A Tanacetum cinerariifolium CYP71 cytochrome P450 enzyme catalyzes the hydroxylation of jasmone to jasmolone, an alcohol moiety of pyrethrin insecticides.
Abstract
Pyrethrins are synthesized by the plant pyrethrum (Tanacetum cinerariifolium), a chrysanthemum relative. These compounds possess efficient insecticidal properties and are not toxic to humans and most vertebrates. Pyrethrum flowers, and to a smaller extent leaves, synthesize six main types of pyrethrins, which are all esters of a monoterpenoid acid moiety and an alcohol moiety derived from jasmonic acid. Here, we identified and characterized the enzyme responsible for the conversion of jasmone, a derivative of jasmonic acid, to jasmolone. Feeding pyrethrum flowers with jasmone resulted in a 4-fold increase in the concentration of free jasmolone as well as smaller but significant proportional increases in free pyrethrolone and all three type I pyrethrins. We used floral transcriptomic data to identify cytochrome P450 genes whose expression patterns were most highly correlated with that of a key gene in pyrethrin biosynthesis, T. cinerariifolium chrysanthemyl diphosphate synthase. The candidate genes were screened for jasmone hydroxylase activity through transient expression in Nicotiana benthamiana leaves fed with jasmone. The expression of only one of these candidate genes produced jasmolone; therefore, this gene was named T. cinerariifolium jasmolone hydroxylase (TcJMH) and given the CYP designation CYP71AT148. The protein encoded by TcJMH localized to the endoplasmic reticulum, and microsomal preparations from N. benthamiana leaves expressing TcJMH were capable of catalyzing the hydroxylation of jasmone to jasmolone in vitro, with a Km value of 53.9 µm. TcJMH was expressed almost exclusively in trichomes of floral ovaries and was induced in leaves by jasmonate.
Pyrethrum (Tanacetum cinerariifolium) is a daisy-like plant in the Asteraceae family that synthesizes a class of insecticidal compounds known as pyrethrins (McLaughlin, 1973). These biodegradable and photolabile pesticides are harmless to most mammals, including humans, and therefore are in demand despite their relatively high price as compared with synthetic analogs, pyrethroids, which generally are less safe to vertebrates and less biodegradable (Jones, 1973; Shafer et al., 2005; DeMicco et al., 2010).
Pyrethrins are esters; the acid moiety is either trans-chrysanthemic acid (in type I pyrethrins) or pyrethric acid (in type II pyrethrins), both of which are monoterpenoids (Head, 1973; Ramirez et al., 2012; Fig. 1). The enzymes involved in the biosynthesis of trans-chrysanthemic acid from two molecules of dimethylallyl diphosphate, a universal precursor of terpenes, have been identified and include trans-chrysanthemyl diphosphate / trans-chrysanthemol synthase (CDS; the original acronym stood for trans-chrysanthemyl diphosphate synthase, but the name of the enzyme was later changed to trans-chrysanthemol synthase when it was shown that, in the presence of low concentrations of dimethylallyl diphosphate, the predominant product is trans-chrysanthemol), alcohol dehydrogenase2 (ADH2), and aldehyde dehydrogenase1 (ALDH1; Rivera et al., 2001; Yang et al., 2014; Xu et al., 2018). Pyrethric acid is derived from trans-chrysanthemic acid by additional oxidation and methylation steps, but the enzymes responsible for these reactions have not yet been reported.
Figure 1.
Proposed biosynthesis pathway of pyrethrin alcohols, known as rethrolones (the compounds in the shaded area), from jasmonic acid (JA). JA is synthesized from linolenic acid in reactions sequentially catalyzed by lipoxygenase (LOX), allene oxide synthase (AOS), allene oxide cyclase (AOC), cis(+)-12-oxo-phytodienoic acid reductase (OPR), and β-oxidation enzymes. JA is converted to jasmone by currently unknown enzymes. As described in this report, the conversion of jasmone to jasmolone is catalyzed by the enzyme jasmolone hydroxylase (JMH). Pyrethrolone and cinerolone are possibly derived from jasmolone in as yet uncharacterized reactions. Finally, each of the three alcohols can be linked to either trans-chrysanthemic acid or pyrethric acid by the enzyme GLIP (GDSL lipase-like protein) to form a total of six types of pyrethrins, called cinerin I, pyrethrin I, and jasmolin I (these three having a trans-chrysanthemic acid moiety) and cinerin II, pyrethrin II, and jasmolin II (with a pyrethric acid moiety).
The alcohol moiety of pyrethrins is either pyrethrolone, cinerolone, or jasmolone, collectively called rethrolones. Based on the results of [1-13C]Glc feeding experiments, Matsuda et al. (2005) concluded that rethrolones are likely made in pyrethrum flowers from jasmone. Jasmone is likely derived from JA (Koch et al., 1997). The synthesis of JA from linolenic acid is well established (Song et al., 1993; Creelman and Mullet, 1997; Tijet and Brash, 2002; Ramirez et al., 2013) and involves the enzymes LOX, AOS, AOC, OPR, and three rounds of β-oxidation (Fig. 1). However, enzymes responsible for the conversion of JA to jasmone, and for the conversion of jasmone to pyrethrolone, cinerolone, or jasmolone, have not yet been reported. The enzyme forming the pyrethrin esters by combining the CoA-activated acid and the alcohol components has been identified as a GDSL lipase-like protein and, accordingly, named GLIP (Kikuta et al., 2012).
As with its precursor JA, jasmone accumulation is induced by wounding, as occurs during herbivory (Loughrin et al., 1995). Both have a repelling effect on insects, such as aphids, and both stimulate the emission of volatile compounds involved in defense, such as monoterpenes (Birkett et al., 2000). Interestingly, although the biosynthesis of pyrethrins is developmentally regulated in the flowers, pyrethrin biosynthesis in leaves is induced by wounding, further suggesting that the synthesis of the alcohol moieties of pyrethrins is tied directly to the JA biosynthetic pathway (Kikuta et al., 2011; Ueda and Matsuda, 2011).
A close inspection of the structures of the alcohols present in pyrethrins and a comparison of these structures with that of jasmone suggest that jasmolone could, in principle, be synthesized from jasmone by a single hydroxylation reaction (Fig. 1). Such hydroxylation reactions often are catalyzed by enzymes belonging to the cytochrome P450 oxidoreductase protein family. Plant genomes typically have a large gene family encoding cytochrome P450 enzymes. P450-mediated hydroxylation occurs on terpenoids, alkaloids, phenylpropanoids, fatty acids, as well as various types of hormones (Mizutani and Ohta, 2010; Zhao et al., 2014). The observation that high levels of pyrethrin biosynthesis occur only in certain parts of the flowers and only during certain developmental stages was used previously to identify candidate genes encoding enzymes responsible for the biosynthesis of trans-chrysanthemic acid (Xu et al., 2018). Here, we used the same strategy to identify a specific gene encoding a P450 cytochrome oxidoreductase that, upon additional experiments, was shown to hydroxylate cis-jasmone to generate jasmolone.
