miR166-overexpressing plants show lower Cd-induced oxidative stress and Cd accumulation than in wild-type plants.
Abstract
MicroRNAs (miRNAs) are 20- to 24-nucleotide small noncoding RNAs that regulate gene expression in eukaryotic organisms. Several plant miRNAs, such as miR166, have vital roles in plant growth, development and responses to environmental stresses. One such environmental stress encountered by crop plants is exposure to cadmium (Cd), an element highly toxic to most organisms, including humans and plants. In this study, we analyzed the role of miR166 in Cd accumulation and tolerance in rice (Oryza sativa). The expression levels of miR166 in both root and leaf tissues were significantly higher in the reproductive stage than in the seedling stage in rice. The expression of miR166 in the roots of rice seedlings was reduced after Cd treatment. Overexpression of miR166 in rice improved Cd tolerance, a result associated with the reduction of Cd-induced oxidative stress in transgenic rice plants. Furthermore, overexpression of miR166 reduced both Cd translocation from roots to shoots and Cd accumulation in the grains. miR166 targets genes encoding the class-III homeodomain-Leu zipper (HD-Zip) family proteins in plants. In rice, HOMEODOMAIN CONTAINING PROTEIN4 (OsHB4) gene (Os03g43930), which encodes an HD-Zip protein, was up-regulated by Cd treatment but down-regulated by overexpression of miR166 in transgenic rice plants. Overexpression of OsHB4 increased Cd sensitivity and Cd accumulation in the leaves and grains of transgenic rice plants. By contrast, silencing OsHB4 by RNA interference enhanced Cd tolerance in transgenic rice plants. These results indicate a critical role for miR166 in Cd accumulation and tolerance through regulation of its target gene, OsHB4, in rice.
Cadmium (Cd) is a nonessential heavy metal harmful to human health. It is widely released into soil, air, and water mainly through industrial and mining effluents and phosphate fertilization (Pinto et al., 2004). The toxic effect of Cd has been a worldwide concern since the outbreak of Itai-Itai disease in the mid-20th century in Japan caused by the daily consumption of Cd-contaminated rice (Horiguchi et al., 1994). For the general population, the major route of Cd exposure is food. Rice grains contaminated with Cd represent a major risk to the health of more than half of the world’s population that depends on rice as a basic staple.
Plants have evolved various molecular mechanisms to respond to heavy metal stress, including microRNA (miRNA)-guided gene regulation at the posttranscriptional level (Ding et al., 2016, 2017; Gielen et al., 2016). miRNAs are a class of endogenous noncoding small RNAs that can base pair their target mRNAs to induce their degradation or repress their translation in eukaryotic organisms. In plants, miRNAs have vital regulatory roles in plant growth, development, and responses to biotic and abiotic stresses, including heavy metal stress (Bartel, 2004; Sunkar et al., 2006; Yang and Chen, 2013; Gu et al., 2017; Noman et al., 2017).
miR166 is a well-conserved miRNA present in all land plants (Floyd and Bowman, 2004; Axtell and Bowman, 2008). miR166 targets the transcripts of the class-III homeodomain-Leu zipper (HD-Zip) transcription factors in plants, including rice (Itoh et al., 2008). miR166-mediated cleavage of HD-Zip III mRNAs has been confirmed by 5′ RACE and degradome sequencing analysis (Liu et al., 2009; Li et al., 2010). miR166 acts as a master regulator of plant development (Schwab et al., 2005). In Arabidopsis (Arabidopsis thaliana), miR166/165 and its targets regulate diverse aspects of plant developmental processes, including shoot apical and lateral meristem formation, leaf polarity, floral development, and vascular patterning of shoot and root (Jung and Park, 2007; Carlsbecker et al., 2010; Chen, 2012; Singh et al., 2014). Recent studies indicate that miR166 also regulates plant responses to abiotic stresses, including drought in Triticum dicoccoides and rice (Oryza sativa; Kantar et al., 2011; Zhang et al., 2018) and mercury treatment in Medicago truncatula (Zhou et al., 2012). Our previous work from microarray assays showed that miR166 was strongly down-regulated in rice by Cd stress (Ding et al., 2011). However, the roles of miR166 and its target genes in Cd accumulation and tolerance in rice have not been established.
In the current study, we performed a comprehensive functional analysis of rice miR166 and its target genes in Cd accumulation and tolerance using both gene expression and transgenic approaches. Our results showed that Cd stress down-regulated the expression of miR166 but up-regulated the expression of its target gene HOMEODOMAIN CONTAINING PROTEIN4 (OsHB4) in rice. In addition, overexpression of miR166 down-regulated OsHB4 expression, decreased Cd-induced oxidative stress, and reduced Cd accumulation in rice grains. By contrast, overexpression of OsHB4 increased both Cd sensitivity and Cd accumulation in the leaves and grains of transgenic rice plants, whereas silencing OsHB4 by RNA interference (RNAi) enhanced Cd tolerance in transgenic rice plants. Based on these results, we conclude that miR166 plays a critical role in Cd accumulation and tolerance through regulation of its HD-Zip target genes in rice.
RESULTS
Cd-Responsive Expression of Rice miR166
We previously reported our investigation of rice miRNA expression patterns under Cd treatment, using a microarray assay with probes complementary to the miRNA sequences of rice from miRBase Release 11.0 (http://microrna.sanger.ac.uk/; Ding et al., 2011). Rice seedlings were treated with 60 μm CdCl2 for 6 h, and small RNAs were isolated from roots and used to screen for differentially expressed miRNAs by microarray analysis. The microarray results showed that the expression of miR166 was substantially decreased under Cd stress (Ding et al., 2011).
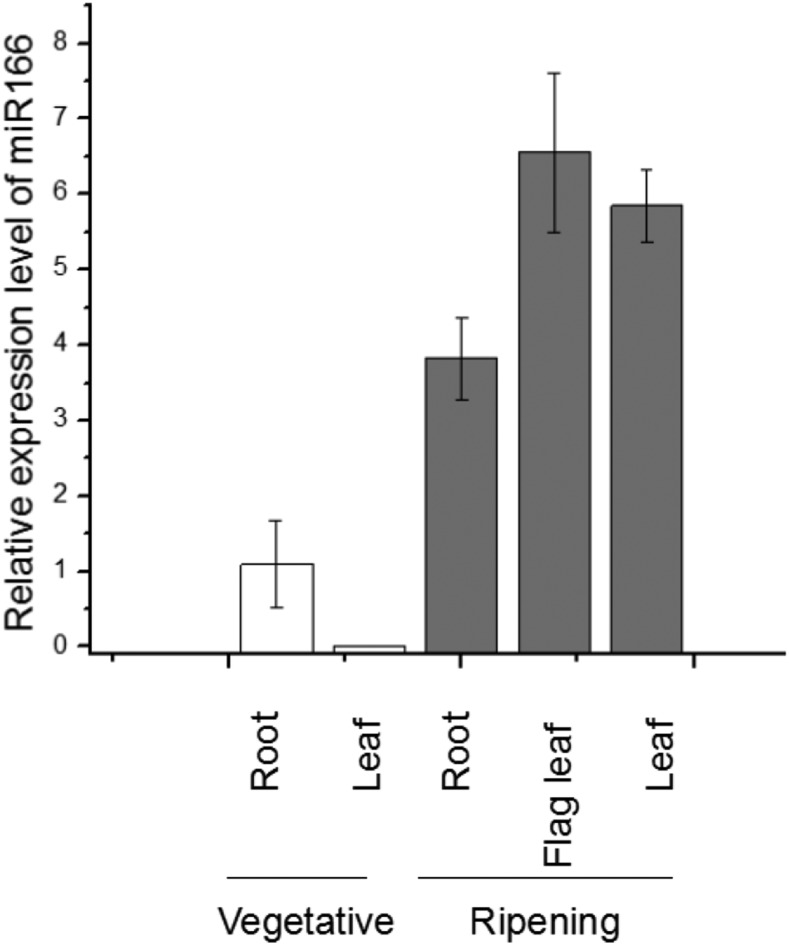
To further analyze miR166, we monitored the expression profile of miR166 in various rice tissues at different growth stages by quantitative real-time PCR (RT-qPCR) analysis. As shown in Figure 1, miR166 was expressed at high levels in flag leaves and leaves during the grain-ripening stage but at relatively low levels in shoots and roots in the vegetative growth stage. These results suggested a role of miR166 in the ripening stage.
