JA preferentially inhibits auxin-induced lateral rooting via stabilization of Aux/IAA repressors.
Abstract
Plant root systems are indispensable for water uptake, nutrient acquisition, and anchoring plants in the soil. Previous studies using auxin inhibitors definitively established that auxin plays a central role regulating root growth and development. Most auxin inhibitors affect all auxin signaling at the same time, which obscures an understanding of individual events. Here, we report that jasmonic acid (JA) functions as a lateral root (LR)-preferential auxin inhibitor in Arabidopsis (Arabidopsis thaliana) in a manner that is independent of the JA receptor, CORONATINE INSENSITIVE1 (COI1). Treatment of wild-type Arabidopsis with either (−)-JA or (+)-JA reduced primary root length and LR number; the reduction of LR number was also observed in coi1 mutants. Treatment of seedlings with (−)-JA or (+)-JA suppressed auxin-inducible genes related to LR formation, diminished accumulation of the auxin reporter DR5::GUS, and inhibited auxin-dependent DII-VENUS degradation. A structural mimic of (−)-JA and (+)-coronafacic acid also inhibited LR formation and stabilized DII-VENUS protein. COI1-independent activity was retained in the double mutant of transport inhibitor response1 and auxin signaling f-box protein2 (tir1 afb2) but reduced in the afb5 single mutant. These results reveal JAs and (+)-coronafacic acid to be selective counter-auxins, a finding that could lead to new approaches for studying the mechanisms of LR formation.
Plant root systems adapt rapidly to environmental changes and enable the plant to survive adverse conditions (Fitter, 1996). Lateral roots extend horizontally from the primary root, contributing to the architecture of the root system. They also help anchor the plant in the soil and facilitate water, micronutrient, and macronutrient uptake (Lynch, 1995). Auxin is a multifunctional plant factor essential to root development, driving gravitropism (Chen et al., 1998; Rashotte et al., 2000; Sukumar et al., 2009), root hair formation (Masucci and Schiefelbein, 1994, 1996; Pitts et al., 1998), elongation and differentiation of root cells (Rahman et al., 2007), and lateral root (LR) formation (Reed et al., 1998; Casimiro et al., 2001; Bhalerao et al., 2002).
Auxin signaling is derived from the interaction between endogenous auxin indole-3-acetic acid (IAA), a TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX PROTEIN (TIR1/AFB) auxin receptor and a family of transcriptional repressors known as AUXIN/IAAs (Aux/IAAs; Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Salehin et al., 2015). In the presence of IAA, the SKP1-CULLIN1-F-BOX (SCF) TIR1/AFB ubiquitin ligase complex binds to Aux/IAAs, which triggers their ubiquitin-mediated degradation of Aux/IAA (Calderon-Villalobos et al., 2010). Most Aux/IAAs contain four conserved domains that contribute to their functional properties. Domain I is a repressor domain (Long et al., 2006; Szemenyei et al., 2008; Causier et al., 2012); domain II is involved in protein stability and is the site of interaction with the TIR1/AFBs and auxin (Gray et al., 2001; Ouellet et al., 2001; Ramos et al., 2001; Dreher et al., 2006); and domains III and IV are responsible for dimerization with other Aux/IAAs and heterodimerization with AUXIN RESPONSE FACTORs (ARFs; Ulmasov et al., 1997). Aux/IAAs themselves bind to and inhibit ARFs, which are DNA-binding transcription factors capable of directing the expression of auxin-responsive genes.
Inhibitors of auxin transport, biosynthesis, and signaling are widely used to block auxin bioactivities. The identification and functional characterization of the membrane proteins involved in polar auxin transport have been greatly facilitated by the use of auxin transport inhibitors such as 1-N-naphthylphthalamic acid (NPA), 2,3,5-triiodobenzoic acid, and gravacin (Casimiro et al., 2001; De Rybel et al., 2009). It was recently reported that l-amino-oxyphenylpropionic acid, l-kynurenine, yucasin, and aminooxy-naphthylpropionic acid were auxin biosynthesis inhibitors (Soeno et al., 2010; He et al., 2011; Nishimura et al., 2014; Narukawa-Nara et al., 2016). In auxin signaling, p-chlorophenoxyisobutyric acid (PCIB), PEO-IAA, and auxinole were reported (Oono et al., 2003; Hayashi et al., 2012). These signaling inhibitors competitively disturb diverse auxin responses, including primary root elongation, root gravitropism, root hair formation, and LR formation.
