EZ-Root-VIS is a free software package that enables the rapid quantification and visual reconstruction of averaged root system architectures for investigating environment-genotype interactions.
Abstract
If we want to understand how the environment has shaped the appearance and behavior of living creatures, we need to compare groups of individuals that differ in genetic makeup and environment experience. For complex phenotypic features, such as body posture or facial expression in humans, comparison is not straightforward because some of the contributing factors cannot easily be quantified or averaged across individuals. Therefore, computational methods are used to reconstruct representative prototypes using a range of algorithms for filling in missing information and calculating means. The same problem applies to the root system architecture (RSA) of plants. Several computer programs are available for extracting numerical data from root images, but they usually do not offer customized data analysis or visual reconstruction of RSA. We developed Root-VIS, a free software tool that facilitates the determination of means and variance of many different RSA features across user-selected sets of root images. Furthermore, Root-VIS offers several options to generate visual reconstructions of root systems from the averaged data to enable screening and modeling. We confirmed the suitability of Root-VIS, combined with a new version of EZ-Rhizo, for the rapid characterization of genotype-environment interactions and gene discovery through genome-wide association studies in Arabidopsis (Arabidopsis thaliana).
Roots play a critical role in soil water and nutrient uptake and, thus, in plant productivity. Root system architecture (RSA) is determined by the relative growth of different parts of the root system as well as frequency of branching and branch angles. RSA is a key determinant of root function and is highly plastic, being controlled by genotype-environment interactions (Yu et al., 2016; Morris et al., 2017; Shahzad and Amtmann, 2017). Optimization of RSA is thus imperative for the improvement of plant resilience, particularly to soil-borne stresses such as soil drought, flooding, nutrient deficiencies, and biotic factors (Julkowska and Testerink, 2015; Lynch, 2015; Rogers and Benfey, 2015). Correlation of root morphology with DNA sequence information allows the identification of the genetic loci that underpin root features, for example, through mutant screens, quantitative trait locus analysis or genome-wide association studies (GWAS). New sequencing technology has vastly increased the amount of available genetic information, and better phenotyping assays are now required to meet the pace of these advancements. Depending on the research question, different phenotyping approaches need to be taken because there is a tradeoff between (1) reflecting realistic environments, (2) controlling environmental factors, (3) obtaining precise information on individual parts of the root systems, and (4) throughput.
Several platforms to obtain root images from plants grown in soil (field, pots, or rhizotrons) are available, together with software to capture and model the overall size and shape of the complex root systems (Lobet et al., 2011; Bucksch et al., 2014; Kalogiros et al., 2016). These studies are complemented by analyses of roots developing on synthetic surfaces, which offer more precise control and manipulation of the root environment. Open source software such as BRAT (Slovak et al., 2014; Satbhai et al., 2017) and RootTrace (Naeem et al., 2011) have been developed to precisely track the early main root growth from scanned images of Arabidopsis (Arabidopsis thaliana) seedlings growing on plates. An intermediate analysis space is occupied by software such as EZ-Rhizo (Armengaud et al., 2009), archiDART (Delory et al., 2016, 2018), RootScape (Ristova et al., 2013), and the commercial WinRhizo (Arsenault et al., 1995). They have facilitated the quantitative assessment of more advanced root systems of Arabidopsis and other dicot plants, including information on lateral root features such as position, length, density, and angle. This information has generated knowledge on branching patterns, rather than just primary root growth or overall shape, and has enabled large-scale phenotyping of Arabidopsis roots to investigate natural variation and responses to multiple environmental cues (Gruber et al., 2013; Kellermeier et al., 2013, 2014; Julkowska et al., 2017).
With a good range of root image analysis software in place, the next challenge is how to process the acquired numerical data in order to extract biologically meaningful and statistically valid results. Averaging across replicates is easy for some simple RSA traits, such as main root (MR) length and lateral root (LR) number, but it is less straightforward for other traits. For example, how should we calculate the mean LR length if the number of LRs differs between replicate plants? We can either average the length of all laterals in each plant before averaging across plants, or we can average individual LRs across replicates. The latter can be done either by the order of appearance (1st, 2nd LR, etc.) or by their position on the main root (e.g. all LRs within a particular sector of the main root). There is no one “correct” solution to this problem; rather, each of the approaches is more or less suited to answer a specific question. For example, the first average provides a measure of overall growth investment into lateral roots, the second provides information on how individual LRs develop, and the last approach best reflects the root shape. It is therefore important that RSA analysis software, in addition to enabling data acquisition, also offers different options for extracting mean root traits and their variance, which can then be used as a numerical input for genetic studies or developmental models.
Another challenge is how to visually represent averaged root systems in a manner that clearly and faithfully reflects differences between genotypes or environmental conditions. Current root image analysis software packages rarely include options to visually reconstruct single-plant or mean RSAs from the measured data. In publications, the individual averaged RSA traits are usually represented in multiple bar graphs. A picture of one “representative” root system is often included, but considering the variability and complexity of RSA, it is unlikely that one individual plant can indeed be representative in all RSA traits. The reader is left with the difficult task of deducing the mean RSA from bar graphs of all individual traits.
To address these problems, we have developed a new application, Root-VIS, which enables calculation and visual reconstruction of mean RSAs from measured data. We exemplify the possibilities offered by Root-VIS using data acquired with a greatly enhanced version of the previously published EZ-Rhizo program. The combined EZ-Root-VIS pipeline provides an easy procedure to capture RSA features of many individual plants and to visualize averaged RSAs for different genotypes under various environments or at different time points. Root-VIS offers a range of choices for how to calculate means and how to visually represent the average root phenotypes. For example, it allows users to calculate means of LR-related traits on a per-plant or a per-branch basis. It provides possibilities to redraw average root architectures from the extracted data or to regenerate root shapes in the form of flag plots, as well as growth-investment or LR length profiles. We validated the utility of the EZ-Root-VIS pipeline for studying root growth dynamics, quantitative genetics, and genotype-environment interactions.
RESULTS
Description of the EZ-Root-VIS Pipeline
The software is a standalone implementation, compatible with all Microsoft Windows operating systems from Windows XP (SP-3) onwards, including Vista, Windows 7, 8/8.1, and 10 (in “classic desktop” mode). Separate versions are available for 32- and 64-bit platforms. The installation package includes Root-VIS and the latest version of the EZ-Rhizo program, which has a number of additions and improvements over the original 2009 release (Armengaud et al., 2009). For example, EZ-Rhizo now accepts input images in any of the standard formats: Windows bitmap (BMP) files, graphics interchange format (GIF), joint photographic experts group compressed images (JPEG), portable network graphics (PNG), or tagged image file format (TIFF) files, and at any resolution. The software presents a typical, Windows-style graphical user interface (GUI; Supplemental Fig. S1A) that will be very familiar to users of the Microsoft Visual Studio development environment. Extensive (and context-sensitive) online help is included with the software (Supplemental Fig. S1B). The software also provides numerous, user-selectable options for controlling the content and style of displays, and the parameters used in the reconstruction and averaging algorithms. These are adjustable via a “settings” property sheet (Supplemental Fig. S1C).
