Hypertrophy of the shoot meristem and increased shoot regeneration capacity in the amp1 mutant depend on the enhanced expression of RAP2.6L, a direct target of miRNA-regulated HD-ZIP III transcription factors in Arabidopsis thaliana.
Abstract
Plants show an indeterminate mode of growth by the activity of organ forming stem cell niches in apically positioned meristems. The correct formation and activity of these meristems are a prerequisite for their adaptive development and also allow the maintenance of organogenesis under adverse circumstances such as wounding. Mutation of the putative Arabidopsis (Arabidopsis thaliana) Glu carboxypeptidase ALTERED MERISTEM PROGRAM1 (AMP1) results in Arabidopsis plants with enlarged shoot apical meristems, supernumerary stem cell pools, and higher leaf formation rate. AMP1 deficiency also causes exaggerated de novo formation of shoot meristems. The activity of AMP1 has been implicated in the control of microRNA (miRNA)-dependent translation; however, it is not known how this function contributes to the shoot meristem defects. Here, we show that the transcription factor RAP2.6L is upregulated in the Arabidopsis amp1 mutant. Overexpression of RAP2.6L in the wild type causes amp1 mutant-related phenotypic and molecular defects, including enhanced shoot regeneration in tissue culture. Conversely, inhibition of RAP2.6L in the amp1 mutant suppresses stem cell hypertrophy and the regenerative capacity. We further provide evidence that RAP2.6L is under direct transcriptional control of miRNA-regulated class III homeodomain-Leu zipper (HD-ZIP III) proteins, key regulators of shoot meristem development, which overaccumulate in the amp1 mutant. Our results reveal a transcription factor module acting downstream of AMP1 in the control of shoot stem cell niche patterning. By positioning the HD-ZIP III/RAP2.6L module downstream of AMP1 function, we provide a mechanistic link between the role of AMP1 in miRNA-mediated translational repression and shoot stem cell specification.
A key requirement for morphogenesis and tissue homeostasis in multicellular organisms is the correct spatiotemporal control of differentiation in the progeny of stem cell populations. Disturbances in this process cause developmental and regenerative defects and are also the basis for cancerous transformation. Plants harbor stem cell niches (SCNs) in apical meristems, which are responsible for the continuous formation of new organs during postembryonic development. This indeterminate mode of growth allows plants to steadily uptake resources and adapt to prevailing environmental conditions. The dome-shaped shoot apical meristem (SAM), which is the source of all shoot organs, consists of different functional zones. Stem cells in the central zone give rise to daughter cells, which undergo transition to a determinate cell fate during the process of organ initiation in the ring-like periphery, called the morphogenetic zone (Sluis and Hake, 2015). Maintenance of this spatial organization is a prerequisite for meristem function and requires the formation of cell identity borders through feedback-regulated communication events (Gaillochet and Lohmann, 2015). Comprehensive characterization of the underlying signaling not only contributes to a mechanistic understanding of plant organogenesis but might also unveil molecular targets to optimize yield traits in crop species (Je et al., 2016).
The stem cell niche in the shoot meristem consists of an apical stem cell pool and an underlying organizing center (OC). The identity of the OC is specified by the expression of the homeodomain transcription factor WUSCHEL (WUS; Mayer et al., 1998). WUS enters the stem cells by symplastic movement to maintain their identity as well as to activate the transcription of CLV3 (Yadav et al., 2011; Daum et al., 2014). The peptide CLV3 in turn diffuses back to more basal regions of the meristem and limits lateral expansion of WUS expression (Brand et al., 2000; Schoof et al., 2000). The position of the WUS expression domain is determined by the establishment of a local response maximum for the hormone cytokinin (Gordon et al., 2009; Chickarmane et al., 2012). Multilayered feedback control between cytokinin, WUS, and CLV3 thus guarantees the dynamic repositioning and size adjustment of the SCN in the shoot meristem (Gaillochet and Lohmann, 2015).
Upon entering the peripheral zone, stem cell descendants undergo transient proliferation and are subsequently incorporated into organ primordia. Suppression of stemness in these cells is a key prerequisite to maintaining the radial integrity of the meristem and is mediated by epigenetic mechanisms since mutants in chromatin assembly and remodeling have been shown to display reactivation of stem cell markers in the morphogenetic zone (Gaillochet and Lohmann, 2015). This process also requires active signaling from the existing SCN as elimination of the primary OC results in the formation of secondary SCNs in the meristem periphery (Loiseau, 1959; Reinhardt et al., 2003). However, the molecular basis of this lateral inhibition mechanism is not known. There is growing evidence that class III homeodomain-Leu zipper (HD-ZIP III) transcription factors contribute to the radial organization of the SAM. HD-ZIP III proteins define apical central identity and proper shoot SCN positioning in the embryo (Roodbarkelari and Groot, 2017) and altered HD-ZIP III activity causes the formation of ectopic stem cell pools in the shoot meristem periphery (Green et al., 2005; Williams et al., 2005; Kim et al., 2008). The function of HD-ZIP III proteins is controlled at several regulatory levels by micro RNAs (miRNAs), microProteins, and potentially also unidentified small molecule signals (Roodbarkelari and Groot, 2017). Moreover, their activity is also modulated by heterodimerization between family members and interaction with other transcription factors (Chandler et al., 2007; Magnani and Barton, 2011). This high level of functional connectivity predestines HD-ZIP IIIs as convergence hubs to integrate spatiotemporal information in the process of SAM patterning. However, how they execute this role at the molecular level is not well known.
Another key regulator of radial SAM organization is the putative Glu carboxypeptidase ALTERED MERISTEM PROGRAM1 (AMP1; Helliwell et al., 2001). Inactivation of this enzyme causes vegetative SAM enlargement and an increased leaf formation rate, which originate at least partially from a strong tendency to generate ectopic SCNs in the SAM periphery (Huang et al., 2015). The observed disintegration of the radial SAM in the Arabidopsis (Arabidopsis thaliana) amp1 mutant could not be attributed to the enhanced cytokinin synthesis found in the mutant and indicated the involvement of a cytokinin-independent mechanism (Huang et al., 2015). Although the biochemical function of AMP1 is not known, it was recently shown to be required for miRNA-mediated translational inhibition at the rough endoplasmic reticulum (Li et al., 2013). However, whether the radial formation of ectopic SCNs in the amp1 mutant is the consequence of overtranslation of miRNA targets involved in SAM control, such as HD-ZIP III transcription factors, remains to be tested.
A key function of AMP1 in the suppression of shoot stem cell identity is also underlined by the high de novo shoot formation capacity of AMP1-defective plants (Chaudhury et al., 1993). Shoot regeneration in tissue culture usually implicates the formation of pluripotent callus cells in explants on auxin-rich callus-induction medium. Callus cells, which have root meristem-like identity, transdifferentiate into WUS-expressing foci on cytokinin-rich shoot induction medium, which subsequently establish functional shoot meristems (Ikeuchi et al., 2016). A number of transcription factors that mediate this transdifferentiation process have been identified, including HD-ZIP III proteins (Kareem et al., 2016). A point mutation affecting the MEHKLA domain of ATHB15/CORONA was reported to trigger shoot regeneration in a cytokinin-independent manner (Duclercq et al., 2011a), and HD-ZIP IIIs directly activate WUS expression in a complex with B-type ARABIDOPSIS RESPONSE REGULATORs during de novo SAM formation (Zhang et al., 2017). Transcription factors belonging to the APETALA2/Ethylene Responsive Factor (AP2/ERF) family also play a prominent role in this process (Ikeuchi et al., 2016). Whereas ENHANCER OF SHOOT REGENERATION1/DORNRÖSCHEN (ESR1/DRN) and ESR2/DRN-LIKE (DRNL) are activated by and subsequently mediate the cytokinin effect (Banno et al., 2001; Matsuo et al., 2011), RAP2.6L has been shown to be required for shoot regeneration in tissue culture without being transcriptionally regulated by cytokinin under these conditions (Che et al., 2006).
To better understand the function of AMP1 in SCN establishment, we screened transcriptome data of the amp1 mutant for potential components acting downstream of the enzyme and identified RAP2.6L to be strongly upregulated in the mutant. Ectopic expression of RAP2.6L in the wild type caused mild amp1-related SAM phenotypes, including an increased meristem size, an elevated leaf formation rate, and altered expression of SAM markers, whereas perturbation of RAP2.6L function in the amp1 mutant suppressed the SAM hypertrophy as well as the increased shoot regeneration capacity. We further show that RAP2.6L is transcriptionally controlled by HD-ZIP III proteins, which directly bind to the RAP2.6L promoter. Based on our findings, we present a model in which AMP1 limits SAM activity by constraining the expression of RAP2.6L through miRNA-dependent translational control of HD-ZIPIII transcription factors.
