The NMT1 and NMT3 phosphoethanolamine N-methyltransferases are required to sustain phosphatidylcholine synthesis above levels needed for growth, development, and reproduction in Arabidopsis thaliana.
Abstract
Phosphatidylcholine (PC) is a major membrane phospholipid and a precursor for major signaling molecules. Understanding its synthesis is important for improving plant growth, nutritional value, and resistance to stress. PC synthesis is complex, involving several interconnected pathways, one of which proceeds from serine-derived phosphoethanolamine to form phosphocholine through three sequential phospho-base methylations catalyzed by phosphoethanolamine N-methyltransferases (PEAMTs). The contribution of this pathway to the production of PC and plant growth has been a matter of some debate. Although a handful of individual PEAMTs have been described, there has not been any in planta investigation of a PEAMT family. Here, we provide a comparative functional analysis of two Arabidopsis (Arabidopsis thaliana) PEAMTs, NMT1 and the little known NMT3. Analysis of loss-of-function mutants demonstrates that NMT1 and NMT3 synergistically regulate PC homeostasis, phase transition at the shoot apex, coordinated organ development, and fertility through overlapping but also specific functions. The nmt1 nmt3 double mutant shows extensive sterility, drastically reduced PC concentrations, and altered lipid profiles. These findings demonstrate that the phospho-base methylation pathway makes a major contribution to PC synthesis in Arabidopsis and that NMT1 and NMT3 play major roles in its catalysis and the regulation of PC homeostasis as well as in plant growth and reproduction.
Phosphatidylcholine (PC) is the major phospholipid of nonplastid membranes in most eukaryotes and is the substrate of numerous fatty acid-modifying enzymes. It is also a major precursor of storage lipid biosynthesis (Ohlrogge and Browse, 1995; Bates et al., 2009; Bates and Browse, 2011) and lipid-based signaling molecules, such as diacylglycerols (DAGs) and phosphatidic acid (PA), that are involved in numerous cellular processes and stress responses in plants (Munnik et al., 1998; Katagiri et al., 2001; Munnik, 2001; Potocký et al., 2003). The choline head group of PC constitutes an essential nutrient for humans and, in some plant species, is the precursor of Gly betaine, which is a compound with protective functions against osmotic stress, high light, or cold stress (Chen and Murata, 2008).
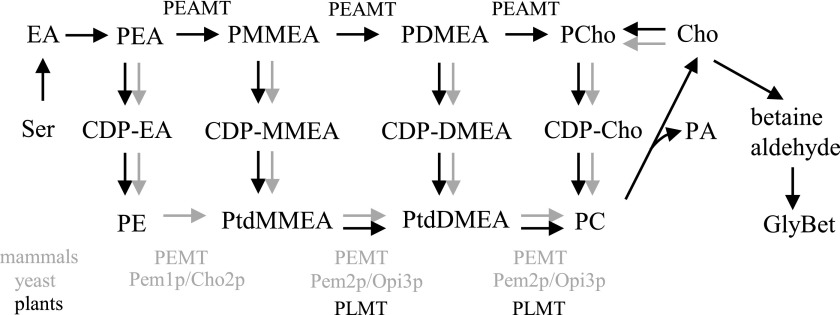
Due to its importance, the synthesis of PC has been studied intensively in a wide range of organisms, including Saccharomyces cerevisiae, mammals (Kennedy and Weiss, 1956; Kodaki and Yamashita, 1987; Summers et al., 1988; Cui et al., 1993; Preitschopf et al., 1993), Caenorhabditis elegans (Palavalli et al., 2006; Brendza et al., 2007), Plasmodium falciparum (Pessi et al., 2005), and plants (Datko and Mudd, 1988a; Bolognese and McGraw, 2000; Mou et al., 2002; Cruz-Ramírez et al., 2004; Jost et al., 2009; Keogh et al., 2009; BeGora et al., 2010). This has revealed the involvement of several pathways, with partial conservation across kingdoms (Fig. 1). One is the cytidine 5′-diphosphocholine (CDP-choline) pathway, commonly referred to as the nucleotide or Kennedy pathway, which starts with the phosphorylation of free choline by choline/ethanolamine kinase (CEK) to yield phosphocholine (PCho). PCho is then coupled to cytidine-5′ triphosphate (CTP) to form CDP-choline through a reaction catalyzed by choline-phosphate cytidyltransferase, and the choline moiety of CDP-choline is transferred to DAG by CDP-choline 1,2-diacylglycerol-cholinephosphotransferase to produce PC. This pathway is highly conserved among eukaryotes. A second pathway in yeast and mammals is the phospholipid methylation pathway, where PC is produced from phosphatidylethanolamine (PE) through three sequential methylations at the phosphatidyl base using S-adenosyl-l-methionine (SAM) as a methyl donor. In yeast, these methylations are catalyzed by two PLMTs (Kodaki and Yamashita, 1987; Summers et al., 1988; McGraw and Henry, 1989; Preitschopf et al., 1993), while in mice, the three transmethylations on PE are performed by a unique enzyme (Bremer and Greenberg, 1961; Ridgway and Vance, 1987; Ridgway et al., 1989; Walkey et al., 1997).
Figure 1.
PC synthesis pathways in plants (black arrows), yeast, and mammals (gray arrows). Cho, Choline; CDP, cytidine diphosphate; DM, dimethyl; EA, ethanolamine; GlyBet, Gly betaine; MM, monomethyl; P, phospho; PEAMT, phosphoethanolamine N-methyltransferase; PEMT, phosphatidylethanolamine N-methyltransferase; PLMT, phospholipid N-methyltransferase; Ptd, phosphatidyl.
PLMT activity has been demonstrated in plants too, in a variety of species, including spinach (Spinacia oleracea), soybean (Glycine max), carrot (Daucus carota), tobacco (Nicotiana tabacum), and recently Arabidopsis (Arabidopsis thaliana) (Marshall and Kates, 1973; Datko and Mudd, 1988b; McNeil et al., 2000; Keogh et al., 2009). Thus far, only enzymes using phosphatidylmonomethylethanolamine (PtdMMEA) and phosphatidyldimethylethanolamine (PtdDMEA) as substrates have been identified; none have been found that can produce PC directly from PE (Keogh et al., 2009). Instead, plants possess an alternative phospho-base methylation pathway that is also present in P. falciparum and C. elegans (Pessi et al., 2005; Palavalli et al., 2006; Brendza et al., 2007) but absent from yeast and mammals (Hitz et al., 1981; Hanson and Rhodes, 1983; Mudd and Datko, 1986; Datko and Mudd, 1988a,b; McGraw and Henry, 1989; Bolognese and McGraw, 2000; Nuccio et al., 2000; Charron et al., 2002; Jost et al., 2009). In this second pathway, also known as the serine decarboxylase-phosphoethanolamine methyltransferase (SDPM) pathway, phosphoethanolamine derived from Ser decarboxylation and ethanolamine phosphorylation (Rontein et al., 2001; Lin et al., 2015) undergoes three sequential phospho-base methylations catalyzed by phosphoethanolamine N-methyltransferase (PEAMT/PMT/NMT) enzymes to form phosphomonomethylethanolamine (PMMEA), phosphodimethylethanolamine (PDMEA), and finally PCho. PCho can then enter the Kennedy pathway to produce PC. Choline is produced either directly from the dephosphorylation of PCho or indirectly from PC hydrolysis by phospholipase D in a reaction also producing the secondary messenger PA. The PMMEA and PDMEA intermediates also can serve as substrates for the nucleotide pathway and be converted to PtdMMEA or PtdDMEA, and then to PC, through phosphatidyl-base methylation using PLMT enzymes (Datko and Mudd, 1988a; Wang and Moore, 1990; Keogh et al., 2009).
The relative contributions of the phospho-base methylation and phosphatidyl-base methylation pathways to the production of PC appear to vary widely among plant species (Hitz et al., 1981; Datko and Mudd, 1988a,b; Williams and Harwood, 1994; McNeil et al., 2000). Regardless of the route used to produce PC, according to the current understanding of phospholipid synthesis in plants, the N-methylation of phosphoethanolamine (PEA) to produce phosphomonoethanolamine is the obligatory entry point to the formation of PC (Datko and Mudd, 1988a; Rhodes and Hanson, 1993; McNeil et al., 2000; Keogh et al., 2009). Given the fundamental importance of PC to the making of plant membranes and signaling, plants defective in the phospho-base pathway, especially in its first step, are expected to exhibit severe defects, and even lethality, unless additional routes for PC synthesis exist (Liu et al., 2015).
The Arabidopsis genome carries three genes encoding PEAMT/PMT/NMT proteins, NMT1 (locus At3g18000), NMT2 (locus At1g48600), and NMT3 (locus At1g73600; Bolognese and McGraw, 2000; BeGora et al., 2010; Lee and Jez, 2017). NMT1 is the best known and only one of the three isoforms to have been studied in any detail in vivo. NMT1 was first identified from the functional complementation of a yeast choline auxotroph mutant and has subsequently been implicated in early root development and temperature-conditional male sterility (Mou et al., 2002; Cruz-Ramírez et al., 2004). Under optimal growth conditions, loss-of-function xpotl1/nmt1 mutants develop into healthy, fully fertile mature plants on soil with no apparent abnormalities. In vivo information on NMT2 and NMT3 is scant, and their disruption in T-DNA mutants was reported to cause no overt phenotype compared with the wild type (BeGora et al., 2010; Lee and Jez, 2017). These studies also describe only a modest (5%–7%) reduction in PC levels in nmt1 seedlings lacking functional NMT1 (Cruz-Ramírez et al., 2004) and no change in the lipid profiles of the nmt2 and nmt3 knockout mutants, except for a 2-fold decrease of one polar lipid species (34:3) in the former (BeGora et al., 2010). These observations suggest high functional redundancy between isoforms, but this remains to be fully examined.
The NMT3 gene has received little attention and was not recovered in the complementation screens of PC synthesis-defective yeast mutants that led to the identification of NMT1 and NMT2. Here, we report the in vivo functional characterization of NMT3 and its overlapping but nonredundant roles with NMT1. Our analysis of nmt1 mnt3 double mutants illuminates novel functions of plant PEAMTs and reveals that the phospho-base methylation pathway makes a major contribution to PC synthesis in Arabidopsis. Together, these results suggest that NMT1 and NMT3 isoforms are of paramount importance in catalyzing that pathway and sustaining homeostatic levels of PC as well as for normal plant development.
RESULTS
NMT3 Catalyzes the Triple Methylation of PEA to PCho in Vivo
Plant PEAMTs have two functional catalytic domains, where the N-terminal domain catalyzes the first methylation of PEA to PMMEA while the two subsequent methylations are catalyzed by the C-terminal domain (Bolognese and McGraw, 2000; Nuccio et al., 2000; Charron et al., 2002; Lee and Jez, 2017). The NMT3 gene encodes a 57.8-kD protein that shares 79% sequence identity with the 56.1-kD PEAMT1/NMT1 (Supplemental Table S1). The C-terminal catalytic domains of the two proteins are identical in each of the four SAM-binding motifs (I, post-I, II, and III) and also identical to the C-terminal motif of the other biochemically characterized PEAMTs from spinach and wheat (Triticum aestivum; Bolognese and McGraw, 2000; Nuccio et al., 2000; Jost et al., 2009). The N-terminal domains, however, present some differences, with single amino acid substitutions in motif I (Leu to Phe), motif II (Gly to Glu), and motif III (Ile to Thr), all nonconservative (Supplemental Fig. S1).
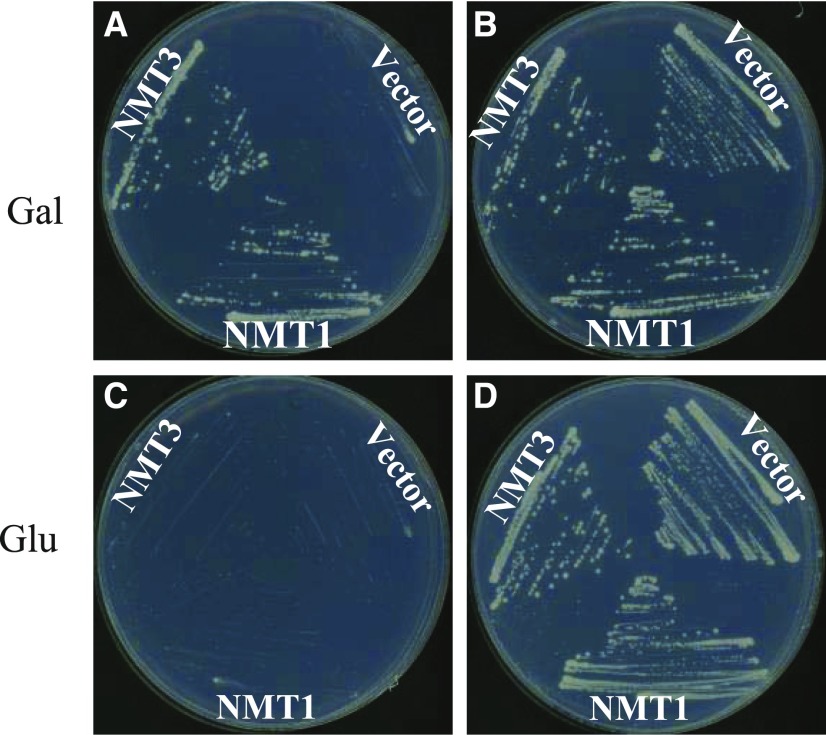
Yeast cells lack PEAMT activity. Instead, they produce PC via the three-step transmethylation of PE into PC, catalyzed by the Pem1p/Cho2p and Pem2p/Opi3p enzymes (Kodaki and Yamashita, 1987; Summers et al., 1988; Preitschopf et al., 1993). Deletion of the two encoding genes abolishes PC synthesis and is lethal unless choline is exogenously provided so that the CDP-choline pathway can be utilized to produce PC (Kodaki and Yamashita, 1987; Summers et al., 1988). To test the capacity of NMT3 to restore the synthesis of PC, we took advantage of the pem1Δ pem2Δ yeast mutant, which lacks functional Pem1p and Pem2p, and examined its ability to grow when expressing the Arabidopsis NMT3 or NMT1 coding sequences under the control of the GAL1-inducible promoter. The empty pYES-DEST52 plasmid was used as a negative control. Transgenic strains were grown in GAL1 promoter-inducing (presence of Gal) or promoter-repressing (presence of Glc) conditions with either the absence (0 mm) or presence (1 mm) of exogenous choline. ProGAL1:NMT1 plasmid transformed in pem1Δ pem2Δ mutants was used as a positive control, as it was shown previously that NMT1 expression can fully rescue pem1Δ pem2Δ growth in the absence of exogenous choline (Bolognese and McGraw, 2000). Transformants carrying the pYES-DEST52 empty vector only grew if supplied with choline under both inducing and repressing conditions (Fig. 2). By contrast, expression of either NMT1 or NMT3 enabled pem1Δ pem2Δ to grow under gene-inducing conditions in the absence of exogenous choline. No growth was observed under repressing conditions (Fig. 2). The assays were repeated in liquid culture, with similar results (Supplemental Fig. S2). These data demonstrate that expression of the Arabidopsis NMT3 protein can suppress the choline auxotrophy of the pem1Δ pem2Δ mutant, similar to NMT1 (Fig. 2; Bolognese and McGraw, 2000).
Figure 2.
NMT3 complements the S. cerevisiae choline-deficient mutant pem1Δ pem2Δ. The pem1Δ pem2Δ mutant yeast strain was transformed with empty vector pYES-DEST52 or the vector containing NMT1 or NMT3 coding sequence. Growth of transformants is shown on minimal medium supplemented with 2% Gal (inducing condition; A and B) or 2% Glc (repressing condition; C and D) and 0 mm choline (A and C) or 1 mm choline (B and D).