In Arabidopsis (Arabidopsis thaliana), perception of jasmonic acid (JA) takes place in a similar manner to that of auxin and involves analogous components, such as jasmonoyl-l-Ile (JA-l-Ile) as the ligand, COI1 (CORONATINE INSENSITIVE1) as the receptor, and jasmonate ZIM-domain (JAZ proteins) as coreceptors (Xie et al., 1998; Devoto et al., 2002; Xu et al., 2002; Chini et al., 2007; Thines et al., 2007; Browse, 2009; Chung and Howe, 2009; Yan et al., 2009; Sheard et al., 2010; Pauwels and Goossens, 2011). In the presence of the JA-l-Ile, JAZ repressor proteins are degraded by the action of the SCFCOI1-E3 ubiquitin ligase complex, releasing MYC2/3/4 transcription factors to trigger the expression of JA-dependent genes (Chini et al., 2007; Fernández-Calvo et al., 2011; Niu et al., 2011). It was recently reported that some jasmonates have bioactivities distinct from the COIl-dependent pathway. For example, 12-oxo-phytodienoic acid and JA-Trp competitively inhibit auxin signaling (Ribot et al., 2008; Staswick, 2009). In this work, we shed new light on the regulation of COI1-independent jasmonate signaling. We show that (−)-JA, (+)-JA and (+)-CFA have a peculiar bioactivity in the inhibition of LR formation, independent of the COI1 signaling pathway.
RESULTS
LR Number Was Reduced by (−)-JA and (+)-JA Independent of COI1
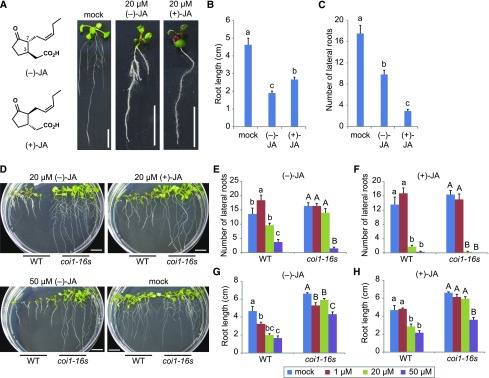
Arabidopsis plants separately treated with both enantiomers of JA exhibited similarly reduced primary root growth compared to mock (Fig. 1, A and B; Asamitsu et al., 2006). Although both (−)-JA and (+)-JA reduced the number of LR, the effect was greater for (+)-JA (Fig. 1C). To investigate whether (+)-JA also induced the JA responses, JA responsive gene expression, JAZ1-GUS degradation, and anthocyanin accumulation were analyzed. Expression of the enzyme ALLENE OXIDE SYNTHASE (AOS) from a COI1-dependent JA biosynthetic gene was induced by both (−)-JA or (+)-JA (Supplemental Fig. S1A; Park et al., 2002a). JAZ1-GUS degradation by either (−)-JA or (+)-JA treatment was rescued by either the proteasome inhibitor MG132, or jarin-1, a selective inhibitor of the JA-amino acid conjugating enzyme JASMONATE RESISTANT1 (Supplemental Fig. S1B; Meesters et al., 2014). Moreover, (−)-JA and (+)-JA both induced anthocyanin accumulation (Supplemental Fig. S1C), but less JA-L-Ile accumulation was observed after (+)-JA treatment compared to (−)-JA treatment, consistent with the low activity of JASMONATE RESISTANT1 for (+)-JA and the stronger binding activity between COI1-JAZ and (+)-JA-l-Ile (Guranowski et al., 2007; Fonseca et al., 2009; Supplemental Fig. S1D).
Figure 1.
