Numerous RLCK VII members modulate plant immune signaling in overlapping and specific manners.
Abstract
Pattern-recognition receptors (PRRs), which consist of receptor kinases (RKs) and receptor-like proteins, sense microbe- and host-derived molecular patterns associated with pathogen infection to trigger immune responses in plants. Several kinases of the 46-member Arabidopsis (Arabidopsis thaliana) receptor-like cytoplasmic kinase (RLCK) subfamily VII play important roles in pattern-triggered immunity, but it is unclear whether different RLCK VII members act specifically or redundantly in immune signaling. Here, we constructed nine higher order mutants of this subfamily (named rlck vii-1 to rlck vii-9) and systematically characterized their immune phenotypes. The mutants rlck vii-5, -7, and -8 had compromised reactive oxygen species production in response to all patterns tested, indicating that the corresponding members are broadly required for the signaling of multiple PRRs. However, rlck vii-4 was defective specifically in chitin-induced reactive oxygen species production, suggesting that RCLK VII-4 members mediate the signaling of specific PRRs. Furthermore, RLCK VII-4 members were required for the chitin-triggered activation of MAPK, demonstrating that these kinases link a PRR to MAPK activation. Moreover, we found that RLCK VII-6 and -8 also were required for RK-mediated root growth. Together, these results show that numerous RLCK VII members are involved in pattern-triggered immune signaling and uncover both common and specific roles of these kinases in plant development and immunity mediated by various RKs.
Plants rely mainly on cell surface-localized receptors, which consist primarily of receptor kinases (RKs) and receptor-like proteins (RLPs), to perceive extracellular signal molecules and modulate growth, development, and immunity. In the absence of a sophisticated adaptive immune system, RKs and RLPs act as pattern-recognition receptors (PRRs) that perceive microbe- and host-derived molecular patterns that are released during pathogen infection and activate plant immunity (Boller and Felix, 2009; Couto and Zipfel, 2016; Li et al., 2016; Tang et al., 2017). For instance, the Arabidopsis (Arabidopsis thaliana) FLAGELLIN SENSING2 (FLS2) and ELONGATION FACTOR-TU (EF-Tu) RECEPTOR (EFR) directly sense the bacterial flagellin epitope flg22 and EF-Tu epitope elf18, respectively (Gómez-Gómez and Boller, 2000; Bauer et al., 2001; Kunze et al., 2004; Zipfel et al., 2006). PEP1 RECEPTOR1 (PEPR1) and PEPR2 recognize plant elicitor peptides (Peps) that are released during pathogen infection (Huffaker and Ryan, 2007; Krol et al., 2010; Yamaguchi et al., 2010). Both CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) and LysM-RK LYSINE MOTIF RECEPTOR KINASE5 bind fungal chitin (Liu et al., 2012; Cao et al., 2014). In addition, RLP23 specifically perceives nlp20, a conserved peptide of Necrosis and Ethylene-Inducing Peptide1-like proteins (NLPs) from bacteria, fungi, and oomycetes (Albert et al., 2015). In addition, RKs regulate plant growth and development by perceiving diverse signals. For example, BRASSINOSTEROID INSENSITIVE1 (BRI1) binds brassinosteroids and, thereby, regulates growth and development (Belkhadir et al., 2006). The phytosulfokine (PSK) receptor perceives a peptide PSK and regulates cellular dedifferentiation and proliferation (Matsubayashi et al., 2002). FERONIA (FER) is required for RAPID ALKALINIZATION FACTOR (RALF)-triggered growth inhibition (Haruta et al., 2014; Stegmann et al., 2017).
Several receptor-like cytoplasmic kinases (RLCKs) have emerged as key players in RK-mediated signaling (Lin et al., 2013; Couto and Zipfel, 2016; Tang et al., 2017; Liang and Zhou, 2018). BOTRYTIS-INDUCED KINASE1 (BIK1) and the closely related PBS1-Like1 (PBL1), belonging to subfamily VII of the RLCKs, mediate pattern-triggered immunity (PTI) by associating directly with FLS2, EFR, CERK1, and PEPR1 (Lu et al., 2010; Zhang et al., 2010; Liu et al., 2013b) and contribute to resistance not only to bacterial and fungal pathogens but also to aphids (Veronese et al., 2006; Lu et al., 2010; Zhang et al., 2010; Lei et al., 2014). Mounting evidence suggests that additional RLCK VII members also contribute to PTI. Both Pseudomonas syringae effector AvrPphB and Xanthomonas campestris effector AvrAC target BIK1 and multiple PBLs belonging to the RLCK VII subfamily to inhibit PTI in plants (Zhang et al., 2010; Feng et al., 2012). PTI-COMPROMISED RECEPTOR-LIKE CYTOPLASMIC KINASE1 (PCRK1) and PCRK2 were shown recently to contribute to PTI and disease resistance to P. syringae (Sreekanta et al., 2015; Kong et al., 2016). Moreover, the involvement of RLCK VII members in RK-mediated signaling is not limited to immunity. CONSTITUTIVE DIFFERENTIAL GROWTH1 (CDG1) associates with BRI1 to regulate brassinosteroid signaling and plant growth (Kim et al., 2011). LOST IN POLLEN TUBE GUIDANCE1 (LIP1) and LIP2, two closely related RLCK VII members, are involved in defensin-like peptide LURE1-induced pollen tube guidance (Liu et al., 2013a), although it remains to be elucidated whether they function downstream of RKs. A recent study showed that RPM1-INDUCED PROTEIN KINASE (RIPK) functions downstream of FER to regulate root growth (Du et al., 2016). The RLCK VII member CAST AWAY (CST) interacts with both HAESA and EVERSHED to negatively regulate floral organ abscission (Burr et al., 2011).
Another important endeavor of plant immunity studies is to understand how PRRs regulate various PTI responses. BIK1 and PBL1 are required for pattern-triggered reactive oxygen species (ROS) production, calcium influx, and callose deposition (Lu et al., 2010; Zhang et al., 2010; Kadota et al., 2014; Li et al., 2014; Ranf et al., 2014). PCRK1 and PCRK2 are required for ROS production, callose deposition, and salicylic acid (SA) accumulation in response to various patterns (Sreekanta et al., 2015; Kong et al., 2016). The activation of MAPK cascades is of profound importance in disease resistance (Meng and Zhang, 2013). Whether RLCK VII members play an important role in pattern-triggered MAPK activation remains poorly understood. The bik1 pbl1 double mutant shows slightly reduced Pep2-triggered MAPK activation but normal flg22-triggered MAPK activation (Feng et al., 2012; Yamada et al., 2016). The pcrk1 pcrk2 double mutant shows a slight reduction in flg22-triggered MAPK activation (Kong et al., 2016). It remains to be tested whether the minor effects of these mutations on MAPK activation are due to functional redundancy among RLCK VII members. In addition, it is unknown whether different RLCK VII members are differentially coopted for distinct PRRs. Furthermore, whether different RLCK VII members regulate different downstream responses has not been tested rigorously.
In an effort to systematically study RLCK VII, we constructed nine higher order mutants and examined their developmental and immune phenotypes triggered by different extracellular signal molecules. These analyses revealed one subgroup of RLCK VII proteins that specifically mediates chitin-triggered immune signaling. We also identified several RLCK VII subgroups that are commonly required for immune signaling triggered by multiple patterns. We categorized a specific subgroup of RLCK VII proteins as crucial components in chitin-triggered MAPK activation. The genetic analysis additionally showed that the RLCK VII members have different roles in PSK- and RALF23-regulated root growth. These results collectively demonstrate both the redundancy and specificity of RLCK VII subgroups in the context of different RKs.
