In legume nodules, selective carbonylation and glycation of proteins occurs during nodule development and may have a role in the regulation of metabolism and senescence.
Abstract
Nitrogen fixation is an agronomically and environmentally important process catalyzed by bacterial nitrogenase within legume root nodules. These unique symbiotic organs have high metabolic rates and produce large amounts of reactive oxygen species that may modify proteins irreversibly. Here, we examined two types of oxidative posttranslational modifications of nodule proteins: carbonylation, which occurs by direct oxidation of certain amino acids or by interaction with reactive aldehydes arising from cell membrane lipid peroxides; and glycation, which results from the reaction of lysine and arginine residues with reducing sugars or their autooxidation products. We used a strategy based on the enrichment of carbonylated peptides by affinity chromatography followed by liquid chromatography-tandem mass spectrometry to identify 369 oxidized proteins in bean (Phaseolus vulgaris) nodules. Of these, 238 corresponded to plant proteins and 131 to bacterial proteins. Lipid peroxidation products induced most carbonylation sites. This study also revealed that carbonylation has major effects on two key nodule proteins. Metal-catalyzed oxidation caused the inactivation of malate dehydrogenase and the aggregation of leghemoglobin. In addition, numerous glycated proteins were identified in vivo, including three key nodule proteins: sucrose synthase, glutamine synthetase, and glutamate synthase. Label-free quantification identified 10 plant proteins and 18 bacterial proteins as age-specifically glycated. Overall, our results suggest that the selective carbonylation or glycation of crucial proteins involved in nitrogen metabolism, transcriptional regulation, and signaling may constitute a mechanism to control cell metabolism and nodule senescence.
Legumes establish nitrogen-fixing symbioses with soil bacteria collectively known as rhizobia. The perception of rhizobial nodulation factors by the host plant induces a cascade of responses that leads to bacterial and root cell differentiation and nodule development (Oldroyd and Downie, 2008). Nodules provide optimal conditions for the expression of bacterial nitrogenase, whose activity contributes fixed nitrogen to ecosystems and croplands (Udvardi and Poole, 2013). These unique symbiotic organs show a high metabolic rate to meet the considerable energy demand of nitrogen fixation. As a result, they produce large amounts of reactive oxygen species (ROS) that need to be kept under control by antioxidant enzymes and metabolites (Becana et al., 2010).
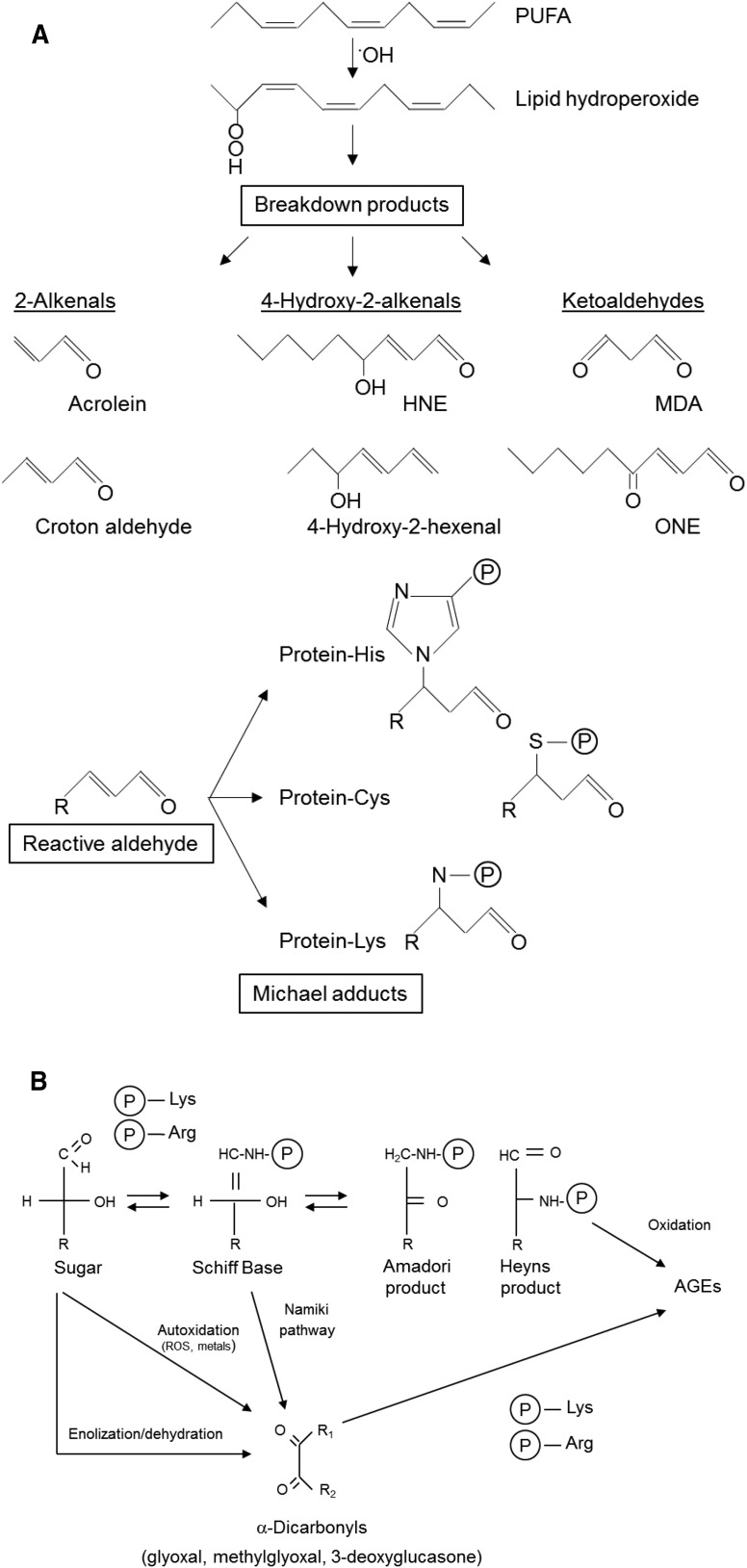
ROS may oxidize proteins reversibly (disulfide bonds, Met sulfoxides) or induce their irreversible modification (carbonylation, advanced glycation). Metal-catalyzed oxidation (MCO) occurs when free Fe2+ or Cu+ reacts with hydrogen peroxide (H2O2) and generates highly reactive hydroxyl radicals through the Fenton reaction (Halliwell, 2006). These radicals can oxidize almost any amino acid side chain, although some of them (Lys, Arg, Pro, and Thr) are particularly susceptible to carbonylation (Møller et al., 2011; Fedorova et al., 2014). Carbonyl groups also may be generated indirectly. In cell membranes, lipid peroxidation takes place when highly reactive ROS, mostly hydroxyl radicals and singlet oxygen, abstract a hydrogen atom from polyunsaturated fatty acids to form lipid hydroperoxides. These molecules are unstable and decompose to generate reactive aldehydes and ketones that form covalent Michael adducts with Arg, Cys, His, and Lys residues (Fig. 1A; Møller et al., 2011; Farmer and Mueller, 2013). Moreover, Arg and Lys residues can react with reducing sugars via the formation of an imine intermediate, generating Amadori and Heyns compounds (Hodge, 1955; Heyns and Noack, 1962). These glycation products are readily oxidized, yielding relatively stable advanced glycation end products (AGEs). Alternatively, AGEs can be formed by reaction with α-dicarbonyls. Glyoxal and methylglyoxal are generated by monosaccharide autooxidation or via the Namiki pathway (oxidation of imine intermediates), whereas 3-deoxyglucasone is a product of nonoxidative enolization and dehydration (Fig. 1B).
Figure 1.
Scheme showing the most common mechanisms of carbonylation and glycation of amino acid residues. A, Polyunsaturated fatty acid (PUFA) oxidation produces unstable lipid hydroperoxides that decompose to secondary products. The Michael addition of the reactive aldehydes to amino acid side chains (mostly His, Cys, and Lys) generates carbonyl derivatives. B, Formation of advanced glycation end products on Arg and Lys residues. HNE, 4-hydroxy-2-nonenal; MDA, malondialdehyde; ONE, 4-oxononenal; P, protein.
Protein carbonylation occurs in animal and plant tissues and contributes to cellular damage caused by stress conditions and age-associated diseases (Berlett and Stadtman, 1997; Höhn et al., 2013; Matamoros et al., 2013; Sun et al., 2014). Recent studies, however, suggest that irreversible protein oxidation might be a major event regulating protein biological function and fate (Lee and Helmann, 2006; Oracz et al., 2007; Winger et al., 2007). The carbonylation of nodule proteins is likely to be important because nodules contain high amounts of Fe proteins, such as nitrogenase, cytochromes, and leghemoglobin (Lb), which are prone to oxidation (Becana et al., 2000).
Protein glycation is well characterized in mammals under several physiological conditions (Spiller et al., 2017). In humans (Homo sapiens), the formation of AGEs accompanies atherosclerosis and diabetes (Höhn et al., 2013). These modifications target mostly long-living proteins, like human lens proteins (Smuda et al., 2015) and collagen (Verzijl et al., 2000), but also plasma proteins (Greifenhagen et al., 2016). Interestingly, the analysis of protein hydrolysates revealed high levels of glycation in plants (Bechtold et al., 2009). This result was confirmed at the proteome level (Bilova et al., 2016), especially under abiotic stress conditions (Paudel et al., 2016). Nevertheless, the investigation of protein carbonylation and advanced glycation in plants is still in its infancy. The identification of oxidized amino acid residues in proteins may provide essential information about the oxidative mechanisms and metabolic pathways involved in the loss of cellular viability that occurs during aging and under stress conditions (Fedorova et al., 2014). Thus, our aim was to gain insight into these processes in vivo using nodulated plants of bean (Phaseolus vulgaris), a major grain legume for direct human consumption (Cavalieri et al., 2011). This legume species also was selected because it was used previously to obtain information about the oxidative stress underlying nodule senescence (Matamoros et al., 2013). Here, we determined the carbonylated and glycated protein profiles of nodules and identified the sites and types of modifications. As a case study, we examined the effects of carbonylation on two key proteins in nodule metabolism, malate dehydrogenase (MDH) and Lb, and provide a detailed quantitative analysis of protein glycation patterns during nodule aging.
RESULTS AND DISCUSSION
Carbonylation Types and Sites in the Nodule Plant Proteome
ROS may regulate cell metabolism by the oxidation of key enzymes and transcription factors. Thus, the sulfur-containing amino acids Cys and Met may be oxidized reversibly to form disulfide bridges and Met sulfoxide, respectively, as part of redox regulatory mechanisms shared by numerous metabolic processes (Tarrago et al., 2009; Couturier et al., 2013). In contrast, higher oxidation states of certain amino acid side chains, including carbonylation, are irreversible and may lead to alterations of protein conformation, inhibition of protein function, and increased susceptibility to degradation (Møller et al., 2011).
Direct oxidation of some amino acids residues (e.g. Lys, Arg, Thr, and Pro) and Michael addition of reactive aldehydes or ketones to Lys, Cys, His, and Arg introduce the carbonyl moiety in proteins. Supplemental Figure S1 shows an overview of the experimental workflow used in this study to identify the carbonylated proteins and the exact modification sites. To avoid artifactual carbonylation during protein extraction, we immediately ground frozen nodules in a buffer containing a mild reducing agent and a metal ion chelator. We digested nodule proteins with trypsin and labeled modified peptides with a carbonyl-specific aldehyde-reactive probe (ARP). Due to the low abundance of modified peptides, the derivatization step was necessary because it facilitated the enrichment of carbonylated peptides by biotin-avidin affinity chromatography and provided specific mass increments in the corresponding spectra, thus significantly improving the identification rates by liquid chromatography-tandem mass spectrometry (LC-MS/MS; Bollineni et al., 2013). This methodology, combined with search engine-assisted protein identification using the mass increments of the different posttranslational modifications (PTMs) as variables (Bollineni et al., 2014a; Griesser et al., 2017), allowed the identification of 238 carbonylated plant proteins. We only considered rank 1 peptides identified with high/medium confidence in at least five biological replicates. To ensure the reliable identification of modified sequences, we randomly selected and manually verified ∼20% of the sequences. Supplemental Figure S2 shows the tandem mass spectra of ARP-labeled peptides containing the different types of carbonyl products analyzed in this study. In each case, the presence of carbonylated residues was verified by the consistent m/z shift of the b and y ions with respect to the theoretical values that would be expected for the unmodified peptides.
