Spatiotemporal analysis of ABA-signaling in plants reveals the dynamics of osmotic stress signaling in roots and identifies novel regulators of ABA-dependent transcriptional programs.
Abstract
The water stress-associated hormone abscisic acid (ABA) acts through a well-defined signal transduction cascade to mediate downstream transcriptional events important for acclimation to stress. Although ABA signaling is known to function in specific tissues to regulate root growth, little is understood regarding the spatial pattern of ABA-mediated transcriptional regulation. Here, we describe the construction and evaluation of an ABSCISIC ACID RESPONSIVE ELEMENT (ABRE)-based synthetic promoter reporter that reveals the transcriptional response of tissues to different levels of exogenous ABA and stresses. Genome-scale yeast one-hybrid screens complemented these approaches and revealed how promoter sequence and architecture affect the recruitment of diverse transcription factors (TFs) to the ABRE. Our analysis also revealed ABA-independent activity of the ABRE-reporter under nonstress conditions, with expression being enriched at the quiescent center and stem cell niche. We show that the WUSCHEL RELATED HOMEOBOX5 and NAC DOMAIN PROTEIN13 TFs regulate QC/SCN expression of the ABRE reporter, which highlights the convergence of developmental and DNA-damage signaling pathways onto this cis-element in the absence of water stress. This work establishes a tool to study the spatial pattern of ABA-mediated transcriptional regulation and a repertoire of TF-ABRE interactions that contribute to the developmental and environmental control of gene expression in roots.
Abscisic acid (ABA) is a plant hormone, widely involved in different biological processes, including plant growth (Humplík et al., 2017), stomatal conductance (Negin and Moshelion, 2016), lateral root development (Duan et al., 2013), and seed dormancy (Leprince et al., 2017), as well as response to water-associated environmental stresses, such as water deficit and osmotic and salt stress (Seo and Koshiba, 2002; Munemasa et al., 2015; Sah et al., 2016). These stresses cause a reduction in the availability of water to the plant, leading to the induction of ABA biosynthesis and subsequent perception by the ABA signaling pathway (Seo and Koshiba, 2002). Reporters that detect the presence and activity of ABA at each step of regulation are needed to study how ABA signaling controls complex physiological responses across the various tissues and organ systems of the plant during stress. Förster resonance energy transfer-based ABA sensors have been developed to study the spatial dynamics of ABA accumulation in root cells (Jones et al., 2014; Waadt et al., 2014). However, the current Förster resonance energy transfer sensors are limited in utility because their binding affinity lies outside the physiological range of ABA levels or expression of the sensors interferes with ABA signaling. ABA may endogenously accumulate to lower levels during environmental stress than occurs after typical exogenous treatment regimes, highlighting the need for reporters that are sensitive to physiologically relevant changes in hormone concentration. Finally, an understanding of the transcriptional regulatory output of a hormone-signaling pathway provides information that complements data obtained from sensors that directly detect hormone levels. Thus, additional reporters are needed to more precisely indicate the spatiotemporal dynamics of ABA and stress signaling.
ABA responses are initiated when the hormone is recognized by a large family of PYL/PYR/RCAR coreceptors and PP2C-type protein phosphatases (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009; Verslues, 2016). When ABA is present, PP2C-type phosphatase activity is suppressed, which frees the SnRK2-type kinases to phosphorylate targets, including ABSCISIC ACID RESPONSIVE ELEMENT (ABRE)-binding transcription factors (TFs), such as the bZIP-type AREBs/ABFs (Fujita et al., 2013). An ABRE sequence from the rice (Oryza sativa) gene RAB16A was the first to be isolated and characterized (Mundy et al., 1990). Synthetic promoters that fuse DNA fragments containing the ABRE with a minimal promoter sequence have the regulatory capacity to cause an increase in GUS expression after ABA treatment (Shen and Ho, 1995). Conservation analysis of these sequences highlights the importance of a core ACGT sequence motif (T/G/C-2A-1C0G0T+1G+2G/T+3C+4; Izawa et al., 1993; Hattori et al., 2002), as well as the G+2 flanking nucleotide for ABA responses (Hattori et al., 2002).
Cis-regulatory element (CRE)-based synthetic promoters (SP), in which a predicted CRE sequence is fused to a minimal promoter (MP) element, represent a powerful tool for validating the regulatory function of CRE sequences (Venter, 2007; Hernandez-Garcia and Finer, 2014). For example, the synthetic promoter DR5 was created by multimerizing a mutated auxin response element seven times and fusing this with a Cauliflower mosaic virus (CaMV) 35S MP (Ulmasov et al., 1995, 1997). DR5 has been widely used as an auxin indicator for tracking auxin signaling events such as those that occur in the root quiescent center (Sabatini et al., 1999) and during morphogenesis (Benková et al., 2003; Friml et al., 2003; Smit and Weijers, 2015), because of its ∼100-fold increased auxin inducibility (Ulmasov et al., 1997). In addition, DR5 was shown to be specifically bound by auxin-responsive ARF TFs but not Aux/IAAs that repress the activity of DR5 in auxin signaling (Ulmasov et al., 1997). While recent studies have led to the development of other reporters that enable a more direct readout of auxin levels in cells, such as DII-based sensors (Brunoud et al., 2012), DR5 is still useful for tracking the final step in auxin-dependent regulation of gene expression.
Here, we present an ABA reporter made from six copies of the ABRE, whose activity recapitulates known centers of ABA signaling. The reporter exhibits sensitivity to physiologically relevant concentrations of ABA and levels of osmotic stress induced by NaCl or mannitol. Genome-scale yeast one-hybrid (Y1H) assays were used to construct an ABRE-TF interaction network. Functional characterization of this network in yeast demonstrated that the recruitment of TFs to ABREs is highly dependent on flanking genomic context and less dependent on the architecture of the synthetic promoter. In addition to the canonical bZIP-type TFs that are known to bind ABRE-like sequences, our ABRE interaction network also identified noncanonical regulators, belonging to the DNA damage response and stem-cell identity pathways, as factors that influence ABRE-mediated expression in plants. We found that WUSCHEL RELATED HOMEOBOX5 (WOX5), a target of auxin signaling that promotes quiescent center (QC) function, is necessary and sufficient to induce expression of the ABRE reporter in this tissue under nonstress conditions. Intriguingly, WOX5 is also necessary for ABA-mediated responses in the QC, suggesting that developmental regulators may be important determinants of the spatial pattern of ABA signaling. Together, our results provide insight and tools for understanding the mechanisms that enable spatially resolved transcriptional programs in plants in response to environmental stress and hormone signaling.
RESULTS
Construction and Testing of an ABRE-Based Reporter
To determine how ABA-dependent gene expression functions within the root under stress conditions, we first performed an analysis of CRE enrichment in salt-stress responsive genes using the tissue-specific and time point-resolved spatiotemporal map we previously generated (O’Connor et al., 2005; Geng et al., 2013). This map examines gene expression across four tissues at six time points during the response to 140 mm NaCl. Our enrichment analysis showed that the ABRE and ABRE-like sequences are highly enriched in the promoters of genes whose expression is induced immediately after salt exposure (Supplemental Fig. S1). These genes were generally induced across all tissues, but to different degrees. These data suggest that ABA-mediated transcriptional responses may be induced broadly in the root during salt stress; however, we cannot exclude the possibility that co-enriched CREs are also responsible for this pattern of activity.
To determine where in the root ABRE-mediated gene expression is induced during stress, we sought to construct an ABRE-based SP reporter that could be studied at the tissue-specific level. Past work has shown that synthetic promoters based on the multimerization of an isolated ABRE sequence can confer ABA responsiveness to a heterologous minimal promoter (Skriver et al., 1991; Vasil et al., 1995); however, their contribution to tissue-level responses was not investigated. To examine this directly, we developed SPs based on ABRE sequences that are present in two well-characterized ABA-responsive genes: RESPONSE TO DEHYDRATION29A (RD29A; Yamaguchi-Shinozaki and Shinozaki, 1993) and ABA INSENSITIVE1 (ABI1; Leung et al., 1994). These two genes are both responsive to salt and osmotic stresses and contain canonical ABRE sequences in their 5′ upstream noncoding regions (Fig. 1). In addition, differences in the ABRE-flanking sequences between ABI1 and RD29A would allow us to examine the effect of sequence context on ABRE activity.
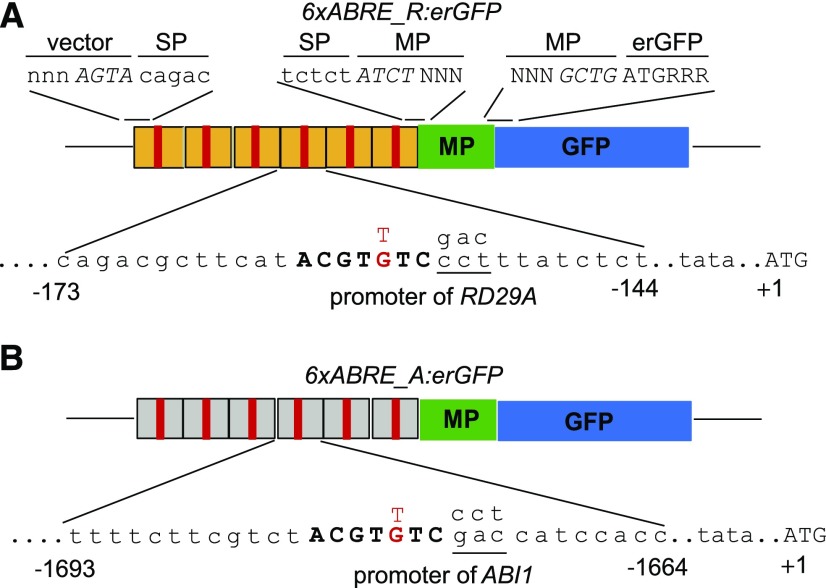
Figure 1.