RESULTS
Expression of RAP2.6L Is Upregulated in the amp1 Mutant
In an effort to better understand the molecular role of AMP1 in shoot meristem function, we screened our recently published amp1 transcriptome data for misregulated factors reported to play a role in SAM patterning and/or activity (Poretska et al., 2016). In this analysis, we found the AP2/ERF transcription factor RAP2.6L to be significantly upregulated in the amp1-13 mutant compared to wild type. Since RAP2.6L is required for shoot regeneration (Che et al., 2006) and the amp1 mutant shows ectopic OC formation in the shoot meristem periphery (Huang et al., 2015), we further investigated the functional relationship between these two factors. To confirm the altered expression of RAP2.6L in the amp1 mutant, we performed reverse transcription-quantitative PCR (qPCR) analysis in the amp1-13 mutant and the amp1 lamp1 double mutant, in which the putative paralog LIKE-AMP1 (LAMP1) is also mutated. RAP2.6L was strongly upregulated in the amp1-13 and amp1-1 lamp1-2 mutants compared to the wild-type Col-0 (Fig. 1A). Similarly, significantly enhanced expression of RAP2.6L was detected in Ler plants bearing the amp1 allele primordia timing (pt; Fig. 1A). Of the closest paralogs of RAP2.6L in the ERF-TF subfamily group X, the transcript levels of RAP2.6 (in the amp1-13 and pt mutants) and ERF112 (in the amp1-13 mutant) were slightly elevated in the amp1 mutant background; however, these changes were not significant under our experimental conditions. (Supplemental Fig. S1, A and B).
Figure 1.
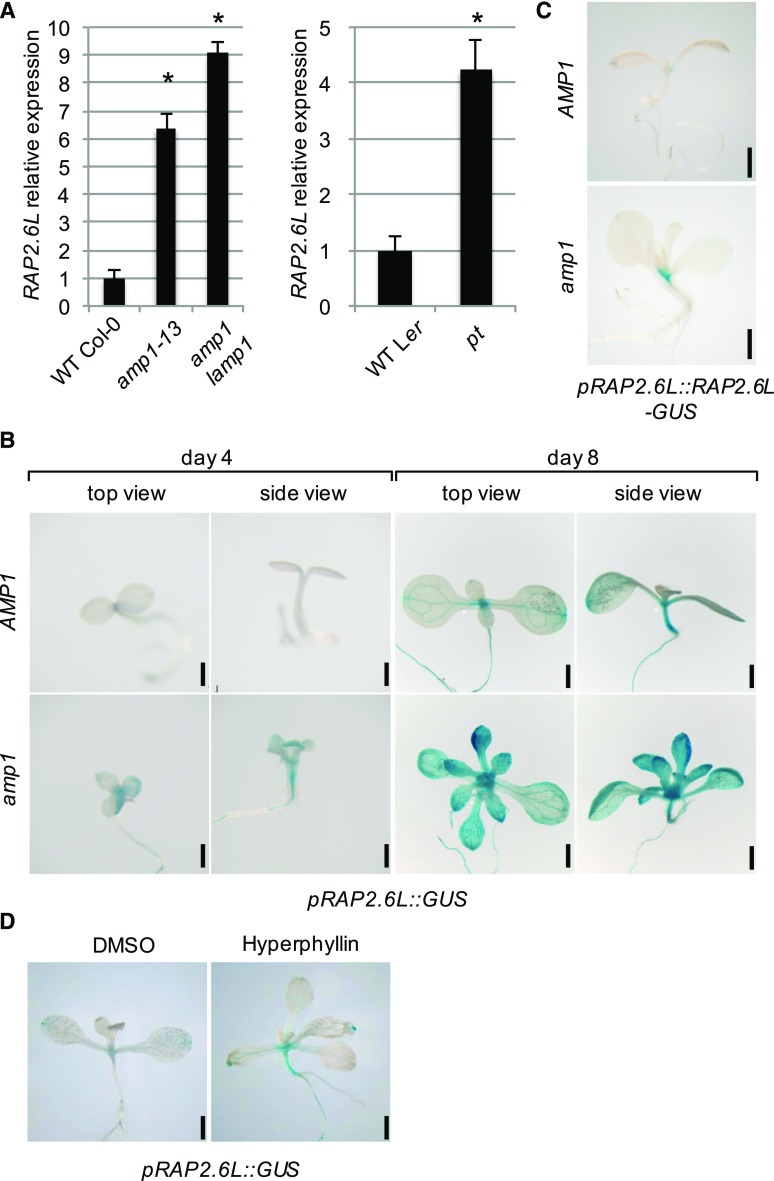
RAP2.6L expression is upregulated in amp1. A, qPCR analysis of RAP2.6L expression in 8-d-old seedlings of the indicated lines. Fold changes compared to the respective wild-type (WT) are shown. Error bars indicate the SE calculated from three biological replicates (at least 30 seedlings per replicate; replicates were grown at the same time on different petri dishes) after normalization to UBIQUITIN-CONJUGATING ENZYME (UBC). Asterisks indicate a significant difference (Student’s two-tailed t test; P < 0.05). B, pRAP2.6L::GUS activity in wild-type Col-0 (WT) and the amp1-1 mutant at 4 and 8 DAG. C, pRAP2.6L::RAP2.6L-GUS activity in wild-type Col-0 (WT) and the amp1-1 mutant at 8 DAG. D, pRAP2.6L::GUS activity in 10-d-old wild-type Col-0 seedlings grown in liquid medium containing either 0.5% DMSO (mock) or 30 μm hyperphyllin. Bars = 1 mm (B–D).
To examine the enhanced expression levels of RAP2.6L in the amp1 mutant at the tissue level, we created the transcriptional GUS reporter pRAP2.6L::GUS. Similar to the RAP2.6L promoter:GUS line (Che et al., 2006), our reporter showed the strongest activity in the shoot apex, the hypocotyl, and the vascular tissues of wild-type seedlings (Fig. 1B), with the single exception that it did not noticeably display the reported expression front in young leaf primordia connected to leaf maturation processes (Che et al., 2006). In 8-d-old amp1-1 seedlings, pRAP2.6L::GUS activity was generally enhanced with the strongest alteration in young leaf primordia, whereas staining was barely visible in the wild-type background (Fig. 1B). This pronounced expression of RAP2.6L in amp1 shoot apices and vascular tissues was already detectable directly after germination in 4-d-old seedlings (Fig. 1B). The elevated transcript levels of RAP2.6L in the amp1 mutant also resulted in an overaccumulation of RAP2.6L since we found a higher activity of pRAP2.6L::RAP2.6L-GUS (Krishnaswamy et al., 2011) in the mutant background compared to the wild type (Fig. 1C). The weak activity of this reporter was only detectable in the areas of strongest expression of the transcriptional pRAP2.6L::GUS reporter, including vascular tissues around the SAM and the base of young leaf primordia. Finally, consistent with an AMP1-dependent control of RAP2.6L expression, pRAP2.6L::GUS activity in the wild type was also induced after treatment with the chemical hyperphyllin (Fig. 1D), which causes amp1-related phenotypic and molecular defects (Poretska et al., 2016).
To test whether the augmented expression of RAP2.6L is caused by the enhanced cytokinin levels previously reported in the amp1 mutant (Nogué et al., 2000), we analyzed the response of pRAP2.6L::GUS activity to short- and long-term treatments with trans-zeatin. None of these exogenous cytokinin treatments affected the pRAP2.6L::GUS staining pattern or intensity in either the wild type or the amp1-1 mutant (Supplemental Fig. S2), indicating that the pronounced expression of RAP2.6L in the amp1 mutant is not caused by the altered homeostasis of this hormone. Taken together, RAP2.6L shows increased and partially ectopic expression in the amp1 mutant in a cytokinin-independent manner, and this altered expression is most prominent in shoot tissues with high AMP1 expression levels such as young leaf primordia (Vidaurre et al., 2007).
Overexpression of RAP2.6L Causes amp1-Related Shoot Phenotypes
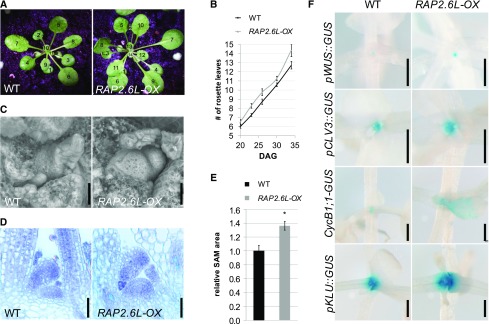
To investigate whether the enhanced expression of RAP2.6L in the amp1 mutant contributes to any of the mutant SAM phenotypes, we analyzed shoot meristem activity parameters in the 35S::RAP2.6L plants (named RAP2.6L-OX throughout this study; Krishnaswamy et al., 2011) grown under short-day conditions. In this line, an obvious increase in true leaf formation became apparent from day 20 on (Fig. 2, A and B). Since the elevated leaf formation in the amp1 mutant correlates with the presence of a hypertrophic SAM, we assessed the meristem dimensions of RAP2.6L-OX seedlings. RAP2.6L-OX SAMs displayed a larger surface area (Fig. 2C) as well as an increased lateral and distal expansion in median longitudinal sections (Fig. 2, D and E). The observed increase in the size of the SAM correlated with enlargement of the OC domain (pWUS::GUS; Fig. 2F) as well as the stem cell pool (pCLV3::GUS; Fig. 2F), which is in its extent comparable to that of early developmental stages of weak amp1 mutant alleles (Huang et al., 2015). Moreover, expression of the mitotic CYCLIN B1;1 (CYCB1;1)::GUS reporter was increased in the shoot meristematic area and in young leaf primordia of the RAP2.6L-OX plants (Fig. 2F), as was found in the faster-forming leaves of the amp1 mutant (Poretska et al., 2016). Notably, we also observed in the RAP2.6L-OX plants a significantly broadened activity of pKLU::GUS, a reporter for the SAM boundary marker CYP78A5/KLUH (Fig. 2F), which controls the rate of leaf formation (Wang et al., 2008) and was prominently upregulated in the amp1 mutant (Helliwell et al., 2001; Griffiths et al., 2011; Poretska et al., 2016).
Figure 2.
