The interaction between the long non-coding RNA PILNCR1 and the microRNA miR399 is important for low Pi tolerance in maize (Zea mays).
Abstract
The regulation of adaptive responses to phosphorus (P) deficiency by the microRNA399 (miR399)/PHOSPHATE2 (PHO2) pathway has been well studied in Arabidopsis (Arabidopsis thaliana) but not in maize (Zea mays). Here, we show that miR399 transcripts are strongly induced in maize by phosphate (Pi) deficiency. Transgenic maize plants that overexpressed MIR399b accumulated excessive amounts of P in their shoots and displayed typical Pi-toxicity phenotypes. We reannotated ZmPHO2 with an additional 1,165 bp of the 5′ untranslated region. miR399-guided posttranscriptional repression of ZmPHO2 was mainly observed in the P-efficient lines. We identified Pi-deficiency-induced long-noncoding RNA1 (PILNCR1) from our strand-specific RNA libraries. Transient expression assays in Nicotiana benthamiana and maize leaf protoplasts demonstrated that PILNCR1 inhibits ZmmiR399-guided cleavage of ZmPHO2. The abundance of PILNCR1 was significantly higher in P-inefficient lines than in P-efficient lines, which is consistent with the abundance of ZmmiR399 transcripts. These results indicate that the interaction between PILNCR1 and miR399 is important for tolerance to low Pi in maize.
Phosphorus (P), the key structural component of nucleic acids, phospholipids, and the energy-carrying molecule ATP, is essential for the normal growth and development of plants. Due to its low availability and slow rate of diffusion in soil, phosphate (Pi) availability is one of the main environmental constraints to crop productivity worldwide (Péret et al., 2011; López-Arredondo et al., 2014). To respond to the dynamic variations of P availability in soils, plants have evolved morphological, physiological, and biochemical adaptations (Péret et al., 2011). These adaptations in response to variable P availability are at least partially dependent on changes in gene expression. With the discovery of noncoding RNAs (ncRNAs), increased attention has been focused on their important roles in posttranscriptional or translational gene regulation.
ncRNAs mainly include small RNAs and long noncoding RNAs (lncRNAs). The small RNAs can be classified as 20- to 24-nucleotide microRNAs (miRNAs), 21-nucleotide transacting small interfering RNAs (siRNAs), ∼24-nucleotide repeat-associated siRNAs, and 21- or 24-nucleotide natural antisense siRNAs. Transcriptome profiling has identified diverse miRNAs, including miR399s, that are involved in responses to Pi deprivation (Hsieh et al., 2009; Pant et al., 2009; Hackenberg et al., 2013). The miR399 family in Arabidopsis (Arabidopsis thaliana) contains six members, and the Arabidopsis miR399 was the first miRNA demonstrated to be involved in Pi-deprivation responses in plants (Fujii et al., 2005; Chiou et al., 2006). The expression of miR399, which is specifically induced by Pi deficiency, regulates Pi homeostasis in Arabidopsis, rice (Oryza sativa), and soybean (Glycine max) by suppressing a ubiquitin-conjugating E2 enzyme, PHOSPHATE2 (PHO2; Fujii et al., 2005; Chiou et al., 2006; Hu et al., 2011; Xu et al., 2013). Further research showed that Pi deficiency-dependent induction of miR399 was significantly suppressed in a phosphate starvation responsive1 (phr1) loss-of-function Arabidopsis mutant (Bari et al., 2006), indicating that PHR1 acts upstream of miR399 in the Pi signaling pathway. PHR1, miR399, and PHO2 define a conserved signaling pathway that responds to Pi deficiency in plants (Bari et al., 2006).
As an important model system for basic biological research, studies in maize (Zea mays) have contributed significantly to our understanding of the regulatory functions of miRNAs (Zhang et al., 2009). The miR399 family in maize, which contains four more members than the family in Arabidopsis, can be divided into six subgroups based on mature miRNA sequences (Zhang et al., 2009). Predicted targets of miR399s in maize mainly include Pi transporters (PHTs) but not AtPHO2 homologs (Zhang et al., 2009; Pei et al., 2013). Thus, the biological roles of miR399 in maize require further investigation.
lncRNAs are typically longer than 200 nucleotides and can be divided into long-intergenic ncRNAs (lincRNAs), intronic ncRNAs (incRNAs), and long noncoding natural antisense transcripts (lncNATs), which are transcribed from the opposite DNA strand to other transcripts and which partly overlap with sense RNA (Chekanova, 2015). In humans, lncRNAs have been demonstrated to be involved in X chromosome inactivation, genomic imprinting, cell cycle regulation, and lineage commitment (Xue et al., 2016). By integrating transcriptomic analysis and high-throughput sequencing data, researchers have identified thousands of lncRNAs in several plant species, such as Arabidopsis, cotton (Gossypium sp.), rice, and maize (Liu et al., 2012; Li et al., 2014; Zhang et al., 2014; Wang et al., 2015a,b). However, the detailed functional identification of plant lncRNAs is still in an early stage. Although only a few lncRNAs have been well characterized in plants, lncRNAs could be important modulators of plant developmental processes, as suggested by research on lincRNA XLOC_057324 (Zhang et al., 2014), lncNAT COOLAIR (Swiezewski et al., 2009), incRNA COLDAIR (Heo and Sung, 2011), and lincRNA HIDDEN TREASURE1 (Wang et al., 2014). Recently, cis-NATPHO1;2 was demonstrated to act as a translational enhancer for its cognate PHO1;2 and to affect Pi homeostasis in rice (Jabnoune et al., 2013), suggesting that lncRNAs also regulate plant adaptive responses to nutrient deprivation.
In this research, we show that ZmMIR399b-overexpressing maize accumulated excessive P in shoots and displayed typical Pi toxicity phenotypes in old leaves. We reannotated ZmPHO2 and found that miR399-guided posttranscriptional repression of ZmPHO2 was observed mainly in the P-efficient lines. Furthermore, we demonstrated that Pi-deficiency-induced lncRNA1 (PILNCR1) could inhibit ZmmiR399-guided cleavage of ZmPHO2 and that the abundance of PILNCR1 was significantly higher in P-inefficient lines than in P-efficient lines. Taken together, our results reveal how lncRNAs help plants adapt to Pi deficiency by interacting with miRNA.
