A second plant succinate dehydrogenase assembly factor is identified for the assembly of SDH1 with SDH2.
Abstract
Succinate dehydrogenase (Complex II; SDH) plays an important role in mitochondrial respiratory metabolism. The SDH complex consists of four core subunits and multiple cofactors, which must be assembled correctly to ensure enzyme function. To date, only an assembly factor (SDHAF2) required for FAD insertion into subunit SDH1 has been identified in plants. Here, we report the identification of Arabidopsis (Arabidopsis thaliana) At5g67490 as a second SDH assembly factor. Knockout of At5g67490 (sdhaf4) did not cause any phenotypic variation in seedlings but resulted in a decrease in both SDH activity and the succinate-dependent respiration rate as well as increased accumulation of succinate. Mass spectrometry analyses revealed stable levels of FAD-SDH1 in sdhaf4, together with increased levels of the FAD-SDH1 assembly factor, SDHAF2, and reduced levels of SDH2 compared with the wild type. Loss of SDHAF4 in sdhaf4 inhibited the formation of the SDH1/SDH2 intermediate, leading to the accumulation of soluble SDH1 in the mitochondrial matrix and reduced levels of SDH1 in the membrane. The increased levels of SDHAF2 suggest that the stabilization of soluble FAD-SDH1 depends on SDHAF2 availability. We conclude that SDHAF4 acts on FAD-SDH1 and promotes its assembly with SDH2, thereby stabilizing SDH2 and enabling its full assembly with SDH3/SDH4 to form the SDH complex.
Mitochondria are involved in a series of cellular processes, including ATP synthesis, respiratory metabolism, and reactive oxygen species (ROS) production (Millar et al., 2011). The electron transport chain (ETC) is embedded in the inner mitochondrial membrane (IMM) and consists of four protein complexes (I–IV). These complexes are essential to the formation of a proton gradient formed by redox reactions along the ETC and the translocation of protons across the IMM, which, in turn, drive the production of ATP via ATP synthase (Complex V; Jacoby et al., 2012). Within the ETC, succinate dehydrogenase (SDH; Complex II) is the smallest complex and forms part of both the ETC and the tricarboxylic acid cycle (Huang and Millar, 2013). It is the only complex within the classical ETC that does not pump protons across the IMM. SDH catalyzes the oxidation of succinate to fumarate and, in doing so, reduces ubiquinone (UQ) to ubiquinol. SDH is a heterotetrameric protein complex, anchored to the IMM by two integral membrane proteins, SDH3 and SDH4, which dimerize to bind a heme and generate the two UQ-binding sites. The SDH3/SDH4 dimer binds to subunit SDH2, which contains three iron-sulfur (Fe-S) clusters and is assembled with the catalytic subunit SDH1. SDH1 contains the succinate-binding site and carries a covalently bound FAD cofactor (Lemire and Oyedotun, 2002; Sun et al., 2005; Huang and Millar, 2013). There are two genes encoding SDH1 in Arabidopsis (Arabidopsis thaliana), SDH1-1 (At5g66760) and SDH1-2 (At2g18450), but the latter is expressed at a very low level (Figueroa et al., 2002).
Comparisons of plant SDH subunit amino acid sequences and purified SDH complexes with those in other organisms showed that the plant SDH complex is divergent in both sequence and composition (Eubel et al., 2003; Huang and Millar, 2013). Although the sequences of the SDH1 and SDH2 subunits are highly conserved across eukaryotes, SDH3 and SDH4 show a high degree of sequence divergence, notably between plants and animals (Burger et al., 1996). Furthermore, the purified plant SDH complex has been reported to contain four additional subunits (SDH5–SDH8) with yet unknown function (Eubel et al., 2003; Millar et al., 2004; Huang et al., 2010). A recent study has suggested that SDH6 and SDH7 act as substitutes for missing helices in SDH3 and SDH4, which are not present in plants but are conserved in other organisms (Schikowsky et al., 2017). For SDH to function, it must be assembled and a series of cofactors must be inserted into the different subunits. Four SDH assembly factors (named SDHAF1–SDHAF4) have been identified in mammals and yeast as being required for the assembly of SDH1 and SDH2 into a soluble intermediate ready for attachment to the membrane-bound SDH3/SDH4 (Ghezzi et al., 2009; Hao et al., 2009; Na et al., 2014; Van Vranken et al., 2014). Three of these proteins have putative orthologs in Arabidopsis (SDHAF1, SDHAF2, and SDHAF4), which have different degrees of sequence conservation with their yeast, Drosophila melanogaster, and mammalian orthologs.
In plants, one of these assembly factors, SDHAF2, has been characterized successfully (Huang et al., 2013). Knockdown of SDHAF2 in Arabidopsis resulted in a significant reduction in the levels of mature SDH complex, as only 50% of assembled SDH holo-complex could be found in sdhaf2 (Huang et al., 2013). Reduced abundances of SDH1 and FAD-bound SDH1 were observed for sdhaf2, indicating the important role played by SDHAF2 in FAD insertion into SDH1 and SDH1 maturation in Arabidopsis (Huang et al., 2013). Previous studies demonstrated that reduced SDH activity and seed set occurred in an SDH1-1 RNA interference line or an SDH1-1/sdh1-1 heterozygous line (León et al., 2007). Interestingly, deficiency of SDH1 in the SDH1-1/sdh1-1 heterozygous line results in an elevated photosynthesis rate and better growth under nitrogen-limiting conditions (Fuentes et al., 2011). An elevated rate of photosynthesis also was observed in tomato SDH2 RNA interference lines with partial SDH dysfunction (Araújo et al., 2011). Additionally, recent studies demonstrated the importance of mature, assembled SDH1 in the salicylic acid-induced stress response (Belt et al., 2017). A mutant carrying a point mutation (dsr1) at the succinate-binding site of SDH1 (Gleason et al., 2011) showed a change in substrate affinity and catalytic efficiency and was, together with sdhaf2, used to measure salicylic acid-induced stress signaling. Due to the kinetic changes in dsr1 and the lower abundance of mature SDH1 in sdhaf2, the stress response was decreased severely in both lines (Belt et al., 2017). Besides the incorporation of FAD into SDH1 via SDHAF2, little is known about the assembly and maturation of SDH1 and its binding to SDH2. As SDH1 maturation is essential for functional mitochondrial metabolism and plant development, it is important to investigate its assembly further by determining the next essential step, the assembly of SDH1 with SDH2.
In yeast, D. melanogaster, and mammalian cells, SDHAF4 (C6orf57 in humans) was shown to bind specifically to the flavinated SDH1 subunit, thereby promoting the assembly of SDH1 with SDH2 after the FAD cofactor was incorporated into SDH1 (Van Vranken et al., 2014). Based on sequence homology, a gene in Arabidopsis (At5g67490, herein named SDHAF4) with a yet unknown function was identified. To investigate the role of At5g67490 and its potential to be a functional equivalent of SDHAF4 for SDH1 assembly in plants, we analyzed a T-DNA insertion line (Landsberg erecta [Ler] background) for SDHAF4 (sdhaf4) to extend the model of the SDH1 assembly pathway in plants, following the insertion of FAD aided by SDHAF2.
RESULTS
SDHAF4 Is Located in Mitochondria and Shares a Conserved Protein Region at the C Terminus
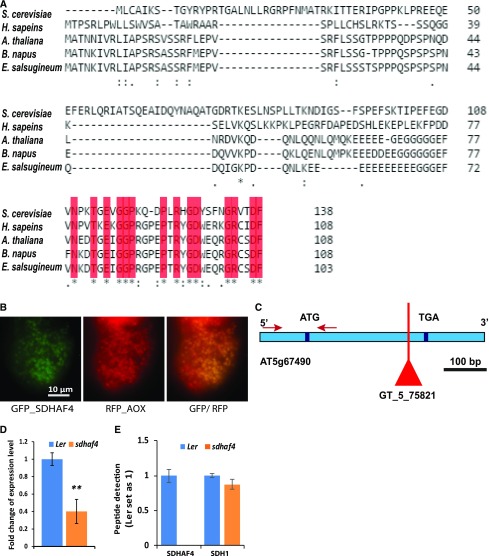
To properly investigate the function of the Arabidopsis gene At5g67490, bioinformatic, localization, genetic, and functional studies needed to be undertaken. Alignment of amino acid sequences for SDHAF4 showed an overall sequence similarity of only 28% between human (Homo sapiens), yeast (Saccharomyces cerevisiae), and plant SDHAF4-like sequences (Arabidopsis, Brassica napus, and Eutrema salsugenium), but a conserved region of approximately 25 amino acids located at the C terminus of each protein was apparent (Fig. 1A; blast.ncbi.nlm.nih.gov and http://www.pantherdb.org). Using subcellular localization information (http://suba.plantenergy.uwa.edu.au/), a mitochondrial targeting sequence was predicted for At5g67490. Consistent with this prediction, At5g67490 also had been reported as an identified unknown protein in a peptide mass spectrometry analysis of Arabidopsis mitochondrial extracts (Taylor et al., 2011). The At5g67490 gene has only one exon and is expressed in all tissues, with the highest expression patterns in cotyledons (http://bar.utoronto.ca). The AtSDHAF4 protein has a calculated molecular mass of approximately 12 kD (www.arabidopsis.org). To further confirm the localization of SDHAF4, GFP was fused to the N terminus of SDHAF4 and expressed transiently in Arabidopsis cell culture. Microscopy imaging of SDHAF4:GFP, as well as mitochondrial ALTERNATIVE OXIDASE1a (AOX1a) fused to red fluorescent protein (AOX-RFP) as a marker, confirmed the mitochondrial localization of SDHAF4 (Fig. 1B). The combined evidence of localization prediction, identification by mass spectrometry, and confirmation by GFP localization provides strong evidence that At5g67490 is a mitochondrial protein.
Figure 1.
SDHAF4 sequences contain a conserved region at the C terminus among different species, the Arabidopsis protein is located within mitochondria, and sdhaf4 is an effective knockout Arabidopsis line at the protein level for SDHAF4. A, Sequence alignment of SDHAF4 protein between S. cerevisiae (YBR269C), human (NM_145267), Arabidopsis (At5g67490), B. napus (XM_013885343), and E. salsugenium (XM_006393894; Clustal Omega). Conserved regions are highlighted in red. Asterisks indicate identical residues, colons indicates conserved substitution, and periods indicate semiconserved substitution. B, GFP_SDHAF4 and RFP_AOX constructs were expressed transiently in Arabidopsis cell culture. GFP localization of SDHAF4 in mitochondria was performed by microscopy imaging. C, Location of the T-DNA insertion in At5g67490 (SDHAF4) within the exon region. Red arrows indicate the binding sites of the primers used for RT-qPCR. D, SDHAF4 gene expression in Ler and sdhaf4. RT-qPCR using primers outside the T-DNA region (red arrows in C) was performed to determine SDHAF4 expression. Expression levels were normalized to actin, and the expression of SDHAF4 in Ler was set as 1. E, SDH1 and SDHAF4 peptides were detected and quantified using mass spectrometry. Mitochondria isolated from Ler and sdhaf4 were used, and protein-specific peptides were used for identification. Samples were normalized to ATP synthase. Shown is the ratio of sdhaf4 to Ler. Error bars indicate se (n = 4). **, P ≤ 0.01 (Student’s t test).