Schematic showing the design of 6xABRE SPs used in this study. A, RD29A version of 6xABRE SP. B, ABI1 version of 6xABRE SP. The 7-bp ABRE sequence (red bar) and gene-specific flanking genomic sequences (orange and gray bars) together constitute the 30-bp SP subunit, which was multimerized six times and fused upstream of a minimal promoter (MP) and a reporter gene. The fifth nucleotide (labeled in red) in the uppercased core ABRE sequence (ACGTGTC; Hattori et al., 2002) was changed from G to T for functional studies using the SP or RD29A full-length promoter context. The three nucleotides that are underlined 3′ of the core motif were swapped between 6xABRE_A and 6xABRE_R SPs to test for effect on recruitment of TFs in the Y1H screens. Above the schematics, the 4-bp overhang sequences used during Golden Gate cloning are indicated in uppercase italics. erGFP, Endoplasmic reticulum-localized GFP.
SPs contained a multimerized SP subunit fused upstream of a MP sequence. This SP subunit was 30 bp in length, with the 7-bp ABRE sequence (ACGTGTC) equidistant from the 5′ and 3′ termini (“Materials and Methods”; Fig. 1). The multimerized promoter sequence from ABI and RD29A were named 6xABRE_A and 6xABRE_R, respectively. The multimerized SP subunits were cloned 5′ of an MP sequence, which was then used to drive expression of an endoplasmic reticulum-localized GFP (erGFP) or GUS reporter gene and transformed into Arabidopsis (Arabidopsis thaliana) Col-0 wild-type plants (“Materials and Methods”; Fig. 1).
We initially characterized an ABRE SP reporter with six copies of the SP subunit fused to the CaMV 35S (−90 to -1) MP and driving the expression of a GUS reporter. These experiments showed that all transgenic lines with visible reporter activity marked cells within the stem cell niche (SCN), QC, and root cap under standard conditions (exemplified in Supplemental Fig. S2). To understand how robust this pattern of expression was, we examined the effect of alternative SP architectures by changing the MP sequence and the number of SP subunit multimers used. Use of the RD29A MP (−54 to +96 bp; Narusaka et al., 2003) or NOS MP (−101 to +4 bp; Michael et al., 2008) in place of the CaMV 35S MP did not have an obvious effect on reporter expression pattern under standard conditions (Supplemental Fig. S3A). Changing the number of multimers of the SP subunit (three, six, and nine repeats) led to quantitatively enhanced activity and a broadening of the expression domain (Supplemental Fig. S3, B and C). These data suggest that the reporter expression pattern conferred by the 6xABRE SP is reproducible but partly influenced by reporter context, as expected.
In order to quantitatively dissect the spatial pattern and compare the conditional spatial pattern changes conferred by these ABRE SPs, the 6xABRE promoter was used to drive expression of erGFP and imaged by confocal microscopy. A quantification strategy was developed using an updated version of CellSeT software (Pound et al., 2012) optimized for ER-localized fluorescence. For the erGFP reporters, the signal from seven different cell types in root tips was measured and quantified throughout the study, unless specified otherwise (Supplemental Fig. S4).
To determine whether the expression of these SP reporters is dependent on the ABRE-core motif, we mutated this sequence in both the 6xABRE_R and 6xABRE_A reporter contexts (Hattori et al., 2002; Fig. 1). In both cases, expression was reduced, particularly in the QC (Supplemental Fig. S5, A and B; Fig. 2, A and B). Reporter expression in the lateral root cap (LRC) and the outer layer of columella was not affected by mutation of the core ABRE sequence (Supplemental Fig. S5, A and B; Fig. 2, A and B), suggesting that this expression domain was likely promoted by ABRE-independent sequences as well.
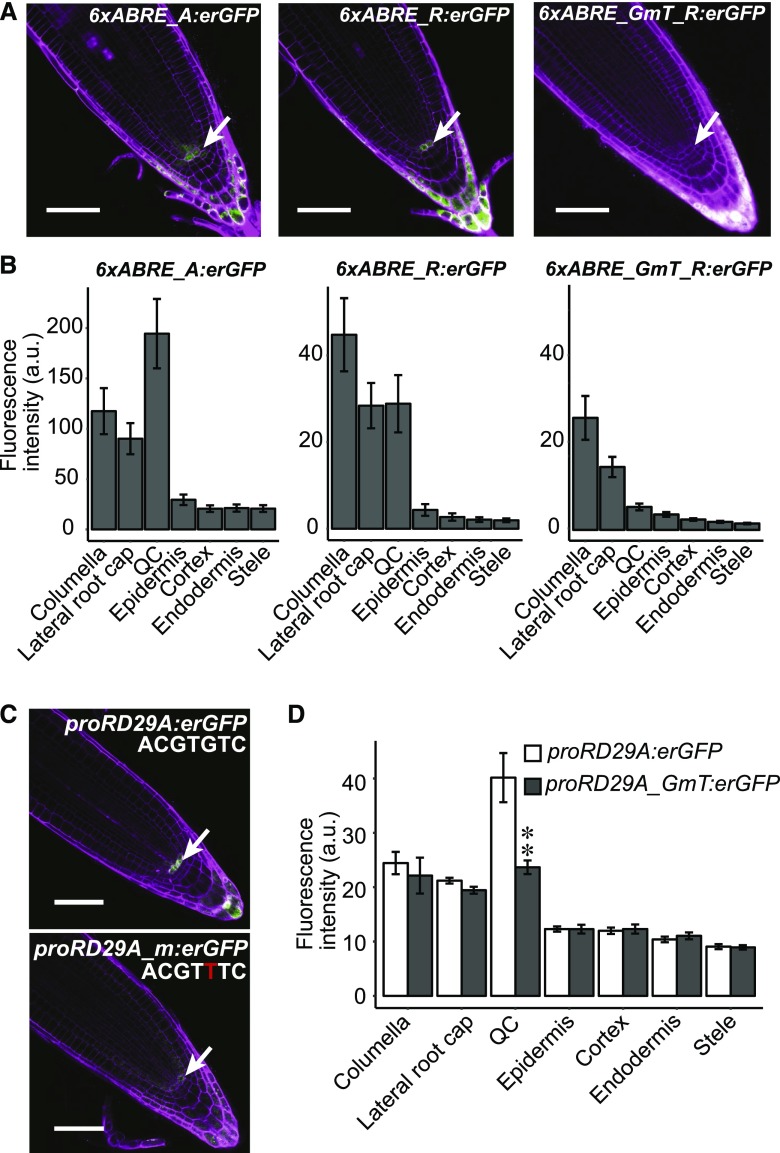
Figure 2.
The ABRE is necessary and sufficient to drive QC expression under standard conditions. A, Confocal images showing expression pattern of 6xABRE_A, 6xABRE_R SP reporters, and mutagenized RD29A version SP, 6xABRE_GmT_R SP, in root tips under standard conditions. B, Bar chart showing the normalized quantification of reporter expression driven by 6xABRE_A SP, 6xABRE_R SP, and 6xABRE_GmT_R SP reporters. Five T1 plants for each reporter were quantified with CellSeT (Pound et al., 2012). C and D, Confocal images of proRD29A:erGFP (C) and ABRE mutated proRD29A_m:erGFP reporter and their quantification in root tips (D). Statistical analysis was conducted on T1 plants (n ≥ 20). Student’s t test significance level, **0.01 < P, *0.01 < P < 0.05. Error bars, se. Scale bars, 50 μm.
The 6xABRE Synthetic Promoter Drives ABA-Inducible Expression in Plants
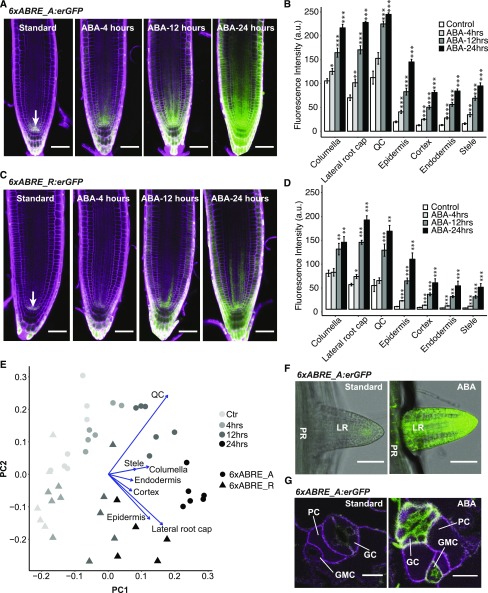
We tested the temporal responsiveness of our 6xABRE SP reporters by treating seedlings with 10 μm ABA for 4, 12, and 24 h. We were able to observe a significant activation of reporter expression after 4 h of treatment in all tissue layers except the QC (Fig. 3, A–D). After longer treatment (from 12 h onward), at least 2-fold activation in all tissues could be observed, which increased with time (Fig. 3, A–D). Principal component analysis (PCA) of the reporter expression patterns showed that the first principal component (PC1) was correlated with the time after ABA treatment, while the second principal component (PC2) distinguished the reporter isoforms (6xABRE_A versus 6xABRE_R; Fig. 3E). Loading plots showed that differential induction of reporter expression in the QC, lateral root cap and epidermis most distinguished the temporal pattern of ABRE reporter activation (Fig. 3E).