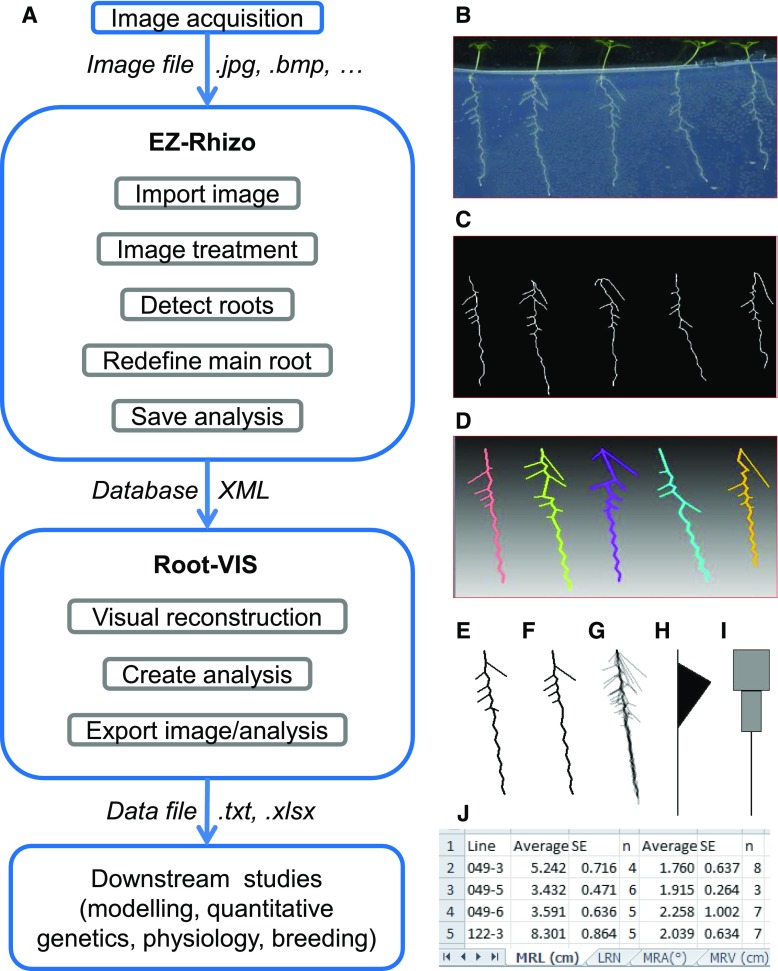
Figure 1A summarizes the workflow of RSA analysis with the EZ-Root-VIS pipeline. Images of plants grown on agar plates in 2D are usually acquired with flatbed scanners (Fig. 1B), but any image capturing device that generates sufficient contrast and resolution is suitable. First, EZ-Rhizo is used to process the images and extract numerical data of RSA features of individual roots (Fig. 1C). After image cropping, conversion into black and white, and background noise filtering, roots are pixelated, skeletonized, edited, and detected as described before (Armengaud et al., 2009). In the new version of EZ-Rhizo, we have included an option to redefine the main root (the original version assumed the longest root path formed the MR and this is also the default setting). The numerical results of the analysis are saved in a user-selectable database, which is now in XML format, allowing easy sharing of data sets between the acquisition and analysis modules. Root-VIS then “reads” the database generated by EZ-Rhizo and displays the files in a database window (left panel in Supplemental Fig. S1A). Prior to averaging RSA data from replicate roots, Root-VIS offers the possibility to draw a visual reconstruction of any individual root in the data set (Fig. 1D), which allows the user to verify the data that were captured by the image acquisition software.
Figure 1.
Overview of EZ-Root-VIS pipeline. A, EZ-Rhizo and Root-VIS software packages provide a convenient analysis pipeline transforming root images into a numerical and statistical data output describing root system architecture. B, A suitable image of Arabidopsis plants growing on a vertical agar plate can be acquired with a flatbed scanner at 200 dpi. C, Skeletonized roots obtained after image processing with EZ-Rhizo provide the basis for quantification of RSA features by EZ-Rhizo. The obtained data are saved in a searchable database. D, Root-VIS reconstructs the individual roots using the data extracted by EZ-Rhizo. E to J, Root-VIS generates visual reconstructions of root system architecture from a user-defined set of individual roots (replicates), including absolute (E) and binned (F) average RSA, alpha blends (G), flag plots (H), and LR profiles (I). The examples shown are based on data from the five individual Arabidopsis plants shown in B. Roots images were taken at 12 DAG. J, For subsequent generation of graphs, statistical analyses, and modeling, the numerical data obtained with Root-VIS are saved and can be displayed in Excel using the XL-Orate plug-in. The screenshot shows an example Excel output from a large data set. Data for each RSA parameter are provided in a separate datasheet. MRL, Main root path length; LRN, lateral root number; MRV, main root vector length.
At the heart of the Root-VIS software lies the concept of the “collection analysis,” in which data extracted from the database are grouped according to the metadata parameters entered during image analysis, including genotype, media type, and plant age. Analyses can be carried out with zero, one, or two of these parameters being variants. The database structure displayed in the analysis window makes it easy to remove or pool individual samples or groups of samples from the analysis (Supplemental Fig. S1B). The averaged root for each group can then be displayed visually using a “reconstruction” algorithm described in more detail in the “Materials and Methods” section. Two types of averaged root systems can be generated: “absolute” and “binned” (Fig. 1, E and F). In the “absolute” mode, lateral roots are averaged according to their order of appearance; in the “binned” mode, the branched zone is divided into sectors (the number of which is equal to the mean number of LRs), and the lengths of LRs in each sector (“bin”) are averaged. The program also provides an option to visualize the variation between individual replicate roots in “alpha blends” (Fig. 1G). Additional visualization options are “flag plots” (Fig. 1H), which represent the overall density and shape of the root system, and LR profiles (Fig. 1I), which represent the size of LRs in different parts of the root system. Flag plots are based on LR angle and LR density as well as the lengths of branched and unbranched zones of the MR as depicted in Supplemental Figure S2. For the LR profiles, the MR length is divided into a user-defined number of sections, and for each section, LR length is displayed as a rectangle of appropriate size. Each of the display modes has a number of suboptions, which the user can select in the “settings” windows. For example, LR profiles can be chosen to show either the sum length of all LRs (LR size [LRS]) or the mean LR length (LRL) in each MR sector. The former provides a measure of total growth investment into a certain part of the root, whereas the latter reflects the local root system width. The different displays are exemplified below using experimental data. We also generated an optional plug-in module, XL-Orate, which allows users to create formatted tables of averaged trait data, including SEs and number of replicates using Microsoft Excel (Fig. 1J; Supplemental Data Set 1), and to rapidly generate bar charts for any data generated with EZ-Rhizo and Root-VIS. The obtained data sets can now be used in a range of downstream studies, including growth models, quantitative genetics, and breeding.
