The two Arabidopsis thaliana one-helix proteins form a trimeric complex with the PSII assembly factor HCF244 and contribute to the synthesis and assembly of the PSII core subunits.
Abstract
The members of the light-harvesting complex protein family, which include the one-helix proteins (OHPs), are characterized by one to four membrane-spanning helices. These proteins function in light absorption and energy dissipation, sensing light intensity, and triggering photomorphogenesis or the binding of chlorophyll and intermediates of chlorophyll biosynthesis. Arabidopsis (Arabidopsis thaliana) contains two OHPs, while four homologs (named high-light-induced proteins) exist in Synechocystis PCC6803. Various functions have been assigned to high-light-induced proteins, ranging from photoprotection and the assembly of photosystem I (PSI) and PSII to regulation of the early steps of chlorophyll biosynthesis, but little is known about the function of the two plant OHPs. Here, we show that the two Arabidopsis OHPs form heterodimers and that the stromal part of OHP2 interacts with the plastid-localized PSII assembly factor HIGH CHLOROPHYLL FLUORESCENCE244 (HCF244). Moreover, concurrent accumulation of the two OHPs and HCF244 is critical for the stability of all three proteins. In particular, the absence of OHP2 leads to the complete loss of OHP1 and HCF244. We used a virus-induced gene silencing approach to minimize the expression of OHP1 or OHP2 in adult Arabidopsis plants and revealed that OHP2 is essential for the accumulation of the PSII core subunits, while the other photosynthetic complexes and the major light-harvesting complex proteins remained unaffected. We examined the potential functions of the OHP1-OHP2-HCF244 complex in the assembly and/or repair of PSII and propose a role for this heterotrimeric complex in thylakoid membrane biogenesis.
Higher plants such as Arabidopsis (Arabidopsis thaliana) express a remarkably versatile family of light-harvesting complex (LHC) proteins, which include the eponymous light-harvesting complex proteins (LHCPs) that form the light-gathering antennae of PSI and PSII (Jansson, 1994). Furthermore, the S subunit of PSII (PsbS), the early light-inducible proteins (ELIPs), stress-enhanced proteins (SEP1 and SEP2), light-harvesting-like3 (SEP3 or LIL3), and the one-helix proteins (referred to as OHPs in plants and as high-light-inducible proteins [Hlips] in cyanobacteria) belong to this family. The common and defining property of all LHC proteins is a hydrophobic, thylakoid membrane-spanning α-helix called the LHC motif, which is involved in chlorophyll binding by the antennal LHCPs (Liu et al., 2004). In principle, all or almost all members of the LHC family should be capable of binding chlorophyll. Indeed, recent publications provide evidence that the Hlips and LIL3 proteins can bind chlorophyll at least in vitro (Mork-Jansson et al., 2015; Staleva et al., 2015; Hey et al., 2017; Shukla et al., 2018).
The various members of the LHC family contain differing numbers of transmembrane-spanning domains. PsbS, which is involved in nonphotochemical quenching (Li et al., 2000), possesses four such domains, two of which exhibit LHC motifs. LHCPs and ELIPs also contain two LHC motifs among their three transmembrane domains. ELIP mRNAs and their protein products are virtually undetectable in unstressed leaf material and accumulate rapidly but transiently upon exposure to elevated light intensities as well as upon the induction of photomorphogenesis in etiolated seedlings (Adamska, 1997; Dhingra et al., 2006). In contrast, SEP-encoding transcripts are detectable even under low light intensities, but, like the ELIP mRNAs, they strongly accumulate upon exposure to elevated light intensities (Heddad and Adamska, 2000). LIL3 has been shown to be functionally connected with chlorophyll synthesis, as loss of both LIL3 isoforms leads to destabilization of the enzymes geranylgeranyl reductase (CHLP) and protochlorophyllide oxidoreductase (POR; Tanaka et al., 2010; Hey et al., 2017). SEPs and LIL3 each contain two transmembrane domains, one of which resembles the LHC motif.
The shortest LHC members in Arabidopsis are OHP1 and OHP2. They contain a single transmembrane domain, which encompasses an LHC motif. Their respective T-DNA insertion mutants are severely compromised in seedling development, and photosynthesis is impaired drastically in young seedlings (Beck et al., 2017). Hlips, the prokaryotic homologs of the OHPs, are the sole representatives of the LHC family in cyanobacteria (Engelken et al., 2010). The Hlips are considered as the ancestors of all eukaryotic members of the LHC family, which presumably evolved from Hlips/OHPs by gene duplication and deletion events (Montané and Kloppstech, 2000; Engelken et al., 2010). The genus Synechocystis encodes four different Hlips (plus the C terminus of ferrochelatase) with a variety of potential functions: as the HliA-D quadruple knockout mutant fails to form trimeric PSI under high-light conditions, a photoprotective function of Hlips for PSI has been proposed (Wang et al., 2008). However, the deletion of Hlips in a PSI-less strain led to oxidative damage of PSII, suggesting that their photoprotective role extends to PSII (Tibiletti et al., 2018). Hlips also have been hypothesized to serve as pigment suppliers during the assembly and repair of PSII (Yao et al., 2007). Furthermore, Hlips have been linked to the control of chlorophyll biosynthesis (Xu et al., 2002). Specifically, those authors suggested that Hlips have a positive effect on the rate-limiting steps at the beginning of chlorophyll biosynthesis.
More recently, Hlips were reported to interact in a putative chlorophyll-binding complex with chlorophyll synthase (CHLG) and the photosystem (PS) assembly factor Ycf39 and, thus, to participate in an early assembly intermediate of PSII complex formation in Synechocystis spp. (Chidgey et al., 2014; Knoppová et al., 2014). CHLG esterifies chlorophyllide with phytyl pyrophosphate, thus producing chlorophyll a for incorporation into the core proteins of PSI and PSII (Schmid et al., 2001). Ycf39 (encoded by the gene slr0399) was proposed as a PSII assembly factor (Ermakova-Gerdes and Vermaas, 1999). The Hlip isoform HliD purified from Synechocystis PCC6803 was found to be associated with chlorophyll a and β-carotene and displayed energy dissipation qualities (Staleva et al., 2015), and homooligomers of HliC were shown to be able to bind pigments in an HliD-less strain (Shukla et al., 2018). In addition to the proposed functions of Hlips in facilitating PS assembly processes, a photoprotective function of HliC and HliD for CHLG also has been reported (Niedzwiedzki et al., 2016).
In contrast to the cyanobacterial Hlips with assigned functions, our knowledge of the function(s) of the Arabidopsis OHPs is limited. OHP1 mRNA was reported to accumulate 2-fold in response to elevated light intensities (Jansson et al., 2000). OHP2 was initially described to cofractionate with PSI-enriched fractions of n-dodecyl β-d-maltoside (DDM)-solubilized thylakoid membranes on Suc density gradients, which prompted the proposal of a role in the photoprotection of PSI (Andersson et al., 2003). Arabidopsis T-DNA insertion (knockout) mutants for each of the two OHP genes revealed drastically impaired development, relative to wild-type seedlings, on Murashige and Skoog medium supplemented with Suc under continuous low-light conditions (Beck et al., 2017). These findings suggested that OHPs play a substantial role in plastid development.
Therefore, we set out to elucidate the function and localization of both Arabidopsis OHP variants using a virus-induced gene silencing (VIGS) approach at a later developmental stage. Interestingly, heterodimer formation of the two OHPs stabilizes both proteins in developing seedlings. Similar to the function of cyanobacterial Hlips, our findings suggest that OHPs interact with PS biogenesis factors and coordinate pigment supply with PS assembly. Multiple assembly factors involved in the de novo biogenesis of PSI or PSII, as well as in PSII repair, are now known (for overview, see Lu, 2016). We show here that OHP2 interacts with HIGH CHLOROPHYLL FLUORESCENCE244 (HCF244; At4g35250), the Arabidopsis homolog of Ycf39, and explore the consequences for the stability and localization of these proteins in thylakoid membranes and their role during the assembly and/or repair of photosynthetic protein complexes.
RESULTS
OHP1 and OHP2 Form a Heterodimer
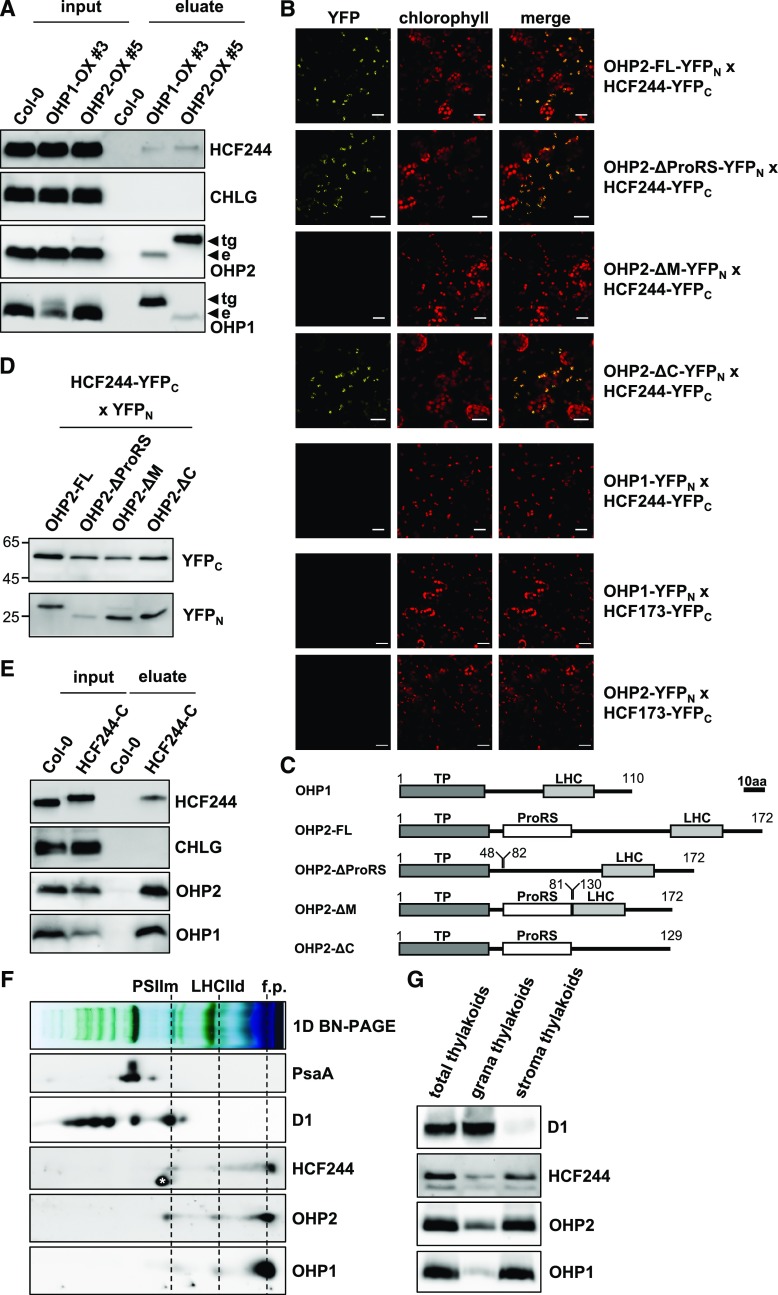
To search for potential interaction partners of the two OHP variants, we performed pull-down experiments with OHP1 and OHP2 fused to a C-terminal HA-strepII tag, which were expressed under the control of the 35S promoter in Arabidopsis wild-type (Columbia-0) plants (hereafter termed OHP1-OX/OHP2-OX lines). These plants accumulated detectable amounts of OHP fusion proteins, whereas the level of the endogenous proteins remained constant. For pull-down assays, thylakoids from the OHP1-OX and OHP2-OX lines were solubilized with 0.5% (w/v) DDM and incubated with streptavidin-coated beads. After washing, proteins bound to the resin were eluted with 10 mm desthiobiotin, fractionated by SDS-PAGE, and analyzed by immunoblotting. Notably, the strepII-tagged OHP in the eluate was always detected together with the other OHP variant (Fig. 1A), but it never coeluted with its endogenous counterpart. These results suggest that neither OHP variant forms homodimers but that one interacts with the other to yield heterodimers.
Figure 1.
OHPs interact with HCF244. A, StrepII tag pull-down analyses using OHPs as bait. Thylakoids from plants expressing OHPs fused to a C-terminal HA-strepII tag under the control of the 35S promoter (OX-lines) were solubilized with 0.5% DDM and incubated with streptavidin-coated beads. After washing and elution with 10 mm desthiobiotin, proteins were separated by Tricine-SDS-PAGE, transferred onto nitrocellulose membranes, and probed with specific antibodies. Endogenous (e) or transgenic OHP species carrying a C-terminal HA-strepII tag (tg) are indicated by arrows marking the respective signals. B, BiFC assays in N. benthamiana leaves. The indicated proteins fused to the N- and C-terminal halves of split YFP were expressed transiently in N. benthamiana leaves. Confocal microscopy on an LSM 800 confocal microscope (Zeiss) was applied to detect yellow fluorescence indicating an interaction. To confirm that the interactions occurred inside the chloroplast, chlorophyll autofluorescence also was recorded. Bars = 20 µm. C, Domain scheme of OHP species used for BiFC assays. The scheme is drawn to scale, and the scale bar represents 10 amino acids (10aa). Numbers indicate amino acids. TP, Transit peptide (predicted); LHC, transmembrane domain harboring the chlorophyll-binding motif; ProRS, Pro-rich sequence. D, Western-blot analysis of full-length (FL) and truncated OHP2 species expressed in N. benthamiana leaves for the BiFC assay. Total protein extracts were separated by SDS-PAGE, transferred onto nitrocellulose membranes, and probed with specific antibodies against the peptide tags. E, StrepII tag pull-down analyses using HCF244 as bait. Thylakoids from plants expressing HA-strepII-tagged HCF244 in the hcf244 mutant background were solubilized with 0.5% DDM, and pulldown was performed as described above. F, Analysis of complex formation of OHPs and HCF244 by BN-PAGE and subsequent second dimension SDS-PAGE. Arabidopsis wild-type thylakoids were solubilized with 1% DDM and separated on BN gels. Lanes of the BN-PAGE gels were denatured and layered on top of acrylamide gels containing 6 m urea or Tricine-SDS gels. After separation, proteins were transferred onto nitrocellulose membranes and probed with specific antibodies. PSIIm, PSII monomer; LHCIId, LHCII dimer; f.p., free proteins. The asterisk next to the HCF244 signal marks a cross reaction of the antibody. G, Subfractionation of thylakoids into grana and stroma fractions by differential centrifugation. Equal amounts of chlorophyll (4 µg) were subjected to SDS-PAGE and probed with specific antibodies.