RESULTS
Construction of Higher Order rlck vii Mutants
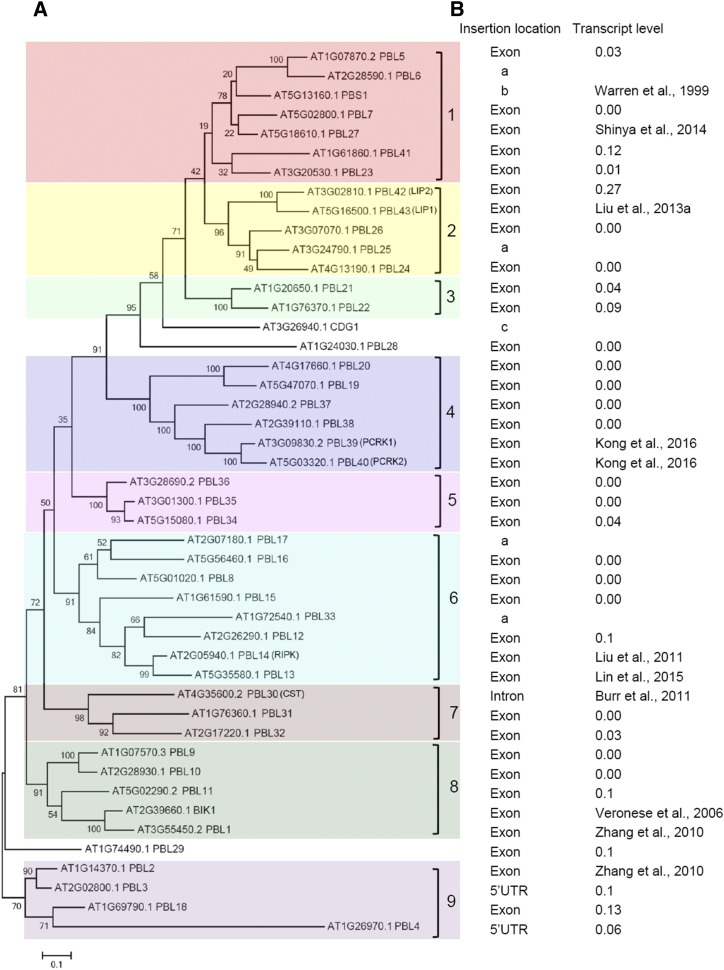
The RLCK subfamily VII contains 46 members in Arabidopsis, including the previously described PBL1 to PBL28, BIK1, PBS1, and CDG1 proteins and the newly named PBL29 to PBL43 (Fig. 1A; Shiu and Bleecker, 2001; Swiderski and Innes, 2001; Muto et al., 2004; Veronese et al., 2006; Zhang et al., 2010; Kim et al., 2011). Among these, PBL43, PBL42, PBL14, PBL30, PBL39, and PBL40 correspond to the previously identified LIP1, LIP2, RIPK, CST, PCRK1, and PCRK2 proteins, respectively (Fig. 1A; Burr et al., 2011; Liu et al., 2011, 2013a; Sreekanta et al., 2015; Kong et al., 2016). The RLCK VII members can be divided into nine subgroups, named RLCK VII-1 to RLCK VII-9 (Fig. 1A), except for CDG1, PBL28, and PBL29, which failed to fall into any subgroup. To study the function of RLCK VII members in RK signaling, we collected Arabidopsis lines containing T-DNA insertions of their corresponding genes from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/). We selected a total of 45 lines with T-DNA insertions in exons, introns, or 5' untranslated regions and confirmed the lines by genotyping. However, no T-DNA insertions were available for PBL6, PBL17, PBL25, and PBL33. To evaluate whether the genes were disrupted by T-DNA insertions, we detected the corresponding gene transcripts using reverse transcription quantitative PCR (RT-qPCR). For further analysis, we selected T-DNA insertion lines representing 40 PBL genes in which the levels of intact transcripts were reduced to approximately 10% or less of wild-type levels (Fig. 1B; Supplemental Fig. S1).
Figure 1.
Phylogenetic clustering of the Arabidopsis RLCK subfamily VII and detailed information on mutant lines. A, Phylogenetic tree of RLCK VII in Arabidopsis. The full-length sequences of RLCK VII members were used to generate the phylogenetic tree using MEGA. B, Summary of RLCK VII mutant lines, including the locations of the T-DNA insertion in the genes (for detailed information, see Supplemental Fig. S1) and the levels of intact RLCK transcripts in the corresponding mutants relative to that in the wild type detected by RT-qPCR (primers are listed in Supplemental Table S2). Under Insertion location, a indicates the absence of any available insertion lines, b indicates a fast neutron mutant, and c indicates a family member not included in our analysis. UTR, Untranslated region.
Considering the high degree of similarity among RLCK VII members, single mutants may or may not exhibit altered phenotypes. We thus combined mutant genes within each subgroup by crossing (Supplemental Table S1). We further introduced pbl6, pbl17, and pbl25 mutations into higher order mutants of RLCK VII-1, -6, and -2, respectively, using the CRISPR-Cas9 method (Supplemental Fig. S2; Supplemental Table S1). Overall, nine higher order mutants were constructed and named rlck vii-1 to rlck vii-9 (Supplemental Table S1).
Roles of Different Subgroups in Signaling Mediated by Different PRRs
To investigate the roles of these RLCK members in plant innate immunity, we collected microarray data sets from ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) and analyzed them for pattern-induced gene expression (Supplemental Fig. S3). More than 20 RLCK VII genes were up-regulated by at least one pattern, and approximately one-third of the genes were induced by two or more patterns (Supplemental Fig. S3), indicating that many members of this subfamily may be involved in PTI.
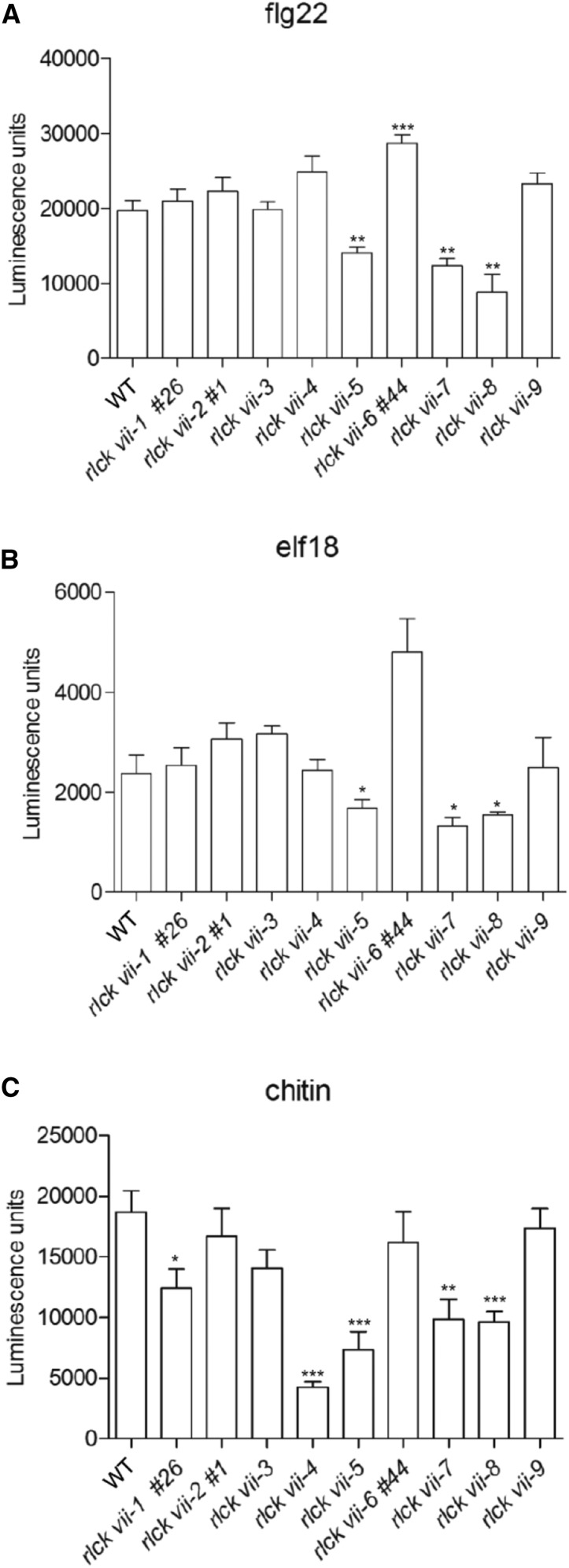
We first examined pattern-triggered ROS production, a robust immune signaling readout, in these higher order mutants. rlck vii-5, -7, and -8 mutants accumulated 40% to 60% less flg22-triggered hydrogen peroxide (H2O2) than did wild-type plants, while the reduced H2O2 triggered by flg22 in rlck vii-1 mutants was observed in only two of four independent experiments (Fig. 2A; Supplemental Fig. S4A). Similarly, elf18- and chitin-induced H2O2 production was reduced significantly in rlck vii-5, -7, and -8 (Fig. 2, B and C; Supplemental Fig. S4, B and C), indicating that RLCK VII-5, -7, and -8 are commonly required for ROS production triggered by multiple patterns. Interestingly, ROS production triggered by chitin, but not other patterns, was nearly abolished in rlck vii-4 and was reduced modestly in rlck vii-1 mutants (Fig. 2C; Supplemental Fig. S4C), indicating that these two subgroups have a specific role in chitin-triggered ROS production. Taken together, these results support the notion that RLCK VII-5, -7, and -8 members are common components acting downstream of multiple PRRs, while RLCK VII-4 and -1 play specific roles downstream of chitin receptors. Interestingly, rlck vii-6 mutants showed higher flg22-triggered H2O2 production than did the wild type (Fig. 2A; Supplemental Fig. S4A). These results suggest that the RLCK VII-6 members have a negative role in flg22-triggered signaling.