Figure 3.
ABA responsiveness of 6xABRE SP reporters. A, Confocal images of 6xABRE_A SP homozygous reporter expression in root tips under 4 , 12, and 24 h of ABA (10 μm) treatment. B, Bar chart showing the averaged fluorescence level of 6xABRE_A SP reporter in various cell types quantified by CellSeT (n ≥ 5). C, Confocal images of 6xABRE_R SP homozygous reporter expression in root tips under 4 h, 12 h, and 24 h of ABA (10 μm) treatment. D, Bar chart showing the averaged fluorescence level of 6xABRE_R SP reporter in various cell types quantified by CellSeT (n ≥ 5). E, Principal component analysis (PCA) with loadings of the expression data from the 6xABRE_A SP and 6xABRE_R SP reporters under 10 μm ABA or no ABA (Ctr) treatment. F, Confocal images of 6xABRE_A SP reporter expression in lateral roots after 3 d 10 μm ABA treatment. G, Confocal images of 6xABRE_A SP reporter expression in leaf epidermis after 24 h ABA treatment. LR, lateral root; PR, primary root; GC, guard cell; GMC, guard mother cell; PC, pavement cell. Scale bars, 50 μm. Student’s t test significance level, *0.01 < P < 0.05, **0.001 < P < 0.01, ***P < 0.001. Error bars, se.
We next tested whether the 6xABRE SP reporters could quantitatively differentiate treatment with varying concentrations of ABA from 1 μm to 25 μm. Surprisingly, both 6xABRE reporters showed strong reporter induction at all ABA concentrations tested, although reporter expression was lowest at 1 µm ABA and highest at 25 µm ABA (Supplemental Fig. S6, A–C). These data suggest that the transcriptional response may be saturated above 1 μm ABA, which is consistent with studies indicating that endogenous ABA concentrations are usually far below the concentrations frequently used.
The broad induction of 6xABRE SP reporter expression in response to ABA treatment suggests that ABA is able to enter into all tissues of the root and that the ABA signal transduction pathway is functional in all tissues of the root. Although all tissues responded, there were differences in the relative magnitude of the changes. However, the overall expression pattern of the reporter was preserved across all treatments with the highest expression in the columella, lateral root cap, and QC. This pattern was also observed across different ABA concentrations and reporter isoforms, suggesting it may reflect an intrinsic regulatory property of the ABRE or of the ABA signaling capacity of these cell types. A reporter construct including the MP, but lacking the 6xABRE cassette, did not show any erGFP induction by ABA treatment, demonstrating that the ABA responsiveness of the 6xABRE SP reporter was due to the multimeric ABRE CRE fragment (Supplemental Fig. S5D).
We also examined reporter expression under standard conditions and in response to ABA in other tissues where ABA signaling is known to play important regulatory roles. Lateral root growth is hypersensitive to ABA treatment (Duan et al., 2013), and we find that this is also reflected in the strong induction of the 6xABRE SP reporter (Fig. 3F). ABA signaling has recently been shown to negatively impact stomatal density by repressing the expression of SPEECHLESS (SPCH), which promotes guard mother cell identity (Tanaka et al., 2013). Interestingly, cells in the stomatal lineage showed a striking induction of 6xABRE SP reporter expression in response to ABA treatment, which was not observed in pavement cells (Fig. 3G). These data suggest that commitment to the stomatal lineage is associated with an enhanced capacity to respond to ABA. ChIP-seq analysis of SPCH shows that ABI1 is a direct target of this TF and that targeted genes are significantly associated with response to ABA (P < 1.4E-17; Lau et al., 2014), suggesting a negative feedback loop between stomatal lineage identity and ABA signaling.
The 6xABRE Synthetic Promoter Drives Stress-Inducible Expression in Plants
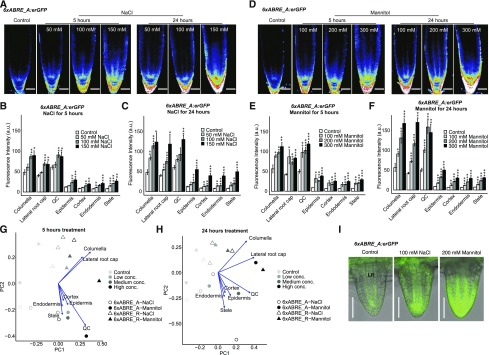
ABA biosynthesis is induced in response to osmotic stresses such as mannitol or NaCl treatment. Past work on salt stress indicated that the induction of ABA signaling is salt concentration dependent; the ABA-responsive proRAB18:GFP reporter is induced in primary roots at concentrations above 100 mm NaCl (Duan et al., 2013). We examined 6xABRE SP reporter expression after 5 or 24 h of treatment with three different concentrations of mannitol or NaCl (Fig. 4, A–F; Supplemental Fig. S6, D–I). PCA of the quantified reporter expression patterns showed that the major source of variation (PC1) separated the data based on the concentration of osmoticum while PC2 separated the two SP reporter isoforms (Fig. 4, G and H). Interestingly, the induction of reporter activity was much more differentiated by osmoticum concentration than ABA treatment (Fig. 4, G and H; Supplemental Fig. S6, A–C), suggesting that ABA signaling was not saturated under the stress conditions used. In general, the mannitol treatment induced SP reporter expression more strongly than NaCl, particularly at the highest concentration tested (150 mm NaCl, 300 mm mannitol; Fig. 4, A–F; Supplemental Fig. S6, D–I). These differences suggest that mannitol is more effective at inducing an ABA-mediated transcriptional response than NaCl. This may be a consequence of the lower membrane permeability of mannitol compared to Na+ and Cl− ions, flux of which across the membrane would reduce the effective difference in solute concentration between the inside and outside of the cell. Consistent with previous findings (Duan et al., 2013), activation of the 6xABRE SP was observed in postemergence stages of lateral root development after 3 d of NaCl or mannitol treatment (Fig. 4I).
Figure 4.
Stress responsiveness of 6xABRE SP reporters. A, Confocal images of 6xABRE_A SP homozygous reporter expression in root tips under three different concentrations of NaCl for short (5 h) and long (24 h) treatment. B and C, Bar charts showing the averaged fluorescence level of 6xABRE_A SP reporter in various cell types under 5 h NaCl (B) or 24 h NaCl (C) treatment, quantified by CellSeT (n ≥ 5). D, Confocal images of 6xABRE_A SP homozygous reporter expression in root tips after treatment with three different concentrations of mannitol for short (5 h) and long (24 h) treatment. E and F, Bar charts showing the averaged fluorescence level of 6xABRE_A SP reporter in various cell types after 5 h mannitol (E) or 24 h mannitol (F) treatment, quantified by CellSeT (n ≥ 5). The same sets of confocal images and bar chart for 6xABRE_R SP reporter are shown in Supplemental Fig. S6, D to I. G and H, PCA with loadings of the expression data from both versions of 6xABRE SP reporters after NaCl or mannitol 5 h (G) or 24 h (H) treatments. I, Confocal images of 6xABRE_A SP reporter expression in lateral roots (LR) after 3 d 100 mm NaCl and 200 mm mannitol treatments. Scale bars, 50 μm. Student’s t test significance level, *0.01 < P < 0.05, **0.001 < P < 0.01, ***P < 0.001. Error bars, se.
As expected, mutating the core ABRE motif in the SP reporters reduced the ABA and NaCl responsiveness of the associated reporters, indicating that the changes in gene expression observed following these treatments were primarily the result of ABRE-mediated gene regulation (Supplemental Fig. S5C). Together, our results demonstrate that the 6xABRE SP reporters generated here are useful tools for quantitatively assessing the ABA-mediated transcriptional response of hormone treatment and abiotic stress.
ABRE Activity under Nonstress Conditions Highlights an ABA-Independent Function
While ABA signaling has been characterized primarily in the context of abiotic stress responses, mutants with defects in ABA biosynthesis show pleiotropic developmental defects such as dwarfism (Nambara et al., 1998), suggesting that the ABA pathway may also be utilized under nonstress conditions. As mentioned earlier, ABRE-mediated reporter expression under standard conditions is highly enriched within the QC and parts of the SCN (Fig. 2, A and B). The QC is the stem cell-organizing center of the root and maintains a population of neighboring undifferentiated cells that later generate the tissue layers of the root. ABA signaling is known to suppress cell division within the QC (Zhang et al., 2010); however, it is not clear whether this results from ABA signaling that is spatially restricted to the QC itself. We confirmed that the ABRE is necessary for QC/SCN expression by mutating a critical base pair in the core motif (Hattori et al., 2002; Figs. 1 and 2, A and B). To test whether the QC/SCN activity of the ABRE was also observed within a native promoter context, we generated plants expressing a proRD29A:erGFP reporter, which includes 891 bp of the promoter region (Yamaguchi-Shinozaki and Shinozaki, 1993). Similar to the 6xABRE_R SP reporter, the full-length RD29A promoter also drove enriched expression in the QC/SCN (Fig. 2, C and D). Mutation of the same nucleotide in the ABRE core motif strongly and specifically reduced QC/SCN expression of RD29A promoter as well (Fig. 2, C and D). Thus, the QC/SCN-enriched expression of the 6xABRE_R SP accurately reflects the partial activity of the endogenous RD29A gene.