RAP2.6L-OX plants show amp1-related vegetative phenotypes. A, Wild-type Col-0 (WT) and RAP2.6L-OX plants grown under short-day conditions for 34 d. Leaves are numbered in the consecutive order of appearance. B, Quantification of rosette leaves in wild-type Col-0 (WT) and RAP2.6L-OX plants at the indicated time points grown under short-day conditions (values represent means ± se; n ≥ 8). C, Scanning electron micrographs of SAM areas of wild-type Col-0 (WT) and RAP2.6L-OX seedlings grown under short-day conditions for 18 d. D, Median longitudinal SAM sections of wild-type Col-0 (WT) and RAP2.6L-OX seedlings grown under short-day conditions for 18 d. E, Quantification of SAM area from median longitudinal sections of wild-type Col-0 (WT) and RAP2.6L-OX seedlings. Normalized values (wild type = 1) are shown (bars represent means ± se; n ≥ 3). Asterisk indicates a significant difference (Student’s two-tailed t test; P < 0.05). F, Comparison of GUS activities of the indicated reporter lines in wild-type and RAP2.6L-OX seedlings grown under short-day conditions. Plants were analyzed at 15 DAG except for pWUS::GUS (18 DAG). Bars = 25 μm (C and D) and 500 μm (F).
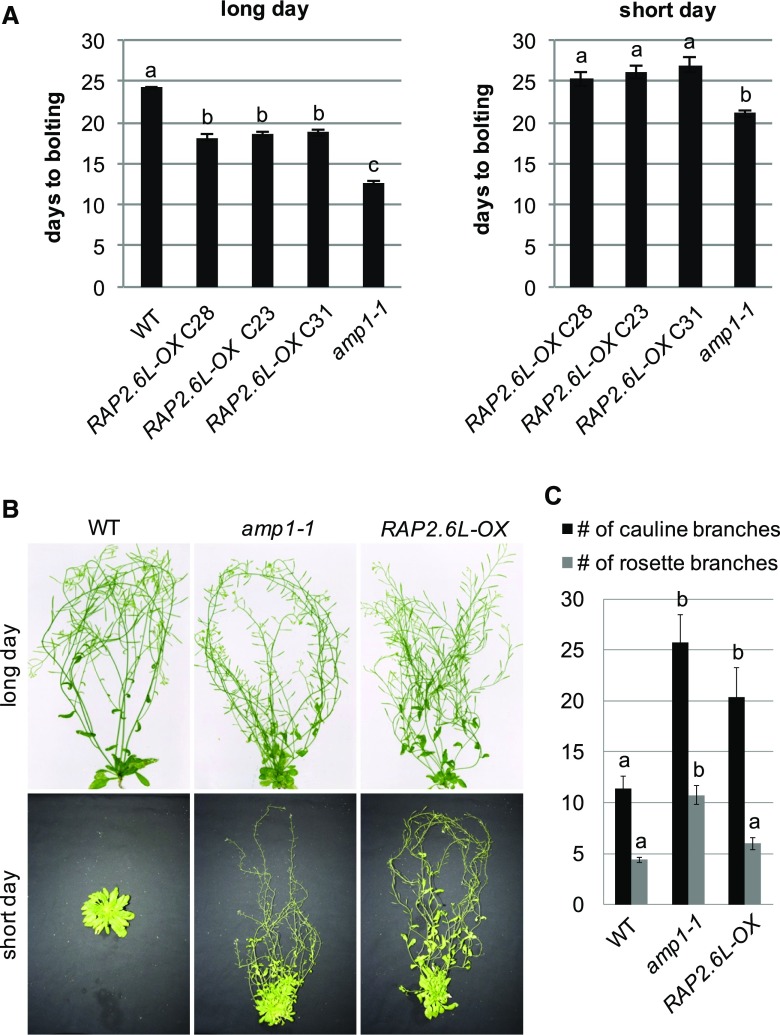
As previously described (Krishnaswamy et al., 2011), RAP2.6L overexpression also provoked a significant reduction in flowering time under long-day conditions, another developmental defect shared with the amp1-1 mutant (Fig. 3A). Moreover, both genotypes underwent floral transition under noninductive short-day conditions in a temporal window of 20 to 27 d after germination (DAG), whereas wild-type plants did not flower under these conditions even at 86 DAG (Fig. 3, A and B). They also produced similar inflorescence structures with a higher number of cauline branches and flower formations starting at a closer distance to the rosette compared to the wild type (Fig. 3, B and C). We conclude that ectopic expression of RAP2.6L causes mild amp1-like phenotypic and molecular alterations in vegetative SAM activity, flowering time, and inflorescence architecture.
Figure 3.
RAP2.6L-OX plants show amp1-related adult phenotypes. A, Quantification of flowering time of the indicated genotypes grown under long-day (left) and short-day conditions (right; wild type not shown since it did not flower in the analyzed period of time). Bars show means ± se; n ≥ 15. Different letters over the error bars indicate significant differences (P < 0.05) as calculated by Duncan’s multiple range test. B, Comparison of wild-type Col-0 (WT), amp1-1, and RAP2.6L-OX shoots grown under long-day (70 DAG) or short-day (86 DAG) conditions. C, Quantification of the rosette branch number and cauline branch number of 70-d-old wild-type Col-0 (WT), amp1-1, and RAP2.6L-OX plants grown under long-day conditions (bars represent means ± se; n ≥ 4). Different letters over the error bars indicate significant differences (P < 0.05) as calculated by Duncan’s multiple range test.
Compromised RAP2.6L Function Suppresses the Enhanced Leaf Formation Rate of the amp1 Mutant
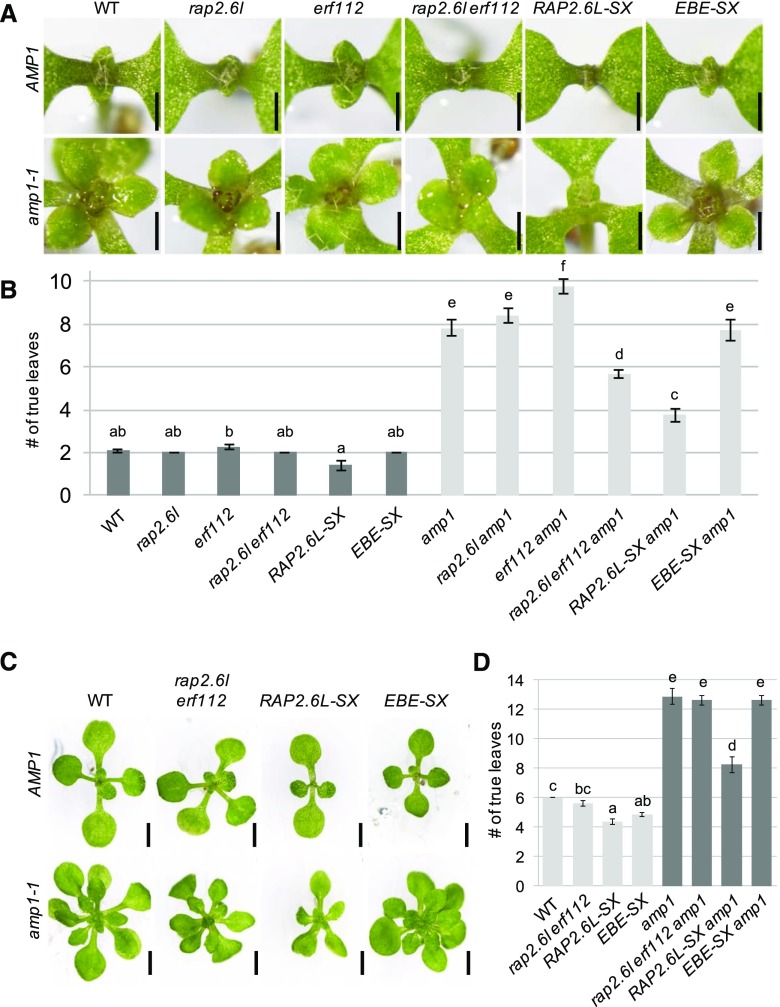
Since the expression of RAP2.6L is deregulated in the amp1 mutant and the RAP2.6L-OX and amp1 plants share similar defects in shoot development, one mode of how AMP1 could restrain SAM activity might be through spatial or temporal limitation of RAP2.6L expression. To this end, we investigated the effect of inactivation of RAP2.6L as well as inactivation of its closest paralogs on shoot meristem function in the wild type and the amp1 mutant. For this purpose, we used T-DNA insertion lines, which were available for RAP2.6L, RAP2.6, and ERF112 (Che et al., 2006). In addition, the transcriptional repression domain SUPERMAN Repression Domain X (SRDX) and the MYC epitope tag were fused to the 35S-driven open reading frames (ORFs) of RAP2.6L and ERF BUD ENHANCER (EBE), resulting in the transgenic lines RAP2.6L-SX and EBE-SX, respectively. Except for a slightly retarded outgrowth of the first pair of true leaves in the loss-of-function single mutant of RAP2.6L, the rap2.6l, rap2.6, and erf112 single mutants did not show a measurable impact on leaf formation in the wild-type or amp1 background (Fig. 4, A and B; Supplemental Fig. S3, A and B). We found a suppressive effect of the rap2.6l erf112 double mutation on the enhanced leaf formation rate of the amp1 mutant in 7-d-old seedlings (Fig. 4, A and B), which disappeared again in later developmental stages (Fig. 4, C and D), whereas the rap2.6l rap2.6 double mutant was aphenotypic in this respect (Supplemental Fig. S3, A and B). The RAP2.6L-SX transgene was even more potent in rescuing the enhanced leaf production of the amp1 mutant at 7 DAG, and this effect persisted throughout the vegetative growth phase. Notably, the relative suppression in leaf number by the RAP2.6L-SX activity was stronger in the amp1 background (7 DAG, 52%; 12 DAG, 36%) than in the wild-type background (7 DAG, 32%; 12 DAG, 27%), supporting a specific genetic interaction rather than an independent effect (Supplemental Fig. S4). Furthermore, although the EBE-SX transgene also affected the leaf formation rate in the wild-type background with increasing efficiency during development, its suppressive effect on the amp1 background was only marginal (Fig. 4, A–D). Taken together, these data suggest that deregulated expression of RAP2.6L in the amp1 mutant contributes to its shortened plastochron.