Figure 2.
Differential roles of RLCK VII subgroups in ROS production. Pattern-triggered ROS production is shown in all higher order mutants compared with the wild type (WT). Leaf strips were treated with flg22 (A), elf18 (B), or chitin (C), and H2O2 production was measured as luminescence. The line numbers (#) refer to alleles created by CRISPR-Cas9. All experiments were repeated at least three times, and data from one representative experiment are shown. Luminescence units refer to arbitrary units recorded by a luminometer. Values are means of the sum of luminescence units collected from all 20 time points over 30 min following the addition of patterns ± sd; n ≥ 6. Significant differences relative to the wild type are indicated by asterisks (*, P < 0.05; **, P < 0.01; and ***, P < 0.001, Student’s t test).
To further assess the functional redundancy of RLCK VII members, we examined chitin-triggered ROS production in both single and higher order mutants of RLCK VII-4, -5, -7, and -8. In general, the rlck vii-4, -5, -7, and -8 higher order mutants were more severely impaired in chitin-triggered ROS production than were the single mutants (Supplemental Fig. S5), indicating the functional redundancy within each clade. No single mutants of RLCK VII-4 members displayed significant ROS defects, indicating that RLCK VII-4 members are completely redundant in chitin-triggered ROS production (Supplemental Fig. S5A). Among the RLCK VII-5, -7, and -8 members, reduced ROS production was observed in pbl34, pbl35, pbl36, pbl31, bik1, and pbl1 single mutants (Supplemental Fig. S5, B–D). These results indicate that the RLCK VII members are functionally redundant both within and across different clades in chitin signaling. Individual members of RLCK VII-5, -7, and -8 do not have equal functions, as pbl35, pbl36, pbl31, and bik1 displayed greater defects than did the other single mutants of genes from the same clades.
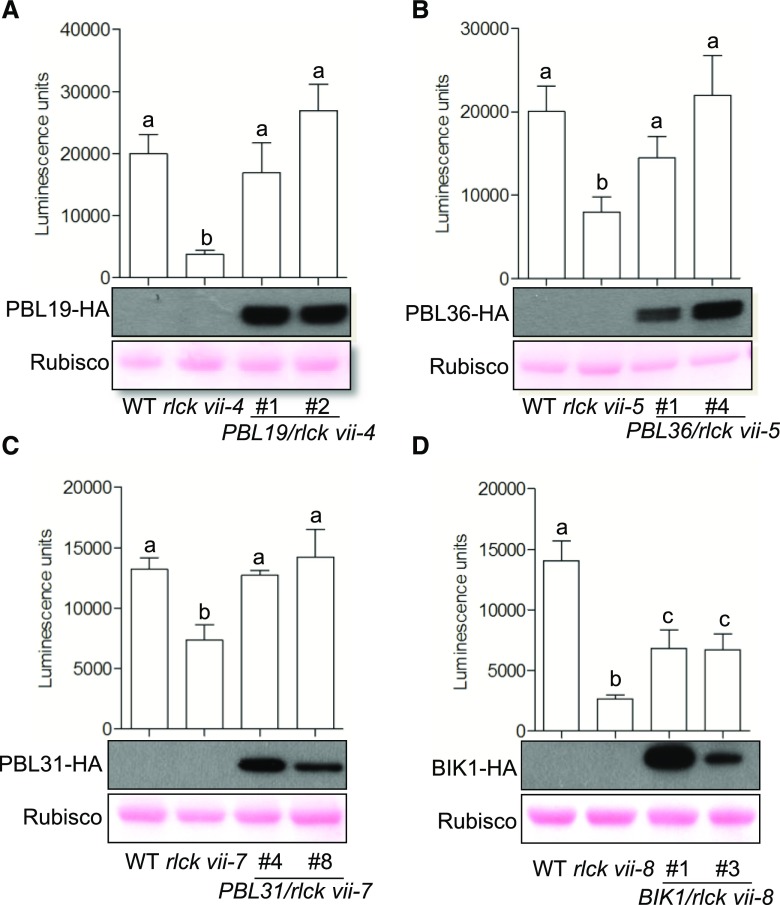
To confirm that these ROS defects were caused by the disruption of these RLCK VII members, we complemented higher order mutants with individual RLCK VII members under the control of their native promoters. For each construct, multiple T1 transgenic plants expressing the transgenes were identified and chitin-induced ROS production was examined (Fig. 3). In each case, all transgenic plants tested showed partial to full restoration of ROS production. The T1 transgenic plant expression of PBL19, PBL36, and PBL31 completely restored ROS production in rlck vii-4, -5, and -7 mutants (Fig. 3, A–C). This was unexpected, since members of these RLCK clades should act in an additive manner. One possible explanation is that the transgenic plants tested had higher expression levels of PBLs than in the wild type. The two T1 transgenic plants carrying BIK1 fused with the hemagglutinin (HA) epitope tag coding sequence partially restored ROS production in the rlck vii-8 mutant, which was expected (Fig. 3D). Thus, these data demonstrate that the RLCK VII-4, -5, -7, and -8 members have important roles in chitin-triggered ROS production.
Figure 3.
Molecular complementation of rlck vii higher order mutants. The expression of PBL19 (A), PBL36 (B), PBL31 (C), and BIK1 (D) driven by their native promoters rescued chitin-triggered ROS production in rlck vii-4, -5, -7, and -8, respectively, compared with the wild type (WT). The indicated constructs were transformed into rlck vii higher order mutants, and two independent T1 transgenic plants for each construct were examined. The line numbers (#) refer to different T1 transgenic plants in each part. The experiment was repeated two times with different T1 transgenic plants and showed similar results. Luminescence units refer to arbitrary units recorded by a luminometer. Four leaves per plant were examined. Values are means of the sum of luminescence units collected from all 20 time points over 30 min following the addition of chitin. Error bars represent sd. Different letters indicate significant differences at P ≤ 0.05 (Student’s t test); n = 4. The gels show PBL accumulation in rlck vii higher order mutants (top) and equal loading, as demonstrated by Ponceau staining of Rubisco (bottom).