Characterization and Phenotypic Analysis of an SDHAF4 T-DNA Insertion Line
We sought to determine whether At5g67490, despite its low sequence identity to SDHAF4 in other organisms, is a second assembly factor for SDH, acting after SDHAF2. If so, we hypothesized that its loss would affect SDH activity and the assembly of SDH1 with SDH2 once FAD was inserted into SDH1. Thus, the aim of this study was to determine the impact of AtSDHAF4 loss on SDH function and the assembly of SDH1 through study of a knockout line. A confirmed knockdown line for SDHAF2 (sdhaf2) also was included in the experiments to determine possible similarities between the two assembly factor mutant lines and to determine the different steps and/or order of these two assembly factors in the maturation of SDH1 and the assembly of SDH1 with SDH2.
To analyze the function of the SDHAF4-like gene in Arabidopsis (At5g67490; SDHAF4), a T-DNA insertion line (GT_5_75821 in the Ler background, hereafter referred to as sdhaf4) from the Nottingham Arabidopsis Stock Centre (http://signal.salk.edu/cgi-bin/tdnaexpress) was obtained and homozygous lines were identified by genotyping (Supplemental Fig. S1). The T-DNA insertion was located within the exon of At5g67490 (Fig. 1C). Reverse transcription quantitative PCR (RT-qPCR) was used to determine the expression level of SDHAF4 in sdhaf4 compared with Ler (Fig. 1D). Primers covering a region outside the T-DNA insertion (Fig. 1C, red arrows) were designed. The expression level of the transcript was reduced about 60% in sdhaf4 (Fig. 1D), showing that, due to the T-DNA insertion, expression from this gene was strongly, but not completely, inhibited. In order to investigate if SDHAF4 protein still accumulates in sdhaf4, multiple reaction monitoring (MRM) was used to detect peptides from SDHAF4 and SDH1 (Fig. 1E). Peptide amount was calculated as the ratio of SDHAF4 or SDH1 in sdhaf4 to Ler. From the results obtained, the abundance of SDH1 was not changed significantly in sdhaf4, while SDHAF4 was not detectable in the mutant, demonstrating that, although transcripts were measurable, SDHAF4 protein did not accumulate (Fig. 1E), indicating an effective knockout of SDHAF4 function in sdhaf4.
We did not observe any change in plant growth or development between sdhaf4 and Ler when they were grown on soil under long-day conditions, which was similar to sdhaf2 and the Columbia-0 (Col-0) wild type (Supplemental Fig. S2). Similarly, there was no difference in root elongation when Ler and sdhaf4 were grown on agar plates (Supplemental Fig. S2), which is in contrast to the short-root phenotype observed in sdhaf2 (Huang et al., 2013).
sdhaf4 Shows Decreased SDH Activity and Succinate-Dependent Respiration
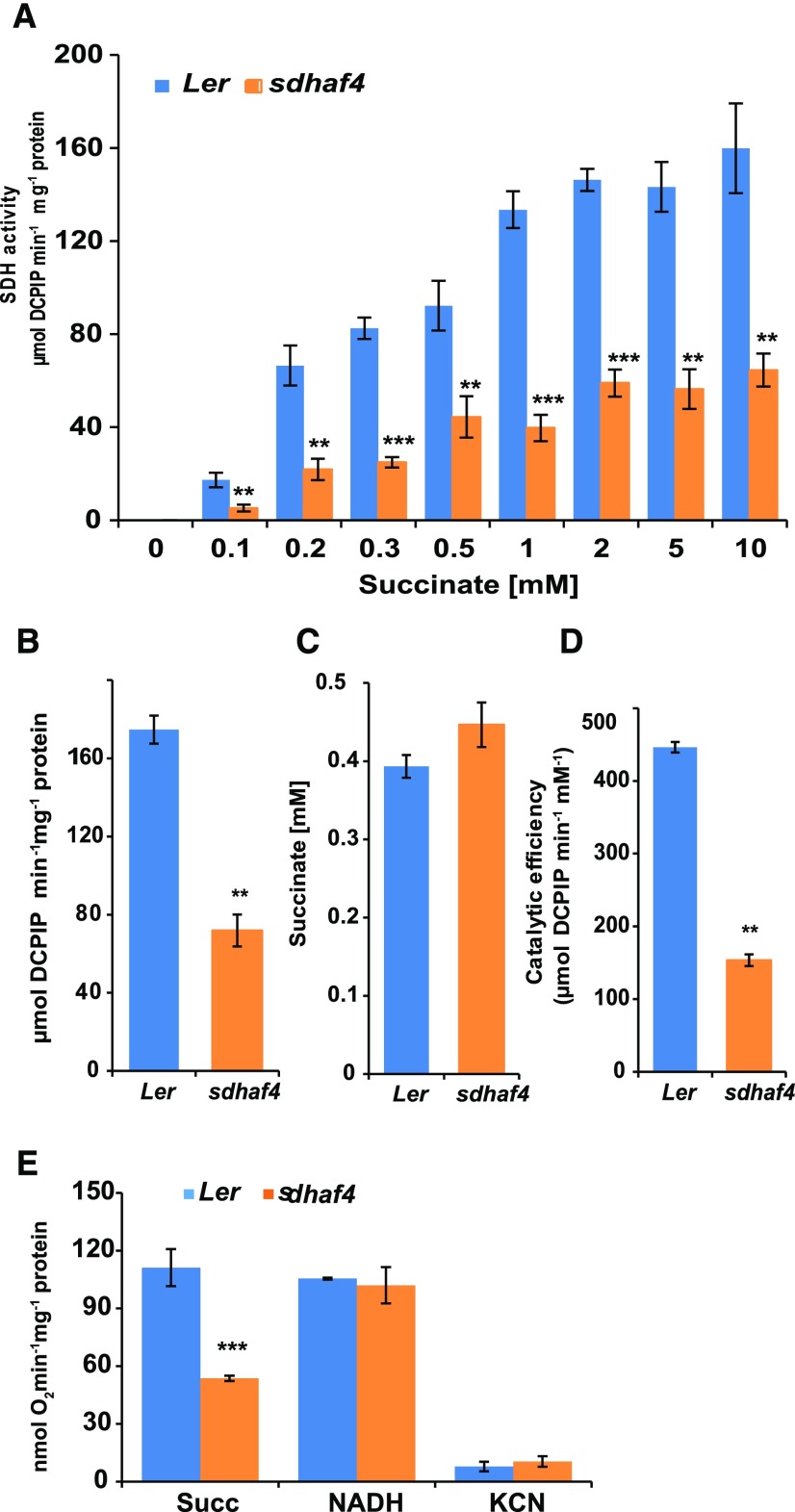
To determine whether knocking out SDHAF4 directly affects SDH function, SDH activity was measured in isolated mitochondria obtained from Ler and sdhaf4 plants (Fig. 2). Phenazine methosulfate (PMS) and 2,6-dichloro-indolephenol (DCPIP) were used to accept electrons generated at the succinate-binding site of SDH. Using a range of 0.1 to 10 mm succinate, a significant decrease in SDH activity was observed in sdhaf4 in comparison with Ler at most substrate concentrations (Fig. 2A). Approximately one-third of the activity seen in Ler could be measured in sdhaf4 (Fig. 2B). The calculated Km value of SDH for succinate (R software; Supplemental Document S1), which describes the substrate concentration necessary to reach half-maximum enzyme velocity, was not significantly different between sdhaf4 and Ler (Student’s t test, P > 0.1; Fig. 2C). Both lines showed a Km of approximately 0.4 mm succinate, indicating that this kinetic property of SDH in Ler is maintained in sdhaf4. Due to its low SDH activity even at high concentrations of succinate, the catalytic efficiency (Vmax/Km) in sdhaf4 was approximately one-third that in Ler (Fig. 2D).
Figure 2.
sdhaf4 shows lower SDH activity and succinate-dependent oxygen consumption compared with Ler. A, SDH activity at different succinate concentrations in the range of 0.1 to 10 mm. B, Kinetic analysis using the Michaelis-Menten formula was performed to determine maximum SDH velocity. C, Km value of SDH for succinate. D, Catalytic efficiency in sdhaf4 and Ler. E, Oxygen consumption in the presence of 5 mm succinate or 1 mm NADH. Error bars indicate se. Student’s t test was performed to determine significant differences between genotypes (n = 4): **, P < 0.01 and ***, P < 0.001.
To determine if succinate-dependent oxygen uptake was altered in sdhaf4, mitochondrial respiration rate was measured using a Clark-type oxygen electrode (Fig. 2E). The succinate-dependent respiration rate in sdhaf4 was only about half the rate in Ler (Fig. 2E), which was very similar to what was observed in sdhaf2 (Huang et al., 2013). To confirm that electron transport through other complexes of the ETC was not affected in sdhaf4, the assay was repeated using NADH as substrate. No significant differences between genotypes could be observed, and both Ler and sdhaf4 showed typical NADH-dependent mitochondrial respiration rates (Fig. 2E). Mitochondria from both lines also showed almost complete sensitivity of their NADH-dependent respiratory rates to cyanide treatment (Fig. 2E). This indicated that AOX capacity was similarly low in both genotypes. Taken together, these data showed that sdhaf4 had a decreased mitochondrial respiration rate when electron flow passed through SDH in a succinate-dependent manner and that no major compensatory changes in other respiratory pathways occurred.