The additional expression of each OHP fusion protein in the OHP1-OX and OHP2-OX lines had no obvious effect on the seedling phenotype (Supplemental Fig. S1, A–C). However, it should be noted that the tagged OHP1 and OHP2 accumulated to levels equivalent to only 10% to 20% of the endogenous protein, although the corresponding transgenic OHP transcripts accumulated strongly. This observation was confirmed in several independent lines (Supplemental Fig. S2, A–C). The steady-state levels of PS core subunits and photosynthetic pigments were not altered in the OHP-OX lines. Likewise, analysis of DDM-solubilized thylakoids on blue native (BN) gels revealed no differences in the amounts of the thylakoid-bound protein complexes between the OHP-OX and control lines (Supplemental Fig. S1, B and C). To exclude that the C-terminal tag impaired OHP function, both the ohp1 and ohp2 T-DNA insertion mutants were complemented with OHP transgenes with their native promoter, which encode HA-tagged variants. Both constructs successfully complemented the mutant phenotype (Supplemental Fig. S3).
OHPs Interact with HCF244
Two Hlips have been found to interact with the putative PSII assembly factor Ycf39 in cyanobacterial extracts (Ermakova-Gerdes and Vermaas, 1999; Chidgey et al., 2014; Knoppová et al., 2014). Strikingly, the OHP2 expression profile shows a strong correlation with that of HCF244, the Arabidopsis homolog of the cyanobacterial protein Ycf39 (Link et al., 2012). Therefore, we used the tagged OHP variants as bait proteins and succeeded in detecting HCF244 in eluates from both OHP1-OX and OHP2-OX thylakoids (Fig. 1A). Note that HCF244 also has been reported to serve similar functions to HCF173 (Schult et al., 2007).
Since cyanobacterial Hlips also were found in complex with CHLG (Chidgey et al., 2014; Knoppová et al., 2014), we also probed our pull-down eluates with an antibody against CHLG. However, no immunoreactive CHLG signal could be detected (Fig. 1A). Moreover, antibodies directed against additional enzymes of chlorophyll synthesis as well as PS core subunits also failed to react with the pull-down eluates. Silver staining of the pull-down eluates revealed that only a few proteins were copurified in detectable amounts along with the bait proteins (Supplemental Fig. S4) and the other OHP variant.
To verify the protein-protein interactions, we performed bimolecular fluorescence complementation (BiFC) experiments with OHP1, OHP2, HCF244, and HCF173. All four were expressed as fusion proteins with the N-terminal and C-terminal halves of split yellow fluorescent protein (i.e. YFPN and YFPC) in Nicotiana benthamiana leaves. Chloroplast-localized YFP fluorescence indicated an OHP2-HCF244 interaction, whereas coexpression of the OHP1 and HCF244 fusion proteins did not reconstitute YFP function (Fig. 1B). Interestingly, YFP signals resulting from the OHP2-HCF244 interaction did not fully overlap with chlorophyll autofluorescence but were visible as speckles inside the chloroplast. In light of previous studies (Zhong et al., 2013), we propose that the speckles are explained by the nucleoid localization of the interaction partners. In this context, it is worth mentioning that the maize (Zea mays) homolog of HCF244 has been found to be localized to nucleoids (Majeran et al., 2012). Neither of the OHP variants bound to HCF173 (Fig. 1B) or to each other in these experiments (data not shown). All combinations also were probed inversely (exchange of the N- and C-terminal halves of YFP) and gave similar results. The expression of the relevant proteins in transfected N. benthamiana cells was confirmed for all tested combinations by western blotting (data not shown).
To identify the segment of OHP2 that interacts with HCF244, OHP2 was dissected into three parts, which were eliminated individually from the full-length protein (Fig. 1C). (1) A Pro-rich region beginning close to the N terminus of the processed OHP2 (∆ProRS; amino acids 49–81) downstream of the putative chloroplast transit peptide (amino acids 1–42); the first seven residues of the mature OHP2 were kept to ensure unimpaired proteolysis of the transit peptide. (2) The C-terminal domain with the LHC motif (ΔC; amino acids 130–172) and (3) the middle region (∆M; amino acids 82–129). Gene constructs encoding truncated OHP2 proteins were fused with the N-terminal and C-terminal halves of split YFP and expressed in combination with HCF244. Deletion of either the Pro-rich region or the C terminus of OHP2 did not prevent the reconstitution of YFP fluorescence, whereas deletion of the middle region eliminated the signal (Fig. 1B). Immunoblotting revealed that all three truncated OHP2 fusion proteins were expressed (Fig. 1D). Thus, the central part of OHP2 at the N-terminal side of the transmembrane domain interacts with HCF244, and the lack of OHP2-∆M-HCF244 interaction cannot be explained by the instability of this deletion variant or the failure of its synthesis (Fig. 1C). The lack of OHP1-OHP2 as well as OHP1-HCF244 interactions in BiFC assays is likely attributable to the split YFP halves, which may hinder the formation of the respective dimers.
The significance of the OHP2-HCF244 interaction was ultimately verified by pull-down assays with HA-strepII-tagged HCF244, which was expressed from a transgene under the control of the 35S promoter in a homozygous hcf244 T-DNA insertion mutant (GK-088C04). The complemented lines (hereafter termed HCF244-C) were phenotypically wild type like (Supplemental Fig. S5), and the HCF244 fusion protein accumulated to a similar level to the endogenous protein in the wild type (Supplemental Fig. S5C). Thylakoid membranes were solubilized and used for strep tag pull-down assays. Probing with specific antibodies revealed that both OHPs were enriched in the eluate from HCF244-C plants (Fig. 1E). Again, pull-down eluates were probed with an antibody against CHLG, but no immunoreactive signal was detected.
To verify the formation of the trimeric OHP1-OHP2-HCF244 complex, we fractionated wild-type thylakoid membranes by BN-PAGE followed by second dimension SDS-PAGE and immunoanalysis of blots with specific antibodies (Fig. 1F). OHP1, OHP2, and HCF244 were found in the low-molecular-weight region at the bottom of the BN gel. In addition, distinct signals for all three proteins also were detected at the level of the monomeric PSII and the LHCII dimer fraction.
Sequential centrifugation was used to separate plastid membranes into stroma and grana thylakoids, and the proteins in these fractions were examined by SDS-PAGE. OHP1, OHP2, and HCF244 were found mainly in the stroma thylakoids (Fig. 1G), confirming the previously reported distribution of OHP2 (Andersson et al., 2003). The predominant assignment of all three proteins to the stroma thylakoids opens the possibility of a physical association with PSI and/or the assembly and repair of PSII.
Inactivation of OHP2 by VIGS Causes a Pale-Green Phenotype in Arabidopsis
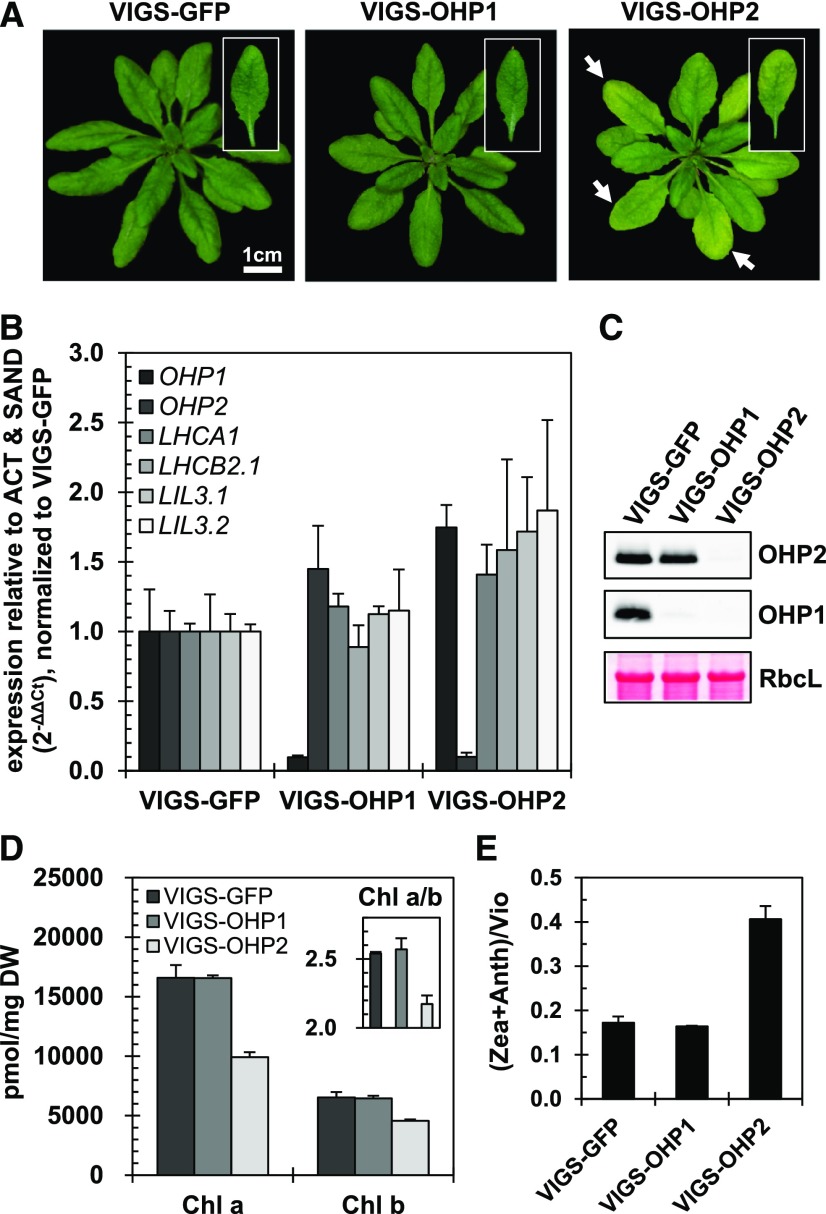
To investigate the functional significance of OHP heterodimerization and the formation of the trimeric OHP1-OHP2-HCF244 complex, we explored the molecular consequences of the impaired expression of both OHP isoforms for chloroplast biogenesis. We first confirmed the recently published phenotype of the T-DNA insertion mutants ohp1 and ohp2 (Beck et al., 2017): homozygous mutant seedlings were nonviable on soil and hardly developed at all on sugar-containing Murashige and Skoog medium (data not shown). However, substantial amounts of leaf material are normally required to elucidate the function and physical interactions of both OHP isoforms in planta. Therefore, we chose the VIGS approach (Dinesh-Kumar et al., 2003; Burch-Smith et al., 2006) to induce the inactivation of OHP1 and OHP2 expression separately in growing Arabidopsis plants. Infiltration with agrobacterial strains containing the T-DNA with the VIGS constructs was performed with 12-d-old seedlings, which had successfully completed the transition from the cotyledon stage to the generation of the first true leaves. By 3 weeks after infiltration with a pTRV2-OHP2 construct, the plants had developed a pale-green leaf phenotype relative to plants infiltrated with a pTRV2-GFP construct (negative control; Fig. 2A). Thus, all experiments were performed with leaf material harvested 3 weeks after infiltration. VIGS of either of the OHP genes reduced expression to only 10% of that seen in VIGS-GFP control plants (Fig. 2B). The expression of the other OHP gene always remained stable, indicating that silencing of one of the genes did not affect the accumulation of the other OHP mRNA. Surprisingly, in contrast to the scarcely viable ohp1 T-DNA insertion mutant, we did not observe any macroscopic change, relative to the controls, in the plants that were infiltrated with the pTRV2-OHP1 construct, even though reverse transcription-quantitative PCR (RT-qPCR) confirmed that levels of OHP1 transcripts were reduced drastically (Fig. 2B).
Figure 2.
VIGS of OHP2 causes a pale-green phenotype in Arabidopsis. A, Phenotypes of 5-week-old VIGS plants (i.e. 3 weeks after infiltration). Pale-green leaves in the VIGS-OHP2 plants are indicated by white arrows. B, RT-qPCR analysis of transcripts of the LHC gene family. OHP transcript levels are reduced specifically in the respective silencing lines. Total RNA was DNase digested and reverse transcribed. RT-qPCR was performed using ACT and SAND genes as references. Data were normalized to the expression in VIGS-GFP lines using the ΔΔCt method (Pfaffl, 2001). Data represent averages of three biological replicates, and error bars represent the sd. C, Western-blot analysis of OHPs showing reduced accumulation of OHPs in the VIGS lines. Total protein extracts were fractionated by SDS-PAGE or Tricine-SDS-PAGE, transferred onto nitrocellulose membranes, and probed with specific antibodies. The large subunit of Rubisco (RbcL) was used as the loading control. D, Chlorophyll (Chl) quantification by HPLC. Pigments were extracted from freeze-dried leaf material in cold alkaline acetone. Extracts were separated by HPLC, and pigments were quantified using pure standards. The inset shows the ratio of chlorophyll a to b species. Data represent averages of four biological replicates, and error bars represent the sd. DW, Dry weight. E, Status of the xanthophyll cycle, expressed as the ratio of violaxanthin (Vio) to antheraxanthin (Anth) and zeaxanthin (Zea). Data represent averages of four biological replicates, and error bars represent the sd.
To assess the specificity and the repercussions of OHP silencing, we examined the expression of genes for other members of the LHC family, namely LHCA1 (At3g54890), LHCB2.1 (At2g05100), LIL3.1 (At4g17600), and LIL3.2 (At5g47110), in the VIGS plants. The transcription of these genes remained wild type like (Fig. 2B). Using a specific antibody raised against OHP1 or OHP2, the loss of the specific protein encoded by the silenced OHP gene was confirmed (Fig. 2C). It is important to emphasize here that the OHP1 content was reduced below the detection limit in VIGS-OHP2 plants, while the OHP2 level was only decreased slightly in VIGS-OHP1 plants. These results are consistent with previous data obtained with the ohp mutants (Beck et al., 2017). Notably, the absence of the respective OHP variant also was observed in young leaves of VIGS-OHP1 and VIGS-OHP2 plants 3 weeks after infiltration (data not shown), although they did not exhibit the reduced pigmentation seen in older leaves of the latter.
HPLC analysis revealed that, in VIGS-OHP2 plants, chlorophyll a and b levels were only about 60% and 70% of the control values, respectively (Fig. 2D), whereas they remained essentially unchanged in VIGS-OHP1 plants. The chlorophyll a/b ratio consequently decreased to 2.17 ± 0.06 in VIGS-OHP2 plants in comparison with 2.54 ± 0.01 in control plants and 2.57 ± 0.08 in VIGS-OHP1 plants (Fig. 2D). The ratio of xanthophylls (zeaxanthin + antheraxanthin)/violaxanthin, representing the status of the xanthophyll cycle pigments, was doubled in VIGS-OHP2 plants compared with control plants (Fig. 2E), indicating that silencing OHP2 induces features of light intensity stress under normal growth light conditions.