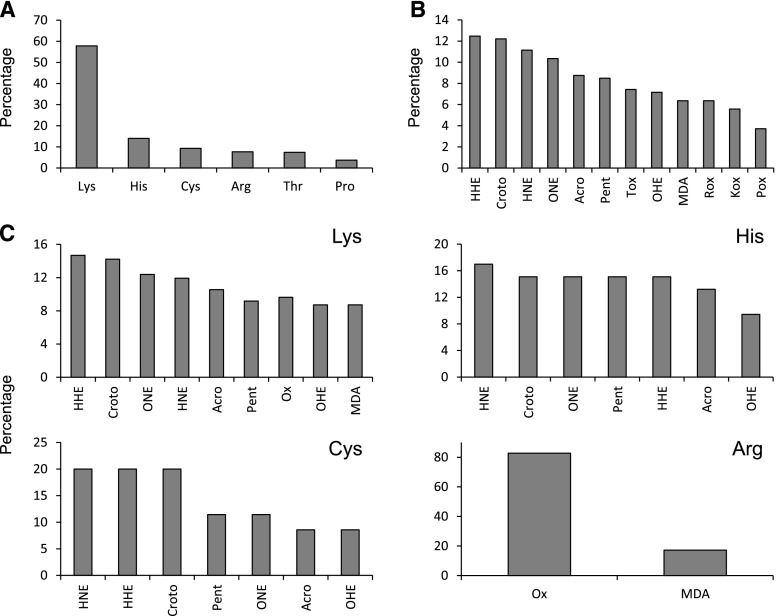
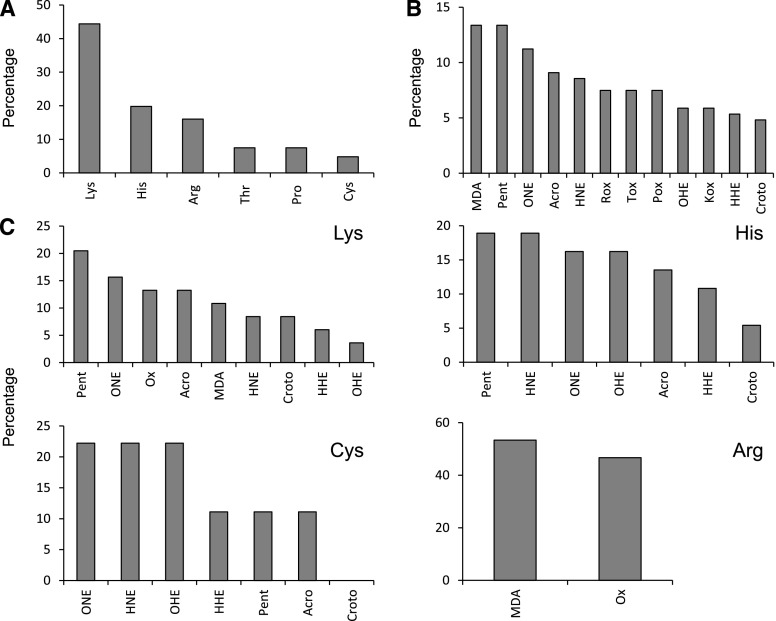
We identified a total of 12 types of carbonylations affecting six amino acid residues (Supplemental Table S1). The most commonly carbonylated amino acid was Lys, which accounted for 58% of all modifications and was present in 66% of the identified carbonylated peptides, followed by His, Cys, Arg, Thr, and Pro, each representing 5% to 15% of the total carbonylated amino acid residues (Fig. 2A). This observation is consistent with the proteomic analyses of yeast and human cells (Møller et al., 2011; Bollineni et al., 2014b; Rao et al., 2018). The reason for the high level of carbonylated Lys is unclear. Tekaia et al. (2002) calculated the amino acid compositions of several organisms. In the Arabidopsis (Arabidopsis thaliana) proteome, the proportion of Lys (6.4%) is similar to that of Arg (5.5%), Thr (5.1%), and Pro (4.7%) and greater than that of His (2.3%) and Cys (1.9%). However, the elevated number of carbonylated Lys residues cannot be explained exclusively by a high abundance of this amino acid in plant proteomes. If the relative abundance of each amino acid is considered, Lys is still the most susceptible amino acid, followed by His and Cys (Supplemental Fig. S3). The number of modification types on Lys residues (nine) also could partially explain its high level of carbonylation. However, His and Cys also can be modified in seven different ways (Supplemental Table S1) and had much lower levels of carbonylation than Lys (Fig. 2A). On the other hand, when only carbonylations via direct oxidation of amino acids were taken into account, Thr was the most susceptible amino acid (25%), followed by Arg (∼20%), Lys (∼20%), and Pro (13%). Similarly, it was reported recently that the side chains of Thr, Lys, and Arg are oxidized predominantly to carbonyl groups in the potato (Solanum tuberosum) tuber mitochondrial proteome (Salvato et al., 2014).
Figure 2.
Carbonylation types and sites in the nodule plant proteome. A, Relative abundance (%) of carbonylated amino acid residues. B, Total modifications. C, Modifications found in Lys, His, Cys, and Arg. The percentages were calculated based on the analysis of 238 proteins. In A, the percentage denotes the proportion of each carbonylated residue with respect to the total number of carbonylated residues (377). In B and C, the percentages denote, respectively, the proportion of each modification with respect to the total number of modifications found in 377 sites and the proportion of each modification on individual residues. Acro, Acrolein Michael addition (MA); Croto, crotonaldehyde MA; HHE, 4-hydroxy-2-hexenal MA; HNE, 4-hydroxy-2-nonenal MA; Kox, Lys oxidation to aminoadipic semialdehyde; MDA, malondialdehyde MA; OHE, 4-oxo-2-hexenal MA; ONE, 4-oxo-2-nonenal MA; Pent, pentenal MA; Pox, Pro oxidation to glutamic semialdehyde; Rox, Arg oxidation to glutamic semialdehyde; Tox, Thr oxidation to 2-amino-3-ketobutyric acid.
Carbonylation sites can be formed by multiple mechanisms that generate various modifications (Møller et al., 2007). Among them, those generated by the formation of adducts with reactive aldehydes derived from the oxidation of polyunsaturated fatty acids were clearly predominant, accounting for 77% of all the modifications. 4-Hydroxy-2-hexenal and crotonaldehyde were the most common types, accounting for ∼12% of the modifications (Fig. 2, B and C). His and Cys also were modified frequently by the addition of 4-hydroxy-2-nonenal, whereas the major carbonylation type affecting Arg was its direct oxidation to glutamic semialdehyde (Fig. 2C). Endogenous oxidized lipids are produced continuously during cell metabolism (Møller et al., 2007). This may be especially true for nodule host cells, which contain large amounts of oxidation-prone symbiosomal membranes (Puppo et al., 1991). Most studies of lipid peroxidation in plants and animals have focused on the toxic effects of lipid-derived carbonyl compounds (Yin et al., 2010). However, the participation of nonenzymatic lipid peroxidation products in signaling mechanisms is increasingly acknowledged (Farmer and Mueller, 2013). Thus, malondialdehyde (MDA) and acrolein induce the expression of stress genes (Vollenweider et al., 2000) and regulate the cell cycle (Farmer and Mueller, 2013). In this regard, the significance of protein carbonylation mediated by lipid peroxidation products as a regulatory mechanism of protein activity in legume nodules merits further research. These compounds are quite stable and could be involved in signaling cascades related to the stress response downstream of ROS. The extent of carbonylation induced by lipid peroxidation products could be regulated by the activities of four carbonyl-scavenging enzymes: aldehyde dehydrogenases, aldo-keto reductases, 2-alkenal reductases, and glutathione transferases. The identification of carbonylated proteins reported below is a necessary first step to gain important information about putative signaling cascades regulated by ROS and lipid peroxidation products.
Identification of Plant Carbonylated Proteins and String Analysis
In this study, we identified 238 oxidized plant proteins containing 377 carbonylation sites in young, mature, and senescent nodules (Supplemental Data Set S1). The initial objective was to identify proteins that undergo carbonylation during the aging process. Most proteins were detected as carbonylated throughout nodule development, and only a few were detected uniquely at a specific stage. It is important to bear in mind that the technique utilized here allowed the identification of oxidized peptides but not their relative quantification. However, a general increase in protein carbonylation levels was reported for senescent nodules (Evans et al., 1999; Matamoros et al., 2013).
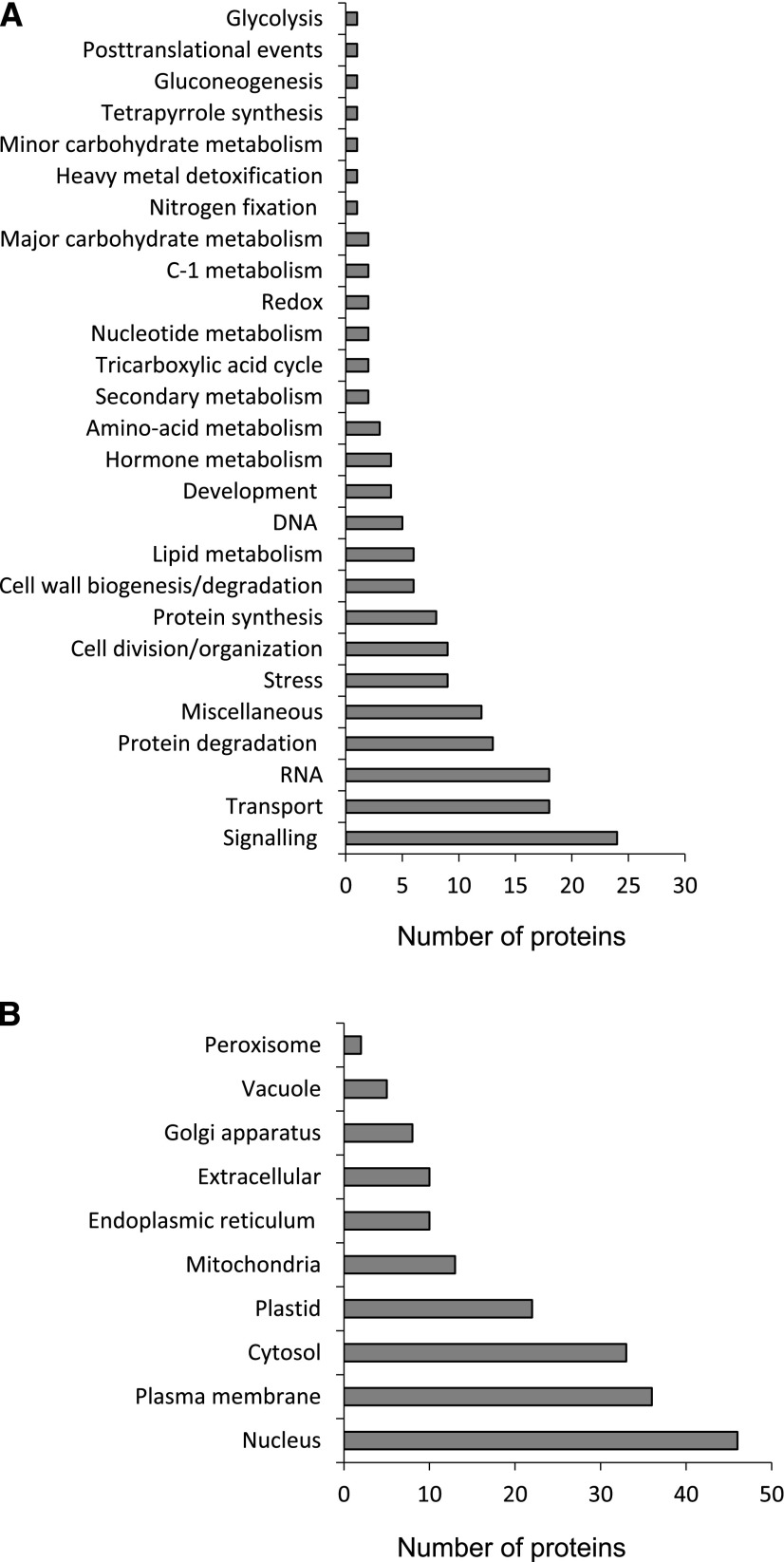
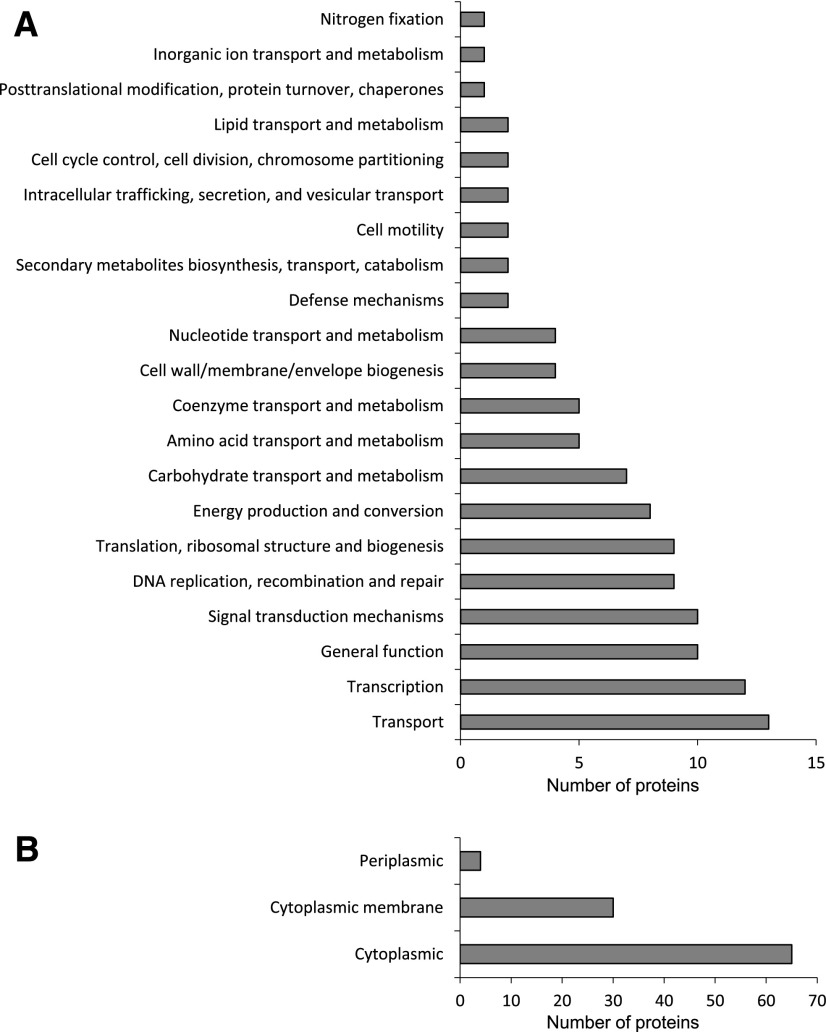
The proteins were classified into 27 functional categories (Fig. 3A) using MapMan (Thimm et al., 2004). A total of 80 proteins could not be assigned to any category because they have unknown functions (21) or are uncharacterized (59). Interestingly, the three categories with a greater number of carbonylated proteins were signaling, transport, and RNA, each of them containing 10% to 15% of all identified proteins. This value is considerably higher than the percentages of proteins belonging to the same categories in the Medicago truncatula nodule proteome: ∼2% for transport and RNA and 7% for signaling, including hormone metabolism (Larrainzar et al., 2007). Our data suggest that the carbonylation of nodule proteins may have some specificity, which is supported by previous work by Winger et al. (2007), who reported some selectivity in the 4-hydroxy-2-nonenal reaction in the mitochondrial proteome, and by Oracz et al. (2007), who showed that the dormancy release is associated with the carbonylation of specific embryo proteins. The implications would be that selective protein oxidation might represent a mechanism by which cell metabolism is regulated and that different modifications would account for specific changes in protein function. In fact, Møller and Sweetlove (2010) proposed that oxidatively modified peptides may act as selective secondary ROS messengers and, thereby, contribute to retrograde signaling.