RESULTS
ZmmiR399 Is Up-Regulated by Pi Deficiency
PHO2 has been experimentally demonstrated to be the target of miR399 in Arabidopsis, rice, soybean, and barley (Hordeum vulgare; Fujii et al., 2005; Hu et al., 2011; Hackenberg et al., 2013; Xu et al., 2013). However, the predicted targets of ZmmiR399 did not include PHO2 or PHO2-like genes, which motivated us to investigate the biological functions of miR399 in maize. We first investigated the responses of ZmmiR399s to Pi stress in hydroponic solutions containing sufficient (250 µm) or limiting (5 µm) Pi. Phenotypic differences between P-sufficient and -deficient maize appeared at 8 d after Pi deficiency treatment. P-deficient maize showed the typical P deficiency phenotype with an accumulation of the purple flavonoid pigment anthocyanin in the old leaves (Supplemental Fig. S1A). Pi deficiency also led to a significant decrease in shoot weight and to an increase in the root-to-shoot weight ratio (Supplemental Fig. S1, B and C).
To test the regulation of ZmmiR399 expression by Pi stress, we conducted a time course experiment with Pi deficiency. Under the Pi-sufficient condition, the expression of ZmmiR399 was undetectable in both shoots and roots of maize (Fig. 1A). Under Pi stress, the level of ZmmiR399 increased substantially in both shoots and roots of maize, and the accumulation of ZmmiR399 was positively correlated with the duration of Pi deficiency (Fig. 1A). To determine whether the response of ZmmiR399 was specific to Pi stress, we also isolated small RNA from maize subjected to nitrogen (N) deficiency. The expression of ZmmiR399 was undetectable in maize under N-sufficient or N-deficient conditions (Supplemental Fig. S2).
Figure 1.
Phenotypes of MIR399b-overexpressing transgenic maize grown in hydroponic solution. A, Small RNA northern blots showing miR399 is up-regulated by Pi deficiency in maize shoots and roots. A 5-µg quantity of small RNA from each sample was loaded per lane and hybridized with a 32P-labeled oligonucleotide probe. U6 was used as a loading control. Numbers below each line of the gel indicate the relative expression of miR399 to U6. nd, Nondetectable; SP and LP, sufficient P and low P, respectively. B, Pi toxicity phenotype of MIR399b-overexpressing transgenic maize (OE2 and OE6) grown in hydroponic solution. Representative seedlings were photographed. The expression level of miR399 in the wild type (WT) and OE were detected by small RNA northern blots. C, P concentrations in shoots and roots of inbred line ZZC01 (wild type) and MIR399b-overexpressing transgenic maize under the Pi-sufficient condition. D, Shoot and root dry weight of inbred line ZZC01 (wild type) and MIR399b-overexpressing transgenic maize under the Pi-sufficient condition. Values in C and D are means ± se (n = 3). Means with the same letter are not significantly different at P < 0.01 according to the LSD test.
Transgenic Maize Plants Overexpressing ZmMIR399b Accumulate P in Shoots
To further investigate the function of ZmMIR399, we chose ZmMIR399b to generate ZmMIR399-overexpressing transgenic maize plants under the constitutive rice ubiquitin promoter. The ZmMIR399b precursor was introduced into the maize inbred line ZZC01 (the genetic background for transgenic maize) through Agrobacterium tumefaciens-mediated transformation of immature embryos. Two independent transgenic events were chosen based on ZmMIR399b expression (Fig. 1B). The responses of transgenic maize overexpressing ZmMIR399b to Pi deficiency were assessed in hydroponic solution. Because ZmPHTs were the potential targets of ZmmiR399, we deduced that the P toxicity phenotypes observed in miR399-overexpressing Arabidopsis and rice should not occur in transgenic maize overexpressing ZmMIR399b. Under the Pi-sufficient condition, ZmMIR399b-overexpressing transgenic maize showed typical P toxicity phenotypes with chlorosis or necrosis at the leaf tips and margins of old leaves (Fig. 1B). The total P concentrations were about two times higher in the old leaves of ZmMIR399b-overexpressing transgenic maize than in the wild type under the Pi-sufficient condition (Fig. 1C). The total P concentration in the wild-type roots was 8.18 mg g−1 dry weight, which was 16%–25% higher than the concentrations in the roots of the transgenic maize (Fig. 1C). In contrast, the shoot and root dry weights of wild-type and ZmMIR399b-overexpressing transgenic maize was similar under the Pi-sufficient condition (Fig. 1D).
When seedlings were transferred to Pi-deficient medium for 3 d, P toxicity phenotypes were not observed in the transgenic maize, but total P concentrations were higher in shoots of transgenic maize than in shoots of the wild type, especially in the ZmMIR399b-overexpressing transgenic maize 2 (OE2) line (Supplemental Fig. S3). These results suggest that ZmmiR399 is important for P homeostasis in maize.
ZmPHO2 Is the Target of ZmmiR399
The P toxicity phenotypes of ZmMIR399b-overexpressing transgenic maize under Pi-sufficient conditions were reminiscent of the phenotypes of the Arabidopsis pho2 mutant (Aung et al., 2006; Bari et al., 2006). This prompted us to analyze the ZmPHO2 transcript levels in ZmMIR399b-overexpressing transgenic maize even though ZmPHO2 was not a predicted target of ZmmiR399. In maize, GRMZM2G381709 and GRMZM2G464572 are the orthologs of Arabidopsis PHO2 (Calderón-Vázquez et al., 2011). We found that the cDNA sequences of GRMZM2G381709 and GRMZM2G464572 were the same as those annotated in MaizeGDB (http://www.maizegdb.org/). Thus, the primer pair we designed would detect the transcripts of both GRMZM2G381709 and GRMZM2G464572 in ZmMIR399b-overexpressing transgenic maize.