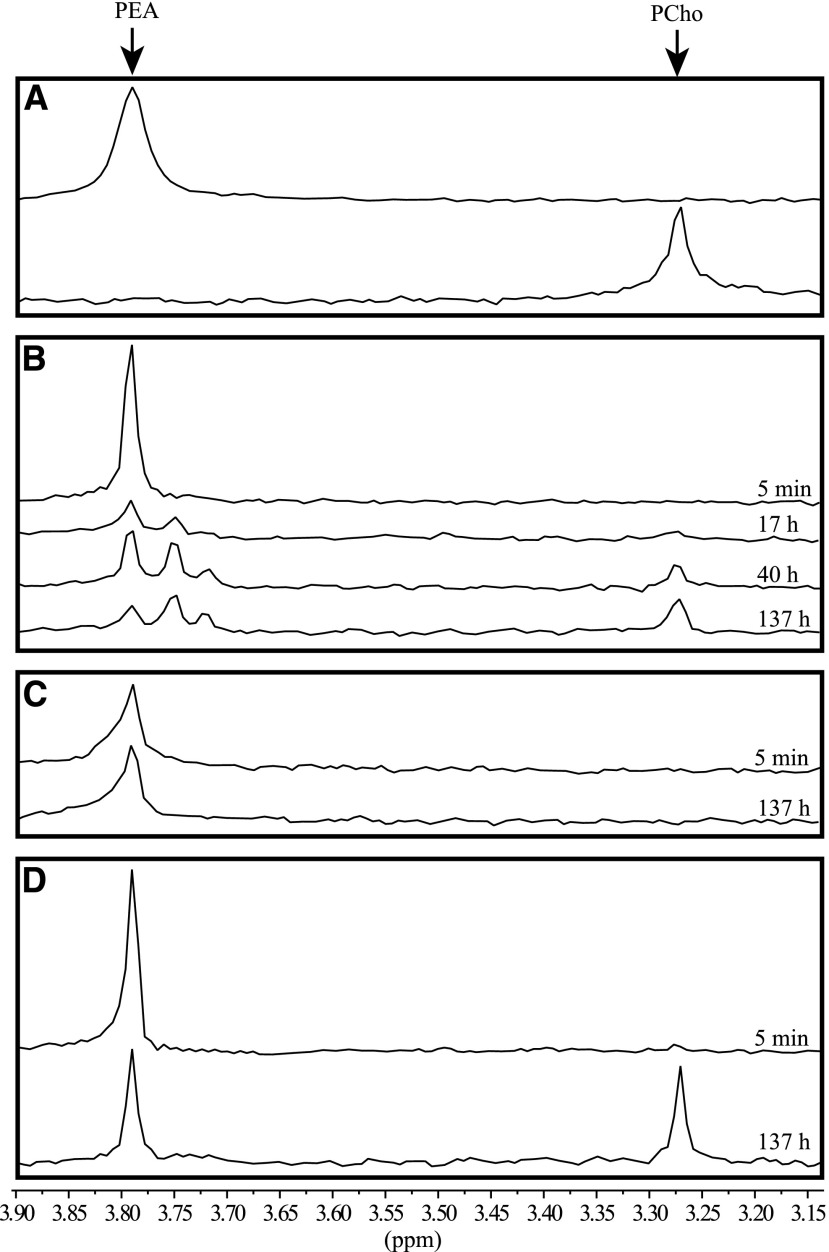
To confirm that NMT3 rescues the PC-deficient yeast mutant by catalyzing the phospho-base methylations of PEA to form PCho and feed it into the CDP-choline pathway for the production of PC, we prepared crude protein extracts from pem1Δ pem2Δ mutant cells expressing pYES-DEST52:NMT3, pYES-DEST52:NMT1, or empty vector and used 31P-NMR to monitor the production of PCho following the addition of PEA substrate and SAM methyl donor to the reaction mixtures. PEA was readily detected in all assays, with a chemical shift of 3.79 ppm with respect to the internal standard methylphosphonate, which matched the shift obtained for the PEA standard in the reaction buffer alone (Fig. 3). For protein extracts of cells harboring the empty vector, the size of that peak remained unchanged throughout the incubation period, and PCho could not be detected at any time point (Fig. 3C). By contrast, for extracts of cells harboring pYES-DEST52:NMT3, the size of the PEA peak declined with time, in parallel to a second compound becoming detectable, with the same chemical shift (3.27 ppm) as observed for the PCho standard, and increasing steadily in relative abundance compared with PEA (0.22, 0.4, and 1.2 PCho:PEA ratios at the 17-, 40-, and 137-h time points, respectively; Fig. 3B). 31P-NMR spectra for the reactions with cells expressing pYES-DEST52:NMT1 showed a similar pattern (Fig. 3D). These data demonstrate that NMT3 acts as a PEAMT enzyme capable of catalyzing the three sequential phospho-base methylations of the SDPM pathway, similar to NMT1. Two intermediate compounds between PEA and PCho could be resolved on spectra from NMT3-expressing cells, most likely corresponding to the PMMEA and PDMEA intermediates. They were not detected in any of the spectra from NMT1-expressing cells, suggesting faster turnover rates of these intermediates than in the reactions catalyzed by NMT3.
Figure 3.
NMT3 has the capacity to catalyze the triple methylation of PEA to PCho. A, 31P-NMR spectra of PEA and PCho standards in HED buffer, pH 8.2. B to D, Time course of PEA methylation reactions catalyzed by protein extracts from yeast cells expressing pYES-DEST52:NMT3 (B), empty vector pYES-DEST52 (C), or pYES-DEST52:NMT1 (D). Shown are 31P-NMR spectra recorded 5 min, 17 h, 40 h, and 137 h after the initiation of the reaction through the addition of substrates PEA and S-adenosyl methionine (B). Only the initial and final spectra, recorded at the 5-min and 137-h time points, respectively, are shown in C and D.
Loss of NMT3 Function in the nmt1 Background Restricts and Alters Primary and Secondary Root Development
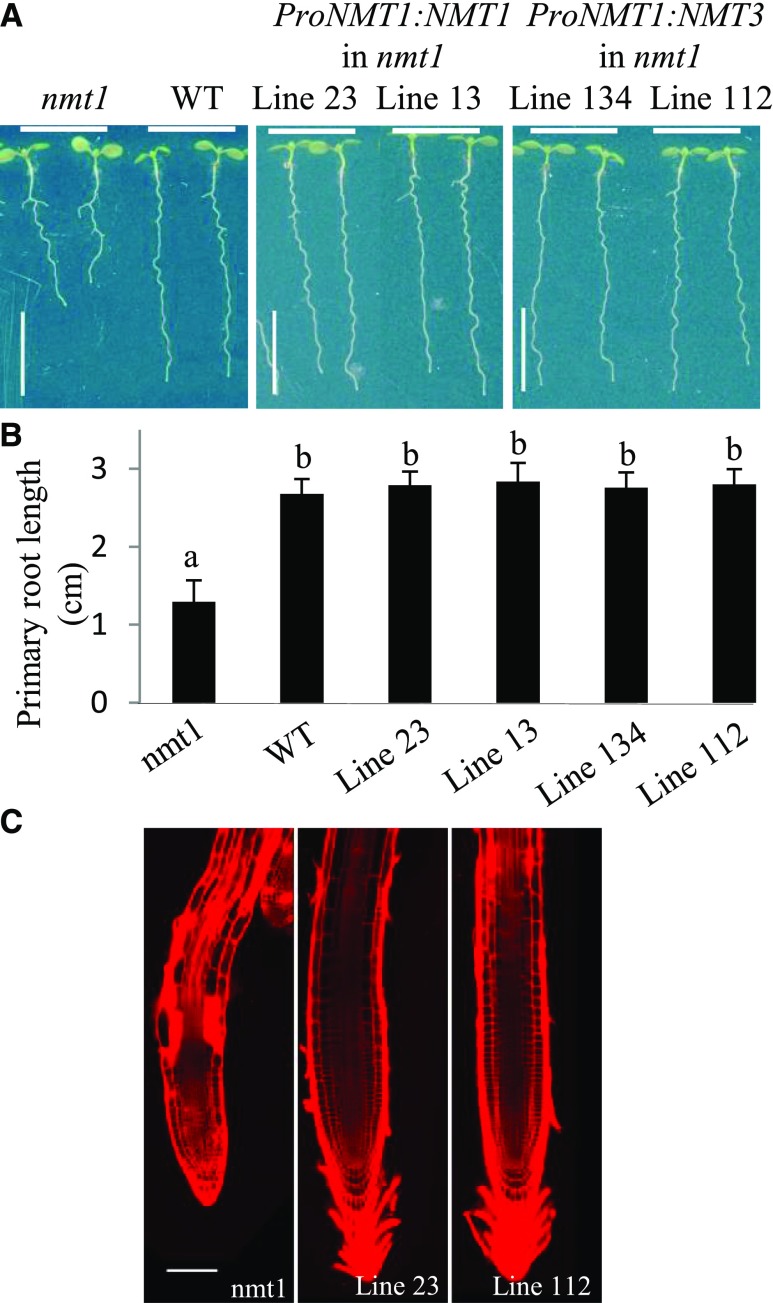
To investigate the in planta biochemical function of NMT3, we expressed the NMT3 gene in the Arabidopsis nmt1 mutant (SALK_036291), which carries a loss-of-function T-DNA insertion in exon 10 (Supplemental Fig. S3) and exhibits a distinctive short-root phenotype (Fig. 4; Cruz-Ramírez et al., 2004). The full-length genomic sequences of NMT3 and NMT1, including 5′ and 3′ untranslated regions, were fused to the 1.9-kb sequence of a putative NMT1 promoter. nmt1 roots were significantly shorter than wild-type roots, as expected (Fig. 4; Cruz-Ramírez et al., 2004). Complementation with ProNMT1:gNMT3 fully recapitulated wild-type root elongation and cellular integrity, similar to ProNMT1:gNMT1, consistent with the shared biochemical function of NMT3 and NMT1 (Fig. 4). This result indicates that NMT3 is a functional homolog of NMT1 in planta.
Figure 4.
NMT3 rescues the short-root phenotype induced by the loss of NMT1 function. A and B, Representative photographs (A) and primary root lengths (B) of 8-d-old seedlings. From left to right: nmt1 mutant, wild type (WT), and four independent transgenic complementation lines expressing ProNMT1:NMT1 (lines 23 and 13) or ProNMT1:NMT3 (lines 134 and 112) in the nmt1 background. Seedlings were grown along vertical plates on 1.2% agar supplemented with one-half-strength Murashige and Skoog medium and 1% Suc. Values in B are means ± se (n = 14–16). Bars topped by different letters denote statistically significant differences in one-way ANOVA (P < 0.001). Bars in A = 1 cm. C, Confocal images of propidium iodide-stained roots of nmt1 and nmt1 expressing ProNMT1:gNMT1 (line 23) or ProNMT1:gNMT3 (line 112). Both transgenes suppress the epidermal defects and extensive cell death caused by NMT1 loss of function (bright red areas). Bar = 100 µm.
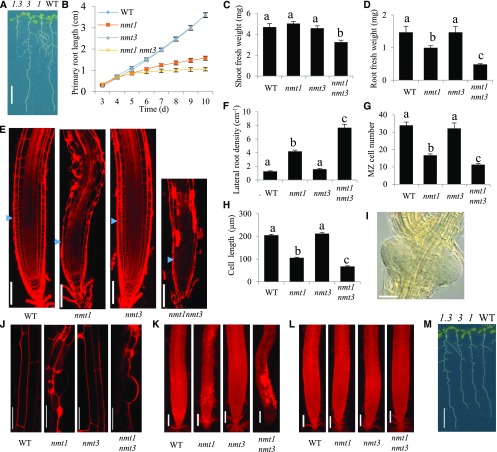
To investigate the physiological functions of NMT3, we obtained the T-DNA insertion line GABI-KAT_109F02 (Rosso et al., 2003), which carries a single loss-of-function T-DNA insertion in NMT3 exon 4 (Supplemental Fig. S3). In contrast to nmt1, homozygous nmt3 seedlings were indistinguishable from the wild type, showing similar overall growth of both shoot and roots, similar primary root elongation rate and lateral branching, and normal root anatomy (Fig. 5, A–F). We crossed nmt3 and nmt1 to generate the nmt1 nmt3 double knockout mutant. nmt1 nmt3 seeds were recovered at the expected frequency (1:16, P < 0.025) and germinated normally. Past day 4, primary root elongation progressed at an extremely low rate, significantly slower than in nmt1 (Fig. 5B). This was due to a reduction of meristem size and number of proliferating cells in nmt1 nmt3 mutants, similar to what has been reported in nmt1 mutants, but of significantly greater magnitude. A reduction of mature cell length also was observed (Fig. 5, G and H). Lateral root density was increased compared with the wild type and nmt1 and nmt3 single mutants (Fig. 5F), partly as a result of reduced mature cell length in the primary root as well as enhanced lateral root initiation. Lateral roots of the nmt1 nmt3 mutant were not only more numerous but often formed in clusters of two to three primordia initiated adjacent or opposite to each other (Fig. 5I). This was never observed in the wild type or either the nmt1 or nmt3 single mutant and suggests an interaction between the SDPM pathway and pathways regulating root branching.
Figure 5.
NMT3 loss of function has, alone, no apparent phenotypic impact but severely impairs vegetative growth and root development in an nmt1 background. A, Overall phenotypes of 12-d-old wild-type (WT), nmt1 (1), nmt3 (3), and nmt1 nmt3 (1.3) seedlings. Bar = 1 cm. B, Kinetics of wild-type, nmt1, nmt3, and nmt1 nmt3 primary root elongation. Values are means ± se (n = 12). C and D, Shoot (C) and root (D) fresh weights of 2-week-old wild-type, nmt1, nmt3, and nmt1 nmt3 seedlings. Values are means ± se (n = 6). E, Confocal optical sections of 10-d-old wild-type, nmt1, nmt3, and nmt1 nmt3 primary roots stained with propidium iodide. Blue arrowheads indicate the boundary between the root meristem and the elongation zone. Bars = 100 µm. F, Lateral root density for 10-d-old wild-type, nmt1, nmt3, and nmt1 nmt3 roots. Values are means ± se (n = 11–12). G, Meristematic cortical cell number in wild-type, nmt1, nmt3, and nmt1 nmt3. Values are means ± se (n = 8). H, Mature cortical cell length in wild-type, nmt1, nmt3, and nmt1 nmt3 roots. Values are means ± se (n = 56–129). I, Example of a lateral root primordia cluster on a 10-d-old nmt1 nmt3 primary root, imaged by differential interference contrast (DIC) light microscopy. Bar = 50 µm. J, Confocal images of epidermal cells in 10-d-old wild-type, nmt1, nmt3, and nmt1 nmt3 primary roots stained with propidium iodide. Bars = 50 µm. K and L, Propidium iodide-stained wild-type, nmt1, nmt3, and nmt1 nmt3 roots grown in the absence (K) or presence (L) of 200 µm choline. Bars = 100 µm. M, Photographs of representative wild-type, nmt1, nmt3, and nmt1 nmt3 seedlings grown on medium supplemented with 200 µm choline. Bar = 1 cm. Bars topped with different letters within each graph (B–D and F–H) denote statistically significant genetic differences by one-way ANOVA (P < 0.05).
The xipotl mutant, which is allelic to the nmt1 mutant used here, was reported to exhibit aberrant epidermal cell morphology and cell death in the root elongation and differentiation zones (Cruz-Ramírez et al., 2004). Similar defects were observed here in nmt1 roots. These defects were severely aggravated in nmt1 nmt3 roots (Fig. 5, E, J, and K), as shown by the widespread penetration of the DNA-binding propidium iodide dye inside numerous epidermal and cortical cells, indicating that these cells were dead (Fig. 5K). The meristem was normal, however, as observed in the nmt1 mutant, but cell death extended closer to the meristem (Fig. 5E). Exogenous choline application completely rescued these phenotypes (Fig. 5, L and M compared with K and A, respectively), demonstrating that they were caused by choline deficiency.
Overall, these results show that, while having no apparent effects on early root development and vegetative growth on its own, the loss of NMT3 in the nmt1 background drastically enhances the nmt1 root defects and also inhibits leaf growth, which is not observed in either of the single mutants. This suggests that NMT1 and NMT3 have synergistic roles in regulating root development and whole plant growth.
The nonadditive, synergistic effects of NMT1 and NMT3 loss of function raised the possibility of a compensatory up-regulation of one PEAMT when the gene encoding the other is disrupted. To examine this, we compared NMT1 and NMT3 gene expression in the nmt3 and nmt1 mutants, respectively, with their expression in the wild type (Supplemental Fig. S3C). NMT1 transcripts were increased about 2- to 3-fold in the shoots and roots of 10-d-old nmt3 plants, compared with their levels in the wild type. This was not sufficient to maintain the overall abundance of PEAMT transcripts as high as in the wild type. In contrast, NMT3 expression remained similar in nmt1 shoots compared with wild-type counterparts and was induced only slightly in the root. These results indicate the coexpression of the two genes, but also organ- and cell type-dependent specificity in their transcriptional regulation.
Specific Expression Patterns of NMT3 and NMT1
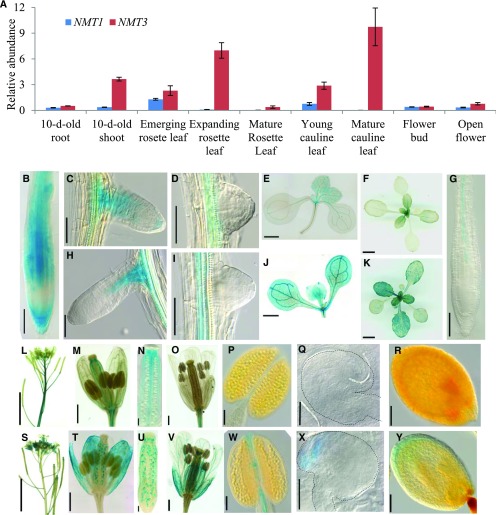
In light of these data and of the ability of NMT3 to correct for the defective root development induced by the loss of NMT1 when driven by ProNMT1, the absence of an apparent phenotype in the nmt3 mutant suggested differences in expression patterns between NMT1 and NMT3. To examine this, we used quantitative real-time PCR to compare the expression profiles of NMT1 and NMT3 in different tissues during vegetative and reproductive development (Fig. 6A). Transcripts of both genes were detected in all tissues tested. However, NMT3 was generally expressed at significantly higher levels than NMT1 and was most abundant in leaves, whether juvenile or adult, expanding or mature, rosette leaves or cauline leaves, while NMT1 was hardly expressed there, consistent with available transcriptomic data (Zimmermann et al., 2004; Winter et al., 2007; Lee and Jez, 2017).