Figure 3.
Functional classification and predicted subcellular localization of plant carbonylated proteins. A, Number of plant carbonylated proteins in different functional categories. Bean nodule proteins were BLASTed against the Arabidopsis proteome. The closest homologs were classified according to MapMan (http://www.gabipd.de/projects/MapMan). B, Predicted subcellular localization of carbonylated proteins. The subcellular localization of the closest homologs was determined according to the SUBA database (Hooper et al., 2017).
The high relative amount of transcription factors and proteins related to gene expression identified in this study is noteworthy and suggests a putative regulatory role of protein carbonylation on gene expression. In a pioneering study, Lee and Helmann (2006) showed that selective protein oxidation provides a regulatory mechanism for peroxide sensing in Bacillus subtilis. The mechanism involves the transcription factor PerR, which controls peroxide-defense genes in response to H2O2. PerR is highly sensitive to MCO driven by bound Fe2+ and H2O2. Oxidized PerR is more susceptible to degradation, triggering the derepression of the PerR regulon. In addition to transcription factors, we identified 16 receptor and protein kinases as highly susceptible to carbonylation (Supplemental Data Set S1). Other categories abundantly represented are protein synthesis and degradation, stress responses (mostly disease resistance proteins), and cell division/organization. Noteworthy, several pentatricopeptide repeat proteins were carbonylated. Although classified within unknown function by MapMan, the pentatricopeptide repeat protein family, which is particularly prevalent in plants, includes many sequence-specific RNA-binding proteins that seem to be involved in all aspects of organelle RNA metabolism, including RNA stability, processing, editing, and translation (Barkan and Small, 2014). Despite differences in the experimental setup and the types of modifications analyzed, numerous carbonylated pentatricopeptide repeat-containing proteins also were detected in the potato mitochondrial proteome (Salvato et al., 2014). Other proteins found as carbonylated in both studies include DNA polymerases, one ATP-dependent RNA helicase, several ribosomal proteins, Ser hydroxymethyltransferase, and succinate dehydrogenase.
To investigate whether carbonylation preferentially affects certain metabolic and/or signaling pathways, we used STRING, a database of physical and functional protein interactions (Szklarczyk et al., 2015). The closest homologs to bean proteins were identified in Arabidopsis and used for the STRING analysis (Supplemental Data Set S1). Two central proteins form major interaction nodes: At5g25930, a receptor-like Leu-rich repeat protein kinase that interacts with several other kinases and three ATP-binding cassette (ABC) transporters; and ABSCISIC ACID-INSENSITIVE1 (At4g26080), a Ser/Thr phosphatase involved in abscisic acid signal transduction that also interacts with several other kinases (Supplemental Fig. S4).
Overall, our results suggest that selective protein carbonylation might constitute a mechanism that regulates gene expression and signaling cascades (e.g. some kinase pathways) in nodule cells.
Nuclear Proteins Are Important Targets of Carbonylation
In this study, we found that many plant proteins involved in the regulation of gene expression and DNA/RNA synthesis and processing were carbonylated. Accordingly, a high proportion of the oxidized proteins were predicted to be nuclear (Fig. 3B). Although the generation of ROS, in particular H2O2, may occur in plant cell nuclei (Ashtamker et al., 2007), the main source of carbonylation of nuclear proteins was reactive aldehydes produced during lipid peroxidation. Unlike ROS, these secondary oxidation products can easily diffuse across membranes and covalently modify nuclear proteins (Farmer and Mueller, 2013). Recently, this and other laboratories have reported the presence in the nucleus of glutathione along with glutathione peroxidase and thioredoxin isoforms, supporting the importance of thiol redox signaling in this cellular compartment (Martí et al., 2009; Vivancos et al., 2010; Gaber et al., 2012; Matamoros et al., 2013, 2015). The occurrence of a variety of carbonylated proteins adds an extra layer of complexity to the redox-dependent regulation in the nucleus.
A high proportion of oxidized proteins was localized in the plasma membrane (Fig. 3B), consistent with the abundance of membranes in nodule infected cells (Puppo et al., 1991). There also were moderate levels of protein oxidation in the cytosol and plastids. Noteworthy, only 13 of the oxidized proteins found in this study were predicted to localize to the mitochondria. This is surprising and, at first sight, seems to be at odds with the general observation that mitochondria are a major source of ROS within the cell (Bartoli et al., 2004; Rhoads et al., 2006; Møller et al., 2007). It should be noted, however, that our results do not provide information on the relative amount of oxidized proteins in each subcellular compartment. Consequently, a different experimental approach should be used to investigate the relative contribution of mitochondria to protein carbonylation in nodule cells. In this context, we have shown that the mitochondria isolated from mature and especially from senescent bean nodules contain significantly higher levels of lipid peroxides and carbonylated proteins than the mitochondria from young nodules (Matamoros et al., 2013).
Carbonylation Inhibits MDH Activity and May Facilitate Lb Cross-Linking
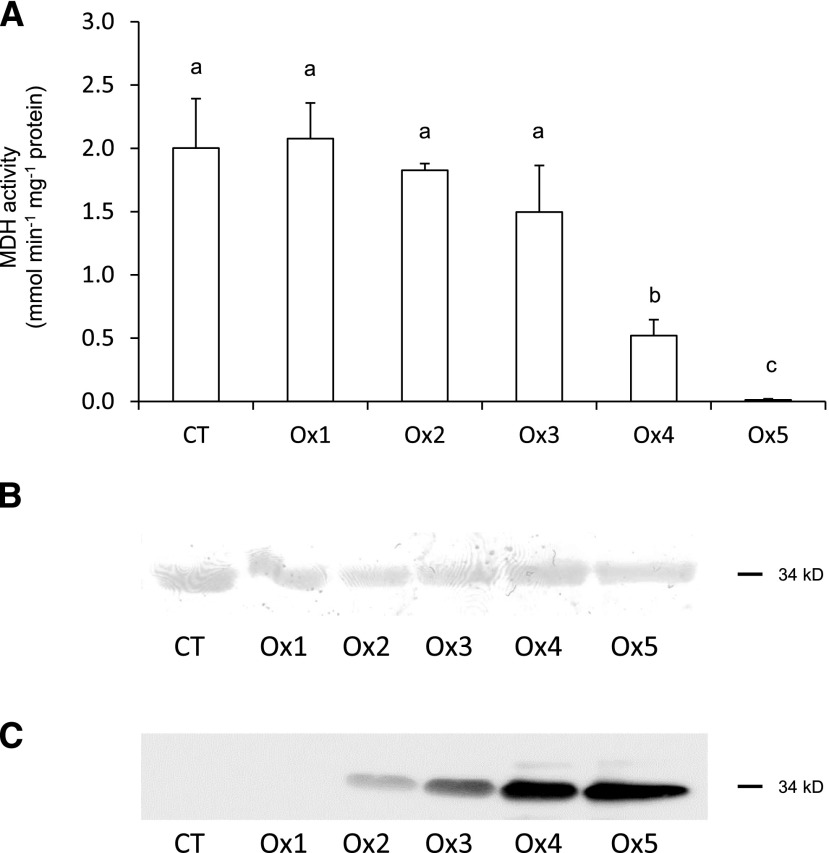
MCO is a chief mechanism for protein oxidation in living organisms (Berlett and Stadtman, 1997; Møller et al., 2011), but little information is available about the effect of specific oxidative modifications on protein structure and function. We have investigated this issue by assessing the in vitro effect of MCO on two key nodule proteins, MDH and Lb. The enzyme MDH catalyzes the reversible oxidation of malate to oxaloacetate and is essential for nitrogen fixation because malate is the primary source of carbon transported to bacteroids (Udvardi and Poole, 2013). We purified bean MDH in recombinant form and incubated the enzyme for 3 to 16 h at 37°C with an MCO system consisting of ascorbate and Fe3+ at various concentrations. Because the incubation times had little effect on the results, we only show the data after 6 h of MCO reaction for simplicity (Fig. 4A). Concentrations up to 100 μm FeCl3 did not significantly affect enzymatic activity, but 200 μm caused 75% inhibition and 300 μm suppressed the activity completely. To ascertain that the activity loss was not caused by protein degradation or by the oxidation of critical Met or Cys residues, we conducted two experiments. First, we exposed recombinant MDH to MCO for 6 h and analyzed the product by SDS-PAGE (Fig. 4B). The incubation with FeCl3 and ascorbate did not cause significant protein degradation or cross-linking. Second, we treated the protein with different concentrations of H2O2. Treatment with H2O2 alone at concentrations up to 10 mm did not inhibit enzyme activity significantly (data not shown), suggesting that protein carbonylation may be responsible for the decrease in activity. This is consistent with the lack of critical residues in bean NAD+-dependent MDHs, which are required for redox regulation and are present in homologous enzymes, such as in the cytosolic and plastid NADP+-dependent MDHs of Arabidopsis (Yoshida and Hisabori, 2016). Moreover, using the anti-dinitrophenylhydrazone antibody, we found that MDH carbonylation was negatively correlated with enzyme activity. Concentrations up to 50 μm FeCl3 had no effect, but higher concentrations increased carbonylation greatly (Fig. 4C). The amino acid residues found to be carbonylated in vivo were Thr-190, Arg-196, and Thr-199 (Supplemental Fig. S5; Supplemental Data Set S1). Of these, Arg-196 is absolutely conserved in all MDHs because its guanidinium group participates in the binding and orientation of the substrate at the active site (Goward and Nicholls, 1994). Therefore, the oxidation of the guanidinium group to glutamic semialdehyde observed in vivo could impair MDH function. However, we cannot entirely rule out that other side chain oxidations not detected in this study contribute to the observed decline in MDH activity. MCO also induced the inhibition of MDH activity in peroxisomal extracts (Nguyen and Donaldson, 2005), although to a lesser extent than in our study.
Figure 4.
Effects of MCO on the activity, protein, and carbonylation of MDH. A, MDH activity after 6 h of MCO at different FeCl3 and ascorbate (ASC) concentrations. CT, Control omitting ASC and FeCl3; Ox1, 2.5 mm ASC, 10 μm FeCl3; Ox2, 12.5 mm ASC, 50 μm FeCl3; Ox3, 25 mm ASC, 100 μm FeCl3; Ox4, 50 mm ASC, 200 μm FeCl3; Ox5, 75 mm ASC, 300 μm FeCl3. Values are means ± se of three replicates. Means denoted by the same letter are not significantly different at P < 0.05 based on Duncan’s multiple range test. B, Coomassie Blue staining of purified MDH after 6 h of MCO at different FeCl3 and ASC concentrations. Each lane was loaded with 5 μg of protein. C, Effects of MCO on MDH carbonylation. The recombinant protein was subjected to MCO, derivatized (4 μg) with 2,4-dinitrophenylhydrazine, and subjected to SDS-PAGE. Immunoblotting was performed using an anti-dinitrophenylhydrazone antibody. B and C show representative gels and blots, respectively, from three independent experiments.
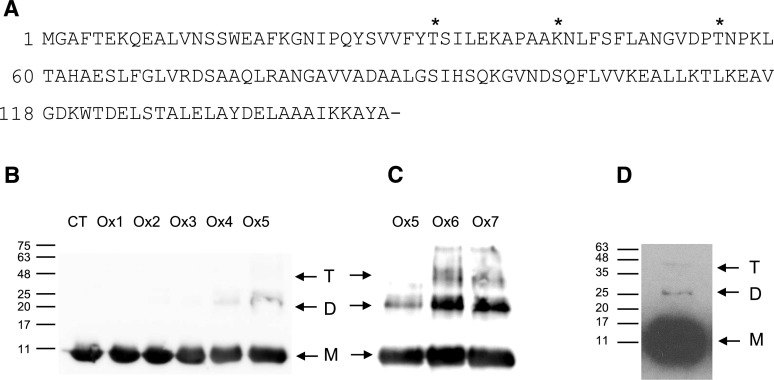
Lb is another critical protein for nodule functioning because it transports oxygen at a low but steady concentration to the symbiosomes, avoiding the irreversible inactivation of nitrogenase (Appleby, 1984; Ott et al., 2005). In addition, Lb may regulate nitric oxide levels within the nodules (Herold and Puppo, 2005; Sánchez et al., 2010; Calvo-Begueria et al., 2018). Developmental and stress-induced nodule senescence are accompanied by decreases in nitrogen fixation and increases in proteolytic activity and ROS (Evans et al., 1999; Groten et al., 2006; Loscos et al., 2008). In turn, Lb levels decline significantly during nodule senescence, although the mechanisms are still poorly known. Soybean (Glycine max) Lbs can be modified by reactive nitrogen species producing in vivo derivatives bearing nitrated hemes (Navascués et al., 2012) and nitro-Tyr residues (Sainz et al., 2015). Also, Lbs undergo cross-linking and dimerization after incubation with H2O2 in vitro, but di-Tyr formation seems not to be involved (Moreau et al., 1995). It also would be plausible that the carbonylation of certain amino acid residues contributes to dimerization via Schiff base formation with Lys residues on another Lb molecule, as observed for other proteins (Madian and Regnier, 2010). We have detected three amino acid modifications in bean nodule Lb: Thr-32 and Thr-55 to 2-amino-3-ketobutyric acid and Lys-42 to aminoadipic semialdehyde (Fig. 5A; Supplemental Data Set S1; Supplemental Fig. S2).
Figure 5.