We then examined whether the QC/SCN expression of the 6xABRE SP reporter was itself ABA dependent under standard conditions. Growth of seedlings on media containing fluridone, which inhibits the carotenoid biosynthetic pathway necessary for ABA production, did not significantly affect SP reporter activity in QC cells (Supplemental Fig. S7, A and B), although it inhibited root growth and caused chlorosis of the cotyledons. The abi1-1 mutation, which leads to dominant suppression of ABA signaling, reduced the ABA response of the 6xABRE reporter in almost all cell types except the QC (Supplemental Fig. S7, C and D). Together, these results suggest that the activity of the 6xABRE reporter under standard conditions may utilize ABA-independent pathways, particularly to promote expression in the QC.
A Genome-Scale Y1H Screen Reveals an ABRE-Centered Transcription Factor Network
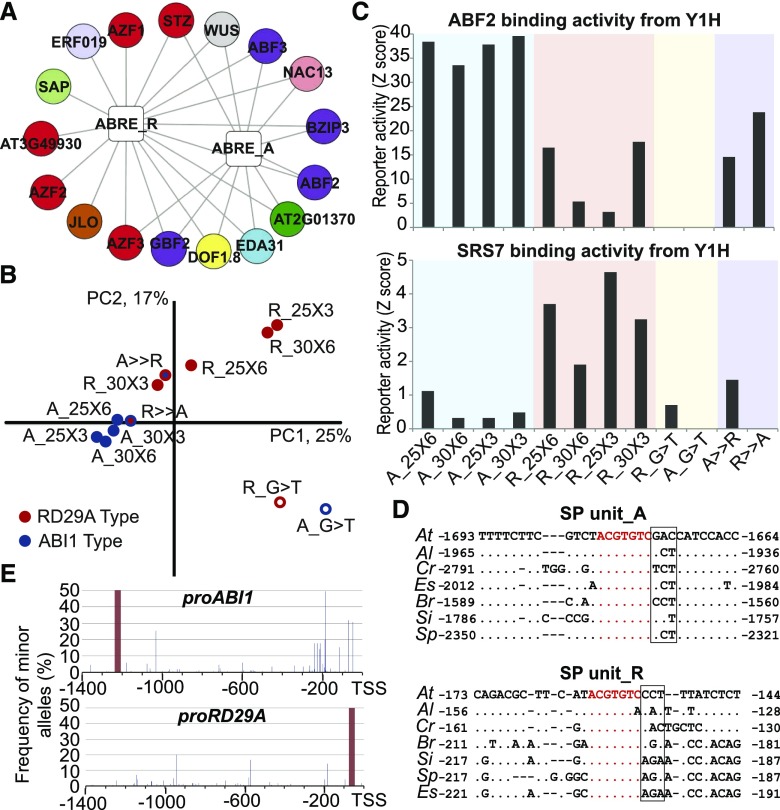
To identify TFs that regulate gene expression through the ABRE, we performed a Y1H screen using a near-complete library of 1,956 putative Arabidopsis TFs (Pruneda-Paz et al., 2014) with different isoforms of ABRE SP (plant MP not included; Supplemental Table S1). Both the ABI1 and RD29A isoforms of the 6xABRE were screened using gLUC59 (Bonaldi et al., 2017) and LacZ reporters (Supplemental Tables S2 and S3). As expected, the Y1H assay identified interactions between these promoters and members of the bZIP class of TFs (ABF2, ABF3, GBF2; Fig. 5A; Supplemental Table S4), which have previously been shown to bind ABRE or ABRE-like sequences and act as transcriptional activators of gene expression during stress responses (Guiltinan et al., 1990; Choi et al., 2000; Uno et al., 2000). ABF3, which interacted with both isoforms of 6xABRE, was expressed in a spatial domain that partially overlapped with the 6xABRE reporters after ABA or NaCl treatment, consistent with ABF3 functioning as an activator of ABRE-dependent gene expression (Supplemental Fig. S8). Interestingly, however, its expression was only faintly detected under standard conditions and was not observed in the QC/SCN (Supplemental Fig. S8), suggesting that other TFs may regulate this domain of expression. Members of the C2H2 TF family (STZ, AZF1, AZF2 and AZF3; Fig. 5A; Supplemental Table S4), which have been shown to act as transcriptional repressors of NaCl and ABA responses (Sakamoto et al., 2000, 2004), were also identified as interactors of the 6xABRE through Y1H. Consistently, similar sets of TFs were also identified as interactors with the 30-bp genomic regions used in the synthetic promoters through ChIP-seq assays (Supplemental Table S5). These data suggest that the ABRE is able to recruit TFs of different classes that may have unique regulatory effects on gene expression.
Figure 5.
Genome-scale Y1H screens reveal TF-CRE interactions and sequence context dependency of TF recruitment. A, TF-ABRE interaction network obtained through Y1H screens (output using both LUC and LacZ reporter platform) visualized using Cytoscape (Shannon et al., 2003). TFs shown in this network have an interaction Z-score > 4 for LUC (Supplemental Table S3) or fold induction > 2 for LacZ (Supplemental Table S2). Square nodes represent 6xABRE SPs. Round colored nodes represent TFs. Edges represent interactions between TFs and CREs; however, the length and thickness of the edges are arbitrary. Colors distinguish TF family (Supplemental Table S4). B, Principal component analysis (PCA) of ABRE SP activities in Y1H assays (output using LUC reporter platform). SP architecture was varied by subunit length (25 or 30 bp), repeat number (three and six times), mutation state of core CRE (G > T change described in Fig. 1), and origin of the 3-bp flanking the 3′ ABRE core sequence (Fig. 1). Red and blue distinguish RD29A (R) and ABI1 (A) isoforms of SPs. ABRE_A with RD29A 3′ 3-bp swap (A >> R, red outline, blue center), ABRE_R with ABI1 3′ 3-bp swap (R >> A, blue outline, red center). Core ABRE-mutated promoters are white filled (A_G > T and R_G > T). The names of the SPs are labeled as follows: isoform _subunit length xsubunit repeat number. C, Y1H-based reporter activity of ABRE_A and ABRE_R isoforms interacting with ABF2 and SRS7 TFs. x axis, ABRE SP isoforms; y axis, Z-scores of reporters’ activity. D, Comparison of ABRE and flanking sequences in the promoter regions of ABI1 and RD29A in different crucifer species. Arabidopsis thaliana (At), Arabidopsis lyrata (Al), and Capsella rubella (Cr) belong to the crucifer lineage I, while Brassica rapa (Br), Sysimbrium irio (Si), Schrenkiella parvula (Sp), and Eutrema salsugineum (Es) are in the crucifer lineage II. Dots indicate nucleotides identical to the Arabidopsis thaliana sequence, and alignment gaps are marked by dashes. Numbers indicate position coordinates starting from the ORF start codon. The core ABRE sequence (ACGTGTC) is colored in red. E, Distribution of SNPs detected among 171 Arabidopsis natural accessions in the upstream 2-kb region of AtABI1 and AtRD29A genes. Dark red bars indicate the respective 30-nt ABRE-containing SP subunit sequence. The frequency of all minor alleles found among the 171 accessions is indicated for each nucleotide position.
Local Sequence Context Affects 6xABRE Activity
We utilized the TF-wide Y1H screen as a method to evaluate how promoter structure and flanking sequence context affect the ability of the ABRE to recruit TFs to a promoter. SP subunits from both ABI1 and RD29A were used to synthesize promoters that varied in the number ABRE SP units multimerized (three or six repeats) or that varied in the length of the repeated SP subunit (25- or 30-bp SP unit; Supplemental Table S1). In addition, the effects of mutating the same critical nucleotide G+2 in the ABRE motif were examined to identify ABRE-independent interactions. In total, 12 different reporter isoforms were tested against all 1,956 TFs, providing a rich dataset to understand the functional significance of SP architecture. PCA of normalized SP reporter activities for all TFs screened separated the ABI1- and RD29A-type isoforms in the first two principal components, regardless of promoter structure (Fig. 5B), which is consistent with the distinct activities of these two reporters in plants (Fig. 3E and Fig. 4, G and H); while the mutated ABRE promoters had activity that was strongly distinguishable from the wild-type SPs (Fig. 5B).