ZmMIR399b overexpression significantly reduced the mRNA levels of GRMZM2G381709/GRMZM2G464572 in maize (Fig. 2A), and similar results were found in different OE lines. To further check the potential relationship between GRMZM2G381709/GRMZM2G464572 and ZmmiR399, we first verified the GRMZM2G381709 and GRMZM2G464572 annotation in maize. 5′ RACE was used, and a 1,276-bp fragment was cloned (Supplemental Fig. S4). An additional 1,165-bp sequence was appended to the originally predicted 5′ UTR of GRMZM2G381709 by MaizeGDB. Further analysis showed that the newly cloned 5′ UTR of GRMZM2G381709 had six ZmmiR399 cleavage sites (Supplemental Fig. S4), suggesting that GRMZM2G381709 could be directly regulated by ZmmiR399. Interestingly, we sequenced 30 clones of ZmPHO2 5′ RACE products, and every fragment belonged to GRMZM2G381709, indicating that there were relatively few or no transcripts of GRMZM2G464572 in maize. GRMZM2G381709 was then designated as ZmPHO2.
Figure 2.
ZmPHO2 is posttranscriptionally regulated by miR399. A, Detection of ZmPHO2 transcripts in MIR399b-overexpressing transgenic maize by RT-qPCR. B and C, Coexpression of various combinations of (B) MIR399b or (C) MIR399bmut and ZmPHO2 expression constructs in N. benthamiana. The expression levels of miR399 were detected by small RNA northern blots. U6 was used as a loading control. Numbers below each line of the gel indicate the relative expression of miR399 to U6. RNA levels were normalized to that of 18S rRNA of tobacco. Values are means ± se (n = 3). Means with the same letter are not significantly different at P < 0.01 according to the LSD test. D, ZmPHO2 mRNA cleavage site detected by 5′ RACE. Numbers indicate the frequency of cleavage at the site. WT, wild type.
We subsequently performed transient coexpression assays in Nicotiana benthamiana using two miR399 constructs, ZmMIR399b and ZmMIR399bmut (six mutations were introduced into the mature sequence of ZmMIR399b). The ZmPHO2 transcripts were significantly decreased when coexpressed with ZmMIR399b (Fig. 2B) but were not affected when coexpressed with ZmMIR399bmut (Fig. 2C). In addition, four of the six predicted target sites in ZmPHO2 could be detected by the 5′ RACE assay using RNA from maize seedlings subjected to Pi deficiency for 6 d (Fig. 2D). These results showed that ZmPHO2 is a target of ZmmiR399 in maize and that the miR399-PHO2 regulatory module is conserved in maize as it is in Arabidopsis and rice.
The Expression Levels of ZmmiR399 and ZmPHO2 in P-Efficient and P-Inefficient Genotypes
We previously reported that, based on RNA-Seq transcriptome analysis, 22 miRNAs belonging to 10 miRNA families, including the miR399 family, were identified as differentially expressed genes between the P-efficient line CCM454 and the P-inefficient line 31778 under Pi-deficient conditions (Du et al., 2016). In this study, we conducted a time course experiment to investigate in detail the expression levels of ZmmiR399 in P-efficient and P-inefficient genotypes. ZmmiR399 was up-regulated by Pi deficiency in both the P-efficient line CCM454 and the P-inefficient line 31778 (Fig. 3A). Unexpectedly, miR399-guided posttranscriptional repression of ZmPHO2 was observed only in line CCM454 (Fig. 3A). In contrast, in line 31778, the transcripts of ZmPHO2 were up-regulated by Pi deficiency even though ZmmiR399 expression was increased (Fig. 3A).
Figure 3.
Analysis of miR399 expression in P-efficient and P-inefficient maize lines. RNA gel blot analysis of miR399 and ZmPHO2 transcripts in (A) maize inbred lines 31778 and CCM454 and (B) in other representative P-efficient and P-inefficient lines. U6 was used as a loading control. RNA levels were normalized to that of ZmTub1. LP, low P. Values are the means ± se (n = 3). In A, means with the same letter are not significantly different at P < 0.01 according to the LSD test. In B, asterisks indicate significant differences between normal-Pi and low-Pi treatments as determined by Student’s t tests (*P < 0.05, **P < 0.01).
To verify these results, we also determined the expression levels of ZmmiR399 in three other P-inefficient lines (FR19, 1538, and JI419) and in three other P-efficient lines (XI14, HAI9-21 and Q1261), which were identified from 560 maize accessions (Du et al., 2016). Consistent with the results for lines 31778 and CCM454, ZmmiR399 was up-regulated by Pi deficiency in both the P-efficient and P-inefficient genotypes (Fig. 3B). Except for inbred line Q1261, in which some nucleotides in the cleavage sites targeted by miR399 were naturally mutated (Supplemental Fig. S5), the transcripts of ZmPHO2 were down-regulated in the P-efficient lines and up-regulated in the P-inefficient lines after Pi deficiency treatment for 7 d (Fig. 3B).
To further verify the relationship between the miR399/PHO2 pathway and low P tolerance in maize, the P-inefficient line Qi205 (Du et al., 2016) was crossed with the P-efficient line CCM454 to generate a segregating population. A total of 100 lines were randomly chosen from the F2:3 family to determine their low P tolerance under a hydroponic condition. We assessed low P tolerance in maize based on their relative shoot fresh weights (Pi-deficient shoot fresh weight versus Pi-sufficient shoot fresh weight; Supplemental Fig. S6A). The 10 most tolerant and 10 most susceptible lines were selected, each accounting for 10% of the tested lines. Another 10 out of the remaining 80 lines were chosen as a control. In all of these lines, ZmmiR399 was up-regulated by Pi deficiency (Supplemental Fig. S6B). In contrast, the down-regulation of ZmPHO2 by ZmmiR399 was mainly observed in P-efficient lines (Supplemental Fig. S6C). These results suggest that miR399-guided posttranscriptional repression of ZmPHO2 increases the tolerance to Pi deficiency in maize.