Roles of Specific Members in MAPK Activation
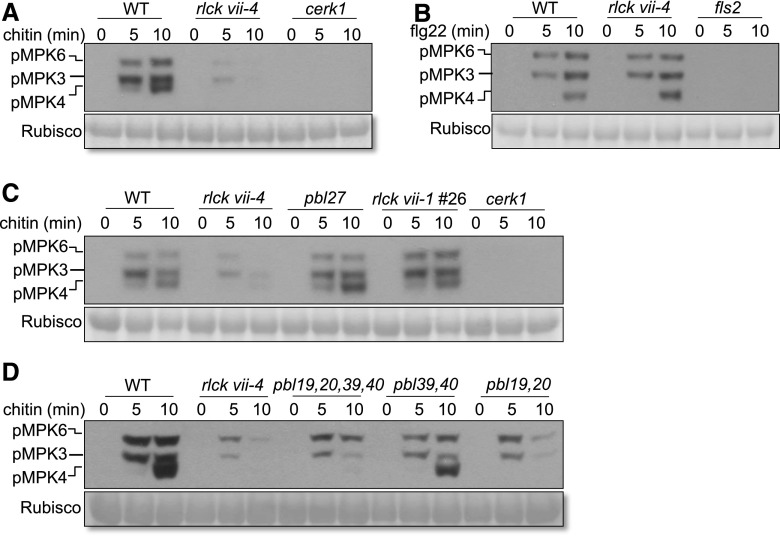
Microbial or plant molecular patterns commonly induce MAPK activation within minutes (Pitzschke et al., 2009; Yamaguchi and Huffaker, 2011). We investigated whether different RLCK VII members are required for MAPK activation in response to different patterns. MAPK activation triggered by chitin was reduced strongly in the rlck vii-4 mutant compared with the wild type (Fig. 4A; Supplemental Fig. S6A). Surprisingly, flg22-, elf18-, and Pep2-triggered MAPK activation were normal in rlck vii-4 mutants (Fig. 4B; Supplemental Figs. S6B and S7), indicating that RLCK VII-4 is required specifically for chitin-triggered MAPK activation. PBL27 was reported previously to be required for chitin-triggered MAPK activation (Shinya et al., 2014). However, we failed to reproduce this result for the same allele, even when we tested the rlck vii-1 quintuple mutant (Fig. 4C; Supplemental Fig. S6C). These results indicated that RLCK VII-4 rather than RLCK VII-1 plays a specific role in chitin-triggered MAPK activation.
Figure 4.
Mutants of RLCK VII-4 are compromised in chitin-triggered MAPK activation. A, Chitin-triggered MAPK activation in the rlck vii-4 mutant compared with the wild type (WT). B, Flg22-triggered MAPK activation in the rlck vii-4 sextuple mutant. C, Chitin-triggered MAPK activation in the pbl27 and rlck vii-1 quintuple mutants. D, Additive effects of RLCK VII-4 members in chitin-triggered MAPK activation. rlck vii-4 sextuple, quadruple (pbl19,20,39,40), and double (pbl39,40 and pbl19,20) mutants were used for this analysis. Seedlings of the indicated genotypes were treated with chitin or flg22 and harvested at the indicated time points. MAPK activation was analyzed by immunoblotting using anti-pERK antibody. cerk1 and fls2 mutated in PRR genes relevant to chitin and flg22 were included as controls, respectively. Equal loading is demonstrated by Ponceau staining of Rubisco (bottom). The results shown are representative of three independent experiments.
To test whether RLCK VII-4 members additively contribute to chitin-triggered MAPK activation, we identified homozygous double (pbl19,20 and pbl39,40) and quadruple (pbl19,20,39,40) mutants and examined their MAPK activation. Lines carrying the pbl19,20 mutations displayed greater defects in chitin-triggered MAPK activation than did lines carrying wild-type PBL19,20, whereas the rlck vii-4 sextuple mutant was nearly unresponsive to chitin (Fig. 4D; Supplemental Fig. S6D). These results showed that RLCK VII-4 members are particularly important for chitin-triggered MAPK activation.
We additionally found that Pep2-triggered MAPK activation was reduced slightly in the triple (bik1 pbl1,11) and rlck ii-8 quadruple mutants, a result consistent with a previous report on the bik1 pbl1 double mutant (Supplemental Fig. S8D; Yamada et al., 2016), while flg22-, elf18-, and chitin-triggered MAPK activation were normal in triple and quadruple mutants of RLCK VII-8 (Supplemental Fig. S8, A–C), which was consistent with our previous report that flg22-induced MAPK activation is normal in bik1 pbl1 mutants (Feng et al., 2012). The MAPK activation in response to flg22 and chitin was unaffected in the other higher order mutants (Supplemental Fig. S9). These results suggest that RLCK VII-4 and RLCK VII-8 link specific PRRs to MAPK activation.
Members of RLCK VII Contribute to Pattern-Triggered Defense Gene Expression and Disease Resistance
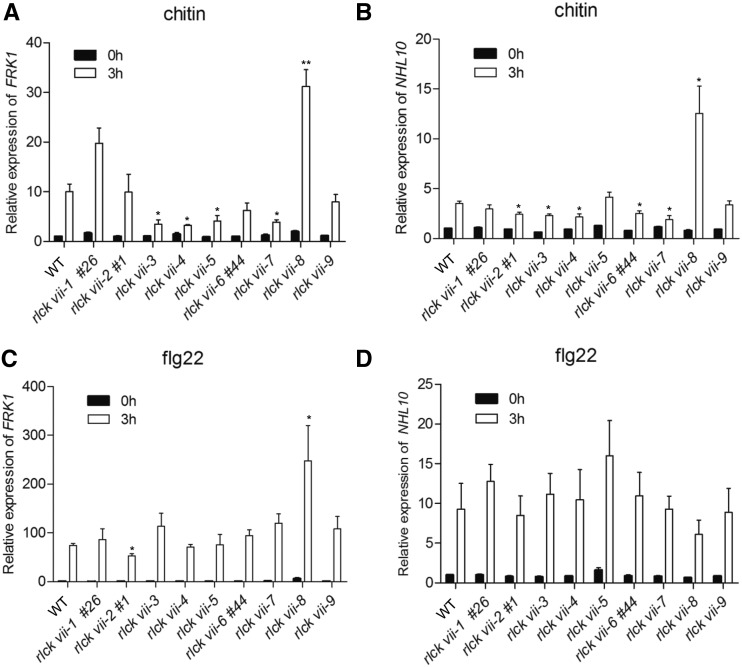
We analyzed defense-related gene expression in the higher order mutants to determine the roles of RLCK VII members in pattern-triggered defense gene expression. The chitin-induced expression of FLG22-INDUCED RECEPTOR-LIKE KINASE1 (FRK1) was reduced to 50% of wild-type levels in rlck vii-3, -4, -5, and -7 higher order mutants (Fig. 5A). The expression of NDR1/HIN1-LIKE10 (NHL10) also was reduced in rlck vii-2, -3, -4, -6, and -7 mutants (Fig. 5B). By contrast, the chitin-induced expression of FRK1 and NHL10 was about 3-fold higher in the rlck vii-8 mutant than in the wild-type (Fig. 5, A and B), indicating that these family members are differentially required for the chitin-triggered expression of FRK1 and NHL10.