RESULTS
Jasmolone Is Produced from Jasmone
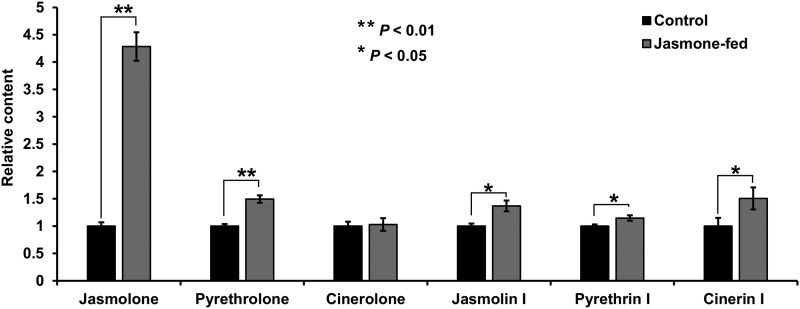
To determine whether jasmone can be a precursor of the pyrethrin alcohols, we fed stage 3 pyrethrum flower buds, the stage at which the rate of pyrethrin biosynthesis is highest (Xu et al., 2018), jasmone by immersing whole flower buds in a 20 μm aqueous jasmone solution for 24 h. Control samples were immersed in water containing no jasmone. After the dipping period, the flower buds were ground and extracted with methyl tert-butyl ether (MTBE), and the extract was analyzed by gas chromatography-mass spectrometry (GC-MS). Extracts from floral buds dipped in the jasmone solution showed a 328% increase in jasmolone concentration and a 49% increase in pyrethrolone concentration relative to the control (both significant at P < 0.01), but the content of free cinerolone did not show any significant change (Fig. 2). In addition, the concentrations of cinerin I, jasmolin I, and pyrethrin I all showed statistically significant increases (P < 0.05). In these experiments, we obtained a jasmolone standard by purifying jasmolin I by HPLC from a commercial preparation of pyrethrins and hydrolyzing it with TcGLIP (Supplemental Fig. S1). Pyrethrolone and cinerolone standards were obtained similarly by purifying type I pyrethrins by HPLC and hydrolyzing them with TcGLIP (Supplemental Fig. S1).
Figure 2.
Relative changes in concentrations of jasmolone, pyrethrolone, and cinerolone, and their respective trans-chrysanthemic acid esters, in flower buds fed with jasmone. The detection of jasmolone, pyrethrolone, and cinerolone was by GC-MS (total ion mode). The detection of jasmolin I, pyrethrin I, and cinerin I was by m/z = 123. Data are presented as means ± sd (n = 3). Statistical analysis was performed using Student’s t-test (two-tailed, two-sample unequal variance).
Characterization of Jasmone Hydroxylase Candidates by Coexpression Analysis and Heterologous Expression
To identify candidate jasmone hydroxylase genes, we performed coexpression analysis of all cytochrome P450 oxidoreductase contigs found in a stage 3 flower bud RNA sequencing (RNA-seq) data set (Xu et al., 2018), representing at least 65 unique genes. The Pearson correlation analysis (Li et al., 2015, 2017) looked for genes whose expression was best correlated with that of TcCDS, the gene encoding the key enzyme in the synthesis of trans-chrysanthemic acid (Rivera et al., 2001; Yang et al., 2014), in all five floral developmental stages as well as in leaves. Of these, the 11 genes with the highest correlation (Table 1; Supplemental Fig. S2) were chosen for further testing.
Table 1. Ranking of the top 11 cytochrome P450 gene candidates by coexpression correlation analysis with the TcCDS gene.
| Cytochrome P450 Genes | Pearson Correlation Coefficient | P (Two Tailed) |
|---|---|---|
| DN144246 | 0.979758 | 0.00002 |
| DN150785 | 0.965354 | 0.00010 |
| DN158750 | 0.960808 | 0.00015 |
| DN133626 | 0.948496 | 0.00033 |
| DN126517 | 0.932317 | 0.00074 |
| DN163032 | 0.921284 | 0.00115 |
| DN97751 | 0.890989 | 0.00298 |
| DN121926 | 0.888259 | 0.00320 |
| DN151943 | 0.850566 | 0.00744 |
| DN15633 | 0.804924 | 0.0160 |
| DN131340 | 0.774698 | 0.0240 |
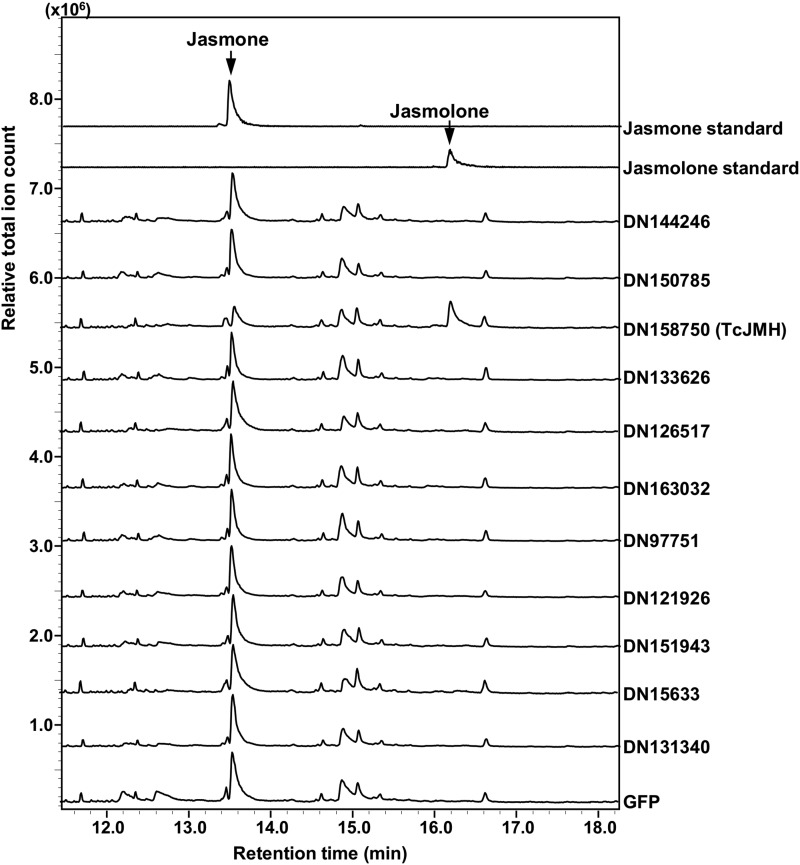
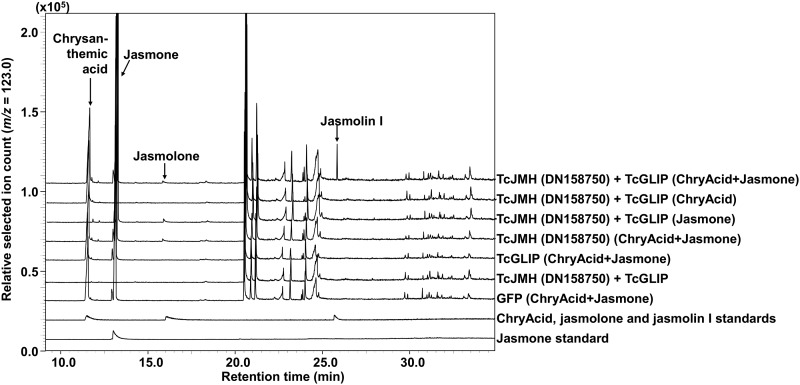
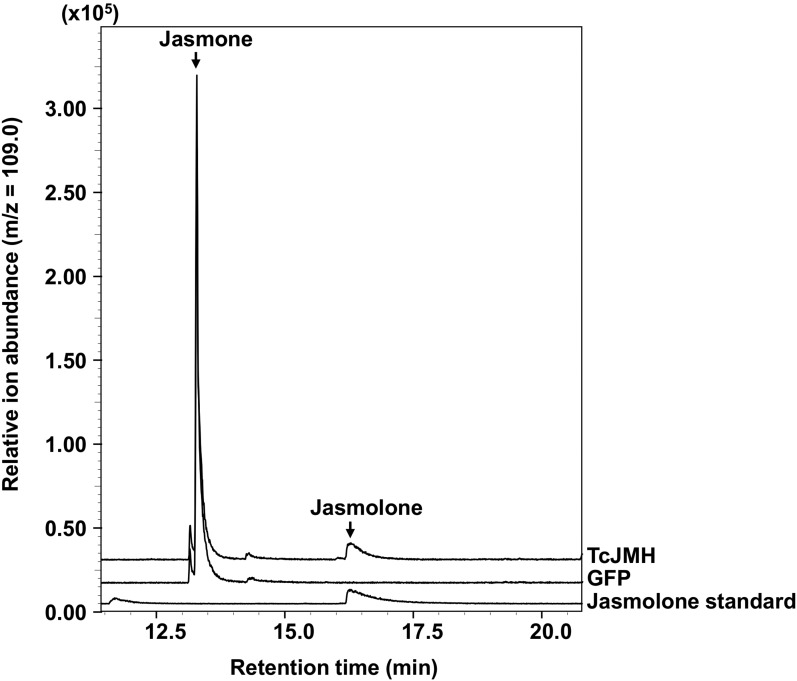
The biochemical activities of the enzymes encoded by these 11 jasmone hydroxylase gene candidates were tested by transient expression in leaves of Nicotiana benthamiana that were simultaneously fed with jasmone. GC-MS analyses of MTBE extracts of the N. benthamiana leaves showed that only plants expressing gene DN158750 produced jasmolone, while plants expressing any of the other 10 pyrethrum candidate cytochrome P450 oxidoreductase genes or GFP did not (Fig. 3). In addition, we also fed N. benthamiana leaves that transiently expressed DN158750 and TcGLIP simultaneously with both jasmone and trans-chrysanthemic acid. Analysis of the leaves of these plants showed the presence of jasmolin I (Fig. 4).