Figure 1.
Expression profiles of miR166 in rice. cDNAs were synthesized from total RNA extracted from various tissues of rice grown in a greenhouse, and the mRNA levels were quantified by RT-qPCR. The data were normalized to β-ACTIN and were shown relative to the vegetative root sample. Error bars represent the se for three independent experiments.
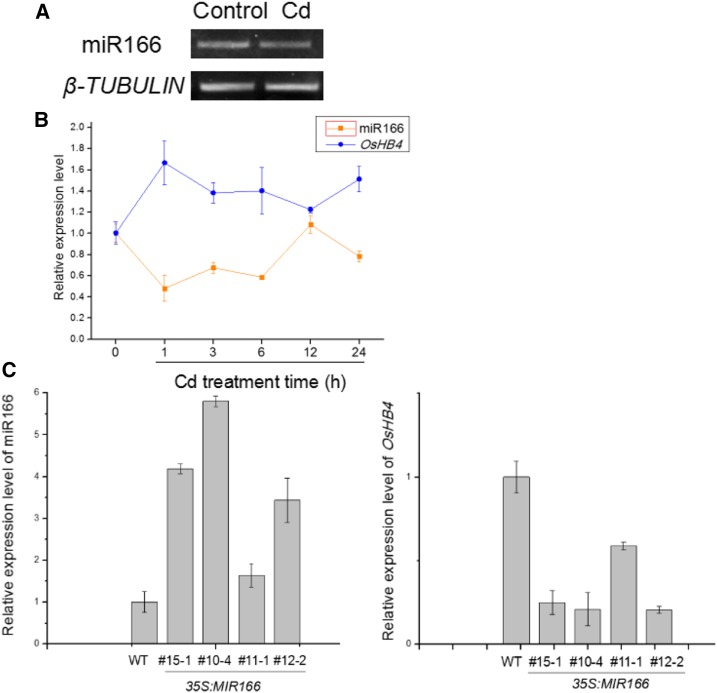
RT-PCR analysis also confirmed findings of the previously reported microarray analysis on the reduction in expression of miR166 after Cd treatment (Fig. 2A). RT-qPCR analysis was then performed to examine the time course in the changes of miR166 expression in response to Cd. In rice roots, miR166 expression was reduced substantially during the first 6 h of Cd treatment (Fig. 2B). We also examined the response of rice class III HD-Zip genes, the targets of miR166, to Cd treatment. There are five closely related class III HD-Zip genes (OsHB1 to OsHB5) in rice that are predicted to be targets of miR166 based on sequence analysis. However, our expression analysis showed that among these, only OsHB4 (Os03g43930) was responsive to Cd. OsHB4 was induced after exposure to Cd (Fig. 2B). Thus, both miR166 and OsHB4 were responsive to Cd stress. Their opposite expression patterns in response to Cd were consistent with the HD-Zip gene being a target of miR166 in plants.
Figure 2.
Expression levels of miR166 and its target gene OsHB4. A, RT-PCR of miR166 precursor from roots under Cd stress for 6 h. B, Detection of miR166 and OsHB4 transcript levels in roots of 1-week-old rice seedlings treated for the indicated times with 60 μm CdCl2 by RT-qPCR. C, Detection of miR166 and OsHB4 gene transcripts in 35S:MIR166 transgenic plant lines by RT-qPCR. Quantifications were normalized to the expression of β-ACTIN. Error bars represent se for three independent experiments.
To further analyze miR166, we overexpressed its precursor in rice to determine the effects of miR166 on the expression of class III HD-Zip genes (Fig. 2C). We placed the miR166 precursor gene behind the cauliflower mosaic virus (CaMV) 35S promoter (35S:MIR166) and transformed it into rice plants. In transgenic rice lines, the transcript levels of mature miR166 increased by 2- to 6-fold over those in wild-type plants (Fig. 2C). We also examined the expression of the class III HD-Zip genes in these 35S:MIR166 lines and found that the transcript levels of only OsHB4 were substantially reduced relative to the levels in wild-type plants. Furthermore, those lines with relatively high levels of miR166 transcripts (lines #15-1, #10-4, and #12-2) displayed relatively strong reduction in the transcript levels of OsHB4 (Fig. 2C). On the other hand, line #11-1 had a lower level of the miR166 transcripts and, at the same time, a higher level of OsHB4 transcripts than other transgenic lines (Fig. 2C). This observation supports that OsHB4 is a major target gene of miR166 and as a result, transcripts of OsHB4 are subjected to miR166-mediated degradation.
Overexpression of miR166 Improved Rice Tolerance to Cd Stress
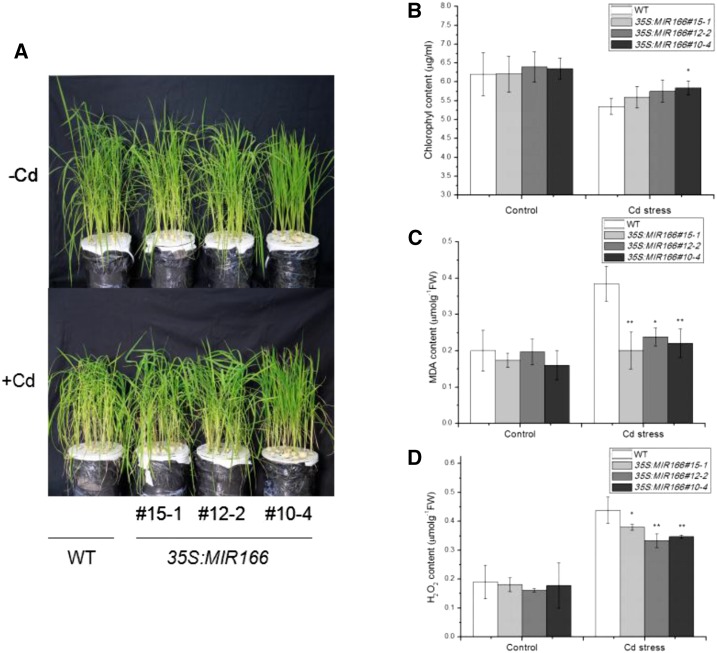
The Cd-responsive expression of both miR166 and its target gene OsHB4 shows that miR166 likely plays a role in the accumulation and tolerance of the heavy metal in plants. To test this possibility, we compared responses to Cd in wild-type plants with those in transgenic 35S:MIR166 lines (#15-1, #10-4, and #12-2) with relatively high miR166 expression levels. The wild-type and transgenic 35S:MIR166 plants were grown for 8 weeks in Yoshida’s culture solution and then treated with 2 μm CdCl2 for 14 d. Although Cd treatment inhibited the growth of both wild-type and transgenic miR166 plants, the extent of growth inhibition in transgenic 35S:MIR166 plants was slightly lower than that in wild-type plants (Fig. 3A). The chlorophyll content in wild-type and transgenic plants was also evaluated. Prior to Cd treatment, wild-type and transgenic 35S:MIR166 lines had similar levels of chlorophyll (Fig. 3B). After Cd treatment, however, the chlorophyll levels in the 35S:MIR166 plants were slightly higher than those in wild-type plants (Fig. 3B). On the other hand, the levels of oxidative stress in the transgenic 35S:MIR166 lines were substantially lower than those in wild-type plants, based on the levels of malonyldialdehyde (MDA), an indicator of oxidized membrane lipids under oxidative stress (Fig. 3C). Under normal growth conditions, there was no significant difference in MDA content between wild-type and transgenic 35S:MIR166 plants (Fig. 3C). Cd treatment increased MDA levels by almost 100% in wild-type plants but by only 20 to 30% in the transgenic 35S:MIR166 lines (Fig. 3C). Thus, the levels of MDA in transgenic 35S:MIR166 lines #15-1, #12-2, and #10-4 were 47.4, 39.5, and 42.1% lower than that in wild-type plants, respectively (Fig. 3C). These results indicated that overexpression of miR166 decreased membrane oxidation under Cd stress.