Loss of OHP2 Is Accompanied by Decelerated Chlorophyll Biosynthesis
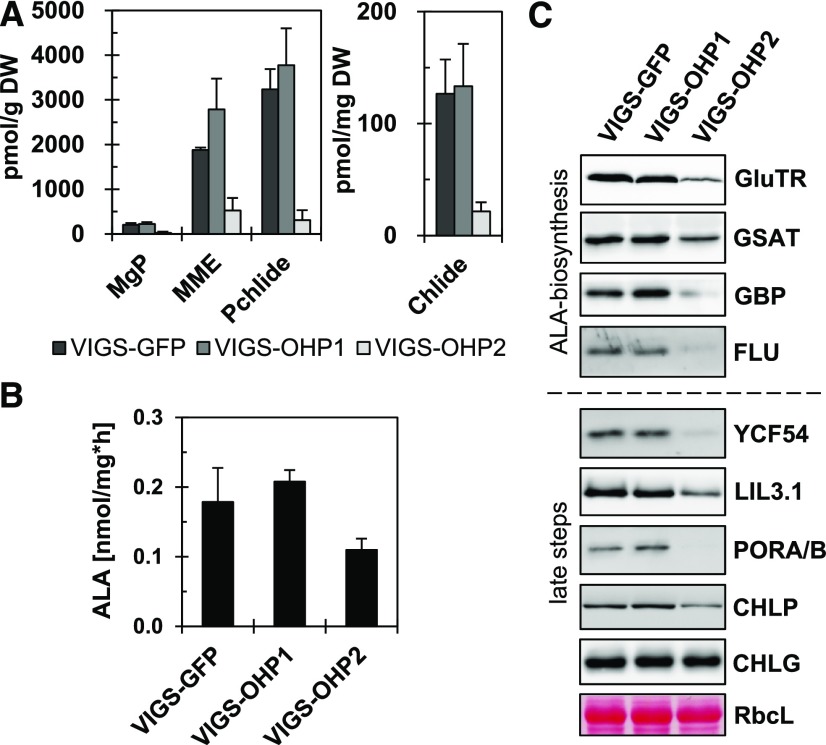
As stated above, multiple functions have been attributed previously to the cyanobacterial Hlips, including a regulatory function in the early steps of chlorophyll biosynthesis to ensure adequate pigment availability (Xu et al., 2002). Therefore, we examined the impact of OHP deficiency on plant chlorophyll biosynthesis. The quantification of chlorophyll precursors by HPLC showed a sharp decrease in the content of all analyzed metabolites in VIGS-OHP2, but not in VIGS-OHP1, plants. Specifically, in VIGS-OHP2 seedlings, the direct chlorophyll precursors protochlorophyllide and chlorophyllide were reduced to 10% and 15% of their control values, respectively. The magnesium porphyrins Mg-protoporphyrin IX and Mg-protoporphyrin IX monomethylester reached only 10% and 25% of control levels, respectively (Fig. 3A).
Figure 3.
Chlorophyll biosynthesis is slowed down in VIGS-OHP2 plants. A, Quantification of intermediates in chlorophyll biosynthesis by HPLC. Pigments were extracted from freeze-dried leaf material in cold alkaline acetone. Extracts were separated by HPLC, and pigments were quantified by comparison with pure standards. DW, Dry weight; MgP, magnesium-protoporphyrin IX; MME, magnesium-monomethylester protoporphyrin IX; Pchlide, protochlorophyllide; Chlide, chlorophyllide. Data represent averages of four biological replicates, and error bars represent the sd. B, Rate of ALA synthesis in VIGS plants. The accumulation of ALA was measured after blocking the subsequent enzyme step by the application of levulinic acid and the conversion of ALA into a pyrrole. Data represent averages of four biological replicates, and error bars represent the sd. C, Western-blot analysis of tetrapyrrole biosynthesis enzymes in the VIGS lines. Total protein extracts were separated by SDS-PAGE, transferred onto nitrocellulose membranes, and probed with specific antibodies. The large subunit of Rubisco (RbcL) served as the loading control.
The flow of intermediates through the chlorophyll biosynthetic pathway is tightly regulated at the level of 5-aminolevulinic acid (ALA) formation (Apitz et al., 2016). It is generally accepted that glutamyl-tRNA reductase (GluTR) mediates the rate-limiting enzymatic step in tetrapyrrole biosynthesis, thus determining the flow of metabolites through the pathway (Tanaka and Tanaka, 2007; Czarnecki and Grimm, 2012). In VIGS-OHP2 plants, the capacity for ALA synthesis was reduced by 50%, but it reached control values in VIGS-OHP1 plants (Fig. 3B). The levels of GluTR, Glu-1-semialdehyde-aminotransferase (GSAT), the GluTR-binding protein (GBP), and FLUORESCENT (FLU) were all reduced noticeably in VIGS-OHP2 plants (Fig. 3C). GBP and FLU are involved in the control of GluTR function (Meskauskiene et al., 2001; Czarnecki et al., 2011). The relative lack of the relevant enzymes probably accounts for the lower rate of ALA synthesis, the reduced ALA supply, and the lower steady-state levels of tetrapyrrole intermediates observed in VIGS-OHP2 plants.
To determine whether the decreased accumulation of tetrapyrrole intermediates was primarily or perhaps exclusively caused by the lower availability of ALA or diminished amounts of downstream enzymes of chlorophyll biosynthesis, whose stability could be affected posttranslationally, we analyzed the steady-state levels of enzymes from the chlorophyll branch using specific antibodies. The levels of the enzymes of chlorophyll biosynthesis farthest downstream (namely POR, CHLP, and YCF54, a subunit of Mg-protoporphyrin IX monomethylester cyclase) were decreased in VIGS-OHP2 plants (Fig. 3C), while only the CHLG content remained similar to the control. The RT-qPCR analysis revealed no significant changes in the expression levels of genes coding for enzymes of chlorophyll biosynthesis (Supplemental Fig. S6). These findings suggest the posttranslational destabilization of many chlorophyll synthesis proteins in response to OHP2 deficiency. In contrast, VIGS-OHP1 plants contained control-like contents of these proteins (Fig. 3C).
Loss of OHP2 Leads to the Specific Destabilization of PSII Core Subunits and PSII Complexes
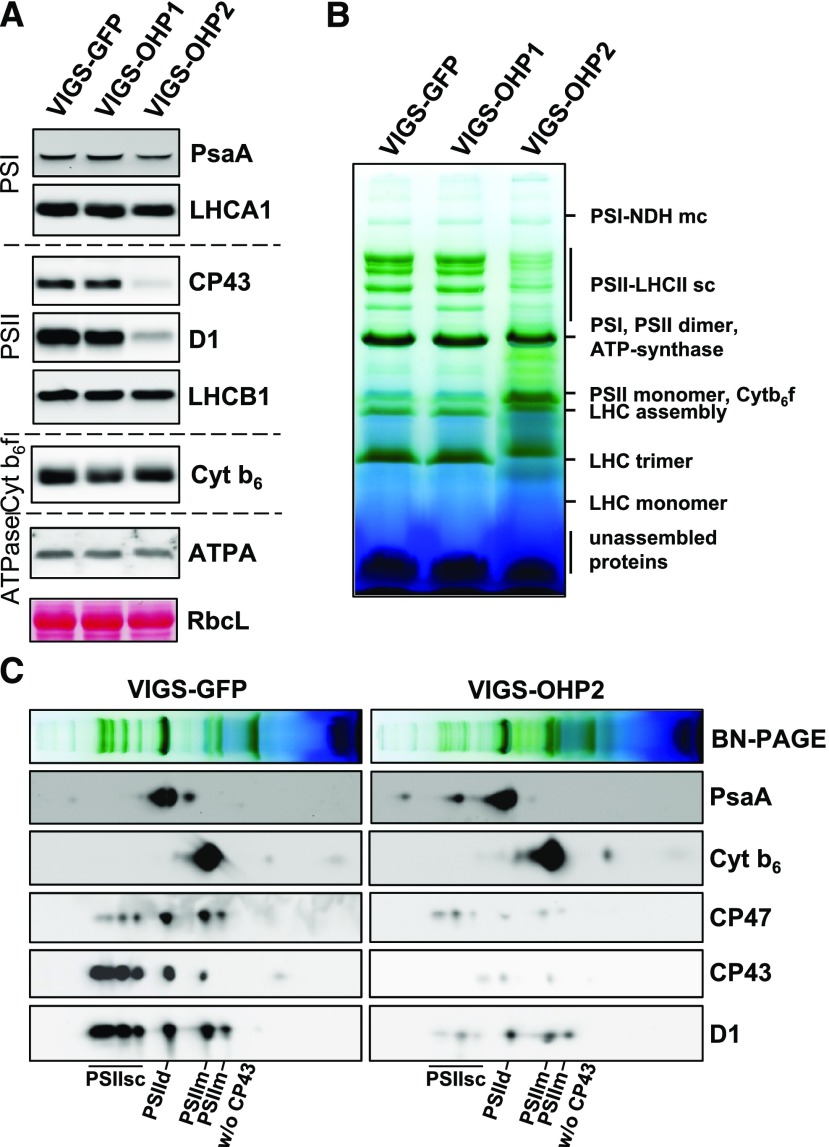
Core subunits of both PSI and PSII were almost absent in homozygous ohp1 seedlings (Beck et al., 2017; Myouga et al., 2018), suggesting a link between OHP1 function and the assembly and/or stabilization of photosynthetic reaction centers. OHP2 was reported previously to be associated with PSI (Andersson et al., 2003). Therefore, we examined the steady-state levels of photosynthetic subunits in the OHP-silenced plants by SDS-PAGE and immunoblotting. Only the core subunits of PSII (D1 and CP43) were greatly diminished in VIGS-OHP2 plants (Fig. 4A), whereas PSI (PsaA), the cytochrome b6f complex (Cyt b6), and ATPase (ATPA) remained unaffected. No significant changes in the levels of these proteins were observed in VIGS-OHP1 plants. In addition, the amounts of LHCA1 and LHCB1 in OHP-silenced lines remained similar to those in the VIGS-GFP control line (Fig. 4A). This combination of stable antennal components and sharply reduced content of PSII core proteins also is compatible with the lower chlorophyll a/b ratio in the VIGS-OHP2 plants.
Figure 4.
PSII core subunits and PSII complexes are destabilized in VIGS-OHP2 plants. A, Western-blot analysis of PS core subunits and the two major LHC proteins LHCA1 and LHCB1 in the VIGS lines. Total proteins were separated on acrylamide gels containing 6 m urea, transferred onto nitrocellulose membranes, and probed with specific antibodies. The large subunit of Rubisco (RbcL) is shown as a loading control. B, BN-PAGE analysis of thylakoid membranes. Thylakoid membranes representing equal amounts of chlorophyll (8 µg) were solubilized with 1% DMM, and photosynthetic complexes were separated on 4.5% to 12% BN gels. C, Fractionation of photosynthetic complexes under denaturing conditions in the second dimension. Lanes from BN gels were denatured and layered on top of acrylamide gels containing 6 m urea. After electrophoresis, proteins were transferred onto nitrocellulose membranes and probed with specific antibodies. PSII-containing complexes are labeled at the bottom edge of the D1 blots: PSIIsc, PSII supercomplexes; PSIId, PSII dimer; PSIIm, PSII monomer.
Photosynthetic protein complexes in the two VIGS-OHP lines and control plants were analyzed by BN-PAGE (samples were normalized with respect to total chlorophyll content) after the solubilization of thylakoids with 1% DDM and separation. No alteration in the pattern of photosynthetic complexes was observed in VIGS-OHP1 extracts, but the relative representation of the abundant complexes was modified extensively in thylakoid membranes of VIGS-OHP2 plants (Fig. 4B). The amounts of PSII supercomplexes were strongly diminished in VIGS-OHP2 plants. Furthermore, monomeric LHCs accumulated, and additional protein bands were visible between the well-resolved main membrane-associated protein complexes in VIGS-OHP2 plants (Fig. 4B).
Complexes that had been separated by BN-PAGE were denatured and subjected to SDS-PAGE to analyze their protein composition. Probing with specific antibodies confirmed the presence of decreased amounts of PSII core subunits in multiple complexes in VIGS-OHP2 extracts, whereas the content and distribution of PSI and cytochrome b6f subunits in different protein complexes resembled those seen in the control (Fig. 4C).
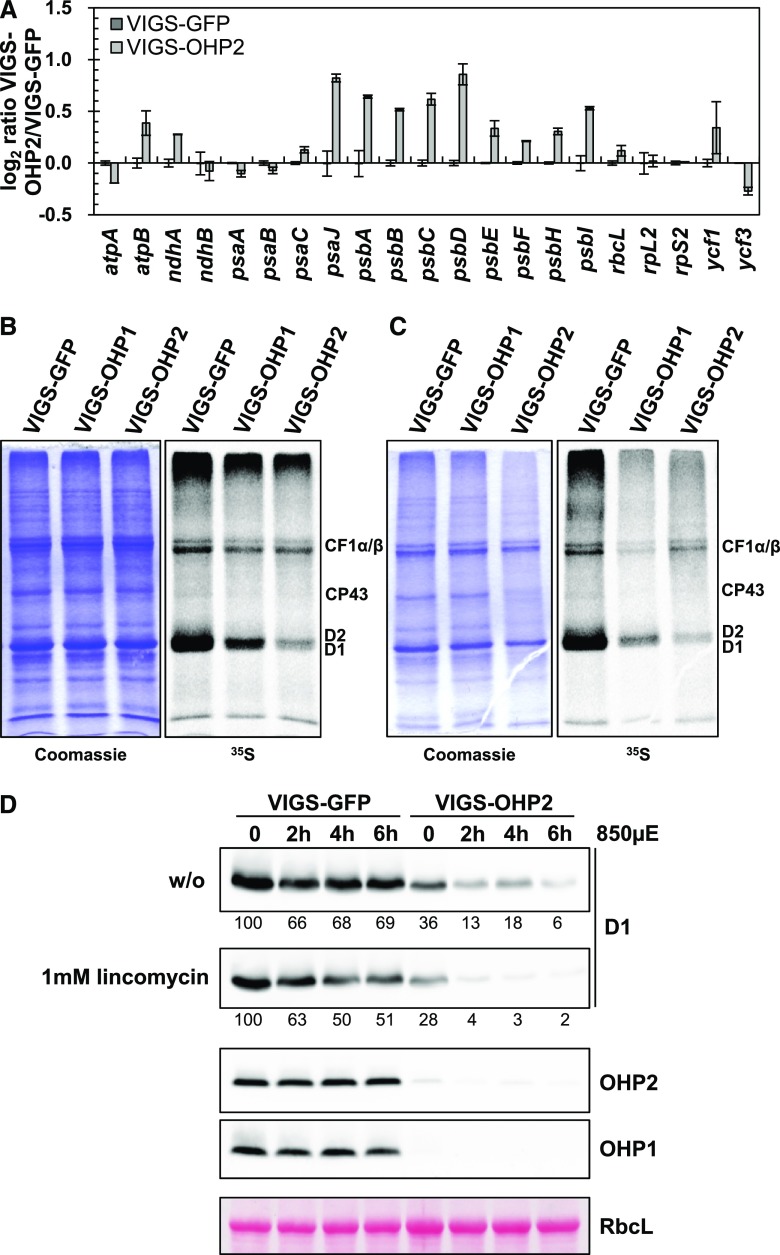
The major subunits of PSI and PSII are encoded in the chloroplast genome, and their genes are transcribed and translated by the plastidic gene expression machinery. Therefore, we explored the regulatory impact of OHP deficiency on plastome gene expression (Fig. 5A). The relative abundances of transcripts of selected plastid-encoded genes were unaltered in VIGS-OHP2 plants relative to the control sample. In contrast, transcripts encoding PSII subunits (e.g. psbA–psbI) were slightly overrepresented. Hence, it can be concluded that the diminished accumulation of plastid-encoded photosynthetic protein subunits in VIGS-OHP2 plants results from posttranscriptional processes.