Figure 6.
NMT1 and NMT3 are differentially expressed and have overlapping but also unique expression patterns. A, Relative NMT1 and NMT3 transcript abundance in vegetative and reproductive organs in the wild type, measured by quantitative reverse transcription PCR analysis and normalized to the abundance of housekeeping genes (AtPDF2, AtAPT1, and AtGAPDH; see “Materials and Methods”). Data shown are means ± se (n = 4 biological replicates). For 10-d-old seedlings, each replicate corresponded to a pool of 15 to 30 seedlings sampled from three different plates; for tissues sampled from soil-grown plants at later stages, each replicate corresponded to one to three individual plants. B to K, GUS staining patterns in vegetative tissues of the wild type expressing ProNMT1:GUS (B–F) or ProNMT3:GUS (G–K). Shown are primary root (B and G), young elongating lateral root (C and H), newly emerged lateral root primordium (D and I), and rosette (E and J) of 10-d-old seedlings grown on agar plates and rosette of 18-d-old plants grown in soil (F and K). Bars = 100 µm in B and G, 50 µm in C, D, H, and I, 1 mm in E and J, and 1 cm in F and K. L to Y, GUS staining patterns in reproductive organs of ProNMT1:GUS (L–R) and ProNMT3:GUS (S–Y) reporter lines. Shown are inflorescence (L and S), stage 12 flowers (M and T), expanded view of ovary (N and U), stage 13 flowers (O and V), mature anthers (P and W), ovules from stage 12 flowers (Q and X), and seeds with embryo at late heart stage (R and Y). The dashed line in each image highlights the contour of the embryo. Bars = 5 mm in L and S, 500 µm in M, T, O, and V, 100 µm in N, U, P, and W, 50 µm in Q and X, and 100 µm in R and Y.
To investigate tissue- and cell-specific expression, we created GUS reporter lines. At the vegetative stage, both ProNMT1:GUS and ProNMT3:GUS lines showed expression in roots, leaves, and the shoot apical meristem, but with differences (Fig. 6, B–K): in ProNMT1:GUS lines, GUS staining was most intense at the root tip, in the columella, and throughout the root elongation zone, but in the mature part of the root it became confined to the vasculature (Fig. 6, B and C; Supplemental Fig. S4, A and B). On the contrary, in ProNMT3:GUS lines, GUS staining was not detectable at the root tip, in either primary or secondary roots, but was present only in the root vasculature and in the connective tissue of young lateral roots to the main root (Fig. 6, G and H; Supplemental Fig. S4C). Furthermore, NMT3 was expressed throughout developing and fully expanded leaf blades (Fig. 6, J and K), while NMT1 expression was high in leaf primordia but faded thereafter, according to an apical-basal gradient mirroring that documented for the cessation of cell division and expansion in developing Arabidopsis leaves (Donnelly et al., 1999), and in mature leaves it was restricted mostly to the vasculature (Fig. 6, E and F). These differences indicate developmental regulation of NMT1 expression during leaf development, while NMT3 expression appears broader and more sustained. GUS expression also was analyzed in reproductive tissues (Fig. 6, L–Y). ProNMT3:GUS inflorescences showed strong staining in sepals, petals, anther filaments, and ovules, specifically at their micropylar end in the region of the synergids. GUS staining also was detected in the seed integuments during embryogenesis. By contrast, NMT1 expression was strong in stems, but in flowers it was confined mostly to carpels, the septum of anthers, and pedicels. These overlapping but unique expression patterns suggested functional specificity between NMT1 and NMT3 in floral development.
Aerial Growth and Reproduction Are Severely Compromised in the nmt1 nmt3 Mutant
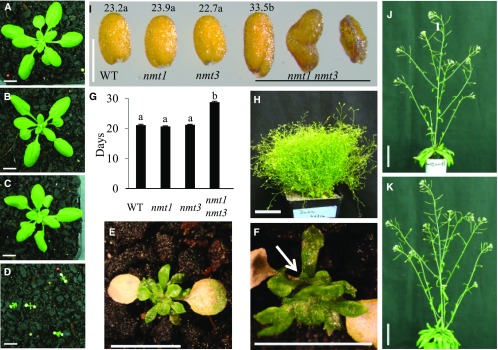
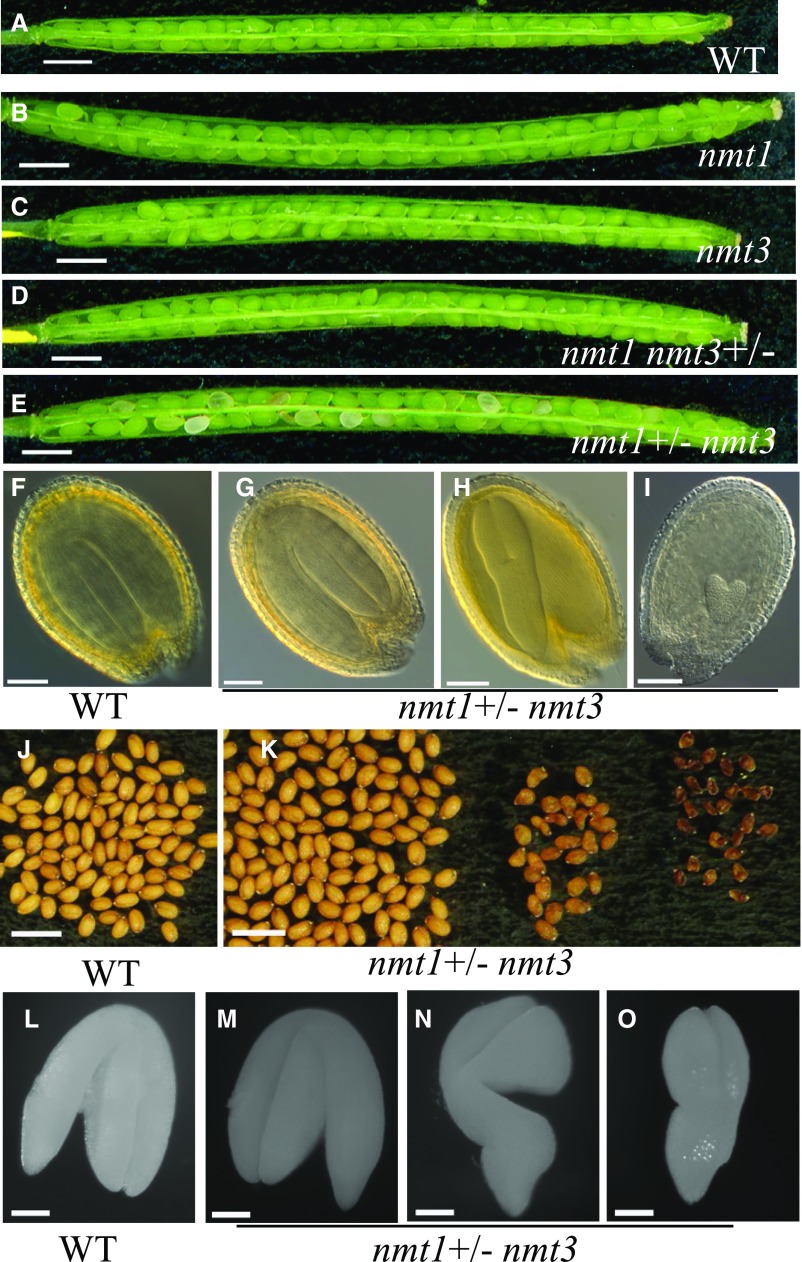
To test for functional specificity between NMT1 and NMT3, nmt3, nmt1, and double nmt1 nmt3 mutants were grown on soil alongside the wild type. nmt1 nmt3 cotyledons were initially normal, but soon they became chlorotic and developed necrosis, whereas leaf emergence was delayed compared with the wild type and single mutants (Fig. 7, A–E; Supplemental Fig. S5, A–H). Leaves hardly expanded, and within less than 3 weeks one or two secondary rosettes were visible (Fig. 7, E and F). Furthermore, wild-type, nmt1, and nmt3 plants formed a single main rosette of similar size and developmental stage (Fig. 7, A–C) and remained indistinguishable during the whole development cycle (Supplemental Fig. S5, I–K), flowering and maturing at the same time (Fig. 7G). Although growing at an extremely reduced pace and showing severe developmental defects, nmt1 nmt3 rosettes switched to reproductive development, with an average 8-d delay compared with the wild type and single mutants (Fig. 7G), and over its lifespan it produced a large number of minute secondary rosettes, many of which also became reproductive, eventually resulting in a bushy, ball-like entanglement of short, thin stems with very reduced cauline leaves and small inflorescences (Figs. 7H and 8A). The vast majority of these were completely sterile. A few formed a very short silique with only a few seeds, half of which aborted early while the other half were either small, wrinkled, and distorted or occasionally full and larger than wild-type or single mutant seeds (Fig. 7I). These seeds represented ∼6% to 8% of the wild-type yield under optimal growth conditions. Continuous choline application restored wild-type rosette and inflorescence morphology, floral development, and fertility (Fig. 7, J and K). Collectively, these results demonstrate that nmt1 nmt3 dwarfism and severely disrupted development are causally related to choline deficiency and that the combined activities of NMT3 and NMT1 in the SDPM pathway play major roles in sustaining Arabidopsis growth, development, and reproduction.
Figure 7.
Loss of NMT3 in an nmt1 background severely compromises aerial growth and reproduction through choline deficiency. A to D, Representative rosettes of 3-week-old soil-grown wild type (A), nmt1 (B), nmt3 (C), and nmt1 nmt3 (D) plants. Bars = 1 cm. E, Enlarged view of a 3-week-old nmt1 nmt3 rosette. Bar = 0.5 cm. F, Side view of E. The white arrow points to a secondary rosette. Bar = 0.5 cm. G, Days to bolting in wild-type (WT), nmt1, nmt3, and nmt1 nmt3 plants under a 16-h photoperiod. Values are means ± se (n = 11–88). Statistical significance tested by one-way ANOVA is denoted by different letters above the bars (P < 0.05). H, Sixteen-week-old reproductive nmt1 nmt3 plant. Bar = 5 cm. I, Photograph of representative wild-type, nmt1, nmt3, and nmt1 nmt3 mature seeds. Wild-type, nmt1, and nmt3 seeds were of similar color, size, and shape. nmt1 nmt3 seeds fell in three visually and morphologically distinct groups, from left to right: full seed, shrunken seed, and aborted seed (see text). Numbers above seeds are mean dry weight per seed (ng; se = 0.9, 1, 0.6, and 0.9, respectively; n = 80–110 seeds). For nmt1 nmt3, only the mean dry weight of the few morphologically normal seeds is shown. Values topped by the same letter were not significantly different by one-way ANOVA (P < 0.05). Bar = 0.5 cm. J and K, Five-week-old nmt1 nmt3 (J) and wild-type (K) inflorescences under choline feeding. Plants were sprayed daily with 2 mm choline, starting from 4 d after cotyledon emergence. Bars = 5 cm.
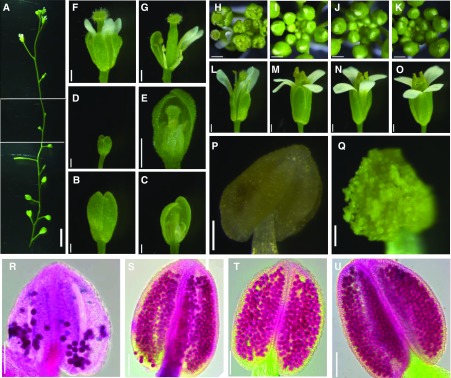
Figure 8.
NMT1 and NMT3 cooperate to control floral organ expansion, anther differentiation and dehiscence, and pollen formation. A, nmt1 nmt3 inflorescence imaged from base to top in three contiguous frames. Bar = 5 mm. B to G, nmt1 nmt3 flowers, intact (B, D, and F) or after dissection (C, E, and G), sampled from the base (B and C) or upper part of the inflorescence (D–G). Bars = 0.5 mm. H to K, Top view of nm1 nmt3 (H), nmt3 (I), nmt1 (J), and wild-type (K) inflorescence. Bars = 0.5 mm. L to O, Open flowers from nmt1 nmt3 (L), nmt3 (M), nmt1 (N), and the wild type (O). nmt1 nmt3 inflorescences comprised very few open and normal flowers, as shown in L (see text); most were abnormal with short sepals, petals, and stamens (F and G) or never opened (B and D). Bars = 0.5 mm. P and Q, nmt1 nmt3 (P) and wild-type (Q) anthers. Bars = 100 µm. R to U, Alexander staining of nmt1 nmt3 (R), nmt3 (S), nmt1 (T), and wild-type (U) anthers. Bars = 100 µm.
Loss of Both NMT3 and NMT1 Compromises the Coordinated Expansion of Floral Organs and Impairs Pollen Formation
To better understand the extensive sterility of nmt1 nmt3 inflorescences, we first examined mature flowers. All nmt1 nmt3 mutants presented the expected whorls of four sepals, four petals, six stamens, and two carpels arranged in a normal phyllotaxy, showing that the determination of floral identity was unaffected. The first few floral buds failed to develop beyond stage 9 or 10 (Smyth et al., 1990) and never opened (Fig. 8, A–C). Some subsequent floral buds higher along the inflorescence had a similar fate, remaining tiny and aborting (Fig. 8, D and E), but most of the later-formed buds developed into flowers with normal-sized sepals and petals and a stigma with normally differentiated papillae (Fig. 8, F, G, and L). However, these buds remained open throughout development due to sepals and petals elongating more slowly than normal relative to the gynoecium and stamens, thus leaving these organs exposed rather than fully enclosed until anthesis, as in the wild type and single nmt1 or nmt3 mutants (Fig. 8, compare H with I–K). In approximately half of these flowers, stamen filaments were abnormally short (Fig. 8G), failing to bring anthers close enough to the stigma for pollination to be possible. In the other half, one or more stamens extended above the stigma as normal (Fig. 8L), but most (89%; n = 200) exhibited pale yellow anthers that were unable to dehisce (Fig. 8, P and Q). In the remaining 11%, one or more bright-yellow anthers developed but, although dehiscent, liberated little pollen. Alexander staining showed that the nmt1 nmt3 lodicules contained substantially fewer mature pollen grains than in wild-type, nmt1, or nmt3 anthers (Fig. 8, R–U). To test whether nmt1 nmt3 pollen was viable, wild-type flowers were hand pollinated with pollen collected from nmt1+/− nmt3 or nmt1 nmt3+/− anthers. The F1 progeny were tested for antibiotic resistance to derive segregation ratios of the nmt1/NMT1 and nmt3/NMT3 alleles. Ratios for each cross were not significantly different from the expected 1:1 ratio for a simple Mendelian dominant trait (P = 0.29 and 0.77; Table I), suggesting similar fertilization success rates by double mutant nmt1 nmt3 pollen compared with nmt3 or nmt1 pollen. Supporting this conclusion, the progeny of self-fertilized nmt1+/− or nmt3+/− hemizygous flowers showed a close to 3:1 segregation ratio of resistance:sensitivity to kanamycin and sulfadiazine, respectively (Table I), showing similar transmission rates of male gametes through nmt1, nmt3, or wild-type pollen. Together, these results demonstrate that the formation of a fully competent male gametophyte does not require local NMT1 or NMT3 expression.
Table I. Segregation of the nmt1 and NMT1 or nmt3 and NMT3 alleles based on antibiotic resistance in the F1 or F2 progeny of crossed or self-pollinated flowers.
P, χ2 test probability values. P > 0.05 indicates no significant difference from a 1:1 segregation ratio (F1 generation) or a 3:1 ratio (F2 generation).
| Cross ♀ ♂ | Generation | nmt1 KanRa | NMT1 KanSa | nmt3 SulRa | NMT3 SulSa | Segregation Ratio | P |
|---|---|---|---|---|---|---|---|
| Wild type nmt3 nmt1+/− | F1 | 154 | 173 | 1:1 | 0.29 | ||
| Wild type nmt1 nmt3+/− | F1 | 206 | 212 | 1:1 | 0.77 | ||
| nmt1+/− nmt1+/− | F2 | 573 | 208 | 3:1 | 0.29 | ||
| nmt3+/− nmt3+/− | F2 | 223 | 67 | 3:1 | 0.46 | ||
| nmt3 nmt1+/− wild type | F1 | 252 | 271 | 1:1 | 0.41 | ||
| nmt1 nmt3+/− wild type | F1 | 246 | 229 | 1:1 | 0.44 |
Numbers of F1 or F2 seedlings resistant or sensitive to kanamycin (KanR and KanS, for the nmt1 and NMT1 alleles, respectively) or to sulfonamide (SulR and SulS, for the nmt3 and NMT3 alleles, respectively).