EZ-Root-VIS Captures Root Growth Dynamics
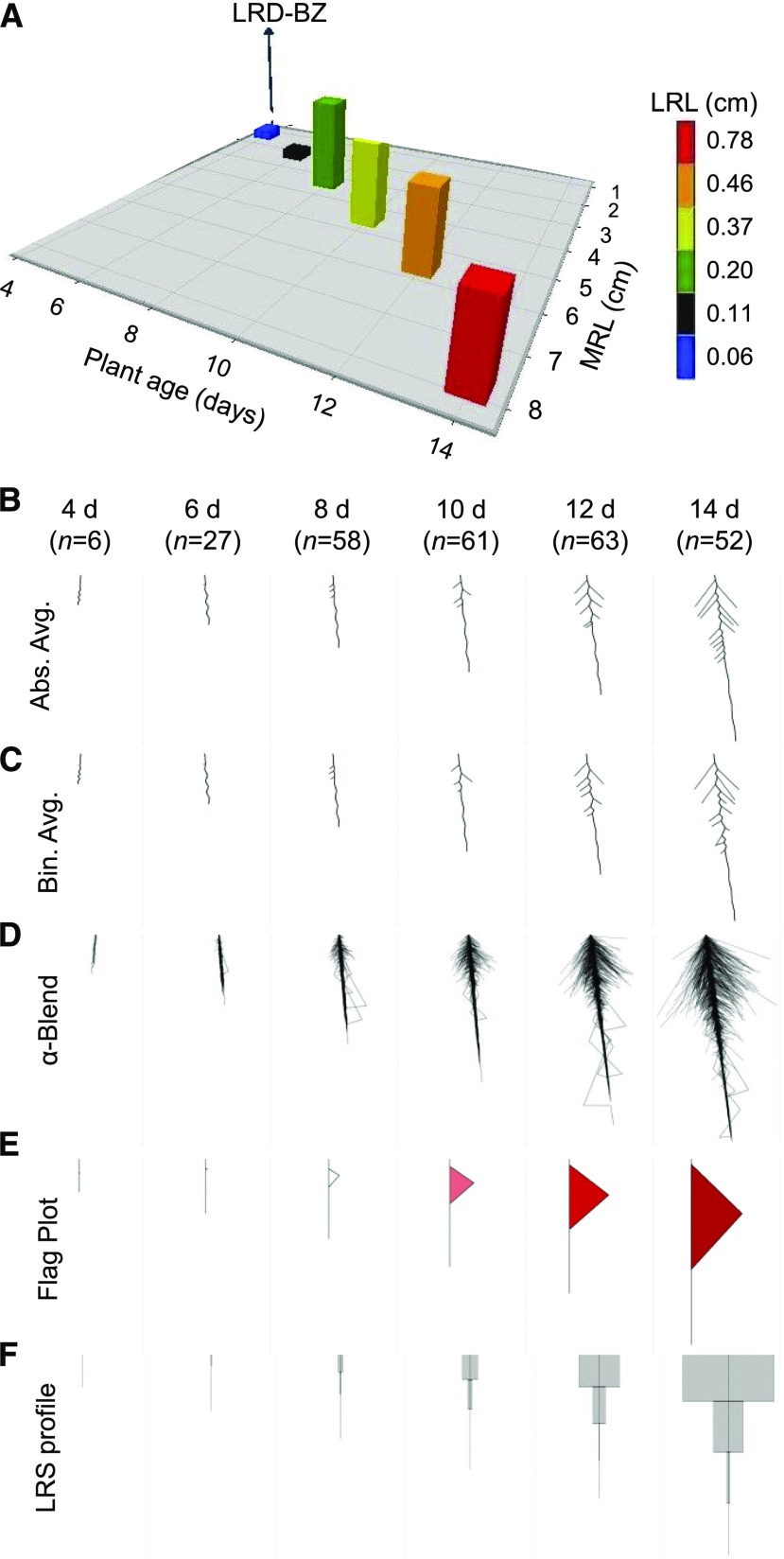
To validate the ability of the EZ-Root-VIS pipeline to capture root system growth, we measured RSA of Arabidopsis Col-0 plants every 2 d over a period of 4 to 14 d after germination (DAG). Quantitative data for 16 root traits (see "Abbreviations") were obtained using EZ-Rhizo. A 3D bar chart generated for three root traits (main root length, LRL, and LR density in the branched zone [LRD-BZ]; Fig. 2A) exemplifies the difficulty of visually representing the complexity of RSA changes in bar graphs; several additional graphs would be required to represent the overall RSA. In contrast, the visual root system reconstructions implemented in Root-VIS accommodate all the root traits and represent averaged RSA (either “absolute” or “binned”) as a single image (Fig. 2, B and C). In this example, the display incorporated bends into the MR path to account for the measured difference between MR path length and MR vector length (straightness). The program accommodates any surplus of path length over vector length across equally sized bends, which are placed at LR emergence sites in the branched zone and at equal distances along the apical zone (see Methods). However, the user should keep in mind that EZ-Rhizo does not determine the number, size, and position of bends, and the displayed bending pattern could look very different from the original images. To avoid overinterpretation of the displayed bending pattern it might be safer to display either path or vector length as a straight line. Therefore, in the settings property sheet, the user can toggle between displaying bends (in the MR and/or LR) or not.
Figure 2.
Visual representation of RSA development over time. A, Change of selected RSA features over time displayed in a conventional bar graph. Plotted are plant age in days after germination (x axis), mean LRD-BZ (y axis), mean main root length (MRL; z axis), and mean LRL (color) of Arabidopsis Col-0 plants growing on control media. Numbers of roots analyzed at each time point are given in B. B to F, Root-VIS offers several options to visualize the RSA of many replicate plants, including absolute (B) and binned (C) average RSA reconstructions, superimposition of normalized RSAs in alpha blends (D), as well as shape reconstructions in the form of flag plots (E) and LRS profiles (F). All Root-VIS reconstructions shown were based on EZ-Rhizo data obtained from Arabidopsis Col-0 plants grown in control conditions. Plant age (in days) and numbers of replicate roots (n) are given in B.
Figure 2D shows the superposition of all reconstructed individual roots as alpha blends, reflecting the variation across replicates. A similar darkness of the blends indicated that the variation of RSA between individual plants did not substantially increase with plant age. The flag plots (Fig. 2E) show how the root system increases in depth, width, and density over time. For this figure, flag plots were produced based on absolute values of MR and LR lengths and angles and on the slope of the linear regression of average LR lengths at the given LR positions (see Supplemental Fig. S2 and “Materials and Methods”). The shade of the flag fill color indicates LR density with darker shades representing higher density. Finally, LRS profiles (Fig. 2F) allow the reader to quickly evaluate the growth investment into LRs in different parts of the root. In this case, the MR was divided into four equal sections (quartiles). The profiles show a sharp increase of mean LRS from 8 DAG onwards, especially in the two basal quartiles of the main root. The data underpinning Figure 2, extracted using XL-Orate (Supplemental Data Set 2), provide a convenient input for root growth models.
EZ-Root-VIS Reveals Genotype-by-Environment Interactions
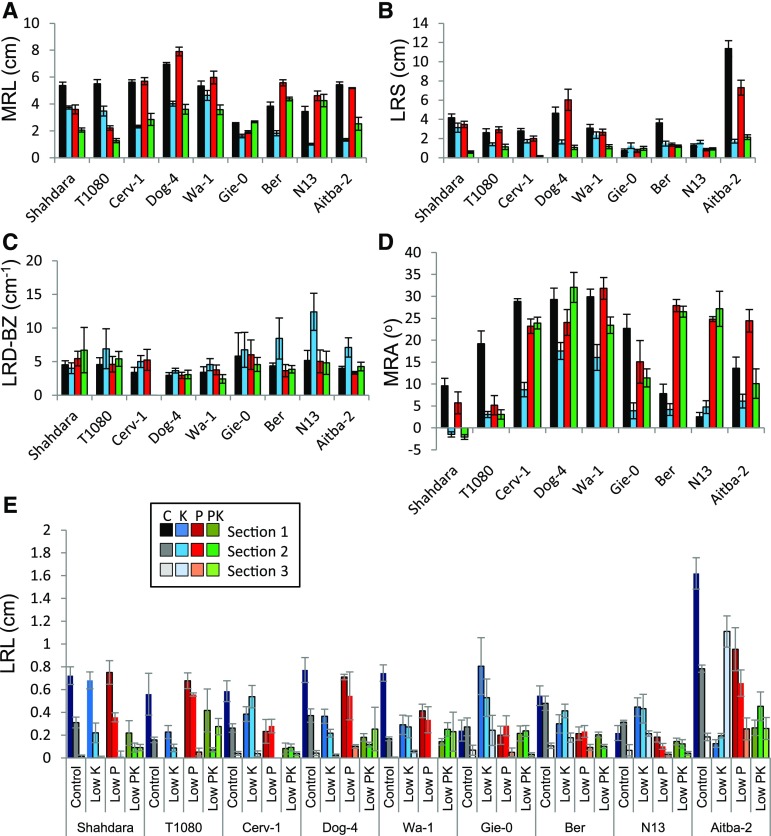
To demonstrate the effectiveness of the pipeline in capturing genotype-by-environment interactions, RSA data were acquired with EZ-Rhizo from replicate plants of nine Arabidopsis accessions, grown in four nutrient media: control, low phosphate (P; 20 µm), low potassium (K; 10 µm), or combined low PK media (Fig. 3; Supplemental Data Set 3; for growth media, see “Materials and Methods”). The visual reconstructions (Fig. 3) generated by Root-VIS facilitate the rapid identification of interesting responses and RSA ideotypes, while Excel-generated bar graphs (Fig. 4) depict means and ses of the individual traits. As reported before (Chevalier et al., 2003; Reymond et al., 2006; Svistoonoff et al., 2007), the Shahdara accession showed a strong inhibition of MR growth in response to P limitation. An even stronger reduction of MR length was found for T1080 despite similar MR length on control media. In most other accessions, e.g. Aitba-2 and Cerv-1, MR growth was not sensitive to P availability, and in the Ber accession, MR growth was even stimulated by decreasing the P supply (Figs. 3A and 4A). By contrast, in Ber, Aitba-2, and Cerv-1, low P resulted in a reduction in the LRS (measured as total lateral root path length; Fig. 4B), particularly in the basal (upper) quartile of the main root (LRS [0.25]; Figs. 3A and 4D). Unlike low P, low K decreased MR length in all accessions tested, albeit to varying extents, and caused a decrease of LRS in most accessions (Fig. 3A). Exceptions to the latter were Gie-0 and N13, which already had very small LRS in control conditions.