Identification of lncRNAs Related to ZmmiR399
To clarify the difference in ZmmiR399-guided regulation of ZmPHO2 between P-efficient and P-inefficient genotypes, we first checked the sequences of the ZmmiR399-binding sites in ZmPHO2 from various maize accessions. Except for the sequences of the P-efficient line Q1261, the sequences of ZmmiR399-binding sites in ZmPHO2 were the same in P-efficient and P-inefficient genotypes (Supplemental Fig. S5). This suggested the presence of another regulatory mechanism affecting miR399-guided repression of ZmPHO2 in maize. One possibility was lncRNAs. To investigate this possibility, we generated strand-specific RNA (ssRNA) libraries from roots and shoots of the P-efficient line CCM454 and the P-inefficient line 31778 that had been subjected to Pi stress for 2 d and 8 d (Du et al., 2016). Using HISAT2 and StringTie (Kim et al., 2015; Pertea et al., 2015), we identified a total of 178,771 directional transcripts from the assembled transcriptome of ssRNA libraries (Supplemental Fig. S7). These reconstructed transcripts were evaluated by FEELnc, an alignment-free program that accurately annotates lncRNAs based on the Random Forest model (Wucher et al., 2017). Finally, 72,429 transcripts containing 46,927 lincRNAs, 2,511 lncNATs, and 1,217 incRNAs were defined as lncRNAs in maize (Supplemental Fig. S7).
Consistent with previous reports (Li et al., 2014; Wang et al., 2015a), the majority (79%) of the identified lncRNAs consisted of a single exon (Fig. 4A). The corresponding lncRNA genes were relatively short with a mean length of 788 bp. Only ∼18% of the lncRNA genes were longer than 1 kb (Fig. 4B). To further confirm the predicted lncRNA genes, we randomly selected 19 lincRNAs to clone using RNA extracted from maize subjected to Pi deficiency stress for 7 d. Sixteen of the 19 randomly selected lincRNAs could be amplified by reverse transcription PCR (RT-PCR; Fig. 4C). The authenticity of these lincRNAs was verified by sequencing. These results demonstrated the reliability of our analysis.
Figure 4.
Identification of maize lncRNAs. A, Distribution of exon number per lncRNA transcript. B, Distribution of exon length density for lncRNAs. C, Validation of randomly selected lncRNAs by RT-PCR. The PCR products were analyzed by the agarose gel electrophoresis.
PILNCR1 Inhibits ZmmiR399-Guided Cleavage of ZmPHO2
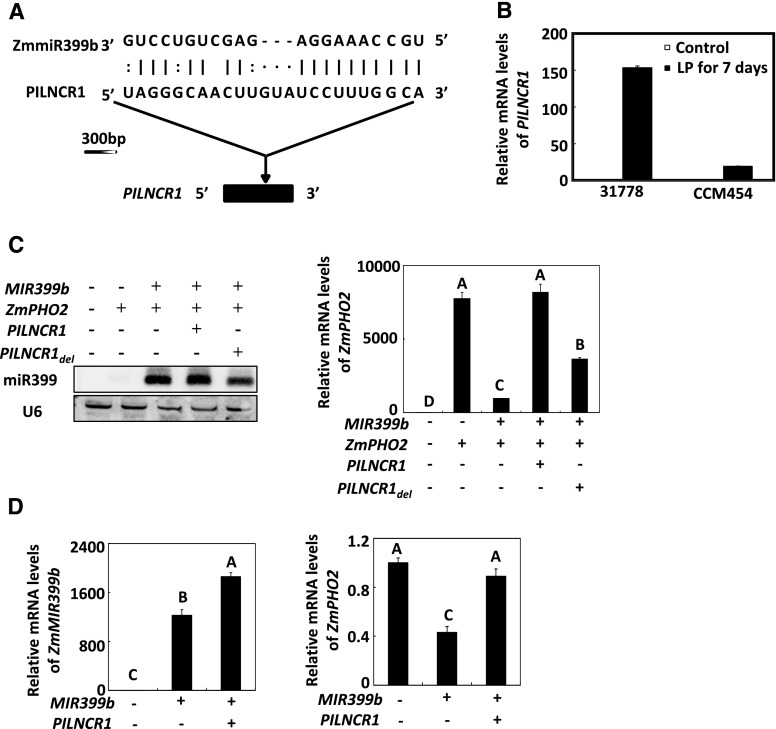
lncRNA819 attracted our attention because it contained a complementary region with ZmmiR399 (Fig. 5A). Using 3′ and 5′ RACE, a 677-bp cDNA clone without an intron was isolated (Supplemental Fig. S8). The expression of lncRNA819 was significantly induced by Pi deficiency in both the P-efficient line CCM454 and the P-inefficient line 31778 (Fig. 5B). Thus, we designated lncRNA819 as PILNCR1.
Figure 5.
Effects of PILNCR1 on ZmmiR399-guided cleavage of ZmPHO2. A, Diagram of the complementary region between PILNCR1 and miR399b in maize. B, RT-qPCR assay of the accumulation of PILNCR1 transcripts in the P-efficient line CCM454 and the P-inefficient line 31778 in response to Pi deficiency. RNA levels were normalized to that of ZmTub1. LP, low P. Values are the means ± se (n = 3). C, Coexpression of various combinations of miR399, ZmPHO2, PILNCR1, and PILNCR1del expression constructs in N. benthamiana. RNA levels were normalized to the expression of the 18S rRNA of tobacco. Error bars represent ses (n = 3). D, Coexpression of various combinations of miR399 and PILNCR1 expression constructs in maize leaf protoplasts. RNA levels were normalized to the expression of ZmTub1. Error bars represent ses (n = 3). In C and D, means with the same letter are not significantly different at P < 0.01 according to the LSD test.