Figure 5.
Synthesis of D1 is impaired in VIGS-OHP2 plants. A, Gene expression analysis of selected plastid-encoded genes. Total RNA was reverse transcribed using random hexamer primers. SAND, ACT1, and AP2M were used as reference genes, and data were analyzed using standard curves. Data represent averages of two biological replicates, and error bars represent the sd. B and C, In vivo labeling assay of thylakoid membrane proteins. Young leaves from VIGS plants (B) or old leaves showing a pale-green phenotype described above (C) were incubated with [35S]Met in the presence of 20 µg mL−1 cycloheximide for 1 h. Crude membranes were isolated and separated by SDS-PAGE. Proteins synthesized during the incubation period were detected autoradiographically. D, In vivo protein stability of D1 in VIGS plants. Leaf discs were incubated without (w/o) or in presence of 1 mm lincomycin in high light (850 µmol photons m−2 s−1) for the times indicated. Total proteins then were separated on acrylamide gels, transferred onto nitrocellulose membranes, and probed with specific antibodies. The large subunit of Rubisco (RbcL) is shown as a loading control. Numbers below the D1 blots indicate relative amounts determined by densitometry.
Finally, we performed in vivo labeling to monitor the production of newly synthesized proteins in intact leaves from VIGS plants. For this experiment, young leaves without phenotypic variation from the control (Fig. 5B) as well as older leaves were used (Fig. 5C). Independent of leaf age, the levels of labeled D1/D2 proteins were lower in VIGS-OHP2 leaves than in control leaves. Quantification of the radioactive signals corresponding to the CF1α/β subunits confirmed that the overall efficiencies of incorporation of [35S]Met in both leaf samples were similar. As exemplified for D1, it is proposed that the relative lack of the plastid-encoded PSII proteins can be explained by impaired synthesis or stability when at least OHP2 is missing. Surprisingly, also in VIGS-OHP1 plants, the synthesis of D1/D2 was reduced, but to a lesser extent than in VIGS-OHP2 plants (Fig. 5, B and C). This points to a general functionality, albeit reduced activity, of the OHP2-HCF244 complex in the absence of OHP1.
To assess the OHP dependency and requirement for in vivo D1 stability and its de novo synthesis, we treated leaves of VIGS-GFP as well as VIGS-OHP2 plants with 1 mm lincomycin, an inhibitor of plastidic protein synthesis, and exposed them to high light (850 µmol photons m−2 s−1). VIGS-OHP2 leaves contained less D1 than VIGS-GFP samples. The control leaf discs lost only one-third of their D1 content within the incubation time (Fig. 5D). This might be explained by a reduced D1 synthesis rate in leaf discs, which cannot compete with the ongoing loss of D1 due to high light-induced photodamage. In contrast, the lack of OHP2 led to an immediate and continuous D1 degradation, starting with an initial loss of two-thirds of the initial D1 amount. A 6-h lincomycin treatment of VIGS-GFP leaves resulted in a 50% drop of D1 and indicates the impaired translation in the chloroplasts. But the D1 content declined rapidly in lincomycin-treated VIGS-OHP2 leaves to nondetectable amounts within 6 h, indicating the requirement of OHP heterodimers during de novo D1 synthesis and the recycling of the PS core complex. The residual D1 levels in VIGS-OHP2 plants seemed to be more susceptible to high-light stress, possibly due to the missing activity of OHP variants, which may exert a photoprotective role on PSII assembly intermediates.
PSII Activity Is Strongly Impaired in VIGS-OHP2 Plants
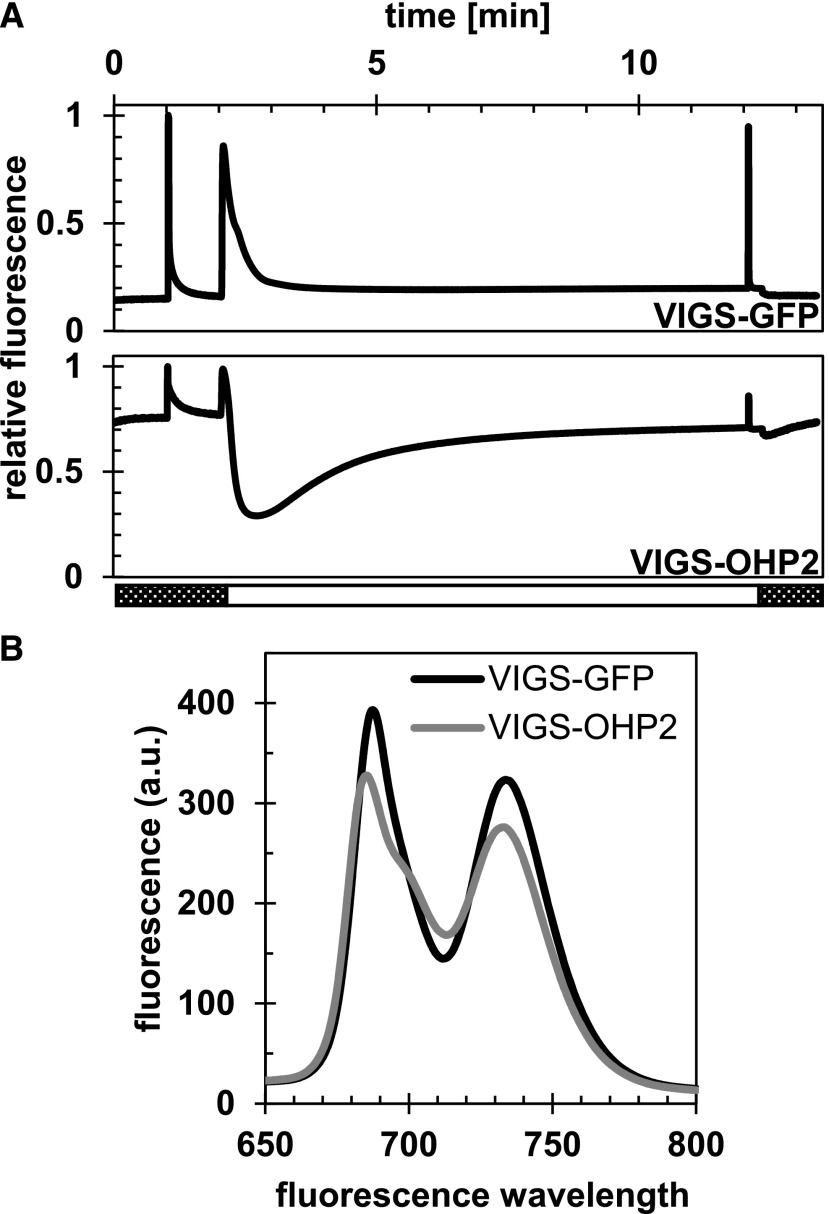
To complement the analysis of the protein composition of photosynthetic complexes, chlorophyll fluorimetric analyses were performed to gain insight into the photosynthetic electron transport in VIGS plants. Using pulse-amplitude modulation (PAM), we analyzed room temperature chlorophyll fluorescence (Fig. 6A; the wild-type-like data for VIGS-OHP1 plants are not shown). Under our growth conditions, the maximum quantum efficiency (Fv/Fm) of VIGS-GFP control and VIGS-OHP1 plants was 0.851 ± 0.001 and 0.846 ± 0.007, respectively. VIGS-OHP2 plants, on the other hand, displayed an extremely low Fv/Fm ratio of 0.182 ± 0.055, which results from a very high ground-level fluorescence of around 0.75, which is 5 times higher than the value for VIGS-GFP control plants (Fig. 6A). When the actinic light was switched on during the PAM measurements, the chlorophyll fluorescence in OHP2-deficient plants declined faster than in the control plants, but it began to rise again after reaching a minimum, before it leveled off to a stable value slightly below the ground fluorescence (Fig. 6A). Consequently, due to the high fluorescence in light, both the quantum efficiency of PSII and photochemical quenching were reduced strongly in VIGS-OHP2 plants. The room temperature chlorophyll fluorescence measurements of VIGS-OHP2 plants under similar experimental conditions yielded a fluorescence curve that resembled that of hcf mutants of group III, which comprises various mutants with defects in PSII (Meurer et al., 1996). In agreement with that report, the VIGS-OHP2 plants clearly displayed a high-chlorophyll-fluorescence phenotype.
Figure 6.
VIGS-OHP2 plants show a high-chlorophyll-fluorescence phenotype. A, Room-temperature chlorophyll fluorescence analysis (PAM). Pulses were set to 6,000 µmol photons m−2 s−1, and Kautsky induction (i.e. actinic light) was performed at a flux of 100 µmol photons m−2 s−1. Curves represent mean values from four biological replicates. B, Low-temperature chlorophyll fluorescence analysis. The chlorophyll concentration was adjusted to 5 µg mL−1 to prevent reabsorption of the emitted light, and fluorescence was recorded at 77K in liquid nitrogen. a.u., arbitrary units.
In summary, these findings demonstrate that silencing of OHP2 in green seedlings strongly impairs PSII formation, whereas the assembly of the other photosynthetic complexes is not affected. To obtain further information about the status of PSI, we subsequently performed 77K fluorescence spectroscopy on leaf extracts using a low concentration of chlorophyll (5 µg mL−1) to prevent reabsorption of the emitted light. The PSI-LHC and PSII-LHC emissions in VIGS-OHP2 plants was decreased to around 85% and 80% of control values, respectively (Fig. 6B). Interestingly, emission maxima from both PSs in VIGS-OHP2 plants showed a blue shift of around 2 nm. Additionally, the PSII-LHC emission spectra of the VIGS-OHP2 extract displayed a shoulder at around 695 to 700 nm, which was not seen in the VIGS-GFP samples and could be assigned to LHCII aggregates that are known to emit fluorescence at 700 nm (Oh et al., 2003; Andrizhiyevskaya et al., 2005). Thus, the VIGS-OHP2 spectrum reflects a potential impairment of energy transfer between the antenna complex and core proteins of PSII. Leaf samples taken from VIGS-OHP1 plants displayed similar emission spectra to the control (data not shown).
Inactivation of OHP2 Expression Destabilizes HCF244
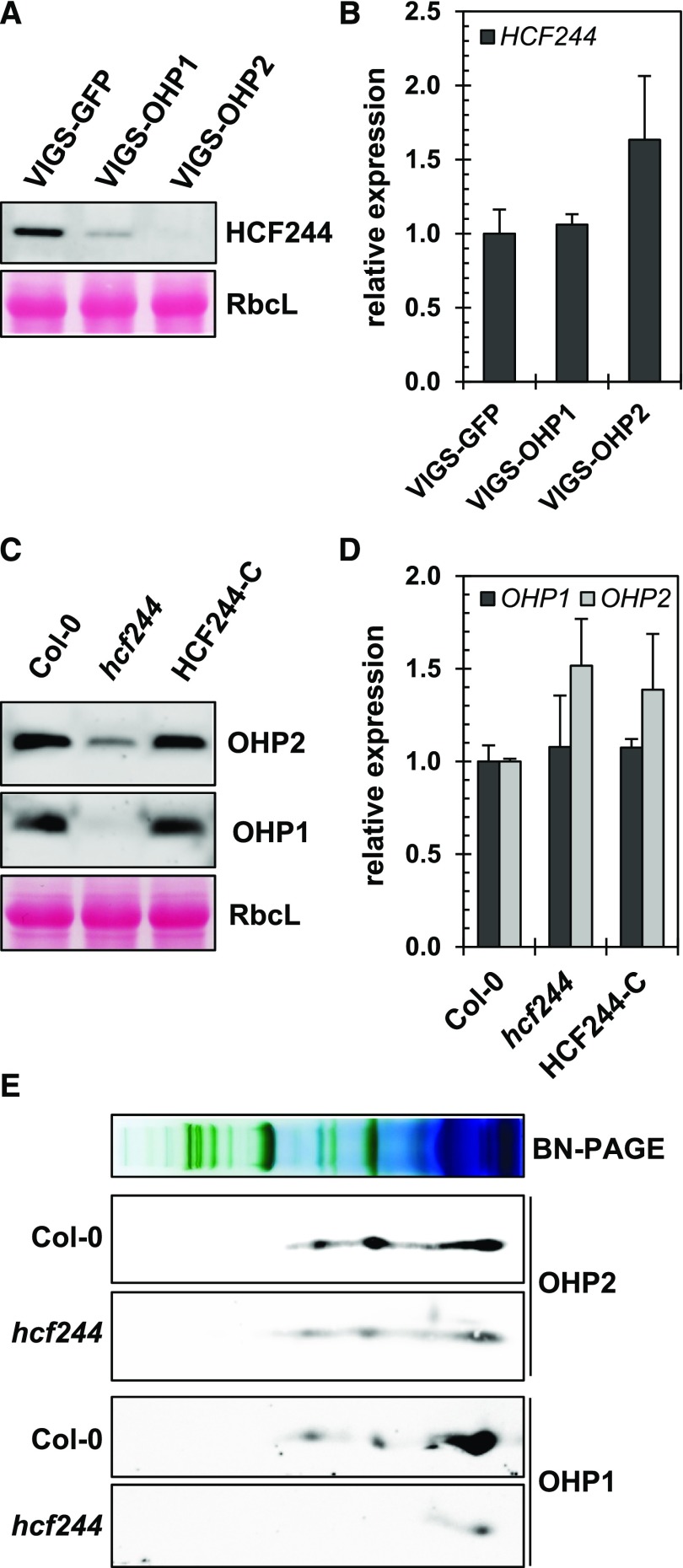
HCF244 is not an integral membrane protein, but it is attached to the stromal side of the thylakoid membrane (Link et al., 2012). As shown above, the stromal part of OHP2 interacts with HCF244 (Fig. 1B), and we hypothesized that OHP2 serves as a membrane anchor for HCF244. To test this notion further, levels of HCF244 mRNA and HCF244 protein in VIGS-OHP2 lines were determined. Strikingly, no HCF244 could be detected (Fig. 7A), although the HCF244 expression level remained unaltered (Fig. 7B). Interestingly, VIGS-OHP1 plants also contained less HCF244 than the wild type, which correlated with the reduced OHP2 content. Nevertheless, the minor decrease in the contents of OHP2 and HCF244 did not perturb thylakoid biogenesis or the assembly of photosynthetic complexes.