Reduction in LR numbers induced by JAs is independent of COI1. A to C, Phenotype (A), the primary root length (B), and LR number (C) of 9-d-old wild-type (Col-0) seedlings grown on 1/2 MS plate containing 20 μm (−)-JA, 20 μm (+)-JA, or water (mock). Values are the means with se (n = 8–10). Different letters indicate significant differences between means (P < 0.05). D, Phenotypes of 10-d-old wild-type (Col-0) and coi1-16s seedlings grown on 1/2 MS plate containing 20 μm (−)-JA, 20 μm (+)-JA, 50 μm (−)-JA, or water (mock). E and F, Number of LR of 10-d-old wild-type (Col-0) and coi1-16s seedlings grown on 1/2 MS plate containing 1, 20, or 50 μm (−)-JA (E), 1, 20, or 50 μm (+)-JA (F), or mock. G and H, The primary root length of 10-d-old wild-type (Col-0) and coi1-16s seedlings grown on 1/2 MS plate containing 1, 20, or 50 μm (−)-JA (G), 1, 20, or 50 μm (+)-JA (H), or mock. Values are the means with se (n = 15–20). Different letters indicate significant differences between means (P < 0.05). Scale bars, 1 cm.
Low concentrations of JA induce COI1-dependent LR formation (Sun et al., 2009, 2011). We also observed a small stimulation of COI1-dependent LR formation upon treatment with a low concentration (1 μm) of (−)-JA, (±)-methyl-JA (MeJA), or (±)-JA, but treatments with either 50 μm (−)-JA or 20 or 50 μm (+)-JA reduced LR numbers in the temperature-dependent COI1 single mutant, coi1-16s, and the COI1 loss-of-function mutant coi1-1 (Fig. 1, D–F; Supplemental Figs. S2 and S3; Reymond et al., 2000; Chen et al., 2013). Slight reductions in primary root length were observed in coi1-16s at higher concentrations of (−)-JA or (±)-JA (Fig. 1, G and H).
ANTHRANILATE SYNTHASE α1 (ASA1), a JA-inducible biosynthetic enzyme of IAA that is associated with LR formation, is induced as a consequence of COI1-JAZ signaling (Sun et al., 2009). Treatment with 50 μm (−)-JA or (+)-JA decreased LR numbers despite induction of ASA1 and the accumulation of IAA in wild type and also decreased LR number with slight ASA1 induction and the accumulation of IAA in coi1-16s (Supplemental Fig. S4). These results suggest that inhibition of LR formation by (−)-JA and (+)-JA is independent of the induction of ASA1, and hence of COI1 regulation.
The Reduction of LR Number Was Not Induced by JA Derivatives
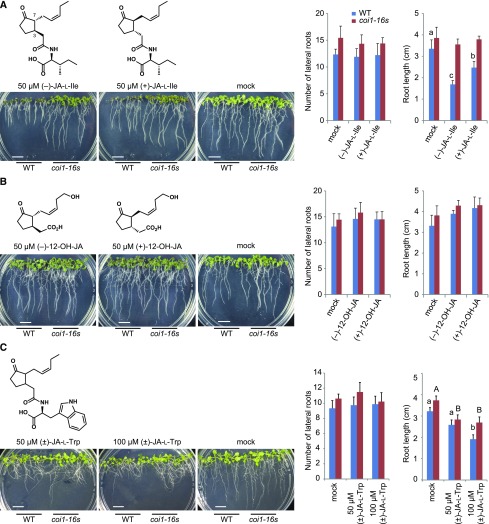
To identify the active form of JA responsible for inhibiting LR formation, we examined the effects of known bioactive JA metabolites in plants, such as (−)-JA-l-Ile for JA response and (−)-12-OH-JA just for tuberization in the potato (Solanum tuberosum; Fonseca et al., 2009; Seto et al., 2009). In contrast to the JA isomers, (−)-JA-l-Ile, (+)-JA-l-Ile, (−)-12-OH-JA, and (+)-12-OH-JA did not inhibit LR formation at 50 μm (Fig. 2, A and B; Supplemental Fig. S5, A and B). It should be emphasized that after treatments with (±)-JA-Trp (JA-Trp), which was reported to be an auxin inhibitor, even at 100 μm, root architectures were distinctly different from those induced by JAs in both wild type and the coi1 mutant (Fig. 2C; Supplemental Fig. S5C; Staswick, 2009).