Figure 3.
Environment- and genotype-dependent growth investment into main root and lateral roots. LRS profiles (A) and average RSA reconstruction (B) of Arabidopsis accessions grown in control, low K, low P, and combined low P and low K (low PK) media. Vertical lines represent the mean main root path length. The main root path was divided into four equally sized sections and lengths of laterals were added within each section. The width of each square represents the mean LRS of three to six replicate plants.
Figure 4.
RSA traits of Arabidopsis accessions grown in different nutrient conditions. Means and ses of selected RSA features of 12-d-old Arabidopsis accessions grown in control (black bars), low K (blue bars), low P (red bars), or low PK (green bars) media. The data were generated by EZ-Rhizo, and means across replicates (n = 3–6 plants) were calculated with Root-VIS for main root length (MRL; A), total LRS (B), LRD-BZ (C), and MRA (D). E, Means and SEs of LRL within four sections of the main root. Data were extracted from LRL profiles generated by Root-VIS and transferred to Excel using the XL-Orate plug-in.
The effects of combined P and K limitation were striking and in many cases could not have been predicted from individual deficiency effects. In accessions experiencing MR inhibition under low P (e.g. Shahdara, T1080), combined PK limitation had an additive inhibitory effect, but in several other accessions (Gie-0, Ber, N13, and Aitba-2), we recorded a complete or partial reversal of the low K-induced MR inhibition by decreasing P (Fig. 4A). In Cerv-1 and Dog-4, MR inhibition by low K was maintained in low PK and dominated over MR insensitivity to low P. In contrast to the genotype-dependent response of MR to single and double deficiency, the LRS of all genotypes was smaller under combined PK limitation than under single deficiencies (Fig. 4B). LRD-BZ was in most accessions stable across treatments (Fig. 4C) with the exception of Ber, N13, and Aitba-2, which showed an increase of LRD-BZ under low K. To separate the effects of LR number and LR length on total LRS, LRL profiles and flag plots were also plotted (Supplemental Fig. S3). Figure 4E shows the extracted data from these profiles as bar graphs.
The reconstructed average RSAs (Fig. 3B) pinpointed additional differences. For example, MR growth was more or less vertical depending on genotype and environment. In control medium, the MR angle (MRA) differed between accessions (Fig. 4D), although the direction was always the same (“positive”; see “Materials and Methods” for sign definition). Under low P, MRA remained similar to control in most accessions, but markedly increased in Ber, N13, and Aitba-2, and decreased in T1080. Under low K, MRAs decreased in all accessions apart from N13. In most accessions the low-K MRA phenotype was reverted back to control or to low-P phenotypes in low PK, with the notable exception of Shahdara.
In summary, the EZ-Root-VIS pipeline enabled the analysis of complex responses of various root architectural traits to either single or combined nutrient limitations. The averaged visual representations allow the fast identification of nutrient interactions and of accessions that show particularly interesting RSA responses. It is remarkable that the relatively small number of accessions and nutrients tested here already encompass a diverse range of combinatorial phenotypic outputs.
EZ-Root-VIS Enables Large-Scale RSA Genetic Studies
To validate the utility of our pipeline for large-scale quantitative genetic studies, we measured RSA of 147 Arabidopsis accessions with EZ-Rhizo and determined means of 16 root traits with Root-VIS for downstream analyses (Supplemental Data Set 4). Five plants per accession were grown on standard medium, and it took one person 13 h to analyze all root images obtained at one time point (12 DAG) using the EZ-Root-VIS pipeline.
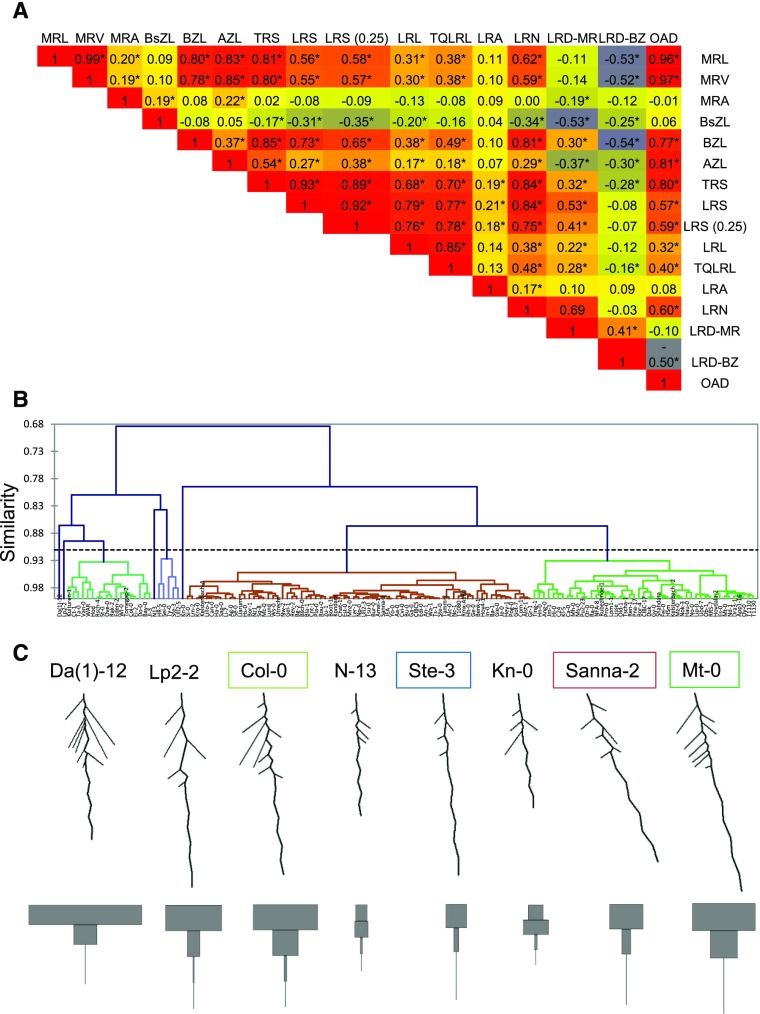
Ten of the analyzed root traits exhibited a normal distribution (Supplemental Fig. S4), but several traits showed a positively skewed trait distribution, including for example LR size and number. Correlation analysis revealed a high degree of interdependence of the RSA traits (Fig. 5A). Only MRA and lateral root angle were relatively independent of other RSA traits, although they showed correlation with the size of MR and LRs, respectively. Surprisingly, MR length was not correlated with the LR density over the MR but was negatively correlated with LRD-BZ. This observation stresses the importance of determining LR density in the branched zone rather than relating the number of LRs to the entire main root. Interestingly, LR density was also negatively correlated with the length of the basal zone (BsZL). In fact, BsZL was negatively correlated with most LR traits, suggesting that the amount of main root growth before emergence of the first LR is an important determinant of other RSA features.