Originally described in tomato (Solanum lycopersicum) and represented in Arabidopsis, several genes containing a complementary region with miR399 constitute the Tomato Phosphate Starvation-induced gene family. The most famous members in this family are INDUCED BY PHOSPHATE STARVATION1 (IPS1) and its close paralog At4, which can affect the cleavage activity of miR399 in Arabidopsis (Franco-Zorrilla et al., 2007). Unlike IPS1 and At4 in Arabidopsis, which encode a conserved peptide (Franco-Zorrilla et al., 2007), PILNCR1 in maize lacks the potential to encode a peptide as predicted by Coding Potential Calculator and Coding-Non-Coding Index software (Sun et al., 2013). In addition, there are three ZmIPS1/ZmAt4 candidates in maize (Calderón-Vázquez et al., 2011). Because of its low confidence as annotated by MaizeGDB, GRMZM2G086179 was not further considered. GRMZM5G843352 and GRMZM2G436295 were transcribed from the same loci in the opposite DNA strands. After the cDNA sequence alignment was analyzed, PILNCR1 showed no homology to GRMZM5G843352 and GRMZM2G436295. Syntenic analysis further demonstrated that PILNCR1 and AtIPS1/AtAT4s were located in nonsyntenic regions (Supplemental Fig. S9). Taken together, these results suggested that the characteristics differed between PILNCR1 and ZmIPS1/ZmAt4. We then performed transient expression assays in N. benthamiana and maize leaf protoplasts to determine whether PILNCR1 could affect the cleavage activity of miR399 in plants.
We tested two PILNCR1 constructs: a full-length PILNCR1 and a construct in which the complementary region with miR399 was deleted (PILNCR1del). These constructs were expressed under the control of the 35S promoter. After 2 d of coexpression in N. benthamiana, RNA was extracted and ZmPHO2 expression was analyzed by reverse-transcription quantitative PCR (RT-qPCR). The efficient reduction of ZmPHO2 mRNA levels by ZmmiR399b was abolished when ZmmiR399b was coexpressed with PILNCR1 (Fig. 5C). However, the ZmPHO2 mRNA level was significantly decreased when ZmmiR399b was coexpressed with PILNCR1del (Fig. 5C), indicating that the complementary region was important for the regulation of ZmmiR399 by PILNCR1. Similar results were obtained when ZmmiR399b was coexpressed with PILNCR1 in maize leaf protoplasts (Fig. 5D). These results suggested that PILNCR1 can efficiently reduce the effects of ZmmiR399.
Expression Patterns of ZmmiR399b and PILNCR1
To further explore the possible relationship between ZmmiR399b and PILNCR1, we examined the expression patterns of ZmmiR399b and PILNCR1 by in situ hybridization. Because of the low abundance of ZmmiR399b and PILNCR1 in maize under Pi-sufficient conditions, both the roots and shoots of maize subjected to Pi deficiency for 7 d were sampled to assess the expression patterns of ZmmiR399b and PILNCR1. ZmmiR399b and PILNCR1 could be detected in roots and shoots of maize, especially in the vascular systems (Fig. 6, A and B). These results suggested that the spatial localizations of ZmmiR399b and PILNCR1 at least partly overlapped.
Figure 6.
Patterns of ZmmiR399b (A) and PILNCR1 (B) accumulation in maize. The roots and shoots of maize plants subjected to low-Pi stress for 7 d were sampled. The corresponding sense probes were used as negative controls. Representative photographs are shown. Scale bars, 10 µm.
PILNCR1 Is Correlated with Low Pi Tolerance in Maize
To further investigate the function of PILNCR1 in low Pi tolerance in maize, we determined the mRNA abundance of PILNCR1 in the P-efficient (CCM454, XI14, HAI9-21, and Q1261) and the P-inefficient lines (31778, FR19, 1538, and JI419) by absolute RT-qPCR. Under Pi-sufficient conditions, the expression of PILNCR1 was low in both P-efficient and P-inefficient lines (Fig. 7). When maize was subjected to Pi deficiency for 7 d, however, the average PILNCR1 abundance was about six times higher in P-inefficient lines than in P-efficient lines (Fig. 7).
Figure 7.
PILNCR1 levels in the leaves of P-inefficient and P-efficient maize accessions. PILNCR1 levels in the leaves of P-inefficient and P-efficient accessions were determined by absolute RT-qPCR. LP, low P. Error bars represent ses (n = 4). Means with the same letter are not significantly different at P < 0.01 according to the LSD test.
DISCUSSION
Although the members and predicted targets of miR399 in maize were clearly different from those in Arabidopsis and rice, this study demonstrated that ZmPHO2 is the authentic target of ZmmiR399 and that the miR399-PHO2 signaling pathway, which responds to Pi deficiency, is also conserved in maize. miR399-guided posttranscriptional repression of ZmPHO2 was observed mainly in the P-efficient lines. From ssRNA libraries of the P-efficient line CCM454 and the P-inefficient line 31778, we identified and characterized the lncRNA PILNCR1, which can inhibit ZmmiR399-guided cleavage of ZmPHO2 in maize. These findings could be useful for the development of maize varieties with low-Pi tolerance.
Conservation of the miR399-PHO2 Signaling Pathway in Maize
The functional significance of miR399 in adaptive responses to Pi limitation has been well explored in Arabidopsis and rice. In Arabidopsis and rice, miR399 is specifically induced by Pi-deficient stress and regulates P homeostasis by suppressing the expression level of PHO2 (Fujii et al., 2005; Chiou et al., 2006; Hu et al., 2011). PHO2 is a ubiquitin-conjugating E2 enzyme that negatively regulates Pi uptake and translocation. Maize contains two AtPHO2 homologs, which share the same cDNA sequences as annotated in MaizeGDB (http://www.maizegdb.org/). 5′ RACE analysis indicated that GRMZM2G381709 is the authentic AtPHO2 homolog in maize. Using stable transgenic maize and transient expression in N. benthamiana, we further demonstrated that ZmPHO2 is posttranscriptionally regulated by miR399.