Figure 5.
FRK1 and NHL10 expression triggered by chitin and flg22 in higher order rlck vii mutants. A and B, Chitin-induced defense gene expression in higher order mutants. C and D, flg22-induced defense gene expression in higher order mutants. Eight-day-old seedlings of the indicated genotypes were treated with chitin or flg22 and harvested at 0 and 3 h. FRK1 and NHL10 transcripts were quantified by RT-qPCR using ACTIN8 as the internal standard. The line numbers (#) refer to alleles created by CRISPR-Cas9. Values are means ± sd of three biological replicates, and significant differences between mutants and the wild type (WT) are indicated by asterisks (*, P < 0.05 and **, P < 0.01, Student’s t test).
Only the rlck vii-2 mutant consistently showed slightly lower expression of flg22-induced FRK1, whereas the rlck vii-8 mutant exhibited higher expression (Fig. 5C). The pcrk1,2 double mutant was reported previously to show a reduction in flg22-triggered FRK1 expression (Kong et al., 2016). However, we failed to observe the reduced FRK1 expression in the rlck vii-4 sextuple mutant. This disparity may be due to the differences in the treatments (i.e. 1 μm flg22 for 4 h versus 0.1 μm flg22 for 3 h) and/or in the materials used. FRK1 and NHL10 expression in response to flg22 were unaffected in most of these higher order mutants (Fig. 5, C and D), suggesting that these RLCK VII subgroups have overlapping roles in this process. Alternatively, these PBL genes may not be required for the activation of FRK1 and NHL10.
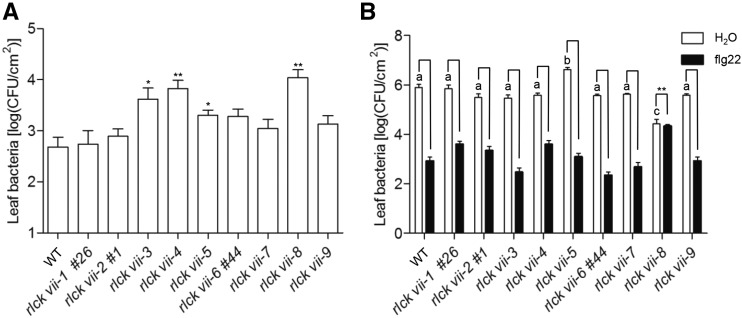
To further determine the functions of these subgroups in pattern-induced resistance to P. syringae, higher order mutants and the wild type were spray inoculated with the nonadapted bacterial pathogen P. syringae hrcC−, which lacks a functional type III secretion system and is unable to inhibit PTI. Three days after inoculation, the bacterial populations in the rlck vii-3, -4, and -8 mutants were approximately 8- to 20-fold greater than in the wild type, with the highest population found in rlck vii-8. The rlck vii-5 mutant also supported a 3-fold greater bacterial population than wild-type plants (Fig. 6A). This is consistent with the previous finding that bik1 single and bik1 pbl1 double mutants were more susceptible than the wild type when spray inoculated with P. syringae hrcC− (Lu et al., 2010; Zhang et al., 2010; Li et al., 2014). The remaining higher order mutants displayed normal disease resistance to this bacterial strain (Fig. 6A). Together, these results indicate that RLCK VII-3, -4, -5, and -8 members play primary roles in resistance to P. syringae.
Figure 6.
Pattern-induced resistance of RLCK VII members to P. syringae. A, Four-week-old plants of the indicated genotypes were spray inoculated with P. syringae hrcC–, and bacterial growth was measured 3 d post inoculation. The line numbers (#) refer to alleles created by CRISPR-Cas9. Values are means of log (colony-forming units [CFU] cm−2 leaf tissue) ± sd; n ≥ 10. Significant differences relative to the wild type (WT) are indicated by asterisks (*, P < 0.05 and **, P < 0.01, Student’s t test). B, Four-week-old plants were pretreated with water or flg22 and infiltrated 1 d later with P. syringae DC3000. The bacterial population was determined 2 d post inoculation. Values are means of log (CFU cm−2 leaf tissue) ± sd. Different letters indicate significant differences between higher order mutants and the wild type in the absence of flg22, and significant differences between higher order mutants and the wild type are indicated by asterisks (**, P < 0.001, Student’s t test); n ≥ 6. The results shown are representative of three independent experiments.
We next performed a flg22 protection assay to determine the roles of these RLCK VII members in pattern-induced resistance to P. syringae DC3000 (Zipfel et al., 2004). In the absence of flg22 treatment, bacterial growth in rlck vii-5 mutant plants was significantly greater than in the wild type (Fig. 6B), suggesting that these plants had a defect in basal resistance unrelated to FLS2 signaling. However, bacterial population numbers in rlck vii-8 plants were the same as in bik1 plants and lower than in the wild type in the absence of flg22 (Fig. 6B; Veronese et al., 2006; Zhang et al., 2010). Flg22 treatment failed to protect rlck vii-8 plants from P. syringae DC3000, consistent with the reported role of BIK1 in pattern-induced resistance to P. syringae DC3000 (Zhang et al., 2010). However, flg22-induced resistance to P. syringae DC3000 in other higher order mutants was comparable to that in the wild type, indicating that RLCK VII-8 members are the most important in FLS2-mediated disease resistance.
Roles of RLCK VII Members in Growth and Development
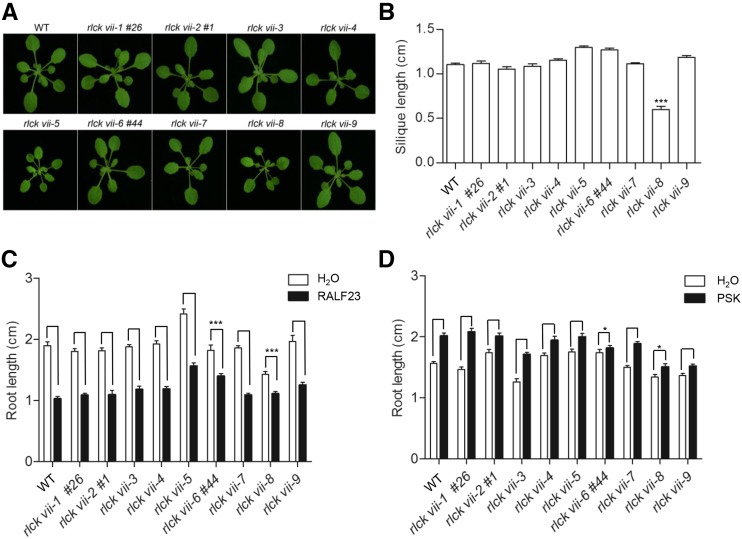
In addition to their roles in plant immunity, RLCK VII members regulate plant growth and development (Liang and Zhou, 2018). An examination of the morphological phenotypes showed that most of the higher order mutants displayed no visible defects in growth and development. rlck vii-5 and -8 were slightly smaller than wild-type plants (Fig. 7A). The phenotype of rlck vii-8 is likely attributable to the bik1 mutation, which was reported to cause increased accumulation of SA and reduced plant size (Veronese et al., 2006). We also examined silique length in these higher order mutants and found that rlck vii-8 mutants had shorter siliques than wild-type plants (Fig. 7B).
Figure 7.