Figure 4.
Compromised function of RAP2.6L suppresses the enhanced leaf formation rate of the amp1 mutant. A, Shoot apices of Col-0 (WT), rap2.6l-1, erf112-1, rap2.6l-1 erf112-1, RAP2.6L-SX, and EBE-SX seedlings in the wild-type background at 7 DAG (top). Shoot apices of the same lines in the amp1-1 background at 7 DAG (bottom). B, Quantification of the number of true leaves of the indicated genotypes at 7 DAG (bars represent means ± se; n ≥ 10). Bars for plant lines in the wild-type background are colored in dark gray, and bars for lines in the amp1-1 background are colored in light gray. Different letters over the error bars indicate significant differences (P < 0.05) as estimated by Duncan’s multiple range test. C, Comparison of Col-0 (WT), rap2.6l-1 erf112-1, RAP2.6L-SX, and EBE-SX seedlings in the wild-type background at 12 DAG (top) and seedlings of the same lines in the amp1-1 background at 12 DAG (bottom). D, Quantification of the number of true leaves of the indicated genotypes at 12 DAG (bars represent means ± se; n ≥ 8). Bars for plant lines in the wild-type background are colored in light gray, and bars for lines in the amp1-1 background are colored in dark gray. Different letters over the error bars indicate significant differences (P < 0.05) as estimated by Duncan’s multiple range test. Bars = 500 μm (A) and 2 mm (C).
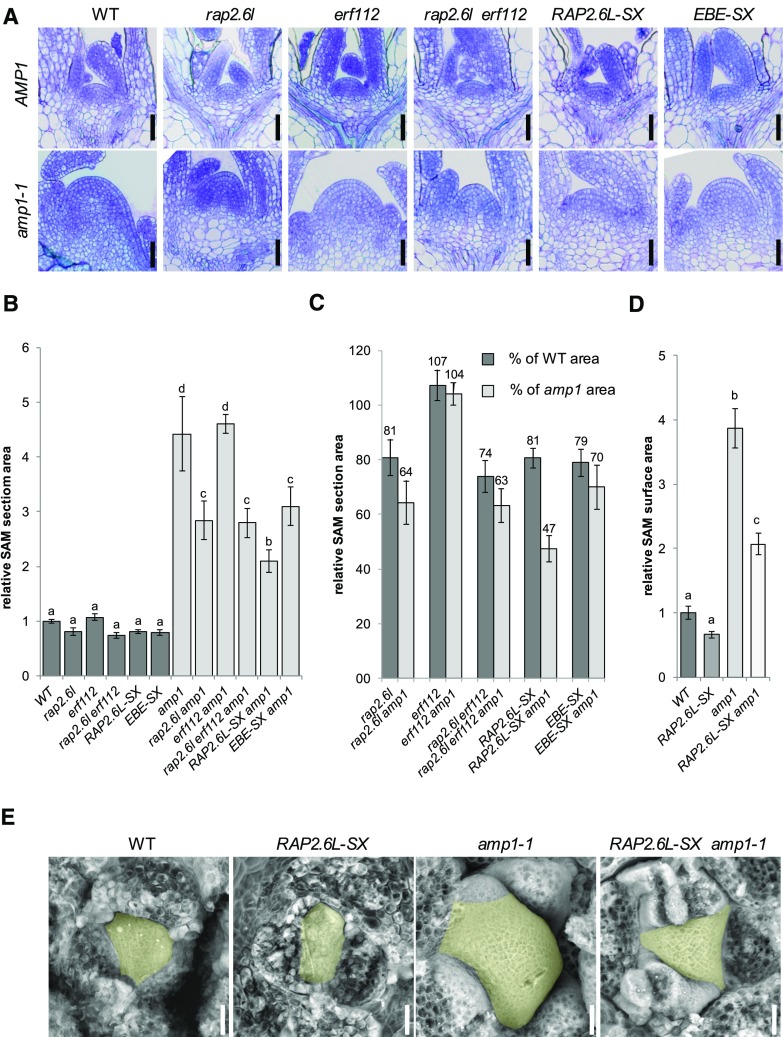
Compromised Function of RAP2.6L Minimizes Overproliferation of the SAM in the amp1 Mutant
To investigate whether the SAM expansion phenotype of the amp1 mutant is also specifically affected by perturbation of the function of RAP2.6L, we analyzed the SAM structures in the generated lines. Determination of the SAM area in median longitudinal shoot sections revealed that all lines except for erf112 showed a slight but not significant reduction in meristem size when in the wild-type background (Fig. 5, A and B). In contrast, the presence of the RAP2.6L-SX transgene reduced the size of the SAM in the amp1 mutant by over 50%. We also found significant, yet less pronounced, suppression of the size of the amp1 mutant SAM in the rap2.6l and rap2.6l erf112 mutants and in the presence of the EBE-SX transgene, whereas the mutation in erf112 was again ineffective in this respect (Fig. 5C). The striking suppressive impact of the RAP2.6L-SX transgene on the meristematic overproliferation of the amp1 mutant was also reflected in a clear reduction of the surface area of the SAM in scanning electron microscopy images, diminishing the visible area from around 400% to 200% compared to wild type (Fig. 5, D and E). As for the analysis of SAM sections, the impact of the RAP2.6L-SX transgene on the surface area of the SAM was only significant in the amp1 mutant, but not in the wild-type background, indicating a specific genetic interaction between RAP2.6L-SX and amp1. Thus, perturbation of the function of RAP2.6L not only suppresses the increased true leaf production of the amp1 mutant but also mitigates its abnormal SAM expansion phenotype.
Figure 5.
Compromised function of RAP2.6L affects SAM size in the amp1 mutant. A, Median longitudinal SAM sections of wild-type Col-0 (WT), rap2.6l-1, erf112-1, rap2.6l-1 erf112-1, RAP2.6L-SX, and EBE-SX plants in the wild-type background at 7 DAG (top) and median longitudinal SAM sections of the same lines in the amp1-1 background at 7 DAG (bottom). B, Quantification of the SAM area from median longitudinal sections of the indicated genotypes at 7 DAG (bars represent means ± se; n ≥ 4). The SAM areas were normalized to that of wild-type Col-0 (wild type = 1). Bars for plant lines in the wild-type background are colored in dark gray, and bars for lines in the amp1-1 background are colored in light gray. Different letters over the error bars indicate significant differences (P < 0.05) as estimated by Duncan’s multiple range test. C, Relative reduction of median SAM section areas by the indicated genotypes compared to wild-type Col-0 (WT) and the amp1-1 mutant based on the data shown in B. Bars represent normalized values ± se; n ≥ 4. Normalized values are shown above the error bars. D, Quantification of SAM surface area from scanning electron micrographs of the indicated genotypes at 7 DAG (bars represent means ± se; n ≥ 3). The SAM areas were normalized to that of wild-type Col-0 (wild type = 1). Different letters over the error bars indicate significant differences (P < 0.05) as estimated by Duncan’s multiple range test. E, Scanning electron micrographs of the SAM of wild-type Col-0 (WT), RAP2.6L-SX, amp1-1, and RAP2.6L-SX amp1-1 seedlings. The SAM areas are highlighted in yellow. Bars = 25 μm (A and E).