Carbonylation of bean nodule Lb. A, Amino acid sequence of Lb in which the carbonylated residues found in vivo are marked with asterisks. B, Immunoblot of purified Lb after 6 h of MCO at different FeCl3 and ascorbate (ASC) concentrations. CT, Control omitting ASC and FeCl3; Ox1, 2.5 mm ASC, 10 μm FeCl3; Ox2, 12.5 mm ASC, 50 μm FeCl3; Ox3, 25 mm ASC, 100 μm FeCl3; Ox4, 50 mm ASC, 200 μm FeCl3; Ox5, 75 mm ASC, 300 μm FeCl3. C, Immunoblot of purified Lb after 6 h of MCO at different FeCl3, ASC, and H2O2 concentrations. Ox5, 75 mm ASC, 300 μm FeCl3; Ox6, 75 mm ASC, 300 μm FeCl3, 1 mm H2O2; Ox7, 75 mm ASC, 300 μm FeCl3, 5 mm H2O2. Gels were loaded with 10 μg of protein per lane. D, Immunoblot of bean nodule extracts. Gels were loaded with 50 μg of protein per lane. For B to D, the apparent molecular masses (kD) of the monomer (M), dimer (D), and tetramer (T) are indicated. The anti-Lb antibody and the secondary antibody were used at dilutions of 1:1,000 and 1:40,000, respectively. Blots are representative of at least three independent experiments.
To further investigate the effects of carbonylation on Lb, we subjected the purified protein to MCO in vitro and studied its changes by SDS-PAGE and immunoblot analyses using a specific Lb antibody (Fig. 5B). The MCO reaction caused aggregation, which was dependent on the concentration of Fe3+ and ascorbate. In samples incubated without Fe3+ and ascorbate (controls) or with up to 100 μm Fe3+ and 25 mm ascorbate, a single band of ∼11 kD was detected. The same results were obtained when Lb was incubated with FeCl3 or ascorbate separately (data not shown). The observed molecular mass was lower than the theoretical value (15.6 kD), consistent with the anomalous migration of Lb by SDS-PAGE reported by Lehtovaara (1978). At higher Fe3+ concentrations (200 μm), a weak band of ∼22 kD, probably an Lb dimer, was observed. The intensity of this band increased at the highest Fe3+ concentration (300 μm), suggesting that the oxidation of certain amino acids by MCO brings about protein aggregation. Dimer formation increased further when H2O2 was included in the reaction mixture with Fe3+ and ascorbate (Fig. 5C). Surprisingly, unlike MDH, we could not detect carbonylated Lb for any treatment (data not shown). This result suggests either that the amount of carbonylated Lb is extremely low or that the epitopes for the antibody recognition are no longer available due to dimerization caused by MCO. Furthermore, the existence of dimers and possibly tetramers in nodule extracts not treated with Fe3+ and ascorbate (Fig. 5D), which are similar to those induced by MCO, can be explained by this mechanism being responsible for Lb oxidation and oligomerization in vivo. Additional studies are required to unambiguously identify the mechanism and the amino acid residues implicated in the formation of Lb oligomers.
Protein Carbonylation in Bacteroids
The main objective of our study was to characterize the oxidation profiles of nodule plant proteins. However, nodule extracts also contained rhizobial proteins due to bacteroid disruption during nodule grinding. To our knowledge, there exists no information in the literature about protein oxidation in nodule bacteroids. To obtain a deeper insight into the role of redox-sensitive irreversible PTMs in nodule physiology, we included in our study the characterization of oxidized proteins from the bacterial symbiont.
Regarding the susceptibility of the amino acid residues to carbonylation, the results were similar to those obtained with plant proteins and other organisms (Møller et al., 2011; Bollineni et al., 2014b; this work). The amino acid most frequently carbonylated was Lys (44% of all modifications), followed by His and Arg (15%–20%), and Thr, Pro, and Cys (less than 10%; Fig. 6A). Likewise, Michael adducts with reactive aldehydes arising from the oxidation of polyunsaturated fatty acids in cell membranes accounted for ∼70% of all the modifications. The most common modifications (10%–15%) of bacterial proteins were caused by MDA, pentenal, and 4-oxononenal (Fig. 6B). This differs from the results observed in plant proteins, where 4-hydroxy-2-hexenal and crotonaldehyde-derived Michael additions were the most common types (Fig. 2B). This could reflect different compositions of polyunsaturated fatty acids in plant and bacterial cell membranes. Remarkably, Arg showed contrasting patterns of oxidation in plant and bacterial proteins (Figs. 2C and 6C). In plant proteins, direct oxidation of Arg to glutamic semialdehyde accounted for 80% of this residue carbonylation, whereas, in the case of bacterial proteins, Arg was carbonylated mostly through Michael addition to MDA.
Figure 6.
Carbonylation types and sites in the nodule bacteroid proteome. A, Relative abundance (%) of carbonylated amino acid residues. B, Total modifications. C, Modifications found in Lys, His, Cys, and Arg. The percentages were calculated based on the analysis of 131 proteins. Abbreviations as in Figure 2.
We identified a total of 134 oxidized peptides and 187 carbonylation sites, corresponding to 131 bacterial proteins from bean nodules (Supplemental Data Set S2). The proteins were classified into 21 functional categories (Fig. 7A) according to the information deposited in the COG of proteins database (Galperin et al., 2015). A total of 20 proteins could not be assigned to any category because they have unknown functions (six) or are uncharacterized (14). The categories with more carbonylated proteins were very similar to those described for plant proteins. Thus, transport, transcription, general function, and signaling categories each included 9% to 12% of all identified proteins. This supports the notion that carbonylation targets specific sets of proteins. Some of the carbonylated proteins in rhizobia have important functions in the symbiosis. For example, one of the transcription factors, FixK, is involved in the activation of the operons required for bacteroid respiration under the microaerobic conditions of nodules and, therefore, is essential for nitrogen fixation (Foussard et al., 1997). The nodulation protein NodU is involved in the synthesis of nodulation factors required for the recognition of plant and bacteria at the early stages of symbiosis (Jabbouri et al., 1995; Oldroyd and Downie, 2008). The predicted subcellular localization of rhizobial proteins was 65% in the cytoplasm, 30% in the plasma membrane, and 5% in the periplasm (Fig. 7B).
Figure 7.
Functional classification and predicted subcellular localization of bacteroid carbonylated proteins. A, Number of bacteroid carbonylated proteins in different functional categories. The proteins were classified according to the Clusters of Orthologous Groups (COG). B, Predicted subcellular localization of carbonylated proteins according to PSORTb version 3.0 (Yu et al., 2010).
Characterization of the Plant Glycated Proteome of Nodules
Reactive aldehydes arising from lipid peroxidation represent the major cause of protein carbonylation. Similarly, the generation of most known AGEs requires oxidative conditions (Fig. 1B; Møller et al., 2011; Höhn et al., 2013). In animal models, the AGE content increases in diverse age-related pathologies (Höhn et al., 2013). However, the current knowledge of protein glycation in plants is limited. This phenomenon was first addressed in the plant kingdom only in the late 2000s (Bechtold et al., 2009), and the AGE-modified proteomes of Arabidopsis and Brassica napus were reported only recently (Bilova et al., 2016).
To characterize the glycated proteome of bean nodules, we used a phenol extraction procedure to isolate the total protein fraction (Isaacson et al., 2006). Because the formation of AGEs in vitro is favored by high temperature and high sugar concentration (Ott et al., 2014), frozen nodules were processed immediately. This approach provided an excellent recovery of even strongly hydrophobic membrane proteins. To reconstitute the dried protein pellets, we employed Progenta Anionic Acid-Labile Surfactant (AALS II), a degradable detergent compatible with LC-MS analysis (Frolov et al., 2017b). Supplementation of this reagent to the shotgun buffer resulted in complete resolubilization of the protein pellets. The protein yields were ∼0.1 to 0.5 mg g−1 fresh weight (Supplemental Table S2), which were adequate for reliable analysis. We separated proteins (5 µg) by SDS-PAGE and stained them with Coomassie Brilliant Blue G250 (Supplemental Fig. S6). The subsequent densitometry analysis indicated reproducible whole-lane average intensities (relative sd = 16.2%). Moreover, the patterns were similar between lanes and age groups (Supplemental Fig. S6). The efficiency of the tryptic digestion was demonstrated by the absence of signals in the corresponding SDS-PAGE analyses (Supplemental Fig. S7). Next, we selected 10 AGEs identified previously in plant proteomes (Bilova et al., 2016) and performed a total of 10 database searches per age group (for settings, see Supplemental Table S3), followed by a multiconsensus analysis. This permitted an efficient reduction of false positive rates, and ∼46% of the hits could be discriminated in this step (data not shown). Consequently, only the most reliable annotations are shown in Supplemental Data Set S3. The accuracy of the identifications was assessed by manual annotation of approximately one-third of the AGE-modified amino acids, based on the characteristic series of b and y ions observed in the MS/MS spectra (Supplemental Figs. S8 and S9).
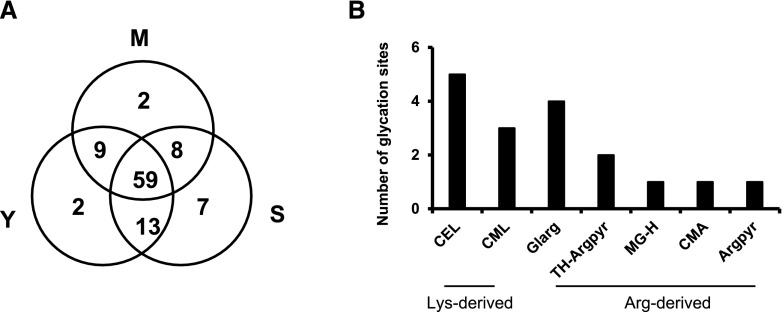
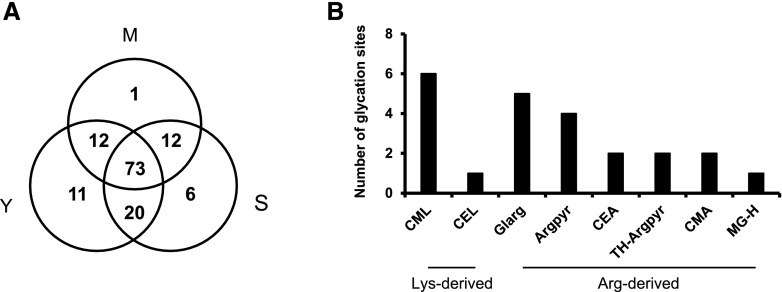
The analysis of the plant glycated proteome from young, mature, and senescent nodules revealed the presence of 100 glycation sites, represented by 80 peptides belonging to 69 proteins (Supplemental Data Set S3). In contrast to previous observations in the Arabidopsis leaf proteome (Bilova et al., 2016), modification sites on Lys and Arg residues were distributed equally in the nodule plant proteome. Thus, Arg-derived AGEs accounted for 52% of the modifications while 48% corresponded to Lys. Nε-(carboxymethyl)lysine (CML), Nε-(carboxyethyl)lysine (CEL), and glyoxal-derived hydroimidazolone (Glarg) were the most common AGEs, accounting for 28%, 19%, and 15% of the modifications, respectively (Supplemental Fig. S10; Supplemental Data Set S3).
The glycated plant proteins were classified into 21 functional categories according to MapMan (Thimm et al., 2004). A total of 15 proteins were uncharacterized and five proteins could not be assigned to any category. The category with the greatest number of glycated proteins was stress response, followed by RNA, protein synthesis, and signaling (Supplemental Fig. S11). This is similar to the results obtained with carbonylated proteins (Fig. 3A). Thus, the selective carbonylation or glycation of key proteins involved in transcriptional regulation, stress response, and signaling might constitute a mechanism to control cell metabolism in response to redox changes. In this study, we identified three prominent glycated proteins required for nitrogen fixation and ammonia assimilation: Suc synthase, Gln synthetase, and Glu synthase. Suc derived from photosynthesis is cleaved by Suc synthase and oxidized further to malate, subsequently transported into the bacteroids, and used as an energy source to sustain nitrogen fixation. The resulting ammonia is released into the cytosol of nodule cells and assimilated into carbon compounds by Gln synthetase and Glu synthase (Udvardi and Poole, 2013). The posttranslational regulation of Suc synthase by redox state (Marino et al., 2008), S-thiolation (Röhrig et al., 2004), and phosphorylation (Duncan and Huber, 2007) has been reported. Likewise, Gln synthetase activity has been shown to be regulated by Tyr nitration (Melo et al., 2011), and both Gln synthetase and Glu synthase are putative targets of S-nitrosylation in Arabidopsis (Hu et al., 2015). Here, we show that the glycation of these three enzymes might constitute an additional mechanism for the regulation of carbon and nitrogen metabolism in legume nodules. Additionally, two proteins involved in the regulation of redox homeostasis in nodule cells, catalase and peptide Met sulfoxide reductase, were found to be glycated in vivo (Supplemental Data Set S3).
The overlap between carbonylated and glycated proteins seems to be quite low. Only four proteins (disease resistance protein [UniProtKB no. V7CU55], nucleotide-binding site Leu-rich repeat defense-type protein [B8R509], cytochrome P450 [V7D0Z1], and phosphatidylinositol/phosphatidylcholine transfer protein [V7ATL9]) were identified as both glycated and carbonylated. This indicates that these two PTMs are selective processes and independently target specific sets of proteins.
Age-Dependent Changes of Glycation in the Nodule Plant Proteome
To our knowledge, there is only one study about the effect of stress conditions on protein glycation in plants, and it shows that osmotic stress causes an increase in protein glycation in Arabidopsis (Paudel et al., 2016). Subsequently, Bilova et al. (2017) reported an age-dependent increase of glycation at specific sites of the Arabidopsis leaf proteome. This points to the existence of glycation hotspots in plants, as shown earlier for humans (Thornalley and Rabbani, 2009). During nodule aging, there is an increase in the concentration of transition metals and ROS production (Becana et al., 2010), which is correlated positively to the rate of glycation (Bilova et al., 2016). This prompted us to investigate the involvement of AGE-related PTMs in nodule senescence using a quantitative approach.
Because data-dependent acquisition (DDA) experiments are prone to undersampling (incomplete coverage of the proteome by MS/MS due to the limitations of the instrument duty cycle; Soboleva et al., 2017), all positive hits, identified in individual age groups, were cross-annotated in other groups. This revealed that the majority of the glycated residues (59) were identified in peptides present in young, mature, and senescent nodules (Fig. 8A). Interestingly, 17 glycated residues were detected in peptides unique to mature and/or senescent nodules, suggesting that those modifications are associated with nodule aging.