Figure 3.
Performance of wild-type and 35S:MIR166 transgenic plants under Cd stress. A, Phenotypic comparison of wild-type (WT) and 35S:MIR166 transgenic rice lines (#10-4, #12-2, and #15-1) under Cd stress. Top: Plants grown for 8 weeks under normal conditions before treatment. Bottom, Eight-week-old plants treated with 2 μm CdCl2 for 14 d. B, Effect of Cd stress on chlorophyll levels. C, Effect of Cd stress on MDA levels. D, Effect of Cd stress on H2O2 content. Asterisks indicate significant differences between wild-type and transgenic plants at *P ≤ 0.05 and **P ≤ 0.01.
Stresses usually cause damage in plants via oxidative stress from increased accumulation of reactive oxygen species, such as hydrogen peroxide (H2O2). Therefore, the H2O2 content in wild-type and transgenic 35S:MIR166 plants was examined. As shown in Figure 3D, under normal conditions, there was no significant difference in the H2O2 content between wild-type and transgenic 35S:MIR166 lines, and after Cd treatment, the H2O2 content increased by 2.3-fold in leaves of wild-type plants but only about 2.0-fold in leaves of 35S:MIR166 lines. As a result, the levels of H2O2 in Cd-treated transgenic 35S:MIR166 lines were significantly lower than those in the Cd-treated wild-type plants (Fig. 3D). Thus, the increased Cd tolerance of miR166-overexpressing rice lines was associated with reduced accumulation of reactive oxygen species and reduced oxidative stress under Cd stress.
Overexpression of miR166 Reduced Cd Translocation in Rice
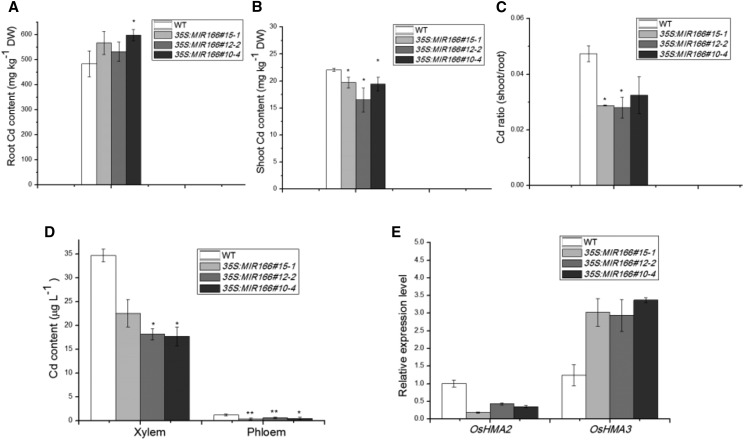
To examine the role of miR166 in rice Cd accumulation, we measured Cd levels in the roots and shoots of rice plants grown hydroponically for 14 d in the presence of 2 μm CdCl2. The Cd content in the roots of transgenic 35S:MIR166 plants was 10%–20% higher than that in the roots of wild-type plants (Fig. 4A). On the other hand, the shoots of 35S:MIR166 plants contained about 15%–30% less Cd than those of wild-type plants (Fig. 4B). Based on the Cd contents in both roots and shoots, we further calculated the Cd translocation factor (the ratio of Cd amount in shoots to that in roots) to examine Cd translocation from roots to above-ground parts. The calculated ratios indicated that the translocation factor in 35S:MIR166 plants was about 40% lower than that in wild-type plants (Fig. 4C), suggesting that overexpression of miR166 led to reduced Cd translocation from roots to shoots in hydroponically grown rice plants.
Figure 4.
Cd content in wild-type and 35S:MIR166 transgenic rice lines (#10-4, #12-2, and #15-1) after exposure to 2 μm CdCl2 for 14 d. A, Cd content in roots. B, Cd content in shoots. C, Ratio of Cd content in shoots to that in roots. D, Cd content in the xylem and phloem sap. E, Relative expression levels of metal transporter genes in the roots of wild-type (WT) and 35S:MIR166 transgenic plants under Cd stress. Vertical bars represent sd of the mean (n = 3). Asterisks indicate mean values are significantly different between wild-type and transgenic plants at *P ≤ 0.05 and **P ≤ 0.01.
To further analyze the effect of miR166 on the root-to-shoot translocation of Cd, we estimated Cd accumulation in the xylem and phloem sap after Cd treatment in hydroponically grown plants. As shown in Figure 4D, the Cd concentrations in the xylem sap were more than 10 times higher than those in the phloem sap in both wild-type and transgenic 35S:MIR166 plants, indicating that Cd root-to-shoot translocation was primarily through xylem. Furthermore, the Cd concentrations in the xylem sap in the 35S:MIR166 plants were 1.5 times lower than those in wild-type plants (Fig. 4D). These results indicate that miR166 plays a negative role in Cd loading to the xylem for root-to-shoot translocation in rice.
To explore possible mechanisms by which miR166 regulates Cd accumulation and translocation, we compared the expression of Cd transporter genes in wild-type and 35S:MIR166 roots using RT-qPCR (Fig. 4E). OsHMA3 is a tonoplast-localized Cd transporter in rice roots that belongs to the heavy metal ATPase (HMA) family. OsHMA3 is mainly expressed in roots, and its overexpression enhances Cd accumulation in roots (Sasaki et al., 2014). Our RT-qPCR analysis revealed that miR166 positively regulated OsHMA3, based on increased levels of OsHMA3 transcripts in 35S:MIR166 roots (Fig. 4E). The increased expression of OsHMA3 in the miR166-overexpressing lines was consistent with increased Cd accumulation in the transgenic rice roots. OsHMA2 is a major Zn/Cd transporter in rice roots (Takahashi et al., 2012). It has been reported that mutations in OsHMA2 restricted Cd translocation (Satoh-Nagasawa et al., 2012). In the 35S:MIR166 plants, in which Cd translocation was also reduced, OsHMA2 expression was decreased (Fig. 4E), which was consistent with the positive role of OsHMA2 in Cd translocation in rice (Satoh-Nagasawa et al., 2012).
