J., Tieleman D.P. Perspective on the Martini model. Chem. Soc. Rev. 2013;42:6801–6822. doi: 10.1039/c3cs60093a. [DOI] [PubMed] [Google Scholar]
- 25.Javanainen M., Martinez-Seara H., Vattulainen I. Excessive aggregation of membrane proteins in the Martini model. PLoS One. 2017;12:e0187936. doi: 10.1371/journal.pone.0187936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller A.N., Long S.B. Crystal structure of the human two-pore domain potassium channel K2P1. Science. 2012;335:432–436. doi: 10.1126/science.1213274. [DOI] [PubMed] [Google Scholar]
- 27.Brohawn S.G., del Mármol J., MacKinnon R. Crystal structure of the human K2P TRAAK, a lipid- and mechano-sensitive K+ ion channel. Science. 2012;335:436–441. doi: 10.1126/science.1213808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong Y.Y., Pike A.C., Carpenter E.P. K2P channel gating mechanisms revealed by structures of TREK-2 and a complex with Prozac. Science. 2015;347:1256–1259. doi: 10.1126/science.1261512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lolicato M., Arrigoni C., Minor D.L., Jr. K2P2.1 (TREK-1)-activator complexes reveal a cryptic selectivity filter binding site. Nature. 2017;547:364–368. doi: 10.1038/nature22988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aryal P., Abd-Wahab F., Tucker S.J. Influence of lipids on the hydrophobic barrier within the pore of the TWIK-1 K2P channel. Channels (Austin) 2015;9:44–49. doi: 10.4161/19336950.2014.981987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levitan I., Singh D., Rosenhouse-Dantsker A. Cholesterol binding to ion channels. Front. Physiol. 2014;5:65. doi: 10.3389/fphys.2014.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brohawn S.G., Campbell E.B., MacKinnon R. Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel. Nature. 2014;516:126–130. doi: 10.1038/nature14013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whorton M.R., MacKinnon R. Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell. 2011;147:199–208. doi: 10.1016/j.cell.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whorton M.R., MacKinnon R. X-ray structure of the mammalian GIRK2-βγ G-protein complex. Nature. 2013;498:190–197. doi: 10.1038/nature12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Y., Cao E., Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mansoor S.E., Lü W., Gouaux E. X-ray structures define human P2X(3) receptor gating cycle and antagonist action. Nature. 2016;538:66–71. doi: 10.1038/nature19367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samways D.S., Li Z., Egan T.M. Principles and properties of ion flow in P2X receptors. Front. Cell. Neurosci. 2014;8:6. doi: 10.3389/fncel.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kasuya G., Fujiwara Y., Nureki O. Structural insights into divalent cation modulations of ATP-gated P2X receptor channels. Cell Rep. 2016;14:932–944. doi: 10.1016/j.celrep.2015.12.087. [DOI] [PubMed] [Google Scholar]
- 39.Kasuya G., Yamaura T., Nureki O. Structural insights into the competitive inhibition of the ATP-gated P2X receptor channel. Nat. Commun. 2017;8:876. doi: 10.1038/s41467-017-00887-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzales E.B., Kawate T., Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 2009;460:599–604. doi: 10.1038/nature08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ballesteros J.A., Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structre-function relations in G protein-coupled receptors. In: Stuart C.S., editor. Methods in Neurosciences. Vol. 25. Academic Press; 1995. pp. 366–428. [Google Scholar]
- 42.Jafurulla M., Rao B.D., Chattopadhyay A. Stereospecific requirement of cholesterol in the function of the serotonin1A receptor. Biochim. Biophys. Acta. 2014;1838:158–163. doi: 10.1016/j.bbamem.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wacker D., Wang C., Stevens R.C. Structural features for functional selectivity at serotonin receptors. Science. 2013;340:615–619. doi: 10.1126/science.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manivet P., Schneider B., Launay J.M. The serotonin binding site of human and murine 5-HT2B receptors: molecular modeling and site-directed mutagenesis. J. Biol. Chem. 2002;277:17170–17178. doi: 10.1074/jbc.M200195200. [DOI] [PubMed] [Google Scholar]