The distinguishable activity of the two ABRE isoforms in the Y1H assay suggested that the flanking sequence is important for defining the strength of interactions with TFs, which is consistent with the previous study on DR5 (Ulmasov et al., 1997). Indeed, TFs such as ABF2 (Kim et al., 2004) or SRS7 (Zawaski et al., 2011) showed differential ability to activate reporter gene expression in yeast depending on whether an ABI1-type or RD29A-type isoform was used (Fig. 5C; Supplemental Table S3). We specifically tested the effect of the flanking sequence by swapping the 3 bp immediately 3′ of the ABRE core sequence between the 6xABRE_R and 6xABRE_A SP isoforms (Fig. 1). PCA of SP activity showed that this 3-bp swap was sufficient to interconvert the TF recruitment activity of the two isoforms, which is also evident from the specific recruitment of ABF2 and SRS7 (Fig. 5, B and C; Supplemental Table S3). The 3-bp flanking sequence 3′ of 6xABRE_A bears similarity to the drought-responsive element-like motif GTCGAC (Yamaguchi-Shinozaki and Shinozaki, 1994; Fig. 1), which is known to act synergistically with the ABRE. Our Y1H data suggest, however, that this drought-responsive element-like motif may also have effects on the recruitment of TFs belonging to other gene families (Supplemental Table S2). This 3-bp flanking sequence showed relatively low conservation in other crucifers compared with Arabidopsis, especially the RD29A type (Fig. 5D), whereas it was highly intraspecifically conserved among the 171 Arabidopsis accessions that we analyzed (Fig. 5E).
Consistent with the greater ability of ABF2 to regulate gene expression through ABRE_A isoforms in yeast, reporter assays in planta showed that the 6xABRE_A SP reporter was able to confer greater responsiveness to stresses (Supplemental Fig. S6, F and I), conditions under which ABF2 is known to regulate gene expression (Kim et al., 2004; Fujita et al., 2005). Consistent with these results, gene expression data from the AtGenExpress consortium revealed that the ABI1 gene was transcriptionally induced by ABA and NaCl treatment more rapidly than RD29A (Supplemental Fig. S9; Kilian et al., 2007). Furthermore, the greater responsiveness of the 6xABRE_A:erGFP reporter under stress conditions provide additional evidence that the ABI1 isoform of the 6xABRE confers enhanced ABA sensitivity.
Developmental and DNA Damage Response Pathways Impinge on ABRE-Dependent Expression
The ABA-independent expression of the 6xABRE SP reporter in the QC and SCN suggests that other TFs, besides the canonical bZIP-type TFs, may regulate expression through the 6xABRE SP (Fig. 5A; Supplemental Table S4). WUSCHEL (WUS), a transcriptional regulator expressed in the shoot apical meristem in a domain functionally homologous with the QC (Laux et al., 1996; Mayer et al., 1998), was also identified as an ABRE-interactor in our Y1H screen. While WUS is not expressed in roots, the closely related gene WOX5 is specifically expressed in the root QC cells and is functionally interchangeable with WUS (Sarkar et al., 2007; Zhou et al., 2015). WOX5 could not have been identified in our original Y1H screens, as it is not present in the set of TFs used in the screen.
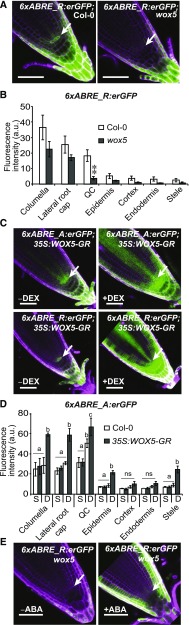
We found that the QC-specific expression of the 6xABRE SP reporter is lost in a wox5 mutant (Sarkar et al., 2007), while other domains of activity are preserved (Fig. 6, A and B). This result is unlikely to be an indirect effect of a change in cell identity, since several other QC markers maintain their expression in the wox5 mutant (Sarkar et al., 2007). Seedlings carrying a DEX-inducible 35S:WOX5-GR construct exhibited strong induction of the 6xABRE reporters throughout the root (Fig. 6, C and D). Consistent with these findings, transient transfection assays in protoplasts showed that the RD29A gene and the 6xABRE_R SP reporter were induced upon expression of WOX5, similar to the effects of ABF3 expression (Supplemental Fig. S10, A–D). Importantly, loss of QC and SCN expression in wox5 was not rescued by exogenous ABA treatment (Fig. 6E), indicating that ABA cannot act independently of WOX5 to induce expression within the QC. WUS was previously shown to bind G-box-like sequences that bear similarity to the ABRE motif (Busch et al., 2010), suggesting WOX5 and WUS regulate meristem-associated gene expression programs through a similar target sequence. One of the direct targets of WOX5 is CYCD3;3, which is repressed in the QC to prevent cell division (Forzani et al., 2014). Published ChIP-seq data reveal that the upstream promoter region of CYCD3;3, where WOX5 binds, is also bound by ABF1/3/4 (Song et al., 2016; Supplemental Fig. S10E). Together, our results identify WOX5 as a regulator of gene expression in the QC and SCN through the ABRE and highlights a shared regulatory mechanism between stress and development.
Figure 6.
ABRE-dependent gene expression is controlled by WOX5. A and B, Confocal images of root tips of 6XABRE_R SP in Col-0 and wox5 mutant backgrounds and averaged fluorescence quantification of T1 plants (n ≥ 7). Student’s t test significance level, **0.001 < P < 0.01. C and D, Confocal images of 6xABRE_A and 6xABRE_R SP reporter expression in 35S:WOX5-GR background root tips, with or without DEX treatment and average fluorescence quantification of F1 seedlings (n ≥ 11). Standard conditions (S) and DEX treatment (D). Significance test based on ANOVA (P < 0.05), and significance groups were labeled with different letters. ns, no significance. E, ABA effect on WOX5 regulated 6xABRE SP reporter. White arrows point to QC. Rotation of root micrographs to align with the diagonal axis of the figure led to apparent cropping of the image. Error bars, se. Scale bars, 50 μm.
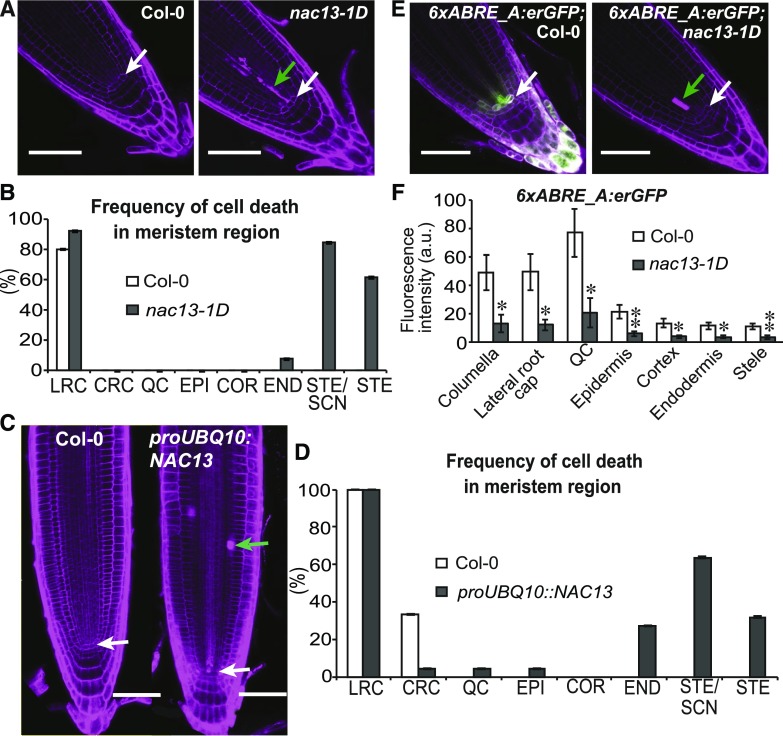
Our Y1H analysis also identified NAC DOMAIN PROTEIN13 (NAC13) as a potential regulator of ABRE-dependent gene expression (Fig. 5A; Supplemental Table S4). Based on published data, NAC13 binds to the promoters of genes that are also bound by ABRE-interacting factors, such as ABF2 (O’Malley et al., 2016; Supplemental Fig. S11A). NAC13 is expressed within the QC and is induced upon DNA damage or associated stresses such as UV irradiation (Supplemental Fig. S11, B and C). We identified a gain-of-function T-DNA mutant allele of NAC13, nac13-1D (Salk_096150C), which caused enhanced expression (Supplemental Fig. S11D) and induced ectopic cell death in the stele, including the stele stem cells (Fig. 7, A and B). Overexpression of NAC13 under the UBQ10 (AT4G05320) promoter also resulted in enhanced cell death, including in other cell types (Fig. 7, C and D), indicating that NAC13 is sufficient to induce programmed cell death (PCD). Cell death was detected using PI (propidium iodide), which exclusively stains the wall when plant cells are alive but stains the interior of the cell after death (Fig. 7; Fulcher and Sablowski, 2009). To be noted, the intense staining of the outer cell layer of the meristem (lateral root cap; Fig. 7, A and C) may be due to the unique composition of the wall when the staining is localized to the wall or due to the normal process of programmed cell death that occurs in this tissue layer, when the interior of the cell is stained. Consistent with our results, NAC13 physically interacts with RADICAL INDUCED CELL DEATH1, which promotes PCD and displays defects in QC development when mutated (O’Shea et al., 2015). Interestingly, the nac13-1D allele suppresses expression of the ABRE reporter (Fig. 7, E and F) as well as the ABRE-containing gene ABI1 (Supplemental Fig. S11E), showing that NAC13 may act as a transcriptional repressor in this context.