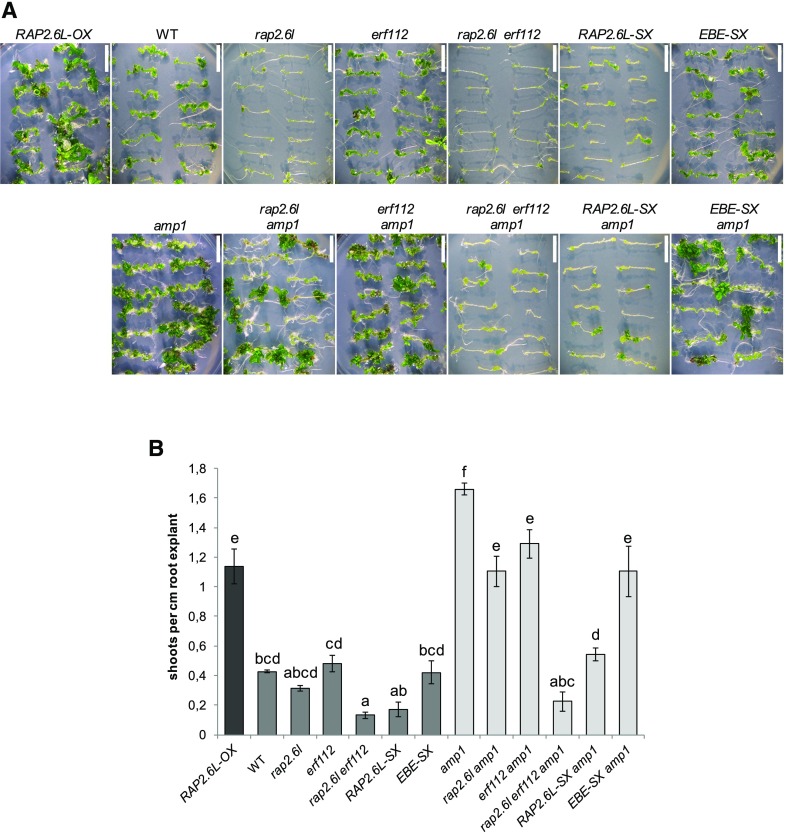
RAP2.6L Mediates the Enhanced Shoot Regeneration Capacity of the amp1 Mutant
RAP2.6L is required for shoot renewal in tissue culture (Che et al., 2006) and the amp1 mutant shows a higher shoot-regenerative capacity (Chaudhury et al., 1993) as well as ectopic shoot stem cell marker expression in the root (Poretska et al., 2016). To determine whether increased expression of RAP2.6L contributes to the elevated de novo SAM formation in the amp1 mutant, we also analyzed the impact of RAP2.6L deficiency on this process. As previously shown (Che et al., 2006), the rap2.6l mutant displayed diminished regeneration of shoots from root explants (Fig. 6, A and B). This defect was further pronounced in the rap2.6l erf112 double mutant and in the RAP2.6L-SX transgenic plants, whereas this process was unaffected in the erf112 mutant and the EBE-SX transgenic plants (Fig. 6, A and B). Intriguingly, overexpression of RAP2.6L in the wild type prominently promoted the shoot de novo formation capacity almost to the same extent as in the amp1-1 mutant. Conversely, the presence of the RAP2.6L-SX transgene or the rap2.6l erf112 mutation suppressed the elevated shoot regeneration of the amp1 mutant to levels similar or even lower than in the wild type. Again here, the effect of the rap2.6l and erf112 mutation and the presence of the EBE-SX transgene were much more subtle in this respect (Fig. 6, A and B). Together, these findings provide evidence that the upregulated expression of RAP2.6L in the amp1 mutant contributes to its enhanced shoot regeneration capacity.
Figure 6.
Compromised function of RAP2.6L affects shoot regeneration in the amp1 mutant. A, Representative root explants of the indicated genotypes after 18 d on shoot induction medium. B, Quantification of shoot regeneration of the indicated genotypes. Bars for plant lines in the wild-type background are colored in dark gray, and bars for lines in the amp1-1 background are colored in light gray. The regenerative capacity was calculated as the number of shoots/cm root explant (bars represent means ± se; n ≥ 30). Different letters over the error bars indicate significant differences (P < 0.05) as estimated by Duncan’s multiple range test. Bars = 10 mm.
RAP2.6L Expression Is Controlled by HD-ZIP III Proteins
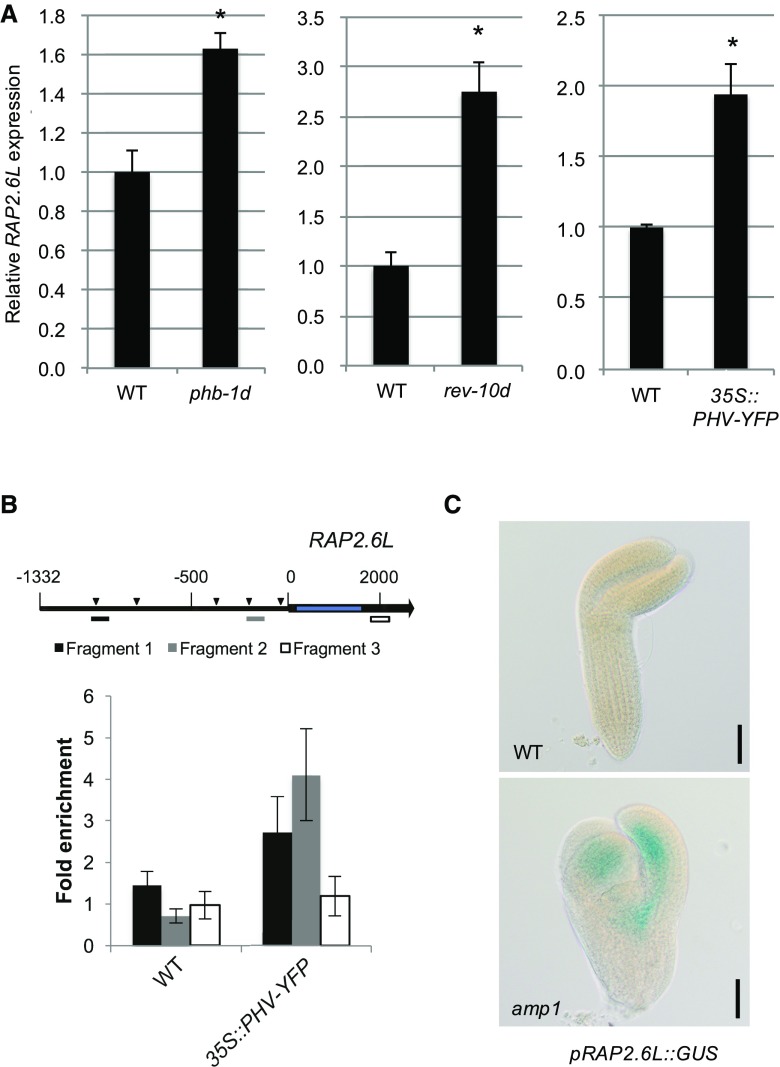
Next, we tried to establish how AMP1 impacts the expression of RAP2.6L at the molecular level. AMP1 is required for the translational inhibition of miRNA targets (Li et al., 2013). Therefore, enhanced activity of the miRNA-controlled components regulating the expression of RAP2.6L might cause its overexpression in the amp1 mutant. We screened the Genevestigator expression database for genetic perturbations affecting the expression of RAP2.6L (Hruz et al., 2008). We detected highly elevated RAP2.6L mRNA levels in embryos of dcl1-15, a miRNA-biosynthesis-deficient mutant (Willmann et al., 2011), consistent with the assumption that RAP2.6L expression is under the control of a miRNA target (Supplemental Fig. S5A). Moreover, this analysis revealed that transcript levels of RAP2.6L are also increased in the 35S:GR-REV* plants (Supplemental Fig. S5B), a line bearing a chemically inducible, miRNA-resistant version of REVOLUTA (Reinhart et al., 2013), suggesting that HD-ZIP III transcription factors, which overaccumulate in the amp1 mutant (Supplemental Fig. S6; Li et al., 2013; Poretska et al., 2016), might affect the expression of RAP2.6L. This was further supported by a significant up-regulation of RAP2.6L mRNA levels in mutants containing miRNA-resistant alleles of either PHABULOSA (phb-1d) or REVOLUTA (rev-10d; Fig. 7A). Similarly, overexpression of a yellow fluorescent protein (YFP)-tagged version of PHAVOLUTA (PHV) promoted the accumulation of RAP2.6L transcripts (Fig. 7A). To test whether the transcription of RAP2.6L is under direct control of HD-ZIP III proteins, we analyzed the RAP2.6L promoter region for the presence of described HD-ZIP III binding motifs. Although the 11-nucleotide inverted palindromic sequence identified by in vitro binding studies of PHV was not present (Sessa et al., 1998), we detected eight AT[G/C]AT repeats representing the inner core of this motif (Fig. 7B), which was defined as the cis-regulatory binding domain of REV based on chromatin immunoprecipitation (ChIP)-seq experiments (Brandt et al., 2012). To assess whether PHV directly binds to the RAP2.6L promoter in vivo, chromatin extracts from wild-type and 35S::PHV-YFP seedlings were immunoprecipitated with an anti-YFP antibody. This analysis revealed that PHV-YFP is associated with two RAP2.6L promoter fragments containing two overlapping AT[G/C]AT repeats each (Fig. 7B). In line with a direct control of RAP2.6L transcription by HD-ZIP III proteins, we found an adaxial/central expression pattern of the pRAP2.6L::GUS reporter in late-stage embryos (Fig. 7C). Moreover, compared to the barely visible reporter activity in the wild type, the signal was strongly enhanced in the amp1 mutant, supporting the model that enhanced HD-ZIP III activity in the amp1 mutant drives the ectopic expression of RAP2.6L. In light of these data, we propose an emerging regulatory module for the control of SAM organization, in which AMP1 constricts stem cell pool homeostasis through limitation of the expression of RAP2.6L via translational control of HD-ZIP III activity.
Figure 7.
RAP2.6L expression is controlled by HD-ZIP III proteins. A, qPCR analysis of RAP2.6L expression in 10-d-old seedlings of phb-1d (left), rev-10d (middle), and 35S::PHV-YFP (right) plants. Normalized means (wild type = 1) are shown. Error bars indicate the se calculated from three biological replicates (at least 30 seedlings per replicate; replicates were grown at the same time on different petri dishes) after normalization to UBC. Asterisks indicate a significant difference (Student’s two-tailed t test; P < 0.05). B, ChIP analysis using 35S::PHV-YFP seedlings. An illustration of the RAP2.6L genomic region is shown (top). The black arrow represents the RAP2.6L transcript region, and the intron region is shown in blue. The transcription start is labeled as 0. Triangles in the promoter region indicate the identified HD-ZIP III binding motifs (AT[G/C]AT). Positions of the genomic fragments analyzed in the ChIP experiment are indicated as differently colored bars. Enrichment of PHV-YFP at the RAP2.6L promoter was determined by qPCR and shown by the ratio between samples treated with antibody and samples treated without antibody. Error bars indicate the se calculated from at least three biological replicates (around 500 seedlings per replicate; replicates were grown at the same time on different petri dishes). C, pRAP2.6L::GUS activity in wild-type (WT) and amp1-1 embryos analyzed by differential interference contrast microscopy. Bars = 50 μm.