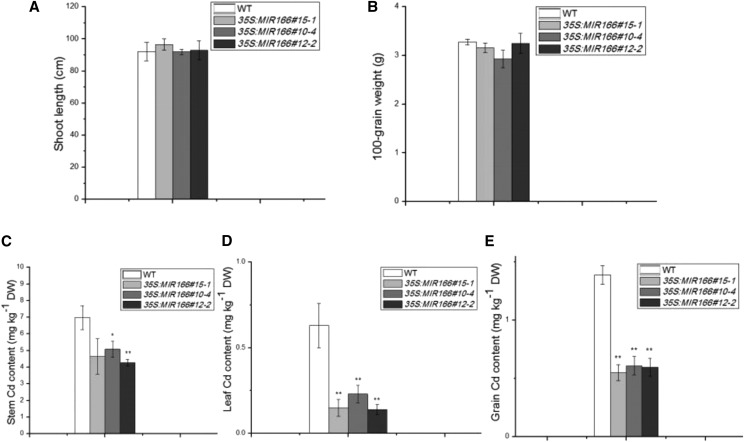
We also analyzed the agronomic traits of and Cd accumulation in soil-grown wild-type and transgenic 35S:MIR166 lines under field conditions in 2013. The experiments were conducted in a restricted experimental field where the soil was artificially contaminated with Cd (average of 3 μg/g). Plants were grown until grain ripening, and both the agronomical traits and Cd content in wild-type and miR166-overexpressing lines were compared. The transgenic 35S:MIR166 plants had normal growth and yield, with no significant difference in plant size, leaf dry weight, and grain weight from wild-type plants (Fig. 5). However, the transgenic plants accumulated about 50% less Cd in the stems and leaves than wild-type plants (Fig. 5, C and D). Moreover, Cd content in the grains of 35S:MIR166 plants was 57% lower than that in wild-type (Fig. 5E). These results indicate that overexpression of miR166 can be explored as an effective way to reduce grain Cd accumulation.
Figure 5.
Agronomic traits and Cd content in miR166-overexpressing lines grown in Cd-containing soil. A, Shoot length. B, Dry weight (DW) of 100 grains. C, Stem Cd concentration. D, Leaf Cd concentration. E, Grain Cd concentration in wild-type (WT) and 35S:MIR166 plants (n = 6). Asterisks indicate significant differences between wild-type and transgenic plants at *P ≤ 0.05 and **P ≤ 0.01.
These measurements showed that in the shoots of soil-grown rice, Cd accumulated primarily in the stem tissues, while the Cd levels in the leaves were generally very low (Fig. 5). In fact, Cd levels were lower in leaves than in grains in both wild-type and transgenic miR166-overexpressing lines (Fig. 5). It appears that a large proportion of the Cd content in the shoots stays in or near the vascular tissues of the stem, where its translocation takes place. Previous studies have also shown that the levels of Cd in the leaves are similar to or even lower than those in the grains in soil-grown rice plants (Shimo et al., 2011; Siebers et al., 2013).
Overexpression of OsHB4 Increased Cd Sensitivity
To determine the role of OsHB4, a target gene of miR166, in Cd accumulation and tolerance, we also generated an OsHB4-overexpression construct driven by the CaMV 35S promoter and introduced it into rice plants using Agrobacterium-mediated transformation. Three transgenic lines (#5, #6, and #11), which contained OsHB4 transcripts at least 25 times higher than in wild-type plants (Fig. 6A), were chosen for further analysis. To evaluate the effect of OsHB4 overexpression, we again treated 8-week-old wild-type and 35S:OsHB4 plants with 2 μm CdCl2 in Yoshida nutrient solution for 14 d. As shown in Figure 6B, more extensive wilting occurred in the transgenic 35S:OsHB4 lines than in wild-type plants after 14 d of Cd treatment (Fig. 6B). In addition, under Cd stress, both the H2O2 and MDA levels were significantly higher in the 35S:OsHB4 lines than in wild-type plants (Fig. 6, C and D). These results indicated that overexpression of OsHB4 reduced plant Cd tolerance.
Figure 6.
Performance of wild-type and 35S:OsHB4 transgenic plants treated with 2 μm CdCl2 for 14 d. A, Detection of OsHB4 mRNA in 35S:OsHB4 transgenic rice. Real-time PCR quantifications were normalized to the expression of β-ACTIN. Error bars represent se (n = 3). B, Phenotypic comparison of wild-type and 35S:OsHB4 transgenic rice lines (#5, #6, and #11) under Cd stress. A representative picture is shown. C, MDA content in plants treated with 2 μm CdCl2 for 14 d. D, H2O2 content in plants treated with 2 μm CdCl2 for 14 d. Asterisks indicate significant differences between wild-type and transgenic plants at *P ≤ 0.05 and **P ≤ 0.01.
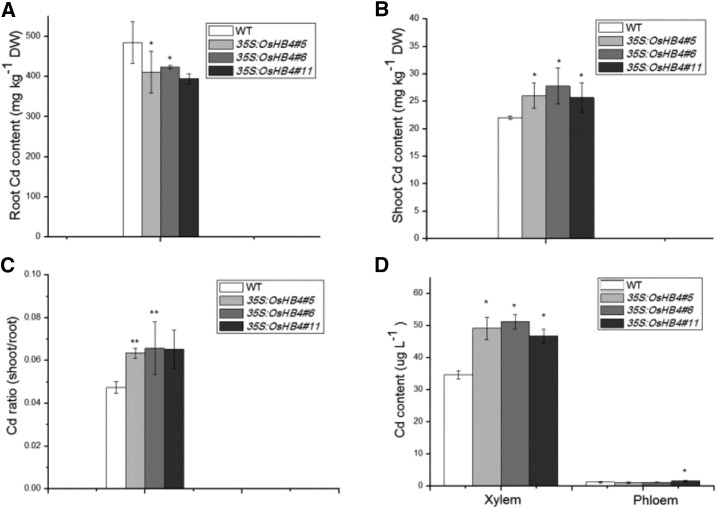
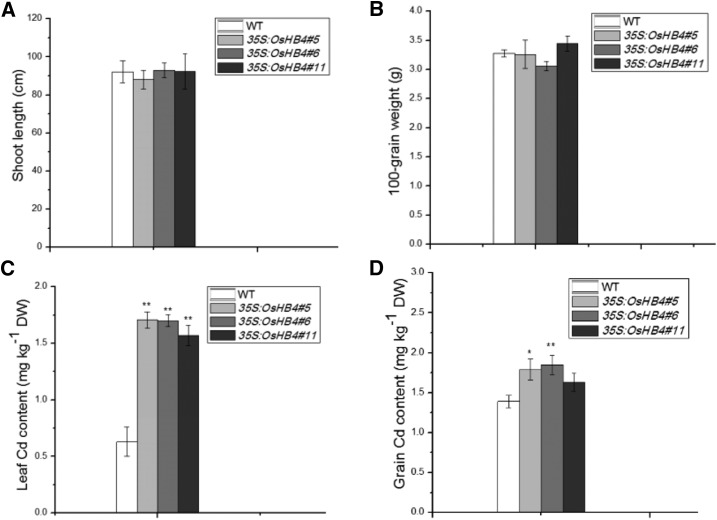
We then analyzed the effect of OsHB4 overexpression on Cd content in hydroponically grown and soil-grown plants (Fig. 7). The 35S:OsHB4 lines had significantly higher Cd levels in the shoots but lower Cd levels in the roots than wild-type plants (Fig. 7). As a result, the Cd translocation factors were significantly higher in the 35S:OsHB4 plants than in wild-type plants (Fig. 7C). Furthermore, Cd concentrations in the xylem sap were 30 to 40% higher in 35S:OsHB4 plants than in wild-type plants (Fig. 7D). These results indicate that OsHB4 plays a positive role in Cd loading to the xylem to promote the root-to-shoot translocation in rice. When grown under field conditions in soil artificially contaminated with Cd, the transgenic 35S:OsHB4 lines grew and developed quite normally (Fig. 8). However, these transgenic plants accumulated more Cd in shoots and grains than wild-type plants (Fig. 8). These results suggest that OsHB4 plays a role opposite to that of miR166 in promoting Cd translocation, thereby increasing Cd accumulation in both rice shoots and grains.
Figure 7.