Morphology and root growth responses of rlck vii higher order mutants. A, Morphology of the wild type (WT) and rlck vii higher order mutants. Plants were grown under 10-h-light/14-h-dark conditions for 4 weeks. The line numbers (#) refer to alleles created by CRISPR-Cas9. The photographs are representative of three independent experiments. B, Silique lengths of the wild type and rlck vii higher order mutants. Values are means ± sd, and significant differences relative to the wild-type are indicated by asterisks (***, P < 0.001, Student’s t test); n = 20. C, Root growth inhibition response to RALF23 in rlck vii higher order mutants. Values are means ± sd, and significant differences between the wild type and higher order mutants are indicated by asterisks (***, P < 0.001, Student’s t test); n = 12. D, Root elongation response to PSK in rlck vii higher order mutants. Values are means ± sd, and significant differences between the wild type and higher order mutants are indicated by asterisks (*, P < 0.05, Student’s t test); n = 12. The results shown are representative of three independent experiments.
We further sought to determine the roles of RLCK VII members in peptide-regulated growth and development by examining the responses of seedlings to RALF23 and PSK (Kutschmar et al., 2009; Stegmann et al., 2017). Root growth inhibition assays showed that rlck vii-6 and -8 had impaired sensitivity to RALF23 compared with the wild type, whereas other higher order mutants did not (Fig. 7C). These results indicated that RLCK VII-6 and -8 may function downstream of FER to regulate root growth. An examination of PSK-induced root growth showed that rlck vii-6 and -8 also were significantly less sensitive than the wild type to PSK, whereas other higher order mutants were comparable to the wild type (Fig. 7D). Together, these results indicated that RLCK VII-6 and -8 members are likely required for RK-regulated plant growth.
DISCUSSION
In this study, we constructed nine higher order mutants of proteins from RLCK subfamily VII and systematically characterized their immune and developmental phenotypes. Various previous reports have shown that single mutants of RLCK VII members have weak phenotypes in PTI (Zhang et al., 2010; Sreekanta et al., 2015; Kong et al., 2016). By contrast, the expression of bacterial effectors that target multiple RLCK VII members leads to strong suppression of PTI (Zhang et al., 2010; Feng et al., 2012), suggesting that these subfamily members have overlapping roles in plant immunity. Given the size of the RLCK VII subfamily, it has been difficult to empirically analyze the redundancy and specificity of its members in different RK signaling pathways and their roles in various downstream responses. The construction and analysis of higher order mutants allowed us to uncover the functions of previously uncharacterized RLCK VII members. We found that several higher order mutants were more severely impaired in chitin-triggered ROS production than the single mutants. Furthermore, multiple higher order mutants displayed impaired PTI responses to each pattern tested, indicating that there is a great degree of functional redundancy within and across different clades of RLCK VII members.
Our data demonstrate that, while the RLCK VII-5, -7, and -8 members are involved in signaling downstream of multiple PRRs, including FLS2, EFR, and CERK1, the RLCK VII-4 members function specifically in chitin-induced immunity. These findings indicate that, although multiple RLCK VII subgroups are commonly employed by different PRRs for immune signaling, one RLCK VII subgroup is recruited by chitin receptors.
The activation of MAPK cascades is critical for the establishment of disease resistance (Meng and Zhang, 2013). However, the molecular link between RKs and MAPK activation remains elusive. There is conflicting evidence regarding whether RLCKs play a major role in MAPK activation. For example, BIK1 and PBL1 have been shown to have a modest role specifically in Pep2-triggered MAPK activation (Yamada et al., 2016), which was confirmed in this study. PCRK1 and PCRK2, which correspond to PBL39 and PBL40 of the RLCK VII-4 subgroup, have been shown previously to play a minor role in flg22-triggered MAPK activation (Kong et al., 2016), but that result was not reproduced in this study. In addition, PBL27 of the RLCK VII-1 subgroup was reported recently to mediate chitin-induced MAPK activation (Shinya et al., 2014), but our extensive analyses failed to provide support for the involvement of PBL27, or any RLCK VII-1 members, in MAPK activation. In these studies, single or double mutants were used for the analyses, which may explain subtle phenotypes that are difficult to reproduce. By using higher order mutants, we have demonstrated that RLCK VII-4 members are profoundly important for chitin-triggered MAPK activation, but they play no detectable role in MAPK activation triggered by other patterns. We were unable to support a role for RLCK VII-1 members in MAPK activation by any patterns, including chitin. These results collectively support the idea that RLCK VII-4 members specifically link the chitin receptors to MAPK activation, whereas RLCK VII-8 members are employed by PEPR1 and PEPR2 for MAPK activation.
BIK1 and PBL1, two members of the RLCK VII-8 subgroup, play positive roles in PTI (Lu et al., 2010; Zhang et al., 2010; Kadota et al., 2014; Li et al., 2014; Ranf et al., 2014). However, the rlck vii-8 quadruple mutant exhibited elevated FRK1 expression in response to chitin and flg22 and increased resistance to P. syringae (Figs. 5, A and C, and 6B). This seems to be at odds with the positive function of BIK1 and PBL1 in PTI. However, bik1 mutants are known to accumulate SA (Veronese et al., 2006), which might have led to the increased defenses in rlck vii-8 higher order mutants. The rlck vii-6 triple mutant exhibited increased ROS production in response to flg22. Interestingly, PBL13, another RLCK VII-6 member, also has been reported to negatively regulate ROS production triggered by flg22 and elf18 (Lin et al., 2015). Whether certain RLCK VII-6 members are negative regulators of ROS production downstream of specific PRRs remains to be examined.
Our data demonstrate that RLCK VII-6 (PBL8, -16, and -17) and RLCK VII-8 specifically link the RALF23 and PSK receptors to regulate root growth in Arabidopsis. Interestingly, a different member of RLCK VII-6, RIPK, has been shown to mediate responses to RALF1 in seedlings (Du et al., 2016), suggesting that RLCK VII-6 may be particularly important for RALF signaling. Moreover, we found that rlck vii-8 mutants also had smaller siliques, which is consistent with the reported role of BIK1 in fertility (Veronese et al., 2006).
Together, our data uncover a large number of RLCK VII members that modulate plant immune signaling. We identified two subgroups of RLCK VII that link PRRs to MAPK activation. This work provides important genetic resources for future studies on plant immunity and growth mediated by diverse RKs. Because RLCK VII members are commonly targeted by multiple pathogen effectors (Zhang et al., 2010; Feng et al., 2012), and several of them have been shown to mediate effector-triggered immunity (Shao et al., 2003; Liu et al., 2011; Guy et al., 2013; Wang et al., 2015), these materials also will be useful in analyses of effector-mediated virulence and effector-triggered immunity.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) plants used in this study included the wild type (Columbia-0), bik1 (Veronese et al., 2006), ripk (Columbia-0; Liu et al., 2011), pbs1-1 (Warren et al., 1999), pbl1 and pbl2 (Zhang et al., 2010), and other PBL T-DNA insertion lines obtained from the Nottingham Arabidopsis Stock Centre (http://www.arabidopsis.info).
The higher order mutants were generated by combining mutant genes within each subgroup. The rlck vii-3, -4, -5, -7, -8, and -9 mutants were generated by crossing homozygous T-DNA lines. Homozygous mutations were identified by genotyping (primers are listed in Supplemental Table S2). Some of the mutations were loosely linked, and these T-DNA lines were first combined to generate homozygous double or triple mutants by crossing, and then these double or triple mutants were crossed to the unlinked T-DNA lines to generate higher order mutants. Two PCRs were performed to identify the homozygous mutation for each gene. The first reaction was carried out with gene-specific primers flanking the T-DNA insertion (left genomic primer and right genomic primer), and the second reaction was performed with a right genomic primer and a T-DNA-specific primer. A homozygous mutation was indicated by a lack of PCR product in the first reaction and the presence of PCR product in the second reaction. For rlck vii-1, -2, and -6 higher order mutants, pbl5,7,27, pbs1-1, pbl24,26, and pbl8,16 homozygous T-DNA mutants were generated before the remaining mutations were introduced using the CRISPR-Cas9 system (Supplemental Fig. S2). The generation and identification of CRISPR-Cas9 mutations are described below.