Figure 8.
Relative abundance of AGE-modified sites in plant proteins from nodules at the three developmental stages. A, Total number of AGE-modified sites identified in plant proteins from young (Y), mature (M), and senescent (S) nodules. B, Number of specific AGE classes identified in glycated peptides differentially abundant in mature and senescent nodules relative to young nodules. Abbreviations as in Table 1.
Label-free quantification of the 100 glycated sites revealed that 11 glycated peptides are differentially abundant in mature and senescent nodules with respect to young nodules (Table 1; Fig. 9; Supplemental Fig. S12). These peptides contained age-dependent glycated sites representing seven individual AGE classes, CEL and Glarg being the most common modifications (Table 1; Fig. 8B). The increase in the amount of glycated peptides can be explained by an age-dependent increment of protein glycation. Alternatively, it could reflect changes in the abundance of specific proteins during nodule development. To clarify this, we compared the alterations in glycation levels at specific sites with changes in the expression of the corresponding proteins (the whole-proteome analysis during bean nodule development will be published elsewhere). For two of the 10 glycated proteins (Gln synthetase and Met-tRNA ligase), we found the corresponding protein-specific nonglycated peptides. For both proteins, the increase in glycation levels was much higher than the increase in protein abundance (Table 1), demonstrating that certain proteins are differentially glycated during nodule development and senescence. The absence of nonmodified peptides for eight glycated proteins was not unexpected. Indeed, this phenomenon was observed in previous studies of the plant glycated proteome (Paudel et al., 2016; Bilova et al., 2017) and could be explained by the low number of trypsin cleavage sites in protein sequences (Guo et al., 2014), short protein half-life (Brings et al., 2017), or higher ionization efficiencies of AGE-modified peptides due to a better charge localization on the imidazolone moiety.
Table 1. Results of the label-free quantification of differentially glycated proteins and the corresponding AGE sites in the plant proteome of bean nodules.
The search was performed by the SEQUEST engine against a combined legume sequence database comprising three legume proteomes. The sequence redundancy of the combined database was eliminated by clustering with the СD-HIT algorithm (Huang et al., 2010), which was used with the sequence identity cutoff set to 1. As far as possible, for each glycated site, the corresponding unique protein-specific nonglycated peptide was used for relative label-free quantification. Unmodified peptides for Gln synthetase PR-1 and Met-tRNA ligase also are shown. For all peptides, the statistical significance of differential expression and glycation (P < 0.05) was confirmed by the Mann-Whitney U test. Argpyr (Argpyrimidine), Nδ-(5-hydroxy-4,6-dimethylpyrimidine-2-yl)-l-ornithine; CEL, Nε-(carboxyethyl)lysine; CMA, Nε-(carboxymethyl)arginine; CML, Nε-(carboxymethyl)lysine; Glarg, glyoxal-derived hydroimidazolone; MG-H, methylglyoxal-derived hydroimidazolone, Nδ-(5- methyl-4-oxo-5-hydroimidazolinone-2-yl)-l-ornithine; TH-Argpyr, tetrahydropyrimidine.
| No. | Protein Annotation | Peptide Sequence | Sites | m/z | zd | tRe | Quantification | ||
|---|---|---|---|---|---|---|---|---|---|
| Protein Name | Accession No.a,b,c | Maturef | Senescentg | ||||||
| fold change | |||||||||
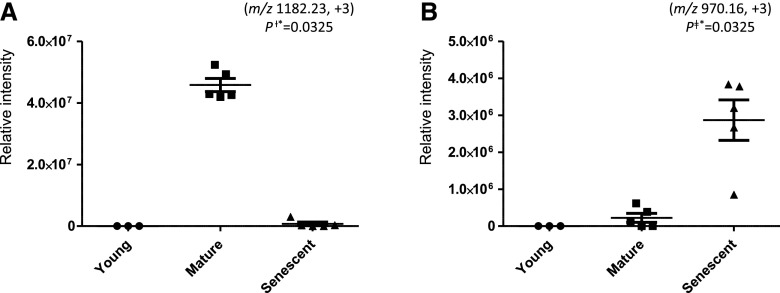
| 1 | Gln synthetase PR-1 | P04770a | 53 WNYDGSSTGQAPGQDSEVIIYPQAIF[CML]DPFR 83 | K79 | 1,182.23 | 3 | 140.63 | ∞ | – |
| 291 RLTGRHETADINTFLWGVANR 311 | 809.76 | 3 | 114.14 | 4.11 | 1.11 | ||||
| 2 | Met-tRNA ligase | V7AE49a | 494 EN[CML]SLCALVMK 504 | K496 | 338.42 | 4 | 70.91 | ∞ | – |
| 334 TISVTEYLNYESGKFSK350 | 655.99 | 3 | 120.71 | 0.59 | 0.08 | ||||
| 655 AQKHPDADALYVEEIDVGEEQTR 677 | 654.06 | 4 | 101.58 | 11.90 | 0.05 | ||||
| 3 | Rho GTPase-activating protein | V7BAK4a | 47 TSLVFFKNDPSALPQ[Glarg]GGEVNLTLGGIDLNNSGSVVV[Glarg] 84 | R62, R84 | 1,013.53 | 4 | 148.79 | 1.42 | 3.65 |
| 4 | Elongation factor 1-α | V7BFQ4a | 180 [CEL]VGYNPEKIPFVPISGFEGDNMIER 204 | K180 | 970.16 | 3 | 134.49 | – | ∞ |
| 5 | rRNA-processing brix domain protein | A0A072VK38b | 5 KG[CML][MG-H]SEIHAK 14 | K7, R8 | 320.68 | 4 | 29.54 | 681.96 | 0.74 |
| 6 | p-Loop nucleoside triphosphate hydrolase superfamily protein, putative | G7I8G1b | 933 TDL[CEL]QHAK 940 | K936 | 338.18 | 3 | 32.76 | 15.63 | 2.97 |
| 7 | Sugar isomerase family protein | Lj0g3v0160179.1c | 122 WVSTRARWLTW[TH-Argpyr] R 134 | R133 | 394.00 | 5 | 69.34 | ∞ | – |
| 8 | rRNA-processing brix domain protein | A0A072VK38b | 5 [CEL]G[CEL][TH-Argpyr] SEIHAK 14 | K5, K7, R8 | 433.24 | 3 | 35.01 | 0.84 | 1.78 |
| 9 | 60S ribosomal protein L6 | B7FN14b | 24 VIEVEGP[Argpyr]GK 33 | R31 | 388.55 | 3 | 68.23 | – | ∞ |
| 10 | Receptor kinase 2 | Lj3g3v0234090.1c | 163 WKQSLLSL[CMA]D[CEL] 173 | R171, K173 | 507.27 | 3 | 99.32 | 24.18 | 0.75 |
| 11 | RNA-binding (RRM/RBD/RNP motif) family protein | G7ZW01b | 39 IVSVDLLWHT[Glarg]GPK[Glarg] 53 | R49, R53 | 465.01 | 4 | 96.47 | ∞ | – |
a–cIndividual proteins were annotated with bean (a), M. truncatula (b), or L. japonicus (c) sequences. d Charge. e Retention time. f Fold change observed in mature plants relative to young plants. g Fold change observed in senescent plants relative to young plants.
Figure 9.
Label-free quantification of glycated peptides of plant proteins from bean nodules at different developmental stages. A, Gln synthetase PR-1. B, Elongation factor 1-α. The statistical significance of differential expression and glycation was confirmed by the Mann-Whitney U test (means ± se of three to five biological replicates; P < 0.05).
Our data show that Gln synthetase is glycated in vivo and that glycation levels depend on the nodule developmental stage (Table 1; Fig. 9A). The modified enzyme was found solely in mature nodules, whereas it was undetectable in young and senescent nodules. Glycation may alter protein structure and function, inducing cross-linking and aggregation. Although the effect of glycation on Gln synthetase activity was not investigated here, this PTM could be involved in the control of nitrogen assimilation in nodules. Additionally, proteins that participate in signaling, RNA processing, and protein synthesis were identified as glycated predominantly in aging nodules, which suggests their involvement in the developmental program that leads to nodule senescence (Table 1; Supplemental Fig. S12). Rho GTPase-activating proteins regulate Rho GTPases that, in turn, are major regulators of signaling pathways in plants (Kawano et al., 2014). They are expressed in legume nodules, and a role in the onset of the nitrogen-fixing symbiosis has been proposed in Lotus japonicus (Ke et al., 2012). Our data show that one Rho GTPase-activating protein is glycated predominantly in aging nodules and, therefore, might be involved in nodule senescence. Moreover, proteins that participate in ribosome biogenesis and protein synthesis (elongation factor 1-α, rRNA-processing brix domain protein, Met-tRNA ligase, and 60S ribosomal protein) were identified as glycated predominantly in mature and senescent nodules (Table 1; Supplemental Fig. S12). The AGE modifications of these proteins could contribute to the decline in nodule function during senescence.
Protein Glycation in the Bacteroids
To complement our study of the glycated nodule proteome, we investigated the protein glycation profiles in the bacterial symbiotic partner. Notably, despite a much lower number of sequence database entries, glycation rates of rhizobial proteins were higher than those observed for the nodule plant proteome. A sequence database search followed by a multiconsensus analysis revealed the presence of 135 glycation sites in 92 peptides from 83 proteins (Supplemental Data Set S4). The proteins were assigned to 18 functional categories based on the COG database (Supplemental Fig. S13). Remarkably, Arg-derived AGEs accounted for 72% of the modifications, whereas only 28% corresponded to Lys modifications (Supplemental Fig. S14). This clearly differs from the results obtained with plant proteins, where Lys and Arg residues were involved in the formation of AGEs at roughly similar levels (Supplemental Fig. S10). The reasons for this discrepancy are not clear but might reflect the different amino acid compositions of plant and bacterial proteins or the metabolic peculiarities of the two symbiotic partners. The most common AGEs detected in bacterial proteins also differed from those found in plants. Nε-(carboxymethyl)arginine (CMA), CML, and argpyrimidine accounted for 28%, 22%, and 19% of the modifications, respectively (Supplemental Data Set S4; Supplemental Fig. S14). Most glycation sites (73) of bacteroid proteins were present throughout nodule development, whereas 19 were unique to peptides from mature and/or senescent nodules, and six were found only in senescent nodules (Fig. 10A). Similar to plant proteins, only three bacteroidal proteins were identified as both carbonylated and glycated, further pointing to the selectivity of both PTMs. These proteins were annotated as alkaline phosphatase (A0A072BWI1), conjugal transfer protein A (A0A072C238), and the cobN subunit of cobaltochelatase (A0A072C0V4).
Figure 10.
Relative abundance of AGE-modified sites in bacteroid proteins from nodules at the three developmental stages. A, Total number of AGE-modified sites identified in bacteroid proteins from young (Y), mature (M), and senescent (S) nodules. B, Number of specific AGE classes identified in glycated peptides differentially abundant in mature and senescent nodules relative to young nodules. Abbreviations as in Table 1 and 2.
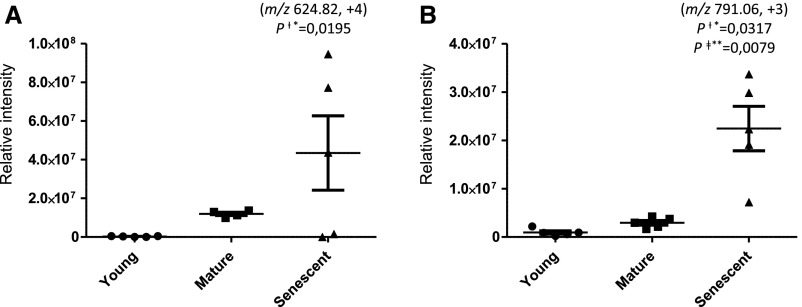
Label-free quantification of the identified glycation sites revealed 18 age-dependent glycated peptides demonstrating differential abundance in individual age groups (Table 2; Fig. 11; Supplemental Fig. S15). These peptides contained eight individual AGEs, with CML and Glarg being the most abundant (Table 2; Fig. 10B). For five of the corresponding proteins, nonglycated peptides also were detected (Table 2). The quantitative analysis revealed the age-dependent glycation of three proteins: Arg biosynthesis bifunctional protein ArgJ, 30S ribosomal protein S10, and monophenol monooxygenase. In contrast, the accumulation of MG-H and/or CMA on Arg-252 in peptidyl dipeptidase could be explained by an age-related increase in protein expression (Table 2). Unfortunately, a reliable conclusion could not be established for the carboxymethylation of Lys-118 in the 30S ribosomal protein S2, because two proteotypic nonglycated peptides showed different quantitative profiles. Nevertheless, the manual interpretation of the corresponding tandem mass spectra confirmed the sequence assignments of these unique peptides (Table 2). Other proteins with predicted differential glycation levels in senescent nodules include two transcriptional regulators, one chaperone, and proteins involved in transport, intracellular trafficking, and energy production and conversion.
Table 2. Results of the label-free quantification of differentially glycated proteins and the corresponding AGE sites in the proteome of bean nodule bacteroids.