DISCUSSION
The putative carboxypeptidase AMP1 has been identified as a crucial determinant of radial SAM organization by suppression of stem cell identity in the morphogenetic zone (Huang et al., 2015). The absence of AMP1 causes disorganized growth patterns and uncoordinated hyperproliferative organ formation in the shoot. Despite the developmental importance of this enzyme, neither its biochemical function nor the regulatory network by which AMP1 controls SAM organization is known. AMP1 limits the translation rate of a broad range of tested miRNA targets (Li et al., 2013). However, whether and how AMP1 controls SAM activity by this mechanism is still elusive. In this work, we showed that the AP2/ERF transcription factor RAP2.6L is strongly upregulated in the absence of AMP1 and that this up-regulation at least partially accounts for the observed phenotypic defects found in the mutant. Moreover, we provide evidence that the ectopic expression of RAP2.6L is driven by the enhanced translation of HD-ZIP III proteins in the amp1 mutant. We propose with RAP2.6L a new regulatory node downstream of AMP1, important for shoot SCN specification.
A role of RAP2.6L in the determination of shoot stem cell identity is supported by earlier studies. RAP2.6L had originally been identified as an essential component of shoot regeneration, since its expression was strongly induced upon transfer of explants to shoot induction medium and its absence severely impaired shoot formation in tissue culture (Che et al., 2006). RAP2.6L is thus a crucial driver of transdifferentiation from root meristem-like callus to newly formed SAMs (Duclercq et al., 2011b). This function is also supported by the finding that upon wounding of inflorescence stems, the expression of RAP2.6L specifically increased at the lower side of the incision zone (Asahina et al., 2011), an area which under certain circumstances, such as shoot decapitation, gives rise to novel SAMs. Moreover, close homologs of RAP2.6L in the Xa group of the AP2/ERF transcription factor family were also shown to be involved in SCN establishment and recovery. Overexpression of EBE/ERF114 not only caused ectopic formation and enhanced outgrowth of axillary shoot meristems, but also increased the size and activity of the vegetative SAM (Mehrnia et al., 2013) similar to the effect of RAP2.6L overexpression described in this study. ERF115 displays an analogous role in the root meristem. Together with PHYTOCHROME A SIGNAL TRANSDUCTION1, ERF115 mediates SCN recovery from differentiated cells in the wounding zone of excised root meristems, and ubiquitous expression of the ERF115/PHYTOCHROME A SIGNAL TRANSDUCTION1 complex causes ectopic stem cell pool formation in the root (Heyman et al., 2016). ERF115 exerts its regenerative capacity at least partially by activating the AP2 transcription factor WOUND INDUCED DEDIFFERENTIATION1, a key player in the wound-induced regeneration processes (Iwase et al., 2011; Heyman et al., 2016). It is still unknown if RAP2.6L also acts through WOUND INDUCED DEDIFFERENTIATION1 or other AP2/ERF proteins involved in shoot regeneration, such as PHLETHORA3/5/7 or ESR1/DRN and ESR2/DRNL.
In light of the phenotypic similarities between RAP2.6L and EBE/ERF114 gain-of-function lines, the question arises whether AMP1 mainly acts through RAP2.6L or also through other members of this ERF subfamily. Although only RAP2.6L among the tested homologs was significantly upregulated in the amp1 mutant in whole-seedling expression analyses, we found that the amp1 shoot defects were more strongly suppressed by the rap2.6l erf112 mutation than by the rap2.6l mutation alone and were most efficiently abrogated by the presence of the RAP2.6L-SX transgene. The RAP2.6L-SX line might affect the activity of additional homologs acting on the same promoters based on its dominant-negative nature. Next to ERF112, RAP2.6 potentially cooperates with RAP2.6L to mediate amp1-related phenotypes, since its expression appeared to be slightly elevated in the amp1 mutant background. Due to the missing suppressive effect of the erf112 and rap2.6 single mutations in the amp1 background, in contrast to the rap2.6l mutation and the strong impact of the RAP2.6L-SX transgene, we postulate that RAP2.6L represents the main target but is functionally escorted by ERF112 and RAP2.6. In contrast, EBE did not show enhanced expression in any of the tested amp1 alleles, and the EBE-SX transgene was much weaker (concerning SAM size) or inefficient (concerning leaf number and shoot regeneration) in suppressing amp1-related shoot phenotypes as compared to the RAP2.6L-SX transgene. Based on these data, it will be interesting to further investigate the specific interaction of ERF112, RAP2.6, and RAP2.6L in shoot SCN replenishment and their functional relationships with AMP1 in this process.
Since AMP1 has been recently positioned in the miRNA-dependent control of translation, we reasoned that the observed induction of RAP2.6L in the amp1 mutant might be caused by excessive translation of miRNA targets affecting its expression. Correspondingly, we found enhanced expression of RAP2.6L in diverse Arabidopsis lines with increased activity of members of the miRNA-controlled family of HD-ZIP III transcription factors and showed that PHV is enriched on the RAP2.6L promoter in vivo. Consistent with a direct control of the expression of RAP2.6L by PHV, we also observed overlapping expression patterns in the embryo. HD-ZIP III transcription factors determine shoot meristem identity during embryogenesis (Smith and Long, 2010), and proper control of their activity is important to maintain the radial symmetry of the SAM (Green et al., 2005; Williams et al., 2005; Kim et al., 2008), making it plausible that their enhanced translation in the amp1 mutant (Li et al., 2013; Poretska et al., 2016) contributes to the ectopic SCN formation. Notably, the enhanced posttranslational activity of HD-ZIP IIIs in the zpr3 zpr4 double mutant provokes the ring-like expansion of the shoot SCN in young seedlings, which is highly reminiscent of what is found in strong amp1 alleles (Kim et al., 2008; Huang et al., 2015). How HD-ZIP III proteins determine shoot meristem identity at the mechanistic level is not well understood, but recent studies revealed a direct impact on key factors such as WUS (Zhang et al., 2017). With RAP2.6L, we identified an additional component acting downstream of HD-ZIP III in the control of shoot meristem patterning. Future genetic analysis should further dissect the functional relevance of this interaction.
Since AMP1 affects the translation of a broad range of tested miRNA targets (Li et al., 2013), the HD-ZIP III/RAP2.6L regulatory cascade is most likely not the only avenue by which AMP1 affects SAM organization. This might be one reason why ectopic expression of RAP2.6L does not fully mimic the amp1 shoot phenotype. Moreover, the enhanced translation of HD-ZIP IIIs in the amp1 mutant potentially misregulates other key determinants of SAM organization such as WUS, which has recently been shown to be under transcriptional control of REV/PHB/PHV in the process of shoot regeneration (Zhang et al., 2017). Nevertheless, the strong suppressive effect of the RAP2.6L-SX transgene on the overall SAM activity of the amp1 mutant shown in this study, compared to the obvious genetic epistasis of amp1 over wus (Huang et al., 2015), supports a prominent role of RAP2.6L downstream of AMP1. Furthermore, a specific functional interaction between AMP1 and HD-ZIP III proteins is supported by the finding that the expression of AMP1 is regulated by REV (Reinhart et al., 2013), potentially representing a negative feedback control mechanism.
We cannot fully exclude the existence of HD-ZIP III-independent modes of RAP2.6L activation in the amp1 mutant. Whereas direct induction by the increased cytokinin levels in the amp1 mutant appears rather unlikely based on the unresponsiveness of RAP2.6L transcription toward exogenous cytokinin application (Supplemental Fig. S2; Che et al., 2006), induction might be caused by the enhanced jasmonic acid (JA) response found in the amp1 mutant (Poretska et al., 2016). RAP2.6L expression is promoted upon application of JA, a hormone synthesized upon wounding. Interestingly, wounding has been shown to be important for the regeneration processes. However, how wounding triggers regeneration at the molecular level and the specific role of JA in this process are not well understood (Ikeuchi et al., 2016). Whether the altered JA response in the amp1 mutant is functionally relevant with respect to the defect in SAM organization and the elevated transcript levels of RAP2.6L are important future questions to be resolved.