Figure 3.
Assays for jasmone hydroxylation activity of the 11 candidate cytochrome P450 oxidoreductases identified in our coexpression analysis. The assays were performed by transiently expressing the candidate genes in N. benthamiana plants and immersing their leaves in a 20 µm solution of jasmone for 24 h, after which the leaves were ground and extracted with MTBE. Detection of the jasmolone in the MTBE extract was by GC-MS (total ion mode). The jasmolone standard was obtained as described in Supplemental Figure S1. Jasmolone was present only in leaves expressing candidate gene DN158750.
Figure 4.
Generation of jasmolin I in leaves of N. benthamiana plants transiently coexpressing TcJMH and TcGLIP and fed with both trans-chrysanthemic acid (ChryAcid) and jasmone.
The DN158750 gene, therefore, was named TcJMH. Phylogenetic analysis of the TcJMH protein indicated that it belongs to the AT clade of the CYP71 family (Supplemental Fig. S3). Accordingly, TcJMH was given the official catalog designation CYP71AT148 by David Nelson (Cytochrome P450 Homepage: http://drnelson.uthsc.edu/cytochromeP450.html; Nelson et al., 1996). The AT clade of CYP71 contains only one other CYP71AT protein with known catalytic function. This protein is CYP71AT146 from Perilla frutescens (Lamiaceae), which catalyzes the hydroxylation of limonene to give perillyl alcohol, which it then further oxidizes to perillaldehyde (Fujiwara and Ito, 2017). The protein sequences of TcJMH and CYP71AT146 are 50% identical. The CYP71AT clade also includes CYP proteins that hydroxylate amino acid-derived aldoximes to nitrile oxides in glucosinolate biosynthesis but are designated as CYP83 proteins for historical reasons (http://drnelson.uthsc.edu/cytochromeP450.html; Supplemental Fig. S3; Bak and Feyereisen, 2001). These proteins show less than 45% sequence identity with TcJMH.
Subcellular Localization of Jasmone Hydroxylase
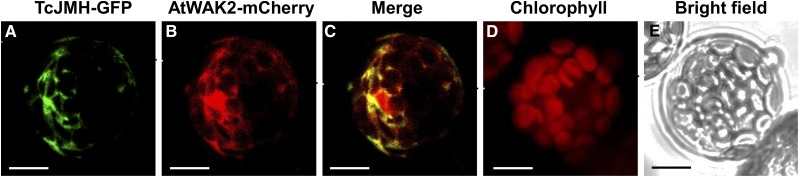
Confocal microscopy of Arabidopsis (Arabidopsis thaliana) protoplasts transiently expressing a TcJMH-GFP fusion gene indicated that the fusion protein produced from this gene localized to the endoplasmic reticulum (ER), since the fluorescent signal from this protein overlapped with the fluorescent signal of a bona fide ER marker (AtWAK2 [At1g21270]; Zhou et al., 2017; Fig. 5).
Figure 5.
Subcellular localization of TcJMH. All images are of the same cell. A, Detection of green fluorescence from the TcJMH-GFP fusion protein. B, Detection of red fluorescence from the ER marker AtWAK2-mCherry fusion protein. C, Overlay of green and red fluorescences from A and B, respectively. D, Detection of red autofluorescence from chloroplasts. E, Bright-field microscopy of the cell under transmitted white light. Bars = 10 µm.
In Vitro Characterization of Jasmone Hydroxylase Activity
To biochemically characterize the enzymatic activity of the putative jasmone hydroxylase, we purified microsomes from N. benthamiana leaves transiently expressing a recombinant TcJMH open reading frame. The microsomal preparation exhibited enzymatic activity capable of converting jasmone to jasmolone (Fig. 6; Supplemental Fig. S4). The Km value for jasmone was calculated as 53.9 ± 13.5 μm (see “Materials and Methods”).
Figure 6.
In vitro conversion of jasmone to jasmolone in microsomes prepared from N. benthamiana leaves transiently expressing TcJMH. After completion of the assay, the solution was extracted with MTBE and the extract was analyzed by GC-MS. For a control, the jasmone hydroxylation assay was conducted with microsomes prepared from N. benthamiana leaves transiently expressing GFP. For details of the enzymatic assay and GC-MS analysis, see “Materials and Methods.”
Tissue-Specific Expression of Jasmone Hydroxylase
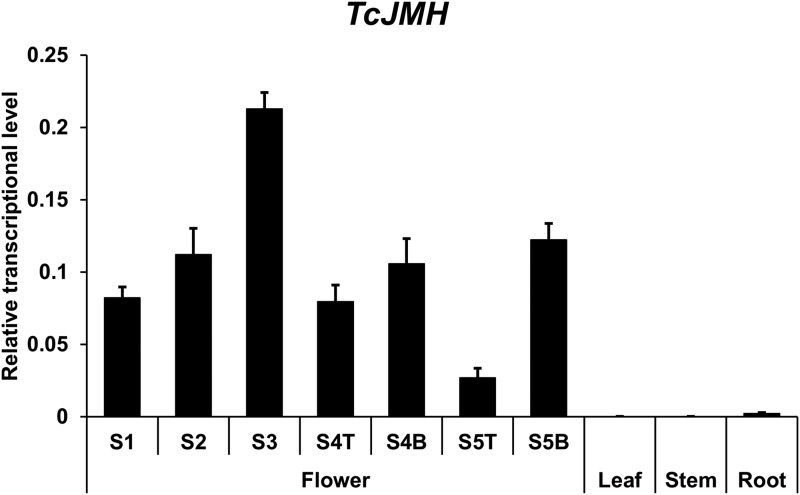
To determine the tissue-specific expression pattern of TcJMH, we performed reverse transcription quantitative PCR (RT-qPCR) analysis of transcripts from the five developmental stages of the flowers (Xu et al., 2018) as well as from leaf, stem, and root tissues (all from plants more than 12 weeks old). For stages 4 and 5, RNA was isolated separately from disk and ray florets. This analysis showed that TcJMH transcripts had little or no presence in leaf, stem, and root tissues (Fig. 7). The abundance of TcJMH transcripts in flowers (Fig. 7) had a similar pattern to that of TcCDS (Xu et al., 2018), peaking at stage 3.