Figure 5.
Use of the EZ-Root-VIS pipeline for large-scale natural variation studies. A, Heat map of pairwise correlations (Pearson correlation coefficient) of 16 RSA traits in 147 Arabidopsis accessions. Plants were grown in control conditions. The correlations between different root features were calculated using means of five plants for each genotype. Stars indicate significance at P < 0.05 (Student’s t test). B, Hierarchical clustering of Arabidopsis accessions by genotype. Calculation of similarity was based on 15 mean RSA traits calculated by Root-VIS. See Supplemental Data Set 5 for the class assignment of the individual accessions. C, Visual reconstructions of RSA and LRS profiles of the central accession of each cluster. Accession names are colored according to B.
Hierarchical clustering was performed to classify the accessions into groups sharing similar RSA based on 15 RSA architecture traits (LR angle was excluded from this analysis due to some negative values). As shown in Figure 5B, the accessions were assigned into eight clusters (Supplemental Data Set 4). Four clusters contained only one accession [Da(1)-12, Lp2-2, N13, and Kn-0], while the largest cluster contained 73 accessions. Average LRS profiles of the central genotype of each cluster (Fig. 5C) indicated different growth investment into MR and LR. Averaged RSA reconstructions revealed differences in MRA. For example, the Sanna-2 and Mt-0 groups exhibited more positively skewed MRs than other groups. Thus, when applied to natural variation studies, Root-VIS quickly identifies root traits that are critical determinants of the overall RSA.
To test if the data extracted with EZ-Root-VIS can facilitate genetic mapping studies, we performed GWA mapping for 16 root traits using an accelerated mixed model implemented in the GWAPP web application (Seren et al., 2012). In total, seven unique locus-specific single nucleotide polymorphisms (SNPs) were found to be associated with various root traits at 5% false discovery rate (FDR; Supplemental Data Set 5). Out of these, one (SNP4 6374071) was associated with TRS, LRS, and LRS (0.25) (Fig. 6A), three were associated with BsZL, and one was associated with lateral root density.
Figure 6.
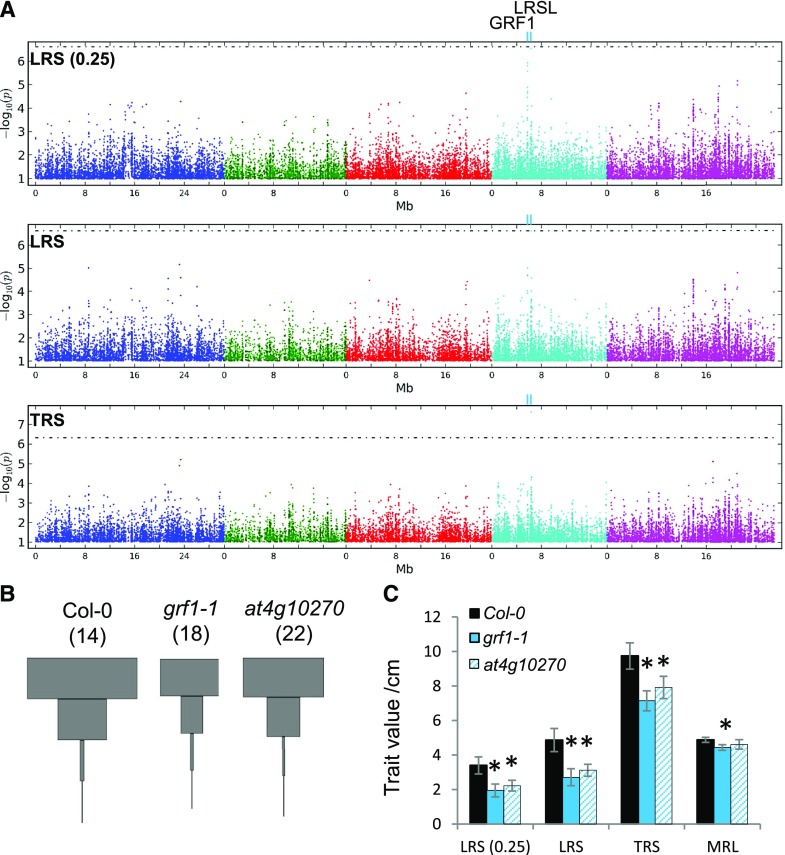
Genome-wide association studies for root system architecture traits. A, Manhattan plots for GWA mapping of three RSA traits; LRS over the basal quartile of the main root [LRS (0.25)], total LRS, and total root size (TRS). The horizontal dotted line corresponds to a 5% FDR threshold. Light-blue ticks labeled “GRF1” and “LRSL” indicate the location of the most significant associations. B,) LRS profiles of Arabidopsis Col-0 wild type and mutant lines. Plants were grown on control media in three independent experiments. The total number of replicate roots contributing to each LRS profile is given in parentheses. C, Means and SEs (n as shown in B) of some RSA traits in wild type Col-0 and mutant lines. Significant differences to the wild type are indicated by asterisks (P < 0.05, Student’s t test).
Forty-seven additional SNPs were identified to be associated with root traits with an adjusted threshold of −log10(P) > 5.0 (Supplemental Data Set 5). One of them resembled SNP4 6374071 insofar as it showed association with LRS (0.25), LRS, and TRS, suggesting a strong contribution of the lateral root size in the basal quartile to the overall root size. We named the two loci Lateral Root Size Locus (LRSL) and General Regulator Factor 1 (GRF1) (Fig. 6A). LRS designates a 5-kb region that harbors four candidate genes (Supplemental Fig. S5B). The GRF1 locus was named after the only gene, AT4G09000 (GRF1), present in a 5-kb window around the strongest associated SNPs for this region (Supplemental Fig. S5A). Interestingly, GRF1 has been reported to be downregulated during LR emergence (Voß et al., 2015), and the expression of one of the candidates in LRSL, AT4G10270, is strongly induced around the time of LR emergence and in emerged LRs (Supplemental Fig. S5, C and D). We therefore investigated root phenotypes of Col-0 T-DNA insertion mutants for these two genes. Indeed, the LRS profiles showed that knockout mutants of GRF1 or AT4G10270 exhibited reductions in LRS (0.25), LRS, and TRS compared to wild-type plants (Fig. 6B), and the extracted data confirmed that the differences were statistically significant (Fig. 6C).
In addition, we characterized the root phenotypes of T-DNA insertion lines of candidate genes underlying 18 other associations [4 significant and 14 between −log10(P) > 5.0 and significant threshold] in the Col-0 or Col-3 backgrounds (Supplemental Data Set 5). In total, for 14 out of the 21 associations, the mutants showed significant phenotypic differences from the wild type in root traits for which associations were initially identified, including MR angle, MR length, LRD-BZ, LR number, BsZL, and apical ZL (Supplemental Fig. S6; Supplemental Data Set 5). Some genes, such as ALF5 and AXR5, were found to contribute to more RSA traits than initially highlighted by the GWAS, suggesting a more general role in root growth and development. Data for all RSA traits in all lines are provided in Supplemental Data Set 6. These results confirm that the EZ-Root-VIS pipeline is a useful tool for fast gene discovery by high-throughput RSA phenotyping.
DISCUSSION
Root-VIS is novel for its ability to average RSA traits using several different algorithms and to visually reconstruct the averaged RSA in several manners. During the development of the software, great effort has been made to create an ergonomically efficient user interface, which is a critical factor in determining the software’s ease of use. Both EZ-Rhizo and Root-VIS present a Windows-style GUI that will be easy to handle for new users. Other features include numerous keyboard accelerators and the option to skip parameter-setting dialogues using the shift key, and an “undo” command to avoid having to restart processing following inadvertent errors. Such features may, at first, seem trivial but, when considering that a typical experiment may involve processing several hundred images per day, they can make an enormous difference to the overall user experience and greatly reduce problems arising from “software fatigue.”