Cd accumulation in wild-type and 35S:OsHB4 transgenic rice lines (#5, #6, and #11) after exposure to 2 μm CdCl2 for 14 d. A, Cd content in roots. B, Cd content in shoots. C, Ratio of Cd content in shoots to that in roots. D, Cd content in the xylem and phloem sap. Vertical bars represent sd of the mean (n = 3). Asterisks indicate mean values are significantly different between wild-type (WT) and transgenic plants at *P ≤ 0.05 and **P ≤ 0.01.
Figure 8.
Agronomic traits and Cd content in OsHB4-overexpressing lines grown in Cd-containing soil. A, Shoot length. B, Dry weight (DW) of 100 grains. C, Leaf Cd concentration. D, Grain Cd concentration in wild-type (WT) and 35S:OsHB4 plants. n = 6; asterisks indicate significant differences between wild-type and transgenic plants at *P ≤ 0.05 and **P ≤ 0.01.
Down-Regulation of OsHB4 Improves Cd Tolerance
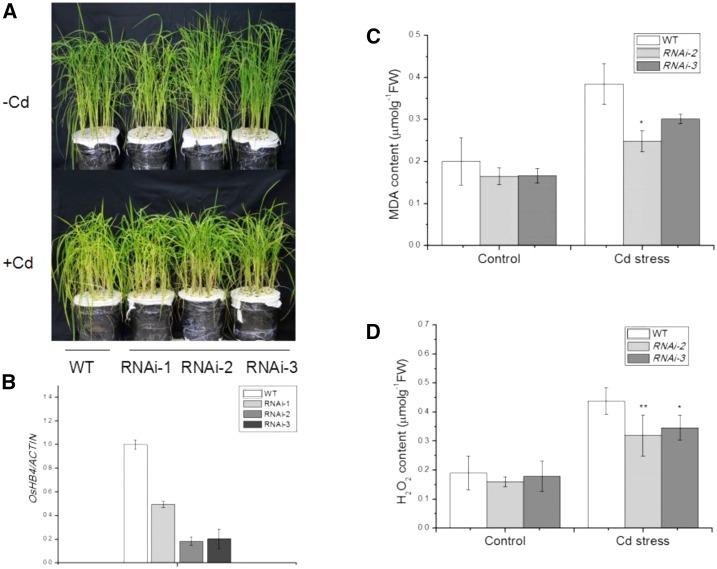
To further investigate the function of OsHB4 in Cd response, we attempted to down-regulate its expression through gene silencing. An RNAi construct for OsHB4 was generated and transformed into rice. Three independent transgenic OsHB4 RNAi lines (#1, #2, and #3) with substantial reduction in the level of OsHB4 transcripts were identified by RT-qPCR analysis (Fig. 9B). Homozygous progeny of the independent RNAi transgenic lines were further analyzed for Cd tolerance.
Figure 9.
Molecular characterization and Cd tolerance analysis of OsHB4 RNAi rice plants. A, Phenotypic comparison of wild-type (WT) and RNA interference (RNAi) transgenic rice lines (-1, -2 and -3) under Cd stress. A, Representative picture taken after Cd treatment for 14 d. B, Reverse0transcription quantitative PCR analysis of the expression of OsHB4 intransgenic rice relative to that of β-ACTIN. Data represent means and se of three replicates. C, MDA content in plants treated with 2 μm CdCl2 for 14 d. D, H2O2 content in plants treated with 2 μm CdCl2 for 14 d. Asterisks indicate significant differences between wild-type and transgenic plants at *P ≤ 0.05 and **P ≤ 0.01.
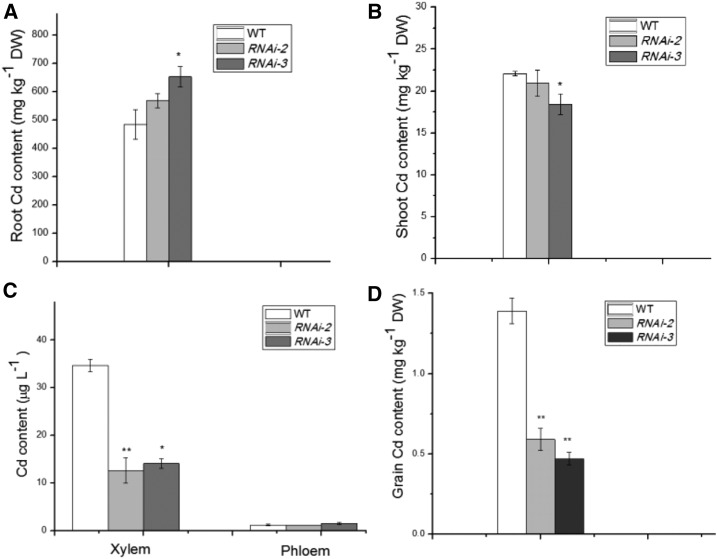
Similar to the transgenic 35S:MIR166 plants, the transgenic OsHB4 RNAi plants displayed improved Cd tolerance. After exposure to 2 μm CdCl2 for 14 d, the MDA and H2O2 levels in the OsHB4 RNAi lines were substantially lower than those in Cd-stressed wild-type seedlings (Fig. 9, C and D). In addition, the OsHB4 RNAi plants accumulated less Cd in shoots but more Cd in roots than wild-type plants (Fig. 10, A and B). Furthermore, Cd concentration in the xylem sap in the OsHB4 RNAi plants was significantly lower than that in wild-type plants (Fig. 10C). When grown in Cd-contaminated soil, OsHB4 RNAi plants accumulated less Cd in the grains than wild-type plants (Fig. 10D). These results indicated that in addition to overexpression of miR166, down-regulation of its target gene OsHB4 could also reduce Cd translocation to grains.
Figure 10.
Cd accumulation in wild-type and OsHB4 RNAi transgenic rice after exposure to 2 μm CdCl2 for 14 d. A, Cd content in roots. B, Cd content in shoots. C, Cd content in the xylem and phloem sap. D, Grain Cd concentration in wild-type (WT) and OsHB4 RNAi plants grown in Cd-containing soil. Asterisks indicate significant differences between wild-type and transgenic plants at *P ≤ 0.05 and **P ≤ 0.01.
DISCUSSION
Heavy metals such as Cd are highly toxic to plants, and plants have evolved a variety of adaptive strategies to reduce Cd toxicity. Over the past several years, a substantial number of studies have revealed that miRNAs play critical roles in the posttranscriptional regulation of plant genes in response to heavy metals. Microarray analysis in Glycine max led to the identification of 26 Cd-responsive miRNAs (Fang et al., 2013). Solexa sequencing in radish (Raphanus sativus) identified 54 known and 16 novel miRNAs that were differentially expressed under Cr stress (Liu et al., 2015). Deep sequencing in the Cd hyperaccumulator Sedum alfredii identified 356 miRNAs, of which 79 miRNAs were differentially expressed under Cd stress (Han et al., 2016). Although an increasing number of heavy metal-responsive miRNAs has been identified, further functional analysis is necessary to establish their roles in heavy metal accumulation and tolerance in plants. Overexpression, mutations, and silencing of miRNAs and their targets have proved useful for analyzing the function of stress-responsive miRNAs and their targeted transcripts in plant stress responses. These molecular genetic approaches have also been used to improve plant tolerance to heavy metals. Zhang et al. (2013) reported that overexpression of miR395 in rapeseed (Brassica napus) reduced Cd-induced oxidative stress and increased the biomass and sulfur levels under Cd exposure in transgenic plants. On the other hand, we have recently reported that overexpression of miR390 in transgenic rice plants increased Cd accumulation and reduced Cd tolerance, indicating that miR390 is a negative regulator of plant adaptive responses to Cd in rice (Ding et al., 2016).