Figure 7.
RT-qPCR analysis of TcJMH transcript abundance in different tissues of pyrethrum plants (T and B represent disk florets and ray florets, respectively, for stages 4 and 5). Data are presented as means ± sd (n = 4).
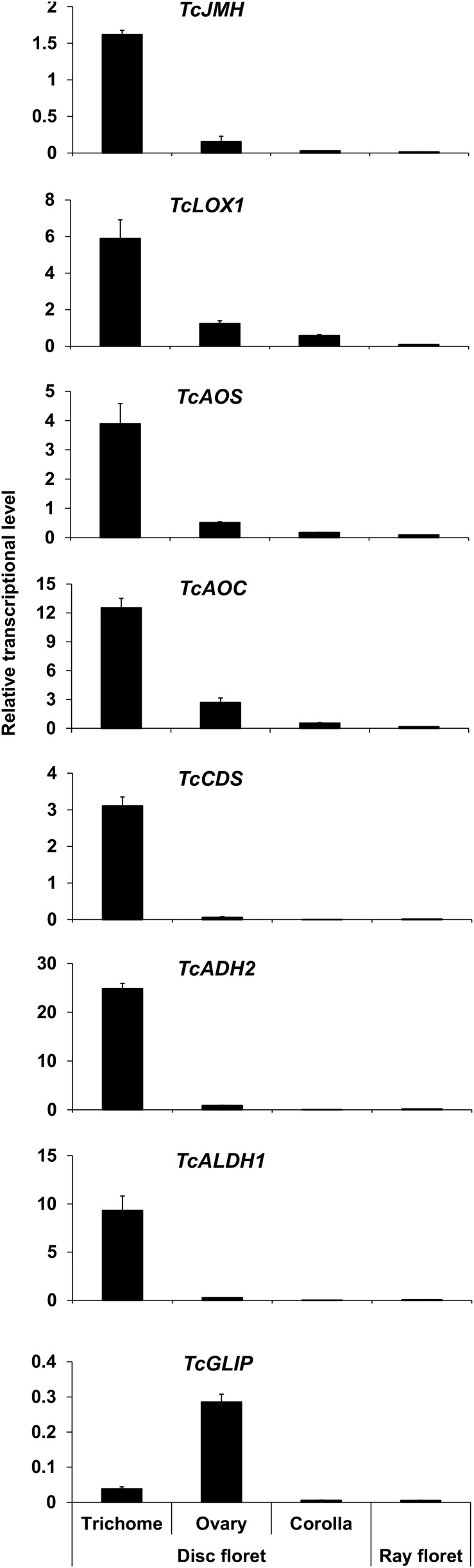
To more specifically identify the part(s) of the flowers where TcJMH is expressed, we performed RT-qPCR analysis of transcripts obtained from trichomes, ovaries, and corollas of disc florets and whole ray florets from stage 3 flowers and compared TcJMH expression patterns with those of TcCDS, TcADH2, and TcADLH1 (all three involved in trans-chrysanthemic acid biosynthesis) as well as several genes involved in JA biosynthesis, including TcLOX1 (Ramirez et al., 2013), TcAOS, and TcAOC (the sequences of the latter two were obtained from the RNA-seq data; Supplemental Fig. S1). We also compared these expression patterns with that of TcGLIP, the final enzyme in the biosynthetic pathway of pyrethrins (Fig. 1). The results of these analyses (Fig. 8) indicated that, in flowers, TcJMH, TcAOS, and TcAOC, as well as TcADH2 and TcADLH1, all are expressed almost exclusively in the trichomes of the ovaries, similar to the previously reported ovarian trichome-specific expression of TcCDS and TcLOX1 (Ramirez et al., 2012, 2013). In contrast, TcGLIP transcripts were found mostly in the nontrichome parts of the ovaries, also consistent with previous reports (Ramirez et al., 2012).
Figure 8.
Transcript abundance of known pyrethrin biosynthetic genes in different parts of stage 3 flowers. Data are presented as means ± sd (n = 3).
Expression of TcJMH in Leaves
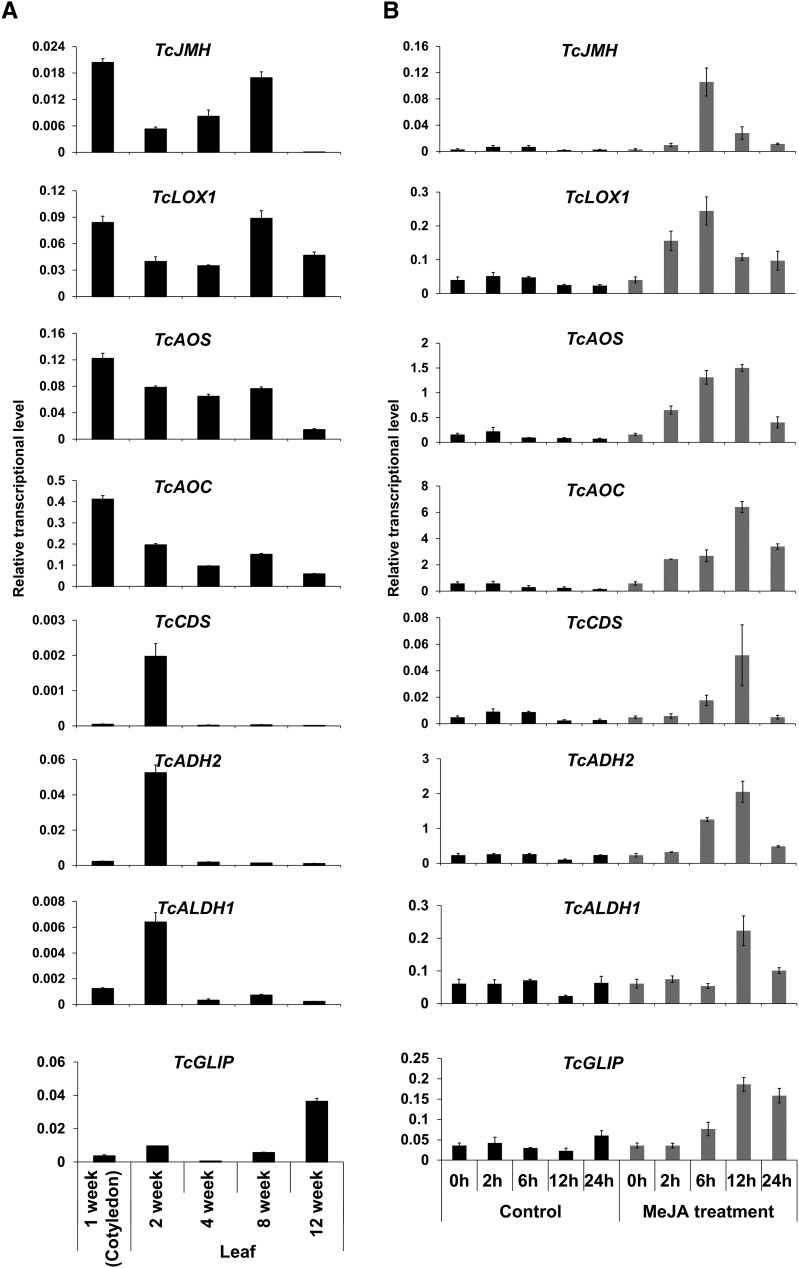
Pyrethrum leaves have been reported to have low levels of pyrethrins compared with flowers (Ueda and Matsuda, 2011). We examined the transcript levels of TcJMH in young leaves of pyrethrum plants of various ages and compared these levels with those of other genes in the pathway (Fig. 9A). Transcript levels of TcJMH, TcLOX1, TcAOS, and TcAOC all were relatively high in cotyledons and decreased in subsequent leaves, although, for TcJMH and TcLOX1, some increase in transcript levels was observed as the plant reached 1 to 3 months of age. The genes involved in trans-chrysanthemic acid biosynthesis showed a different pattern, having low levels of transcripts in cotyledons, peaking at the age of 2 weeks, and then declining again. TcGLIP transcripts showed a third pattern, being low for most of the age range studied here but finally increasing when the plant reached the age of 3 months.