Figure 2.
Effects of known bioactive JA metabolites on LR number and the primary root length. A, Phenotype, LR number, and the primary root length of 9-d-old wild-type (Col-0) and coi1-16s seedlings grown on 1/2 MS plate containing 50 μm (−)-JA-l-Ile, 50 μm (+)-JA-l-Ile, or 0.05% ethanol (mock). B, Phenotype, LR number, and the primary root length of 9-d-old wild-type (Col-0) and coi1-16s seedlings grown on 1/2 MS plate containing 50 μm (−)-12-OH-JA, 50 μm (+)-12-OH-JA, or water (mock). C, Phenotype, LR number, and the primary root length of 9-d-old wild-type (Col-0) and coi1-16s seedlings grown on 1/2 MS plate containing 50 or 100 μm (±)-JA-l-Trp or 0.1% DMSO (mock). Scale bars, 1 cm. Values are the means with se (n = 9–10). Different letters indicate significant differences between means (P < 0.05).
JA Suppressed the Genes Related to Auxin-Inducible LR Formation
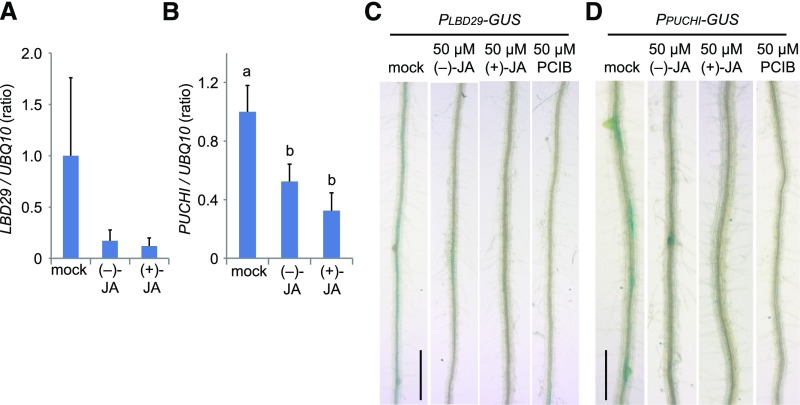
To reveal the molecular mechanism for JA-mediated inhibition of LR formation, (−)-JA- and (+)-JA-induced gene expression profiles were analyzed using a 44K microarray. We identified different classes of JA-responsive genes by two criteria: first, expression was unaffected by 50 μm MeJA, which is converted to JA and then to bioactive jasmonoyl-Ile in plants (independent of COI1-JAZ signaling; Browse, 2009); and second, expression level was changed >2.5-fold by (+)-JA compared to (−)-JA treatment. Three transcription factor genes, LATERAL ORGAN BOUNDARIES DOMAIN (LBD)29, PUCHI, and GATA23, all known to be auxin-inducible genes and related to PIN-FORMEDs regulation leading to LR formation, were dramatically down-regulated in response to JA treatment (Supplemental Table S1; Hirota et al., 2007; Okushima et al., 2007; De Rybel et al., 2010). Quantitative reverse transcription (RT)-PCR revealed that only PUCHI was downregulated by 50 μm (−)-JA or (+)-JA in the roots, although the expression levels of PUCHI and LBD29 were considerably reduced (Fig. 3, A and B). Consistent with the microarray data, GUS reporter expression around LR primordia in PLBD29-GUS and PPUCHI-GUS was suppressed by 50 μm (−)-JA or (+)-JA, as well as by the antiauxin-like compound, p-chlorophenoxyisobutyric acid (PCIB; Fig. 3, C and D; Oono et al., 2003; Biswas et al., 2007).
Figure 3.