The search was performed by the SEQUEST engine against a sequence database of Rhizobium leguminosarum bv phaseoli. As far as possible, for each glycated site, the corresponding unique protein-specific nonglycated peptide was used for relative label-free quantification. Unmodified peptides for Arg biosynthesis bifunctional protein ArgJ, 30S ribosomal proteins S2 and S10, peptidyl-dipeptidase, and monophenol monooxygenase are shown. For all peptides, the statistical significance of differential expression and glycation (P < 0.05) was confirmed by the Mann-Whitney U test. CEA, Nε-(carboxyethyl)arginine. Other abbreviations as in Table 1.
| No. | Protein Annotation | Peptide Sequence | Sites | m/z | z | tR | Quantification | ||
|---|---|---|---|---|---|---|---|---|---|
| Protein Name | Accession No. | Maturea | Senescentb | ||||||
| fold change | |||||||||
| 1 | Arg biosynthesis bifunctional protein ArgJ | A0A072C5E0 | 35 NRTDVLMMVFD[CML]PAAVAGVFTR 56 | K46 | 624.82 | 4 | 137.91 | 40.53 | 182.82 |
| 331 IVMAVGK 337 | 359.22 | 2 | 52.50 | 1.68 | 0.21 | ||||
| 155 AIMTTDTYPK 164 | 570.78 | 2 | 65.13 | 0.02 | 0.02 | ||||
| 2 | 30S ribosomal protein S2 | A0A072C5T7 | 116 LR[CML]LDEILGGEAQGFTKK 133 | K118 | 516.04 | 4 | 90.41 | 3.52 | 0.56 |
| 108 TISNSIQR 115 | 459.75 | 2 | 40.72 | 5.76 | 0.30 | ||||
| 55 ALQVVSDTVAR 65 | 579.82 | 2 | 73.25 | – | 0.19 | ||||
| 3 | 30S ribosomal protein S10 | A0A072C6R8 | 10 L[CEL]AFDHR 16 | K11 | 320.17 | 3 | 67.85 | 18.75 | 44.64 |
| 17 ILDASTREIVSTAKR 31 | 553.98 | 3 | 73.49 | 1.46 | 1.02 | ||||
| 4 | Peptidyl-dipeptidase | A0A072BWW2 | 248 AWVA[CMA]GENGGETDNRAVIR 266 | R252 | 533.01 | 4 | 69.75 | 8.43 | 9.34 |
| 248 AWVA[MG-H]GENGGETDNRAVIR 266 75 | R252 | 482.76 | 2 | 108.18 | 0.20 | 5.31 | |||
| 75 VSALFWNK 82 | |||||||||
| 227 SIIEPFLTFSER 238 | 719.88 | 2 | 138.27 | 6.67 | 14.72 | ||||
| 5 | Monophenol monooxygenase | A0A072BUN7 | 462 VYGGRGQTDGEAIDLQLIPVGSGAG[CML]PGAVAPAKLEIAIVSA 503 | K487 | 1,368.74 | 3 | 138.16 | ∞ | – |
| 462VYGGRGQTDGEAIDLQLIPVGSGAGKPGAVAPA[CML]LEIAIVSA 503 | K495 | 530.29 | 2 | 113.58 | 0.19 | 3.65 | |||
| 248 IWATWNLR 255 | |||||||||
| 6 | Uncharacterized protein | A0A072C7M7 | 186 NVQDSV[Glarg]ALLSGVVR 200 | R192 | 413.98 | 4 | 109.24 | 5.92 | 193.92 |
| 7 | Anti-sigma factor transcriptional activator protein | A0A072C686 | 42 LVSSLELLAGEALE[CMA]SPEAAIADR 65 | R56 | 642.84 | 4 | 63.078 | 3.59 | 0.36 |
| 8 | Type VI secretion system baseplate subunit TssK | A0A072BS04 | 50 LN[Argpyr]ELLGLGK 59 | R52 | 398.24 | 3 | 97.12 | – | ∞ |
| 9 | ABC transporter substrate-binding protein | A0A072BSX7 | 213 LMSGDVALVEDPRMIEALEML[Argpyr][Argpyr]LSNYAEPLEK 245 | R234, R235 | 1,322.34 | 3 | 142.63 | – | ∞ |
| 10 | Integrase | A0A072BY45 | 68 LDRLA[Glarg]SVADLMGITE[Glarg]LR 86 | R73, R84 | 571.05 | 4 | 118.65 | 13.03 | 0.79 |
| 11 | GntR family transcriptional regulator | A0A072C3K4 | 24 IVGMIADGTFPVNE[Glarg]LPSETR 44 | R38 | 586.30 | 4 | 105.32 | 3.72 | 0.09 |
| 12 | ABC transporter ATP-binding protein | A0A072BVM6 | 41 HPWQAGFAIGATLVASTFQLMIP[Glarg] 64 | R64 | 712.17 | 5 | 110.60 | 0.30 | 1.94 |
| 13 | α/β-Hydrolase fold protein | A0A072CKM0 | 82 GL[CML]AVNVFAMSWGGTVAGAYAAR 104 | K84 | 791.06 | 3 | 139.83 | 3.11 | 23.70 |
| 14 | Cytochrome bc biosynthesis ABC transporter ATP-binding protein | A0A072BTW6 | 307 GALDLG[Argpyr]TLLAAR 319 | R313 | 352.45 | 4 | 77.35 | 69.96 | 27.83 |
| 15 | Uncharacterized protein | A0A072C8Y3 | 24 IGELL[TH-Argpyr] 29 | R29 | 422.74 | 2 | 56.70 | 2.88 | 0.13 |
| 16 | Molecular chaperone protein DnaJ | A0A072BWF0 | 7 YFD[TH-Argpyr] IR 12 | R10 | 338.50 | 3 | 63.58 | 0.08 | 53.40 |
| 17 | NADPH quinone reductase | A0A072BTA3 | 22 LA[CML]ALVRFAPPELEMSEISFK 42 | K24 | 812.10 | 3 | 133.53 | 2.36 | 24.76 |
| 18 | FAD-dependent l-sorbose dehydrogenase | A0A072C6X7 | 207 KNLTV[CEA]TGA[CEA] 216 | R212, R216 | 420.57 | 3 | 34.45 | 9.31 | 0.01 |
Fold change observed in mature plants relative to young plants.
Fold change observed in senescent plants relative to young plants.
Figure 11.
Label-free quantification of glycated peptides of bacteroid proteins from bean nodules at different developmental stages. A, Arg biosynthesis bifunctional protein ArgJ. B, α/β-Hydrolase fold protein. The statistical significance of differential expression and glycation was confirmed by the Mann-Whitney U test (means ± se of five biological replicates; P < 0.05).
In summary, we have used novel methodological approaches to identify numerous proteins susceptible to irreversible oxidative modifications (carbonylation and advanced glycation) and to characterize their modification sites in bean nodules. The effect of carbonylation on two key nodule proteins was investigated. In terms of relative abundance, high numbers of proteins involved in the stress response, the regulation of gene expression, and intracellular signaling were detected as carbonylated or glycated. Moreover, several proteins were identified as glycated specifically in aging nodules. We propose that these two redox-sensitive, stable PTMs are important in the regulation of nodule metabolism and senescence.
MATERIALS AND METHODS
Biological Material
Bean (Phaseolus vulgaris ‘Contender’) seeds were surface sterilized with 70% (v/v) ethanol and germinated in pots containing a perlite:vermiculite (1:1, v/v) mixture. After 7 d, seedlings were inoculated with Rhizobium leguminosarum bv phaseoli strain 3622 and were grown on a nutrient solution containing 0.25 mm NH4NO3 in a controlled-environment chamber (Loscos et al., 2008). This small concentration of combined nitrogen in the nutrient solution does not inhibit nodulation and favors plant growth. Nodules were harvested from plants at three different developmental stages (days after germination): young (∼28 d, late vegetative stage), mature (∼40 d, late flowering-early fruiting stage), and senescent (∼53 d, fully developed pods). Nodules were kept at −80°C until use.
Expression, Purification, and Assay of Recombinant MDH
The nodule cDNA sequence encoding bean NAD+-dependent MDH was PCR amplified using PfuUltra II DNA polymerase (Agilent) and primers (forward, 5′-CACCATGGAGGCAACTGCAGGAGC-3′; and reverse, 5′-TTATTTTCTGATGAAGTCTA-3′) compatible with pET200 directional TOPO expression kits (Invitrogen). After transformation by heat shock at 42°C for 30 s, protein expression was induced in Escherichia coli BL21 (DE3) by the addition of 1 mm isopropyl-β-d-thiogalactopyranoside for 4 h at 37°C. Bacteria were harvested by centrifugation, resuspended in 50 mm potassium phosphate buffer (pH 8) containing 300 mm NaCl and 40 mm imidazole, and sonicated six times for 30 s each. Extracts were cleared by centrifugation, and supernatants were loaded onto HiTrap chelating HP Ni-affinity columns (GE Healthcare Life Sciences). The His-tagged proteins were eluted with buffer containing 250 mm imidazole, desalted, and concentrated by ultrafiltration. MDH activity was measured at 25°C by following the decrease in A340 due to NADH oxidation. The reaction mixture contained 50 mm potassium phosphate buffer (pH 7), 1 mm oxaloacetic acid, and 0.2 mm NADH.
Lb Purification and Immunoblotting
Lb was purified from bean nodules. Briefly, nodules (200 mg) were ground at 0°C in a mortar with 400 μL of 50 mm Tris-HCl (pH 8) supplemented with complete EDTA-free protease inhibition cocktail (Sigma). After centrifugation, the proteins were separated on 12.5% (w/v) native polyacrylamide gels, and the red band of Lb was excised and eluted from the gel by incubation with 10 mm ammonium bicarbonate for 16 h at 4°C. The protein was then concentrated by ultrafiltration.
Immunoblotting was performed following standard protocols. The primary antibody was raised against bean Lb and the secondary antibody was goat anti-rabbit IgG horseradish peroxidase conjugate (Sigma). The antibodies were used at dilutions of 1:1,000 and 1:40,000, respectively. Immunoreactive proteins were detected by chemiluminescence (Thermo Scientific).
MCO and Detection of Carbonylated Proteins
MCO was performed as described by Requena et al. (2001) with some modifications. Briefly, proteins were dissolved at 1 mg mL−1 in oxidation buffer (50 mm HEPES, pH 7.4, 100 mm KCl, and 10 mm MgCl2) and incubated with different concentrations of ascorbate (12.5–75 mm) and FeCl3 (10–300 μm) for 6 h at 37°C with shaking at 700 rpm in the dark. The reaction was terminated by adding 1 mm EDTA, removing the excess reagents by ultrafiltration, and subsequent freezing at −80°C. Similarly, proteins were incubated with different concentrations of H2O2 (0–10 mm) in oxidation buffer supplemented with 1 mm EDTA under the same conditions as for MCO. Carbonylated proteins were detected using the OxyBlot Protein Oxidation Detection Kit (Millipore) following the manufacturer’s protocol.
Identification of Carbonylated Proteins and Modified Sites
Proteins were extracted from 200 mg of frozen nodules with 500 μL of a medium containing 30 mm MOPS (pH 7.2), 1 mm EDTA, 10 mm KH2PO4, 2% (w/v) polyvinylpolypyrrolidone, and 0.14 m β-mercaptoethanol. After centrifugation, the supernatants were dialyzed in 25 mm ammonium bicarbonate containing 0.14 m β-mercaptoethanol. For trypsin digestion, 100 μg of protein were denatured with 1% (w/v) deoxycholate, reduced with 5 mm Tris-(2-carboxyethyl)-phosphine at 60°C for 30 min (with shaking at 550 rpm), and alkylated with 10 mm iodoacetamide at 37°C for 30 min in the dark. Digestion was carried out with 2 μg of Trypsin Gold (Promega) for 16 h at 37°C (with shaking at 550 rpm). The resulting peptides were labeled at 27°C for 2 h with 10 mm O-(biotinylcarbazoylmethyl)hydroxylamine (ARP; Gerbu Biotechnik) in 1% (v/v) formic acid. Excess ARP was removed by solid-phase extraction using Oasis HLB cartridges (Waters). Peptides were eluted with 70% (v/v) acetonitrile and 0.5% (v/v) formic acid and dried under vacuum. The ARP-labeled tryptic peptides (100 μL in PBS) were added to spin columns containing 200 μL of monomeric avidin agarose slurry (Thermo Scientific). After washing with 1 mL of PBS, 1 mL of 10 mm NaH2PO4, and 2 mL of 50 mm ammonium bicarbonate in 20% (v/v) methanol, the column was rinsed with 1 mL of water, and the purified peptides were eluted with 30% (v/v) acetonitrile containing 0.4% (v/v) formic acid, vacuum concentrated, and stored at −80°C. Samples were analyzed on a nano-Acquity ultra-performance liquid chromatography (UPLC) device (Waters) coupled online to an LTQ Orbitrap XL ETD mass spectrometer equipped with a nano-electrospray ionization (ESI) source (Thermo Scientific). Eluent A was aqueous formic acid (0.1%, v/v) and eluent B was formic acid (0.1%, v/v) in acetonitrile. Affinity-enriched peptides were loaded onto the trap column (nanoAcquity symmetry C18; internal diameter 180 µm, length 20 mm, and particle diameter 5 µm) at a flow rate of 10 µL min−1. Peptides were separated on a BEH 130 column (C18-phase; internal diameter 75 µm, length 100 mm, and particle diameter 1.7 µm) with a flow rate of 0.4 µL min−1 using the following gradient: 3% to 9% (2.1 min), 9.9% (1.9 min), 17.1% (10 min), 18% (0.5 min), 20.7% (0.2 min), 22.5% (3.1 min), 25.6% (3 min), 30.6% (5 min), 37.8% (2.8 min), and finally to 81% eluent B (2 min). The transfer capillary temperature was set to 200°C and the tube lens voltage to 120 V. An ion spray voltage of 1.5 kV was applied to a PicoTip online nano-ESI emitter (New Objective). Precursor ion survey scans were acquired with an Orbitrap mass spectrometer (resolution of 60,000) for an m/z range of 400 to 2,000. The collision-induced dissociation-tandem mass spectra (isolation width 2, activation Q 0.25, normalized collision energy 35%, and activation time 30 ms) were recorded in DDA mode for the six most intense signals in each survey scan with dynamic exclusion for 60 s using Xcalibur software (version 2.0.7).
The acquired MS/MS spectra were searched against the UniProtKB bean database using the SEQUEST search engine (Proteome Discoverer 1.4; Thermo Scientific), allowing up to two missed cleavages and a mass tolerance of 10 ppm for precursor ions and 0.8 D for product ions. Search results were filtered for rank 1 (ARP-labeled samples), high confidence, and score versus charge states corresponding to XCorr/z 2.0/2, 2.25/3, 2.5/4, 2.75/5. Only peptides identified by MS/MS in at least five biological replicates were considered, and ∼20% of MS/MS spectra were assigned manually to confirm identifications by database search. The resulting set of modified proteins was annotated manually based on their Gene Ontology terms provided by UniProtKB. The BLAST algorithm (Altschul et al., 1990) was used to select the closest Arabidopsis (Arabidopsis thaliana) homolog for each bean protein. This was a necessary first step to take advantage of the bioinformatic resources developed for Arabidopsis research.