Figure 9.
Transcript abundance of pyrethrin biosynthetic genes in pyrethrum leaves. A, Transcript abundance of known pyrethrin biosynthetic genes in leaves of plants of different ages. B, Transcript abundance of known pyrethrin biosynthetic genes induced in leaves of 2-week-old plants by MeJA. Data are presented as means ± sd (n = 3).
JA, from which jasmolone is derived, is a defense hormone known to induce the expression of defense genes. JA production itself is induced by wounding (Creelman and Mullet, 1997), and it was reported that pyrethrin biosynthesis, as well as TcGLIP expression, in leaves increase upon wounding (Ueda and Matsuda, 2011; Kikuta et al., 2012). Therefore, it was of interest to determine if JA induced the transcription of TcJMH and the other genes in the pyrethrin pathway. The results of JA (applied as methyl jasmonate [MeJA]) induction experiments (Fig. 9B) indicate that TcJMH, TcLOX1, TcAOS, TcAOC, TcCDS, TcADH2, TcADLH1, and TcGLIP all showed similar induction patterns, with transcript levels peaking within 6 to 12 h after the application of jasmonate and then declining within 24 h.
DISCUSSION
Jasmolone Is Synthesized from Jasmone in Flowers of T. cinerariifolium
The conversion of jasmone to jasmolone theoretically requires a single hydroxylation reaction (Fig. 1). We observed that feeding T. cinerariifolium flowers with jasmone increased the concentration of free jasmolone in the flowers 4.3-fold, suggesting that jasmone is indeed a precursor of jasmolone in these flowers. The concentration of free pyrethrolone also showed a statistically significant increase, albeit by a smaller factor (1.5-fold). We observed no change in the concentration of free cinerolone. However, the concentration of all type I pyrethrins, those derived from the esterification of trans-chrysanthemic acid with these three alcohols, showed statistically significant increases (Fig. 2). (The concentrations of the class II pyrethrins [esters of pyrethric acid with these alcohols] were too low to be measured accurately.) These results indicate that jasmolone is derived from jasmone. These results also support the conclusion that pyrethrolone is derived from jasmolone, possibly by a single desaturation reaction (Fig. 1). The biosynthesis of cinerolone is less clear. While it is possible that the alcohol side chain shortening to give cinerolone occurs at the free alcohol stage, the observation that the concentration of free cinerolone did not increase upon feeding with jasmone but cinerin I concentration did increase raises the possibility that the alcohol side chain shortening occurs after the ester is formed. It is important to note that the expected increases in the concentrations of the free alcohols versus the final pyrethrin products depend on actual initial pool sizes and the properties of enzymes catalyzing downstream steps in the pathway, including the affinity of the ester-forming enzyme, TcGLIP, for the various alcohols. Such information regarding the enzymes involved, some of which are still unknown, has not yet been reported, and we could not determine absolute concentrations of free alcohols for lack of available standards of known concentrations for calibration. Therefore, it is not possible at present to give a definitive explanation for why jasmone feeding led to a much greater increase in the free jasmolone pool than in the pool of free pyrethrolone (typically, pyrethrin I is found at higher levels than jasmolin I; Matsuda et al., 2005) and why the free cinerolone pool showed no increase.
Coexpression Analysis Combined with Heterologous in Planta Expression and in Vitro Biochemical Assay Identified T. cinerariifolium CYP71AT148 as Jasmone Hydroxylase
Based on the hypothesis that the hydroxylation reaction that converts jasmone to jasmolone is catalyzed by a cytochrome P450 oxidoreductase, we searched our transcriptome database (Xu et al., 2018) for transcripts of genes encoding proteins present in stage 3 flowers, the floral stage in which pyrethrin biosynthesis peaks (Ramirez et al., 2012). Transcripts of at least 65 unique genes were identified based on this criterion, and the 11 top-ranked transcripts for coexpression correlation with TcCDS, which encodes a key enzyme in pyrethrin biosynthesis, were selected for further analysis. It should be noted that additional steps in the biosynthesis of pyrethrin precursors may be catalyzed by cytochrome P450 oxidoreductase, for example, in the synthesis of pyrethric acid.
These 11 gene candidates (Table 1) were transiently expressed in N. benthamiana leaves that were also fed with jasmone (Fig. 3). Leaves expressing gene DN158750 showed the presence of jasmolone; expression of the other 10 genes did not. DN158750, therefore, was named TcJMH. Subsequent in vitro assays with partially purified recombinant TcJMH showed that it catalyzes the hydroxylation of jasmone to jasmolone, with a Km value for jasmone of 53.9 µm.
Expression analysis of TcJMH by RT-qPCR using RNA from different tissues and from different stages of development of T. cinerariifolium flowers indicated that TcJMH is expressed specifically in flowers, and peak levels of transcripts coincided with previously reported peak levels of pyrethrin biosynthesis (Ramirez et al., 2012; Xu et al., 2018). Subcellular localization showed that TcJMH, like many other cytochrome P450 oxidoreductases, is localized to the ER (Fig. 5). More importantly, our data (Fig. 8) indicated that TcMJH is expressed specifically in trichomes, as had been shown previously for TcCDS and TcLOX1 (Ramirez et al., 2012, 2013). We also showed that TcAOS, TcAOC, TcADH2, and TcALDH1 exhibit trichome-specific expression (Fig. 8). Thus, all currently known genes involved in the biosynthesis of both the monoterpene acid and the linolenic acid-derived alcohol moieties of pyrethrins are expressed specifically in the trichomes.
The Role of TcGLIP in Pyrethrin Biosynthesis
As reported previously (Ramirez et al., 2012), we also observed that TcGLIP was not expressed substantially in trichomes but instead showed the highest level of transcript accumulation in the ovaries, suggesting that the assembly of the pyrethrin esters from the acid and alcohol components occurs there. Kikuta et al. (2012) reported that TcGLIP used the activated CoA ester of trans-chrysanthemic acid to link it to the alcohol moiety but did not report whether TcGLIP also can form the ester directly from trans-chrysanthemic acid. When we transiently expressed both TcJMH and TcGLIP in N. benthamiana and fed the leaves with both jasmone and trans-chrysanthemic acid, jasmolin I was formed (Fig. 4), further confirming that the product of TcJMH activity on jasmone is jasmolone. In addition, the formation of jasmolin I in these N. benthamiana leaves suggests that TcGLIP may be able to catalyze the formation of the pyrethrin esters by using the nonactivated acid, although it also is possible that N. benthamiana leaves have an endogenous CoA ligase activity that works on trans-chrysanthemic acid. Both of these possibilities remain to be tested.