In this study, we discovered that expression of miR166 was significantly reduced under Cd exposure (Fig. 2). Transgenic rice plants overexpressing miR166 displayed improved growth under Cd stress, unlike wild-type (Fig. 3). Increased growth of the miR166-overexpressing rice plants was associated with significant increase in the chlorophyll content and attenuation in Cd-induced oxidative damage in rice (Fig. 3). Furthermore, overexpression of miR166 reduced root-to-shoot Cd translocation rate (Fig. 4). The 35S:MIR166 plants had substantially lower Cd concentrations in the xylem sap than the wild-type plants (Fig. 4D). It has been well established that root-to-shoot Cd translocation via the xylem is the major and common physiological process determining Cd accumulation in shoots and grains of rice (Uraguchi et al., 2009). These results suggest that miR166 plays a role in Cd loading to the xylem and regulates root-to-shoot translocation in rice. Indeed, overexpression of miR166 changed the expression pattern of OsHMA metal transporter genes with important roles in the transport and homeostasis of metals in plants (Sasaki et al., 2014). The previous finding that mutations in OsHMA2 restrict Zn/Cd translocation (Satoh-Nagasawa et al., 2012) show that OsHMA2 encodes a major Zn/Cd transporter that promotes Zn/Cd root-to-shoot transport. Our discovery that overexpression of miR166 reduces both the expression of OsHMA2 and root-to-shoot translocation in the transgenic rice plants (Fig. 4) is consistent with this role of OsHMA2. OsHMA3, on the other hand, is a tonoplast-localized Cd transporter that promotes vacuolar compartmentalization of Cd, thereby restricting its root-to-shoot translocation (Uraguchi et al., 2009; Sasaki et al., 2014). Overexpression of OsHMA3 also increased Cd tolerance in rice, likely due to increased vacuolar sequestration of Cd in the roots (Sasaki et al., 2014). Likewise, in 35S:MIR166 plants, we observed increased expression of OsHMA3 (Fig. 4E). Thus, the increased Cd tolerance of and decreased Cd translocation in the 35S:MIR166 plants could be attributable at least in part to the opposite effects on the expression of OsHMA2 and OsHMA3.
A key finding of this study is the substantial reduction of leaf and grain Cd contents in the transgenic miR166-overexpressing rice lines grown in soil containing a low Cd concentration (Fig. 5), making miR166 a potential candidate for genetic engineering to reduce Cd content in rice grains. The reduced Cd content in the leaves and grains of transgenic miR166-overexpressing lines likely resulted from the reduction in the root-to-shoot translocation of the heavy metal. Expression of miR166 in leaves and flag leaves during the grain-ripening stage was substantially higher than that in leaves during the vegetative growth stage (Fig. 1). Since the grain-ripening stage is a critical period for grain Cd accumulation (Arao et al., 2009), the increased expression of miR166 in leaves and flag leaves during the grain-ripening stage may help reduce Cd transport into rice grains. Intriguingly, miR166 expression was reduced by Cd treatment (Fig. 2). The physiological significance of Cd-responsive reduction in miR166 expression is unclear but may be related to the other roles of this miRNA in plant growth and development under stress conditions.
The biological functions of miRNAs are mediated through negative regulation of their specific targets. In rice, miR166 targets genes encoding the class III HD-Zip, as shown by sequencing analysis and confirmation by 5′ RACE and degradome sequencing (Liu et al., 2009; Li et al., 2010). In this study, we found that miR166 and OsHB4, one of the HD-Zip genes, exhibited opposite expression patterns in rice roots after treatment with Cd (Fig. 2B). OsHB4 and miR166 also showed negatively correlated expression patterns in miR166-overexpression lines (Fig. 2C). These results confirmed that OsHB4 was targeted by miR166. In rice and Arabidopsis, about 50% of all homeobox proteins belong to the HD-Zip superfamily (Chan et al., 1998; Jain et al., 2008). HD-Zip transcription factors regulate the enlargement of shoot apical meristem and vascular development (Prigge et al., 2005; Williams et al., 2005). There are also HD-Zip family genes that are responsive to various stress conditions, including drought, salt, and abscisic acid (Agalou et al., 2008; Zhang et al., 2012). Shin et al. (2004) reported that Athb-12, an HD-Zip protein from Arabidopsis, increased salt tolerance in yeast by promoting sodium exclusion. Although some members of the HD-Zip gene family have been implicated in the regulation of development and stress responses, the functions of most HD-Zip genes are still unknown. Based on our comprehensive analysis, we have provided strong evidence for a critical role of a rice HD-Zip gene, OsHB4, as an important target of miR166 in the regulation of Cd accumulation and tolerance in rice. Expression of OsHB4 in Cd-treated rice seedlings was negatively regulated by miR166 (Fig. 2). The transgenic plants overexpressing OsHB4 displayed phenotypes opposite to those of transgenic plants overexpressing miR166, including changes in Cd tolerance, Cd concentrations in the xylem sap, Cd root-to-shoot translocation, and Cd content in the grains. In contrast, silencing of OsHB4 improved Cd tolerance and reduced Cd translocation to grains (Figs. 9 and 10). These phenotypes of OsHB4-silenced rice plants were similar to those of 35S:MIR166 plants (Fig. 10). These results further support the notion that OsHB4 is a critical target of miR166 in the regulation of Cd translocation and tolerance in rice. As a transcription factor, OsHB4’s role in Cd transport and tolerance is likely through regulation of other genes, including OsHMA2 and OsHMA3, which affect various aspects of Cd uptake, sequestration, translocation, and detoxification in rice. Future studies should identify the direct target genes of OsHB4 and associated molecular and biochemical processes in order to establish the miR166/OsHB4-mediated pathway of plant-adaptive responses to this toxic heavy metal.
MATERIALS AND METHODS
Plant Growth Conditions, Seedling Cultivation, and Stress Treatment
Seeds of rice cultivar Zhonghua 11 (Oryza sativa subsp. japonica) were sterilized with 10% sodium hypochlorite for 20 min, rinsed three times with distilled water, and soaked for 2 d at 37°C in the dark. The seeds were then transferred onto moist filter paper and cultured at 30°C in the dark for 1 d. The germinated seeds were then transferred to Yoshida nutrient solution under a 13 h light (29°C)/11 h dark (22°C) photoperiod. For Cd treatment, 1-week-old seedlings were transferred to plastic containers containing Yoshida nutrient solution with appropriate concentrations of CdCl2. Seedlings that were not treated with Cd were used as controls. After Cd treatment, rice tissues were collected separately and immediately frozen in liquid nitrogen for further analysis.
RT-qPCR Assay
Total RNA collected from rice roots after exposure to 60 μm CdCl2 for 0, 1, 3, 6, 12, and 24 h was separately isolated using Trizol reagent (Invitrogen) and treated with RNase-free DNase I (TaKaRa) to remove contaminated DNA. Real-time PCRs were carried out using a Rotor-Gene Q machine (parameters, 95°C for 1 min, followed by 45 cycles of 95°C for 10 s, 58°C for 15 s, and 72°C for 15 s). All reactions were performed in triplicate. Quantification of gene expression was performed using the comparative Ct method. The β-ACTIN gene was chosen as the internal control. The primer pairs for RT-qPCR analysis of these genes were as follows: β-ACTIN forward, 5′-GCCGTCCTCTCTCTGTATGC-3′; reverse, 5′-GGGGACAGTGTGGCTGAC-3′; miR166, stem-loop RT primer, 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAGGGAA-3′; forward, 5′-GTCGGACCAGGCTTCA-3′; reverse, 5′-TGCGTGTCGTGGAGTC-3′; OsHB4 forward, 5′-ATGTCAAGAACCTCCCCAG-3′; reverse, 5′-CACACGCAAAAATCACTCA-3′; OsHMA2 forward, 5′-TGGCACCACAAAAGGCTATT-3′; reverse, 5′-CACCGTCGATTGGAATGACT-3′; OsHMA3 forward, 5′-TGGGGATCTTCATCAAGGGT-3′; reverse, 5′-AACGAGTCGATGCTGAACTC-3′.