The systemic movement of miR399 is an early response to Pi deficiency in plants. The transcripts of PHO2 in roots can be suppressed by the shoot-to-root movement of mature miR399, which led to Pi overaccumulation in Arabidopsis shoots (Lin et al., 2008; Pant et al., 2008). Here, we also found that ZmmiR399b is expressed in the vascular systems of maize and that more P was translocated from roots to shoots in ZmMIR399b-overexpressing transgenic maize than in the wild-type. These results indicate that the miR399-PHO2 signaling pathway and its response to Pi deficiency are conserved among plant species and that this pathway is important for modulating P homeostasis in maize as it is in Arabidopsis and rice.
The Interaction between PILNCR1 and miR399 in Maize
The results of transient coexpression assays in N. benthamiana and maize protoplasts demonstrated that PILNCR1 can reduce the effects of miR399 in maize and that the reduction depends on complementary regions in PILNCR1 and miR399. This is reminiscent of the function of the IPS1 and At4 in Arabidopsis (Franco-Zorrilla et al., 2007). PILNCR1 is not ZmIPS1 because (1) PILNCR1 lacks the potential to encode a peptide, (2) PILNCR1 shows no homology to ZmIPS1/ZmAt4 candidates, and (3) syntenic analysis shows that PILNCR1 is not related to AtIPS1 or AtAt4s.
In micrografting experiments with wild-type Arabidopsis and pho2 mutant seedlings, the Pi content of shoots was high when the roots were derived from the pho2 mutant, but not when the shoots of the pho2 mutant were grafted onto wild-type roots (Pant et al., 2008), suggesting that shoots contain other regulators to modulate Pi homeostasis. The following evidence from this study indicates that PILNCR1 should be a good regulator candidate to modulate Pi homeostasis because (1) the spatial localizations of PILNCR1 and miR399 overlapped when maize was subjected to Pi deficiency; (2) consistent with the induction of miR399 expression by Pi deficiency, PILNCR1 abundance increased under low-Pi stress; (3) when maize was subjected to Pi deficiency for 7 d, the average PILNCR1 abundance was six times higher in P-inefficient lines than in P-efficient lines, but the average miR399 level was only 2.5 times higher in P-inefficient lines than in P-efficient lines; (4) compared with the high level of induction of ZmmiR399 by Pi deficiency, the ZmPHO2 transcripts of some P-inefficient inbred lines were not down-regulated after Pi deficiency for 7 d. In humans, research increasingly indicates that coding genes are not the only targets regulated by miRNAs and that different RNAs compete with each other to interact with miRNAs (Cesana et al., 2011; Hansen et al., 2013). Thus, we propose a possible model for the cofunction between PILNCR1 and ZmmiR399 in maize under Pi-deficient conditions (Fig. 8). The relatively high amount of PILINCR1 in P-inefficient lines could efficiently mask the PHO2-targeting miR399 and impair the miR399-guided cleavage of PHO2, which would lead to PHO2 accumulation under Pi deficiency and therefore reduce the tolerance to low Pi in maize. Of course, ZmPHO2 might be deferentially regulated between the maize lines in a way that was to some extent miR399/PILNCR1 independent, such as the allelic variation in cis elements. The hypothesis requires further investigation.
Figure 8.
Proposed model of the role of the PILNCR1-miR399 regulatory module in determining the tolerance of maize to low Pi. Under Pi deficiency, the relatively high amount of PILINCR1 in P-inefficient lines could efficiently impair the miR399-guided cleavage of PHO2, which would lead to PHO2 accumulation and therefore reduce the tolerance to low Pi in maize. The red and blue symbols represent the corresponding mRNA levels in P-inefficient and P-efficient maize accessions, respectively.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of uniform size were surface sterilized in 3% NaOCl for 20 min. After they were washed with distilled water, the seeds were soaked in a saturated CaSO4 solution with continuous aeration for 6 h and were then germinated in coarse quartz sand at room temperature until two leaves were visible. After their endosperms were removed, seedlings were transferred to 3-L pots supplied with modified half-strength Hoagland’s nutrient solution for 2 d and then supplied with full-strength Hoagland’s nutrient solution containing either 250 µm PO43− or 5 µm PO43− to generate Pi-sufficient or Pi-deficient conditions. In the low Pi treatment, KCl was added to maintain the same concentration of potassium in both treatments. The basic nutrients in hydroponic solutions were similar to those described by Du et al. (2016). The seedlings were grown in a growth chamber with 14-h light/10-h dark and with a 28°C/22°C day/night temperature regime.
Identification of lncRNAs
The clean reads of 24 ssRNA libraries from shoots and roots of the P-efficient line CCM454 and the P-inefficient line 31778 that had been subjected to Pi stress for 2 and 8 d were produced as described by Du et al. (2016). These clean reads were then mapped to the maize (Zea mays) B73 reference genome (AGP v3.27) using the alignment program HISAT2 (version 2-2.0.1; Kim et al., 2015). The transcriptome was separately assembled using the StringTie program (Pertea et al., 2015). The Ballgown procedure was applied to estimate the transcript expression levels (Pertea et al., 2016). lncRNAs were identified from the assembled transcriptomes by the FEELnc program (Wucher et al., 2017).
Generation of Transgenic Maize
The maize transformation vector (pCUB) was constructed using the rice (Oryza sativa) ubiquitin promoter. The selectable marker cassette for pCUB contained the Streptomyces hygroscopicus phosphinothricin acetyltransferase gene (bar) under the control of a Cauliflower mosaic virus 35S promoter. A 497-bp fragment surrounding the MIR399b sequence including the fold-back structure was amplified from genomic DNA with the primers listed in Supplemental Table S1. The amplified fragments were introduced into the pCUB vector using the BamHI restriction site by In-Fusion reaction (Clontech Laboratories). The plasmid was electroporated into Agrobacterium tumefaciens EHA105.