Finally, it remains unclear how enhanced RAP2.6L expression in amp1 triggers the expansion of SCN identity in the peripheral zone of the meristem as well as the enhanced ability for shoot regeneration in root explants. A potential downstream component is CUP-SHAPED COTYLEDON2, a transcription factor required for embryonic SAM formation and shoot regeneration in tissue culture, whose expression has been shown to depend on RAP2.6L (Che et al., 2006). Another future research direction might be to analyze whether RAP2.6L feeds back on HD-ZIP III activity by physical interaction. The AP2/ERF transcription factors ESR1/DRN and ESR2/DRNL bind to HD-ZIP IIIs and cooperate in shoot patterning during embryogenesis (Chandler et al., 2007). It will be interesting to see whether RAP2.6L has a related competence to modulate HD-ZIP III function in the control of SAM development.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotypes Columbia (Col-0) and Landsberg erecta (Ler) were used in this study. amp1-1 (Col-0; N8324), amp1-13 (Col-0; N522988), pt (Ler; N235), lamp1-2 (Col-0; SM_3.22750), rap2.6-2 (N863006), rap2.6l-1 (N656482), erf112-1 (N563727), and phb-1d (N3761) were ordered from the Nottingham Arabidopsis Stock Centre (http://www.arabidopsis.info), and genotypes of rap2.6-2, rap2.6l-1, and erf112-1 were verified by PCR genotyping (Supplemental Fig. S7). 35S::PHV-YFP was described previously (Poretska et al., 2016). The 35S::RAP2.6L lines (C23, C28, and C31; named RAP2.6L-OX throughout this study) and pRAP2.6L::RAP2.6L:GUS were kindly provided by Nat Kav (Krishnaswamy et al., 2011). Of the RAP2.6L overexpression lines, C28 was used throughout this study unless indicated otherwise. rev-10d was obtained from Stephan Wenkel. CycB1;1::CycB1;1-GUS (in Col-0) was provided by John Celenza (DiDonato et al., 2004), pCLV3::GUS, and pWUS::GUS (in Ler) were received from Thomas Laux (Gross-Hardt et al., 2002), and pKLU::GUS (in Col-0) was donated by Michael Lenhard (Anastasiou et al., 2007). Combinations of mutants and reporter lines were generated by crossing individual lines, and genotypes were verified phenotypically and by PCR genotyping (see primers in Supplemental Table S1).
Arabidopsis seeds were surface sterilized with 70% ethanol containing 0.05% SDS for 3 min and rinsed with 96% ethanol for 1 min.
The seeds were plated on half-strength Murashige and Skoog (MS) medium (Duchefa) containing 0.7% (w/v) agar (Duchefa) and 1% (w/v) Suc or in soil. After stratification (4°C for 48 h), seeds were transferred to the incubator set to long-day conditions (16 h of 80 µmol s−1 m−2 light/8 h dark) or short-day conditions (8 h of 80 µmol s−1 m−2 light/16 h dark). For the hyperphyllin treatment, seeds were germinated in liquid half-strength MS medium containing 1% (w/v) Suc in the presence or absence of the drug and analyzed at 10 DAG.
Gene Constructs and Transformation
PCR was performed with proofreading thermostable polymerase (Thermo Fisher Scientific), and all clones were confirmed by sequencing. To create 35S::RAP2.6L-MYC-SRDX (RAP2.6L-SX) and 35S::EBE-MYC-SRDX (EBE-SX), the RAP2.6L and EBE ORFs were amplified from cDNA (Col-0) by PCR with the primer pairs RAP2.6L ORF F (EcoRV)/RAP2.6L ORF R (NotI) and EBE ORF F (EcoRV)/EBE ORF R (NotI), respectively. The fragment was subcloned into pGEM-T Easy (Promega). Subsequently, in a previously created pGWR8-CES-MYC-SRDX (Poppenberger et al., 2011), the CES ORF was removed by a double digestion with EcoRV and NotI to generate the pGWR8-MYC-SRDX backbone. The RAP2.6L and EBE ORFs were then transferred via EcoRV and NotI into pGWR8-MYC-SRDX to generate RAP2.6L-SX and EBE-SX, respectively.
To create pRAP2.6L::GUS, a 1,487-bp genomic sequence upstream of the RAP2.6L ORF was amplified with primers AP2.6proF(PstI) and AP2.6proR(BamHI) and subcloned into pGEM-T Easy (Promega). The fragment was excised using PstI and BamHI and ligated into pPZP-GUS-1 (Diener et al., 2000), resulting in pRAP2.6L::GUS.
Plants were transformed using the floral-dip method, at least 10 independent transformants were generated for each line, and the resulting T2 lines were confirmed for single transgene insertion sites based on the 3:1 segregation of the selection marker and propagated for further analysis.
GUS Staining
Seedlings were put into the freshly made GUS staining buffer (100 mm sodium phosphate buffer, 10 mm EDTA, 0.5 mm K3Fe(CN)6, 0.5 mm K4Fe(CN)6, Triton X-100 [0.1% v/v], and 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide). The seedlings were incubated at 37°C for a duration of time depending on the reporter strength. After staining, the seedlings were dehydrated with 70% ethanol. Seedlings were analyzed using a stereomicroscope (SZX10; Olympus) equipped with a digital camera (DP26; Olympus).
Histology
The histological analysis was performed as previously described (De Smet et al., 2004). Seedlings were grown under long-day conditions for 8 d or short-day conditions for 18 d and fixed overnight at 4°C in formaldehyde-acetic acid-alcohol (5% [v/v] formaldehyde, 5% [v/v] acetic acid, and 50% [v/v] ethanol). After fixation, samples were dehydrated in a graded ethanol series (2 h each in 30%, 50%, 70%, and 96% ethanol) and embedded with Technovit 7100 (Heraus Kulzer) according to the manufacturer’s instructions. A series of 5- to 7-μm-thick longitudinal sections was made with a Leica RM 2065 microtome. Sections were transferred to microscopic slides (Marienfeld), stained for 5 min in 0.02% aqueous Toluidine blue O (Sigma-Aldrich), and rinsed with water. Subsequently, the stained sections were analyzed with a microscope (BX-61; Olympus).
Scanning Electron Microscopy
Seedlings were fixed in formaldehyde-acetic acid-alcohol (50% ethanol, 10% acetic acid, and 5% formaldehyde) overnight at 4°C and then dehydrated through a graded ethanol series up to 96% ethanol and supercritical point-dried (CPD300; Leica Microsystems). Dried seedlings were dissected and mounted on conductive adhesive tabs (PLANO) under a stereomicroscope (SZX10; Olympus). Samples were subsequently examined using a T-3000 tabletop scanning electron microscope (Hitachi).
Leaf Number Analysis
The shoot apex area of seedlings was examined under the stereomicroscope (2× magnification) at the indicated developmental stages and the number of visible leaves was recorded.
Regeneration Capacity Assay
The regeneration capacity assay was performed based on a previous protocol (Che et al., 2006). In brief, Arabidopsis seedlings were grown for 7 d on 0.5× MS medium under long-day conditions. Root segments of 1 to 1.5 cm were cut, transferred to Gamborg’s B5 medium (Sigma-Aldrich) supplemented with 0.5 g/L MES (pH set to 5.7 with KOH), 2.2 μm 2,4-dichlorophenoxyacetic acid, 0.2 μm kinetin, and 0.8% agarose (callus induction medium), and incubated under constant light for 4 d. The root explants were then transferred to shoot induction medium (Gamborg’s B5 medium containing 0.5 g/L MES [pH set to 5.7 with KOH], 5.0 μm isopentenyladenine, 0.9 μm indole acetic acid, and 0.8% agarose) and incubated under constant light for 18 d. Regenerated shoots were counted and the number of regenerated shoots per cm explant was calculated.
Reverse Transcription-qPCR
About 50 mg of seedling material was collected, shock-frozen in liquid nitrogen, and homogenized with a Retsch mill (Verder Scientific). The total RNA was extracted with the E.N.Z.A. Plant RNA Mini Kit (OMEGA Bio-Tek) and treated with DNase I (Thermo Fisher Scientific). The first-strand cDNA was synthesized using the extracted RNA as template with the RevertAid first-strand cDNA synthesis kit (Thermo Fisher Scientific). qPCR was performed with an Eppendorf Realplex Mastercycler using SensiFAST SYBR Lo-ROX Mix (Bioline) and specific primers (Supplemental Table S1) for the mRNAs of interest. Data were normalized to UBC (AT5G25760) and measured in at least three technical replicates.
Western Blotting
Western blotting was performed as previously described (Poretska et al., 2016).
ChIP Assays
One gram of 10-d-old plants was submerged in 1% formaldehyde solution, and a vacuum was applied for 10 min. Cross linking was quenched by adding 125 mm Gly, and a vacuum was applied for 5 min. After rinsing two times with double-distilled water, the plant material was ground to a fine powder in liquid nitrogen, and chromatin preparation was performed as previously described (Poppenberger et al., 2011). For immunoprecipitation, anti-GFP VHH agarose beads (Chromotek) and agarose beads as control were used. Washing of the beads and elution of the DNA-protein complex were performed as described previously (Kaufmann et al., 2010). DNA purification was performed with the MinElute PCR purification kit (Qiagen, Venlo, NL). For qPCR analysis, primers specific for the desired promoter region (listed in Supplemental Table S1) were used. Purified DNA fragments were used for establishing standard curves for quantification.
Accession Numbers
Sequence data from this article can be found in the GenBank data library under accession numbers AT3G54720 (AMP1), AT5G19740 (LAMP1), AT5G13330 (RAP2.6L), AT1G43160 (RAP2.6), AT2G33710 (ERF112), AT5G61890 (EBE), AT5G07310 (ERF115), AT5G25760 (UBC), AT2G34710 (PHB), AT5G60690 (REV), AT2G17950 (WUS), AT2G27250 (CLV3), AT1G13710 (KLU), and AT4G37490 (CYCB1;1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression analysis of ERF transcription factors closely related to RAP2.6L.
Supplemental Figure S2. RAP2.6L expression is not affected by cytokinin.
Supplemental Figure S3. Effect of compromised RAP2.6 function on the leaf formation rate and shoot regeneration.
Supplemental Figure S4. Effect of compromised RAP2.6L function on the leaf formation rate of the amp1 mutant.
Supplemental Figure S5. Expression of RAP2.6L is increased in the dcl1 mutant and the 35S::GR-REV* transgenic plants.
Supplemental Figure S6. PHV::YFP overaccumulates in the amp1 and amp1 lamp1 mutants.
Supplemental Figure S7. Genotyping of the rap2.6-2, rap2.6l-1, and erf112-1 mutants.