Figure 7.
ABRE-dependent gene expression is controlled by NAC13. A, Confocal images of Col-0 and nac13-1D (Salk_096150C) root tips. Dead cells are detected by intense propidium iodide (PI) staining (magenta), green arrow. QC cells, white arrow. B, Frequency of cell death in the lateral root cap (LRC), columella root cap (CRC), QC, epidermis (EPI), cortex (COR), endodermis (ENDO), stele stem cell niche (STE SCN), and distal stele (STE). Five and 13 roots were analyzed for Col-0 and nac13-1D mutant, respectively. C, Confocal images of Col-0 and proUBQ10:NAC13. Dead cells are detected by intense PI staining, green arrow. QC cells, white arrow. D, Frequency of cell death in different cell types of root tip. Abbreviations are the same as those in B. A total of 6 and 22 roots were analyzed for Col-0 and proUBQ10:NAC13 T2 generation plants, respectively. E and F, Confocal image of 6xABRE_A:erGFP SP reporter expression in Col-0 and nac13-1D mutant background root tips and quantification of T1 plants (n ≥ 6). Student’s t test significance level, **0.001 < P < 0.01. Error bars, se. Scale bars, 50 μm.
DISCUSSION
Tissue-specific changes in gene expression are the most common form of transcriptional regulation in response to environmental stresses (Dinneny et al., 2008; Iyer-Pascuzzi et al., 2011); however, our understanding of how stress-signaling pathways contribute to this regulatory complexity is poor. In this study, extensive characterization of ABRE-based reporters allowed us to provide a deep understanding of the regulatory potential of this sequence in plant tissues. Similar to the auxin response element, which confers response to hormonal and developmental cues, the ABRE confers spatially defined and environmentally regulated patterns of expression that are elicited through diverse upstream signaling pathways. Osmotic stress-induced and ABA-responsive expression demonstrates the efficacy of the 6xABRE reporter as a useful tool for characterizing the spatial pattern of ABA-dependent gene expression. We validated the robustness of this reporter by testing different isoforms and reporter architectures and provided an analysis of promoter activity using genome-scale Y1H screens. Using these approaches, we found that noncanonical ABA-independent pathways can also regulate gene expression through the ABRE, highlighting the ABRE as a regulatory hub. This convergence of ABA-independent and -dependent pathways provides a mechanism to explain how gene expression patterns might exhibit tissue specificity and environmental responsiveness using the same DNA regulatory code. This hypothesis requires further testing, however, particularly to examine the interaction of the noncanonical ABRE-interacting TFs in an endogenous genomic context. Furthermore, determining the precise DNA-binding site preference of these TFs will allow for a better understanding of how the ABRE motif is able to recruit non-bZIP-type TFs.
A Reporter to Study ABA-Mediated Transcriptional Regulation
The 6xABRE reporter generated here provides a tool to study the spatial patterning of stress responses in plants. The initial analysis of the reporter presented here has already revealed important novel patterns of ABA-responsive gene expression. Specific cell types in roots, lateral roots, and the leaf epidermis show enhanced response to ABA treatment suggesting that their capacity to sense or respond to this hormone may be enhanced, relative to neighboring tissues. We also showed that WOX5 is necessary for this enhanced responsivity in the QC, and it will be interesting for future studies to determine what other developmental regulators contribute to the spatial pattern of ABA response elsewhere.
The 6xABRE provides information on the spatial pattern of ABA-mediated transcriptional regulation. The reporter expression pattern observed is robust to the architecture of the reporter, suggesting that it is not an artifact of the specific design features we have chosen. Certainly, it may be that in certain biological contexts, reporters with other synthetic promoter architectures would be more optimal than the 6xABRE predominantly used here; for example, a 3xABRE reporter might provide greater dynamic range when examining responses to elevated levels of exogenous ABA.
The sequence context of the ABRE core motif has a clear effect on the expression level of the associated reporter. This is likely an unavoidable consequence of the impact that the flanking genomic sequence has on the binding of TFs to the core motif, since TFs such as ABF2 are influenced by these sequence differences. Our comparison of the 6xABRE_A and 6xABRE_R provides information on how sequence context affects the pattern of activity in planta and in yeast. While the pattern of expression was similar between these two isoforms, overall responsiveness to ABA and osmotic stress varied substantially. Such differences were consistent across all assays, suggesting an overall lower ability of the 6xABRE_R to recruit TFs involved in the ABA response, which is consistent with the reduced ability of ABF2 to act through this sequence in yeast. The 6xABRE_A is likely the most useful for future studies focused on the function of ABA signaling in mediating a transcriptional response to environmental cues.
WOX5 Promotes ABA Responsiveness in the SCN
The longevity of root growth is dependent on the maintenance of the SCN, which feeds the meristem and root cap with a continuous supply of daughter cells. Environmental stress can cause the production of stress hormones such as ethylene, ABA, and brassinosteroids, which affect the division rates of cells within the niche (Ortega-Martínez et al., 2007; Zhang et al., 2010; Vilarrasa-Blasi et al., 2014). Upon DNA damage, the stem cells that surround the quiescent center undergo PCD, while the QC cells proliferate to regenerate the niche after a period of recovery (Fulcher and Sablowski, 2009). Suppression of cell division within the QC is thought to be important for allowing these cells to avoid the effects of DNA-damaging chemicals and environmental stimuli. Upon recovery, however, such quiescence must be released to allow for re-establishment of the niche.
Previous work has shown that ABA is necessary to promote two QC-associated functions: suppression of cell cycle activity in the QC and suppression of differentiation in the neighboring stem cell layers (Zhang et al., 2010). ABA performs the latter function together with WOX5, with neither alone being sufficient to suppress differentiation. In the work we described here, we found that the ABRE CRE is an important integrator for these two pathways. The activity of the ABRE is dependent on WOX5 and ABA independent under nonstress conditions. ABA treatment can induce expression within the QC and SCN; however, these effects are dependent on WOX5.
We propose a model in which the sensitivity of cells to ABA is facilitated by developmental regulators such as WOX5. The case may be similar in the leaf epidermis, where entry into the guard cell lineage leads to enhanced ABA responsiveness, perhaps as a consequence of SPCH activity. Part of the molecular mechanism involved in sensitizing cells to ABA may involve the direct interaction of proteins such as WOX5 with promoters containing ABREs and changes in chromatin state (Sang et al., 2012; Pi et al., 2015). ABA-mediated induction of the 6xABRE reporter expression is WOX5 independent in the epidermis and root cap and may rely upon other developmental regulators or the epidermal-enriched expression of ABA-dependent TFs such as ABF3.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was the parent strain transformed with all synthetic promoter reporter constructs, the proABF3:erGFP reporter, and full-length and mutated promoters of the RD29A erGFP reporter, UBQ10:NAC13 and 35S:WOX5-GR. Mutants of nac13-1D (Salk_096150C) and wox5-1 (SALK_038262; Sarkar et al., 2007) are in the Col-0 background, and the abi1-1 mutant is in the Landsberg eracta (Ler) background.
Growth Conditions
Seeds were surface sterilized, and seedlings were grown as previously described (Geng et al., 2013). For salt treatment and hormone treatments, 5-d-postgermination seedlings were transferred to standard media supplemented with various concentrations of sodium chloride (NaCl; Sigma-Aldrich), mannitol (Sigma-Aldrich), ABA (Sigma-Aldrich), 10 μm fluridone (Sigma-Aldrich), and 1 μm DEX (Sigma-Aldrich). Germination and growth of seedlings was performed in a Percival CU41L4 incubator at a constant temperature of 22°C with long-day lighting conditions (16 h light and 8 h dark). Petri plates were sealed with two layers of Parafilm (Alcan Packaging).
Bioinformatic Analysis: Sequence Variation in Genomic Sequences Flanking the ABRE
SNP information was obtained for 171 Arabidopsis genotypes sequenced by the Salk Institute for the 1001 Genomes Project (1001 Genomes Consortium, 2016). Allele frequency for each genomic position was counted using a customized script. Promoter regions of seven crucifer species in Lineage I and II (Haudry et al., 2013) were searched for the ABRE consensus sequences tested in this study. Alignments were generated using Muscle (Edgar, 2004) and further manually adjusted.
Synthetic Promoter Pesign
For all constructs, unless specifically indicated, a 30-bp window of sequence was used and multimerized six times. SPs containing three, six, or nine repeats of 25 or 30 bp were used (Supplemental Table S1). Mutation of the ABRE SP was performed as previously described (Hattori et al., 2002). Multimerized sequences were fused upstream of an MP to generate core promoter sequences. A modified fragment of the CaMV 35S domain A was used (−90 to −1 bp; Benfey and Chua, 1990; Lam et al., 1990) as our MP. The RD29A MP sequence (−54 bp to +96 bp region; Narusaka et al., 2003) and NOS MP (−101 bp to +4 bp; Michael et al., 2008) were also tested. Promoter sequences were synthesized using gene synthesis services (GeneArt, Thermo Fisher, and Genscript).