Figure 7.
OHP1, OHP2, and HCF244 are required for mutual stabilization. A, Western-blot analysis of HCF244 in OHP-VIGS plants. Total proteins were separated on acrylamide gels, transferred onto nitrocellulose membranes, and probed with specific antibodies. The large subunit of Rubisco (RbcL) is shown as a loading control. B, RT-qPCR analysis of HCF244 transcripts in OHP-VIGS plants. Total RNA was DNase digested and reverse transcribed. RT-qPCR was performed using ACT and SAND genes as references. Data represent averages of three biological replicates, and error bars represent the sd. C, Western-blot analysis of OHPs in the hcf244 mutant and HCF244-C complementation lines. D, RT-qPCR analysis of OHP transcripts in the hcf244 mutant and HCF244-C complementation lines. Data represent averages of three biological replicates, and error bars represent the sd. E, Analysis of the localization of OHP2 in the absence of HCF244 by BN-PAGE and subsequent second dimension SDS-PAGE. Thylakoids were solubilized with 1% DDM and separated on BN gels. Lanes of the BN-PAGE gel were denatured and layered on top of Tricine-SDS gels. After separation, proteins were transferred onto nitrocellulose membranes and probed with specific antibodies. Col-0, Ecotype Columbia-0.
As all HCF244 comigrates with both OHPs in wild-type samples (Fig. 1F), we concluded that free HCF244 is unstable. This idea is consistent with the observation that HCF244-C lines accumulate markedly increased amounts of HCF244 transcripts, but no additional HCF244 is detectable (Supplemental Fig. S4, C and D). Thus, it is likely that HCF244 requires parallel accumulation of OHP2 for its stability. Although a central role of OHP2 for the stability of both HCF244 and OHP1 is suggested, it cannot be excluded that a dimeric OHP2-HCF244 complex is at least partially functional without OHP1, as depletion of OHP1 by VIGS had no influence on plant growth, which, in turn, indicates that the stability of HCF244 depends solely on OHP2.
A T-DNA insertion in the first exon of HCF244 generates a strong knockdown mutant, which expresses the HCF244 transcript at only 10% of wild-type levels (Supplemental Fig. S4D). The concomitant decrease in HCF244 content resulted in pale-green, growth-retarded plants, which accumulated less chlorophyll and lower amounts of the PSII core proteins CP43 and D1 than the wild type, while the contents of LHCPs and the other photosynthetic complexes were not affected (Supplemental Fig. S5, A–F). These findings confirm previous observations showing that PSII assembly and stability are compromised in hcf244 mutants (Link et al., 2012). Comparison of their phenotypical and biochemical modifications underlines the resemblances between VIGS-OHP2 and hcf244 seedlings (Figs. 2 and 4; Supplemental Fig. S5) and supports the suggested functional link between HCF244 and OHPs in ensuring PSII assembly and stability. The levels of both OHPs were strongly decreased in the hcf244 mutant, although the expression of their mRNAs was not changed (Fig. 7, C and D). Hence, the similarity of the phenotypes of hcf244 and VIGS-OHP2 plants correlates with the mutual stabilization and common function of HCF244 and the two OHP variants.
Apart from the lower content of the OHP variants in the hcf244 mutant, their correlation to the thylakoid membrane complexes in the absence of HCF244 was examined. Purified thylakoid membranes of the wild type and the hcf244 mutant were subjected to BN-PAGE and second dimension Tricine-SDS-PAGE as described above and probed with OHP antibodies (Fig. 7E). Interestingly, in the absence of HCF244 (and OHP1; i.e. in the hcf244 mutant), OHP2 still assembled in the same Mr complexes as in wild-type samples. The OHP-specific immune signals are in agreement with those in Figure 1F and correspond to the HCF244-containing protein complexes. These complexes correlate with the monomeric PSII, LHCII dimer, and the free protein range (Fig. 1F). As equal amounts of thylakoid membranes were loaded onto BN-PAGE in this experiment (normalized to the chlorophyll concentration), the decreased OHP2 content in the hcf244 mutant (Fig. 7C) also is reflected by the weaker OHP2 immune signals in Figure 7E. In agreement with Figure 7C, OHP1 in hcf244 thylakoids was virtually undetectable, despite some trace amounts in the free protein range.
In contrast to HCF244, no direct homolog of plant HCF173 exists in Synechocystis PCC6803. To investigate the impact of HCF173 on the stability of HCF244 and the OHPs, we also silenced HCF173 and HCF244 by VIGS (Supplemental Fig. S7). VIGS-HCF244 plants phenotypically resembled the hcf244 mutant (Supplemental Figs. S5 and S7), and VIGS-HCF173 plants showed a less drastic macroscopic phenotype in comparison with the HCF173 knockout mutant but still exhibited a reduction in chlorophyll content to 85% of that in the control (Supplemental Fig. S7, A and B). The latter result agrees with earlier findings showing that the hcf173 mutant is less severely affected than its hcf244 counterpart (Schult et al., 2007; Link et al., 2012). Transcript analysis by RT-qPCR revealed a decrease of the targeted HCF173 and HCF244 transcripts to 10% to 15% compared with the control, whereas the content of other analyzed transcripts was unchanged (Supplemental Fig. S7C). As observed in the hcf244 mutant line, the amounts of core subunits of PSII as well as both OHP variants were strongly diminished in VIGS-HCF244 plants. Steady-state levels of PSII subunits also were lower in VIGS-HCF173 plants than in the control, but the content of both OHP variants as well as HCF244 remained essentially unaltered in these plants (Supplemental Fig. S7D). A reduced accumulation of PSII subunits has been reported for hcf173 lines; therefore, our results are in accordance with published data (Schult et al., 2007; Link et al., 2012).
Our studies indicate that the stability of each of the three proteins, the two OHP variants and HCF244, at least partially depends on the presence of the other two components. Unfortunately, a lack of one of the three proteins consequently also compromised the stability of the other two proteins. Thus, it remained difficult to explore the individual contribution of each protein to the cotranslational or posttranslational assembly of chlorophyll-binding proteins in the thylakoid membrane. We proposed an experimental approach to explore the function of the OHP heterodimer in the presence of a nonfunctional mutant of HCF244, which has lost its activity but still provides the scaffold function for OHP accumulation and stabilization.
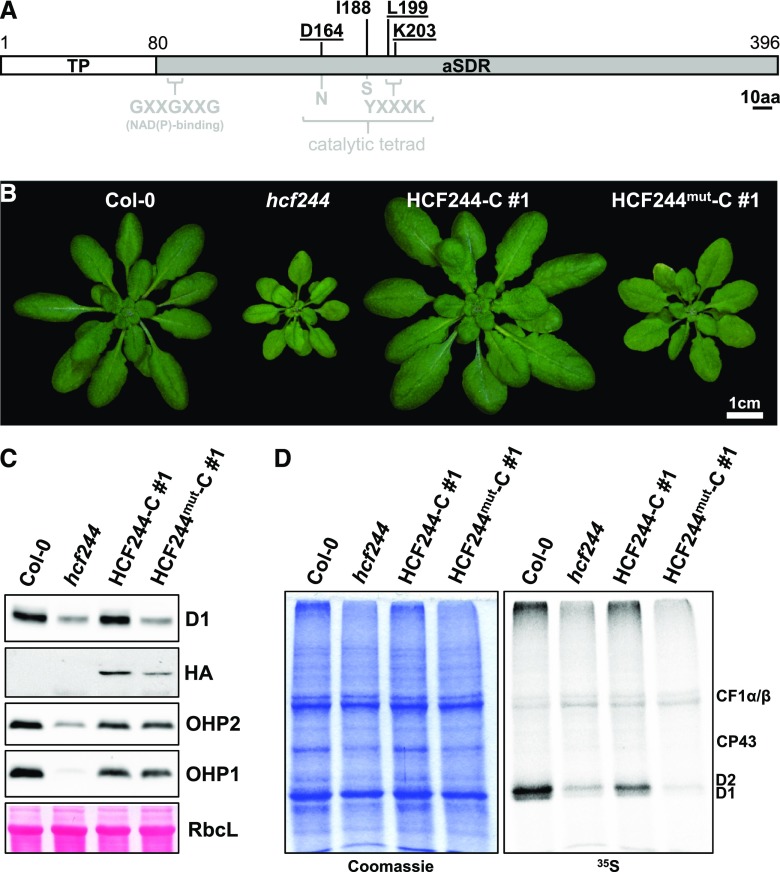
HCF244 belongs to subgroup 5 of atypical short-chain dehydrogenases (SDRs), which only partially possess the conserved amino acid residues known to be responsible for catalytic activity in typical SDR proteins. For instance, the usual catalytic tetrad N-S-Y-K of SDRs (Filling et al., 2002; Hwang et al., 2005) was modified to D-I-L-K in HCF244 (Link et al., 2012), and a catalytic activity of HCF244, if any, is not known. We nonetheless substituted the amino acids Asp-164, Leu-199, and Lys-203 (as part of the former catalytic tetrad) by Ala residues and overexpressed the mutated protein in the hcf244 background (Fig. 8A). Plants expressing this construct (hereafter termed HCF244mut-C) resembled the mutant phenotype despite a slight increase in leaf size and pigmentation (chlorophyll a accumulated to 80% of the wild-type level in HCF244mut-C plants compared with 60% in the hcf244 mutant), indicating failure of the mutated protein to fully compensate for the loss of endogenous HCF244 (Fig. 8B). This is in contrast to the overexpression of the wild-type protein in the HCF244-C line, which successfully complemented the mutant phenotype (Fig. 8B; Supplemental Fig. S5). In both complementation lines, comparable levels of tagged HCF244 accumulated (Fig. 8C). In accordance with the macroscopic phenotype, the content of D1 did not recover in the HCF244mut-C line, although the protein levels of OHPs were comparable to those in the HCF244-C line and, thereby, were almost wild type like (Fig. 8C). Analysis of the in vivo labeling activity revealed that the HCF244mut-C line exhibited strongly reduced D1/D2 synthesis, as in the hcf244 mutant (Fig. 8D).
Figure 8.
Amino acid substitutions in HCF244 impair the activity but not the mutual stabilization of OHPs. A, Schematic representation of HCF244 indicating the essential amino acids of SDR proteins (displayed in gray) and their counterparts in HCF244 (displayed in black). Underlined residues have been exchanged to Ala residues in the HCF244mut-C line. The scale bar represents 10 amino acids (10aa). B, Phenotypes of 5-week-old mutant plants grown under short-day conditions. C, Western-blot analysis of HCF244-C lines. Total proteins were separated on acrylamide gels, transferred onto nitrocellulose membranes, and probed with specific antibodies. The large subunit of Rubisco (RbcL) is shown as a loading control. D, In vivo labeling assay of thylakoid membrane proteins. Young leaves were incubated with [35S]Met in the presence of 20 µg mL−1 cycloheximide for 1 h. Crude membranes were isolated and separated by SDS-PAGE. Proteins synthesized during the incubation period were detected autoradiographically. Col-0, Ecotype Columbia-0.
DISCUSSION
Within the LHC family, cyanobacterial Hlips and plant OHPs share the single hydrophobic, membrane-spanning LHC motif, but the sequences on either side of it show only weak similarity to each other or to those of other LHC-like proteins. Hence, their functions in thylakoid membrane biogenesis do not necessarily need to match completely. To elucidate their in planta function, we tagged Arabidopsis OHPs to determine their localization and identify their interaction partners, and we analyzed the impact of their inactivation on the assembly and stability of photosynthetic complexes.
Requirement for Synchronous Accumulation of Both OHP Variants
Interaction studies based on BiFC in N. benthamiana leaves, and strepII tag pull-down assays using Arabidopsis thylakoid membranes from OHP1-OX/OHP2-OX seedlings, were employed to screen for interactors with both OHPs in Arabidopsis (Fig. 1). When used as bait, each OHP bound the other variant (Fig. 1A). Interestingly, the endogenous OHP variant was never copurified together with its tagged version. Thus, we conclude that OHPs do not form homodimers in Arabidopsis but act in a functional complex as a heterodimer. These results are consistent with the mutual interaction of cyanobacterial HliD and HliC, which was shown to be required for the energy quenching of bound chlorophyll in Synechocystis spp. (Staleva et al., 2015). However, HliC was able to form pigment-binding homooligomers in HliD-less strains (Shukla et al., 2018).
Deletion of either of the OHP genes leads to severe physiological defects, indicating that one OHP variant cannot compensate for the loss of the other. This is corroborated by the finding that overexpression of one OHP variant fails to rescue the knockout of the other OHP gene (Beck et al., 2017). Moreover, the functional deficiency of one OHP variant reduces the stability of the other (Fig. 2C), which indicates that each depends on the other for its stability and argues for the stoichiometrically balanced accumulation of both OHPs in wild-type plants. However, it is worth mentioning that silencing OHP1 in adult plants, which resulted in a decrease in the OHP1 level below the immunological detection limit, did not strongly modify the OHP2 content. In marked contrast, OHP2 silencing resulted in the disappearance of OHP1, without affecting the transcription of the latter’s coding gene (Fig. 2, B and C). These findings are in agreement with data reported for the corresponding ohp knockout mutants. Here, OHP1 was undetectable in ohp2 seedlings, while ohp1 seedlings clearly contained OHP2, albeit in reduced amounts (Beck et al., 2017). Therefore, it is suggested that OHP1 is indispensable only in very early stages of Arabidopsis seedling development, whereas older plants may tolerate the loss of OHP1 under normal growth conditions. Conversely, OHP2 seems to provide an essential function from the early germination phase onward. Moreover, the stability of OHP1 depends on OHP2 during plant development, while OHP1 is less critical for the maintenance of OHP2 stability. However, it is likely that additional factors are responsible for ensuring the stable accumulation of OHP2.