Characterization of the Glycated Nodule Proteome
Frozen nodules (∼250 mg) were ground in a Mixer Mill MM 400 with 3-mm-diameter stainless steel balls (Retsch) at a vibration frequency of 30 Hz (2 × 1 min). The total protein fraction was prepared using phenol extraction as described (Frolov et al., 2017a). The proteins were reconstituted in 100 µL of shotgun buffer (8 m urea, 2 m thiourea, and 0.2% (w/v) AALS II in 100 mm Tris-HCl, pH 7.5). Protein concentrations were determined by the 2D Quant kit (GE Healthcare), and the results were cross-validated by SDS-PAGE as described by Greifenhagen et al. (2016). Protein (30 µg) was diluted with shotgun buffer without AALS II to obtain a final volume of 100 µL and reduced with 10 µL of 50 mm Tris-(2-carboxyethyl)-phosphine for 30 min at 37°C. The samples were cooled to room temperature, alkylated with 11 µL of 100 mm iodoacetamide in AALS II-free shotgun buffer in darkness for 60 min at 4°C, and diluted 8-fold with 50 mm ammonium bicarbonate. Trypsin (0.4 µg µL−1 in 50 mm ammonium bicarbonate) was added sequentially in 1:20 and 1:50 enzyme:substrate ratios, and incubations were performed at 37°C for 5 and 18 h, respectively. Afterward, 10% (v/v) aqueous trifluoroacetic acid (UV grade) was added to a final concentration of 1% (v/v), and the digests were incubated at 37°C for 20 min to eliminate AALS II. The efficiency of the tryptic digestion was determined by SDS-PAGE. The resulting peptides were desalted by solid-phase extraction using in-house-prepared stage tips (i.e. polypropylene pipette tips [200 µL] filled with six layers of C18 reverse-phase material [Empower SPE discs]).
The peptides were reconstituted in 3% (v/v) acetonitrile in 0.1% (v/v) aqueous trifluoroacetic acid, and aliquots equivalent to 500 ng were loaded on an Acclaim PepMap 100 C18 trap column (300 µm × 5 mm, 3 µm particle size) during 15 min at 30 µL min−1. Separation was performed at 300 nL min−1 on an Acclaim PepMap 100 C18 column (75 µm × 250 mm, particle size 2 µm) using an Ultimate 3000 RSLC nano-HPLC system coupled online to a Q Exactive Q-Orbitrap mass spectrometer via a nano-ESI source equipped with a 30-µm i.d., 40-mm-long steel emitter (Thermo Scientific). The eluents A and B were 0.1% (v/v) aqueous formic acid and 0.08% (v/v) formic acid in acetonitrile, respectively. The peptides were eluted with linear gradients: from 1% to 35% (90 min) followed by 35% to 85% eluent B (5 min). The column was washed for 11 min and reequilibrated with 1% eluent B for 10 min. The nano-LC-MS analysis was performed in the positive ion mode using the mass spectrometer settings listed in Supplemental Table S4 and relied on the data-dependent acquisition experiments comprising a survey Orbitrap-MS scan and dependent Q-Orbitrap-MS/MS scans for the five most abundant signals (at certain retention times) with charge states ranging from 2 to 6. Peptide identification and protein annotation were performed by the SEQUEST search engine (Proteome Discoverer 2.1) against a combined database containing protein sequences of bean, Lotus japonicus, and Medicago truncatula using the settings listed in Supplemental Table S5. The search against the R. leguminosarum database (www.uniprot.org) relied on the same search machine and software.
Characterization of the glycated proteome relied on the raw data acquired as described in the previous section. For this, SEQUEST searches (Proteome Discoverer 1.4) were run against the legume and rhizobial sequence databases shown in Supplemental Table S5. Ten search templates, containing both AGEs and oxidative modifications, were designed (Supplemental Table S3). The false positive hits were excluded by a multiconsensus search with false discovery rate < 0.01. The sequence identity was confirmed by manual interpretation for the annotations with the lowest XCorr values. The identifications were cross-annotated between experimental groups by exact m/z values and retention times. For this, the corresponding extracted ion chromatograms were integrated at specific m/z values by LCquan 2.8 software. The significance of intergroup differences was assessed by the Mann-Whitney U test (Prism 5.0) after data normalization by median. The functional annotation of age-dependently glycated proteins relied on the MapMan software (http://mapman.gabipd.org).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers XP_007159190 (MDH) and XP_007144265 (Lb).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Overview of the experimental workflow followed for the identification of the carbonylated and glycated peptides and of their modification sites in nodule proteins.
Supplemental Figure S2. Tandem mass spectra of the ARP-derivatized modified peptides.
Supplemental Figure S3. Relative abundance of carbonylated amino acid residues considering the amino acid composition of the Arabidopsis proteome.
Supplemental Figure S4. STRING analysis of carbonylated proteins.
Supplemental Figure S5. Amino acid sequences of NAD+-MDHs from vascular plants.
Supplemental Figure S6. SDS-PAGE of individual protein samples isolated from nodules of young, mature, and senescent bean plants.
Supplemental Figure S7. SDS-PAGE of tryptic digests of the total protein fraction from nodules of young, mature, and senescent bean plants.
Supplemental Figure S8. Tandem mass spectra of AGE-modified tryptic peptides corresponding to the bean nodule plant proteins shown in Table 1.
Supplemental Figure S9. Tandem mass spectra of AGE-modified tryptic peptides corresponding to the bean nodule bacteroid proteins shown in Table 2.
Supplemental Figure S10. Number of glycation sites in each of the AGE classes detected in the plant proteome of bean nodules by nano-UPLC-LIT-Orbitrap-DDA-MS/MS.
Supplemental Figure S11. Number of glycated plant proteins from bean nodules in each functional category.
Supplemental Figure S12. Label-free quantification of glycated peptides of plant proteins from bean nodules at different developmental stages.
Supplemental Figure S13. Number of glycated bacteroid proteins in different functional categories.
Supplemental Figure S14. Number of glycation sites in each of the AGE classes detected in the proteome of bean nodule bacteroids by nano-UPLC-LIT-Orbitrap-DDA-MS/MS.
Supplemental Figure S15. Label-free quantification of glycated peptides of bacteroid proteins from bean nodules at different developmental stages.
Supplemental Table S1. Amino acid modifications generated by direct oxidation or Michael addition of reactive aldehydes or ketones.
Supplemental Table S2. Protein yields and total protein intensities per lane calculated for individual samples separated by SDS-PAGE.
Supplemental Table S3. Settings for the SEQUEST database search used for the identification of glycation sites in plant and bacteroid proteins from bean nodules.
Supplemental Table S4. Instrument settings applied for ESI-Orbitrap-MS experiments.
Supplemental Table S5. Combined database used for the SEQUEST search engine comprising individual sequence databases.
Supplemental Data Set S1. Plant proteins from bean nodules carbonylated in vivo.
Supplemental Data Set S2. Bacteroid proteins from bean nodules carbonylated in vivo.
Supplemental Data Set S3. Plant proteins from bean nodules glycated in vivo.
Supplemental Data Set S4. Bacteroid proteins from bean nodules glycated in vivo.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Irene Orera and Giuseppe Lattanzio (Unidad de Proteómica de los Servicios Científico-Técnicos del Centro de Investigación Biomédica de Aragón-Instituto Aragonés de Ciencias de la Salud) for assistance with sample preparation. The Proteomics Unit is a member of ProteoRed ISCIII. Thanks also are due to Dennis Westphal for help with proteomic data analysis and Carmen Pérez-Rontomé for figure editing. We are most grateful to Ian Max Møller (Aarhus University), David Dalton (Reed College), and two anonymous reviewers for helpful comments on the article.
Footnotes
This work was supported by the Ministerio de Economía y Competitividad-Fondos Europeos de Desarrollo Regional (grant AGL2014-53717-R to M.A.M. and M.B.), by the German Research Society (grant FR3117/2-1 to A.F. and A.K.), by the Russian Foundation for Basic Research (grant 18-016-00190 to A.F.), by the Deutsche Forschungsgemeinschaft (grant FE-1236/3-1 to M.F.), and by the European Regional Development Fund (European Union and Free State Saxony; grants 100146238 and 100121468 to M.F.).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Appleby CA. (1984) Leghemoglobin and Rhizobium respiration. Annu Rev Plant Physiol 35: 443–478 [Google Scholar]
- Ashtamker C, Kiss V, Sagi M, Davydov O, Fluhr R (2007) Diverse subcellular locations of cryptogein-induced reactive oxygen species production in tobacco Bright Yellow-2 cells. Plant Physiol 143: 1817–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415–442 [DOI] [PubMed] [Google Scholar]
- Bartoli CG, Gómez F, Martínez DE, Guiamet JJ (2004) Mitochondria are the main target for oxidative damage in leaves of wheat (Triticum aestivum L.). J Exp Bot 55: 1663–1669 [DOI] [PubMed] [Google Scholar]
- Becana M, Dalton DA, Moran JF, Iturbe-Ormaetxe I, Matamoros MA, Rubio MC (2000) Reactive oxygen species and antioxidants in legume nodules. Physiol Plant 109: 372–381 [Google Scholar]
- Becana M, Matamoros MA, Udvardi M, Dalton DA (2010) Recent insights into antioxidant defenses of legume root nodules. New Phytol 188: 960–976 [DOI] [PubMed] [Google Scholar]
- Bechtold U, Rabbani N, Mullineaux PM, Thornalley PJ (2009) Quantitative measurement of specific biomarkers for protein oxidation, nitration and glycation in Arabidopsis leaves. Plant J 59: 661–671 [DOI] [PubMed] [Google Scholar]
- Berlett BS, Stadtman ER (1997) Protein oxidation in aging, disease, and oxidative stress. J Biol Chem 272: 20313–20316 [DOI] [PubMed] [Google Scholar]
- Bilova T, Lukasheva E, Brauch D, Greifenhagen U, Paudel G, Tarakhovskaya E, Frolova N, Mittasch J, Balcke GU, Tissier A, et al. (2016) A snapshot of the plant glycated proteome: structural, functional, and mechanistic aspects. J Biol Chem 291: 7621–7636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilova T, Paudel G, Shilyaev N, Schmidt R, Brauch D, Tarakhovskaya E, Milrud S, Smolikova G, Tissier A, Vogt T, et al. (2017) Global proteomic analysis of advanced glycation end products in the Arabidopsis proteome provides evidence for age-related glycation hot spots. J Biol Chem 292: 15758–15776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollineni RC, Fedorova M, Hoffmann R (2013) Qualitative and quantitative evaluation of derivatization reagents for different types of protein-bound carbonyl groups. Analyst (Lond) 138: 5081–5088 [DOI] [PubMed] [Google Scholar]
- Bollineni RC, Fedorova M, Blüher M, Hoffmann R (2014a) Carbonylated plasma proteins as potential biomarkers of obesity induced type 2 diabetes mellitus. J Proteome Res 13: 5081–5093 [DOI] [PubMed] [Google Scholar]
- Bollineni RC, Hoffmann R, Fedorova M (2014b) Proteome-wide profiling of carbonylated proteins and carbonylation sites in HeLa cells under mild oxidative stress conditions. Free Radic Biol Med 68: 186–195 [DOI] [PubMed] [Google Scholar]
- Brings S, Fleming T, Freichel M, Muckenthaler MU, Herzig S, Nawroth PP (2017) Dicarbonyls and advanced glycation end-products in the development of diabetic complications and targets for intervention. Int J Mol Sci 18: 984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo-Begueria L, Rubio MC, Martínez JI, Pérez-Rontomé C, Delgado MJ, Bedmar EJ, Becana M (2018) Redefining nitric oxide production in legume nodules through complementary insights from electron paramagnetic resonance spectroscopy and specific fluorescent probes. J Exp Bot 69: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri A, Merchant A, van Volkenburgh E (2011) Why not beans? Funct Plant Biol 38: 3–6 [DOI] [PubMed] [Google Scholar]
- Couturier J, Chibani K, Jacquot JP, Rouhier N (2013) Cysteine-based redox regulation and signaling in plants. Front Plant Sci 4: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan KA, Huber SC (2007) Sucrose synthase oligomerization and F-actin association are regulated by sucrose concentration and phosphorylation. Plant Cell Physiol 48: 1612–1623 [DOI] [PubMed] [Google Scholar]
- Evans PJ, Gallesi D, Mathieu C, Hernández MJ, de Felipe M, Halliwell B, Puppo A (1999) Oxidative stress occurs during soybean nodule senescence. Planta 208: 73–79 [Google Scholar]
- Farmer EE, Mueller MJ (2013) ROS-mediated lipid peroxidation and RES-activated signaling. Annu Rev Plant Biol 64: 429–450 [DOI] [PubMed] [Google Scholar]
- Fedorova M, Bollineni RC, Hoffmann R (2014) Protein carbonylation as a major hallmark of oxidative damage: update of analytical strategies. Mass Spectrom Rev 33: 79–97 [DOI] [PubMed] [Google Scholar]
- Foussard M, Garnerone AM, Ni F, Soupène E, Boistard P, Batut J (1997) Negative autoregulation of the Rhizobium meliloti fixK gene is indirect and requires a newly identified regulator, FixT. Mol Microbiol 25: 27–37 [DOI] [PubMed] [Google Scholar]
- Frolov A, Bilova T, Paudel G, Berger R, Balcke GU, Birkemeyer C, Wessjohann LA (2017a) Early responses of mature Arabidopsis thaliana plants to reduced water potential in the agar-based polyethylene glycol infusion drought model. J Plant Physiol 208: 70–83 [DOI] [PubMed] [Google Scholar]
- Frolov A, Didio A, Ihling C, Chantzeva V, Grishina T, Hoehenwarter W, Sinz A, Smolikova G, Bilova T, Medvede S (2017b) The effect of simulated microgravity on the Brassica napus seedling proteome. Funct Plant Biol 45: 440–452 [DOI] [PubMed] [Google Scholar]