Construct Generation
A Multisite Gateway cloning strategy was used for all GUS reporter genes generated in this study, while the Golden Gate cloning strategy was used to generate reporter genes with erGFP (Engler et al., 2009; Emami et al., 2013). The GUS coding sequence was amplified from the pBI101 vector and cloned into the D-TOPO vector that contains the L1/L2 Gateway recombination sites (Invitrogen). For Golden Gate cloning, the pSE7 binary vector was used together with pCR-BluntII-TOPO-MP and pCR-BluntII-TOPO-erGFP. For Y1H assays, the SP sequences were directly cloned into the pY1-PGA59(MCS) vector (Bonaldi et al., 2017) with BsaI-mediated Golden Gate cloning (Emami et al., 2013). All of the designed sequences are available in Supplemental Table S1.
The promoter of the ABF3 gene was amplified by PCR from genomic DNA introducing BsaI sites and cloned into the pCR-BluntII TOPO vector, while for cloning the RD29A promoter, SapI sites were added. Golden Gate cloning was used to generate transcriptional fusions between the promoter and erGFP sequences within pSE7 or pMCY2 (Emami et al., 2013). To mutate the promoter sequences, PCR-based site-directed mutagenesis was used to introduce a 1-bp mutation into the core ABRE sequence, changing it from “ACGTG” to “ACGTT.” All of the primers used are listed in Supplemental Table S6.
To overexpress the NAC13 and WOX5 coding sequences in protoplasts, the UBQ10 promoter or 35S CaMV promoter was cloned upstream of these coding sequences using the same Golden Gate cloning strategy described above. All coding sequences were amplified from cDNA synthesized from Col-0 plant mRNA. All of the primers used are listed in Supplemental Table S6.
Fluorescence Microscopy
For confocal microscope imaging, roots were stained in 10 μg/mL PI (dissolved in water) solution for up to 5 min prior to imaging, except when roots were grown on media supplemented with NaCl or fluridone where a FM4-64 solution (Invitrogen) was used for staining. A Leica SP5 or SP8 point-scanning confocal microscope was used for imaging. For defining the spatial expression pattern of synthetic promoter reporters, at least 15 T1 transgenic seedlings were imaged; while for all other experiments, at least 10 roots were imaged for each condition and genotype. Reporter activity was quantified using CellSeT (Pound et al., 2012), and significance was determined using a two-tailed Student’s t test with a P value threshold of 0.05 or two-way ANOVA test as indicated in the figure legends.
GUS Analysis
Quantification of GUS reporter expression was performed using Fiji (Schindelin et al., 2012). Images were taken under bright field mode and transformed into 16-bit grayscale, and the mean value of the regions specified in the apical meristem, distant meristem, and maturation regions were measured. The mean value of a region in the root having no GUS signal was also measured to perform a background subtraction normalization.
Y1H Screening and Data Analysis
The integration of SP:LacZ, SP:HIS3, and SP:LUC constructs into the YM4271 yeast genome, selection on medium lacking His and uracil, and autoactivation were performed as previously described (Pruneda-Paz et al., 2009). The transgene-positive yeast strains were used for Y1H screening against the genome-scaled AD-TF library containing about 1,956 transcription factors (Pruneda-Paz et al., 2014), and interactions were monitored using the activity of LacZ or gLUC59. OD600 was measured to quantify yeast growth and luminescence (LUC) or OD420 (β-galactosidase) was measured to quantify reporter gene activity. A minimum OD600 of 0.12 was used as a cutoff before further consideration of the sample. Reporter activity was calculated as follows: (1) each luminescence or OD600 was corrected by subtracting the average luminescence or OD600 background from each plate; (2) reporter activity was calculated as (luminescence [or OD420 for lacZ]*1000)/(OD600 × cell volume × reaction time). Reaction time was designated as 1 when the reporter was luciferase; (3) the relative reporter activation was determined using the empty plasmid control for each 384-well plate and Z-score calculated for all TFs per synthetic promoter (Supplemental Table S3). Z-scores were used for the PCA using MeV (Howe et al., 2010) and network generation with Cytoscape (Shannon et al., 2003).
Transient Assays in Protoplasts
Protoplasts were isolated from the leaf mesophyll of plants homozygous for the 6xABRE_A:GUS or 6xABRE_R:GUS reporters using the PEG-calcium method (Yoo et al., 2007). For ABA treatment, 50 μm ABA was added to the WI solution (0.5 m mannitol, 4 mm MES, and 20 mm KCl) prior to the last incubation step. pSE7 was used as a negative control plasmid for the samples without any TF overexpression. mRNA was isolated from the transformed protoplasts using the RNeasy Plant Mini Kit (Qiagen). cDNA was synthesized using the iScript Supermix cDNA synthesis kit (Bio-Rad). Reverse transcription-quantitative PCR was performed using a Roche LightCycler with SensiFAST SYBR Kit (Bioline). Three biological replicates and two technical replicates were performed, and the expression was normalized to an internal control gene (AT4G37830). The expression level was calculated using the delta CT method (Schmittgen and Livak, 2008).
Root Phenotyping
The nac13-1D mutant (Salk_096150C) has a T-DNA inserted in the promoter region of the NAC13 gene (AT1G32870). Expression level of the associated gene was tested by reverse transcription-quantitative PCR on the same plants as were used for phenotypic analysis (Supplemental Fig. S11D). Seeds of wild-type Col-0, nac13-1D, and UBQ10:NAC13 mutants were germinated and grown on standard medium for 5 d, and the root tips were imaged on a Leica SP5 or SP8 confocal microscope after being stained with 10 μg/mL PI for 5 min. PI staining intensity facilitates the identification of dead cells, as it is membrane impermeable in live cells, whereas the dye incorporates readily into dead cells. The number of plants showing dead cells in the LRC, columella root cap, QC, epidermis, cortex, endodermis, stele in stem cell niche, and stele in maturation zone were quantified, and the percentage of roots exhibiting cell death was calculated.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers ABI1, AT4G26080, NP_194338.1, and NM_118741.3; RD29A, AT5G52310, NP_200044.1, and NM_124610.3; WOX5, AT3G11260, NP_187735.2, and NM_111961.4; NAC13, AT1G32870, NP_001323024.1, and NM_001333020.1; ABF2, AT1G45249, NP_849777.1, and NM_179446.5; ABF3, AT4G34000, NP_001320130.1, and NM_001342246.1; and SRS7, AT1G19790, NP_001031069.1, and NM_001035992.4.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The identification of known elements enriched with spatiotemporal salt responsiveness map in Arabidopsis root tip.
Supplemental Figure S2. Expression pattern of ABRE SPs with GUS reporter.
Supplemental Figure S3. Effects of MP sequence and SP unit repeat number on the expression of ABRE-containing SPs.
Supplemental Figure S4. Quantification strategy for the 6xABRE SP reporters in this study.
Supplemental Figure S5. Mutation analysis of the ABRE core sequence in synthetic promoters.
Supplemental Figure S6. Responsiveness of 6xABRE SP reporters to gradient of ABA and stresses in the primary root tip.
Supplemental Figure S7. ABA does not significantly affect ABRE SP expression in QC.
Supplemental Figure S8. Expression pattern of ABF3.
Supplemental Figure S9. Temporal expression pattern of RD29A and ABI1 under salt and ABA treatment.
Supplemental Figure S10. Potential connection between WOX5 and ABA signaling through ABRE or ABRE contained CYCD3;3.
Supplemental Figure S11. Involvement of NAC13 in regulating ABA-induced genes.
Supplemental Table S1. Information of SP reporters tested in this study.
Supplemental Table S2. Fold induction by TF-AD library on ABRE-bait in yeast one hybrid screening using the LacZ reporter.
Supplemental Table S3. Z-score of induction by TF-AD library on the CRE-bait in the yeast one hybrid screening using the LUC reporter.
Supplemental Table S4. TF-ABRE network obtained from Y1H for Figure 5A.
Supplemental Table S5. TFs showing high affinity with the 30-bp genomic regions used in 6xABRE synthetic promoters.
Supplemental Table S6. Primers used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank members of the Dinneny lab for careful review of this manuscript.
Footnotes
Funding of the portion of this work performed at the Temasek Lifesciences Laboratory was provided by the Singapore National Research Foundation. Research performed at the Carnegie Institution for Science was supported by the Carnegie Institution for Science Endowment Fund and a National Science Foundation grant (MCB-1157895) awarded to J.R.D.
Articles can be viewed without a subscription.