The nmt1 nmt3 Mutant Is Defective in Female Gametophyte Development
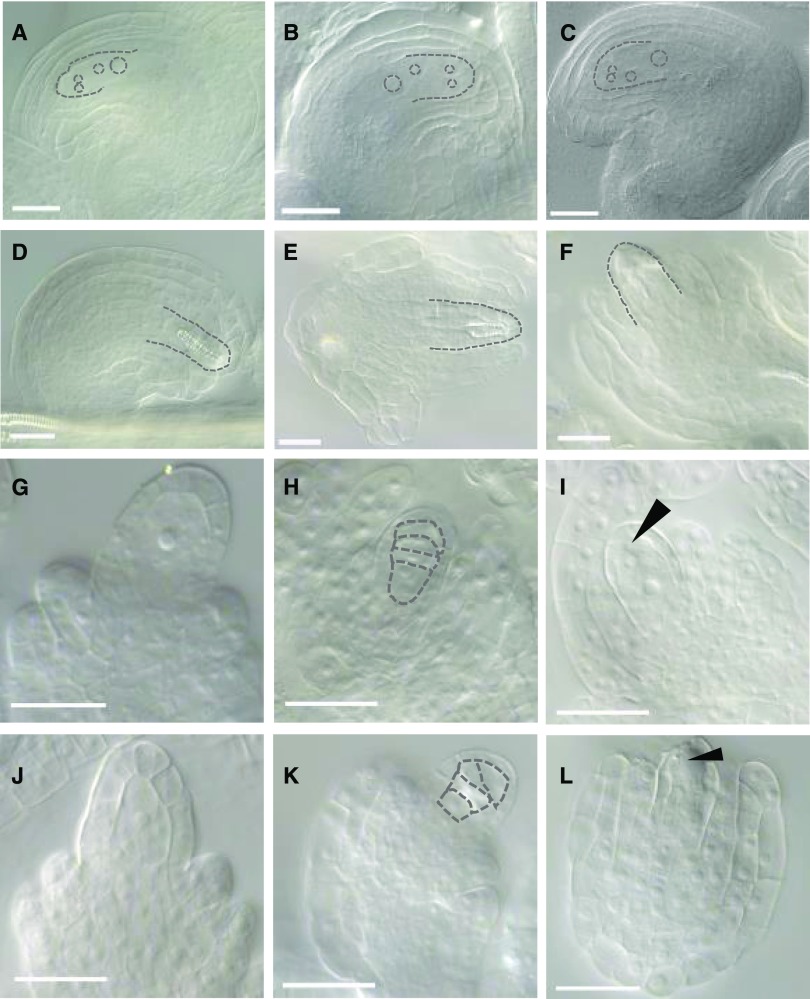
Given the overlapping but unique expression domains of NMT3 and NMT1 in female organs (Fig. 6), the apparently normal pistils and stigmas in many nmt1 nmt3 flowers, apart from the oldest ones, were intriguing and prompted us to examine ovule viability. To that end, we pollinated these normal-looking nmt1 nmt3 stigmata with wild-type pollen. Less than 10% of the crosses (3:53) gave rise to a silique, and these yielded two to three seeds at most. To determine whether this was the result of defective ovules and/or impaired fertilization, ovules were carefully excised from the ovaries of mature mutant and wild-type flowers (stage 13). Wild-type or nmt1 and nmt3 ovules had all reached the FG7 mature stage, showing a four-celled embryo sac surrounded by fully developed integuments (Christensen et al., 1997; Fig. 9, A–C). In stark contrast, in nmt1 nmt3 siliques, stage FG7 ovules were rare. Instead, most ovules showed an abnormal shape and anatomy with an amorphous or highly divided finger-like structure visible in the embryo sac and disorganized integuments that failed to enclose the nucellus (Fig. 9, D–F). This was not seen in either nmt1 or nmt3 ovules, which were indistinguishable from the wild type (Fig. 9, A–C). To determine the onset of these abnormalities, nmt1 nmt3 ovules were dissected at a range of earlier stages. MMC determination and enlargement appeared normal (Fig. 9, G and J), and the enlarged MMC gave rise to a tetrad of haploid megaspores as in the wild type (Fig. 9, H and K). However, all four megaspores generally degenerated (Fig. 9L), instead of normally three (Fig. 9I). To better understand the relative importance of NMT3 and NMT1 in female gametophyte development, we pollinated nmt1 nmt3+/− and nmt3 nmt1+/− flowers (i.e. hemizygous for NMT3 and NMT1, respectively, but homozygous for the other NMT) with wild-type pollen. No significant deviation from 100% female transmission rate was observed in either cross (Table I). This result reveals that NMT1 and NMT3 are both haplosufficient in a double mutant gynoecium for the normal development of an nmt1 nmt3 double mutant haploid egg cell and that the abnormal development of the female gametophyte frequently seen in nmt1 nmt3 flowers is of sporophytic origin. Taken together, our data demonstrate redundant functions of NMT1 and NMT3 in female megaspore selection and ovule integument outgrowth. The semisterility of nmt1 nmt3 inflorescences shows that these functions are not totally penetrant, similar to NMT1 and NMT3 functions at the earlier stages of floral development.
Figure 9.
NMT1 and NMT3 redundantly control ovule and female gametophyte development through nontotally penetrant functions. A to F, Mature ovules, FG7 stage: the wild type (A), nmt1 (B), nmt3 (C), and nmt1 nmt3 (D–F). nmt1 nmt3 ovules with fully enclosing integuments were rare (at most one in 10 to 20 of the several hundred ovules examined, across multiple siliques). . G and J, MMC of wild-type (G) and nmt1 nmt3 (J) ovules. H and K, Tetrad of haploid megaspores in wild-type (H) and nmt1 nmt3 (K) ovules. I and L, Female gametophyte in the wild type (I) and nmt1 nmt3 (L) at stage FG1. The arrowhead in I points to the functional megaspore. No megaspore has been retained in L, as in the vast majority (at least 90%) of the ovules examined that developed normally up to that stage. In A to F, dashed lines highlight the contours of the four-celled embryo sac (A–C), an abnormal structure within the nucellus of nmt1 nmt3 ovules (D and E), and an abnormally patterned and incompletely enclosed nmt1 nmt3 ovule (F). In H and K, dashed lines highlight the megaspore tetrad in wild-type (H) and nmt1 nmt3 (K) ovules. Bars = 25 nm.
NMT1 and NMT3 Play Synergistic But Unequal Roles in Embryogenesis and Seed Development
The high proportion of aborted seeds in nmt1 nmt3 siliques indicated a high frequency of developmental impairment postfertilization, during seed development. To investigate this, we sampled fully elongated siliques of self-fertilized nmt1 nmt3+/− and nmt1+/− nmt3 alongside single mutant and wild-type siliques 10 d after pollination (i.e. at the end of wild-type embryogenesis). Wild-type, nmt1, nmt3, and nmt1 nmt3+/− siliques were filled with green, swollen seeds of mostly uniform size, whereas nmt1+/− nmt3 siliques contained a mix of broadly three seed phenotypes (Fig. 10, A–E): wild-type-looking seeds; seeds of wild-type size but paler green; or small, flatter, brown seeds. The last two classes together represented about one-quarter of the total number of seeds (449:1,943), indicating that they likely corresponded to nmt1 nmt3 segregants. Upon clearing, pale green and white seeds revealed torpedo and heart stage embryos, respectively, contrasting with the fully developed, folded embryos enclosed in the dark green seeds sampled from the same silique or from single mutant or wild-type siliques (Fig. 10, F–I).
Figure 10.
Lack of functional NMT3 and NMT1 impairs embryogenesis and seed development. A to E, Ten-day-old wild-type (WT; A), nmt1 (B), nmt3 (C), nmt1 nmt3+/− (D), and nmt1+/− nmt3 (E) siliques. Bars = 1 mm. F to I, Developing seeds excised from 10-d-old siliques, at the end of embryogenesis, in the wild type (F), nmt1+/− nmt3 class I seeds (turgid, bright green seeds; see text; G), class II, pale green seeds (H), and class III, white seeds (I). Bars = 100 µm. J to K, Mature desiccated seeds from wild-type (J) and nmt1+/− nmt3 (K) siliques. Three visually distinct phenotypes are present in the latter, from left to right: full, golden seeds; shrunken seeds; and aborted seeds. Bars = 1 mm. L to O, Embryos excised from mature desiccated seeds of wild-type (L) or nmt1+/− nmt3 (M) full seeds and shrunken seeds (N and O). As for floral development, NMT1 and NMT3 function during embryogenesis, and embryo morphogenesis had high but incomplete penetrance. Bars = 100 µm.
To determine whether embryonic development had been arrested in those paler seeds or might only be proceeding at a reduced pace, we dissected siliques at full maturity. Wild-type, nmt1, nmt3, or nmt1 nmt3+/− siliques yielded seeds of uniform size and normal golden color, as expected (Fig. 10J; Supplemental Fig. S6), while nmt1+/− nmt3 siliques contained three clearly distinct seed groups: smooth, oblong wild-type-like seeds (class I); smaller seeds, often of darker color and with a pointed or distorted shape at the chalazal end (class II); or even smaller, shriveled, and dark brown seeds (class III; Fig. 10K). Class I seeds contained a normal fully developed embryo that completely filled the seed cavity (Fig. 10M). Class II seeds had embryos of varied but always reduced size and a morphology typical of late-torpedo to walking stick embryonic stages, with a bulging radicle (Fig. 10, N and O). No embryo could be found in class III seeds, implying very precocious abortion during early embryogenesis. These data suggest that the loss of both NMT1 and NMT3 function caused slower or prematurely arrested seed development and disturbed differentiation of the embryo basal domain. The relative frequencies of type I (78%), II (10%), or III seeds (12%; n = 1,362 seeds) suggested that classes II and III corresponded to homozygous nmt1 nmt3 segregants (expected frequency, 25%). Supporting this, class I seeds readily germinated and produced normal seedlings. By contrast, although class II showed very high germination rates (90%; n = 370), they produced seedlings with a typical nmt1 nmt3 very-short-root phenotype. PCR genotyping of a random subset (n = 87 seeds) systematically returned a homozygous nmt1 nmt3 genotype, and all class III seeds failed to germinate. Together, these results demonstrate synergistic but unequal roles of NMT1 and NMT3 as positive regulators of embryogenesis and embryo morphogenesis. NMT3 is haplosufficient in an nmt1 gynoecium for an nmt1 nmt3 zygote to give rise to a fully mature and viable embryo, developing at normal pace, whereas NMT1 is haploinsufficient in an nmt3 gynoecium for seed development to proceed to completion and is only partly haplosufficient for seed viability.
Loss of Both NMT1 and NMT3 Severely Compromised PC Synthesis
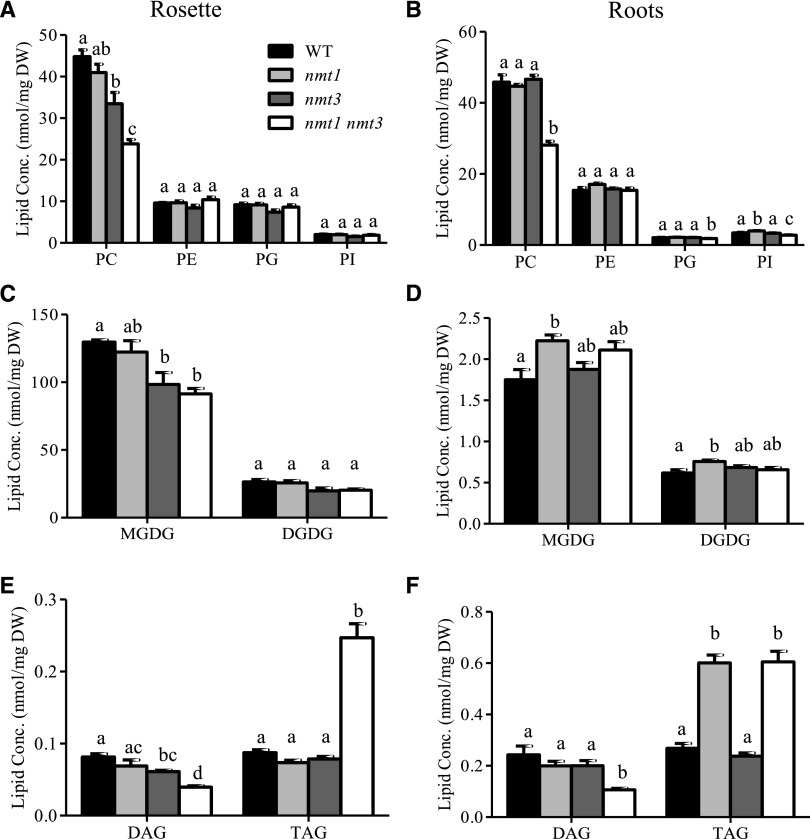
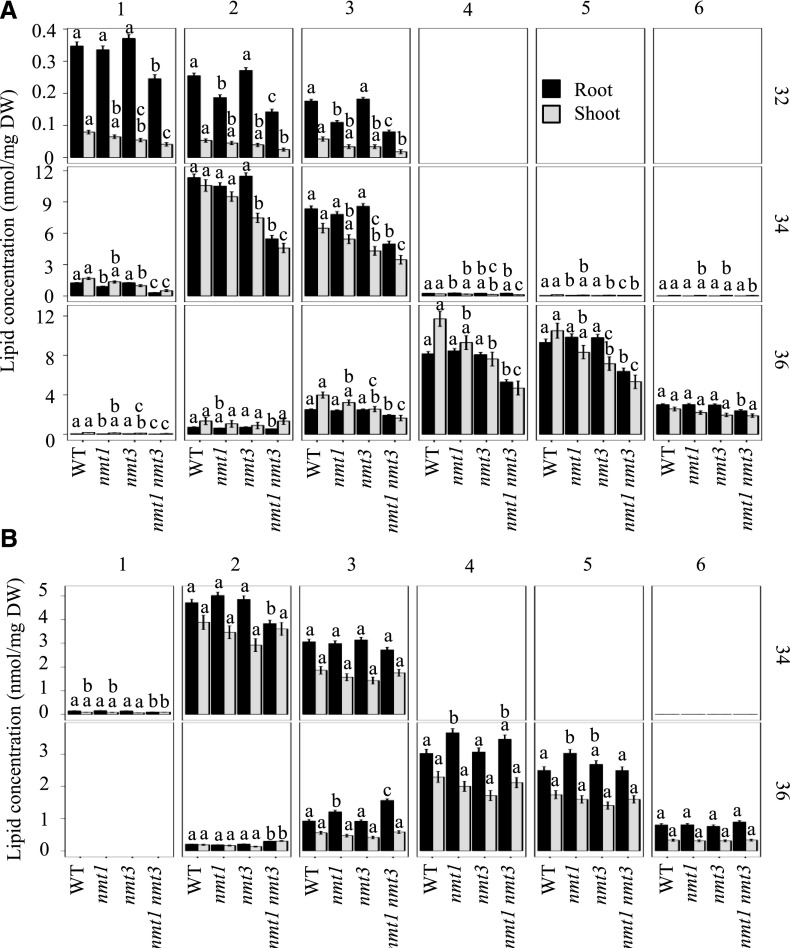
The nmt1 nmt3 seedlings were barely able to grow and produced only a few seeds. To better understand how this was achieved, we conducted lipid-profiling experiments on young roots and rosettes. PC represents the major class of polar lipids in wild-type roots and leaves (Fig. 11, A and B; Devaiah et al., 2006). Loss of NMT1 reduced the abundance of the minor C32:2 and C32:3 species in roots, and there was a trend for small reductions in C36:3, C36:4, and C36:5 in rosettes, but these effects were relatively too small to be reflected in overall organ PC content (Fig. 11). Loss of NMT3 also left root PC content unaffected but caused a 17% decrease in shoot PC content due to a significant reduction of most C32, C34, and C36 molecular species (Fig. 12A). These decreases were amplified in the diminutive double mutant, especially for the 34:2 and 36:4 species, and all PC molecular species were now decreased significantly in roots too (Fig. 12), resulting in large decreases of overall PC content (39% and 47% in roots and shoot, respectively; Fig. 11, A and B). These data support the shared biochemical function of the two enzymes in the SDPM pathway and their importance for PC synthesis. They also show that each enzyme can compensate in part for the other, but not equally, adding evidence for functional specificity between the two PEAMTs.
Figure 11.