Assembly of the SDH Holo-Complex Is Decreased in sdhaf4
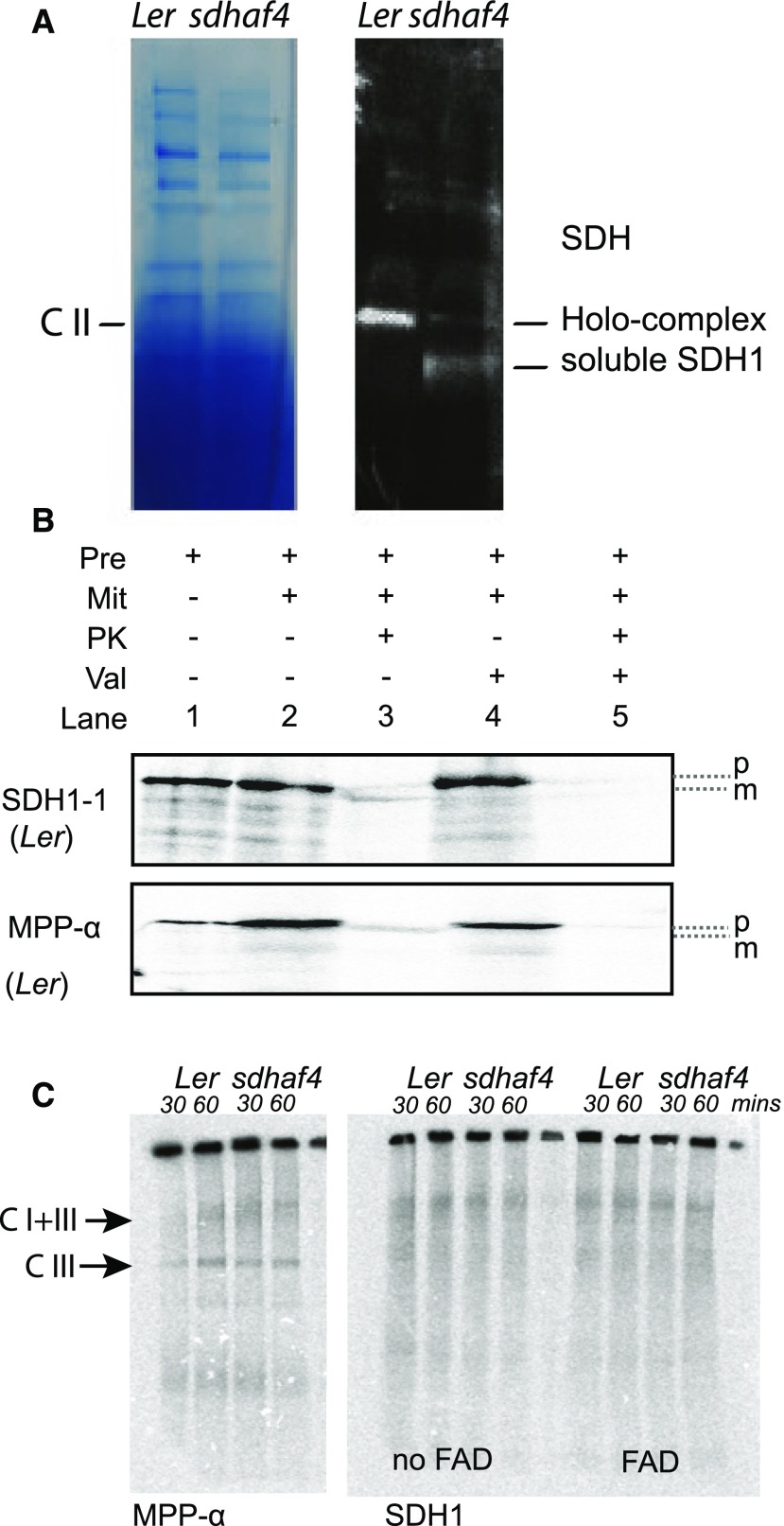
To determine if SDHAF4 is required for SDH1 assembly in plants, mitochondria from Ler and sdhaf4 were separated by blue native (BN) PAGE (Fig. 3). Anti-SDH1 antibodies were used to detect SDH1 protein (Fig. 3A). Ler showed a strong SDH1 signal at the expected size of the SDH holo-complex (∼170 kD). In the case of sdhaf4, two distinct bands were detected. One was about the same size as the immunoreactivity in Ler, representing the SDH holo-complex, but it showed only a weak signal compared with Ler. A second lower band, which was absent in Ler, was detected in sdhaf4 with a strong signal. This lower band likely represents soluble SDH1 (∼70 kD) that was not incorporated into the holo-complex (Fig. 3A). This is consistent with SDHAF4 being required for the assembly of SDH1 into the SDH holo-complex.
Figure 3.
Less SDH1 is incorporated into the SDH holo-complex, and it accumulates as a soluble protein in sdhaf4. A, SDH1 antibody was used to detect SDH1 abundance in whole mitochondria samples from Ler and sdhaf4 loaded on a BN gel and blotted on a PVDF membrane. Two bands were detected, potentially representing SDH holo-complex and soluble SDH1 protein, as indicated. B, Import of [35S]Met-labeled SDH1 and MPP-α into Ler mitochondria analyzed by SDS-PAGE. Pre, Precursor; Mit, mitochondria; PK, proteinase K; Val, valinomycin (inhibitor for import across the IMM); p, precursor protein; m, mature protein. C, [35S]Met-labeled SDH1 and MPP-α imported into Ler and sdhaf4 mitochondria for 30 and 60 min and separated by BN-PAGE. C I+III, ComplexI+III; C I, Complex I.
To test this further, the import of radiolabeled SDH1 into mitochondria was performed in an attempt to visualize directly the SDH1 assembly process (Fig. 3B). MPP-α subunit was used as a control. Radiolabeled 35S precursor proteins were imported into mitochondria and samples were separated by SDS-PAGE, demonstrating that both SDH1 and MMP-α could be imported and were protease protected in mitochondria isolated from Ler (Fig. 3B). However, when BN-PAGE was used to separate proteins in their native state after 30 or 60 min of incubation, the assembly of SDH could not be observed in either Ler or sdhaf4 (Fig. 3C). Exogenous FAD was added to determine if it would improve SDH1 assembly, without success (Fig. 3C). As a control, MPP-α subunit was imported in parallel experiments and showed successful radiolabeling of Complex III and the Supercomplex I and III in both Ler and sdhaf4 (Fig. 3C), demonstrating that the import and assembly of ETC components into these mitochondria could be successful, but not for SDH1 (Fig. 3C). It might be possible that, in the case of SDH1 assembly, additional substrates and/or conditions in the mitochondria are necessary that are still unknown.
Protein Abundance of SDH2 and SDHAF2 Is Altered in sdhaf4 Mitochondria
The results noted above showed that the loss of functional SDHAF4 resulted in decreased SDH activity and less succinate-dependent respiration. Based on the model in yeast, it was hypothesized that SDHAF4 would act as an assembly factor of the flavinated SDH1 and would be important for the assembly of an SDH1/SDH2 intermediate. Although the SDH1 import did not work to provide evidence in support of this, western-blot analysis did show a second lower molecular weight band in sdhaf4, potentially indicating soluble SDH1. To further investigate this, the abundance of gene-specific tryptic peptides derived from SDHAF4 as well as SDHAF2, and the subunits SDH1, SDH2, SDH6, and SDH7, were obtained from isolated mitochondria using MRM analysis. Samples of mitochondrial proteins from sdhaf4, Ler, sdhaf2, and Col-0 were analyzed to further investigate alterations in SDHAF2 and SDHAF4 abundance when SDH1 is reduced in abundance.
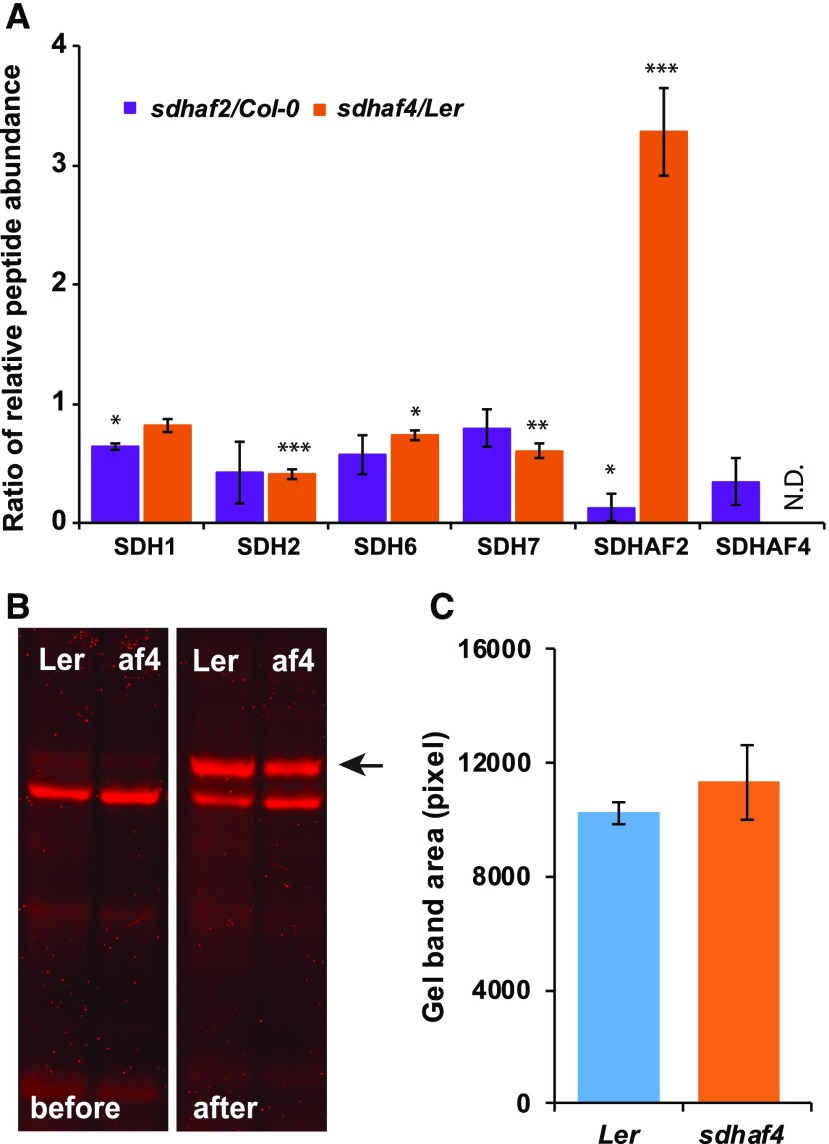
Comparing protein abundance in sdhaf4 and Ler, we found that the abundance of SDH1 was not altered in sdhaf4 (Fig. 4A), consistent with Figure 1E. Interestingly, SDH2 abundance was reduced significantly by half in sdhaf4 compared with Ler (Fig. 4A). The reduced abundance of SDH2 is consistent with the hypothesis that SDHAF4 acts as an assembly factor for SDH and acts during the incorporation of SDH2 into SDH1/SDH2 assembly intermediates. If this hypothesis is true, the loss of SDHAF4 should result in a reduction in the amount of SDH2 and a change in stoichiometry of SDH1 to SDH2. In sdhaf2, SDH1 abundance was decreased, indicating that, without SDHAF2 and without the inserted FAD, SDH1 was not stable enough to accumulate, leading to decreased amounts of SDH1 (Fig. 4A). The abundances of SDH6 and SDH7 peptides were reduced slightly in sdhaf4 but not in sdhaf2 (Fig. 4A). As they are likely to be replacements of the helices missing from SDH3 and SDH4 (Schikowsky et al., 2017), SDH6 and SDH7 can be used as representations for subunits SDH3 and SDH4, as specific peptides for these subunits could not be identified by mass spectrometry. Nevertheless, the lack of severe consequences in the stability or abundance of SDH6 and SDH7 indicated that the assembly of the SDH membrane arm might occur independently from SDH1/SDH2.
Figure 4.
Abundances of SDH2 and SDHAF2, but not FAD-bound SDH1, are altered in sdhaf4. A, MRM was used to detect peptides from SDH subunits and assembly factors. Shown are the ratios of peptides (mutant to wild type) based on whole mitochondria protein samples (50 µg; n = 3). N.D. indicates not detected in sdhaf4. Error bars indicate se. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 (Student’s t test for sdhaf2:Col-0 or sdhaf4:Ler). B, A FAD-bound protein assay was performed to compare FAD binding with SDH1 in sdhaf4 (af4) and Ler. Mitochondrial proteins (10 µg) were separated by SDS-PAGE, following a gel incubation in 10% (v/v) acetic acid for 30 min. FAD fluorescence scans were performed before and after the acetic acid treatment using Typhoon Trio Laser (Amersham Biosciences) and filters Cy5 (670 bp) and Cy3 (580 bp). The FAD-bound SDH1 band became visible after acetic acid incubation (marked with a black arrow). C, Quantification of FAD-bound SDH1 bands on gels, with three replicates.