References
- 1001 Genomes Consortium (2016) 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Chua NH (1990) The Cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250: 959–966 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bonaldi K, Li Z, Kang SE, Breton G, Pruneda-Paz JL (2017) Novel cell surface luciferase reporter for high-throughput yeast one-hybrid screens. Nucleic Acids Res 45: e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, et al. (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106 [DOI] [PubMed] [Google Scholar]
- Busch W, Miotk A, Ariel FD, Zhao Z, Forner J, Daum G, Suzaki T, Schuster C, Schultheiss SJ, Leibfried A, et al. (2010) Transcriptional control of a plant stem cell niche. Dev Cell 18: 849–861 [DOI] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, and Benfey PN (2008). Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Duan L, Dietrich D, Ng CH, Chan PMY, Bhalerao R, Bennett MJ, Dinneny JR (2013) Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25: 324–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami S, Yee M-C, Dinneny JR (2013) A robust family of Golden Gate Agrobacterium vectors for plant synthetic biology. Front Plant Sci 4: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Gruetzner R, Kandzia R, Marillonnet S (2009) Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4: e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forzani C, Aichinger E, Sornay E, Willemsen V, Laux T, Dewitte W, Murray JAH (2014) WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Curr Biol 24: 1939–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park S-Y, Cutler SR, Sheen J, Rodriguez PL, Zhu J-K (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Yoshida T, Yamaguchi-Shinozaki K (2013) Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol Plant 147: 15–27 [DOI] [PubMed] [Google Scholar]
- Fulcher N, Sablowski R (2009) Hypersensitivity to DNA damage in plant stem cell niches. Proc Natl Acad Sci USA 106: 20984–20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Wu R, Wee CW, Xie F, Wei X, Chan PMY, Tham C, Duan L, Dinneny JR (2013) A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 25: 2132–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiltinan MJ, Marcotte WR Jr., Quatrano RS (1990) A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250: 267–271 [DOI] [PubMed] [Google Scholar]
- Hattori T, Totsuka M, Hobo T, Kagaya Y, Yamamoto-Toyoda A (2002) Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol 43: 136–140 [DOI] [PubMed] [Google Scholar]
- Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ, Forczek E, Joly-Lopez Z, Steffen JG, Hazzouri KM, et al. (2013) An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet 45: 891–898 [DOI] [PubMed] [Google Scholar]
- Hernandez-Garcia CM, Finer JJ (2014) Identification and validation of promoters and cis-acting regulatory elements. Plant Sci 217-218: 109–119 [DOI] [PubMed] [Google Scholar]
- Howe E, Holton K, Nair S, Schlauch D, Sinha R, Quackenbush J (2010) MeV: MultiExperiment Viewer. In Ochs MF, Casagrande JT, Davuluri RV, eds, Biomedical Informatics for Cancer Research. Springer US, Boston, MA, pp 267–277 [Google Scholar]
- Humplík JF, Bergougnoux V, Van Volkenburgh E (2017) To stimulate or inhibit? That is the question for the function of abscisic acid. Trends Plant Sci 22: 830–841 [DOI] [PubMed] [Google Scholar]
- Iyer-Pascuzzi AS, Jackson T, Cui H, Petricka JJ, Busch W, Tsukagoshi H, Benfey PN (2011) Cell identity regulators link development and stress responses in the Arabidopsis root. Dev Cell 21: 770–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Foster R, Chua NH (1993) Plant bZIP protein DNA binding specificity. J Mol Biol 230: 1131–1144 [DOI] [PubMed] [Google Scholar]
- Jones AM, Danielson JA, Manojkumar SN, Lanquar V, Grossmann G, Frommer WB (2014) Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife 3: e01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kim S, Kang JY, Cho DI, Park JH, Kim SY (2004) ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J 40: 75–87 [DOI] [PubMed] [Google Scholar]
- Lam E, Kano-Murakami Y, Gilmartin P, Niner B, Chua NH (1990) A metal-dependent DNA-binding protein interacts with a constitutive element of a light-responsive promoter. Plant Cell 2: 857–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Davies KA, Chang J, Adrian J, Rowe MH, Ballenger CE, Bergmann DC (2014) Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 345: 1605–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jürgens G (1996) The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122: 87–96 [DOI] [PubMed] [Google Scholar]
- Leprince O, Pellizzaro A, Berriri S, Buitink J (2017) Late seed maturation: drying without dying. J Exp Bot 68: 827–841 [DOI] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al. (2008) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy J, Yamaguchi-Shinozaki K, Chua NH (1990) Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc Natl Acad Sci USA 87: 1406–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S, Hauser F, Park J, Waadt R, Brandt B, Schroeder JI (2015) Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr Opin Plant Biol 28: 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Kawaide H, Kamiya Y, Naito S (1998) Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol 39: 853–858 [DOI] [PubMed] [Google Scholar]
- Narusaka Y, Narusaka M, Seki M, Fujita M, Ishida J, Nakashima M, Enju A, Sakurai T, Satou M, Kamiya A, et al. (2003) Expression profiles of Arabidopsis phospholipase A IIA gene in response to biotic and abiotic stresses. Plant Cell Physiol 44: 1246–1252 [DOI] [PubMed] [Google Scholar]
- Negin B, Moshelion M (2016) The evolution of the role of ABA in the regulation of water-use efficiency: From biochemical mechanisms to stomatal conductance. Plant Sci 251: 82–89 [DOI] [PubMed] [Google Scholar]
- O’Connor TR, Dyreson C, Wyrick JJ (2005) Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413 [DOI] [PubMed] [Google Scholar]
- O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR (2016) Cistrome and Epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea C, Kryger M, Stender EGP, Kragelund BB, Willemoës M, Skriver K (2015) Protein intrinsic disorder in Arabidopsis NAC transcription factors: transcriptional activation by ANAC013 and ANAC046 and their interactions with RCD1. Biochem J 465: 281–294 [DOI] [PubMed] [Google Scholar]
- Ortega-Martínez O, Pernas M, Carol RJ, Dolan L (2007) Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317: 507–510 [DOI] [PubMed] [Google Scholar]
- Park S-Y, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, Groot E, Laux T (2015) Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev Cell 33: 576–588 [DOI] [PubMed] [Google Scholar]
- Pound MP, French AP, Wells DM, Bennett MJ, Pridmore TP (2012) CellSeT: novel software to extract and analyze structured networks of plant cells from confocal images. Plant Cell 24: 1353–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA (2009) A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Nagel DH, Kang SE, Bonaldi K, Doherty CJ, Ravelo S, Galli M, Ecker JR, Kay SA (2014) A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell Reports 8: 622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Sah SK, Reddy KR, Li J (2016) Abscisic acid and abiotic stress tolerance in crop plants. Front Plant Sci 7: 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H, Araki T, Meshi T, Iwabuchi M (2000) Expression of a subset of the Arabidopsis Cys(2)/His(2)-type zinc-finger protein gene family under water stress. Gene 248: 23–32 [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol 136: 2734–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Silva-Ortega CO, Wu S, Yamaguchi N, Wu M-F, Pfluger J, Gillmor CS, Gallagher KL, Wagner D (2012) Mutations in two non-canonical Arabidopsis SWI2/SNF2 chromatin remodeling ATPases cause embryogenesis and stem cell maintenance defects. Plant J 72: 1000–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Seo M, Koshiba T (2002) Complex regulation of ABA biosynthesis in plants. Trends Plant Sci 7: 41–48 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Ho TH (1995) Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 7: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriver K, Olsen FL, Rogers JC, Mundy J (1991) cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc Natl Acad Sci USA 88: 7266–7270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit ME, Weijers D (2015) The role of auxin signaling in early embryo pattern formation. Curr Opin Plant Biol 28: 99–105 [DOI] [PubMed] [Google Scholar]
- Song L, Huang SC, Wise A, Castanon R, Nery JR, Chen H, Watanabe M, Thomas J, Bar-Joseph Z, Ecker JR (2016) A transcription factor hierarchy defines an environmental stress response network. Science 354: aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Nose T, Jikumaru Y, Kamiya Y (2013) ABA inhibits entry into stomatal-lineage development in Arabidopsis leaves. Plant J 74: 448–457 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ (1995) Composite structure of auxin response elements. Plant Cell 7: 1611–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil V, Marcotte WR Jr., Rosenkrans L, Cocciolone SM, Vasil IK, Quatrano RS, McCarty DR (1995) Overlap of Viviparous1 (VP1) and abscisic acid response elements in the Em promoter: G-box elements are sufficient but not necessary for VP1 transactivation. Plant Cell 7: 1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter M. (2007) Synthetic promoters: genetic control through cis engineering. Trends Plant Sci 12: 118–124 [DOI] [PubMed] [Google Scholar]
- Verslues PE. (2016) ABA and cytokinins: challenge and opportunity for plant stress research. Plant Mol Biol 91: 629–640 [DOI] [PubMed] [Google Scholar]
- Vilarrasa-Blasi J, González-García M-P, Frigola D, Fàbregas N, Alexiou KG, López-Bigas N, Rivas S, Jauneau A, Lohmann JU, Benfey PN, et al. (2014) Regulation of plant stem cell quiescence by a brassinosteroid signaling module. Dev Cell 30: 36–47 [DOI] [PubMed] [Google Scholar]
- Waadt R, Hitomi K, Nishimura N, Hitomi C, Adams SR, Getzoff ED, Schroeder JI (2014) FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. eLife 3: e01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1993) Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236: 331–340 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zawaski C, Kadmiel M, Ma C, Gai Y, Jiang X, Strauss SH, Busov VB (2011) SHORT INTERNODES-like genes regulate shoot growth and xylem proliferation in Populus. New Phytol 191: 678–691 [DOI] [PubMed] [Google Scholar]
- Zhang H, Han W, De Smet I, Talboys P, Loya R, Hassan A, Rong H, Jürgens G, Paul Knox J, Wang M-H (2010) ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J 64: 764–774 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, Tarr PT, Yan A, Kay SA, Meyerowitz EM (2015) Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517: 377–380 [DOI] [PMC free article] [PubMed] [Google Scholar]