- Gaber A, Ogata T, Maruta T, Yoshimura K, Tamoi M, Shigeoka S (2012) The involvement of Arabidopsis glutathione peroxidase 8 in the suppression of oxidative damage in the nucleus and cytosol. Plant Cell Physiol 53: 1596–1606 [DOI] [PubMed] [Google Scholar]
- Galperin MY, Makarova KS, Wolf YI, Koonin EV (2015) Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43: D261–D269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goward CR, Nicholls DJ (1994) Malate dehydrogenase: a model for structure, evolution, and catalysis. Protein Sci 3: 1883–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greifenhagen U, Frolov A, Blüher M, Hoffmann R (2016) Plasma proteins modified by advanced glycation end products (AGEs) reveal site-specific susceptibilities to glycemic control in patients with type 2 diabetes. J Biol Chem 291: 9610–9616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesser E, Vemula V, Raulien N, Wagner U, Reeg S, Grune T, Fedorova M (2017) Cross-talk between lipid and protein carbonylation in a dynamic cardiomyocyte model of mild nitroxidative stress. Redox Biol 11: 438–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groten K, Dutilleul C, van Heerden PDR, Vanacker H, Bernard S, Finkemeier I, Dietz KJ, Foyer CH (2006) Redox regulation of peroxiredoxin and proteinases by ascorbate and thiols during pea root nodule senescence. FEBS Lett 580: 1269–1276 [DOI] [PubMed] [Google Scholar]
- Guo X, Trudgian DC, Lemoff A, Yadavalli S, Mirzaei H (2014) Confetti: a multiprotease map of the HeLa proteome for comprehensive proteomics. Mol Cell Proteomics 13: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B. (2006) Reactive species and antioxidants: redox biology is a fundamental theme of aerobic life. Plant Physiol 141: 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Puppo A (2005) Oxyleghemoglobin scavenges nitrogen monoxide and peroxynitrite: a possible role in functioning nodules? J Biol Inorg Chem 10: 935–945 [DOI] [PubMed] [Google Scholar]
- Heyns K, Noack H (1962) Die umsetzung von D-fructose mit L-lysine und L-arginin und deren beziehung zu nichtenzymatischen bräunungsreaktionen. Chem Ber 95: 720–727 [Google Scholar]
- Hodge JE. (1955) The Amadori rearrangement. Adv Carbohydr Chem 10: 169–205 [DOI] [PubMed] [Google Scholar]
- Höhn A, König J, Grune T (2013) Protein oxidation in aging and the removal of oxidized proteins. J Proteomics 92: 132–159 [DOI] [PubMed] [Google Scholar]
- Hooper CM, Castleden IR, Tanz SK, Aryamanesh N, Millar AH (2017) SUBA4: the interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res 45: D1064–D1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Huang X, Chen L, Sun X, Lu C, Zhang L, Wang Y, Zuo J (2015) Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol 167: 1731–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Niu B, Gao Y, Fu L, Li W (2010) CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26: 680–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson T, Damasceno CM, Saravanan RS, He Y, Catalá C, Saladié M, Rose JK (2006) Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat Protoc 1: 769–774 [DOI] [PubMed] [Google Scholar]
- Jabbouri S, Fellay R, Talmont F, Kamalaprija P, Burger U, Relić B, Promé JC, Broughton WJ (1995) Involvement of nodS in N-methylation and nodU in 6-O-carbamoylation of Rhizobium sp. NGR234 nod factors. J Biol Chem 270: 22968–22973 [DOI] [PubMed] [Google Scholar]
- Kawano Y, Kaneko-Kawano T, Shimamoto K (2014) Rho family GTPase-dependent immunity in plants and animals. Front Plant Sci 5: 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke D, Fang Q, Chen C, Zhu H, Chen T, Chang X, Yuan S, Kang H, Ma L, Hong Z, et al. (2012) The small GTPase ROP6 interacts with NFR5 and is involved in nodule formation in Lotus japonicus. Plant Physiol 159: 131–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrainzar E, Wienkoop S, Weckwerth W, Ladrera R, Arrese-Igor C, González EM (2007) Medicago truncatula root nodule proteome analysis reveals differential plant and bacteroid responses to drought stress. Plant Physiol 144: 1495–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Helmann JD (2006) The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440: 363–367 [DOI] [PubMed] [Google Scholar]
- Lehtovaara P. (1978) Anomalous migration of leghaemoglobin on sodium dodecyl sulphate/polyacrylamide-gel electrophoresis. Biochem J 169: 251–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscos J, Matamoros MA, Becana M (2008) Ascorbate and homoglutathione metabolism in common bean nodules under stress conditions and during natural senescence. Plant Physiol 146: 1282–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madian AG, Regnier FE (2010) Proteomic identification of carbonylated proteins and their oxidation sites. J Proteome Res 9: 3766–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino D, Hohnjec N, Küster H, Moran JF, González EM, Arrese-Igor C (2008) Evidence for transcriptional and post-translational regulation of sucrose synthase in pea nodules by the cellular redox state. Mol Plant Microbe Interact 21: 622–630 [DOI] [PubMed] [Google Scholar]
- Martí MC, Olmos E, Calvete JJ, Díaz I, Barranco-Medina S, Whelan J, Lázaro JJ, Sevilla F, Jiménez A (2009) Mitochondrial and nuclear localization of a novel pea thioredoxin: identification of its mitochondrial target proteins. Plant Physiol 150: 646–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoros MA, Fernández-García N, Wienkoop S, Loscos J, Saiz A, Becana M (2013) Mitochondria are an early target of oxidative modifications in senescing legume nodules. New Phytol 197: 873–885 [DOI] [PubMed] [Google Scholar]
- Matamoros MA, Saiz A, Peñuelas M, Bustos-Sanmamed P, Mulet JM, Barja MV, Rouhier N, Moore M, James EK, Dietz KJ, et al. (2015) Function of glutathione peroxidases in legume root nodules. J Exp Bot 66: 2979–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo PM, Silva LS, Ribeiro I, Seabra AR, Carvalho HG (2011) Glutamine synthetase is a molecular target of nitric oxide in root nodules of Medicago truncatula and is regulated by tyrosine nitration. Plant Physiol 157: 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller IM, Sweetlove LJ (2010) ROS signalling: specificity is required. Trends Plant Sci 15: 370–374 [DOI] [PubMed] [Google Scholar]
- Møller IM, Jensen PE, Hansson A (2007) Oxidative modifications to cellular components in plants. Annu Rev Plant Biol 58: 459–481 [DOI] [PubMed] [Google Scholar]
- Møller IM, Rogowska-Wrzesinska A, Rao RSP (2011) Protein carbonylation and metal-catalyzed protein oxidation in a cellular perspective. J Proteomics 74: 2228–2242 [DOI] [PubMed] [Google Scholar]
- Moreau S, Davies MJ, Puppo A (1995) Reaction of ferric leghemoglobin with H2O2: formation of heme-protein cross-links and dimeric species. Biochim Biophys Acta 1251: 17–22 [DOI] [PubMed] [Google Scholar]
- Navascués J, Pérez-Rontomé C, Gay M, Marcos M, Yang F, Walker FA, Desbois A, Abián J, Becana M (2012) Leghemoglobin green derivatives with nitrated hemes evidence production of highly reactive nitrogen species during aging of legume nodules. Proc Natl Acad Sci USA 109: 2660–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Donaldson RP (2005) Metal-catalyzed oxidation induces carbonylation of peroxisomal proteins and loss of enzymatic activities. Arch Biochem Biophys 439: 25–31 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Oracz K, El-Maarouf Bouteau H, Farrant JM, Cooper K, Belghazi M, Job C, Job D, Corbineau F, Bailly C (2007) ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. Plant J 50: 452–465 [DOI] [PubMed] [Google Scholar]
- Ott C, Jacobs K, Haucke E, Navarrete Santos A, Grune T, Simm A (2014) Role of advanced glycation end products in cellular signaling. Redox Biol 2: 411–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott T, van Dongen JT, Günther C, Krusell L, Desbrosses G, Vigeolas H, Bock V, Czechowski T, Geigenberger P, Udvardi MK (2005) Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr Biol 15: 531–535 [DOI] [PubMed] [Google Scholar]
- Paudel G, Bilova T, Schmidt R, Greifenhagen U, Berger R, Tarakhovskaya E, Stöckhardt S, Balcke GU, Humbeck K, Brandt W, et al. (2016) Osmotic stress is accompanied by protein glycation in Arabidopsis thaliana. J Exp Bot 67: 6283–6295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puppo A, Herrada G, Rigaud J (1991) Lipid peroxidation in peribacteroid membranes from French-bean nodules. Plant Physiol 96: 826–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RSP, Zhang N, Xu D, Møller IM (2018) CarbonylDB: a curated data-resource of protein carbonylation sites. Bioinformatics (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requena JR, Chao CC, Levine RL, Stadtman ER (2001) Glutamic and aminoadipic semialdehydes are the main carbonyl products of metal-catalyzed oxidation of proteins. Proc Natl Acad Sci USA 98: 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads DM, Umbach AL, Subbaiah CC, Siedow JN (2006) Mitochondrial reactive oxygen species: contribution to oxidative stress and interorganellar signaling. Plant Physiol 141: 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhrig H, John M, Schmidt J (2004) Modification of soybean sucrose synthase by S-thiolation with ENOD40 peptide A. Biochem Biophys Res Commun 325: 864–870 [DOI] [PubMed] [Google Scholar]
- Sainz M, Calvo-Begueria L, Pérez-Rontomé C, Wienkoop S, Abián J, Staudinger C, Bartesaghi S, Radi R, Becana M (2015) Leghemoglobin is nitrated in functional legume nodules in a tyrosine residue within the heme cavity by a nitrite/peroxide-dependent mechanism. Plant J 81: 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvato F, Havelund JF, Chen M, Rao RSP, Rogowska-Wrzesinska A, Jensen ON, Gang DR, Thelen JJ, Møller IM (2014) The potato tuber mitochondrial proteome. Plant Physiol 164: 637–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez C, Gates AJ, Meakin GE, Uchiumi T, Girard L, Richardson DJ, Bedmar EJ, Delgado MJ (2010) Production of nitric oxide and nitrosylleghemoglobin complexes in soybean nodules in response to flooding. Mol Plant Microbe Interact 23: 702–711 [DOI] [PubMed] [Google Scholar]
- Smuda M, Henning C, Raghavan CT, Johar K, Vasavada AR, Nagaraj RH, Glomb MA (2015) Comprehensive analysis of Maillard protein modifications in human lenses: effect of age and cataract. Biochemistry 54: 2500–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboleva A, Schmidt R, Vikhnina M, Grishina T, Frolov A (2017) Maillard proteomics: opening new pages. Int J Mol Sci 18: 2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller S, Li Y, Blüher M, Welch L, Hoffmann R (2017) Glycated lysine-141 in haptoglobin improves the diagnostic accuracy for type 2 diabetes mellitus in combination with glycated hemoglobin HbA1c and fasting plasma glucose. Clin Proteomics 14: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Lu L, Liu L, Liu W, Yu Y, Liu X, Hu Y, Jin C, Lin X (2014) Nitrate reductase-mediated early nitric oxide burst alleviates oxidative damage induced by aluminum through enhancement of antioxidant defenses in roots of wheat (Triticum aestivum). New Phytol 201: 1240–1250 [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43: D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrago L, Laugier E, Rey P (2009) Protein-repairing methionine sulfoxide reductases in photosynthetic organisms: gene organization, reduction mechanisms, and physiological roles. Mol Plant 2: 202–217 [DOI] [PubMed] [Google Scholar]
- Tekaia F, Yeramian E, Dujon B (2002) Amino acid composition of genomes, lifestyles of organisms, and evolutionary trends: a global picture with correspondence analysis. Gene 297: 51–60 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Thornalley PJ, Rabbani N (2009) Highlights and hotspots of protein glycation in end-stage renal disease. Semin Dial 22: 400–404 [DOI] [PubMed] [Google Scholar]
- Udvardi M, Poole PS (2013) Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol 64: 781–805 [DOI] [PubMed] [Google Scholar]
- Verzijl N, DeGroot J, Thorpe SR, Bank RA, Shaw JN, Lyons TJ, Bijlsma JW, Lafeber FP, Baynes JW, TeKoppele JM (2000) Effect of collagen turnover on the accumulation of advanced glycation end products. J Biol Chem 275: 39027–39031 [DOI] [PubMed] [Google Scholar]
- Vivancos PD, Dong Y, Ziegler K, Markovic J, Pallardó FV, Pellny TK, Verrier PJ, Foyer CH (2010) Recruitment of glutathione into the nucleus during cell proliferation adjusts whole-cell redox homeostasis in Arabidopsis thaliana and lowers the oxidative defence shield. Plant J 64: 825–838 [DOI] [PubMed] [Google Scholar]
- Vollenweider S, Weber H, Stolz S, Chételat A, Farmer EE (2000) Fatty acid ketodienes and fatty acid ketotrienes: Michael addition acceptors that accumulate in wounded and diseased Arabidopsis leaves. Plant J 24: 467–476 [DOI] [PubMed] [Google Scholar]
- Winger AM, Taylor NL, Heazlewood JL, Day DA, Millar AH (2007) The cytotoxic lipid peroxidation product 4-hydroxy-2-nonenal covalently modifies a selective range of proteins linked to respiratory function in plant mitochondria. J Biol Chem 282: 37436–37447 [DOI] [PubMed] [Google Scholar]
- Yin L, Mano J, Wang S, Tsuji W, Tanaka K (2010) The involvement of lipid peroxide-derived aldehydes in aluminum toxicity of tobacco roots. Plant Physiol 152: 1406–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Hisabori T (2016) Adenine nucleotide-dependent and redox-independent control of mitochondrial malate dehydrogenase activity in Arabidopsis thaliana. Biochim Biophys Acta 1857: 810–818 [DOI] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, et al. (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26: 1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]