Both OHPs Interact with HCF244
In the BiFC assay, YFP fluorescence was restored when OHP2 was coexpressed with HCF244, whereas no OHP1-HCF244 interaction was detected (Fig. 1B), which is most likely due to sterical hindrance caused by the split YFP tag. Deletion studies enabled the interaction site of OHP2 with HCF244 to be delimited to the middle region at the N-terminal side of the transmembrane helix (Fig. 1, B and C), which most likely projects into the stroma. In addition, no interaction of the OHPs with HCF173, a factor with a comparable function to HCF244 in thylakoid biogenesis, was detected. Regardless of whether OHP2 or OHP1 served as the bait, HCF244 was always captured in pull-down approaches. Consequently, both OHPs were found in the eluates of pull-down assays performed with tagged HCF244 protein (Fig. 1E). HCF244 has been reported to associate with the thylakoid membrane via an interacting protein and to be protected from protease action by its putative binding partner (Link et al., 2012). It is tentatively proposed here that OHP2 functions as the membrane anchor for HCF244.
Our data are in agreement with the interaction of the cyanobacterial HliD with the HCF244 homolog Ycf39, which is one of the many assembly factors involved in the de novo biogenesis of PSI or PSII as well as in the PSII repair cycle (Lu, 2016). The cyanobacterial Ycf39 has been demonstrated to bind directly to HliD but also to CHLG (Knoppová et al., 2014). In contrast, Arabidopsis CHLG could not be detected in either OHP or HCF244 eluates (Fig. 1, A and E). However, as CHLG is an intricately folded binding partner that contains seven transmembrane helices, it is not entirely excluded that its detergent-solubilized form has lost the ability to interact stably with OHPs or HCF244 under in vitro conditions.
OHP1, OHP2, and HCF244 Form a Trimeric Complex and Mutually Stabilize Each Other
Notably, in OHP1- and OHP2-silenced plants, the steady-state level of HCF244 was drastically reduced (VIGS-OHP1) and scarcely detectable (VIGS-OHP2), respectively (Fig. 7A). Conversely, the hcf244 mutant contained decreased amounts of OHP2, while OHP1 was undetectable (Fig. 7C). We suggest that the simultaneous accumulation of the OHP isoforms and HCF244 ensures their mutual stabilization.
The formation of a trimeric protein complex (OHP1-OHP2-HCF244) was verified by the separation of solubilized proteins from wild-type thylakoid membranes by BN-PAGE followed by SDS-PAGE in the second dimension and immunoblotting with specific antibodies. OHP1, OHP2, and HCF244 comigrated in at least three areas of the BN gel, which correspond to protein complexes with different Mr values (Fig. 1F). All three proteins were most abundant in the low-molecular-weight region of the BN gel and also were detected at the level of monomeric PSII and at the position of the LHCII dimer fraction. Dimeric LHCIIs were recently proposed to represent a special assembly form of LHCII possessing high rates of chlorophyll excitation quenching (Janik et al., 2017). Although the three proteins largely accumulated in the low-molecular-weight range, which might reflect single proteins dissociated from the complex during solubilization, the formation and migration as a trimeric 60-kD complex is not entirely excluded. In our studies, OHP1 migrated differently in BN-PAGE and associated to different protein complexes than has been reported recently (Myouga et al., 2018). However, Myouga et al. (2018) used plants expressing a 10×-myc-tagged OHP1 for BN-PAGE analyses, whereas we intended to detect endogenous OHP1. Thus, it cannot be excluded that the oversized peptide tag relative to the low Mr of OHP1 affects the electrophoretic mobility and/or the potential interaction capacities in comparison with an untagged endogenous OHP1.
As stated above, OHP1 was undetectable in the hcf244 mutant, but a significant amount of OHP2 still accumulated (Fig. 7C). BN-PAGE and second dimension SDS-PAGE analyses of the hcf244 thylakoid membranes revealed that the residual OHP2 assembled to the same molecular mass complexes in BN-PAGE as in wild-type samples (Fig. 7E). It is likely that the OHP1-OHP2-HCF244 complex is coupled to other complexes by the interaction of OHP2 with other unknown binding partners and that this interaction is not abolished in the absence of OHP1 and HCF244. It is proposed that its intrinsic stability predestines OHP2 to serve as a starting point for the assembly of the OHP1-OHP2-HCF244 complex (see below).
HCF244 comigrated with both OHPs. Thus, it is proposed that HCF244 is bound mainly to the OHP heterodimer, while unbound HCF244 is less stable. This idea is further supported by the analysis of HCF244 transcripts in HCF244-C lines, where it accumulated some 40-fold relative to the level in the wild type, although no such accumulation of its protein product was observed (Supplemental Fig. S5, B and C). We suggest that the excess HCF244 is unstable in the absence of additional OHP2. In contrast to HCF244, the loss of HCF173 has no effect on the stability of either OHP or HCF244 (Supplemental Fig. S7). VIGS of HCF244 or HCF173 phenocopied the respective T-DNA insertion mutants, including the specific destabilization of PSII core subunits (Supplemental Fig. S7, C and D). However, silencing of HCF173 did not affect the levels of HCF244 or the OHP variants (Supplemental Fig. S7D). OHP1 has been reported previously to interact with HCF244 as well as HCF173 (Myouga et al., 2018). However, we cannot detect a functional connection between the role of HCF173 and that of the trimeric OHP1-OHP2-HCF244 complex in PSII biogenesis. These observations also are compatible with the inability of HCF173 to interact with OHP variants in BiFC (Fig. 1B), although sterical hindrance between the split YFP halves and the tagged proteins preventing an interaction cannot be excluded.
VIGS-OHP2 Plants Display a Pale-Green Phenotype
In this study, we successfully applied the VIGS system to investigate the physiological and biochemical consequences of Arabidopsis OHP deficiency. In contrast to the ohp1 and ohp2 lines, which are deficient in these proteins from germination and early greening stages on, VIGS of OHP expression was first imposed on 2-week-old seedlings, which apparently passed the critical stages of development. The VIGS approach was employed in order to obtain sufficient amounts of leaf material for in-depth functional analysis of both OHP isoforms. Three weeks after the induction of OHP silencing, the steady-state levels of the OHP transcripts and proteins had decreased markedly (Fig. 2, B and C). Interestingly, only the complete loss of OHP2 perturbed plant growth and pigmentation, resulting in pale-green leaves with only 60% to 70% of the chlorophyll content in the control (Fig. 2, A and D). This phenotype differs strikingly from that of the growth-arrested ohp1 and ohp2 mutants, which did not grow on medium without an additional carbon source. The decrease in chlorophyll accumulation in the ohp1 and ohp2 plants is reflected in the abnormal pigmentation of VIGS-OHP2 lines.
Analysis of chlorophyll biosynthesis in VIGS-OHP2 plants revealed a decrease in both steady-state levels of chlorophyll biosynthetic enzymes as well as chlorophyll precursors, whereas no changes in chlorophyll metabolism were observed in VIGS-OHP1 plants (Fig. 3, A and C). VIGS-OHP2 leaves exhibited a 50% decrease in the ALA synthesis rate. This is in agreement with data from Synechocystis strains lacking all Hlips, which exhibited a drastic decrease in ALA synthesis (Yao et al., 2012). These defects in chlorophyll synthesis and photosynthesis in VIGS-OHP2 plants were not caused by altered transcriptional control of the corresponding genes (Supplemental Fig. S6). However, it remains unclear how OHP2 deficiency interferes with chlorophyll metabolism. A reduced demand for chlorophyll for the photosynthetic apparatus will attenuate chlorophyll synthesis by feedback control at the rate-limiting step of ALA synthesis (Shalygo et al., 2009). Thus, it is suggested that, in response to OHP2 silencing, this mechanism attenuates ALA synthesis, which reduces metabolic flow, and results in lower steady-state levels of chlorophyll precursors and ultimately in a decreased rate of chlorophyll synthesis, which reduces the risk of photooxidative damage by free chlorophylls.
The decreased metabolic flow and reduced content of enzymes of chlorophyll biosynthesis are consistent with previous data obtained from cyanobacterial and plant mutants deficient in other representatives of the LHC protein family. Thus, decreased levels of LIL3.1 are accompanied by the destabilization of CHLP and POR, indicating that the stability of these proteins depends on their mutual physical interaction (Hey et al., 2017).
OHP2 Is Required for the Accumulation of PSII Core Subunits
The function of Hlips has been associated with their interactions with proteins involved in pigment synthesis and the delivery of pigments to proteins during the assembly of PS complexes. As OHP deficiency negatively affects the synthesis of chlorophyll, an impact on the biogenesis of thylakoid membranes and the assembly process of chlorophyll-binding proteins of the photosynthetic complexes also was expected. Moreover, as Ycf39 interacts and likely cooperates mutually with cyanobacterial Hlips, it is likely that OHPs also are involved in the PS assembly processes. The decreased chlorophyll a/b ratio in VIGS-OHP2 plants (Fig. 2D) points to a negative impact of OHP2 deficiency on the stability and/or biogenesis of PS core subunits rather than LHCs (Fig. 4A). Interestingly, VIGS-induced OHP2 deficiency sharply reduces steady-state levels of the PSII core subunits D1 and CP43, whereas PSI core subunits and the major LHC proteins LHCA1 and LHCB1 remain essentially unchanged (Fig. 4A).
In agreement with these findings, PSII complex formation was impaired, as was demonstrated by the analysis of DDM-solubilized thylakoid membranes on BN-PAGE gels. PSII supercomplexes were strongly diminished, and monomeric LHCs accumulated (Fig. 4B). Immunological analyses of 2D gel blots with specific antibodies against PS core subunits confirmed the OHP-mediated reduction of PSII, as the accumulation of CP47, CP43, and D1 was strongly decreased (Fig. 4C). In addition, the functional deficit of PSII was confirmed by room-temperature and 77K fluorescence spectroscopy (Fig. 6). VIGS-OHP2 plants showed a high-chlorophyll-fluorescence phenotype (Fig. 6A), which characterizes several PSII mutants (Meurer et al., 1998; Armbruster et al., 2010; Zhang et al., 2011). Hence, these results suggest that OHP2 is involved mainly in the maintenance of adequate amounts of PSII core subunits but does not influence LHC protein stability.
As LHC levels remained stable in VIGS-OHP2 plants, while the amounts of PSII core proteins decreased, it is hypothesized that much of the available LHCII cannot couple to the PSII core antenna and the reaction center. Consequently, excess light energy cannot be effectively quenched, and increased fluorescence emission from uncoupled LHCIIs would account for the elevated dark-level fluorescence in VIGS-OHP2 plants (Fig. 6A). This notion is corroborated by the 77K fluorescence emission spectra, as a blue shift of the PSII emission peak (emanating from LHCII) is indicative of increased LHCII emission (Fig. 6B).
The changes observed in protein levels of PSII core subunits in VIGS-OHP2 plants are not due to decreased plastid transcript levels. On the contrary, transcripts encoding PSII subunits were increased compared with the control (Fig. 5A). In vivo labeling assays with [35S]Met revealed a strongly decreased synthesis of D1/D2 in VIGS-OHP2 plants (Fig. 5, B and C), suggesting the involvement of OHP2 together with OHP1 and HCF244 in the synthesis and accumulation of the stable PSII core proteins. It is proposed that the reduced accumulation of the plastid-encoded proteins is a consequence of the impaired cotranslational assembly of plastid-encoded proteins of PSII with their pigments and cofactors during integration into the thylakoid membrane when OHP variants and HCF244 are missing. Interestingly, also in VIGS-OHP1 lines, a slight decrease in the synthesis of D1/D2 was observed (Fig. 5, B and C). This indicates that the OHP2-HCF244 dimer is principally active without OHP1, but OHP1 seems to be required for the complete activity of the complex during PSII assembly. As VIGS-OHP1 lines showed, under moderate growth conditions, only this weak phenotype, the action of an OHP2-HCF244 complex seems to be sufficient for adequate D1 accumulation. Future studies on the functions of the OHP variants will include growth under abiotic stress conditions.
OHPs Play a Role in PSII Biogenesis/Repair Together with HCF244
Effective knockdown of HCF244 expression results in pale-green and growth-retarded seedlings, which exhibit strongly reduced polysomal loading of the plastid-encoded psbA mRNA (Link et al., 2012). These results suggested that HCF244 is important for the initiation of D1 synthesis. In accordance with these data, coexpression analyses using the ATTED-II database (Aoki et al., 2016) reveal a strong correlation between OHP2 and HCF244 expression profiles. This also holds true for HCF173 (At1g16720), which codes for a factor with a similar function to HCF244 during translational initiation on the psbA transcript (Schult et al., 2007). Indeed, the hcf244 hcf173 double mutant shows an additive effect (Link et al., 2012), in agreement with the functional independence of HCF244 and HCF173 described above.
Although the exact molecular function of HCF244 is not clear, it is undoubtedly required for PSII biogenesis (Link et al., 2012). Membrane tethering of HCF244 via the OHP1-OHP2 heterodimer would enable it to link the synthesis of D1 with the attachment of cofactors and pigments and its integration into the thylakoid membrane. The cotranslational insertion of pigments is required to stabilize D1 (He and Vermaas, 1998). As both OHPs are equipped with the conserved amino acids of the LHC motif (Engelken et al., 2010), it is likely that they are able to bind chlorophyll, presumably as a dimer. Therefore, the OHP heterodimer could functionally connect the synthesis of D1 directly to the subsequent delivery of chlorophylls (Fig. 9). In addition, the OHP heterodimer could serve as a photoprotectant for newly synthesized D1 and/or PSII assembly intermediates. Future experiments should investigate the effects on VIGS-OHP1 plants of exposure to increasing light intensities for prolonged periods in order to check their susceptibility to light stress.
Figure 9.