Arabidopsis plants were grown at 22°C in a greenhouse with a 10-h-light/14-h-dark photoperiod. Four- to 5-week-old plants were used for oxidative burst detection and the bacterial infection assay.
CRISPR-Cas9-Mediated Mutations
To generate mutations in PBL6, PBL17, and PBL25 using the CRISPR-Cas9 system, corresponding single guide RNAs were cloned into a CRISPR-Cas9 vector (Xing et al., 2014) and transformed into plants by Agrobacterium tumefaciens-mediated transformation.
To genotype these mutated lines, total DNA was extracted from T1 transgenic plants, and fragments containing the target sites were amplified by PCR using gene-specific primers (Supplemental Table S2). The PCR products were digested with appropriate restriction enzymes (HhaI, HphI, and BstUI for PBL6, PBL17, and PBL25, respectively), and plant lines carrying completely or partially undigested bands were selected. The PCR products of homozygous T2 lines, for which restriction enzyme treatment produced no digested bands, were sequenced to confirm mutation.
Phylogenetic Analysis
The full-length sequence of each RLCK VII member was aligned using MUSCLE in MEGA (Kumar et al., 2016), and this alignment was used to generate a phylogenetic tree with the neighbor-joining method (Saitou and Nei, 1987). The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965). The bootstrap analyses were conducted with 500 replicates. The numbers associated with each branch represent the bootstrap support.
Molecular Complementation
The indicated full-length genes were amplified from wild-type genomic DNA and subcloned into a modified pCAMBIA1300 binary vector that harbors an HA epitope tag, to generate PBL-HA constructs. These constructs were then introduced into the indicated higher order mutants using an A. tumefaciens-mediated floral dip transformation method (Clough and Bent, 1998).
RNA Isolation and Gene Expression Analysis
Eight-day-old seedlings grown on one-half-strength Murashige and Skoog (MS) medium were sprayed with 0.1 μm flg22 or 200 μg mL−1 chitin in deionized water containing 0.01% (v/v) Silwet L-77. For each treatment, eight to 15 plants were harvested for RNA isolation. To detect PBL transcripts in pbl mutants, RNA was extracted from untreated 8-d-old seedlings.
Total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen), and 2 μg of total RNA was reverse transcribed using a Maxima First Strand cDNA Synthesis Kit (Thermo Scientific). Subsequently, RT-qPCR was performed using a SYBR Premix Ex TaqII Kit (Takara). Primers used are listed in Supplemental Table S2.
Oxidative Burst
The leaves of 5-week-old plants were sliced into 1-mm strips and incubated in deionized water on a 96-well plate for 10 to 12 h and then treated with 0.1 μm flg22, 0.1 μm elf18, or 200 μg mL−1 chitin in 200 μL of buffer (20 μm Luminol and 1 μg mL−1 horseradish peroxidase) as described (Zhang et al., 2007). Luminescence was recorded immediately with an integration time of 0.5 s using a GloMax 96 Microplate Luminometer (Promega). Data from a total of 20 time points were collected for each sample over the course of 30 min, and the sum of the total luminescence units measured was used as a measurement of total ROS production. All experiments were repeated two to four times.
MAPK Activity Assay
Eight-day-old seedlings grown on one-half-strength MS medium were sprayed with 0.1 μm flg22, 0.1 μm elf18, 0.1 μm Pep2, or 200 μg mL−1 chitin (containing 0.01% Silwet L-77). About 10 seedlings were harvested at 0, 5, and 10 min after each treatment and ground thoroughly in liquid nitrogen. Protein was extracted from these seedlings with extraction buffer (50 mm HEPES, pH 7.5, 150 mm KCl, 1 mm EDTA, 0.5% Triton X-100, 1 mm DTT, and 1× protease inhibitors [Roche]), and the phosphorylation of MAPKs was detected by anti-pERK (Cell Signaling) immunoblotting. All experiments were repeated at least three times.
Bacterial Growth Assay
Pseudomonas syringae DC3000 and the hrcC– mutant were cultured in King’s medium B (King et al., 1954) containing 50 mg L−1 rifampicin at 28°C. For the flg22 protection assay, 5-week-old plants were preinfiltrated with 1 μm flg22 or deionized water and then, 1 d later, infiltrated with 1 × 106 CFU mL−1 P. syringae DC3000. The bacterial populations were determined after 2 d as described previously (Katagiri et al., 2002). Two leaf discs of 0.6 cm in diameter from different leaves served as one replicate, and each data point represented at least six replicates. For spray inoculation, plant leaves were sprayed with P. syringae mutant hrcC– (Yuan and He, 1996) at 5 × 108 CFU mL−1 containing 0.017% Silwet L-77. The bacterial populations were determined after 3 d. All experiments were repeated at least three times.
Root Growth Responses to RALF23 or PSK Peptide
For the RALF23-induced root growth inhibition assay, seedlings were germinated, grown vertically on one-half-strength MS medium for 4 d, and then transferred to 600 μL of one-half-strength MS liquid medium containing 1 μm RALF23 on a 24-well plate and incubated at 22°C with mild shaking for 3 d. For the PSK-triggered root growth assay, seedlings were germinated and grown vertically on one-half-strength MS medium containing 1 μm PSK for 7 d. All experiments were repeated at least three times.
Accession Numbers
Sequence data from this article can be found in the TAIR database (https://www.arabidopsis.org): At2G28590 (PBL6), At1G07870 (PBL5), At5G02800 (PBL7), At5G18610 (PBL27), AT5G13160 (PBS1), AT1G61860 (PBL41), AT3G20530 (PBL23), AT4G13190 (PBL24), AT3G24790 (PBL25), AT3G07070 (PBL26), AT5G16500 (PBL43), AT3G02810 (PBL42), AT1G76370 (PBL22), AT1G20650 (PBL21), AT3G26940 (CDG1), AT1G24030 (PBL28), AT5G03320 (PBL40), AT3G09830 (PBL39), AT2G39110 (PBL38), AT2G28940 (PBL37), AT5G47070 (PBL19), AT4G17660 (PBL20), AT5G15080 (PBL34), AT3G01300 (PBL35), AT3G28690 (PBL36), AT1G72540 (PBL33), AT2G26290 (PBL12), AT5G35580 (PBL13), AT2G05940 (PBL14), AT1G61590 (PBL15), AT2G07180 (PBL17), AT5G56460 (PBL16), AT5G01020 (PBL8), AT2G17220 (PBL32), AT1G76360 (PBL31), AT4G35600 (PBL30), AT2G28930 (PBL10), AT1G07570 (PBL9), AT5G02290 (PBL11), AT3G55450 (PBL1), AT2G39660 (BIK1), AT1G74490 (PBL29), AT1G26970 (PBL4), AT1G69790 (PBL18), AT2G02800 (PBL3), and AT1G14370 (PBL2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Gene structures of PBL genes and locations of T-DNA insertions.
Supplemental Figure S2. CRISPR-Cas9-mediated mutations of PBL6, PBL17, and PBL25.
Supplemental Figure S3. Expression profiles of RLCK VII members in response to different molecular patterns.
Supplemental Figure S4. Replicate ROS production data for Figure 2.
Supplemental Figure S5. Chitin-triggered ROS production in single mutants.
Supplemental Figure S6. Replicate blots for Figure 4.
Supplemental Figure S7. RLCK VII-4 members are not required for elf18- and Pep2-triggered MAPK activation.
Supplemental Figure S8. MAPK activation of RLCK VII-8 mutants.
Supplemental Figure S9. MAPK activation triggered by flg22 and chitin.
Supplemental Table S1. List of mutant lines and higher order mutants.