TcMJH and Other Pyrethrin Biosynthetic Genes Are Induced in Leaves by JA
Previous reports indicated that pyrethrin biosynthesis in pyrethrum leaves is induced by wounding (Ueda and Matsuda, 2011; Kikuta et al., 2012). Consistent with such reports, changes in transcript levels of TcMJH and other known genes of pyrethrin biosynthesis in pyrethrum leaves that were exposed to MeJA vapor showed a typical JA induction pattern (Sasaki et al., 2001), peaking within 6 to 12 h of the beginning of exposure and then declining within 24 h (Fig. 9B). It is important to note that neither in the wounding experiments nor in our MeJA induction experiments were trichomes of leaves examined directly; instead, whole leaves were analyzed. Nonetheless, the observation that some constitutive level of TcMJH expression was observed in cotyledons (Fig. 9A), which lack trichomes, suggests that TcMJH expression in leaves may not occur exclusively, or even mainly, in trichomes. The fact that JA is both an inducer of pyrethrin biosynthesis as well as a precursor substrate may impose additional complications in the regulation of its biosynthesis, and such possible complications also remain to be investigated.
TcJMH, a Key Enzyme in the Biosynthesis of the Alcohol Moiety of Pyrethrins, Is a Member of the CYP71 Family
JA is a ubiquitous plant hormone, and jasmone, a volatile compound, also has been detected in the floral scent of multiple species, such as jasmine (Jasminum officinale), tea (Camila sinensis), and orange (Citrus sinensis; Ruzicka and Pfeiffer, 1933; Yamanishi et al., 1965; Chung, 2012). Jasmolone and its related alcohols pyrethrolone and cinerolone (i.e. rethrolones), on the other hand, have been reported so far only from Tanacetum spp. Therefore, it appears that the ability to synthesize these alcohols from jasmone evolved within this genus (Head, 1973; Ramirez et al., 2012). Here, we showed that the first step in the rethrolone biosynthetic pathway is catalyzed by TcJMH, an enzyme that belongs to the cytochrome P450 oxidoreductase family. TcJMH falls into the CYP71 family of the extensive CYP superfamily. More specifically, it falls into the CYP71AT clade, which also encompasses the CYP83 clade. While the CYP83 enzymes catalyze the hydroxylation of aldoximes, they are not closely related to TcJMH, and even the most similar protein in the CYP71AT clade to TcJMH with a known enzymatic activity, the terpene hydroxylase CYPAT146 from P. frutescens (Lamiaceae; Supplemental Fig. S3), is only 50% identical to TcJMH. The lack of closely related CYP proteins with known functions currently precludes a more detailed reconstruction of the molecular steps involved in the evolution of TcJMH from another CYP enzyme with a different function. However, the availability of TcJMH will facilitate the biochemical analysis of closely related CYP genes and proteins in Asteraceae and, thus, ultimately allow the determination of the origin and extent of the rethrolone pathway in plants.
Ramirez et al. (2012) showed that the biosynthesis of pyrethrins begins in the trichomes of the ovaries and ends in the pericarp. The association of TcMJH with the ER also indicates that the biosynthetic pathway of pyrethrins not only involves different cells but also different compartments within the same cell, as the initial formation of trans-chrysanthemol by TcCDS occurs in the plastids (Yang et al., 2014), while the synthesis of the trans-chrysanthemic acid likely occurs in the cytosol (Xu et al., 2018). It would be of interest to identify the cellular and subcellular locations of all other enzymes in the pathway. Even with the limited information available now, it is likely that additional proteins involved in the transport of various intermediates are required for pyrethrin biosynthesis.
MATERIALS AND METHODS
Plant Growth Condition
Pyrethrum (Tanacetum cinerariifolium) plants were grown on soil in the greenhouse at 30°C during the 12-h light period and 24°C during the 12-h dark period. Nicotiana benthamiana plants were grown on soil in the greenhouse at a constant 28°C temperature with a 16-h light period and an 8-h dark period.
Jasmone Feeding of Tissues
Feeding experiments were carried out as described previously (Li et al., 2015, 2017) with the few modifications noted below. Pyrethrum flowers and leaves of N. benthamiana were immersed in 20 mL of aqueous solution of 20 μm jasmone, and control samples were immersed in pure water. The samples were vacuumed for 10 min, then incubated at room temperature for 24 h. The samples were then rinsed with water twice, extra water was removed with filter paper, and the samples were extracted with 10 mL of MTBE overnight. The samples were then centrifuged, and the top MTBE phase was collected and vacuumed to condense the extract to a final volume of 0.5 mL for GC-MS analysis. Cinerolone, jasmolone, and pyrethrolone peaks eluting from the GC column were identified by comparing published information about the identification of these compounds from pyrethrins hydrolyzed by TcGLIP (Kikuta et al., 2012; Supplemental Fig. S1).
RNA-Seq Data Analysis
P450 genes were obtained by searching the pyrethrum RNA-seq database with the term Cytochrome P450. Retrieved sequences of contigs were checked for the presence of a complete open reading frame, and only the ones with complete open reading frames were considered as P450 candidates. The sequences of AOS and AOC were obtained by BLAST search of the pyrethrum RNA-seq database (Xu et al., 2018) using Arabidopsis (Arabidopsis thaliana) AOS and AOC as respective queries.
Coexpression Analysis
Coexpression analysis was performed by comparing the transcript abundance of TcCDS and cytochrome P450 candidates using the Pearson correlation method to quantitate the similarity of their expression, as described by Li et al. (2015). The software for this analysis was SPSS statistics 22 (https://www.ibm.com/support/knowledgecenter/en/SSLVMB_22.0.0/com.ibm.spss.statistics_22.kc.doc/pv_welcome.html).
GC-MS Analysis
GC-MS analysis was performed using a GCMS-QP2010 SE apparatus (Shimadzu) with an Rxi-5Sil MS column. The column temperature was programmed as follows: 50°C hold for 3 min, 50°C to 160°C at 10°C min−1 and hold for 3.5 min, and 160°C to 300°C at 10°C min−1 and hold for 6 min. Total time for the program was 37.5 min.
Commercial pyrethrum extract (Sigma-Aldrich; 82670-25G) containing more than 50% (w/v) pyrethrins was used for the separation of pyrethrins. The separation procedure was based on Matsuda et al. (2005) with minor modifications. Pyrethrum extract was diluted 1:20 by acetonitrile, and 20 μL was loaded on a SUPELCOSI LC-18 column in a Waters HPLC machine and eluted at 0.5 mL min−1 constant rate with 65% (v/v) acetonitrile (in water) as the mobile phase. Fractions were collected every 0.5 min, and injections were repeated seven times to obtain a sufficient amount of pyrethrins. Acetonitrile was removed by evaporation in a vacuum centrifuge concentrator, and the remaining material was extracted with 500 μL of MTBE for GC-MS analysis. For fractions containing pyrethrins, MTBE was dried in a vacuum centrifuge concentrator, and the compound was redissolved in 100 μL of DMSO for further hydrolysis. Ten μL of purified pyrethrins was treated with 40 μL of purified GLIP (4 μg) overnight at room temperature to generate rethrolones. Reaction products were extracted with 100 μL of MTBE for GC-MS analysis.