We validated our pipeline to capture fairly complex RSA of 12-d-old Arabidopsis plants in multiple contexts, including time series experiments, large-scale natural variation, and genotype-environment interaction studies. We provide the options to extract means based on order (“absolute”) or local position of the lateral (“binned”). It was noted that depending on how much LR number varies between individual plants, the different averaging methods can lead to about 10% variation in means determined for LR-related traits. Such variation can be crucial when handling a large number of genotypes since the phenotypic spectrum across accessions is continuous with relatively small differences between the individual accessions. We do not consider one of the averaging methods to be superior to the other; instead, we provide the user with both options, allowing them to consider which approach is particularly suitable to answer their questions.
We exemplified the utility of the EZ-Root-VIS pipeline to represent the plastic responses of various Arabidopsis accessions to environmental cues, with a particular focus on LRS profiles. Indeed, this analysis helped us to identify extreme and interesting genotypes in terms of their sensitivity to P or K limitation and combined deficiency. Under the low-P conditions applied here, none of the Arabidopsis accessions showed preferential growth investment into upper (basal) lateral roots despite the widespread view that low-P adapted RSA ideotypes preferentially explore residual P in the topsoil (Lynch and Brown, 2001). This suggests that the accessions have not evolved in a P-limited environment or that they only extend laterals if they perceive an actual P gradient. Our pipeline also revealed that interactive effect of K and P limitation modulate RSA in a genotype-specific manner that could not have been predicted from measuring RSA under single deficiency or in one accession only.
We further validated the usefulness of the EZ-Root-VIS pipeline for facilitating GWAS on RSA. A total of 56 loci controlling various root traits could be mapped at a cutoff threshold of –log10(P) > 5.0. We adjusted the threshold to this level for two reasons: first, SNPs in genes encoding known regulators affecting root traits such as IAA1/AXR5 and ALF5 were associated with LRD-BZ and main root length with a –log10(P) > 5.0 and <5% FDR significance threshold, respectively. Second, some associations that are not significant at 5% FDR, yet above –log10(P) > 5.0, have been shown to be false negatives because multiple testing correction methods are overly conservative (Meijón et al., 2014; Müller et al., 2016; Kalladan et al., 2017). The vast majority of the loci identified here did not contain any previously known RSA regulators. Indeed, the follow-up characterization of Col-0 knockout mutants pointed to several genes with functions in RSA development, including AT4G09000 (GRF1), AT4G10270, AT4G10570 (UBP9), AT4G01550 (NTM2), AT3G29300, AT5G51560, and AT3G04260 (PTAC3). We also found natural variation in genes that had previously been shown to affect root traits, such as AT4G14560 (IAA1/AXR5), AT5G22300 (NIT4), and AT3G23560 (ALF5; Supplemental Data Sets 5 and 6). These findings provide an excellent starting point for understanding the genetics of previously unresolved traits, such as LRS (0.25) and MR angle, and the role of RSA in plant adaptation to their natural habitats. Studying these traits can also provide insights into fundamental biological processes. For example, MR angle studies are expected to advance our understanding of plagiogravitropism, cell wall integrity (Van der Does et al., 2017), and voltage sensing (Kellermeier et al., 2014), as Arabidopsis mutants impaired in cellulose biosynthesis or membrane depolarization show altered MR angle.
Approaches such as GWAS become highly time and cost-effective if data for multiple traits can be obtained from the same experiment. Using the EZ-Root-VIS pipeline, it is currently possible to generate data for 20 root traits (more can be added) over a mapping population comprising about 150 accessions in the course of a few days. While the throughput is lower than for some other software packages (e.g. BRAT; Slovak et al., 2014), the RSAs quantified by EZ-Root-VIS can be more complex and hence more traits can be extracted. Importantly, the visual reconstructions of averaged root systems replace the need to rely on “representative” example pictures and enable fast integration of the measured individual RSA traits into meaningful root system shapes, which can be considered a breakthrough in the field of plant root biology. In publications, provision of averaged root system images rather than representative images is critical for interpretation and follow-up studies and it may also improve the reproducibility of results between various labs. Visual root shape reconstruction can also aid breeding efforts aimed at improving plant resilience to soil-borne stresses, facilitating the selection of desired root ideotypes from large phenotypic data sets.
EZ-Rhizo and Root-VIS have been developed over several years, and the versions released with this article have already been beta-tested by volunteer researchers. The software therefore represents a community effort, and we will continue to make improvements in response to suggestions and requests from the academic community.
MATERIALS AND METHODS
Software Development
The EZ-Rhizo and Root-VIS applications, and their various plug-in modules (collectively packaged as The Rhizo-II Root Biometrics Suite), were developed on, and for, the Microsoft Windows operating systems, in the C++ language, using Microsoft Visual Studio 2010/2017. The GUI was implemented using objects derived from the Microsoft Foundation Class library, and interaction with the XML database is built on code from the CMarkup (V-11.5) C++ XML Parser (www.firstobject.com). The software package can be downloaded from http://www.psrg.org.uk/download/Rhizo-64.msi (for 64-bit platforms) or http://www.psrg.org.uk/download/Rhizo-32.msi (for 32-bit platforms); it is free of charge, but we ask users to register their installation.
The XL-Orate plug-in is an “application extension module” (dynamic link library), which is activated via the “Plug-Ins” command in the “File” menu. It was also written in C++ and developed using the same tools and coding strategy as for the main programs. It requires installation of the Microsoft Excel software on the user’s computer and interacts with Excel directly (without user involvement) through programmed automation.
Root-VIS was developed to be used in conjunction with EZ-Rhizo. Nevertheless, in principle, any data set can be analyzed with Root-VIS so long as it is presented in the correct format and contains all the data required for the visual reconstructions. To facilitate data input from other sources, Root-VIS is already compatible with RSML (Lobet et al., 2015), and procedures for enabling Root-VIS to read those data files are currently under development.
Visual Reconstructions
The Root-VIS program uses a subtle, smart algorithm to reconstruct visually representative forms, (optionally) adding “bends” to roots based on the available information (the length-to-vector ratio, and the number, position, and orientation of lateral roots). We implemented five types of average visual reconstructions of root systems in Root-VIS, named Absolute Averages, Binned Averages, Alpha Blends, Flag Plots, and LR Profiles. The information available from the database, for a main root, comprises its path length (the total length of the root, from base to apex), its vector length (the straight-line distance between base and apex), the angle of its vector from vertical (positive angles being clockwise when looking at the root from the back of the plate), and the number of lateral roots. Lengths are expressed in centimeters and angles are in degrees. For a lateral root, the information also includes its position (the distance along its parent root from its parent’s base).
Adding Bending Patterns
Roots are usually not straight and therefore any visualization has to accommodate the difference between path length and vector length. However, the precise bending patterns are currently not quantified by EZ-Rhizo. We implemented different visualization options in Root-VIS consisting of either straight lines representing path or vector length, or added bends. For roots that have no subroot, bends can be added in a predetermined number, equally spaced along the vector’s axis, with the displacement of each point from the axis being calculated by the application of Pythagoras’ theorem. However, for roots with one or more subroots, a correlation has been demonstrated between the bending of a root and the emergence of a lateral root and that lateral roots will preferentially form at bends and follow an alternating directional pattern (Laskowski et al., 2008). We therefore decided to place bends along the vector, not at equal intervals, but rather at points corresponding to the position of each lateral root, with the direction of the off-axis displacement being determined by the sign of the branch angle. If two consecutive lateral roots branch in the same direction, Root-VIS adds an extra bend point between them, being displaced in the direction opposite to that of the two real subroots. Finally, we need to allow for a degree of bending in the apical part of the root: For this, we distribute the total amount of added “bend” between the branched and apical zones according to the relative lengths of these two root segments and allow the user to specify how many (equally spaced) bends to add in the apical zone. Clearly, the described procedure does not recreate the original bending pattern but applies existing evidence and user’s choices to fill in missing information. While the displayed bending pattern will not be faithful, the approach allowed us to correctly display both path and vector length in one picture.