Profiles of major lipids in wild-type (WT), nmt1, nmt3, and nmt1 nmt3 seedlings. Lipids were analyzed separately for the aerial parts and roots of 12-d-old seedlings. A and B, Concentrations of phospholipids PC, PE, phosphatidylglycerol (PG), and phosphatidylinositol (PI). C and D, Concentrations of galactolipids monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG). E and F, DAG and triacylglycerol (TAG) concentrations. Shown are means ± se (n = 4–6 pools of 36–100 seedlings). One-way ANOVA followed by Bonferroni’s test was used for statistical analysis of genotypic differences for each lipid in root or shoot tissue. Bars topped by different letters denote statistically different values (P < 0.05). DW, Dry weight.
Figure 12.
PC and PE molecular species in the wild type (WT) and mutants lacking NMT1 or NMT3 function, or both. Molecular species of PC (A) and PE (B) are shown in roots and shoots of 12-d-old wild-type, nmt1, nmt3, and nmt1 nmt3 plants, graphed as facet plots with the total carbons as vertical facets and the number of double bonds as horizontal facets. Bars show means ± se (n = 4–6 biological replicates, each corresponding to a pool of 36–100 seedlings, separating shoots and roots). Bars topped by different letters denote statistically significant differences among genotypes by one-way ANOVA run for each tissue, followed by Bonferroni’s pairwise test (P < 0.05). DW, Dry weight.
Lipid Remodeling and TAG Accumulation in Seedlings Lacking NMT1 and NMT3 Function
PE is the other major phospholipid of plant membranes. Remarkably, in nmt1 nmt3 mutants, the PE content and proportions of molecular species remained similar to those in the wild type (Figs. 11, A and B, and 12). PG and PI concentrations showed little to no variations either (Fig. 11; Supplemental Fig. S7). By contrast, MGDG content was altered significantly, with a 21% increase in roots and a 29% decrease in shoots (Fig. 11, C and D). These shifts largely reflected the effects of the nmt1 and nmt3 mutations, respectively, fitting with the predominant expression of NMT1 in roots and of NMT3 in leaves. Interestingly, the overall abundance of DGDG was little affected (Fig. 11), but loss of NMT1 and NMT3 induced compositional modifications, with a general decrease of 34-carbon fatty acid species and an opposite increase in 36-carbon chains in roots, as well as decreases of both C34 and C36 species in shoots, but more limited (Supplemental Figs. S8 and S9). Collectively, these data demonstrate significant lipid remodeling in nonplastidic and plastidic membranes of the nmt1 nmt3 mutant, with differences between aerial and root tissues and synergistic roles of the two NMTs in both tissues.
DAG is the substrate for PC and PE synthesis and is a branch point between the synthesis of membrane lipids or TAG storage lipids, prompting us to investigate whether the significantly reduced capacity of the nmt1 nmt3 mutant to synthesize PC had an impact on DAG and TAG concentrations (Fig. 11, E and F). We observed a consistent trend for reduced DAG in the three mutants; reductions were highly significant in the double mutant (56% and 51% decreases in roots and shoot, respectively). On the contrary, TAG contents were increased significantly (125% and 183% in roots and shoots, respectively; Fig. 11, E and F; Supplemental Fig. S10). In roots, this was brought about by the nmt1 mutation, while in shoots, TAG concentrations were increased only in the double mutant but unaffected in the two single mutants. These data demonstrate that DAG is preferentially channeled toward the synthesis of storage lipids in the nmt1 nmt3 seedlings, and provide further evidence of functional specificity between NMT1 and NMT3.
TAG is derived from DAG via the acylation of acyl-CoA at the sn-3 position by diacylglycerol acyltransferase (DGAT) or the acylation of PC at the sn-2 position by phosphatidylcholine diacylglycerol acyltransferase (PDAT). Overexpression of DGAT1 results in the accumulation of TAG in vegetative tissue (Bouvier-Navé et al., 2000; Kelly et al., 2013). To investigate whether the increased TAG contents of nmt1 nmt3 roots and shoots were due to the enhanced expression of biosynthesis genes, we measured DGAT1 and PDAT1 transcript abundance. Only DGAT1 expression showed a significant but relatively modest reduction (∼20%; Supplemental Fig. S11). The accumulation of TAG reflects the balance between synthesis and breakdown. Therefore, we quantified the expression of the TAG lipase-encoding genes SUGAR-DEPENDENT1 (SDP1), SDP1-LIKE (SDP1L), and ADIPOSE TRIGLYCERIDE LIPASE (ATGL; Eastmond, 2006; Kelly et al., 2011, 2013), and also CGI58, which encodes an enzyme reported to have lipase, phospholipase, and lysophosphatidic acid acyltransferase activities (Ghosh et al., 2009) and has been suggested to be involved in lipid turnover and signaling (James et al., 2010; Park et al., 2013), as well as PEROXISOME ABC TRANSPORTER1 (PXA1), whose encoded protein acts in fatty acid transport in peroxisomes (De Marcos Lousa et al., 2013). No differential gene expression was found except for small increases and decreases in ATGL and CGI58L expression, respectively, in roots (Supplemental Fig. S11). These data suggest that the increased TAG content of nmt1 and nmt1 nmt3 tissues is mostly posttranscriptionally controlled.
DISCUSSION
NMT3 Is a Functional Homolog of NMT1 But Is Not Totally Redundant Physiologically
Our data provide biochemical and genetic evidence that the NMT3 gene encodes a PEAMT with the in vivo capacity to catalyze the triple transmethylation of PEA into PCho. First, expression of Arabidopsis NMT3 alleviates the choline auxotrophy of the PC-deficient yeast strain pem1Δ pem2Δ. Second, supply of PEA to crude protein extracts from these transgenic yeast cells enables the de novo synthesis of PCho. Third, expression of NMT3 in the short-root nmt1 knockout mutant restores normal root development when expressed under the control of the NMT1 promoter. Fourth, supplementation of choline, which can be converted directly to PCho, the end product of the phospho-base methylation pathway, by choline kinase, recovers the severe growth and developmental defects of the nmt1 nmt3 double mutant in roots, leaves, and inflorescences. Altogether, these results identify NMT3 as a functional PEAMT in vivo, with the same biochemical function as the previously characterized NMT1. However, the detection of two intermediate peaks between PEA and PCho in our NMR spectra from protein reaction mixtures containing NMT3, but their absence in spectra from NMT1-expressing cells, suggest faster turnover rates of the PMMEA and PDMEA intermediates in the presence of NMT1 than of NMT3 enzymes in vivo. This interpretation is supported by a recent in vitro study showing lower values of kcat/Km in NMT3 (Lee and Jez, 2017). In our earlier in vitro comparison of the wheat TaPEAMT1 and TaPEAMT2 enzymes, we found that, while sharing the same biochemical function, the two PEAMTs differed in their intrinsic catalytic activity and affinities for PEA and SAM substrates and also in their sensitivity to feedback inhibition by the end products PCho and choline (Jost et al., 2009). NMT1 expression is known to be sensitive to feedback inhibition by choline, both in vitro and in planta. The mechanisms are not fully understood, but evidence suggests that it involves a small open reading frame upstream of the gene (Tabuchi et al., 2006; Eastmond et al., 2010; Alatorre-Cobos et al., 2012; Craddock et al., 2015). NMT3 has a small open reading frame inserted in its first intron but none in its upstream regulatory region (Supplemental Fig. S3). This difference may explain why the lack of NMT3 leads to an up-regulation of NMT1 expression while the converse is not observed (Supplemental Fig. S3). It will be interesting to investigate this further and, more broadly, the regulation of the two proteins under a range of physiologically relevant conditions, especially as the regulation of PEAMTs appears to involve interactions with various metabolites, including stress signaling molecules such as PA or some phosphoinositides (Jost et al., 2009).
The acquisition of functional diversity among closely related isoforms within gene families occurs through modifications of protein function but also commonly through alterations of expression patterns. Fitting with this, the NMT3 and NMT1 genes present partly overlapping but also specific expression patterns in roots, leaves, and reproductive organs (Fig. 6). These overlaps and differences in expression partly account for the type and magnitude of developmental defects induced by the loss of one or the other NMT, or both: from nondetectable defects in the nmt3 root where NMT1 activity can compensate for loss of NMT3 in the vasculature, being also expressed there (Fig. 6), and furthermore sensitive to choline feedback regulation (Tabuchi et al., 2006; Eastmond et al., 2010; Alatorre-Cobos et al., 2012); to severe defects in the nmt1 root meristem and elongation zone, two physically small regions but ones of locally high demand for PC, in which, furthermore, NMT3 is not expressed (Fig. 6); to catastrophic defects in the double mutant nmt1 nmt3 root, reflecting the compounded losses of NMT activity in the root tip (NMT1) and the vasculature (NMT1 and NMT3).
However, differences in the expression patterns between the two NMTs alone cannot account for the observed mutant phenotypes. In the inflorescence, for example, NMT1 and NMT3 are expressed in mostly distinct domains (Fig. 6; https://bar.utoronto.ca/eplant/), yet the single mutant nmt1 or nmt3 flowers present no apparent floral or embryogenic defect. Extracellular and intercellular choline transport is well documented in animals, where it plays a major role in cell-cell communication, development, and neurological pathologies (Ueland, 2011). Little is known about choline transport in plants. However, an Arabidopsis homolog of the human choline transporter-like protein CTL (CHER1) was shown recently to be involved in choline transport and required for the maintenance of PCho and choline levels within roots and leaves (Dettmer et al., 2014; Kraner et al., 2017). Choline also has long been known to be present in xylem sap (Maizel et al., 1956; Martin and Tolbert, 1983). In addition, the fact that exogenously provided choline or PCho can rescue the root defects of the nmt1 mutant or the allelic xpotl1 (Fig. 5; Cruz-Ramírez et al., 2004) and also the multiple defects of the nmt1 nmt3 mutant uncovered here, from roots to leaves and inflorescences, also implies the existence of long-distance choline/PCho movement within the plant, correcting for local endogenous deficiencies in PC synthesis. As a whole, these observations suggest that the formation of some fertile flowers in the nmt1 and nmt3 mutants could be enabled by choline transport from domains where the residual NMT is expressed to domains where the locally expressed NMT has been rendered nonfunctional, such as from carpels to ovules in nmt1 ovaries or from the stem vasculature to sepals, petals, and stamens in nmt1 flowers. Supporting an important role of choline/PCho transport to mitigate the consequences of reduced or absent local PC synthesis, the nmt1 nmt3 female gametophyte develops normally if NMT1 or NMT3 expression is present in the enclosing sporophytic tissues but not when it is absent, and the nonvascularized root tip is the only root domain presenting severe defects when the activity of the only one of the two NMTs to be expressed there (NMT1) is disrupted.
NMT1 and NMT3 Control Phase Transition and Floral Development
The nmt1 nmt3 mutant is a powerful tool for analyzing the physiological functions of PEAMTs and identifying novel PEAMT and PC functions in multiple aspects of Arabidopsis development. Lack of functional NMT1 and NMT3 causes a significant delay in floral initiation and flowering time, revealing a role of these PEAMTs in the control of phase transition. The mechanisms are unknown; however, in a recent study of plants with modified PECT1, and hence modified ability to produce PE, flowering time was found to correlate positively with the PC-PE ratio in the shoot apical meristem in a dose-dependent manner (Nakamura et al., 2014). Further investigation showed that PC binds to the florigen FT in vitro and that the positive effect of PC on flowering requires FT. Here, we report that PC levels were lowered drastically in nmt1 nmt3 shoot and roots. Both the NMT1 and NMT3 genes are strongly expressed in the shoot apical meristem (Fig. 6; Waese et al., 2017; https://bar.utoronto.ca/eplant/), making it very likely that PC levels were decreased severely there too. In addition, our lipidomic data show decreases in most PC molecular species, including those that appear to preferentially bind to FT (Nakamura et al., 2014). Altogether, this suggests that the delayed flowering in nmt1 nmt3 might be explained by a disruption of the FT-mediated signaling via reduced availability of PC.
Once floral initiation eventually occurred, numerous flowers were initiated. Many aborted early, and some survived but showed a range of defects. The slow-growing and never-closing sepals and petals, shorter stamen filaments, and reduced outgrowth of ovule integuments that they exhibited are all consistent with the structural role of PC for membrane synthesis, cellular proliferation, and growth. It is interesting, however, that growth inhibition was relatively stronger in sepals and petals than in the pistil. The underlying causes could relate to cell or organ specificity in membrane lipid composition and/or in lipid-signaling networks. There is mounting evidence for a broad spatiotemporal diversity in lipid composition of Arabidopsis floral organs, including in some lipid species that have an important signaling role, such as PA (Nakamura, 2015). Our lipid profiling of nmt1 nmt3 vegetative tissues shows significant perturbations of the relative proportions of the major membrane lipids and constitutive molecular species (Figs. 11 and 12; Supplemental Figs. S7–S9). The nmt1 nmt3 floral phenotypes identified here, and their noncomplete penetrance, suggest that profound and spatially variable compositional modifications likely take place in the inflorescence too, with variations among organs and cell types.
While many nmt1 nmt3 flowers showed a general failure of organs to develop and expand normally, a few were morphologically normal and indistinguishable from the wild type, produced viable pollen, yet were most often infertile. We found defective megaspore selection and retention of a functional megaspore in most female gametophytes (more than 90%). The MMC went through meiosis and gave rise to four haploid megaspores, as normal (Christensen et al., 1997), but all megaspores degenerated, instead of only three in the wild type. That infertility is of sporophytic origin, since one functional copy of either NMT1 or NMT3 in the sporophytic tissues surrounding the embryo sac was sufficient to rescue nmt1 nmt3 ovule fertilization (Table I). Specification of the functional megaspore requires communication between megaspores and sporophytic tissues (Bencivenga et al., 2011; Chevalier et al., 2011; Figueiredo and Köhler, 2016). The nature of the signals is not well understood, but cytokinin-dependent signals are involved (Cheng et al., 2013), and a role for auxin also has been proposed (Pagnussat et al., 2009). The molecular controls of megaspore selection also remain elusive, apart from the recent discovery of an important role of the arabinogalactan protein AGP18 in megaspore selection and viability (Demesa-Arévalo and Vielle-Calzada, 2013). Here, we uncover two other players, the NMT1 and NMT3 PEAMTs. It will be fascinating to determine whether PC or related metabolites and signaling molecules are involved directly in the communication between female gametophytic and sporophytic cells or act indirectly through controlling integument outgrowth and cellular membrane conformation or the choline-dependent differentiation of plasmodesmata (Kraner et al., 2017).
In this context of signaling between gametophytic and sporophytic cells, it is notable that NMT1 has been implicated in ovule targeting, a process controlled by complex intersignaling between the pollen tube and the maternal surrounding tissues and extracellular matrix; this was ascribed to a role in pollen tube growth and navigation within the pistil (Qin et al., 2009) through unknown mechanisms. We conducted a series of genetic crosses to test pollen viability and pollen-borne defects in transmitting male gametes. The results give no indication of defective ovule targeting by nmt1 or nmt3 pollen. nmt1 nmt3 pollen was produced in much reduced amounts compared with the wild type and single mutants but, when used to pollinate wild-type stigmas, showed normal male gamete transmission rates (Table I). The discrepancy between the two studies could be due to differences in growth conditions, such as temperature or nutrition, that have the potential to interact with NMTs in the control of membrane properties and the environment of the pollen tube within the pistil, and thus its navigation toward the ovule and egg cell (Higashiyama and Takeuchi, 2015). This supports a need for future investigation under a range of conditions, as a temperature-conditional male sterility has been reported in an NMT1-silencing line (Mou et al., 2002), and should include examining the possibility that, in the homozygous nmt1 nmt3 double mutant, female sporophytic defects may influence the male gametophyte, as observed in some female gametophytic mutants (Huck et al., 2003), especially given the strong and specific expression of NMT3 in the synergid area during late ovule development (Fig. 6).