Vector Construction and Rice Transformation
For generation of 35S:MIR166 constructs, a 299-bp fragment surrounding the miRNA sequence that includes the foldback structure of miR166 was amplified from rice genomic DNA using the following primers: miR166 forward 5′-CGGGGTACCCTTTCTCTGCTTTGGTGGTT-3′ and reverse 5′-AACTGCAGTTCCGATGGATTTCCTGGAC-3′. The amplified fragments were digested and subcloned into KpnI and PstI sites in pCAMBIA1300-35S-NOS between the CaMV 35S promoter and the NOS terminator. The hygromycin phosphotransferase gene (hpt) was used as a selectable marker. To generate 35S:OsHB4 constructs, the OsHB4 (Os03g43930) open reading frame was amplified by RT-PCR with the forward primer 5′-GGGGTACCGGAGAAGGGTCGACGGGGATG-3′ and reverse primer 5′-CGGGATCCGACGAATGACCAGTTGACGAAC-3′. The PCR products were first cloned into pMD-19T vector (TaKaRa) and verified by sequencing. Then, the OsHB4 open reading frame was released by digestion with KpnI and BamHI and subcloned into pCAMBIA1300. To generate the RNAi constructs, an OsHB4 gene fragment was PCR-amplified using gene-specific primers (5′-GGGGATCCGAGCTCAGGCAGGACAGGTCGTCG-3′ and 5′-CGACTAGTGGTACCCCTTGCGCTGCTTCTCG-3′) and cloned into pCAMBIA1300-35SI-X by forward and reverse insertions.
Cultivars of japonica rice Zhonghua 11 were used for transformation. Transgenic lines were generated by cocultivation of rice calli with Agrobacterium tumefaciens strain EHA105, which contained the constructs (Hiei et al., 1994). Transgenic rice seedlings were selected using 50 mg/L hygromycin. All the putative T0 transgenic plants were screened using PCR analysis with genomic DNA from their leaves. The PCR was performed with hpt-specific primers in a 20-µL reaction system. Expression levels of miR166 and OsHB4 in transgenic rice plants were examined using RT-qPCR. The transformation vector also contains a GUS reporter gene, and GUS staining was also used to confirm the transgenic plants (Jefferson, 1987).
Effect of Cd Stress on Rice Seedling Growth
To determine the role of miR166 on rice response to Cd stress, wild-type and 35S:MIR166 seedlings were cultured with Yoshida nutrient solution for 8 weeks. After exposure to 2 μm CdCl2 for 14 d, phenotypes of the plants were examined by photographing plants, and the chlorophyll, MDA, H2O2, and Cd contents were also determined. All experiments were performed in triplicate. Data points were presented as the mean ± sd of three replications. Significant differences among the wild-type and transgenic lines were analyzed using Student’s t test.
The chlorophyll content was determined according to the method of Knudson et al. (1977). In brief, 0.1 g fresh rice leaf was extracted in 2 mL 95% ethanol for 36 h in the dark, and the extracted solution was analyzed spectrophotometrically. The H2O2 content was measured using a Hydrogen Peroxide Assay Kit (Nanjing Jiancheng). Leaf tissues (0.5 g) were ground into powder using liquid nitrogen and homogenized in 0.9 mL of precooled 50 mm phosphate buffer (pH 7.0). The homogenate was centrifuged at 10,000g for 10 min, and the supernatants were used for the H2O2 assay. Absorbance of the supernatant was recorded at 405 nm.
The MDA content was measured as described by Heath and Packer (1968), with slight modification. In brief, rice shoots of 0.1 g were ground in 1 mL of 10% (w/v) trichloroacetic acid. After centrifugation at 12,000g for 10 min at 4°C, the supernatants were collected, and 2 mL of the supernatant fraction was mixed with 2 mL 0.6% TBA solution. The mixtures were heated at 95°C for 30 min and then cooled quickly in an ice bath. The resulting mixtures were centrifuged at 10,000g for 10 min, and the absorbance of the supernatants was measured at 450, 532, and 600 nm.
Determination of Cd Content
After treatment with 2 μm CdCl2 for 14 d, roots of rice seedlings were immersed in 20 mm disodium ethylenediamine tetra-acetic acid for 20 min and then rinsed three times with deionized water. The roots and shoots were dried at 105°C for 2 h and then at 70°C to achieve a constant weight. In total, 200 mg of dry plant material was treated with 6 mL HNO3 at 120°C with a Multiwave until they were completely digested. Concentrations of Cd were then determined using an atomic absorption spectrometer (AA-7000; Shimadzu). Xylem sap exuded from the cut surface was collected by trapping it into a 1.5-mL plastic vial filled with a small piece of cotton for 1 h after cutting the shoots at 3 cm above the roots. The amount of collected sap was weighed. Cd concentration in the sap was quantified using atomic absorption spectrometry.
Soil Growing of Rice Plants and Analysis of Cd Content
Soil for the experiments was collected from the surface layer (0–20 cm) of an unpolluted paddy field in Hangzhou, China. The soil was sieved using a 2-mm grid sieve and air dried. Fertilizers were added to the soil and incubated for 2 weeks before Cd treatment, which was added by manually mixing in a CdCl2 solution. To ensure Cd homogeneity, the soil was mixed every 5 d for 30 d. Before planting, the Cd level was determined to be about 3 μg per gram of soil.
For analysis of Cd content in soil-grown rice plants, wild-type, miR166, and OsHB4 transgenic rice seeds were surface-sterilized and germinated in deionized water for 3 d. Two-week-old rice seedlings were transplanted to a plastic rectangular pot filled with Cd-containing soil (∼3 μg/g) in May, 2013 at China Jiliang University in Hangzhou, China. The pots were set up outdoors, with a glass shelter to protect from rainfall and pests. Intermittent flooding was introduced throughout cultivation to enhance Cd availability in the soil. Plants were harvested in September and washed thoroughly with tap water and then distilled water after grain ripening. The plant samples were then divided into stems, leaves, and grains. After harvesting, the fresh weights were recorded, and the plants were dried at 70°C. Then, the dry weights were recorded and the plant materials were ground using a stainless steel grinder. For grain Cd analysis, dried grain samples were hulled and milled into powder. Brown rice (0.3 g) was treated with 6 mL HNO3 at 120°C with a Multiwave until they were completely digested. The Cd content of the grains was quantified by an atomic absorption spectrometer (AA-7000; Shimadzu).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AK103284.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Footnotes
This study was supported by the Natural Science Foundation of Zhejiang Province (LR17C130001), the National Natural Science Foundation of China (31771698), the Key Research and Development Project of Zhejiang Province (2015C03020-4), and the National Key Research and Development Program of China (2017YFD0801104).