Expression of LBD29 and PUCHI in seedlings treated with JAs. A and B, Quantitative RT-PCR data of LBD29 (A) and PUCHI (B) in 4-d-old wild type (Col-0) roots grown on 1/2 MS plate containing 50 μm (−)-JA, 50 μm (+)-JA, or water (mock). Values are the means with sd (n = 3). Different letters indicate significant differences between means (P < 0.05). C and D, GUS localizations in PLBD29-GUS (C) and PPUCHI-GUS (D) in roots. Four-day-old seedlings were transferred to 1/2 MS plate containing 50 μm (−)-JA, 50 μm (+)-JA, 50 μm PCIB, or 0.04% dimethyl sulfoxide (mock) in the absence or presence of 0.1 μm IAA and grown for an additional 3 d. Scale bars, 0.5 mm.
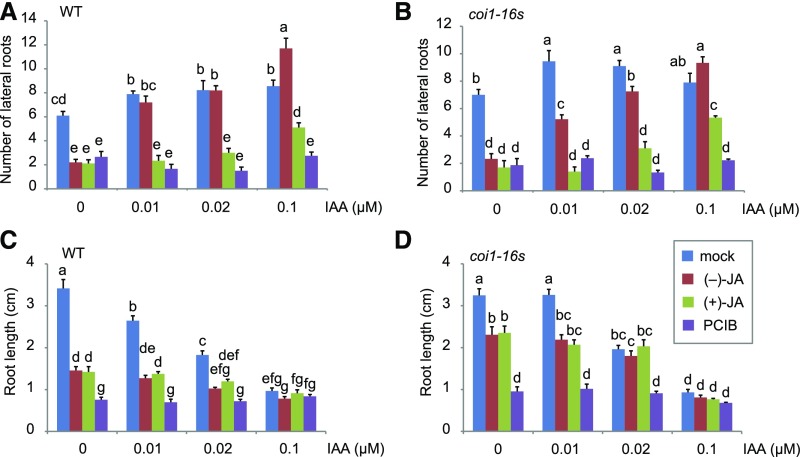
These expression changes may explain phenotypes of suppression of LR formation by the treatment of (−)-JA and (+)-JA under LR promotion by auxin signaling. Therefore, the effects for LR number were analyzed by the cotreatment with IAA and JAs. Cotreatment with various concentrations of IAA reduced the inhibitory effects of (−)-JA and (+)-JA on LR formation in a concentration-dependent manner in wild type, coi1-16s, and coi1-1 mutants, with a slight reduction of the primary root length (Fig. 4, A–D; Supplemental Fig. S6).
Figure 4.
Effects by the cotreatment of JAs and IAA on LR number and the primary root length. A and B, LR number of wild-type (Col-0; A) and coi1-16s (B). C and D, The primary root length of wild type (Col-0; C) and coi1-16s (D). Four-day-old seedlings were transferred to 1/2 MS plate containing 0.04% DMSO (mock), 50 μm (−)-JA, 50 μm (+)-JA, or 50 μm PCIB in the absence or presence of 0.1 μm IAA and grown for an additional 3 d. Values are the means with se (n = 9–10). Different letters indicate significant differences between means (P < 0.05).