There was a statistically significant 3-fold increase in SDHAF2 levels in sdhaf4 (Fig. 4A). It could be hypothesized that, due to the loss of SDHAF4, SDHAF2 may have accumulated in order to keep SDH1 stable as a soluble protein or that SDHAF2 was simply stabilized by the availability of its product, FAD-SDH1. Depending on where SDHAF4 acts, one might expect to see the same effect in sdhaf2, with the accumulation of SDHAF4 in order to keep SDH1 as stable as possible. However, from the results obtained, sdhaf2 plants did not show any increase in SDHAF4 abundance; in fact, it was reduced by half compared with Col-0 (Fig. 4A). The fact that SDHAF2 accumulated in sdhaf4, but not the reverse, is consistent with the hypothesis that SDHAF2 acts upstream of SDHAF4 in the assembly pathway of SDH. Together, these findings are consistent with the hypothesis that SDHAF4 abundance is dependent on the presence of SDH1, which is reduced in sdhaf2. SDHAF2, on the other hand, is highly abundant in sdhaf4, consistent with a role for SDHAF2 in SDH1 maturation and/or stability upstream of SDHAF4.
In previous work, the sdhaf2 line was shown to have lower levels of FAD-bound SDH1 protein (Huang et al., 2013). As SDHAF4 is suggested to act after SDHAF2, following the incorporation of FAD into SDH1, sdhaf4 should not show any differences in FAD binding compared with Ler. To test this hypothesis, the FAD-bound protein assay developed by Bafunno et al. (2004) was performed, which reveals FAD-SDH1 after gel acidification (Fig. 4B). Based on gel band area comparisons calculated in ImageJ (Fig. 4, B and C), no differences could be detected in the fluorescence of the acetic acid-dependent protein bands from Ler and sdhaf4, showing that FAD insertion into SDH1 was unaltered in sdhaf4. This further strengthens the hypothesis that SDHAF4 acts upon the already flavinated SDH1 subunit in plants.
SDH1 Protein Accumulates in the Soluble Fraction of sdhaf4 Mitochondria
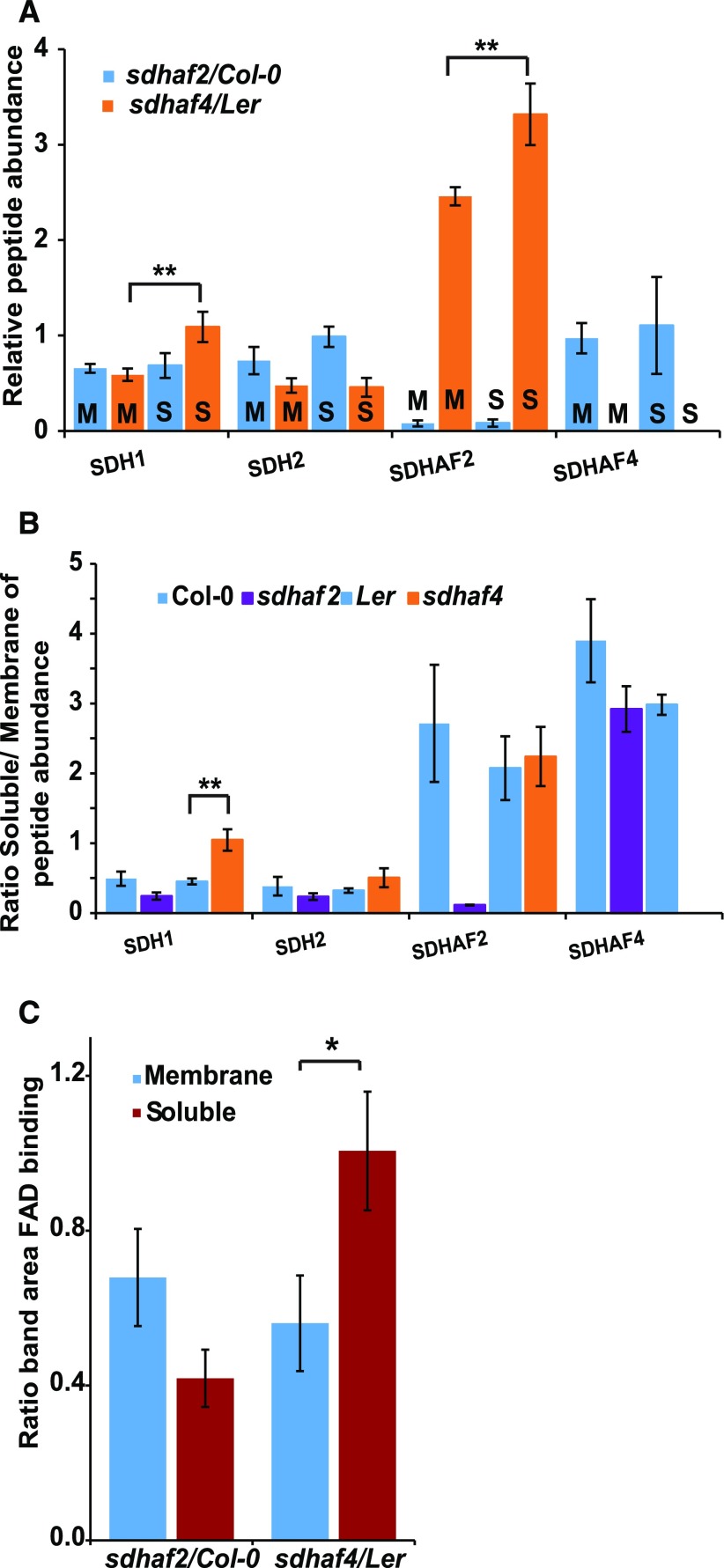
The sdhaf4 line displayed decreased amounts of SDH2 and increased amounts of SDHAF2, whereas the abundances of SDH1 as well as SDH6 and SDH7 were largely unaltered (Fig. 4A). FAD binding in SDH1 was unaltered (Fig. 4, B and C), but two bands for SDH1 were identified by native BN-PAGE (Fig. 3A). Based on these results, it was hypothesized that, in the case of sdhaf4, SDH1 may be stable and accumulate as a soluble protein; however, it is unable to attach to SDH2 and less assembled into the holo-complex II enzyme. To test this idea, the MRM assays were repeated, but mitochondrial samples were separated into soluble and membrane fractions. To compare possible differences in SDH components and SDH assembly factors, peptide detection of SDH1 and SDH2, as well as the assembly factors SDHAF2 and SDHAF4, was performed in sdhaf4, Ler, sdhaf2, and Col-0 (Fig. 5). To compare differences between the mutant lines, relative peptide abundance was calculated as the ratio of abundance in the mutant to that in the respective wild-type background (Fig. 5A). The results showed that, within the membrane samples, the abundance of SDH1 and SDH2 was reduced by half that in the two assembly factor mutants (Fig. 5A), indicating that there was less membrane-assembled SDH holo-complex present. However, comparing soluble fractions, sdhaf4 showed significantly higher amounts of SDH1 (Student’s t test, P = 0.04) compared with its matched membrane sample, with about the same amount of SDH1 protein as in Ler, while sdhaf2 showed no such difference (Fig. 5A). There was no SDH2 protein accumulation in the soluble fraction in sdhaf4; rather, it was about half the amount compared with Ler (Fig. 5A). In agreement with MRM analysis of whole mitochondria (Fig. 4A), the SDHAF2 protein accumulated in both fractions in sdhaf4; however, this accumulation was significantly higher in the soluble fraction (Fig. 5A).
Figure 5.
SDH1 accumulates as a soluble FAD-bound protein in sdhaf4. A, Peptide abundance in soluble (S) and membrane (M) mitochondrial protein fractions compared between mutant lines and the wild type (n = 3). B, Ratio of protein abundance in soluble fractions to that in membrane fractions within each genotype (n = 3). C, Comparison of the gel band area of FAD-bound SDH1 between mutant lines and the wild type from soluble and mitochondrial membrane fractions (n = 4). *, P ≤ 0.08 and **, P ≤ 0.05, by Student’s t test. Error bars indicate se.
Col-0 and Ler showed half as much SDH1 and SDH2 protein in the soluble fraction compared with their respective membrane fractions. In sdhaf2, approximately one-quarter of the SDH1 and SDH2 were in the soluble mitochondrial fraction (Fig. 5B). In contrast, sdhaf4 had about the same amount of SDH1 protein in its soluble mitochondrial fraction compared with its membrane fraction (Fig. 5B), a significantly higher soluble-to-membrane ratio than in Ler. These findings are in agreement with the hypothesis that SDHAF4 acts on flavinated SDH1 and promotes the assembly of SDH1 with SDH2. Due to the knockout of SDHAF4, SDH1 assembly with SDH2 appeared to be inhibited, resulting in the accumulation of soluble SDH1 and the degradation of SDH2. The SDH1 assembly factors SDHAF2 and SDHAF4 were significantly more abundant in the soluble fraction of Col-0 and Ler, indicating that they are present mostly as soluble proteins in mitochondria.
To test whether SDH1 accumulated as a soluble FAD-bound protein in sdhaf4, the FAD-binding assays also were repeated and quantified using soluble and membrane mitochondrial samples (Fig. 5C; Supplemental Fig. S3). The FAD-bound SDH1 contents in soluble and membrane fractions were compared between genotypes and are shown as the ratio of mutant to wild-type FAD-bound SDH1 (Fig. 5C). sdhaf2 showed reduced FAD binding, particularly in the soluble fraction, which is in agreement with the MRM findings and previous studies in sdhaf2 (Huang et al., 2013). In contrast, sdhaf4 showed a similar amount of FAD-bound protein in the soluble fraction to that in Ler (Fig. 5C). This provides additional evidence that SDH1 accumulated in the soluble fraction as a FAD-bound protein in sdhaf4, suggesting that SDHAF4 is important for SDH1 assembly.
Having a high amount of free flavinated SDH1 in the soluble fraction of mitochondria isolated from sdhaf4 plants might theoretically lead to a higher SDH activity or ROS production in that fraction. Therefore, to check for either scenario, SDH activity was measured in soluble and membrane fractions of mitochondria isolated from Ler, sdhaf4, sdhaf2, and Col-0 (Supplemental Fig. S4). Based on total protein amount, no increased activity in the soluble fraction from sdhaf4 could be observed; instead, the opposite was demonstrated (Supplemental Fig. S4A). The soluble fraction in all genotypes showed less DCPIP-dependent SDH activity than in the membrane fraction. Similar results were obtained from ROS detection in soluble and membrane fractions using DCFDA as a fluorescent dye (Supplemental Fig. S4B). Soluble mitochondrial samples showed less than half the rate of ROS production of membrane samples (Supplemental Fig. S4B). A possible explanation could be that soluble FAD-SDH1 by itself is only able to achieve partial functional electron transfer to PMS-DCPIP, while Complex II in membranes containing Fe-S clusters and the UQ site have a higher efficiency for electron transfer to PMS-DCPIP.