N. benthamiana Transient Expression
The vector pEAQ-HT was used for transient expression in N. benthamiana, and the procedure was done as reported previously (Sainsbury et al., 2009). Primers used in the construction of the vectors are listed in Supplemental Table S1. pEAQ-HT constructs carrying specific genes were mobilized into Agrobacterium tumefaciens strain GV3101. A. tumefaciens cells were incubated in liquid Luria-Bertani medium and grown overnight to an OD value of 1, then centrifuged at 5,000 rpm for 10 min to remove the Luria-Bertani medium. The cells were resuspended in an equal volume of MMA buffer containing 10 mm MES, pH 6.8, 10 mm MgCl2, and 100 μm acetosyringone. Cell suspensions were injected into leaves of 4-week-old N. benthamiana from the abaxial side using a 3-mL syringe. After 3 d, the leaves were harvested for the feeding experiment, microsome preparation, or protein purification.
Microsomal preparations were performed according to the procedures outlined by Schaller (2017) with minor modifications. Eight milliliters of homogenization buffer without MgCl2 (50 mm Tris-HCl, pH 8.2, 2 mm EDTA, 20% (v/v) glycerol, 1 mm DTT, and 1:100 Sigma-Aldrich protease inhibitor cocktail for plant cell and tissue extracts) was added to 8 g of N. benthamiana leaves. The leaves were next ground with mortar and pestle on ice and filtered through a 250-μm mesh. The filtrate (around 12 mL) was centrifuged at 5,000 rpm for 5 min. The supernatant was ultracentrifuged in a swing rotor at 100,000 rpm for 45 min to pellet the membranes. The supernatant was discarded, and the pellet was resuspended with 0.6 mL of resuspension buffer (50 mm Tris-HCl, pH 8.2, 2 mm EDTA, 10% (w/v) Suc, 1 mm DTT, and 1:100 Sigma-Aldrich protease inhibitor cocktail for plant cell and tissue extracts) for enzyme assays.
TcGLIP protein was expressed in N. benthamiana leaves and purified by His-tag affinity. Fifty N. benthamiana seedlings were infiltrated with A. tumefaciens, and their leaves were collected after 3 d. All leaves were ground in liquid nitrogen and extracted in 100 mL of binding buffer (50 mm NaH2PO4, pH 8, 300 mm NaCl, and 10 mm imidazole), centrifuged (5,000 rpm, 10 min) to remove debris, and the supernatant was saved and passed through a filter paper. Ni-NTA agarose (300 μL) was added to the lysate and let stand for 1 h. Next, the agarose was washed with 30 mL of wash buffer (50 mm NaH2PO4, pH 8, 300 mm NaCl, and 20 mm imidazole) in a Qiagen 1-mL column, and the protein was eluted with 1 mL of elution buffer (50 mm NaH2PO4, pH 8, 300 mm NaCl, and 250 mm imidazole). Purified protein was analyzed by SDS-PAGE to determine purity, and the concentration was measured with Bio-Rad protein assay dye reagent (500-0006).
Analysis of the Km Value of TcJMH
JMH reactions were carried out in 50 µL of aqueous solution containing 100 mm Tris (pH 7.5), 30 μL of protein (microsomal preparation), varying concentrations (0/25/50/100/150/200 µm) of jasmone, and 300 µm NADPH. After a 2-h incubation, products were extracted with MTBE for GC-MS analysis and plotted to verify enzyme saturation (Supplemental Fig. S4). The Km value was calculated by the hyperbolic regression analysis method with software Hyper32 (http://hyper32.software.informer.com/). Data are presented as means ± sd (n = 3).
RT-qPCR of Floral and Leaf Transcripts
RNA was extracted with an Omega plant RNA kit (catalog no. R6827-02). The reverse transcription reaction was performed with a Thermo Fisher high-capacity cDNA reverse transcription kit (catalog no. 4368814). RT-qPCR was performed using Thermo Fisher Power SYBR Green PCR master mix (catalog no. 4367659). For tissue-specific expression analysis, T. cinerariifolium glyceraldehyde 3-phosphate dehydrogenase, TcActin7, and TcTubulin3 were used as internal reference genes. Primers used are listed in Supplemental Table S1.
Trichome isolation from pyrethrum flowers was performed according to Ramirez et al. (2012). Disc flowers at stage 3 were cut in the middle, corollas were removed, and the ovaries were placed in a 50-mL tube filled with approximately 20 mL of liquid nitrogen. The tube was vortexed for 1 min, then liquid nitrogen was added and the tube was vortexed again, for a total of 10 repeats. Trichomes were collected by filtering the slurry through 150-μm mesh and collecting the flow through.
MeJA Induction of Pyrethrin Biosynthetic Genes in Leaves
For MeJA induction, 2-week-old pyrethrum seedlings growing on one-half-strength Murashige and Skoog medium were placed in an air-tight container. A 1-cm × 1-cm 3MM filter paper soaked with MeJA was placed inside the container for the indicated time, after which the first two true leaves were collected and used for RNA extraction.
Subcellular Localization
The vector used for subcellular localization analysis was pEZS-NL. Protoplasts were prepared from Arabidopsis leaves, and transformation and confocal microscopy were performed as described previously (Xu et al., 2013). Primers used in the construction of the vectors are listed in Supplemental Table S1.
Phylogenetic Analysis
Amino acid sequences of CYP71AT subfamily proteins were provided by David Nelson (http://drnelson.uthsc.edu/cytochromeP450.html) and by BLASTP search in the National Center for Biotechnology Information. Phylogenetic analysis was conducted by MEGA7 using the maximum likelihood method based on the JTT matrix-based model (Kumar et al., 2016).
Accession Numbers
The sequence data in this study can be obtained from the National Center for Biotechnology Information with the following GenBank accession numbers: TcJMH, MG189934; DN144246, MG874675; DN150785, MG874676; DN133626, MG874677; DN126517, MG874678; DN163032, MG874679; DN97751, MG874680; DN121926, MG874681; DN151943, MG874682; DN15633, MG874683; DN131340, MG874684; TcAOS, MH397471; and TcAOC, MH397472.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of rethrolones.
Supplemental Figure S2. cDNA and protein sequences of the 11 cytochrome P450 oxidoreductases, TcAOS, and TcAOC identified in this study.
Supplemental Figure S3. A maximum likelihood phylogenetic tree of TcJMH and related sequences.
Supplemental Figure S4. Enzyme saturation curve for in vitro assays for the conversion of jasmone to jasmolone by purified ER preparations from N. benthamiana leaves expressing TcJMH.
Supplemental Table S1. Primers used in this investigation.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dr. Haiyang Xu for helpful discussions concerning the analysis of the T. cinerariifolium cytochrome P450 enzymes. We also thank Dr. David Nelson for naming TcJMH and providing CYP71AT subfamily sequences for phylogenetic analysis.
Footnotes
This work was supported by the National Science Foundation collaborative research grant 1565355 to E.P.
Articles can be viewed without a subscription.