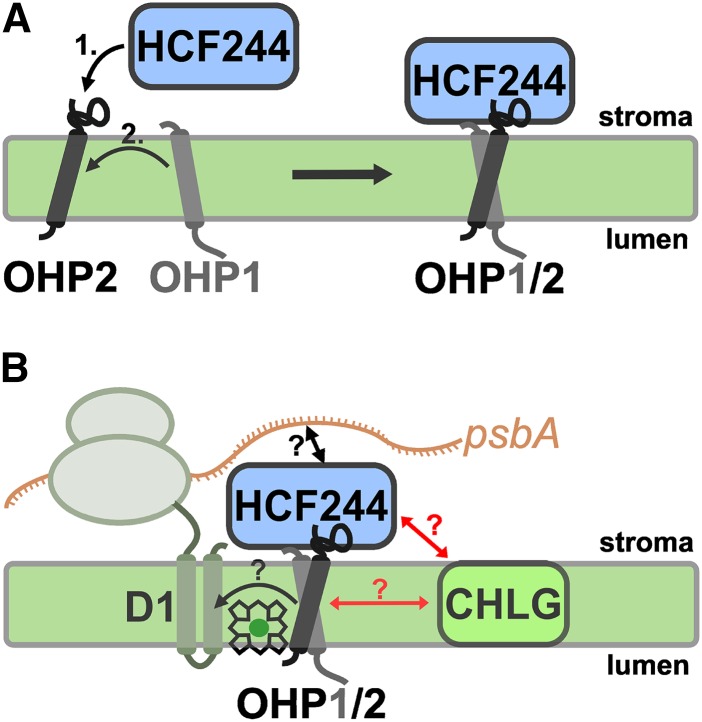
Working model for the assembly and function of the OHP1-OHP2-HCF244 heterotrimeric complex. A, The formation of the heterotrimer begins with the association of HCF244 to OHP2. The N-terminal end of OHP2 is facing the stroma and involved in binding HCF244. This dimer already possesses partial activity, and dimer formation is required for the subsequent binding of OHP1. The heterotrimeric complex is formed when OHP1 associates with OHP2 in the membrane and, furthermore, with the membrane-associated HCF244. HCF244 is required to stably hold OHP1 in the trimeric complex. B, In our model, the OHP heterodimer is involved in the delivery of chlorophylls to newly synthesized D1 proteins, whereas HCF244 is involved in the translation initiation of psbA. A stable interaction or complex formation of either OHPs or HCF244 with CHLG is unlikely (indicated by red arrows), although a transient and/or weak interaction cannot be excluded.
It should be noted that the two OHP isoforms and HCF244 comigrate, inter alia, with monomeric PSII on BN-PAGE gels (Fig. 1F). Monomeric PSII is known to be the intermediate stage in PSII protein assembly and the D1 repair cycle (Rokka et al., 2005; Nixon et al., 2010; Herbstová et al., 2012). Moreover, OHP2 was initially reported to be enriched in stroma lamellae relative to grana thylakoids (Andersson et al., 2003), and this observation was confirmed in this study (Fig. 1G). As the assembly and repair of PSII take place in the stroma thylakoids, the localization of OHP2 together with OHP1 and HCF244 in the stroma thylakoids supports a role for the heterotrimer in the assembly/repair process of PSII complexes.
To disentangle the molecular function of HCF244 from the specific function of the OHP heterodimer remains challenging. However, as HCF244 belongs to a group of atypical SDRs, we substituted three of the four residues forming the catalytic tetrad of typical SDRs (Fig. 8A) and expressed it into the hcf244 knockout mutant. As a result, this mutant HCF244 protein did not compensate entirely for the loss of the endogenous HCF244 (Fig. 8, B and D). However, the mutant HCF244 still acts as a scaffold to stabilize wild-type-like amounts of both OHPs. Therefore, it is clear that HCF244 function must precede the action of OHPs.
CONCLUSION
We provide evidence that OHP2 interacts directly with OHP1 and HCF244 in planta to form a trimeric complex. Balanced accumulation of all three proteins is required for the stability of this complex, while OHP2 appears to provide the basic scaffold. If OHP2 is missing, both OHP1 and HCF244 are destabilized, while deficiency of HCF244 or OHP1 has less impact on the amounts of OHP2. Thus, apart from its role in the trimeric complex, OHP2 probably also interacts with other membrane-bound complexes and couples the trimeric complex with other proteins. In our model, assembly begins with monomeric OHP2, either free or already bound to other complexes. Then, HCF244 interacts with the stromal part of OHP2 (Fig. 9A). HCF244-OHP2 already possesses (partial) activity, as shown in the in vivo labeling experiments in VIGS-OHP1 plants (Fig. 5, B and C). The presence of HCF244 is required for the subsequent association of OHP1 to the complex, which interacts with OHP2 in the membrane and, furthermore, with the membrane-associated HCF244. HCF244 thereby acts as a scaffold, tethering together both OHP variants, and stabilizes the OHP heterodimer. On the other hand, the association of OHP1 also has a stabilizing effect on HCF244. It is likely that the full activity of the complex in the PS assembly can be achieved only when OHP1 is added (Fig. 9B). However, the questions of which conditions substantially require OHP1 and why OHP1 is degraded in the absence of OHP2 and HCF244 need to be addressed in the future.
While induced OHP1 silencing does not perturb chloroplast function in 12-d-old seedlings, the strongly growth-retarded phenotype of the ohp1 mutant indicates that OHP1 is indispensable in the initial phase of germination. However, the interdependent function of the two OHP variants is supported by the observation that neither ohp mutant could be complemented by the expression of the other OHP gene under the control of the 35S promoter (Beck et al., 2017). Thus, the presence of the OHP heterodimer is essential during the critical, early developmental stage and most probably also is relevant under stressful environmental conditions.
Future work will focus on the chlorophyll-binding properties of OHPs. Hlip dimers with bound chlorophyll a and β-carotene act in energy dissipation (Staleva et al., 2015). It is likely that OHPs also bind chlorophylls and/or carotenes as dimers. As the homodimer formation of OHPs could not be detected in the thylakoid membrane, pigments are likely to be bound by the OHP1-OHP2 heterodimer. To what extent the putative chlorophyll-binding capability is essential for the function of the heterotrimeric complex will be analyzed by characterizing plants expressing OHPs with mutated chlorophyll-binding motifs.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Generation of Transgenic Lines
Arabidopsis (Arabidopsis thaliana) plants were grown on soil in a growth chamber (100 µmol photons m−2 s−1, 8-h photoperiod, 20°C). The hcf244 mutant line was obtained from the Nottingham Arabidopsis Stock Centre (GK-088C04), and the homozygosity of the T-DNA insertion was confirmed by PCR using specific primers (Supplemental Table S1). For the overexpression of OHPs, both OHP1 and OHP2 cDNAs were amplified with specific primers from Arabidopsis total cDNA generated by RT-PCR (Supplemental Table S1) and cloned into a pCAMstrepII vector that was created as follows. The 2.2-kb T-DNA region of pXCS-HA-strepII (Witte et al., 2004) was amplified using Phusion polymerase (New England Biolabs) and primers pXCSfrag_fw (5′-CGTTTAAACAGCTTGCATGCCGGTCG-3′) and pXCSfrag_rev (5′-AGTTTAAACTGAAGGCGGGAAAC-3′). The PCR product was ligated into pJET (Thermo Scientific) and sequenced to confirm sequence identity. Digestion by PmeI yielded the blunt-ended 2.2-kb T-DNA fragment. A 6.3-kb fragment containing the vector backbone of the binary vector pCAMBIA3301 was amplified from pCAMBIA3301 using primers pCAM33_RBout (5′-GTTTAAACTATCAGTGTTTGACAGG-3′) and pCAM33_LBout (5′-GTTTAAACAATTCAGTACATTAAAAACGTCCG-3′). The product was cloned into pJET and subsequently cut out by PmeI. Both PmeI fragments were ligated to create the 8.5-kb pCAMstrepII. The orientation of the T-DNA was verified by restriction enzyme analysis. The vector was transformed subsequently into Arabidopsis wild-type plants (Columbia-0) via Agrobacterium tumefaciens-mediated transformation. For complementation of the hcf244 mutant, HCF244 cDNA was amplified with specific primers from Arabidopsis total cDNA generated by RT-PCR (Supplemental Table S1) and cloned into the pCAMstrepII vector. The mutated HCF244 sequence was generated by overlap-extension PCR and cloned into the pCAMstrepII vector. The vectors were transformed subsequently into hcf244 mutant plants as described above. For complementation of the ohp mutants, an ∼2-kb genomic fragment of each OHP gene (consisting of a 1-kb upstream region, the coding sequence, and a 0.5-kb downstream region) was expanded with an HA tag fused to the coding sequence by overlap-extension PCR using specific primers (Supplemental Table S1), and the whole fragment was cloned subsequently into the pCAMBIA3301 vector. The vector was transformed subsequently into the T-DNA insertion mutants as described above.
VIGS Assay
Arabidopsis plants used for the VIGS assay were grown on soil in a growth chamber (100 µmol photons m−2 s−1, 16-h photoperiod, 20°C). The tobacco rattle virus-based vectors pTRV1 and pTRV2 were used for the VIGS assay (Liu et al., 2002). OHP1 and OHP2 cDNAs were amplified from Arabidopsis total cDNA generated by RT-PCR as described above (Supplemental Table S1) and cloned into the pTRV2 vector. A pTRV2-GFP vector was employed as the negative control. Infiltration of A. tumefaciens cells transformed with the pTRV plasmids was performed according to Burch-Smith et al. (2006).
Pigment Extraction and HPLC
For pigment extraction, leaf material was harvested 2 h after daybreak and ground in liquid nitrogen. Subsequently, the powder was freeze dried and the pigments were extracted with ice-cold, alkaline acetone (acetone:0.2 n NH4OH, 9:1). Pigments were separated by HPLC and quantified using pure standards.
Determination of the ALA Synthesis Rate
ALA synthesis rates were measured according to Mauzerall and Granick (1956). Briefly, detached leaves or leaf discs were incubated in 40 mm levulinic acid and 50 mm Tris-HCl, pH 7.2, for 3 to 4 h under growth light conditions. Then, the leaf material was ground in liquid nitrogen and resuspended in 50 mm KPO4 buffer, pH 6.8. After centrifugation, 4 volumes of the supernatant was mixed with 1 volume of ethyl acetoacetate, boiled for 10 min, and mixed 1:1 with modified Ehrlich reagent. The ALA concentration was then determined photometrically and quantified by comparison with an authentic standard.
Preparation of Total Leaf Protein, and SDS-PAGE and Immunoblot Analyses
Total leaf proteins were extracted from leaf material, ground in liquid nitrogen by resuspension in sample buffer (100 mm Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 200 mm DTT, and 0.01% Bromophenol Blue), followed by incubation at 95°C for 10 min. SDS-PAGE was performed on 12% SDS-PAGE gels according to Laemmli (1970), and equal amounts of protein were loaded for the different samples. In order to separate the protein components of photosynthetic complexes, 6 m urea was added to the separating gel as required. For the detection of OHPs, protein extracts were fractionated on 10% Tricine-SDS gels (Schägger, 2006). In either case, proteins were transferred subsequently onto nitrocellulose membranes and probed with specific antibodies. For signal detection, the Clarity Western ECL Blotting Substrate (Bio-Rad) was used.
Thylakoid Extraction and BN-PAGE
Thylakoid membranes were extracted according to Järvi et al. (2011), except that leaves were first homogenized in buffer containing 450 mm sorbitol, 20 mm Tricine-KOH, pH 8.4, 10 mm EDTA, and 0.1% BSA. For storage, thylakoids were resuspended in 25BTH20G buffer (25 mm BisTris, pH 7, and 20% glycerol) and stored at −80°C. BN-PAGE was performed according to Peng et al. (2008) on 4% to 12.5% BN gels.
Subfractionation of Thylakoids
Thylakoids were subfractionated into grana and stroma thylakoids by differential centrifugation according to Baena-González et al. (1999). Briefly, isolated thylakoids were solubilized with 0.2% digitonin at a chlorophyll concentration of 0.2 µg µL−1 in fractionation buffer (100 mm sorbitol, 50 mm HEPES, pH 7.5, and 10 mm MgCl2) and centrifuged for 5 min at 4,500g. The supernatant was centrifuged at 40,000g for 30 min to collect the grana stacks and further at 140,000g for 90 min to obtain the stroma lamellae. Membranes (equal amounts of chlorophyll) were then subjected to SDS-PAGE.
In Vivo Radioactive Labeling of Thylakoid Proteins
Radioactive labeling of thylakoid proteins was performed according to Meurer et al. (1998) with modifications. Leaves from VIGS plants were incubated in 200 µL of reaction buffer (1 mm KH2PO4, pH 6.3, 0.1% Tween 20, and 20 µg mL−1 cycloheximide) for 30 min, and then 30 µCi of [35S]Met (Hartmann Analytic) in 100 µL of reaction buffer was added (0.1 µCi µL−1 final concentration). After a brief vacuum infiltration, leaves were incubated in ambient light for 60 min. Leaves then were homogenized in PBS buffer and centrifuged, and the pellet was resuspended into SDS-PAGE sample buffer. SDS-PAGE was carried out as described above.
Gene Expression Analyses
Total RNAs were extracted from leaf material as described previously (Oñate-Sánchez and Vicente-Carbajosa, 2008). Subsequently, 1-µg aliquots of RNA were digested with DNase, and cDNA synthesis was performed with RevertAid RT enzyme (Thermo) according to the manufacturer’s instructions. Gene expression was analyzed by RT-qPCR, which was carried out in a CFX 96 real-time system (Bio-Rad) using 2× SensiMixSYBR (Bioline). Primers used for qPCR analysis are listed in Supplemental Table S2. ACT2 (At3g18780) and SAND (At2g28390) were used as reference genes, and normalization was performed by the ΔΔCt method (Pfaffl, 2001). Expression analysis of plastid-encoded genes was done in a LightCycler 480 II system (Roche) using LightCycler 480 SYBR Green I (Roche). In this case, 18S-rRNA (At3g41768), ACT1-1 (At2g37620), and AP2M (At5g46630) were used as reference genes, and data were analyzed with standard curves.
Chlorophyll Fluorescence Measurements
PAM room-temperature fluorescence was measured with an FMS2 Portable Pulse Modulated Chlorophyll Fluorometer (Hansatech). In general, the protocol described by Meurer et al. (1996) was followed, except that the actinic light was adjusted to growth light conditions (100 µmol photons m−2 s−1). For 77K spectroscopy, leaf material was homogenized in buffer containing 0.4 m sorbitol and 50 mm Tricine, pH 7.8, and the chlorophyll concentration was adjusted to 10 µg mL−1. Subsequently, the suspension was mixed with 1 volume of 80% glycerol, filled into a capillary, and frozen in liquid nitrogen. Chlorophyll fluorescence was then measured with an F-7000 fluorescence spectrophotometer (Hitachi).
Expression and Purification of Recombinant OHP1 and OHP2 Proteins, and Antibody Production
Coding sequences of both OHP genes without the putative transit peptides were amplified from cDNA with primers listed in Supplemental Table S1 and cloned into the pET22b vector (Novagen). Protein expression was carried out at 37°C for 3 h in Rosetta 2 cells (Novagen) grown in 2YT medium containing 1% Glc and was induced by adding 1 mm IPTG (Sigma). Recombinant proteins were solubilized in the presence of 8 m urea and purified on HisPur Ni-NTA Resin (Thermo) following the manufacturer’s instructions. After purification, proteins were dialyzed into buffer containing 50 mm KPO4, pH 7.6, 5% glycerol, and 0.8 m urea. Immunization of rabbits and antibody purification were performed by BioGenes (Berlin).