Averaged RSA: Absolute and Binned Modes
The Root-VIS program provides two different methods for calculating the averaged RSA of a given data set, which we call “absolute averaging” and “binned averaging.” For absolute averages, first, the mean parameters (length, vector, and angle) for the main root are determined by dividing the totals for all plants by the number of samples; for the angle parameter, averages are formed using the absolute values (i.e. not including the sign) and the modal sign is then given to the calculated mean. Next, the median number of lateral roots is calculated. Then, for each lateral root, in order of appearance, their means are calculated (in the same way as for the main root, with the addition of the position parameter). If a plant in the data set has less than the median number of lateral roots, zeros are given as the four parameter values for any “missing” roots, and the divisor for the parameter totals for those lateral roots is reduced by one. Since LRs are numbered from base to apex in EZ-Rhizo any “missing” lateral roots will always be at the tip of the parent root.
For the binned averaging algorithm, first, each main root within the data set is split into three sections: the basal zone, the branched zone, and the apical zone as described (Armengaud et al., 2009). The path and vector lengths are then established for each of these zones and accumulated across all of the roots, and the means calculated. By adding each mean, the total mean path length and mean vector length of the root are determined. Next, the mean angle and median number of laterals are calculated as for absolute averages. However, to calculate the mean parameters for each lateral root, we first split the branched zone into a number of sections, or bins, equal to the mean number of lateral roots. The lateral roots for each sample in the data set are allocated to one of these bins according to their position. The mean values for LR length, vector, and angle are then calculated for each bin (as before) and for visualization they are positioned at the midpoint of the bin section. This averaging algorithm takes into account that the order of appearance does not necessarily reflect where a LR is placed; for example, the 4th lateral root in one plant could be located near the top of the main root, whereas in another it could be positioned in the middle. In the visual reconstruction the representative average LR is placed at the halfway point of the respective parent root segment.
Alpha Blends
The RSA reconstructions do not include an indication of the amount of variance between samples in each data set (although SEs are included in the numerical output). The alpha blend option allows the reconstructions of the individual roots to be overlaid in a semitransparent fashion, thus indicating the degree of variance across the data set. By default, when creating alpha blends, the vector angles of the main roots are normalized to the average vector angle for the samples included. Additional user-selectable options also allow for the MR path lengths and/or vector lengths to be normalized.
Flag Plots
The various components of a flag plot are indicated in Supplemental Figure S2; the flagpole is divided into three sections, the lengths of which are proportional to the lengths of each of the root zones (basal, branched, and apical); the angles between the upper and lower edges of the flag and the pole represent the mean insertion angles of the LRs (upper edge) and the slope of the linear regression between the mean length of the LRs and their position on the main root. Alternatively, the length of the upper edge can represent the average length of the LRs, in which case the angle between the lower edge and the flagpole will not represent any directly measured RSA data. The density of the flag’s fill color represents the average lateral root density—the value of which is normalized (i.e. scaled to a value between 0 and 1) within the given set of plots.
The software provides a number of options to control how to draw flag plots, including the selections described above, selections for root path or vector data, and choice of fill color.
LR Profiles
The fact that EZ-Rhizo determines the position of each LR allows simple visualization of local differences in LR growth. To construct the LR profiles, the MR is divided into a user-defined number of sectors (2–10) and the LR data within each sector is represented as a rectangle over the sector. The width of the rectangle can be chosen to represent either the total LR size (added lengths of all LRs in the sector, LRS profile) or the mean length of the LRs in the sector (LRL profile).
Plant Material, Growth Conditions, and Root Image Processing
Arabidopsis (Arabidopsis thaliana) accessions from the RegMap panel (Horton et al., 2012), and homozygous T-DNA insertion mutant lines for candidate genes and their corresponding wild types, Col-0 or Col-3, were obtained from the Nottingham Arabidopsis Stock Centre.
Seeds were surface sterilized in absolute ethanol for 1 min and then washed for 5 min with a solution containing 2.8% (w/v) sodium hypochlorite and 0.1% (v/v) Tween 20, followed by five washes with sterile distilled water. Seeds were incubated at 4°C for 5 to 7 d. The sterilized and stratified seeds were sown on vertical plates with minimal media (Kellermeier et al., 2014) containing 1% (w/v) agar (Formedium), 0.5% (w/v) Suc, and 0.2 m MES/Tris, pH 5.6. For nutrient limitation experiments, concentrations of K and P were lowered to 10 and 20 µm, respectively, and the osmotic potential of these media was adjusted as described before (Kellermeier et al., 2014). Plants were grown in a growth chamber at 60% relative humidity and 20°C, with 9 h of light (120 µE m−2 s−1). Images of each plate were acquired using a flatbed scanner at 200 dpi as previously described (Kellermeier and Amtmann, 2013). The RSA of each plant was quantified from the images using EZ-Rhizo as described before (Armengaud et al., 2009), facilitated by some new software functions described in the Help files of the latest version of EZ-Rhizo package released with this publication.
Statistical Tests
Averaged root trait data obtained from Root-VIS using the XL-Orate plug-in were used for statistical analyses unless otherwise stated. Trait frequency distribution and Shapiro-Wilk test of uniformity, correlation, and agglomerative hierarchical clustering analyses were performed using XLSTAT software. Genome-wide association mapping was conducted using GWAPP web interface (https://gwas.gmi.oeaw.ac.at/) with 250k SNP data and the accelerated mixed-model algorithm method (Seren et al., 2012). Statistically significant differences between wild-type and mutant plants was assessed using a Student’s t test (*P < 0.05) using Microsoft Excel.
Abbreviations
MRL, Main root path length; MRV, main root vector length; MRA, main root angle; BsZL, basal zone length (from hypocotyl to the first lateral root); BZL, branched zone length; AZL, apical zone length; TRS, total root system; LRS, total lateral root path length; LRS (0.25), total lateral root path length in the top quartile of main root; LRL, average lateral root path length; LRL (0.25), average lateral root path length in the upper quartile of main root; LRN, number of lateral roots; LRD-MR, lateral root density over the entire main root path (LRN/MRL); LRD-BZ, lateral root density in the branched zone of the main root (LRN/BZL); LRA, lateral root angle; OAD, overall depth.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT4G09000 (GRF1), AT4G10270 (in locus LRS), AT4G10570 (UBP9), AT4G01550 (NTM2), AT3G29300 (in locus LRDB), AT5G51560 (in locus LRK-LRN), AT3G04260 (PTAC3), AT4G14560 (IAA1/AXR5), AT5G22300 (NIT4), and AT3G23560 (ALF5). See also Supplemental Data Sets 5 and 6.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Screenshots of the Root-VIS software.
Supplemental Figure S2. Visual explanation of the data incorporated into a Root-VIS flag plot.
Supplemental Figure S3. Root-VIS representations of average RSAs of nine Arabidopsis accessions in four different nutrient conditions.
Supplemental Figure S4. Frequency distribution of RS traits in Arabidopsis accessions.
Supplemental Figure S5. Genes and transcript profiles for two RSA associations.
Supplemental Figure S6. Candidate gene validation
Supplemental Data Set 1. Numerical data for average RSA traits, number of replicate plants, and se underpinning the roots shown in Figure 1, E and F.
Supplemental Data Set 2. Numerical data for average RSA traits, number of replicate plants, and se underpinning the roots shown in Figure 2.