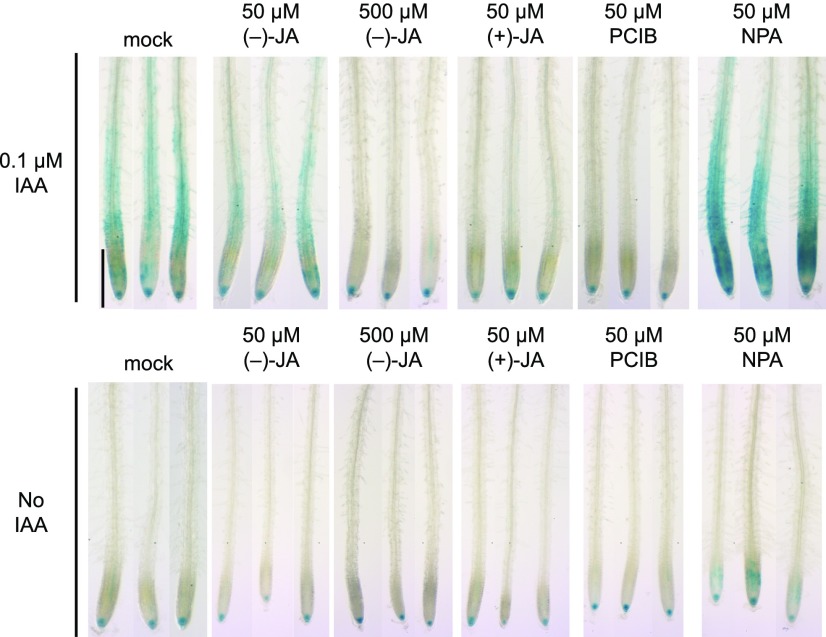
To assess the effects of JA on auxin signaling directly, auxin-responsive gene expression was analyzed using the DR5::GUS reporter line (Ulmasov et al., 1997). (−)-JA at a concentration of 50 μm inhibited IAA-induced DR5::GUS expression, but 50 μm (+)-JA completely eliminated the response, as did 50 μm PCIB and 500 μm (−)-JA treatments (Fig. 5). Polar auxin transport is essential for LR formation, and the auxin transport inhibitor NPA inhibits LR formation (Casimiro et al., 2001). In contrast to the inhibition of DR5::GUS expression by JA and PCIB, NPA enhanced DR5::GUS expression (Fig. 5). This result suggests that (−)-JA might reduce auxin signaling, but not auxin transport.
Figure 5.
Expression patterns of the DR5::GUS reporter line in roots of seedlings treated with JAs. Expression patterns of DR5::GUS in roots. Five-day-old seedlings were incubated in 1/2 MS liquid medium with 0.04% DMSO (mock) or indicated compounds for 6 h before development. Scale bar, 0.5 mm.
JAs Stabilized Aux/IAA
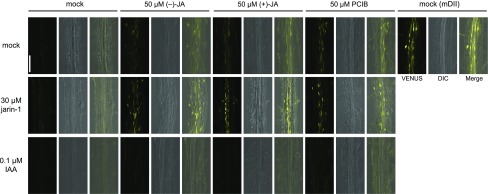
To reveal whether (−)-JA targets the auxin-signaling cascade, further analysis was performed using the DII-VENUS reporter line as an auxin sensor (Brunoud et al., 2012). Degradation of DII-VENUS fusion protein is accelerated by IAA via the SCFTIR1/AFB pathway. The auxin signal is repressed by PCIB, indicated by the accumulation of DII-VENUS in the root maturation zone and the root tip (Fig. 6; Supplemental Fig. S7). Similarly, 50 μm (−)-JA or (+)-JA treatment caused the accumulation of DII-VENUS fluorescence in the root maturation zone (Fig. 6). This fluorescence was enhanced by cotreatment with jarin-1, which suppresses the conversion of JA to JA-Ile and increased JA concentration, and diminished by cotreatment with IAA in both the maturation zone and the root tip (Fig. 6; Supplemental Figure S7; Meesters et al., 2014). These data are consistent with the loss of SCFTIR1/AFB activity and, hence, auxin signaling in the root maturation zone where LRs elongate (Fig. 1).
Figure 6.
DII-VENUS fluorescence in seedlings treated with JAs. Laser scanning confocal micrographs of DII-/mDII-VENUS at the root maturation zone. Left, fluorescent image of DII-VENUS (false-colored yellow). Center, DIC image. Right, merge of DII-VENUS and DIC. Seedlings were incubated in 1/2 MS liquid medium with 0.05% DMSO (mock), 50 μm (−)-JA, 50 μm (+)-JA, or 50 μm PCIB in mock, 30 μm jarin-1, or 0.1 μm IAA for 3 h. Scale bar, 0.1 mm.