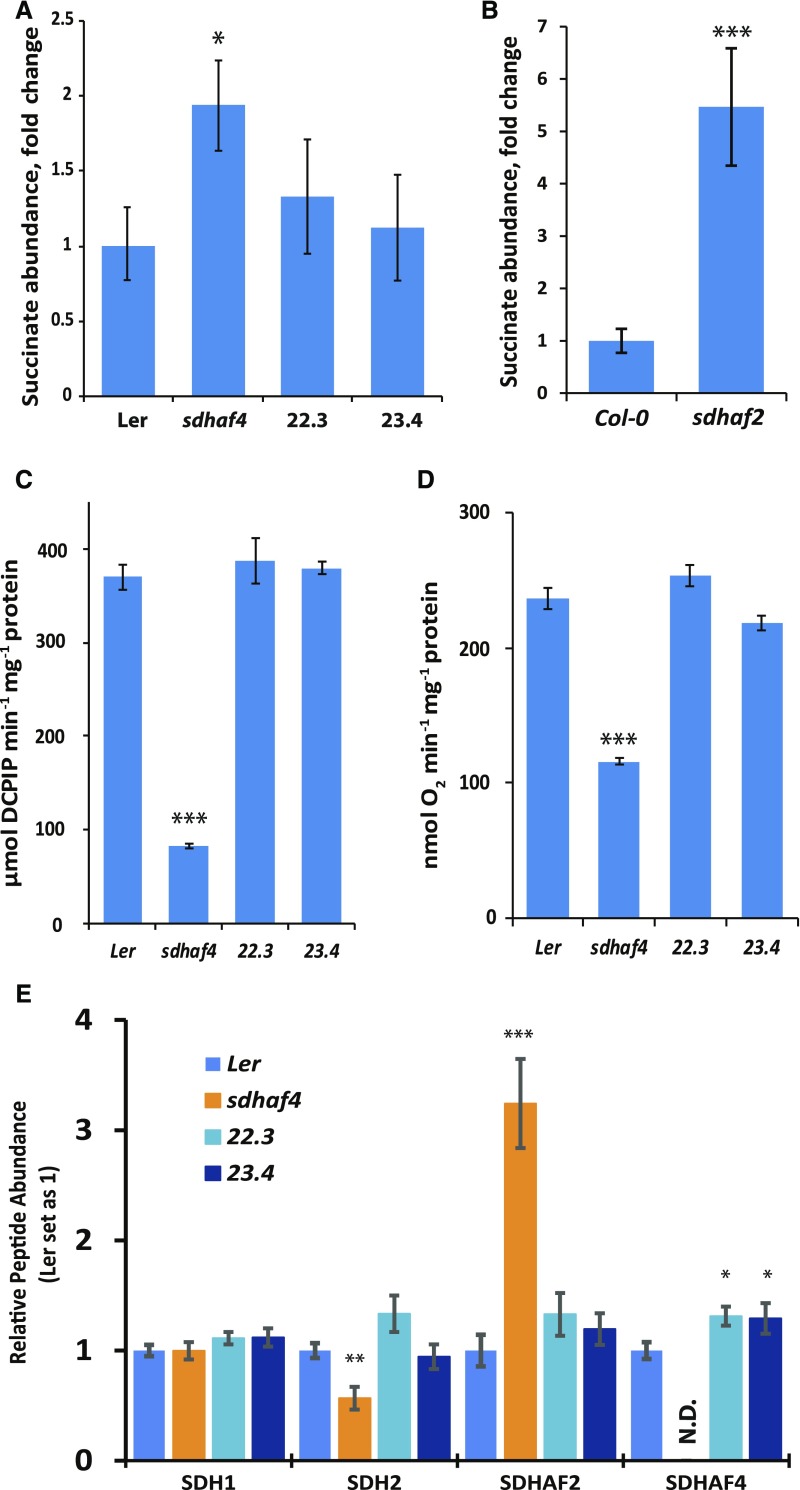
sdhaf4 Shows an Increase in Succinate, and Its SDH Dysfunction Can Be Genetically Complemented
Consistent with the inhibition of SDH activity (Fig. 2), sdhaf4 showed a 2-fold increase in the abundance of succinate in whole plant extracts of 10-d-old seedlings compared with Ler (n = 4; P = 0.015; Fig. 6A). To confirm that the succinate accumulation was caused by the loss of SDHAF4, 35S:SDHAF4 complementation lines were designed and two independent lines (22.3 and 23.4) were selected and analyzed together with sdhaf4 (Fig. 6). Both complementation lines showed similar amounts of succinate compared to Ler. As observed in previous studies, sdhaf2 showed a 5-fold accumulation of succinate compared with Col-0 in analogous assays (Fig. 6B). In order to confirm that the accumulation of succinate was due to the dysfunction of SDH in sdhaf4, mitochondria were isolated and SDH enzymatic activity and succinate-dependent respiration rate in both complementation lines were measured side by side with Ler and sdhaf4. Both lines showed similar levels of SDH enzymatic activity and succinate-dependent respiration rate to the wild type in all measurements, in contrast to sdhaf4, which showed only one-quarter of the SDH enzymatic activity (Fig. 6C) and half the respiration rate (Fig. 6D) of Ler. We also measured the protein levels of SDH subunits SDH1 and SDH2 and SDH assembly factors SDHAF2 and SDHAF4 in isolated mitochondria of two complemented lines (22.3 and 23.4), sdhaf4, and Ler (Fig. 6E). There were no differences in SDH1 abundance among the four lines (Fig. 6E). Both complemented lines showed similar levels of SDH2 and SDHAF2 compared with Ler (Fig. 6E). Furthermore, both complemented lines had significantly higher levels of SDHAF4 protein than that in Ler (Fig. 6E), presumably due to the overexpression of SDHAF4 using the 35S promoter. Therefore, the 35S:SDHAF4 complementation restored the defects in sdhaf4 (Fig. 6), and we concluded that the knockout of At5g67490 is responsible for its SDH dysfunction.
Figure 6.
35S-SDHAF4 complements SDH activity in sdhaf4. A, Fold change of succinate abundance in sdhaf4 and two independent complementation lines (22.3 and 23.4) compared with Ler (n = 4). B, Fold change of succinate abundance in sdhaf2 compared with Col-0 (n = 4). C, SDH enzyme activity at 10 mm succinate. D, Oxygen consumption in the presence of 5 mm succinate. E, Protein peptide detection of SDH subunits SDH1 and SDH2 and SDH assembly factors SDHAF2 and SDHAF4 (n = 4). Error bars indicate se; N.D., not detected. Student’s t test was used to determine significant differences between sdhaf4 and Ler complementation lines: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
DISCUSSION
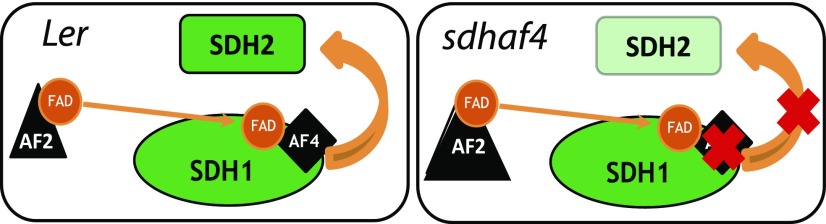
The functional assembly of SDH1 into the SDH protein complex is crucial for its activity, as SDH1 is the catalytic site for the oxidization of succinate to fumarate. Matured SDH1 must then assemble with SDH2, forming the SDH1/SDH2 intermediate that then docks with the SDH3/SDH4/SDH6/SDH7 membrane subcomplex. SDHAF2 was identified previously as an assembly factor of SDH in Arabidopsis (Huang et al., 2013) and shown to be involved in inserting the FAD cofactor into SDH1. Here, an Arabidopsis gene analogous to SDHAF4 in humans was studied, and our analysis of an At5g67490 T-DNA knockout line (sdhaf4) provided evidence that AtSDHAF4 is important for SDH2 stability and SDH1/SDH2 intermediate assembly (Fig. 7). The results presented in this study thus extend our understanding of the machinery of SDH assembly in plants and mechanistically confirm the putative role attributed to At5g67490 based on sequence comparison alone.
Figure 7.
Schematic diagram of FAD insertion into SDH1 and the assembly of SDH1 with SDH2. Left, As a first step in SDH1 assembly, SDHAF2 (AF2) is required to insert the FAD cofactor into SDH1. SDHAF4 (AF4) likely binds to the FAD site in SDH1 and promotes the assembly of SDH1 with SDH2. Right, Loss of SDHAF4 causes the decreased assembly of SDH1 with SDH2, leading to the degradation of SDH2. Loss of AF4 also induces the accumulation of AF2, presumably improving the stability of SDH1-FAD.
The results obtained from our studies of At5g67490 are broadly consistent with the model that SDHAF4 acts directly at the FAD-bound cofactor in SDH1 (Van Vranken et al., 2014). Knockout of this gene decreased SDH activity and succinate-dependent respiration and increased succinate content in plant tissues. Complementation with 35S:SDHAF4 restored not only succinate levels in the mutant but also mitochondrial SDH activity and the respiration rate to the levels observed in Ler (Figs. 2 and 6). No significant changes in SDH1 abundance or FAD-bound cofactor on the whole mitochondria level were observed in sdhaf4 compared to Ler (Fig. 4), demonstrating that, in contrast to sdhaf2, the amounts of available SDH1 and SDH1-FAD were not reduced in sdhaf4. In contrast, SDH2 abundance was reduced by half, indicating that the assembly of SDH1 with SDH2 was likely disrupted (Fig. 7). Furthermore, a mismatch in the stoichiometry of SDH1 and SDH2 occurred in sdhaf4. This is in agreement with studies performed in yeast, where knockout of SDHAF4 caused a decrease in SDH2 abundance (Van Vranken et al., 2014). Altogether, this provides further evidence of SDH2 stability being dependent on SDH1 availability and maturation across eukaryotes (Fig. 7). Interestingly, in D. melanogaster, SDHAF4 deletion demonstrated that dSDHAF4 is necessary to maintain SDH1 stability (Van Vranken et al., 2014).
When we compared mitochondrial membrane and soluble fractions, an accumulation of soluble flavinated SDH1 was observed in sdhaf4 (Fig. 5), a phenomenon consistent with reports in other eukaryotes (Van Vranken et al., 2014). Membrane-bound SDH1 and SDH2 abundance was halved in sdhaf4 compared with Ler (Fig. 5), which indicates that there was a reduced amount of membrane-bound SDH holo-complex in the mutant. This is in agreement with studies in yeast and D. melanogaster, which also showed decreased steady-state levels of the SDH holo-complex (Van Vranken et al., 2014). In addition, SDHAF2 abundance increased about 3-fold in sdhaf4, likely to prevent the destabilization of SDH1. In the case of sdhaf2, SDHAF4 did not accumulate but rather decreased in abundance (Fig. 4). Together, these results indicate that SDHAF2 acts upstream of SDHAF4 (Fig. 7). A high availability of nonassembled SDH1 protein in the soluble fraction provides evidence that SDHAF4 is involved in SDH1/SDH2 assembly in plants as described for animals (Van Vranken et al., 2014). It most likely acts on the FAD-bound cofactor in SDH1 (Van Vranken et al., 2014), following FAD insertion by SDHAF2 (Hao et al., 2009; Huang et al., 2013). SDHAF4 binds to SDH1 independently of any other core subunit and SDHAF2 (Van Vranken et al., 2014). In addition, the interaction of SDHAF4 and SDH1 is dependent on the covalent binding of FAD to SDH1, as yeast SDHAF4 failed to interact with SDH1 if the FAD cofactor was missing (Van Vranken et al., 2014). Furthermore, BN-PAGE analysis revealed that SDHAF4 formed a stable subcomplex with SDH1 that was not associated with the SDH holo-complex and that accumulated if SDH2 was not present (Van Vranken et al., 2014). This is in agreement with the decreased abundance of the SDH holo-complex in sdhaf4 and the formation of a slightly smaller subcomplex of soluble SDH1 (Fig. 3). In addition, the levels of SDH6 and SDH7, which are likely replacements of helices missing from SDH3 and SDH4 (Schikowsky et al., 2017), were not altered as drastcally as SDH2 in sdhaf2 or sdhaf4 (Fig. 4), indicating that the assembly of SDH1 and SDH2 occurred independently of SDH3 and SDH4.