NMT1 and NMT3 Function Controls Seed Development and Viability
A number of Arabidopsis mutants disrupted immediately upstream of NMTs in the SDPM pathway or coding for enzymes of the CDP-choline pathway show embryonic defects. Thus, plants lacking SCD1 or CEK4 activity for the conversion of Ser to ethanolamine and phosphorylation of the latter to PEA exhibit arrested embryo development (Lin et al., 2015; Yunus et al., 2016; http://www.seedgenes.org/NomarskiImages?alleleSymbol=emb+1075). Disruption of PECT1, the rate-limiting enzyme for the synthesis of PE, causes embryo abortion before the octant stage (Mizoi et al., 2006). Disruption of the AAPT1 and AAPT2 enzymes that catalyze the last steps of PC and PE synthesis also leads to embryo defects (Liu et al., 2015). Embryonic defects thus appear to be a general response to PC deficiency or decrease in PC/PE. In keeping with this, disruption of NMT1 and NMT3 in the nmt1 nmt3 mutant causes widespread, but not completely penetrant, embryo abortion or premature developmental arrest (Fig. 10). The synthesis of lipids in plants occurs through two interdependent pathways: the prokaryotic pathway, which takes place in the chloroplast and is responsible for the synthesis of the MGDG and DGDG galactolipids as well as of sulfolipids, and the eukaryotic pathway, which refers to reactions involved in the synthesis of lipids in the endoplasmic reticulum, including PC, PE, phosphatidylserine (PS), and PI (Ohlrogge and Browse, 1995; Li-Beisson et al., 2010, 2013). There is intense trafficking of lipids between the two pathways (Somerville and Browse, 1996; Benning et al., 2006; Benning, 2009). Much reduced PC synthesis in nmt1 nmt3 may make embryogenesis more highly dependent on lipids produced through the prokaryotic pathway and, thus, critically restricted, especially as only MGDG is a bilayer-forming lipid potentially able to act as a surrogate lipid for membrane synthesis (Li-Beisson et al., 2013). In developing Arabidopsis seeds, undifferentiated plastids only begin to develop into chloroplasts and increase in numbers at the torpedo stage (i.e. after embryogenesis is complete and the embryo has entered the morphogenetic phase; Hsu et al., 2010). This may explain why nmt1 nmt3 seed abortion occurs predominantly during early embryogenesis. The next phases of embryo morphogenesis and maturation are phases of high demand for PC to sustain active cell proliferation and expansion and then the formation of oil bodies through the coalescence of TAG molecules. TAGs are the primary storage compounds in the Arabidopsis seed, and their synthesis primarily utilizes PC-derived DAG (Bates and Browse, 2011). The high frequency among the few nmt1 nmt3 viable seeds of small, wrinkled seeds with incompletely expanded embryos not filling the seed cavity, but normal embryo development and seed viability in the single mutants (Fig. 10; Supplemental Fig. S6) shows that at least one of the two NMTs is required to fulfill the high embryo demand for PC during embryo morphogenesis and maturation. Interestingly, as observed for floral development, NMT1 and NMT3 functions on embryogenesis and seed abortion show incomplete penetrance, pointing to spatially or developmentally regulated modulating factors that it will be intriguing to elucidate.
NMT1 and NMT3 as Potential Targets for Modulating Lipid Homeostasis and Biomass Nutritional Value?
In the absence of choline supplementation, the combined disruption of NMT1 and NMT3 catalysis of the methylation pathway rendered the plant barely able to acquire autotrophy, maintain some growth, and reproduce. This indicates that PC synthesis was decreased close to the minimum critical PC level permissive of survival and that the observed 40% to 50% decrease of tissue PC concentration is close to the threshold beyond which membrane assembly, cell viability, and the generation of even a few viable progeny are no longer possible. Remarkably, PE homeostasis remained unaltered in these seedlings. Since PC and PE synthesis use PEA as a common substrate (Fig. 1), this implies a negative feedback regulation by the PE product on its biosynthetic pathway. Similarly, when PE synthesis was inhibited in the aapt1 aapt2 double mutant, PC concentrations were unaffected (Liu et al., 2015). This indicates separate controls of the two branches where each branch ensures that its product is maintained constant. One may surmise that the net effect is to buffer variations of the PC-PE ratio toward maintaining membrane function.
Nevertheless, the relative abundance of the two phospholipids was altered significantly in the nmt1 nmt3 mutant (38% and 42% decreases in PC/PE ratio in roots and shoot, respectively) and also in the nmt1 roots (10% decrease in PC/PE ratio), while showing no significant change in nmt3 tissues. Membrane lipid composition plays an important role in membrane fluidity and stability, and thus in membrane-mediated signaling and trafficking. Some phospholipids increase fluidity, such as PC, while others decrease it, such as PE (van den Brink-van der Laan et al., 2004; Fajardo et al., 2011). A decrease in PC/PE ratio and consequent increase in membrane rigidity occur during cold acclimation (Sutoh et al., 2010), and membrane fluidity has been postulated to be a primary signal in stress sensing, such as cold or osmotic stress sensing (Orvar et al., 2000; Los and Murata, 2004). Acyl chain length and saturation, which also are important determinants of membrane physical properties, also were altered in the nmt1 nmt3 mutant, and to some degree in the single mutants, but differentially depending on lipid and organ (roots or leaves), indicating nontotally redundant metabolic functions of the two NMTs. These differences may partly reflect the differential expression of the two isoforms in roots and photosynthetic organs, as glycerolipids synthesized through the prokaryotic pathway in the chloroplast have predominantly 16:0 fatty acids at the sn-2 position of the glycerol backbone, while those synthesized in the eukaryotic pathway mostly have 18-carbon unsaturated fatty acids. It will be of great interest to explore the impact of NMT1- and NMT3-mediated alterations of the relative PC and PE contents, and also galactolipids, on membrane and cellular functions under changing and challenging environmental conditions.
Disruption of NMT1 and NMT3 led to a 2- to 4-fold increase in major TAG molecular species, indicating that the two PEAMTs are actively involved in the regulation of lipid storage in Arabidopsis. Enhanced TAG accumulation upon the disruption of PC synthesis appears to be a feature common to various eukaryotes. Silencing of PMT1 in C. elegans increases TAG levels in lipid droplets (Li et al., 2011). The interruption of PC synthesis in yeast, mouse liver, and human cells also has been shown to enhance TAG storage (Malanovic et al., 2008; Walker et al., 2011). These findings are in agreement with the tight interconnection between the synthesis of phospholipids and TAGs in plants (Bates et al., 2009; Bates and Browse, 2011; Chapman and Ohlrogge, 2012). Together with PA, DAG is the common precursor for the synthesis of TAG and PC. When PC synthesis is inhibited, DAG may be diverted into TAG synthesis. Supporting this, concomitant with increased TAG concentrations, nmt1 nmt3 seedlings exhibited decreased DAG content (Fig. 11). Interestingly, however, TAG content also was elevated in nmt1 roots, where overall PC content did not differ significantly from that measured in the wild type (Fig. 11; Supplemental Fig. S10). However, in those roots, the C32:2 and C32:3 PC species were present at 20% to 30% lower concentrations than in wild-type and nmt3 roots (Fig. 12). It is possible that some PC molecular species, although minor in terms of their contribution to the total PC pool, may play a monitoring role in directing DAG to storage or membrane lipid synthesis. It is also possible that high local PCho and PC deficiency at the nmt1 root tip played a role. Plant oils are an important natural resource to meet the increasing demands of food, feed, biofuel, and industrial applications. Nonseed plant biomass represents the most abundant feedstock source on earth. Therefore, engineering TAG accumulation in nonseed tissues has important implications. Modulating PEAMT activity in concert with other regulatory enzymes regulating lipid homeostasis may be a promising avenue for manipulating TAG content in plants and improving their energy and nutritional value.
Possible Origin of Residual PC Synthesis in the Absence of Functional NMT1 and NMT3
In mammals and C. elegans, disruption of PC synthesis is lethal unless choline is provided exogenously (Li et al., 2005; Palavalli et al., 2006; Brendza et al., 2007). In P. falciparum, loss of PMT caused extreme growth and survival defects but was not lethal in the absence of choline supplementation; this was ascribed to some residual choline in the human erythrocytes (Witola et al., 2008). A triple nmt1 nmt2 nmt3 mutant will be instrumental in establishing whether the SDPM pathway is as critical in Arabidopsis and whether NMT2 activity can fully account for residual PC in the double nmt1 nmt3 mutant. Contrary to an earlier report (BeGora et al., 2010), a recent enzymatic study described the in vitro capacity of NMT2 to catalyze the conversion of PEA to PCho (Lee and Jez, 2017). Unlike in mammalian and yeast cells, no methylation activity over PE has been detected in plant cells (Marshall and Kates, 1973; Datko and Mudd, 1988a; McNeil et al., 2000). Moreover, the recently cloned PMLTs from Arabidopsis and soybean can only methylate PtdMMEA and PtdDMEA but not PE (Keogh et al., 2009). Under the current understanding of phospholipid synthesis in plants, this makes the phospho-base methylation of PEA by NMT enzymes absolutely necessary for the production of any PC at all in Arabidopsis cells. However, Liu et al. (2015) recently proposed a novel methylation pathway that might indirectly convert PE to PC in Arabidopsis. LysoPE derived from PE is methylated to lysoPC, which is then converted to PC by lysophosphatidylcholine acyltransferase or by lysophosphatidylcholine transacylase. The presence of such a pathway needs further investigation. In mammalian cells, PS is synthesized by base-exchange reactions in which the choline moiety of PC or the ethanolamine moiety of PE is replaced by l-Ser catalyzed by phosphatidylserine synthase (PSS; Kuge and Nishijima, 2003; Vance, 2008; Vance and Tasseva, 2013). These reactions are reversible, so that PC and PE also can be formed from PS. Purified PSS1 from Chinese hamster (Cricetulus griseus) can catalyze choline base exchange with the Ser moiety of PS to PC (Tomohiro et al., 2009). Arabidopsis contains a single PSS1 homologous to mammalian enzymes, and it has been suggested to have ethanolamine and Ser base-exchange activity but to lack choline base-exchange activity (Yamaoka et al., 2011). Even if PSS1 can catalyze the choline base exchange, this pathway could not then contribute to net PC synthesis in Arabidopsis, unless de novo synthesis of the free choline base does not totally rely on the SDPM methylation pathway, as currently thought. Characterization of a triple nmt1 nmt2 nmt3 mutant will help in definitely proving whether PC synthesis absolutely requires PEAMT activity.
In summary, the relative contribution of the phospho-base methylation pathway to de novo PC biosynthesis in plants has remained a pending question. Our data establish that it is of paramount importance and provide the first comparative functional analysis of two PEAMT family members in a plant species. With its dramatically altered phenotypes and phospholipid profiles, the nmt1 nmt3 mutant provides a powerful tool for further dissecting the critical regulatory roles of the methylation pathway and its products in lipid metabolism, signaling, and development in plants. High-resolution lipidomic analysis and lipid fluxomics coupled with high-resolution cellular, molecular, and developmental analyses, under varying environmental conditions, will greatly assist progress in the mechanistic understanding of PEAMT isoform functional specificity in different cell and organ types.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used as the wild type. T-DNA insertion lines nmt1 (SALK_036291) and SAIL_22_D10 were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org/abrc/); nmt3 (GABI-Kat_109F02) was obtained from GABI-Kat (Rosso et al., 2003; https://www.gabi-kat.de/). Genotyping primers are listed in Supplemental Table S2. Primers 1988, 1989, and 1987 were used to genotype for the presence of T-DNA insertion in nmt1. Primers 3157, 3158, and 2681 were used to genotype SAIL_22_D10. Primers 451, 452, and 455 were used to genotype for the presence of the T-DNA insertion in nmt3. Primer sequences are shown in Supplemental Table S2.
Double mutants nmt1 nmt3, nmt1+/− nmt3, and nmt1 nmt3+/− were created via crossing nmt1 and nmt3, and the F2 progeny were screened on medium supplemented with 40 mg L−1 kanamycin for separating seedlings carrying the nmt1 or NMT1 allele or 5 mg L−1 sulfadiazine for separating seedlings carrying the nmt3 or NMT3 allele.
For seedlings grown on plates, seeds were surface sterilized with (0.12% [v/v] sodium hypochlorite and 90% [v/v] ethanol for 5 min, washed once with 100% ethanol, and dried) and sown on one-half-strength Murashige and Skoog salt medium (Murashige and Skoog, 1962) with 1% (w/v) Suc and 1.2% (w/v) agar (Bacto). After 2 to 4 d of stratification at 4°C in the dark, plates were placed vertically in a controlled growth chamber set at 21°C and providing 120 ± 10 μmol m−2 s−1 light intensity for 10 h of light/14 h of dark unless specified otherwise.
For plants grown in soil, seeds were stratified at 4°C for 2 to 4 d, sown directly on the soil surface, and grown in controlled growth chambers set as above except for the photoperiod, which was raised to 16 h.
Transgenic Plants
NMT1 and NMT3 genomic DNA sequences were amplified by PCR using primer pairs 1764/1765 and 1768/1769, respectively (Supplemental Table S2). The amplified PCR products were cloned into pDONR207 via BP reaction according to the manufacturer’s instructions (ThermoFisher Scientific), resulting in pDONR207:NMT1gDNA and pDONR207:NMT3gDNA. The NMT1 promoter used to drive NMT1 and NMT3 expression was amplified by PCR using primer pair 1793/1794. The PCR product was cloned into pDONRP4P1r via BP reaction, resulting in pDONRp4p1r:NMT1Pro1. To facilitate the fusion of NMT1 promoter with the NMT1 or NMT3 gene, we first created a multisite Gateway vector, pWHC100, by replacing the attB1 site of pMDC99 (Curtis and Grossniklaus, 2003) with the attB4 site. A ccdB cassette was amplified from pMDC99 with primers 1795/1796 and digested with KpnI and SacI, followed by ligation to pMDC99 digested by KpnI and SacI, yielding pWHC100 vector. NMT1Pro1:gNMT1 and NMT1Pro1:gNMT3 fusions were generated by recombining pDONR207:NMT1gDNA and pDONR207:NMT3gDNA, respectively, with pDONRP4P1r:NMT1Pro1 into the multisite destination vector pWHC100, to yield the final pWHC100:NMT1Pro1:NMT1gDNA and pWHC100:NMT1Pro1:NMT3gDNA vectors.
To generate the ProNMT1:GUS and ProNMT3:GUS promoter fusion, a 1.9-kb fragment spanning −1,465 to +473 bp of NMT1 and a 5.6-kb fragment from −3,488 to +2,162 bp of NMT3 were amplified by primers 704/705 and 708/709, respectively. The amplified 1.9-kb fragment of NMT1 was digested with EcoRV and BamHI restriction enzymes and transferred to the EcoRV and BamHI sites of pENTR1A, resulting in pENTR1A:ProNMT1 plasmid. The amplified 5.6-kb fragment of NMT3 was digested with XhoI and BamHI and ligated to the pENTR1A vector BamHI and XhoI sites, resulting in pENTR1A:ProNMT3. The entry vectors were recombined with binary vector pMDC163 (Curtis and Grossniklaus, 2003) via LR reaction according to the manufacturer’s instruction to create expression vectors pMDC163:ProNMT1 and pMDC163:ProNMT3.
The expression vectors were transformed into Agrobacterium tumefaciens (strain GV3101) via electroporation. The A. tumefaciens-mediated floral dip method (Davis et al., 2009) was used for Arabidopsis transformation. NMT1Pro:NMT1gDNA and NMT1Pro:NMT3gDNA constructs were introduced into the nmt1 mutant background. NMT1Pro:GUS and NMT3Pro:GUS constructs were transferred into the Columbia-0 background.
Southern-Blot Analysis
Genomic DNA isolated from the nmt3 mutant using the CTAB method was digested with several restriction endonuclease enzymes and separated on 0.8% (w/v) agarose with 0.5× TBE buffer. The gel was blotted onto a nylon membrane and hybridized with a [32P]dCTP-labeled probe. The ampicillin probe was isolated by PCR with primers 648 AmpU (5′-ATGTATCCGCTCATGAGACAATAACCCT-3′) and 649 AmpL (5′-TTACCAATGCTTAATCAGTGAGGCACCTA-3′) using the Rediprime II random prime labeling system (Amersham Biosciences). Following hybridization, the membrane was washed twice (2× SSC, 0.1% SDS and 0.2× SSC, 0.1% SDS, at 65°C for 10–20 min) and analyzed by Bio-Rad autoradiography.
Transgenic Yeasts
To generate ProGal:NMT1 and ProGal:NMT3 plasmids, the coding regions of NMT1 and NMT3 were amplified by PCR using primer pairs 2178/2193 and 2211/2212, respectively, and cloned into pDONR207 using Gateway technology (ThermoFisher Scientific). These clones were transferred to pYES-DEST52 vector (ThermoFisher Scientific), resulting in pYES-DEST52:NMT1 and pYES-DEST52:NMT3, respectively. These vectors as well as the empty vector pYES-DEST52 were transformed into Saccharomyces cerevisiae pem1Δ pem2Δ (MATα hisΔ3I leu2Δ0 ura3Δ0 pem1::Kanr -pem2::Kanr; Pessi et al., 2005).
For spotting assays, transformed yeast single colonies were first grown on liquid YNB medium with 2% (w/v) Glc, 1 mm choline, and amino acids His (20 µg mL−1), Leu (30 mg mL−1), and Met (20 µg mL−1) at 28°C, at 200 rpm agitation, until OD600 reached about 1. Yeast cells were harvested by centrifugation at 10,000g for 5 min followed by three rinses with sterile water. OD600 was adjusted to 1 and 0.1 with sterile water. Ten microliters of yeast cells was then spotted on solid YNB medium supplemented with amino acids as above and 2% Glc or 2% Gal, as indicated. Choline (1 mm) was supplemented as indicated. Plates were incubated at 37°C for 2 to 6 d.