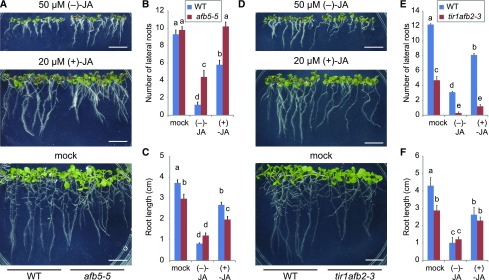
To elucidate the relationships between TIR1/AFB1–5 receptors and JA, we analyzed tir1afb2-3 and afb5-5 mutants. Their treatment with (−)-JA or (+)-JA caused a decrease in the number of LR in tir1afb2-3 compared with wild type, whereas afb5-5 resisted (−)-JA and (+)-JA and no reduction in LR formation was observed (Fig. 7).
Figure 7.
Effects of JAs on afb5 and tir1afb2 mutant lines compared to wild type. A, Phenotype of 9-d-old wild-type (WT; Col-0) or afb5-5 seedlings grown on 1/2 MS plate containing 50 μm (−)-JA, 20 μm (+)-JA, or water (mock). B and C, LR number (B) and the primary root length (C) in 9-d-old wild-type (Col-0) or afb5-5 seedlings grown on 1/2 MS plate containing 50 μm (−)-JA, 20 μm (+)-JA, or water (mock). D, Phenotype of 9-d-old wild-type (Col-0) or tir1afb2-3 seedlings grown on 1/2 MS plate containing 50 μm (−)-JA, 20 μm (+)-JA, or water (mock). E and F, LR number (E) and the primary root length (F) in 9-d-old wild-type (Col-0) or tir1afb2-3 seedlings grown on 1/2 MS plate containing 50 μm (−)-JA, 20 μm (+)-JA, or water (mock). Values are the means with se (n = 9–10). Different letters indicate significant differences between means (P < 0.05). Scale bars, 1 cm.
Coronafacic Acid Was an LR-Preferential Auxin Inhibitor
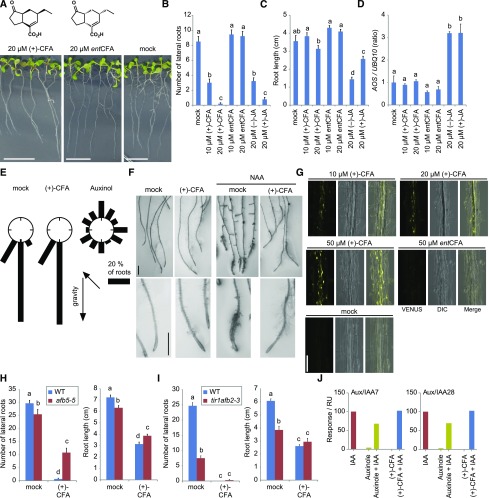
Jasmonic acids are converted to JA-l-Iles, which induce COI1-dependent responses, and so coi1 mutants are necessary to analyze JA treatment-induced LR formation (Supplemental Fig. S1). Accordingly, we sought an LR-preferential auxin inhibitor that does not induce COI1-dependent JA responses. Coronatine is a phytotoxin produced by Pseudomonas syringae and known to function as a structural mimic of (+)-7-iso-JA-l-Ile in planta. The structure of coronatine consists of coronafacic acid (+)-CFA (an analog of MeJA) and coronamic acid, joined by an amide bond between the acid group of CFA and the amino group of coronamic acid, and so (+)-CFA could be considered as a structural mimic of (+)-7-iso-JA (Supplemental Fig. S8; Egoshi et al., 2016). Treatment of wild type with (+)-CFA dramatically suppressed LR formation, with only slight effects on primary root growth (Fig. 8, A–C). Enantiomeric (−)-CFA did not noticeably affect LR numbers (Fig. 8, A–C). Gene expression of AOS was not observed after treatment with either (+)-CFA or (−)-CFA, suggesting that (+)-CFA can indeed mimic JA, without inducing a COI1-dependent JA response (Fig. 8D). (+)-CFA treatment did not affect root gravitropism or auxin-inducible root hair formation (Fig. 8, E and F). Treatment with 10 μm (+)-CFA induced accumulation of DII-VENUS fluorescence (Fig. 8G), but treatment with (−)-CFA did not. As was found for the JA treatments, (+)-CFA decreased LR numbers in tir1afb2-3 compared with wild type, whereas afb5-5 resisted (+)-CFA-induced reductions in LR formation (Fig. 8, H and I). We also tested whether (+)-CFA cross-reacted with auxin receptors using surface plasmon resonance (SPR) assays; however, (+)-CFA exhibited neither auxin-like nor antiauxin-like activity, when binding to either AFB5-Aux/IAA7 or 28 (Fig. 8J; Supplemental Fig. S9).