Supplemental Table S1. List of oligos used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank John Chelenza, Nat Kav, Thomas Laux, Michael Lenhard, and Stephan Wenkel for providing published material. We thank Pablo Albertos, Brigitte Poppenberger, and Wilfried Rozhon for critical reading of the manuscript. We also thank Julius Mayer for helping with SAM sectioning and Irene Ziegler for technical assistance.
Footnotes
This work was supported by the Austrian Science Fund (P19935 to T.S.), by an APART fellowship from the Austrian Academy of Sciences (11300 to T.S.), by the German Federal Ministry of Education and Research (031A327 and 031B0554 to T.S.), and by a PhD fellowship from the China Scholarship Council (to S.Y.). S.Y. was a member of the Technical University of Munich Graduate School.
Articles can be viewed without a subscription.
References
- Anastasiou E, Kenz S, Gerstung M, MacLean D, Timmer J, Fleck C, Lenhard M (2007) Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev Cell 13: 843–856 [DOI] [PubMed] [Google Scholar]
- Asahina M, Azuma K, Pitaksaringkarn W, Yamazaki T, Mitsuda N, Ohme-Takagi M, Yamaguchi S, Kamiya Y, Okada K, Nishimura T, et al. (2011) Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proc Natl Acad Sci USA 108: 16128–16132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno H, Ikeda Y, Niu QW, Chua NH (2001) Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 13: 2609–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619 [DOI] [PubMed] [Google Scholar]
- Brandt R, Salla-Martret M, Bou-Torrent J, Musielak T, Stahl M, Lanz C, Ott F, Schmid M, Greb T, Schwarz M, et al. (2012) Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J 72: 31–42 [DOI] [PubMed] [Google Scholar]
- Chandler JW, Cole M, Flier A, Grewe B, Werr W (2007) The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 134: 1653–1662 [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Letham S, Craig S, Dennis ES (1993) Amp1: a mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J 4: 907–916 [Google Scholar]
- Che P, Lall S, Nettleton D, Howell SH (2006) Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol 141: 620–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chickarmane VS, Gordon SP, Tarr PT, Heisler MG, Meyerowitz EM (2012) Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc Natl Acad Sci USA 109: 4002–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Medzihradszky A, Suzaki T, Lohmann JU (2014) A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc Natl Acad Sci USA 111: 14619–14624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Chaerle P, Vanneste S, De Rycke R, Inzé D, Beeckman T (2004) An easy and versatile embedding method for transverse sections. J Microsc 213: 76–80 10.1111/j.1365-2818.2004.01269.x [DOI] [PubMed] [Google Scholar]
- DiDonato RJ, Arbuckle E, Buker S, Sheets J, Tobar J, Totong R, Grisafi P, Fink GR, Celenza JL (2004) Arabidopsis ALF4 encodes a nuclear-localized protein required for lateral root formation. Plant J 37: 340–353 [DOI] [PubMed] [Google Scholar]
- Diener AC, Li H, Zhou W, Whoriskey WJ, Nes WD, Fink GR (2000) Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 12: 853–870 10.1105/tpc.12.6.853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclercq J, Assoumou Ndong YP, Guerineau F, Sangwan RS, Catterou M (2011a) Arabidopsis shoot organogenesis is enhanced by an amino acid change in the ATHB15 transcription factor. Plant Biol (Stuttg) 13: 317–324 [DOI] [PubMed] [Google Scholar]
- Duclercq J, Sangwan-Norreel B, Catterou M, Sangwan RS (2011b) De novo shoot organogenesis: from art to science. Trends Plant Sci 16: 597–606 [DOI] [PubMed] [Google Scholar]
- Gaillochet C, Lohmann JU (2015) The never-ending story: from pluripotency to plant developmental plasticity. Development 142: 2237–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM (2009) Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci USA 106: 16529–16534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KA, Prigge MJ, Katzman RB, Clark SE (2005) CORONA, a member of the class III homeodomain leucine zipper gene family in Arabidopsis, regulates stem cell specification and organogenesis. Plant Cell 17: 691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J, Barrero JM, Taylor J, Helliwell CA, Gubler F (2011) ALTERED MERISTEM PROGRAM 1 is involved in development of seed dormancy in Arabidopsis. PLoS One 6: e20408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Hardt R, Lenhard M, Laux T (2002) WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev 16: 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Chin-Atkins AN, Wilson IW, Chapple R, Dennis ES, Chaudhury A (2001) The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell 13: 2115–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman J, Cools T, Canher B, Shavialenka S, Traas J, Vercauteren I, Van den Daele H, Persiau G, De Jaeger G, Sugimoto K, et al. (2016) The heterodimeric transcription factor complex ERF115-PAT1 grants regeneration competence. Nat Plants 2: 16165. [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Pitorre D, Poretska O, Marizzi C, Winter N, Poppenberger B, Sieberer T (2015) ALTERED MERISTEM PROGRAM1 suppresses ectopic stem cell niche formation in the shoot apical meristem in a largely cytokinin-independent manner. Plant Physiol 167: 1471–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Ogawa Y, Iwase A, Sugimoto K (2016) Plant regeneration: cellular origins and molecular mechanisms. Development 143: 1442–1451 [DOI] [PubMed] [Google Scholar]
- Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, et al. (2011) The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol 21: 508–514 [DOI] [PubMed] [Google Scholar]
- Je BI, Gruel J, Lee YK, Bommert P, Arevalo ED, Eveland AL, Wu Q, Goldshmidt A, Meeley R, Bartlett M, et al. (2016) Signaling from maize organ primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nat Genet 48: 785–791 [DOI] [PubMed] [Google Scholar]
- Kareem A, Radhakrishnan D, Sondhi Y, Aiyaz M, Roy MV, Sugimoto K, Prasad K (2016) De novo assembly of plant body plan: a step ahead of Deadpool. Regeneration (Oxf) 3: 182–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Østerås M, Farinelli L, Krajewski P, Angenent GC (2010) Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat Protoc 5: 457–472 [DOI] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Lee M, Lee I, Park HY, Seo PJ, Jung JH, Kwon EJ, Suh SW, Paek KH, et al. (2008) HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell 20: 920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy S, Verma S, Rahman MH, Kav NN (2011) Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol Biol 75: 107–127 [DOI] [PubMed] [Google Scholar]
- Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, et al. (2013) MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153: 562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau JE. (1959) Observations et experimentation sur la phyllotaxie et le fonctionnement du sommet vegetatif chez quelques Balsaminacees. Ann Sci Nat Bot 11: 1–214 [Google Scholar]
- Magnani E, Barton MK (2011) A per-ARNT-sim-like sensor domain uniquely regulates the activity of the homeodomain leucine zipper transcription factor REVOLUTA in Arabidopsis. Plant Cell 23: 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo N, Makino M, Banno H (2011) Arabidopsis ENHANCER OF SHOOT REGENERATION (ESR)1 and ESR2 regulate in vitro shoot regeneration and their expressions are differentially regulated. Plant Sci 181: 39–46 [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- Mehrnia M, Balazadeh S, Zanor MI, Mueller-Roeber B (2013) EBE, an AP2/ERF transcription factor highly expressed in proliferating cells, affects shoot architecture in Arabidopsis. Plant Physiol 162: 842–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogué F, Hocart C, Letham DS, Dennis ES, Chaudhury AM (2000) Cytokinin synthesis is higher in the Arabidopsis amp1 mutant. Plant Growth Regul 32: 267–273 [Google Scholar]
- Poppenberger B, Rozhon W, Khan M, Husar S, Adam G, Luschnig C, Fujioka S, Sieberer T (2011) CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J 30: 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poretska O, Yang S, Pitorre D, Rozhon W, Zwerger K, Uribe MC, May S, McCourt P, Poppenberger B, Sieberer T (2016) The small molecule hyperphyllin enhances leaf formation rate and mimics shoot meristem integrity defects associated with AMP1 deficiency. Plant Physiol 171: 1277–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Frenz M, Mandel T, Kuhlemeier C (2003) Microsurgical and laser ablation analysis of interactions between the zones and layers of the tomato shoot apical meristem. Development 130: 4073–4083 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Liu T, Newell NR, Magnani E, Huang T, Kerstetter R, Michaels S, Barton MK (2013) Establishing a framework for the Ad/abaxial regulatory network of Arabidopsis: ascertaining targets of class III homeodomain leucine zipper and KANADI regulation. Plant Cell 25: 3228–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roodbarkelari F, Groot EP (2017) Regulatory function of homeodomain-leucine zipper (HD-ZIP) family proteins during embryogenesis. New Phytol 213: 95–104 [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Sessa G, Steindler C, Morelli G, Ruberti I (1998) The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol Biol 38: 609–622 [DOI] [PubMed] [Google Scholar]
- Sluis A, Hake S (2015) Organogenesis in plants: initiation and elaboration of leaves. Trends Genet 31: 300–306 [DOI] [PubMed] [Google Scholar]
- Smith ZR, Long JA (2010) Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464: 423–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaurre DP, Ploense S, Krogan NT, Berleth T (2007) AMP1 and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development 134: 2561–2567 [DOI] [PubMed] [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E, Weigel D (2008) Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20: 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC (2005) Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132: 3657–3668 [DOI] [PubMed] [Google Scholar]
- Willmann MR, Mehalick AJ, Packer RL, Jenik PD (2011) MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol 155: 1871–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV (2011) WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev 25: 2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TQ, Lian H, Zhou CM, Xu L, Jiao Y, Wang JW (2017) A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29: 1073–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]