Immature embryos of the maize inbred line ZZC01, which was used as the wild type, were inoculated with EHA105 harboring the appropriate plasmid as described previously (Frame et al., 2006). Transformants were selected with gradually increasing concentrations of bialaphos. Independent transgenic events with a single copy or a low number of copies were self-pollinated or crossed with ZZC01 to produce T1 seeds. T2 plants with confirmed transgene expression were self-pollinated to generate T3 seeds. T3 homozygous lines were used for all experiments.
RNA Analysis
Total RNA was extracted from inbred lines and transgenic maize with Trizol reagent (Invitrogen). The enrichment, fractionation, and detection of miR399 were performed as previously described (Li et al., 2008).
To quantify ZmmiR399, stem-loop RT-qPCR was performed as previously described (Chen et al., 2005). For RT-PCR, first-strand cDNA was synthesized using SuperScript III First-Strand Synthesis Supermix (Invitrogen). Specific primers were used to detect the expression levels of ZmPHO2, ZmMIR399b, and PILNCR1. Primer efficiencies were measured and calculated. qPCR was carried out in an ABI 7500 system (Applied Biosystems) using the SYBR Premix Ex Taq (perfect real time) kit (TaKaRa Biomedicals). The PCR products were loaded on 1% agarose gels and were checked after staining with ethidium bromide. Each experiment was replicated three times. The comparative Ct method was applied. The specific primers of ZmPHO2 are listed in Supplemental Table S1.
Absolute Analysis of PILNCR1 Abundance
To determine the absolute abundance of PILNCR1 in leaves of P-inefficient and P-efficient inbred lines, the full-length of PILNCR1 was amplified and then subcloned into the pMD 18-T vector (TaKaRa). A series of the plasmid dilutions (from 10−1 to 10−8 ng) were prepared to generate the standard curve for absolute quantification. The absolute copy numbers of each sample were calculated on the basis of the corresponding Ct values and standard curve. Each experiment was replicated four times.
5′ RACE
RNA ligase-mediated RACE was performed using the GeneRacer Kit (Invitrogen). Total RNA was extracted from Pi-deficient roots of the inbred line CCM454 by the Trizol method. The GeneRacer RNA Oligo adaptor was directly ligated to the total RNA without calf intestinal phosphatase treatment. The GeneRacer oligo dT was then used for cDNA synthesis. Initial PCR was carried out using the GeneRacer 5′ primer and gene-specific outer primer. Nested PCR was carried out using the initial PCR reaction products diluted 50 times as template and the GeneRacer 5′ nested primer and gene-specific inner primer. The sequences of the gene-specific primers are listed in Supplemental Table S1. The PCR products were cloned into pMD18-T (TaKaRa Biomedicals) and sequenced.
In Situ Hybridization
In situ hybridization for miR399 was performed as described by Chen (2004). For the PILNCR1 probe, full-length PILNCR1 was amplified and cloned into the pGEM-T-easy vector (Promega). T7 or SP6 RNA polymerases were used for in vitro transcription to generate the sense or antisense probes, respectively. The corresponding procedures were previously described (Zhang et al., 2007).
Transient Expression in Nicotiana benthamiana
Site-directed mutagenesis was performed to generate ZmMIR399bmut with the QuickChange II site-directed mutagenesis kit (Stratagene). The fragments were sequenced to ensure that only the desired mutations were introduced. Full-length cDNA sequences of PILNCR1, PILNCR1del, ZmPHO2, and ZmMIR399b including the fold-back structure were amplified with the primers listed in Supplemental Table S1. To generate the corresponding expression plasmids, the amplified fragments were cloned into the pCPB vector using the BamHI restriction site by In-Fusion reaction. The plasmids were electroporated into A. tumefaciens GV3101 and were transiently expressed in N. benthamiana epidermal cells as described by Li et al. (2008). Leaves were harvested 2 d after the infiltration and were subjected to RNA analysis as described above.
Maize Protoplast Preparation and Transfection
Maize leaf protoplasts were isolated as previously described by Chen et al. (2016). In brief, young leaves of maize seedlings were cut into approximately 0.5-mm strips along the veins. The strips were incubated in an enzyme solution (1.5% Cellulase R10, 0.5% macerozyme R10, 0.4 m mannitol, 20 mm KCl, 20 mm MES pH 5.7, 10 mm CaCl2, 0.1% BSA, and 5 mm β-mercaptoethanol) for 6 h in the dark with gentle shaking. After digestion, an equal volume of W5 solution (125 mm CaCl2, 154 mm NaCl, 5 mm KCl, 5 mm Glc, and 0.03% MES pH 5.7) was added. The protoplasts were collected by centrifugation at 100g for 2 min and were resuspended in MMG solution (15 mm MgCl2, 0.1% MES pH 5.7, and 0.4 m mannitol) to obtain a final concentration of 2 × 106 cells mL−1.
For transformation, approximately 25 μg of plasmid DNA was mixed with 500 μL of protoplasts and incubated in PEG4000 solution (45% PEG4000, 0.8 m mannitol, and 1 m CaCl2) for 20 min in the dark. A 1-mL volume of W5 solution was added to end the transformation. The transfected protoplasts were incubated at 28°C for 18 h to allow RNA expression. Total RNA was extracted from transfected protoplasts with Trizol reagent (Invitrogen) as described above.
Determination of Total P Content
Transgenic maize plants were separated into shoots and roots. After they were subjected to 105°C for 30 min, the samples were dried at 65°C for 5 d and then milled to a fine powder. The weighed samples were then digested in 5 mL of H2SO4-H2O2 at 300°C until the solution became clear. The total P contents were determined by the vanadomolybdate method.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: miR399b (zma-MIR399b), ZmPHO2 (GRMZM2G3817019), ZmIPS1 (GRMZM5G843352, GRMZM2G436295), AtIPS1 (AT3G09922), and AtAT4 (AT5G03545).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypic responses of maize inbred line B73 to P stress.
Supplemental Figure S2. The responses of miR399 to N stress.
Supplemental Figure S3. P concentrations in shoots and roots of inbred line ZZC01 (wild type) and MIR399b-overexpressing transgenic maize under Pi-deficient conditions.