- 45.Kikkawa H., Isogaya M., Kurose H. The role of the seventh transmembrane region in high affinity binding of a β 2-selective agonist TA-2005. Mol. Pharmacol. 1998;53:128–134. doi: 10.1124/mol.53.1.128. [DOI] [PubMed] [Google Scholar]
- 46.Barak L.S., Ménard L., Caron M.G. The conserved seven-transmembrane sequence NP(X)2,3Y of the G-protein-coupled receptor superfamily regulates multiple properties of the β 2-adrenergic receptor. Biochemistry. 1995;34:15407–15414. doi: 10.1021/bi00047a003. [DOI] [PubMed] [Google Scholar]
- 47.Ghanouni P., Schambye H., Kobilka B.K. The effect of pH on beta(2) adrenoceptor function. Evidence for protonation-dependent activation. J. Biol. Chem. 2000;275:3121–3127. doi: 10.1074/jbc.275.5.3121. [DOI] [PubMed] [Google Scholar]
- 48.Bhattacharya S., Vaidehi N. Differences in allosteric communication pipelines in the inactive and active states of a GPCR. Biophys. J. 2014;107:422–434. doi: 10.1016/j.bpj.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berstein G., Haga T., Ichiyama A. Effect of the lipid environment on the differential affinity of purified cerebral and atrial muscarinic acetylcholine receptors for pirenzepine. Mol. Pharmacol. 1989;36:601–607. [PubMed] [Google Scholar]
- 50.Albert A., Alexander D., Boesze-Battaglia K. Cholesterol in the rod outer segment: a complex role in a “simple” system. Chem. Phys. Lipids. 2016;199:94–105. doi: 10.1016/j.chemphyslip.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 51.Mason W.T., Abrahamson E.W. Phase transitions in vertebrate and invertebrate photoreceptor membranes. J. Membr. Biol. 1974;15:383–392. doi: 10.1007/BF01870096. [DOI] [PubMed] [Google Scholar]
- 52.Wootten D., Simms J., Sexton P.M. Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc. Natl. Acad. Sci. USA. 2013;110:5211–5216. doi: 10.1073/pnas.1221585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siu F.Y., He M., Stevens R.C. Structure of the human glucagon class B G-protein-coupled receptor. Nature. 2013;499:444–449. doi: 10.1038/nature12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fukuda J., Suzuki G., Ohta H. Identification of a novel transmembrane domain involved in the negative modulation of mGluR1 using a newly discovered allosteric mGluR1 antagonist, 3-cyclohexyl-5-fluoro-6-methyl-7-(2-morpholin-4-ylethoxy)-4H-chromen-4-one. Neuropharmacology. 2009;57:438–445. doi: 10.1016/j.neuropharm.2009.06.017. [DOI] [PubMed] [Google Scholar]
- 55.Woldegiorgis G., Shrago E. Adenine nucleotide translocase activity and sensitivity to inhibitors in hepatomas. Comparison of the ADP/ATP carrier in mitochondria and in a purified reconstituted liposome system. J. Biol. Chem. 1985;260:7585–7590. [PubMed] [Google Scholar]
- 56.Kimura Y., Kioka N., Ueda K. Modulation of drug-stimulated ATPase activity of human MDR1/P-glycoprotein by cholesterol. Biochem. J. 2007;401:597–605. doi: 10.1042/BJ20060632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clay A.T., Lu P., Sharom F.J. Interaction of the P-glycoprotein multidrug transporter with sterols. Biochemistry. 2015;54:6586–6597. doi: 10.1021/acs.biochem.5b00904. [DOI] [PubMed] [Google Scholar]
- 58.Shintre C.A., Pike A.C., Carpenter E.P. Structures of ABCB10, a human ATP-binding cassette transporter in apo- and nucleotide-bound states. Proc. Natl. Acad. Sci. USA. 2013;110:9710–9715. doi: 10.1073/pnas.1217042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pin J.P., Galvez T., Prézeau L. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol. Ther. 2003;98:325–354. doi: 10.1016/s0163-7258(03)00038-x. [DOI] [PubMed] [Google Scholar]
- 60.Wang C., Wu H., Stevens R.C. Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs. Nat. Commun. 2014;5:4355. doi: 10.1038/ncomms5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.