Supplemental Data Set 3. Means and SE of RSA traits of different accessions in four nutrient conditions (data underpinning Figs. 3 and 4 and Supplemental Fig. S3).
Supplemental Data Set 4. Means of RSA traits of 147 Arabidopsis accessions under control conditions and hierarchical clustering of accessions based on root traits.
Supplemental Data Set 5. Description of the GWAS data set.
Supplemental Data Set 6. Means and SEs of root system architecture traits of mutants of candidate genes identified through GWAS (data underpinning Fig. 6 and Supplemental Fig. S6).
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Geraldine Goldie, who carried out preliminary consultation and programming as part of a student project at the University of Glasgow. We also thank Mike Blatt for his support in the development of this software.
Footnotes
The work was funded by the Biotechnology and Biological Sciences Research Council (BB/N018508/1) and by the Gatsby Charitable Trust (Sainsbury studentship to F.K.).
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Armengaud P, Zambaux K, Hills A, Sulpice R, Pattison RJ, Blatt MR, Amtmann A (2009) EZ-Rhizo: integrated software for the fast and accurate measurement of root system architecture. Plant J 57: 945–956 [DOI] [PubMed] [Google Scholar]
- Arsenault J, Pouleru S, Messier C, Guay R (1995) WinRHlZO, a root-measuring system with a unique overlap correction method. HortScience 30: 906 [Google Scholar]
- Bucksch A, Burridge J, York LM, Das A, Nord E, Weitz JS, Lynch JP (2014) Image-based high-throughput field phenotyping of crop roots. Plant Physiol 166: 470–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier F, Pata M, Nacry P, Doumas P, Rossignol M (2003) Effects of phosphate availability on the root system architecture: large-scale analysis of the natural variation between Arabidopsis accessions. Plant Cell Environ 26: 1839–1850 [Google Scholar]
- Delory BM, Baudson C, Brostaux Y, Lobet G, du Jardin P, Pagès L, et al. (2016) archiDART: an R package for the automated computation of plant root architectural traits. Plant Soil 398: 351–365 [Google Scholar]
- Delory BM, Li M, Topp CN, Lobet G (2018) archiDART v3.0: A new data analysis pipeline allowing the topological analysis of plant root systems. F1000 Res 7: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber BD, Giehl RFH, Friedel S, von Wirén N (2013) Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol 163: 161–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton MW, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjálmsson BJ, et al. (2012) Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet 44: 212–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julkowska MM, Testerink C (2015) Tuning plant signaling and growth to survive salt. Trends Plant Sci 20: 586–594 [DOI] [PubMed] [Google Scholar]
- Julkowska MM, Koevoets IT, Mol S, Hoefsloot H, Feron R, Tester MA, Keurentjes JJB, Korte A, Haring MA, de Boer G-J, Testerink C (2017) Genetic Components of Root Architecture Remodeling in Response to Salt Stress. Plant Cell 29: 3198–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalladan R, Lasky JR, Chang TZ, Sharma S, Juenger TE, Verslues PE (2017) Natural variation identifies genes affecting drought-induced abscisic acid accumulation in Arabidopsis thaliana. Proc Natl Acad Sci USA 114: 11536–11541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalogiros DI, Adu MO, White PJ, Broadley MR, Draye X, Ptashnyk M, Bengough AG, Dupuy LX (2016) Analysis of root growth from a phenotyping data set using a density-based model. J Exp Bot 67: 1045–1058 [DOI] [PubMed] [Google Scholar]
- Kellermeier F, Amtmann A (2013) Phenotyping jasmonate regulation of root growth. Methods Mol Biol 1011: 25–32 [DOI] [PubMed] [Google Scholar]
- Kellermeier F, Chardon F, Amtmann A (2013) Natural variation of Arabidopsis root architecture reveals complementing adaptive strategies to potassium starvation. Plant Physiol 161: 1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermeier F, Armengaud P, Seditas TJ, Danku J, Salt DE, Amtmann A (2014) Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 26: 1480–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Marée AFM, Scheres B (2008) Root system architecture from coupling cell shape to auxin transport. PLoS Biol 6: e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobet G, Pagès L, Draye X (2011) A novel image-analysis toolbox enabling quantitative analysis of root system architecture. Plant Physiol 157: 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobet G, Pound MP, Diener J, Pradal C, Draye X, Godin C, Javaux M, Leitner D, Meunier F, Nacry P, Pridmore TP, Schnepf A (2015) Root system markup language: toward a unified root architecture description language. Plant Physiol 167: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch JP. (2015) Root phenes that reduce the metabolic costs of soil exploration: opportunities for 21st century agriculture. Plant Cell Environ 38: 1775–1784 [DOI] [PubMed] [Google Scholar]
- Lynch JP, Brown KM (2001) Topsoil foraging - An architectural adaptation of plants to low phosphorus availability. Plant Soil 237: 225–237 [Google Scholar]
- Meijón M, Satbhai SB, Tsuchimatsu T, Busch W (2014) Genome-wide association study using cellular traits identifies a new regulator of root development in Arabidopsis. Nat Genet 46: 77–81 [DOI] [PubMed] [Google Scholar]
- Morris EC, Griffiths M, Golebiowska A, Mairhofer S, Burr-Hersey J, Goh T, von Wangenheim D, Atkinson B, Sturrock CJ, Lynch JP, et al. (2017) Shaping 3D root system architecture. Curr Biol 27: R919–R930 [DOI] [PubMed] [Google Scholar]
- Müller LM, Lindner H, Pires ND, Gagliardini V, Grossniklaus U (2016) A subunit of the oligosaccharyltransferase complex is required for interspecific gametophyte recognition in Arabidopsis. Nat Commun 7: 10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem A, French AP, Wells DM, Pridmore TP (2011) High-throughput feature counting and measurement of roots. Bioinformatics 27: 1337–1338 [DOI] [PubMed] [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T (2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29: 115–125 [DOI] [PubMed] [Google Scholar]
- Ristova D, Rosas U, Krouk G, Ruffel S, Birnbaum KD, Coruzzi GM (2013) RootScape: a landmark-based system for rapid screening of root architecture in Arabidopsis. Plant Physiol 161: 1086–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers ED, Benfey PN (2015) Regulation of plant root system architecture: implications for crop advancement. Curr Opin Biotechnol 32: 93–98 [DOI] [PubMed] [Google Scholar]
- Satbhai SB, Setzer C, Freynschlag F, Slovak R, Kerdaffrec E, Busch W (2017) Natural allelic variation of FRO2 modulates Arabidopsis root growth under iron deficiency. Nat Commun 8: 15603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seren Ü, Vilhjálmsson BJ, Horton MW, Meng D, Forai P, Huang YS, Long Q, Segura V, Nordborg M (2012) GWAPP: a web application for genome-wide association mapping in Arabidopsis. Plant Cell 24: 4793–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzad Z, Amtmann A (2017) Food for thought: how nutrients regulate root system architecture. Curr Opin Plant Biol 39: 80–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slovak R, Göschl C, Su X, Shimotani K, Shiina T, Busch W (2014) A scalable open-source pipeline for large-scale root phenotyping of Arabidopsis. Plant Cell 26: 2390–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- Van der Does D, Boutrot F, Engelsdorf T, Rhodes J, McKenna JF, Vernhettes S, Koevoets I, Tintor N, Veerabagu M, Miedes E, et al. (2017) The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PLoS Genet 13: e1006832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voß U, Wilson MH, Kenobi K, Gould PD, Robertson FC, Peer WA, Lucas M, Swarup K, Casimiro I, Holman TJ, et al. (2015) The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nat Commun 6: 7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Gutjahr C., Li C.J., Hochholdinger F. (2016) Genetic control of lateral root formation in cereals. Trends Plant Sci 21: 951-961 [DOI] [PubMed] [Google Scholar]