BiFC Analysis
OHP1, OHP2, HCF244, and HCF173 cDNAs were amplified from total cDNA using specific primers carrying attB sites (Supplemental Table S1) and cloned into pDEST-GW-VYNE/-VYCE vectors (Gehl et al., 2009) via pDON207 using the Gateway system (Thermo Fisher). Truncated OHP2 sequences were generated by overlap-extension PCR with specific primers (Supplemental Table S1). Plasmids were transformed into A. tumefaciens strain GV2260, and BiFC experiments were performed as described previously (Hey et al., 2017). YFP fluorescence was recorded on an LSM 800 confocal microscope (Zeiss).
StrepII Tag Pull-Down Assay
Isolated thylakoids were solubilized with 1% DDM in pull-down buffer (50 mm Tris, pH 8, 0.5 mm EDTA, 1 mm NaCl, and 10% glycerol) and incubated at 4°C for 2 h with streptavidin-coated beads (Strep-Tactin Macroprep; IBA). After intensive washing, proteins were eluted with 10 mm desthiobiotin (Sigma) and concentrated on Corning Spin-X UF concentrators (3K MWCO; Sigma). Proteins then were fractionated on acrylamide gels and detected by immunoblotting.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under accession numbers At5g02120 (OHP1), At1g34000 (OHP2), At4g35250 (HCF244), and At1g16720 (HCF173).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization of the OHP-OX lines.
Supplemental Figure S2. Expression levels of HA-strepII-tagged OHP species in the OHP-OX lines.
Supplemental Figure S3. Complementation of the ohp1 and ohp2 mutants with HA-tagged OHP variants.
Supplemental Figure S4. Silver staining of the pull-down eluates.
Supplemental Figure S5. Characterization of the hcf244 mutant and the HCF244-C complementation line.
Supplemental Figure S6. Transcript analysis of tetrapyrrole biosynthesis genes in VIGS plants.
Supplemental Figure S7. Characterization of VIGS-HCF244/VIGS-HCF173 lines.
Supplemental Table S1. Primers used for cloning and genotyping.
Supplemental Table S2. Primers used for RT-qPCR analysis.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Peter Westhoff and Karin Meierhoff for kindly providing α-HCF244 antiserum. Furthermore, we thank Peng Wang for providing the TRV1 and TRV2 vectors and Heiko Lokstein and Josephine Herbst for valuable discussions.
Footnotes
This work was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft) to B.G. (Gr936 14-1, subproject of the DFG-Research Unit FOR2092).
Articles can be viewed without a subscription.
References
- Adamska I. (1997) ELIPs: light-induced stress proteins. Physiol Plant 100: 794–805 [Google Scholar]
- Andersson U, Heddad M, Adamska I (2003) Light stress-induced one-helix protein of the chlorophyll a/b-binding family associated with photosystem I. Plant Physiol 132: 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrizhiyevskaya EG, Chojnicka A, Bautista JA, Diner BA, van Grondelle R, Dekker JP (2005) Origin of the F685 and F695 fluorescence in photosystem II. Photosynth Res 84: 173–180 [DOI] [PubMed] [Google Scholar]
- Aoki Y, Okamura Y, Tadaka S, Kinoshita K, Obayashi T (2016) ATTED-II in 2016: a plant coexpression database towards lineage-specific coexpression. Plant Cell Physiol 57: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apitz J, Nishimura K, Schmied J, Wolf A, Hedtke B, van Wijk KJ, Grimm B (2016) Posttranslational control of ALA synthesis includes GluTR degradation by Clp protease and stabilization by GluTR-binding protein. Plant Physiol 170: 2040–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U, Zühlke J, Rengstl B, Kreller R, Makarenko E, Rühle T, Schünemann D, Jahns P, Weisshaar B, Nickelsen J, et al. (2010) The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell 22: 3439–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Barbato R, Aro EM (1999) Role of phosphorylation in the repair cycle and oligomeric structure of photosystem II. Planta 208: 196–204 [Google Scholar]
- Beck J, Lohscheider JN, Albert S, Andersson U, Mendgen KW, Rojas-Stütz MC, Adamska I, Funck D (2017) Small one-helix proteins are essential for photosynthesis in Arabidopsis. Front Plant Sci 8: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Liu Y, Dinesh-Kumar SP (2006) Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol 142: 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidgey JW, Linhartová M, Komenda J, Jackson PJ, Dickman MJ, Canniffe DP, Koník P, Pilný J, Hunter CN, Sobotka R (2014) A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. Plant Cell 26: 1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki O, Grimm B (2012) Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J Exp Bot 63: 1675–1687 [DOI] [PubMed] [Google Scholar]
- Czarnecki O, Hedtke B, Melzer M, Rothbart M, Richter A, Schröter Y, Pfannschmidt T, Grimm B (2011) An Arabidopsis GluTR binding protein mediates spatial separation of 5-aminolevulinic acid synthesis in chloroplasts. Plant Cell 23: 4476–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra A, Bies DH, Lehner KR, Folta KM (2006) Green light adjusts the plastid transcriptome during early photomorphogenic development. Plant Physiol 142: 1256–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar SP, Anandalakshmi R, Marathe R, Schiff M, Liu Y (2003) Virus-induced gene silencing. Methods Mol Biol 236: 287–294 [DOI] [PubMed] [Google Scholar]
- Engelken J, Brinkmann H, Adamska I (2010) Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily. BMC Evol Biol 10: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermakova-Gerdes S, Vermaas W (1999) Inactivation of the open reading frame slr0399 in Synechocystis sp. PCC 6803 functionally complements mutations near the Q(A) niche of photosystem II: a possible role of Slr0399 as a chaperone for quinone binding. J Biol Chem 274: 30540–30549 [DOI] [PubMed] [Google Scholar]
- Filling C, Berndt KD, Benach J, Knapp S, Prozorovski T, Nordling E, Ladenstein R, Jörnvall H, Oppermann U (2002) Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J Biol Chem 277: 25677–25684 [DOI] [PubMed] [Google Scholar]
- Gehl C, Waadt R, Kudla J, Mendel RR, Hänsch R (2009) New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol Plant 2: 1051–1058 [DOI] [PubMed] [Google Scholar]
- He Q, Vermaas W (1998) Chlorophyll a availability affects psbA translation and D1 precursor processing in vivo in Synechocystis sp. PCC 6803. Proc Natl Acad Sci USA 95: 5830–5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddad M, Adamska I (2000) Light stress-regulated two-helix proteins in Arabidopsis thaliana related to the chlorophyll a/b-binding gene family. Proc Natl Acad Sci USA 97: 3741–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbstová M, Tietz S, Kinzel C, Turkina MV, Kirchhoff H (2012) Architectural switch in plant photosynthetic membranes induced by light stress. Proc Natl Acad Sci USA 109: 20130–20135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey D, Rothbart M, Herbst J, Wang P, Müller J, Wittmann D, Gruhl K, Grimm B (2017) LIL3, a light-harvesting complex protein, links terpenoid and tetrapyrrole biosynthesis in Arabidopsis thaliana. Plant Physiol 174: 1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CC, Chang YH, Hsu CN, Hsu HH, Li CW, Pon HI (2005) Mechanistic roles of Ser-114, Tyr-155, and Lys-159 in 3alpha-hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni. J Biol Chem 280: 3522–3528 [DOI] [PubMed] [Google Scholar]
- Janik E, Bednarska J, Sowinski K, Luchowski R, Zubik M, Grudzinski W, Gruszecki WI (2017) Light-induced formation of dimeric LHCII. Photosynth Res 132: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson S. (1994) The light-harvesting chlorophyll a/b-binding proteins. Biochim Biophys Acta 1184: 1–19 [DOI] [PubMed] [Google Scholar]
- Jansson S, Andersson J, Kim SJ, Jackowski G (2000) An Arabidopsis thaliana protein homologous to cyanobacterial high-light-inducible proteins. Plant Mol Biol 42: 345–351 [DOI] [PubMed] [Google Scholar]
- Järvi S, Suorsa M, Paakkarinen V, Aro EM (2011) Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem J 439: 207–214 [DOI] [PubMed] [Google Scholar]
- Knoppová J, Sobotka R, Tichy M, Yu J, Konik P, Halada P, Nixon PJ, Komenda J (2014) Discovery of a chlorophyll binding protein complex involved in the early steps of photosystem II assembly in Synechocystis. Plant Cell 26: 1200–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Li XP, Björkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Link S, Engelmann K, Meierhoff K, Westhoff P (2012) The atypical short-chain dehydrogenases HCF173 and HCF244 are jointly involved in translational initiation of the psbA mRNA of Arabidopsis. Plant Physiol 160: 2202–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W (2004) Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428: 287–292 [DOI] [PubMed] [Google Scholar]
- Lu Y. (2016) Identification and roles of photosystem II assembly, stability, and repair factors in Arabidopsis. Front Plant Sci 7: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, Watkins KP, Barkan A, van Wijk KJ (2012) Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol 158: 156–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauzerall D, Granick S (1956) The occurrence and determination of delta-amino-levulinic acid and porphobilinogen in urine. J Biol Chem 219: 435–446 [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J, Meierhoff K, Westhoff P (1996) Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198: 385–396 [DOI] [PubMed] [Google Scholar]
- Meurer J, Plücken H, Kowallik KV, Westhoff P (1998) A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J 17: 5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montané MH, Kloppstech K (2000) The family of light-harvesting-related proteins (LHCs, ELIPs, HLIPs): was the harvesting of light their primary function? Gene 258: 1–8 [DOI] [PubMed] [Google Scholar]
- Mork-Jansson AE, Gargano D, Kmiec K, Furnes C, Shevela D, Eichacker LA (2015) Lil3 dimerization and chlorophyll binding in Arabidopsis thaliana. FEBS Lett 589: 3064–3070 [DOI] [PubMed] [Google Scholar]
- Myouga F, Takahashi K, Tanaka R, Nagata N, Kiss AZ, Funk C, Nomura Y, Nakagami H, Jansson S, Shinozaki K (2018) Stable accumulation of photosystem II requires ONE-HELIX PROTEIN1 (OHP1) of the light harvesting-like family. Plant Physiol 176: 2277–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiedzki DM, Tronina T, Liu H, Staleva H, Komenda J, Sobotka R, Blankenship RE, Polívka T (2016) Carotenoid-induced non-photochemical quenching in the cyanobacterial chlorophyll synthase-HliC/D complex. Biochim Biophys Acta 1857: 1430–1439 [DOI] [PubMed] [Google Scholar]
- Nixon PJ, Michoux F, Yu J, Boehm M, Komenda J (2010) Recent advances in understanding the assembly and repair of photosystem II. Ann Bot 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MH, Moon YH, Lee CH (2003) Increased stability of LHCII by aggregate formation during dark-induced leaf senescence in the Arabidopsis mutant, ore10. Plant Cell Physiol 44: 1368–1377 [DOI] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J (2008) DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes 1: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Shimizu H, Shikanai T (2008) The chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis. J Biol Chem 283: 34873–34879 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokka A, Suorsa M, Saleem A, Battchikova N, Aro EM (2005) Synthesis and assembly of thylakoid protein complexes: multiple assembly steps of photosystem II. Biochem J 388: 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H. (2006) Tricine-SDS-PAGE. Nat Protoc 1: 16–22 [DOI] [PubMed] [Google Scholar]
- Schmid HC, Oster U, Kögel J, Lenz S, Rüdiger W (2001) Cloning and characterisation of chlorophyll synthase from Avena sativa. Biol Chem 382: 903–911 [DOI] [PubMed] [Google Scholar]
- Schult K, Meierhoff K, Paradies S, Töller T, Wolff P, Westhoff P (2007) The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell 19: 1329–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalygo N, Czarnecki O, Peter E, Grimm B (2009) Expression of chlorophyll synthase is also involved in feedback-control of chlorophyll biosynthesis. Plant Mol Biol 71: 425–436 [DOI] [PubMed] [Google Scholar]
- Shukla MK, Llansola-Portoles MJ, Tichy M, Pascal AA, Robert B, Sobotka R (2018) Binding of pigments to the cyanobacterial high-light-inducible protein HliC. Photosynth Res 137: 29–39 [DOI] [PubMed] [Google Scholar]
- Staleva H, Komenda J, Shukla MK, Šlouf V, Kaňa R, Polívka T, Sobotka R (2015) Mechanism of photoprotection in the cyanobacterial ancestor of plant antenna proteins. Nat Chem Biol 11: 287–291 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Tanaka A (2007) Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol 58: 321–346 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Rothbart M, Oka S, Takabayashi A, Takahashi K, Shibata M, Myouga F, Motohashi R, Shinozaki K, Grimm B, et al. (2010) LIL3, a light-harvesting-like protein, plays an essential role in chlorophyll and tocopherol biosynthesis. Proc Natl Acad Sci USA 107: 16721–16725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibiletti T, Rehman AU, Vass I, Funk C (2018) The stress-induced SCP/HLIP family of small light-harvesting-like proteins (ScpABCDE) protects photosystem II from photoinhibitory damages in the cyanobacterium Synechocystis sp. PCC 6803. Photosynth Res 135: 103–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Jantaro S, Lu B, Majeed W, Bailey M, He Q (2008) The high light-inducible polypeptides stabilize trimeric photosystem I complex under high light conditions in Synechocystis PCC 6803. Plant Physiol 147: 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte CP, Noël LD, Gielbert J, Parker JE, Romeis T (2004) Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol Biol 55: 135–147 [DOI] [PubMed] [Google Scholar]
- Xu H, Vavilin D, Funk C, Vermaas W (2002) Small Cab-like proteins regulating tetrapyrrole biosynthesis in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 49: 149–160 [DOI] [PubMed] [Google Scholar]
- Yao D, Kieselbach T, Komenda J, Promnares K, Prieto MA, Tichy M, Vermaas W, Funk C (2007) Localization of the small CAB-like proteins in photosystem II. J Biol Chem 282: 267–276 [DOI] [PubMed] [Google Scholar]
- Yao DCI, Brune DC, Vavilin D, Vermaas WFJ (2012) Photosystem II component lifetimes in the cyanobacterium Synechocystis sp. strain PCC 6803: small Cab-like proteins stabilize biosynthesis intermediates and affect early steps in chlorophyll synthesis. J Biol Chem 287: 682–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhou G, Liu B, Kong Y, Chen N, Qiu Q, Yin H, An J, Zhang F, Chen F (2011) HCF243 encodes a chloroplast-localized protein involved in the D1 protein stability of the Arabidopsis photosystem II complex. Plant Physiol 157: 608–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Zhou W, Wang H, Ding S, Lu Q, Wen X, Peng L, Zhang L, Lu C (2013) Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell 25: 2925–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]