It was proposed that SDHAF4 blocks the production of ROS during the assembly of SDH1 to SDH2 (Van Vranken et al., 2014). Therefore, the high abundance of free flavinated SDH1 in sdhaf4 might be expected to have toxic effects. ROS could be generated by solvent-accessible FAD, which may oxidize succinate to fumarate independently of the SDH complex. This would lead to the reduction of FAD, which would be autooxidized by molecular oxygen and result in the formation of superoxide (Messner and Imlay, 2002; Guzy et al., 2008). However, measurement of SDH activity via DCPIP reduction as well as ROS measurements using DCFDA as a fluorescent dye in soluble and membrane fractions of mitochondria isolated from sdhaf4 showed no increase in SDH activity or ROS accumulation in the soluble fractions (Supplemental Fig. S4). In fact, in sdhaf4, SDH activity and ROS production rates were lower in the soluble mitochondrial samples compared with the membrane samples (Supplemental Fig. S4), indicating that, in Arabidopsis at least, free FAD-SDH1 is not as efficient as membrane Complex II in promoting succinate oxidation. These results are in contrast to previous findings, where deletion of SDHAF4 resulted in a high sensitivity to oxidative stress induced by hyperoxia in D. melanogaster and the accumulation of ROS in yeast (Van Vranken et al., 2014), indicating that the assembly of SDH1 and its catalytic capability may be regulated differently in plants. Studies to date on SDH disagree with regard to the site of autooxidation responsible for ROS production. Some studies point to the FAD site within SDH (Imlay, 1995; Messner and Imlay, 1999, 2002), while others indicate ubisemiquinone and Fe-S centers to be the responsible sites (Guo and Lemire, 2003; Liang and Patel, 2004; Huang and Lemire, 2009). Studies in mammalian cells also showed that knockdown of SDH2, but not SDH1, caused an increase in ROS production (Ishii et al., 1998; Guzy et al., 2008). In Arabidopsis, it seems that FAD is not a major site responsible for ROS production (Supplemental Fig. S4). It is possible that Fe-S clusters or the UQ site form the reactive site, or that it is formed by a combination of FAD and Fe-S centers that are present in the membrane Complex II (Supplemental Fig. S4).
Despite its role in SDH assembly, it is clear that SDHAF4 is not essential for SDH function in Arabidopsis, as low enzyme activity and respiration rate could still be achieved in sdhaf4 (Fig. 2) and there was no apparent growth penalty or phenotype associated with the complete loss of the SDHAF4 protein (Supplemental Fig. S2). This is in contrast with the knockout of SDHAF2, which is embryo lethal in Arabidopsis, indicating that, while the SDHAF2 protein is essential for SDH1 flavination in plants (Huang et al., 2013), the SDHAF4 protein is not essential for FAD-SDH1 assembly with SDH2. Combined with this study, the observation that, in other mutant lines, partial dysfunction of the SDH complex can occur without phenotypic growth variation, or can result in the inhibition of root elongation or even better growth performance with enhanced photosynthesis rate (León et al., 2007; Araújo et al., 2011; Fuentes et al., 2011; Gleason et al., 2011; Huang et al., 2013), indicates that the complicated physiological roles of SDH depend on the level or site of dysfunction and, presumably, also downstream signaling.
D. melanogaster lacking SDHAF4 had a more severe phenotype than the plant mutants, with D. melanogaster showing reduced SDH activity and destabilization of both SDH1 and SDH2 as well as a much more significant decrease in abundance of the SDH holo-complex (Van Vranken et al., 2014). Yeast and mammalian cells, on the other hand, were able to maintain 40% to 50% of wild-type SDH activity and showed a similar level of mature SDH complex assembly (Van Vranken et al., 2014) to that observed in Arabidopsis (Fig. 2). While yeast mutants showed relatively unaltered levels of SDH1, D. melanogaster mutants had a highly reduced abundance of SDH1, which was potentially responsible for the almost complete loss of holo-complex in those lines (Van Vranken et al., 2014). Yeast and mammalian cells had functional SDH in the absence of SDHAF4 (Van Vranken et al., 2014). Similarly, Arabidopsis also was able to maintain SDH1 levels in the form of soluble protein and maintained some SDH activity (Figs. 2 and 5A), suggesting that either additional assembly factors exist in plants or that the process of SDH1/SDH2 intermediate assembly occurs spontaneously at a low rate without the requirement for this assembly factor.
MATERIALS AND METHODS
Plant Lines
Seeds of Arabidopsis (Arabidopsis thaliana) ecotype Col-0 (Lamb et al., 1989) and Ler, as well as previously described sdhaf2, were used within this study. Seeds of a T-DNA insertion line (GT_5_75821) were obtained from the Nottingham Arabidopsis Stock Centre. Plants were screened for homozygous insertion using standard PCR-based methods, the primers for which are listed in Supplemental Figure S1.
Growth of Arabidopsis Plants on Soil
Seeds of Arabidopsis lines were sown on a 1:3:1 (v/v) perlite:shamrock compost:vermiculite soil mix and covered with a transparent acrylic hood. After 3 d of stratification in the dark at 4°C, plants were transferred to a growth chamber with controlled long-day conditions (16 h of light/8 h of dark, light intensity of 200 µmol m−2 s−1, relative humidity of 70%, and 22°C day/17°C night).
Growth of Hydroponic Arabidopsis Plants
Arabidopsis seeds were washed in 70% (v/v) ethanol for 2 min and in sterilization solution (5% [v/v] bleach and 0.1% [v/v] Tween 20) for 5 min with periodic shaking. Seeds were washed five times in sterile water before being dispensed into 250-mL plastic vessels containing 80 mL of MS medium (one-half-strength Murashige and Skoog medium without vitamins, one-half-strength Gamborg B5 vitamin solution, 5 mm MES, and 2.5% [w/v] Suc, pH 5.7). Hydroponic cultures were grown under long-day conditions (described above) with shaking at 220 rpm for 2 weeks.
Isolation of Mitochondria from Hydroponic Cultures
Mitochondria were isolated from 2-week-old hydroponically grown Arabidopsis plants based on the method described by Millar et al. (2001) with slight modifications. Plant materials were homogenized in grinding buffer (0.3 m Suc, 25 mm tetrasodium pyrophosphate, 1% [w/v] polyvinylpyrrolidone [PVP]-40, 2 mm EDTA, 10 mm KH2PO4, 1% [w/v] BSA, and 20 mm ascorbic acid, pH 7.5) using mortar and pestle for 2 to 5 min, twice. The homogenate was filtered through four layers of Miracloth and centrifuged at 2,500g for 5 min at 4°C. The supernatant was centrifuged at 14,000g for 20 min at 4°C, and the resulting pellet was resuspended in Suc wash buffer (0.3 m Suc, 0.1% [w/v] BSA, and 10 mm TES, pH 7.5). Resuspended tissue material was carefully layered over a 35-mL PVP-40 gradient (30% (v/v) Percoll and 0%–4% (w/v) PVP). The gradient was centrifuged at 40,000g for 40 min at 4°C. The mitochondrial band was collected and washed three times in Suc wash buffer without BSA at 20,000g for 20 min at 4°C, and aliquots of isolated mitochondrial protein were stored at −80°C.
Determination of Root Growth
Arabidopsis seeds were washed in 70% (v/v) ethanol for 2 min and in sterilization solution (5% [v/v] sodium hydrochlorite and 0.1% [v/v] Tween 20) for 5 min with periodic shaking. Seeds were washed five times in sterile water before individual seeds were transferred onto 100-mm2 square petri dishes containing MS medium (one-half-strength Murashige and Skoog medium without vitamins, one-half-strength Gamborg B5 vitamin solution, 5 mm MES, 0.8% [w/v] agar, and 2.5% [w/v] Suc, pH 5.8). Plates were wrapped in aluminum foil and kept at 4°C for 48 h before being transferred into a growth chamber set up with controlled conditions (16/8-h light/dark period with a light intensity of 100–125 µmol m−2 s−1 at 22°C). Over a period of 2 weeks, root length development was documented and root area was calculated using ImageJ.
Designing the SDHAF4-GFP Construct
Gateway technology (Thermo Fisher) was used to create the GFP construct. Full-length SDHAF4 genomic sequence was amplified by PCR with primers containing attB adapters: forward primer, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCACCATGGCGACGAACAACATCGTACG-3′; and reverse primer, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCGAAATCAGAGCATCGACCACGTTG-3′. PCR fragments were loaded onto a 1.5% (w/v) agarose gel, followed by gel purification (Qiagen). Purified DNA of SDHAF4 was cloned into pDONR201 and subsequently the GFP vector (pDest-CGFP) under the control of the 35S promoter of Cauliflower mosaic virus using the BP and LR reaction kit (Thermo Fisher), followed by transformation into Escherichia coli DH5α (Carrie et al., 2008). Plasmid isolation using the Plasmid Midi Kit (Qiagen) was performed as described by the manufacturer.
Arabidopsis Transient Transformation Using Gold Particle Bombardment
As a mitochondrial marker, red fluorescent protein was fused to Glycine max alternative oxidase (RFP-AOX). Arabidopsis suspension cell culture was used for transient transformation. Arabidopsis cell culture (2–3 mL) was placed onto sterile Whatman filter paper and placed onto cell culture MS plates containing mannitol as osmoticum for 2 h prior to transformation. To 50 µL of washed gold (1 µm size; Bio-Rad), 5 µg of DNA (SDHAF4-GFP or AOX-RFP) was added and mixed. During mixing, 50 µL of 2.5 m CaCl2 and 20 µL of 100 mm spermidine were added one after the other. Tubes were vortexed and centrifuged for 30 s, and the supernatant was removed. A total of 140 µL of 70% (v/v) ethanol was added and mixed, and the supernatant was removed, followed by adding 140 µL of 100% ethanol, vortexing, and removal of the supernatant. Finally, 56 µL of 100% ethanol was added. Seven macrocarriers (Bio-Rad) were prepared for each, and precipitated gold was resuspended before 8 µL was transferred onto the center of each macrocarrier.
Arabidopsis cell culture was transformed using the PDS-1000 system according to the manufacturer’s instructions (Bio-Rad). Gold particles were fired onto cells under vacuum at an approximate pressure of 1,300 bar. Cells were kept at 22°C in the dark for 12 to 24 h before GFP and RFP were visualized at 100× magnification with a BX61 Olympus microscope using excitation wavelengths at 460/480 nm (GFP) or 535/555 nm (RFP) and emission wavelengths of 495 to 540 nm (GFP) or 570 to 625 nm (RFP). Images were captured using CellR imaging software as described previously (Carrie et al., 2009).