Yeast Protein Extraction and Measurement of PEA Methylation Activity by 31P-NMR
S. cerevisiae pem1Δ pem2Δ cells harboring pYESDEST52, pYES-DEST52:NMT1, or pYESDEST52:NMT3 were grown in liquid YNB medium with 2% Gal, 1 mm choline, and amino acids His (20 µg mL−1), Leu (30 mg mL−1), and Met (20 µg mL−1) at 28°C, at 200 rpm agitation, until OD600 reached about 1. Yeast crude protein extracts were obtained as reported previously (BeGora et al., 2010). Extraction buffer containing 100 mm HEPES-KOH (pH 8.02), 1 mm Na2EDTA, and 5 mm DTT (HED buffer) was prepared in heavy water. Yeast cells were pelleted by centrifuging at 3,500g for 10 min. Cell pellets were rinsed once with HED buffer before disruption by glass beads in 400 µL of HED buffer. Cell debris were removed by centrifugation at 12,000g for 10 min. The protein concentrations in these cell-free crude extracts were determined by Bradford assay (Bradford, 1976). A total of 2.8 mg of total proteins was desalted using Zeba Spin Desalting Columns, 40K MWCO, according to the manufacturer’s instructions (ThermoFisher Scientific). 31P-NMR was used to monitor PEA conversion into PCho following initiation of the reaction by adding 5 mm PEA (Sigma-Aldrich; catalog no. P0503) and 15 mm SAM (Sigma-Aldrich; catalog no. A7007) substrates and 5 mm methylphosphonate (Sigma-Aldrich; catalog no. 289868) as an internal standard (Rueppel and Marvel, 1976) to the desalted proteins in a final volume of 400 µL in HED buffer (time zero).
31P-NMR spectra of these reaction mixtures, and of PEA and PCho (Sigma-Aldrich; catalog no. P0378) standards (5 mm in 400 µL of HED buffer), were recorded at 162 MHz on a Bruker Avance-400 instrument using a 5-mm probe operating at 25°C. All FIDS (free induction decay) outputs were accumulated across 400 ppm, using 64 iterations, collecting 32K data points with a pulse delay of 5 s. The decoupler was turned on during acquisition. All spectra were referenced to the chemical shift of the internal standard methylphosphonate at pH 8.2 (23.29 ppm).
Reverse Transcription Quantitative PCR
Total RNA was extracted from various tissues using TRI Reagent (Sigma-Aldrich). mRNA was isolated from total RNA using oligo(dT)25 magnetic beads (ThermoFisher Scientific) and reverse transcribed into cDNA using MMLV reverse transcriptase (Promega) in a total volume of 20 µL containing 6.25 ng µL−1 oligo(dT)15 (Promega) and 1.25 ng µL−1 random hexamers (ThermoFisher Scientific). Reverse transcription was carried out at 37°C for 30 min, 42°C for 30 min, and 50°C for 2 min. A total of 2.5 µL of 10× diluted cDNA was used for quantitative PCR in a total volume of 10 µL with FastStart Universal SYBR Green Master Mix (Roche) using the Applied Biosystems ViiA 7 Real-Time PCR System. A standard thermal profile was used: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Gene expression was expressed relative to the geometric mean of APT1, PDF2, and GAPDH reference genes. The primers used for reverse transcription quantitative PCR are listed in Supplemental Table S3.
Microscopy Analysis
For GUS staining, whole seedlings, rosette, flowers, and stems and roots were incubated in acetone for 20 min on ice and then rinsed briefly in GUS staining solution without X-Gluc (50 mm sodium phosphate buffer, pH 7, 10 mm potassium ferrocyanide, 10 mm potassium ferricyanide, and 10 mm EDTA), followed by transfer to GUS staining solution containing 1 mg mL−1 X-Gluc. Vacuum was immediately applied for 15 min twice. Seedlings were incubated at 37°C for 1.5 h in GUS staining solution, except for ProNMT1:GFP/GUS roots (16 h); rosettes, floral organs, and stem were incubated in GUS staining solution supplemented with 0.1% Triton X-100 for 16 h. After GUS staining, chlorophyll was removed using 70% ethanol. Samples were examined by DIC light microscopy using a Leica DM6000 microscope fitted with a Leica DFC 310FX camera (Leica Instruments). Eighteen-day-old rosettes were scanned with an Epson perfection v600 scanner. Florets and stems were photographed using a Leica M205 FA stereomicroscope with a Leica DFC 550 color camera (Leica Instruments).
For analysis of root anatomy, roots were briefly stained in 10 µm propidium iodide and observed by confocal or light microscopy, using a Leica TCS SP8 or Leica DM6000 microscope, respectively. Mature cell length was determined as the average length of six to 10 contiguous cells in the mature zone of the primary root on eight to 12 replicates per genotype. The boundary between meristem and elongation zone was determined from the profiles of cell length with respect to distance from the quiescent center, as classically done. In the meristem, average cell length does not change with position and individual cell lengths vary between l (just before the cell divides) and l/2 (just after division). The end of the meristem was determined as the position where cell lengths started to increase sharply, and the boundary between elongation and mature zones was determined as the position where cell lengths were not increasing anymore.
For ovule observation, pistils were slit open longitudinally using a stereomicroscope and fixed in 50% ethanol and 10% acetic acid for 2 h at room temperature or 4°C overnight, followed by clearing in chloral hydrate:glycerol:water (20 g:1 mL:6 mL). Hoyer’s solution containing choral hydrate:glycerol:water:gum arabic (40 g:2 mL:12 mL:5 g) was used for mounting the cleared ovules. DIC images were taken with a Leica DM6000 microscope fitted a Leica DFC 310FX camera (Leica Instruments).
To determine the embryogenic stage of developing seeds, embryos were cleared and mounted in Hoyer’s solution. DIC images were taken as described above.
For pollen staining, anthers were collected and immediately fixed in Carnoy’s solution (ethanol:chloroform:acetic acid [v/v/v], 6:3:1) and kept at 4°C until samples were examined. Pollen was stained with a modified version of Alexander’s stain (Peterson et al., 2010). Stained pollen grains were observed and photographed as described above.
Lipid Extraction and Quantification
Total lipids were extracted from freeze-dried tissues as described previously (Petrie et al., 2012). Samples were first homogenized in 300 µL of methanol using TissueLyser II followed by a brief centrifugation and addition of 600 µL of chloroform. After homogenization and brief centrifugation, 300 µL of KCl was added, followed by homogenization. The mixture was then centrifuged at 12,000g for 5 min, and the lower phase was collected. The remaining phase was extracted again through the addition of 500 µL of chloroform and then homogenized and centrifuged for 5 min at 12,000g. The organic phase was collected and pooled with the phase collected earlier and dried under a nitrogen flow.
Liquid Chromatography-Mass Spectrometry Analysis
Chloroform extracts of lipids from plant tissue were dried under nitrogen and resuspended in chloroform to 100 mg mL−1, then diluted to 1 mg mL−1 in butanol:methanol (1:1, v/v) and analyzed by liquid chromatography-mass spectrometry, based on previously described methods (Reynolds et al., 2015). Briefly, a Waters BEH C8 (100 mm × 2.1 mm, 2.7 µm) was used to separate the lipids on an Agilent 1290 series liquid chromatograph and 6490 triple quadrupole liquid chromatograph-mass spectrometer with Jet Stream ionization with a binary gradient flow rate of 0.2 mL min−1. The mobile phases were as follows: water:acetonitrile (10:90, v/v) with 10 mm ammonium formate and 0.2% formic acid (A); and water:acetonitrile:isopropanol (5:15:80, v/v/v) with 10 mm ammonium formate and 0.2% formic acid (B). The gradient was held at 1% mobile phase B for 2 min before being raised to 20% B at 5 min, 60% B at 8 min, 70% B at 12 min, and final increase to 90% B at 14 min, before reequilibration to 1% B. The ionization conditions were as follows: nebulizer at 45 p.s.i., gas temperature and flow at 250°C and 14 L min−1, sheath gas and flow at 250°C and 11 L min−1, and capillary and nozzle voltages at 3,000 and 1,000 V. Time-dependent multiple reaction monitoring lists were developed after initial screening of pooled samples on an Agilent quadrupole time of flight device using the same gradient and ionization conditions. The positive ion ammonium adducts of MGDG, DGDG, DAG, and TAG lipid species were fragmented using a collision energy of 28 V for all lipid classes except DAG, where a collision energy of 14 V was used. Multiple reaction monitoring lists were based on the lipid species detected by quadrupole time of flight and the neutral loss of the following major fatty acids: 16:0, 16:3, 18:0, 18:1, 18:2, and 18:3. The molecular anions [M–H]− of PE, PI, and PG were fragmented with a collision energy of 36 V, and the carboxylate anions of the acyl chains were used to identify lipid species and acyl chain composition (Pulfer and Murphy, 2003). The positive ion PC and lysoPC species hydrogen adducts were quantified by the characteristic 184 m/z phosphatidyl head group.
External standards were purchased from Avanti Polar Lipids (PE 34:1 16:0/18:1, PG 36:1 18:0/18:1, PI 34:2 16:0/18:2, PC 36:2 18:1/18:1, MGDG 34:6 16:3/18:3, DGDG 34:3 16:0/18:3, DAG 36:2 18:1/18:1, and TAG 54:3 18:1/18:1/18:1) and standard curves ranging from 0.001 to 100 µg mL−1 used to quantify all lipid species within the class. Peak integration and analysis was conducted using Agilent Quantitative Analysis software and exported for further analysis using the dplyr and ggplot2 packages from R using RStudio (Horton and Kleinman, 2015).
Accession Numbers
Sequence data for this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: NMT1 (At3g18000), NMT2/PMEAMT (At1g48600), NMT3 (At1g73600), APT1 (At1g27450), PDF2/PP2AA3 (At1g13320), and GAPDH (AT1G13440).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of Arabidopsis NMT3 with NMT1 and NMT2 and other previously characterized plant NMT/PEAMT enzymes.
Supplemental Figure S2. Complementation of the choline auxotrophic yeast pem1Δ pem2Δ mutant by NMT3.
Supplemental Figure S3. Characterization of the nmt1 and nmt3 T-DNA lines, SALK_036291 and GABI-KAT_109F02, respectively.
Supplemental Figure S4. NMT1 promoter activity at the root tip and along the whole root up to the rosette in ProNMT1:GUS and PproNMT3:GUS reporter lines.
Supplemental Figure S5. NMT1 and NMT3 interact to control aerial development.
Supplemental Figure S6. Visual phenotypes of mature seeds.
Supplemental Figure S7. PG and PI molecular species in wild-type, nmt1, nmt3, and nmt1 nmt3 seedlings.
Supplemental Figure S8. MGDG molecular species in wild-type, nmt1, nmt3, and nmt1 nmt3 seedlings.
Supplemental Figure S9. DGDG molecular species in wild-type, nmt1, nmt3, and nmt1 nmt3 seedlings.
Supplemental Figure S10. TAG molecular species in wild-type, nmt1, nmt3, and nmt1 nmt3 seedlings.
Supplemental Figure S11. Transcript abundance of genes involved in TAG biosynthesis and degradation and in lipid turnover.
Supplemental Table S1. Percentage identity values among the three Arabidopsis NMT proteins and the characterized PEAMT enzymes from spinach and bread wheat.
Supplemental Table S2. Cloning and genotyping primers.
Supplemental Table S3. Primers used for reverse transcription quantitative PCR.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dr. Choukri Ben Mamoun (Yale University) for the gift of the pem1Δ pem2Δ yeast mutant strain.
Footnotes
This research work was supported by the Australian National University, Grains Research and Development Corporation grants ANU0014 and ANU00023 (J.M.), and a postgraduate scholarship from the Ministry of Science, Research, and Technology of Iran (H.S.).
Articles can be viewed without a subscription.
References
- Alatorre-Cobos F, Cruz-Ramírez A, Hayden CA, Pérez-Torres CA, Chauvin AL, Ibarra-Laclette E, Alva-Cortés E, Jorgensen RA, Herrera-Estrella L (2012) Translational regulation of Arabidopsis XIPOTL1 is modulated by phosphocholine levels via the phylogenetically conserved upstream open reading frame 30. J Exp Bot 63: 5203–5221 [DOI] [PubMed] [Google Scholar]
- Bates PD, Browse J (2011) The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J 68: 387–399 [DOI] [PubMed] [Google Scholar]
- Bates PD, Durrett TP, Ohlrogge JB, Pollard M (2009) Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol 150: 55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BeGora MD, Macleod MJ, McCarry BE, Summers PS, Weretilnyk EA (2010) Identification of phosphomethylethanolamine N-methyltransferase from Arabidopsis and its role in choline and phospholipid metabolism. J Biol Chem 285: 29147–29155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencivenga S, Colombo L, Masiero S (2011) Cross talk between the sporophyte and the megagametophyte during ovule development. Sex Plant Reprod 24: 113–121 [DOI] [PubMed] [Google Scholar]
- Benning C. (2009) Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu Rev Cell Dev Biol 25: 71–91 [DOI] [PubMed] [Google Scholar]
- Benning C, Xu C, Awai K (2006) Non-vesicular and vesicular lipid trafficking involving plastids. Curr Opin Plant Biol 9: 241–247 [DOI] [PubMed] [Google Scholar]
- Bolognese CP, McGraw P (2000) The isolation and characterization in yeast of a gene for Arabidopsis S-adenosylmethionine:phospho-ethanolamine N-methyltransferase. Plant Physiol 124: 1800–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier-Navé P, Benveniste P, Oelkers P, Sturley SL, Schaller H (2000) Expression in yeast and tobacco of plant cDNAs encoding acyl CoA:diacylglycerol acyltransferase. Eur J Biochem 267: 85–96 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bremer J, Greenberg DM (1961) Methyl transfering enzyme system of microsomes in the biosynthesis of lecithin (phosphatidylcholine). Biochim Biophys Acta 46: 205–216 [Google Scholar]
- Brendza KM, Haakenson W, Cahoon RE, Hicks LM, Palavalli LH, Chiapelli BJ, McLaird M, McCarter JP, Williams DJ, Hresko MC, et al. (2007) Phosphoethanolamine N-methyltransferase (PMT-1) catalyses the first reaction of a new pathway for phosphocholine biosynthesis in Caenorhabditis elegans. Biochem J 404: 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Ohlrogge JB (2012) Compartmentation of triacylglycerol accumulation in plants. J Biol Chem 287: 2288–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron JBF, Breton G, Danyluk J, Muzac I, Ibrahim RK, Sarhan F (2002) Molecular and biochemical characterization of a cold-regulated phosphoethanolamine N-methyltransferase from wheat. Plant Physiol 129: 363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TH, Murata N (2008) Glycinebetaine: an effective protectant against abiotic stress in plants. Trends Plant Sci 13: 499–505 [DOI] [PubMed] [Google Scholar]
- Cheng CY, Mathews DE, Schaller GE, Kieber JJ (2013) Cytokinin-dependent specification of the functional megaspore in the Arabidopsis female gametophyte. Plant J 73: 929–940 [DOI] [PubMed] [Google Scholar]
- Chevalier É, Loubert-Hudon A, Zimmerman EL, Matton DP (2011) Cell-cell communication and signalling pathways within the ovule: from its inception to fertilization. New Phytol 192: 13–28 [DOI] [PubMed] [Google Scholar]
- Christensen CA, King EJ, Jordan JR, Drews GN (1997) Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex Plant Reprod 10: 49–64 [Google Scholar]
- Craddock CP, Adams N, Bryant FM, Kurup S, Eastmond PJ (2015) PHOSPHATIDIC ACID PHOSPHOHYDROLASE regulates phosphatidylcholine biosynthesis in Arabidopsis by phosphatidic acid-mediated activation of CTP:PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE activity. Plant Cell 27: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A, López-Bucio J, Ramírez-Pimentel G, Zurita-Silva A, Sánchez-Calderon L, Ramírez-Chávez E, González-Ortega E, Herrera-Estrella L (2004) The xipotl mutant of Arabidopsis reveals a critical role for phospholipid metabolism in root system development and epidermal cell integrity. Plant Cell 16: 2020–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z, Vance JE, Chen MH, Voelker DR, Vance DE (1993) Cloning and expression of a novel phosphatidylethanolamine N-methyltransferase: a specific biochemical and cytological marker for a unique membrane fraction in rat liver. J Biol Chem 268: 16655–16663 [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datko AH, Mudd SH (1988a) Phosphatidylcholine synthesis: differing patterns in soybean and carrot. Plant Physiol 88: 854–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datko AH, Mudd SH (1988b) Enzymes of phosphatidylcholine synthesis in lemna, soybean, and carrot. Plant Physiol 88: 1338–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AM, Hall A, Millar AJ, Darrah C, Davis SJ (2009) Streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marcos Lousa C, van Roermund CWT, Postis VLG, Dietrich D, Kerr ID, Wanders RJA, Baldwin SA, Baker A, Theodoulou FL (2013) Intrinsic acyl-CoA thioesterase activity of a peroxisomal ATP binding cassette transporter is required for transport and metabolism of fatty acids. Proc Natl Acad Sci USA 110: 1279–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demesa-Arévalo E, Vielle-Calzada JP (2013) The classical arabinogalactan protein AGP18 mediates megaspore selection in Arabidopsis. Plant Cell 25: 1274–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Ursache R, Campilho A, Miyashima S, Belevich I, O’Regan S, Mullendore DL, Yadav SR, Lanz C, Beverina L, et al. (2014) CHOLINE TRANSPORTER-LIKE1 is required for sieve plate development to mediate long-distance cell-to-cell communication. Nat Commun 5: 4276. [DOI] [PubMed] [Google Scholar]