References
- Bak S, Feyereisen R (2001) The involvement of two p450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol 127: 108–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkett MA, Campbell CAM, Chamberlain K, Guerrieri E, Hick AJ, Martin JL, Matthes M, Napier JA, Pettersson J, Pickett JA, et al. (2000) New roles for cis-jasmone as an insect semiochemical and in plant defense. Proc Natl Acad Sci USA 97: 9329–9334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MS. (2012) Volatile compounds of the hallabong (Citrus kiyomi × Citrus ponkan) blossom. Food Sci Biotechnol 21: 285–290 [Google Scholar]
- Creelman RA, Mullet JE (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48: 355–381 [DOI] [PubMed] [Google Scholar]
- DeMicco A, Cooper KR, Richardson JR, White LA (2010) Developmental neurotoxicity of pyrethroid insecticides in zebrafish embryos. Toxicol Sci 113: 177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Ito M (2017) Molecular cloning and characterization of a Perilla frutescens cytochrome P450 enzyme that catalyzes the later steps of perillaldehyde biosynthesis. Phytochemistry 134: 26–37 [DOI] [PubMed] [Google Scholar]
- Head SW. (1973) Composition of pyrethrum extract and analysis of pyrethrins. In Casida JE, ed, Pyrethrum. Academic Press, New York, pp 25–53 [Google Scholar]
- Jones GDG. (1973) Pyrethrum production. In Casida JE, ed, Pyrethrum. Academic Press, New York, pp 17–22 [Google Scholar]
- Kikuta Y, Ueda H, Nakayama K, Katsuda Y, Ozawa R, Takabayashi J, Hatanaka A, Matsuda K (2011) Specific regulation of pyrethrin biosynthesis in Chrysanthemum cinerariaefolium by a blend of volatiles emitted from artificially damaged conspecific plants. Plant Cell Physiol 52: 588–596 [DOI] [PubMed] [Google Scholar]
- Kikuta Y, Ueda H, Takahashi M, Mitsumori T, Yamada G, Sakamori K, Takeda K, Furutani S, Nakayama K, Katsuda Y, et al. (2012) Identification and characterization of a GDSL lipase-like protein that catalyzes the ester-forming reaction for pyrethrin biosynthesis in Tanacetum cinerariifolium: a new target for plant protection. Plant J 71: 183–193 [DOI] [PubMed] [Google Scholar]
- Koch T, Bandemer K, Boland W (1997) Biosynthesis of cis-jasmone: a pathway for the inactivation and the disposal of the plant stress hormone jasmonic acid to the gas phase? Helv Chim Acta 80: 838–850 [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang F, Chang Y, Zhao T, Schranz ME, Wang G (2015) Nicotinate O-glucosylation is an evolutionarily metabolic trait important for seed germination under stress conditions in Arabidopsis thaliana. Plant Cell 27: 1907–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang F, Wu R, Jia L, Li G, Guo Y, Liu C, Wang G (2017) A novel N-methyltransferase in Arabidopsis appears to feed a conserved pathway for nicotinate detoxification among land plants and is associated with lignin biosynthesis. Plant Physiol 174: 1492–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughrin JH, Manukian A, Heath RR, Tumlinson JH (1995) Volatiles emitted by different cotton varieties damaged by feeding beet armyworm larvae. J Chem Ecol 21: 1217–1227 [DOI] [PubMed] [Google Scholar]
- Matsuda K, Kikuta Y, Haba A, Nakayama K, Katsuda Y, Hatanaka A, Komai K (2005) Biosynthesis of pyrethrin I in seedlings of Chrysanthemum cinerariaefolium. Phytochemistry 66: 1529–1535 [DOI] [PubMed] [Google Scholar]
- McLaughlin GA. (1973) History of pyrethrum. In Casida JE, ed, Pyrethrum. Academic Press, New York, pp 3–15 [Google Scholar]
- Mizutani M, Ohta D (2010) Diversification of P450 genes during land plant evolution. Annu Rev Plant Biol 61: 291–315 [DOI] [PubMed] [Google Scholar]
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, et al. (1996) P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 6: 1–42 [DOI] [PubMed] [Google Scholar]
- Ramirez AM, Stoopen G, Menzel TR, Gols R, Bouwmeester HJ, Dicke M, Jongsma MA (2012) Bidirectional secretions from glandular trichomes of pyrethrum enable immunization of seedlings. Plant Cell 24: 4252–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez AM, Yang T, Bouwmeester HJ, Jongsma MA (2013) A trichome-specific linoleate lipoxygenase expressed during pyrethrin biosynthesis in pyrethrum. Lipids 48: 1005–1015 [DOI] [PubMed] [Google Scholar]
- Rivera SB, Swedlund BD, King GJ, Bell RN, Hussey CE Jr, Shattuck-Eidens DM, Wrobel WM, Peiser GD, Poulter CD (2001) Chrysanthemyl diphosphate synthase: isolation of the gene and characterization of the recombinant non-head-to-tail monoterpene synthase from Chrysanthemum cinerariaefolium. Proc Natl Acad Sci USA 98: 4373–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka L, Pfeiffer M (1933) Über asminriechstoffe. I. Die Konstitution des Jasmons. Helv Chim Acta 16: 1208–1214 [Google Scholar]
- Sainsbury F, Thuenemann EC, Lomonossoff GP (2009) pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J 7: 682–693 [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Asamizu E, Shibata D, Nakamura Y, Kaneko T, Awai K, Amagai M, Kuwata C, Tsugane T, Masuda T, et al. (2001) Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res 8: 153–161 [DOI] [PubMed] [Google Scholar]
- Schaller GE. (2017) Isolation of endoplasmic reticulum and its membrane. Methods Mol Biol 1511: 119–129 [DOI] [PubMed] [Google Scholar]
- Shafer TJ, Meyer DA, Crofton KM (2005) Developmental neurotoxicity of pyrethroid insecticides: critical review and future research needs. Environ Health Perspect 113: 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WC, Funk CD, Brash AR (1993) Molecular cloning of an allene oxide synthase: a cytochrome P450 specialized for the metabolism of fatty acid hydroperoxides. Proc Natl Acad Sci USA 90: 8519–8523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijet N, Brash AR (2002) Allene oxide synthases and allene oxides. Prostaglandins Other Lipid Mediat 68-69: 423–431 [DOI] [PubMed] [Google Scholar]
- Ueda H, Matsuda K (2011) VOC-mediated within-plant communications and nonvolatile systemic signals upregulate pyrethrin biosynthesis in wounded seedlings of Chrysanthemum cinerariaefolium. J Plant Interact 6: 89–91 [Google Scholar]
- Xu H, Zhang F, Liu B, Huhman DV, Sumner LW, Dixon RA, Wang G (2013) Characterization of the formation of branched short-chain fatty acid:CoAs for bitter acid biosynthesis in hop glandular trichomes. Mol Plant 6: 1301–1317 [DOI] [PubMed] [Google Scholar]
- Xu H, Moghe GD, Wiegert-Rininger K, Schilmiller AL, Barry CS, Last RL, Pichersky E (2018) Coexpression analysis identifies two oxidoreductases involved in the biosynthesis of the monoterpene acid moiety of natural pyrethrin insecticides in Tanacetum cinerariifolium. Plant Physiol 176: 524–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanishi T, Kiribuchi T, Mikumo Y, Sato H, Ohmura A, Mine A, Kurata T (1965) Studies on the flavor of green tea. Part VI. Neutral fraction of the essential oil of tea leaves. Agric Biol Chem 29: 300–306 [Google Scholar]
- Yang T, Gao L, Hu H, Stoopen G, Wang C, Jongsma MA (2014) Chrysanthemyl diphosphate synthase operates in planta as a bifunctional enzyme with chrysanthemol synthase activity. J Biol Chem 289: 36325–36335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YJ, Cheng QQ, Su P, Chen X, Wang XJ, Gao W, Huang LQ (2014) Research progress relating to the role of cytochrome P450 in the biosynthesis of terpenoids in medicinal plants. Appl Microbiol Biotechnol 98: 2371–2383 [DOI] [PubMed] [Google Scholar]
- Zhou F, Wang CY, Gutensohn M, Jiang L, Zhang P, Zhang D, Dudareva N, Lu S (2017) A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice. Proc Natl Acad Sci USA 114: 6866–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]