References
- Agalou A, Purwantomo S, Overnäs E, Johannesson H, Zhu X, Estiati A, de Kam RJ, Engström P, Slamet-Loedin IH, Zhu Z, et al. (2008) A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol Biol 66: 87–103 [DOI] [PubMed] [Google Scholar]
- Arao T, Kawasaki A, Baba K, Mori S, Matsumoto S (2009) Effects of water management on cadmium and arsenic accumulation and dimethylarsinic acid concentrations in Japanese rice. Environ Sci Technol 43: 9361–9367 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Bowman JL (2008) Evolution of plant microRNAs and their targets. Trends Plant Sci 13: 343–349 [DOI] [PubMed] [Google Scholar]
- Bartel DP. (2004) MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. (2010) Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RL, Gago GM, Palena CM, Gonzalez DH (1998) Homeoboxes in plant development. Biochim Biophys Acta 1442: 1–19 [DOI] [PubMed] [Google Scholar]
- Chen X. (2012) Small RNAs in development—insights from plants. Curr Opin Genet Dev 22: 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chen Z, Zhu C (2011) Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). J Exp Bot 62: 3563–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Ye Y, Jiang Z, Wang Y, Zhu C (2016) MicroRNA390 is involved in cadmium tolerance and accumulation in rice. Front Plant Sci 7: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Wang Y, Jiang Z, Wang F, Jiang Q, Sun J, Chen Z, Zhu C (2017) MicroRNA268 overexpression affects rice seedling growth under cadmium stress. J Agric Food Chem 65: 5860–5867 [DOI] [PubMed] [Google Scholar]
- Fang X, Zhao Y, Ma Q, Huang Y, Wang P, Zhang J, Nian H, Yang C (2013) Identification and comparative analysis of cadmium tolerance-associated miRNAs and their targets in two soybean genotypes. PLoS One 8: e81471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd SK, Bowman JL (2004) Gene regulation: Ancient microRNA target sequences in plants. Nature 428: 485–486 [DOI] [PubMed] [Google Scholar]
- Gielen H, Remans T, Vangronsveld J, Cuypers A (2016) Toxicity responses of Cu and Cd: the involvement of miRNAs and the transcription factor SPL7. BMC Plant Biol 16: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q, Chen Z, Yu X, Cui W, Pan J, Zhao G, Xu S, Wang R, Shen W (2017) Melatonin confers plant tolerance against cadmium stress via the decrease of cadmium accumulation and reestablishment of microRNA-mediated redox homeostasis. Plant Sci 261: 28–37 [DOI] [PubMed] [Google Scholar]
- Han X, Yin H, Song X, Zhang Y, Liu M, Sang J, Jiang J, Li J, Zhuo R (2016) Integration of small RNAs, degradome and transcriptome sequencing in hyperaccumulator Sedum alfredii uncovers a complex regulatory network and provides insights into cadmium phytoremediation. Plant Biotechnol J 14: 1470–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125: 189–198 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Horiguchi H, Teranishi H, Niiya K, Aoshima K, Katoh T, Sakuragawa N, Kasuya M (1994) Hypoproduction of erythropoietin contributes to anemia in chronic cadmium intoxication: Clinical study on Itai-itai disease in Japan. Arch Toxicol 68: 632–636 [DOI] [PubMed] [Google Scholar]
- Itoh J, Hibara K, Sato Y, Nagato Y (2008) Developmental role and auxin responsiveness of Class III homeodomain leucine zipper gene family members in rice. Plant Physiol 147: 1960–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP (2008) Genome-wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice. FEBS J 275: 2845–2861 [DOI] [PubMed] [Google Scholar]
- Jefferson R. (1987) Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol Biol Report 5: 387–405 [Google Scholar]
- Jung JH, Park CM (2007) MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta 225: 1327–1338 [DOI] [PubMed] [Google Scholar]
- Kantar M, Lucas SJ, Budak H (2011) miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 233: 471–484 [DOI] [PubMed] [Google Scholar]
- Knudson LL, Tibbitts TW, Edwards GE (1977) Measurement of ozone injury by determination of leaf chlorophyll concentration. Plant Physiol 60: 606–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R (2010) Transcriptome-wide identification of microRNA targets in rice. Plant J 62: 742–759 [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhang YC, Wang CY, Luo YC, Huang QJ, Chen SY, Zhou H, Qu LH, Chen YQ (2009) Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett 583: 723–728 [DOI] [PubMed] [Google Scholar]
- Liu W, Xu L, Wang Y, Shen H, Zhu X, Zhang K, Chen Y, Yu R, Limera C, Liu L (2015) Transcriptome-wide analysis of chromium-stress responsive microRNAs to explore miRNA-mediated regulatory networks in radish (Raphanus sativus L.). Sci Rep 5: 14024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noman A, Fahad S, Aqeel M, Ali U, Amanullah, Anwar S, Baloch SK, Zainab M (2017) miRNAs: Major modulators for crop growth and development under abiotic stresses. Biotechnol Lett 39: 685–700 [DOI] [PubMed] [Google Scholar]
- Pinto AP, Mota AM, de Varennes A, Pinto FC (2004) Influence of organic matter on the uptake of cadmium, zinc, copper and iron by sorghum plants. Sci Total Environ 326: 239–247 [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE (2005) Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Ma JF (2014) Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J Exp Bot 65: 6013–6021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Nagasawa N, Mori M, Nakazawa N, Kawamoto T, Nagato Y, Sakurai K, Takahashi H, Watanabe A, Akagi H (2012) Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol 53: 213–224 [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Shimo H, Ishimaru Y, An G, Yamakawa T, Nakanishi H, Nishizawa NK (2011) Low cadmium (LCD), a novel gene related to cadmium tolerance and accumulation in rice. J Exp Bot 62: 5727–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Koo YD, Lee J, Lee HJ, Baek D, Lee S, Cheon CI, Kwak SS, Lee SY, Yun DJ (2004) Athb-12, a homeobox-leucine zipper domain protein from Arabidopsis thaliana, increases salt tolerance in yeast by regulating sodium exclusion. Biochem Biophys Res Commun 323: 534–540 [DOI] [PubMed] [Google Scholar]
- Siebers N, Siangliw M, Tongcumpou C (2013) Cadmium uptake and subcellular distribution in rice plants as affected by phosphorus: soil and hydroponic experiments. J Soil Sci Plant Nutr 13: 833–844 [Google Scholar]
- Singh A, Singh S, Panigrahi KC, Reski R, Sarkar AK (2014) Balanced activity of microRNA166/165 and its target transcripts from the class III homeodomain-leucine zipper family regulates root growth in Arabidopsis thaliana. Plant Cell Rep 33: 945–953 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18: 2051–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R, Ishimaru Y, Shimo H, Ogo Y, Senoura T, Nishizawa NK, Nakanishi H (2012) The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ 35: 1948–1957 [DOI] [PubMed] [Google Scholar]
- Uraguchi S, Mori S, Kuramata M, Kawasaki A, Arao T, Ishikawa S (2009) Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J Exp Bot 60: 2677–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC (2005) Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132: 3657–3668 [DOI] [PubMed] [Google Scholar]
- Yang ZM, Chen J (2013) A potential role of microRNAs in plant response to metal toxicity. Metallomics 5: 1184–1190 [DOI] [PubMed] [Google Scholar]
- Zhang JS, Zhang H, Srivastava AK, Pan YJ, Bai JJ, Fang JJ, Shi HZ, Zhu JK (2018) Knock-down of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development. Plant Physiol 176: 2082–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LW, Song JB, Shu XX, Zhang Y, Yang ZM (2013) miR395 is involved in detoxification of cadmium in Brassica napus. J Hazard Mater 250-251: 204–211 [DOI] [PubMed] [Google Scholar]
- Zhang S, Haider I, Kohlen W, Jiang L, Bouwmeester H, Meijer AH, Schluepmann H, Liu CM, Ouwerkerk PB (2012) Function of the HD-Zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Mol Biol 80: 571–585 [DOI] [PubMed] [Google Scholar]
- Zhou ZS, Zeng HQ, Liu ZP, Yang ZM (2012) Genome-wide identification of Medicago truncatula microRNAs and their targets reveals their differential regulation by heavy metal. Plant Cell Environ 35: 86–99 [DOI] [PubMed] [Google Scholar]