Design of 35S:SDHAF4 Complementation Lines
Complementation of the sdhaf4 mutant line (GT_5_75821) was achieved using the full-length At5g67490 cDNA. The resulting sequence was cloned into Gateway pDonr Zeo vector (Invitrogen), sequenced, and recombined into binary vector pB2GW7 containing the cauliflower mosaic virus 35S promoter (Invitrogen). This construct was transformed into Agrobacterium tumefaciens and used to transform homozygous sdhaf4 mutants by the floral dip method (Mara et al., 2010). Transformed plants were selected on agar plates containing Basta (5 mg mL−1).
Metabolite Extraction and Gas Chromatography-Mass Spectrometry Data Analysis of Arabidopsis Leaves
Two-week-old plant leaves grown on MS petri dishes under long-day conditions were collected in 2-mL microfuge tubes and snap frozen in liquid nitrogen. Stainless grinding balls were added, and leaf tissue was homogenized twice using a mixer mill (MM 301; Retsch) at a frequency 20 Hz for 1 min each. Metabolite extraction medium (20 mL of HPLC-grade methanol, 2 mL of fresh MilliQ water, and 1 mL of Ribitol [0.2 mg mL−1]; 150 µL per 10 mg of leaf material) was added to samples, mixed, and incubated at 65°C for 20 min. Samples were centrifuged at 20,000g for 10 min, and 60 µL of supernatant was transferred into verex insert tubes (6 mm diameter; Phenomenex) and set in 2-mL tubes. Samples were vacuum dried and derivatized before analysis on an Agilent GC/MSD system (Agilent Technologies). Data preprocessing and statistical analysis were performed using MetabolomeExpress software (version 1.0; http://www.metabolome-express.org) as described previously (Carroll et al., 2010).
Quantitative RT-PCR to Determine SDHAF4 Gene Expression
Ler and sdhaf4 plants were grown on soil under long-day conditions for 3 weeks. RNA isolation was performed using the RNAeasy extraction kit (Qiagen) following the manufacturer’s instructions. Three micrograms of DNA-free RNA was used for cDNA synthesis using the reverse transcriptase provided in the SuperScript III kit (Invitrogen). One microliter of 25-fold diluted reverse transcriptase reaction was used for the quantitative RT-PCR. Samples were loaded onto 384-well plates and mixed with 4 µL of SYBR Green I Master Mix (Roche Diagnostics). Samples were analyzed using the LightCycler 480 Roche real-time PCR system as described previously (de Longevialle et al., 2008). Primers used for quantitative RT-PCR were as follows: SDHAF4-forward, 5′-TGTTAGGCCTAGCTCCTGATG-3′; SDHAF4-reverse, 5′-ACTGGAATAACAAGATCACCAG-3′; Actin-forward, 5′-GAAGATCAAGATCATTGCTCCT-3′; and Actin-reverse, 5′-TACTCTGCTTGCTGATCCA-3′.
SDH Activity and Kinetic Analysis
SDH activity was measured spectrophotometrically by following the reduction of DCPIP at 600 nm using succinate as the substrate. Arabidopsis mitochondria (50 µg) isolated from the wild type and sdhaf4 were used in 1 mL of reaction medium (50 mm potassium phosphate, pH 7.4, 0.1 mm EDTA, 0.1% [w/v] BSA, 10 mm potassium cyanide, 0.12 mm DCPIP, and 1.6 mm PMS). To calculate SDH activity, an extinction coefficient of 21 mm−1 cm−1 at 600 nm for DCPIP was used. Calculations of the Michaelis-Menten kinetic constants Km and Vmax were performed using R software (script can be found in Supplemental Document S1).
Measurement of Oxygen Uptake Using a Clark Electrode
Oxygen consumption was measured using a Clark oxygen electrode (Hansatech Instruments). Arabidopsis mitochondria (100 µg) isolated from the wild type and sdhaf4 were used, and oxygen uptake was measured as described previously (Huang et al., 2013) in the presence of either 5 mm succinate or 1 mm NADH, in both cases with the addition of 1 mm cyanide.
Measurement of Flavin-Protein Binding
Isolated mitochondrial protein samples were separated on an SDS mini-gel (Mini-PROTEAN, any kD; Bio-Rad). Ten micrograms of mitochondrial protein was mixed with β-mercaptoethanol and 2× SDS buffer (8% [w/v] SDS, 125 mm Tris-HCl, 20% [v/v] glycerol, and 0.01% [w/v] Bromphenol Blue) in a 1:4 (v/v) ratio, and samples were incubated at 95°C for 3 min before being loaded onto the mini-gel. The gel run was set to a constant voltage of 200 V for 20 min. Flavin-protein binding was measured as described by Bafunno et al. (2004). After electrophoresis, the gel was incubated in 10% acetic acid solution and scanned before and after treatment using a Typhoon Trio Laser Imager (Amersham Biosciences) and Cy5 (670 bp) and Cy3 (580 bp) filters. Bound FAD can be measured based on the gel band area that becomes visible after acetic acid treatment. The maximum fluorescence intensity was reached at pH 3.2 to 3.5. ImageJ was used to determine the band area between different genotypes.
Measurement of ROS from Isolated Mitochondria Using DCFDA
Mitochondria isolated from Ler and sdhaf4 were separated into membrane and soluble fractions by freeze/thawing three times followed by centrifugation at 20,000g for 10 min. DCFDA measurements were performed as described previously (Belt et al., 2017). In brief, fresh mitochondria from soluble and membrane fractions (15 µg) were transferred in 50 µL of buffer (0.3 m Suc, 5 mm KH2PO4, 10 mm TES, 10 mm NaCl, 2 mm MgSO4, and 0.1% [w/v] BSA, pH 7.2). DCFDA was diluted to 10 µm, a final volume of 50 µL in the same buffer solution together with the individual substrates. Both solutions were transferred and mixed on a 96-well plate to a final volume of 100 µL. Fluorescence with excitation/emission spectra of 480/529 nm was measured over 10 min, and the slope was calculated.
BN-PAGE and Western-Blot SDH1-1 Detection
A precast gradient native gel (4.5%–16%; Invitrogen) was used to load mitochondria from wild-type and sdhaf4 samples. Thirty micrograms of mitochondrial pellet was resuspended in 10 µL of solubilization buffer (30 mm HEPES, pH 7.4, 150 mm potassium acetate, 10% (v/v) glycerol, and 0.5 g of digitonin per 10 mL) per 100 µg of mitochondrial protein. Samples were incubated on ice for 20 min, followed by a second centrifugation for 20 min at 18,300g at 4°C. Supernatant was transferred into a new 1.5-mL microfuge tube, and 1 µL of 5% (w/v) Serva Blue G per 20 µL of solubilization buffer was added. Gel electrophoresis was run at 150 V for 2 to 3 h following the manufacturer’s protocol.
For western-blot transfer, the protein gel was soaked in transfer buffer (25 mm Tris-HCl, pH 7.6, 192 mm Gly, 20% (v/v) methanol, and 0.03% (w/v) SDS) for 30 min before assembly in a Hoefer semiphor semidry blotting apparatus. Blot transfer was set up for 0.8 mA cm−2 membrane area, and voltage was limited to 100 V. After blot transfer, the membrane was incubated in blocking solution (3 mL of 10× blocking solution [Sigma] and 27 mL of 1× TBS Tween buffer [1.5 m NaCl and 100 mm Tris-HCl, pH 7.4]) at room temperature for 1 h before incubation with primary SDH1-1 antibody (Peters et al., 2012; SDH1-1 antibody was kindly provided by Hans-Peter Braun) in a 1:1,000 dilution in TBS Tween buffer overnight at 4°C on a shaker. The membrane was washed in TBS Tween solution three times before incubation with the secondary antibody (anti-rabbit IgG; Sigma) for 1 h at room temperature. Washing steps in TBS Tween buffer were repeated, and signal was detected using 3 mL of western-blot detection substrates in a 1:1 ratio (Clarity Western Blotting Substrates; Bio-Rad).
Mitochondrial Protein in Vitro Import
Mitochondrial protein import assays were carried out as described previously (Wang et al., 2012). [35S]Met-labeled precursor proteins of MPP-α (At1g51980) and SDH1-1 (At5g66760) were synthesized using rabbit reticulocyte TNT in vitro transcription/translation lysate (Promega). Briefly, 100 μg of freshly isolated mitochondria was incubated in 180 μL of import master mix (0.3 m Suc, 50 mm KCl, 10 mm MOPS, pH 7.4, 5 mm KH2PO4, 0.1% [w/v] BSA, 1 mm MgCl2, 1 mm Met, 200 mm ADP, 750 mm ATP, 5 mm succinate, 5 mm DTT, 5 mm NADH, and 1 mm GTP) with or without the addition of valinomycin. Radiolabeled precursor protein (10 µL) was added, and the import reaction was initiated by incubation at 26°C with gentle rocking for 30 min. To one-half of each reaction, 3.2 µg of proteinase K was added, and reactions were incubated on ice for 30 min. Proteolysis was inhibited with 1 µL of 100 mm PMSF. The mitochondria were pelleted by centrifugation at 20,000g for 5 min at 4°C, and the mitochondrial pellet was resuspended with SDS-PAGE sample buffer and resolved on 12% Tris acrylamide gels. For BN-PAGE import assays, 30 µg of mitochondria was inoculated with 10 µL of radiolabeled protein, and the samples were incubated for 30 and 60 min. Mitochondria were pelleted as above, and the pellets were resuspended in 5% (w/v) digitonin buffer and resolved on precast gradient native gels (4.5%–16%; Invitrogen). The gels were Coomassie Blue stained, dried, and exposed to a BAS TR2040 phosphor image screen for 24 h. The phosphor-imaging plates were scanned using the Typhoon 5 (GE Healthcare).
MRM Sample Preparation and Analysis
Fifty to 100 µg of isolated mitochondrial protein extract (see “Isolation of Mitochondria from Hydroponic Cultures”) per plant line was used for MRM analysis. Whole mitochondria protein samples as well as membrane and soluble fractions were used. To separate soluble and membrane mitochondrial fractions, whole mitochondria samples were frozen and thawed three times with vortexing in between. To separate fractions, samples were centrifuged at 20,000g for 20 min at 4°C. Supernatant was transferred into a new tube (soluble fraction), and the pellet was resuspended in Suc wash buffer (membrane fraction). Proteins were acetone precipitated overnight at −20°C. After the removal of acetone, dried pellets were resuspended in buffer containing 7 m urea, 2 m thiourea, 50 mm NH4HCO3, and 10 mm DTT. Samples were solubilized at room temperature with shaking at 500 rpm. Iodacetamide (25 mm) was added, and samples were incubated for 45 min at room temperature in the dark before being diluted to a final concentration of less than 1 m urea using 50 mm NH4HCO3. Trypsin (0.8 µg µL−1) was added to the samples in a 1:20 ratio of trypsin to protein content, and samples were incubated overnight at 37°C, followed by acidification using 0.1% (v/v) formic acid. Peptides from trypsin-digested protein extracts from soluble and membrane fractions were analyzed by triple-quadrupole mass spectrometry as described previously (Huang et al., 2013). The following peptide sequences were used to quantify protein abundance: AVIELENYGLPFSR, SMTMEIR, and SSYTIVDHTYDAVVVGAGGAGLR for SDH1; NEMDPSLTFR for SDH2; FMEWWER and LSFFENYTR for SDH6; ALLAEDASLR for SDH7; AAAGQPWVR for SDHAF2; and YGDWEQR for SDHAF4.