Supplemental Figure S4. The 5′ UTR sequence of ZmPHO2 obtained by 5′ RACE.
Supplemental Figure S5. Alignment of the miR399 binding sites of ZmPHO2 in P-efficient and P-inefficient accessions.
Supplemental Figure S6. The expression levels of ZmmiR399b and ZmPHO2 in the F2:3 generation of the CCM454 × Qi205 cross.
Supplemental Figure S7. The pipeline for the systematic identification of lncRNA in maize.
Supplemental Figure S8. The sequence of PILNCR1 obtained by RACE.
Supplemental Figure S9. Comparative maps of PILNCR1 and AtIPS1/AtAT4s between maize and Arabidopsis.
Supplemental Table S1. Oligos used as primers in the experiment.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dr. Keke Yi (Institute of Agricultural Resources and Regional Planning, Chinese Academy of Agricultural Sciences) for useful comments. We also thank Dr. Hong Wang (Institute of Agricultural Resources and Regional Planning, Chinese Academy of Agricultural Sciences) for excellent technical support in the elemental assay.
Footnotes
This work was supported by the National Key Research and Development Program of China (grant no. 2016YFD0101002), the Ministry of Agriculture of China for Transgenic Research (grant no. 2018ZX0800988B), and the Agricultural Science and Technology Innovation Program of CAAS to W.X.L.
References
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón-Vázquez C, Sawers RJ, Herrera-Estrella L (2011) Phosphate deprivation in maize: genetics and genomics. Plant Physiol 156: 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147: 358–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA. (2015) Long non-coding RNAs and their functions in plants. Curr Opin Plant Biol 27: 207–216 [DOI] [PubMed] [Google Scholar]
- Chen X. (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yi Q, Cao Y, Wei B, Zheng L, Xiao Q, Xie Y, Gu Y, Li Y, Huang H, et al. (2016) ZmbZIP91 regulates expression of starch synthesis-related genes by binding to ACTCAT elements in their promoters. J Exp Bot 67: 1327–1338 [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL (2006) Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Wang K, Xu C, Zou C, Xie C, Xu Y, Li WX (2016) Strand-specific RNA-Seq transcriptome analysis of genotypes with and without low-phosphorus tolerance provides novel insights into phosphorus-use efficiency in maize. BMC Plant Biol 16: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame BR, McMurray JM, Fonger TM, Main ML, Taylor KW, Torney FJ, Paz MM, Wang K (2006) Improved Agrobacterium-mediated transformation of three maize inbred lines using MS salts. Plant Cell Rep 25: 1024–1034 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- Hackenberg M, Shi BJ, Gustafson P, Langridge P (2013) Characterization of phosphorus-regulated miR399 and miR827 and their isomirs in barley under phosphorus-sufficient and phosphorus-deficient conditions. BMC Plant Biol 13: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388 [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Hsieh LC, Lin SI, Shih ACC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151: 2120–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Zhu C, Li F, Tang J, Wang Y, Lin A, Liu L, Che R, Chu C (2011) LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol 156: 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu Q, Poirier Y (2013) A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 25: 4166–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Eichten SR, Shimizu R, Petsch K, Yeh CT, Wu W, Chettoor AM, Givan SA, Cole RA, Fowler JE, et al. (2014) Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol 15: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20: 2238–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ (2008) Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH (2012) Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Arredondo DL, Leyva-González MA, González-Morales SI, López-Bucio J, Herrera-Estrella L (2014) Phosphate nutrition: improving low-phosphate tolerance in crops. Annu Rev Plant Biol 65: 95–123 [DOI] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR (2009) Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol 150: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Jin Z, Li K, Yin H, Wang J, Yang A (2013) Identification and comparative analysis of low phosphate tolerance-associated microRNAs in two maize genotypes. Plant Physiol Biochem 70: 221–234 [DOI] [PubMed] [Google Scholar]
- Péret B, Clément M, Nussaume L, Desnos T (2011) Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci 16: 442–450 [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11: 1650–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y (2013) Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res 41: e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Wang H, Niu QW, Wu HW, Liu J, Ye J, Yu N, Chua NH (2015a) Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J 84: 404–416 [DOI] [PubMed] [Google Scholar]
- Wang M, Yuan D, Tu L, Gao W, He Y, Hu H, Wang P, Liu N, Lindsey K, Zhang X (2015b) Long noncoding RNAs and their proposed functions in fibre development of cotton (Gossypium spp.). New Phytol 207: 1181–1197 [DOI] [PubMed] [Google Scholar]
- Wang Y, Fan X, Lin F, He G, Terzaghi W, Zhu D, Deng XW (2014) Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc Natl Acad Sci USA 111: 10359–10364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucher V, Legeai F, Hédan B, Rizk G, Lagoutte L, Leeb T, Jagannathan V, Cadieu E, David A, Lohi H, et al. (2017) FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res 45: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Liu Q, Chen L, Kuang J, Walk T, Wang J, Liao H (2013) Genome-wide identification of soybean microRNAs and their targets reveals their organ-specificity and responses to phosphate starvation. BMC Genomics 14: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Hennelly S, Doyle B, Gulati AA, Novikova IV, Sanbonmatsu KY, Boyer LA (2016) A G-rich motif in the lncRNA Braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage. Mol Cell 64: 37–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chia JM, Kumari S, Stein JC, Liu Z, Narechania A, Maher CA, Guill K, McMullen MD, Ware D (2009) A genome-wide characterization of microRNA genes in maize. PLoS Genet 5: e1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Madi S, Borsuk L, Nettleton D, Elshire RJ, Buckner B, Janick-Buckner D, Beck J, Timmermans M, Schnable PS, et al. (2007) Laser microdissection of narrow sheath mutant maize uncovers novel gene expression in the shoot apical meristem. PLoS Genet 3: e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YC, Liao JY, Li ZY, Yu Y, Zhang JP, Li QF, Qu LH, Shu WS, Chen YQ (2014) Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol 15: 512. [DOI] [PMC free article] [PubMed] [Google Scholar]