Supplemental Table S2. Primers used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Qi-Jun Chen for providing the CRISPR-Cas9 vector and Yufei Yu for assistance in drawing PBL structures.
Footnotes
This work was funded by the National Natural Science Foundation of China (31370293 to F.F.), by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDB11020200), and by The State Key Laboratory of Plant Genomics (SKLPG2016A-18 to J.-M.Z.).
References
- Albert I, Böhm H, Albert M, Feiler CE, Imkampe J, Wallmeroth N, Brancato C, Raaymakers TM, Oome S, Zhang H, et al. (2015) An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat Plants 1: 15140. [DOI] [PubMed] [Google Scholar]
- Bauer Z, Gómez-Gómez L, Boller T, Felix G (2001) Sensitivity of different ecotypes and mutants of Arabidopsis thaliana toward the bacterial elicitor flagellin correlates with the presence of receptor-binding sites. J Biol Chem 276: 45669–45676 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Wang X, Chory J (2006) Arabidopsis brassinosteroid signaling pathway. Sci STKE 2006: cm5. [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Burr CA, Leslie ME, Orlowski SK, Chen I, Wright CE, Daniels MJ, Liljegren SJ (2011) CAST AWAY, a membrane-associated receptor-like kinase, inhibits organ abscission in Arabidopsis. Plant Physiol 156: 1837–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, Stacey G (2014) The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3: e03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537–552 [DOI] [PubMed] [Google Scholar]
- Du C, Li X, Chen J, Chen W, Li B, Li C, Wang L, Li J, Zhao X, Lin J, et al. (2016) Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis. Proc Natl Acad Sci USA 113: E8326–E8334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng F, Yang F, Rong W, Wu X, Zhang J, Chen S, He C, Zhou JM (2012) A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485: 114–118 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Guy E, Lautier M, Chabannes M, Roux B, Lauber E, Arlat M, Noël LD (2013) xopAC-triggered immunity against Xanthomonas depends on Arabidopsis receptor-like cytoplasmic kinase genes PBL2 and RIPK. PLoS ONE 8: e73469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Ryan CA (2007) Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc Natl Acad Sci USA 104: 10732–10736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, et al. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell 54: 43–55 [DOI] [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He SY (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. The Arabidopsis Book 1: e0039, doi/10.1199/tab.0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Guan S, Burlingame AL, Wang ZY (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell 43: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44: 301–307 [PubMed] [Google Scholar]
- Kong Q, Sun T, Qu N, Ma J, Li M, Cheng YT, Zhang Q, Wu D, Zhang Z, Zhang Y (2016) Two redundant receptor-like cytoplasmic kinases function downstream of pattern recognition receptors to regulate activation of SA biosynthesis. Plant Physiol 171: 1344–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, Postel S, Arents M, Jeworutzki E, Al-Rasheid KA, et al. (2010) Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem 285: 13471–13479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutschmar A, Rzewuski G, Stührwohldt N, Beemster GTS, Inzé D, Sauter M (2009) PSK-α promotes root growth in Arabidopsis. New Phytol 181: 820–831 [DOI] [PubMed] [Google Scholar]
- Lei J, A Finlayson S, Salzman RA, Shan L, Zhu-Salzman K (2014) BOTRYTIS-INDUCED KINASE1 modulates Arabidopsis resistance to green peach aphids via PHYTOALEXIN DEFICIENT4. Plant Physiol 165: 1657–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, et al. (2014) The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338 [DOI] [PubMed] [Google Scholar]
- Li L, Yu Y, Zhou Z, Zhou JM (2016) Plant pattern-recognition receptors controlling innate immunity. Sci China Life Sci 59: 878–888 [DOI] [PubMed] [Google Scholar]
- Liang X, Zhou JM (2018) Receptor-like cytoplasmic kinases: central players in plant receptor kinase-mediated signaling. Annu Rev Plant Biol 69: 267–299 [DOI] [PubMed] [Google Scholar]
- Lin W, Ma X, Shan L, He P (2013) Big roles of small kinases: the complex functions of receptor-like cytoplasmic kinases in plant immunity and development. J Integr Plant Biol 55: 1188–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ZJ, Liebrand TW, Yadeta KA, Coaker G (2015) PBL13 is a serine/threonine protein kinase that negatively regulates Arabidopsis immune responses. Plant Physiol 169: 2950–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Lin ZJ, Coaker G (2011) A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhong S, Guo X, Hao L, Wei X, Huang Q, Hou Y, Shi J, Wang C, Gu H, et al. (2013a) Membrane-bound RLCKs LIP1 and LIP2 are essential male factors controlling male-female attraction in Arabidopsis. Curr Biol 23: 993–998 [DOI] [PubMed] [Google Scholar]
- Liu T, Liu Z, Song C, Hu Y, Han Z, She J, Fan F, Wang J, Jin C, Chang J, et al. (2012) Chitin-induced dimerization activates a plant immune receptor. Science 336: 1160–1164 [DOI] [PubMed] [Google Scholar]
- Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, Zhou JM (2013b) BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci USA 110: 6205–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y, Ogawa M, Morita A, Sakagami Y (2002) An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296: 1470–1472 [DOI] [PubMed] [Google Scholar]
- Meng X, Zhang S (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51: 245–266 [DOI] [PubMed] [Google Scholar]
- Muto H, Yabe N, Asami T, Hasunuma K, Yamamoto KT (2004) Overexpression of constitutive differential growth 1 gene, which encodes a RLCKVII-subfamily protein kinase, causes abnormal differential and elongation growth after organ differentiation in Arabidopsis. Plant Physiol 136: 3124–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A, Schikora A, Hirt H (2009) MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Ranf S, Eschen-Lippold L, Fröhlich K, Westphal L, Scheel D, Lee J (2014) Microbe-associated molecular pattern-induced calcium signaling requires the receptor-like cytoplasmic kinases, PBL1 and BIK1. BMC Plant Biol 14: 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233 [DOI] [PubMed] [Google Scholar]
- Shinya T, Yamaguchi K, Desaki Y, Yamada K, Narisawa T, Kobayashi Y, Maeda K, Suzuki M, Tanimoto T, Takeda J, et al. (2014) Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J 79: 56–66 [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekanta S, Bethke G, Hatsugai N, Tsuda K, Thao A, Wang L, Katagiri F, Glazebrook J (2015) The receptor-like cytoplasmic kinase PCRK1 contributes to pattern-triggered immunity against Pseudomonas syringae in Arabidopsis thaliana. New Phytol 207: 78–90 [DOI] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C (2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289 [DOI] [PubMed] [Google Scholar]
- Swiderski MR, Innes RW (2001) The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J 26: 101–112 [DOI] [PubMed] [Google Scholar]
- Tang D, Wang G, Zhou JM (2017) Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29: 618–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, Mengiste T (2006) The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Roux B, Feng F, Guy E, Li L, Li N, Zhang X, Lautier M, Jardinaud MF, Chabannes M, et al. (2015) The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 18: 285–295 [DOI] [PubMed] [Google Scholar]
- Warren RF, Merritt PM, Holub E, Innes RW (1999) Identification of three putative signal transduction genes involved in R gene-specified disease resistance in Arabidopsis. Genetics 152: 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Yamashita-Yamada M, Hirase T, Fujiwara T, Tsuda K, Hiruma K, Saijo Y (2016) Danger peptide receptor signaling in plants ensures basal immunity upon pathogen-induced depletion of BAK1. EMBO J 35: 46–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A (2011) Endogenous peptide elicitors in higher plants. Curr Opin Plant Biol 14: 351–357 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA (2010) PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, He SY (1996) The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. J Bacteriol 178: 6399–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, et al. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185 [DOI] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L (1965) Bryson V, Vogel H.J, eds, Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins, Academic Press, New York, pp 97–166 [Google Scholar]