A 6495 triple-quadrupole mass spectrometer (Agilent Technologies) regulated by MassHunter Workstation Data Acquisition software (version B.07.01, build 7.1.7112.0; Agilent Technologies) was used, and MRM data were analyzed in Skyline (version 3.5.0.9319, MacCoss Laboratory, University of Washington [https://skyline.gs.washington.edu]) by integrating peak areas for each quantifier ion. To correct for differences, the sum of all detected peptides in each run was normalized to the sum of ATP synthase peptides.
Accession Number
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number At5g67490 (Q84WS5) for SDHAF4.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Genotyping of the T-DNA insertion line of At5g67490.
Supplemental Figure S2. Plant growth and development in soil and on plates.
Supplemental Figure S3. SDS-PAGE of FAD-bound SDH1 from soluble and membrane protein fractions.
Supplemental Figure S4. SDH activity and ROS production are not increased in the soluble mitochondrial fraction of sdhaf4.
Supplemental Document S1. R script version 3.3.1 was used for the calculation of SDH Km and Vmax.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Ricarda Fenske for MRM analysis.
Footnotes
This work was supported by the facilities of the Australian Research Council (ARC) Centre of Excellence Program (CE140100008). S.H. and M.M. were funded as ARC Australian Future Fellows (FT130101338 and FT13010012, respectively). O.V.A. was supported by ARC Discovery Projects (DP160103573), Swedish Research Council (VR 2017003854), Crafoord Foundation (20170862), and Carl Trygger Foundation (CTS17-487). K.B. holds a SIRF (Scholarship for International Research Fees) and UIS (University International Stipend) from the University of Western Australia.
Articles can be viewed without a subscription.
References
- Araújo WL, Nunes-Nesi A, Osorio S, Usadel B, Fuentes D, Nagy R, Balbo I, Lehmann M, Studart-Witkowski C, Tohge T, et al. (2011) Antisense inhibition of the iron-sulphur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture. Plant Cell 23: 600–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafunno V, Giancaspero TA, Brizio C, Bufano D, Passarella S, Boles E, Barile M (2004) Riboflavin uptake and FAD synthesis in Saccharomyces cerevisiae mitochondria: involvement of the Flx1p carrier in FAD export. J Biol Chem 279: 95–102 [DOI] [PubMed] [Google Scholar]
- Belt K, Huang S, Thatcher LF, Casarotto H, Singh KB, Van Aken O, Millar AH (2017) Salicylic acid-dependent plant stress signaling via mitochondrial succinate dehydrogenase. Plant Physiol 173: 2029–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Lang BF, Reith M, Gray MW (1996) Genes encoding the same three subunits of respiratory complex II are present in the mitochondrial DNA of two phylogenetically distant eukaryotes. Proc Natl Acad Sci USA 93: 2328–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Kuehn K, Duncan O, Barthet M, Smith PM, Eubel H, Meyer E, Day DA, Millar AH, et al. (2008) Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett 582: 3073–3079 [DOI] [PubMed] [Google Scholar]
- Carrie C, Kühn K, Murcha MW, Duncan O, Small ID, O’Toole N, Whelan J (2009) Approaches to defining dual-targeted proteins in Arabidopsis. Plant J 57: 1128–1139 [DOI] [PubMed] [Google Scholar]
- Carroll AJ, Badger MR, Millar AH (2010) The MetabolomeExpress Project: enabling web-based processing, analysis and transparent dissemination of GC/MS metabolomics datasets. BMC Bioinformatics 11: 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Longevialle AF, Hendrickson L, Taylor NL, Delannoy E, Lurin C, Badger M, Millar AH, Small I (2008) The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J 56: 157–168 [DOI] [PubMed] [Google Scholar]
- Eubel H, Jänsch L, Braun HP (2003) New insights into the respiratory chain of plant mitochondria: supercomplexes and a unique composition of complex II. Plant Physiol 133: 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P, Léon G, Elorza A, Holuigue L, Araya A, Jordana X (2002) The four subunits of mitochondrial respiratory complex II are encoded by multiple nuclear genes and targeted to mitochondria in Arabidopsis thaliana. Plant Mol Biol 50: 725–734 [DOI] [PubMed] [Google Scholar]
- Fuentes D, Meneses M, Nunes-Nesi A, Araújo WL, Tapia R, Gómez I, Holuigue L, Gutiérrez RA, Fernie AR, Jordana X (2011) A deficiency in the flavoprotein of Arabidopsis mitochondrial complex II results in elevated photosynthesis and better growth in nitrogen-limiting conditions. Plant Physiol 157: 1114–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi D, Goffrini P, Uziel G, Horvath R, Klopstock T, Lochmüller H, D’Adamo P, Gasparini P, Strom TM, Prokisch H, et al. (2009) SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat Genet 41: 654–656 [DOI] [PubMed] [Google Scholar]
- Gleason C, Huang S, Thatcher LF, Foley RC, Anderson CR, Carroll AJ, Millar AH, Singh KB (2011) Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proc Natl Acad Sci USA 108: 10768–10773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Lemire BD (2003) The ubiquinone-binding site of the Saccharomyces cerevisiae succinate-ubiquinone oxidoreductase is a source of superoxide. J Biol Chem 278: 47629–47635 [DOI] [PubMed] [Google Scholar]
- Guzy RD, Sharma B, Bell E, Chandel NS, Schumacker PT (2008) Loss of the SdhB, but not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol Cell Biol 28: 718–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, Devilee P, Cremers CW, Schiffman JD, Bentz BG, et al. (2009) SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science 325: 1139–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Lemire BD (2009) Mutations in the C. elegans succinate dehydrogenase iron-sulfur subunit promote superoxide generation and premature aging. J Mol Biol 387: 559–569 [DOI] [PubMed] [Google Scholar]
- Huang S, Millar AH (2013) Succinate dehydrogenase: the complex roles of a simple enzyme. Curr Opin Plant Biol 16: 344–349 [DOI] [PubMed] [Google Scholar]
- Huang S, Taylor NL, Narsai R, Eubel H, Whelan J, Millar AH (2010) Functional and composition differences between mitochondrial complex II in Arabidopsis and rice are correlated with the complex genetic history of the enzyme. Plant Mol Biol 72: 331–342 [DOI] [PubMed] [Google Scholar]
- Huang S, Taylor NL, Ströher E, Fenske R, Millar AH (2013) Succinate dehydrogenase assembly factor 2 is needed for assembly and activity of mitochondrial complex II and for normal root elongation in Arabidopsis. Plant J 73: 429–441 [DOI] [PubMed] [Google Scholar]
- Imlay JA. (1995) A metabolic enzyme that rapidly produces superoxide, fumarate reductase of Escherichia coli. J Biol Chem 270: 19767–19777 [PubMed] [Google Scholar]
- Ishii N, Fujii M, Hartman PS, Tsuda M, Yasuda K, Senoo-Matsuda N, Yanase S, Ayusawa D, Suzuki K (1998) A mutation in succinate dehydrogenase cytochrome b causes oxidative stress and ageing in nematodes. Nature 394: 694–697 [DOI] [PubMed] [Google Scholar]
- Jacoby RP, Li L, Huang S, Pong Lee C, Millar AH, Taylor NL (2012) Mitochondrial composition, function and stress response in plants. J Integr Plant Biol 54: 887–906 [DOI] [PubMed] [Google Scholar]
- Lamb CJ, Lawton MA, Dron M, Dixon RA (1989) Signals and transduction mechanisms for activation of plant defenses against microbial attack. Cell 56: 215–224 [DOI] [PubMed] [Google Scholar]
- Lemire BD, Oyedotun KS (2002) The Saccharomyces cerevisiae mitochondrial succinate:ubiquinone oxidoreductase. Biochim Biophys Acta 1553: 102–116 [DOI] [PubMed] [Google Scholar]
- León G, Holuigue L, Jordana X (2007) Mitochondrial complex II is essential for gametophyte development in Arabidopsis. Plant Physiol 143: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang LP, Patel M (2004) Iron-sulfur enzyme mediated mitochondrial superoxide toxicity in experimental Parkinson’s disease. J Neurochem 90: 1076–1084 [DOI] [PubMed] [Google Scholar]
- Mara C, Grigorova B, Liu Z (2010) Floral-dip transformation of Arabidopsis thaliana to examine pTSO2:β-glucuronidase reporter gene expression. J Vis Exp 1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner KR, Imlay JA (1999) The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J Biol Chem 274: 10119–10128 [DOI] [PubMed] [Google Scholar]
- Messner KR, Imlay JA (2002) Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J Biol Chem 277: 42563–42571 [DOI] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giegé P, Leaver CJ (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127: 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Eubel H, Jänsch L, Kruft V, Heazlewood JL, Braun HP (2004) Mitochondrial cytochrome c oxidase and succinate dehydrogenase complexes contain plant specific subunits. Plant Mol Biol 56: 77–90 [DOI] [PubMed] [Google Scholar]
- Millar AH, Whelan J, Soole KL, Day DA (2011) Organization and regulation of mitochondrial respiration in plants. Annu Rev Plant Biol 62: 79–104 [DOI] [PubMed] [Google Scholar]
- Na U, Yu W, Cox J, Bricker DK, Brockmann K, Rutter J, Thummel CS, Winge DR (2014) The LYR factors SDHAF1 and SDHAF3 mediate maturation of the iron-sulfur subunit of succinate dehydrogenase. Cell Metab 20: 253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters K, Niessen M, Peterhänsel C, Späth B, Hölzle A, Binder S, Marchfelder A, Braun HP (2012) Complex I-complex II ratio strongly differs in various organs of Arabidopsis thaliana. Plant Mol Biol 79: 273–284 [DOI] [PubMed] [Google Scholar]
- Schikowsky C, Senkler J, Braun HP (2017) SDH6 and SDH7 contribute to anchoring succinate dehydrogenase to the inner mitochondrial membrane in Arabidopsis thaliana. Plant Physiol 173: 1094–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Huo X, Zhai Y, Wang A, Xu J, Su D, Bartlam M, Rao Z (2005) Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 121: 1043–1057 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Millar AH (2011) The Arabidopsis thaliana 2-D gel mitochondrial proteome: refining the value of reference maps for assessing protein abundance, contaminants and post-translational modifications. Proteomics 11: 1720–1733 [DOI] [PubMed] [Google Scholar]
- Van Vranken JG, Bricker DK, Dephoure N, Gygi SP, Cox JE, Thummel CS, Rutter J (2014) SDHAF4 promotes mitochondrial succinate dehydrogenase activity and prevents neurodegeneration. Cell Metab 20: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Carrie C, Giraud E, Elhafez D, Narsai R, Duncan O, Whelan J, Murcha MW (2012) Dual location of the mitochondrial preprotein transporters B14.7 and Tim23-2 in complex I and the TIM17:23 complex in Arabidopsis links mitochondrial activity and biogenesis. Plant Cell 24: 2675–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]