- Devaiah SP, Roth MR, Baughman E, Li M, Tamura P, Jeannotte R, Welti R, Wang X (2006) Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry 67: 1907–1924 [DOI] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ. (2006) SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18: 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Quettier AL, Kroon JTM, Craddock C, Adams N, Slabas AR (2010) Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell 22: 2796–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo VA, McMeekin L, LeBlanc PJ (2011) Influence of phospholipid species on membrane fluidity: a meta-analysis for a novel phospholipid fluidity index. J Membr Biol 244: 97–103 [DOI] [PubMed] [Google Scholar]
- Figueiredo DD, Köhler C (2016) Bridging the generation gap: communication between maternal sporophyte, female gametophyte and fertilization products. Curr Opin Plant Biol 29: 16–20 [DOI] [PubMed] [Google Scholar]
- Ghosh AK, Chauhan N, Rajakumari S, Daum G, Rajasekharan R (2009) At4g24160, a soluble acyl-coenzyme A-dependent lysophosphatidic acid acyltransferase. Plant Physiol 151: 869–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Rhodes D (1983) 14C tracer evidence for synthesis of choline and betaine via phosphoryl base intermediates in salinized sugarbeet leaves. Plant Physiol 71: 692–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama T, Takeuchi H (2015) The mechanism and key molecules involved in pollen tube guidance. Annu Rev Plant Biol 66: 393–413 [DOI] [PubMed] [Google Scholar]
- Hitz WD, Rhodes D, Hanson AD (1981) Radiotracer evidence implicating phosphoryl and phosphatidyl bases as intermediates in betaine synthesis by water-stressed barley leaves. Plant Physiol 68: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton NJ, Kleinman K (2015) Using R and RStudio for Data Management, Statistical Analysis and Graphics. Chapman and Hall/CRC Press, Boca Raton, FL [Google Scholar]
- Hsu SC, Belmonte MF, Harada JJ, Inoue K (2010) Indispensable roles of plastids in Arabidopsis thaliana embryogenesis. Curr Genomics 11: 338–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck N, Moore JM, Federer M, Grossniklaus U (2003) The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130: 2149–2159 [DOI] [PubMed] [Google Scholar]
- James CN, Horn PJ, Case CR, Gidda SK, Zhang D, Mullen RT, Dyer JM, Anderson RGW, Chapman KD (2010) Disruption of the Arabidopsis CGI-58 homologue produces Chanarin-Dorfman-like lipid droplet accumulation in plants. Proc Natl Acad Sci USA 107: 17833–17838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost R, Berkowitz O, Shaw J, Masle J (2009) Biochemical characterization of two wheat phosphoethanolamine N-methyltransferase isoforms with different sensitivities to inhibition by phosphatidic acid. J Biol Chem 284: 31962–31971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T, Takahashi S, Shinozaki K (2001) Involvement of a novel Arabidopsis phospholipase D, AtPLDdelta, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J 26: 595–605 [DOI] [PubMed] [Google Scholar]
- Kelly AA, Quettier AL, Shaw E, Eastmond PJ (2011) Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Plant Physiol 157: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, van Erp H, Quettier AL, Shaw E, Menard G, Kurup S, Eastmond PJ (2013) The sugar-dependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Plant Physiol 162: 1282–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EP, Weiss SB (1956) The function of cytidine coenzymes in the biosynthesis of phospholipides. J Biol Chem 222: 193–214 [PubMed] [Google Scholar]
- Keogh MR, Courtney PD, Kinney AJ, Dewey RE (2009) Functional characterization of phospholipid N-methyltransferases from Arabidopsis and soybean. J Biol Chem 284: 15439–15447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaki T, Yamashita S (1987) Yeast phosphatidylethanolamine methylation pathway: cloning and characterization of two distinct methyltransferase genes. J Biol Chem 262: 15428–15435 [PubMed] [Google Scholar]
- Kraner ME, Link K, Melzer M, Ekici AB, Uebe S, Tarazona P, Feussner I, Hofmann J, Sonnewald U (2017) Choline transporter-like1 (CHER1) is crucial for plasmodesmata maturation in Arabidopsis thaliana. Plant J 89: 394–406 [DOI] [PubMed] [Google Scholar]
- Kuge O, Nishijima M (2003) Biosynthetic regulation and intracellular transport of phosphatidylserine in mammalian cells. J Biochem 133: 397–403 [DOI] [PubMed] [Google Scholar]
- Lee SG, Jez JM (2017) Conformational changes in the di-domain structure of Arabidopsis phosphoethanolamine methyltransferase leads to active-site formation. J Biol Chem 292: 21690–21702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Na K, Lee HJ, Lee EY, Paik YK (2011) Contribution of sams-1 and pmt-1 to lipid homoeostasis in adult Caenorhabditis elegans. J Biochem 149: 529–538 [DOI] [PubMed] [Google Scholar]
- Li Z, Agellon LB, Vance DE (2005) Phosphatidylcholine homeostasis and liver failure. J Biol Chem 280: 37798–37802 [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2010) Acyl-lipid metabolism. The Arabidopsis Book 8: e0133, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2013) Acyl-lipid metabolism. The Arabidopsis Book 11: e0161, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Liu YC, Nakamura Y (2015) The choline/ethanolamine kinase family in Arabidopsis: essential role of CEK4 in phospholipid biosynthesis and embryo development. Plant Cell 27: 1497–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang G, Wang X (2015) Role of aminoalcoholphosphotransferases 1 and 2 in phospholipid homeostasis in Arabidopsis. Plant Cell 27: 1512–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los DA, Murata N (2004) Membrane fluidity and its roles in the perception of environmental signals. Biochim Biophys Acta 1666: 142–157 [DOI] [PubMed] [Google Scholar]
- Maizel JV, Benson AA, Tolbert NE (1956) Identification of phosphoryl choline as an important constituent of plant sap. Plant Physiol 31: 407–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanovic N, Streith I, Wolinski H, Rechberger G, Kohlwein SD, Tehlivets O (2008) S-Adenosyl-L-homocysteine hydrolase, key enzyme of methylation metabolism, regulates phosphatidylcholine synthesis and triacylglycerol homeostasis in yeast: implications for homocysteine as a risk factor of atherosclerosis. J Biol Chem 283: 23989–23999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall MO, Kates M (1973) Biosynthesis of phosphatidyl ethanolamine and phosphatidyl choline in spinach leaves. FEBS Lett 31: 199–202 [DOI] [PubMed] [Google Scholar]
- Martin BA, Tolbert NE (1983) Factors which affect the amount of inorganic phosphate, phosphorylcholine, and phosphorylethanolamine in xylem exudate of tomato plants. Plant Physiol 73: 464–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw P, Henry SA (1989) Mutations in the Saccharomyces cerevisiae opi3 gene: effects on phospholipid methylation, growth and cross-pathway regulation of inositol synthesis. Genetics 122: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil SD, Nuccio ML, Rhodes D, Shachar-Hill Y, Hanson AD (2000) Radiotracer and computer modeling evidence that phospho-base methylation is the main route of choline synthesis in tobacco. Plant Physiol 123: 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi J, Nakamura M, Nishida I (2006) Defects in CTP:PHOSPHORYLETHANOLAMINE CYTIDYLYLTRANSFERASE affect embryonic and postembryonic development in Arabidopsis. Plant Cell 18: 3370–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Wang X, Fu Z, Dai Y, Han C, Ouyang J, Bao F, Hu Y, Li J (2002) Silencing of phosphoethanolamine N-methyltransferase results in temperature-sensitive male sterility and salt hypersensitivity in Arabidopsis. Plant Cell 14: 2031–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd SH, Datko AH (1986) Phosphoethanolamine bases as intermediates in phosphatidylcholine synthesis by lemna. Plant Physiol 82: 126–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T. (2001) Phosphatidic acid: an emerging plant lipid second messenger. Trends Plant Sci 6: 227–233 [DOI] [PubMed] [Google Scholar]
- Munnik T, Irvine RF, Musgrave A (1998) Phospholipid signalling in plants. Biochim Biophys Acta 1389: 222–272 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nakamura Y. (2015) Function of polar glycerolipids in flower development in Arabidopsis thaliana. Prog Lipid Res 60: 17–29 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Andrés F, Kanehara K, Liu YC, Dörmann P, Coupland G (2014) Arabidopsis florigen FT binds to diurnally oscillating phospholipids that accelerate flowering. Nat Commun 5: 3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccio ML, Ziemak MJ, Henry SA, Weretilnyk EA, Hanson AD (2000) cDNA cloning of phosphoethanolamine N-methyltransferase from spinach by complementation in Schizosaccharomyces pombe and characterization of the recombinant enzyme. J Biol Chem 275: 14095–14101 [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J (1995) Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvar BL, Sangwan V, Omann F, Dhindsa RS (2000) Early steps in cold sensing by plant cells: the role of actin cytoskeleton and membrane fluidity. Plant J 23: 785–794 [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Alandete-Saez M, Bowman JL, Sundaresan V (2009) Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324: 1684–1689 [DOI] [PubMed] [Google Scholar]
- Palavalli LH, Brendza KM, Haakenson W, Cahoon RE, McLaird M, Hicks LM, McCarter JP, Williams DJ, Hresko MC, Jez JM (2006) Defining the role of phosphomethylethanolamine N-methyltransferase from Caenorhabditis elegans in phosphocholine biosynthesis by biochemical and kinetic analysis. Biochemistry 45: 6056–6065 [DOI] [PubMed] [Google Scholar]
- Park S, Gidda SK, James CN, Horn PJ, Khuu N, Seay DC, Keereetaweep J, Chapman KD, Mullen RT, Dyer JM (2013) The α/β hydrolase CGI-58 and peroxisomal transport protein PXA1 coregulate lipid homeostasis and signaling in Arabidopsis. Plant Cell 25: 1726–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi G, Choi JY, Reynolds JM, Voelker DR, Mamoun CB (2005) In vivo evidence for the specificity of Plasmodium falciparum phosphoethanolamine methyltransferase and its coupling to the Kennedy pathway. J Biol Chem 280: 12461–12466 [DOI] [PubMed] [Google Scholar]
- Peterson R, Slovin JP, Chen C (2010) A simplified method for differential staining of aborted and non-aborted pollen grains. Int J Plant Biol 1: 66–69 [Google Scholar]
- Petrie JR, Vanhercke T, Shrestha P, El Tahchy A, White A, Zhou XR, Liu Q, Mansour MP, Nichols PD, Singh SP (2012) Recruiting a new substrate for triacylglycerol synthesis in plants: the monoacylglycerol acyltransferase pathway. PLoS ONE 7: e35214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocký M, Eliás M, Profotová B, Novotná Z, Valentová O, Zárský V (2003) Phosphatidic acid produced by phospholipase D is required for tobacco pollen tube growth. Planta 217: 122–130 [DOI] [PubMed] [Google Scholar]
- Preitschopf W, Lückl H, Summers E, Henry SA, Paltauf F, Kohlwein SD (1993) Molecular cloning of the yeast OPI3 gene as a high copy number suppressor of the cho2 mutation. Curr Genet 23: 95–101 [DOI] [PubMed] [Google Scholar]
- Pulfer M, Murphy RC (2003) Electrospray mass spectrometry of phospholipids. Mass Spectrom Rev 22: 332–364 [DOI] [PubMed] [Google Scholar]
- Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA, Palanivelu R (2009) Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds KB, Taylor MC, Zhou XR, Vanhercke T, Wood CC, Blanchard CL, Singh SP, Petrie JR (2015) Metabolic engineering of medium-chain fatty acid biosynthesis in Nicotiana benthamiana plant leaf lipids. Front Plant Sci 6: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D, Hanson AD (1993) Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu Rev Plant Physiol Plant Mol Biol 44: 357–384 [Google Scholar]
- Ridgway ND, Vance DE (1987) Purification of phosphatidylethanolamine N-methyltransferase from rat liver. J Biol Chem 262: 17231–17239 [PubMed] [Google Scholar]
- Ridgway ND, Yao Z, Vance DE (1989) Phosphatidylethanolamine levels and regulation of phosphatidylethanolamine N-methyltransferase. J Biol Chem 264: 1203–1207 [PubMed] [Google Scholar]
- Rontein D, Nishida I, Tashiro G, Yoshioka K, Wu WI, Voelker DR, Basset G, Hanson AD (2001) Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme. J Biol Chem 276: 35523–35529 [DOI] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Rueppel ML, Marvel JT (1976) 1H and 31P NMR spectra of substituted methylphosphonic acids with indirect determination of 31P shifts. Org Magn Reson 8: 19–20 [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Browse J (1996) Dissecting desaturation: plants prove advantageous. Trends Cell Biol 6: 148–153 [DOI] [PubMed] [Google Scholar]
- Summers EF, Letts VA, McGraw P, Henry SA (1988) Saccharomyces cerevisiae cho2 mutants are deficient in phospholipid methylation and cross-pathway regulation of inositol synthesis. Genetics 120: 909–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutoh K, Sanuki N, Sakaki T, Imai R (2010) Specific induction of TaAAPT1, an ER- and Golgi-localized ECPT-type aminoalcoholphosphotransferase, results in preferential accumulation of the phosphatidylethanolamine membrane phospholipid during cold acclimation in wheat. Plant Mol Biol 72: 519–531 [DOI] [PubMed] [Google Scholar]
- Tabuchi T, Okada T, Azuma T, Nanmori T, Yasuda T (2006) Posttranscriptional regulation by the upstream open reading frame of the phosphoethanolamine N-methyltransferase gene. Biosci Biotechnol Biochem 70: 2330–2334 [DOI] [PubMed] [Google Scholar]
- Tomohiro S, Kawaguti A, Kawabe Y, Kitada S, Kuge O (2009) Purification and characterization of human phosphatidylserine synthases 1 and 2. Biochem J 418: 421–429 [DOI] [PubMed] [Google Scholar]
- Ueland PM. (2011) Choline and betaine in health and disease. J Inherit Metab Dis 34: 3–15 [DOI] [PubMed] [Google Scholar]
- Vance JE. (2008) Phosphatidylserine and phosphatidylethanolamine in mammalian cells: two metabolically related aminophospholipids. J Lipid Res 49: 1377–1387 [DOI] [PubMed] [Google Scholar]
- Vance JE, Tasseva G (2013) Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim Biophys Acta 1831: 543–554 [DOI] [PubMed] [Google Scholar]
- van den Brink-van der Laan E, Killian JA, de Kruijff B (2004) Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim Biophys Acta 1666: 275–288 [DOI] [PubMed] [Google Scholar]
- Waese J, Fan J, Pasha A, Yu H, Fucile G, Shi R, Cumming M, Kelley LA, Sternberg MJ, Krishnakumar V, et al. (2017) Eplant: visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell 29: 1806–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Jacobs RL, Watts JL, Rottiers V, Jiang K, Finnegan DM, Shioda T, Hansen M, Yang F, Niebergall LJ, et al. (2011) A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell 147: 840–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkey CJ, Donohue LR, Bronson R, Agellon LB, Vance DE (1997) Disruption of the murine gene encoding phosphatidylethanolamine N-methyltransferase. Proc Natl Acad Sci USA 94: 12880–12885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Moore TS (1990) Phosphatidylcholine biosynthesis in castor bean endosperm: purification and properties of cytidine 5′-triphosphate:choline-phosphate cytidylyltransferase. Plant Physiol 93: 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M, Harwood JL (1994) Alternative pathways for phosphatidylcholine synthesis in olive (Olea europaea L.) callus cultures. Biochem J 304: 463–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witola WH, El Bissati K, Pessi G, Xie C, Roepe PD, Mamoun CB (2008) Disruption of the Plasmodium falciparum PfPMT gene results in a complete loss of phosphatidylcholine biosynthesis via the serine-decarboxylase-phosphoethanolamine-methyltransferase pathway and severe growth and survival defects. J Biol Chem 283: 27636–27643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y, Yu Y, Mizoi J, Fujiki Y, Saito K, Nishijima M, Lee Y, Nishida I (2011) PHOSPHATIDYLSERINE SYNTHASE1 is required for microspore development in Arabidopsis thaliana. Plant J 67: 648–661 [DOI] [PubMed] [Google Scholar]
- Yunus IS, Liu YC, Nakamura Y (2016) The importance of SERINE DECARBOXYLASE1 (SDC1) and ethanolamine biosynthesis during embryogenesis of Arabidopsis thaliana. Plant J 88: 559–569 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]