Figure 8.
The effects of CFA on the wild type. A–C, Phenotype of 8-d-old wild-type (Col-0) seedlings grown on 1/2 MS plate containing 20 μm (+)-/(−)-CFA or 0.05% ethanol (mock). LR number (B), the primary root length (C), and expression patterns of AOS (D) in 8-d-old wild-type (Col-0) seedlings grown on 1/2 MS plate containing 10 μm or 20 μm (+)-CFA, 10 μm or 20 μm (−)-CFA, 20 μm (−)-JA, 20 μm (+)-JA, or 0.05% ethanol (mock). Values are the means with se (n = 18–20). Different letters indicate significant differences between means (P < 0.05). Scale bars, 1 cm. E, Effects of (+)-CFA on root gravitropism. Five-day-old seedlings were vertically grown in the first gravity direction (first g) with 50 μm (+)-CFA or 5 μm auxinole and then rotated at an angle of 135° (second g) and cultured for an additional 24 h. The angles were grouped into 30° classes and plotted as circular histograms (n = 40). F, Root hair growth of 5-d-old Col-0 with 50 μm (+)-CFA or 0.05% ethanol (mock) with/without 0.5 μm naphthalene1-acetic acid (NAA) for 2 d. Scale bars, 1 mm. G, Laser scanning confocal micrographs of DII-VENUS at root maturation zone. Five-day-old seedlings were incubated in 1/2 MS liquid medium with 10, 20, or 50 μm (+)-CFA or 50 μm (−)-CFA or 0.05% ethanol (mock) for 3 h. Scale bars, 0.1 mm. H, LR number and primary root length in 12-d-old wild-type (Col-0) or afb5-5 seedlings grown on 1/2 MS plate containing 50 μm (+)-CFA or 0.05% ethanol (mock). I, LR number and primary root length in 11-d-old wild-type (Col-0) or tir1afb2-3 seedlings grown on 1/2 MS plate containing 50 μm (+)-CFA or 0.05% ethanol (mock). Values are the means with se (n = 9–10). Different letters indicate significant differences between means (P < 0.05). J, Surface plasmon resonance assays of AFB5 auxin and antiauxin activities using (+)-CFA. All sensorgram data are normalized to the signal from 5 μm IAA (red, 100%). Auxinole is used as a control for antiauxin activity, using 50 μm auxinole. (+)-CFA were added at 50 μm, and with 5 μm IAA for the antiauxin assay. The SPR chip was coated with biotinylated Aux/IAA7-DII or Aux/IAA28-DII peptide and binding was recorded at the end of the association phase.
DISCUSSION
(−)-JA and (+)-JA have been implicated in the inhibition of LR formation in Arabidopsis, independent of COI1 signaling (Fig. 1). Both (−)-JA and (+)-JA stabilize DII-VENUS, indicating that their activity stabilizes domain II of Aux/IAAs and therefore suppresses the expression of auxin-inducible genes related to LR formation, such as LBD29 and PUCHI (Figs. 3, 6, and 7). Furthermore, (+)-CFA was found to be an active structural mimic of (−)-JA, establishing it as a preferential-auxin inhibitor for LR formation (Fig. 8).
Bioactivities of JA
JAZ degradation, induction of JA-responsive genes, anthocyanin accumulation, chlorophyll accumulation, and growth defects are also observed by the treatment of high JA concentration, as well as low JA concentration and wounding stress (Lorenzo et al., 2004; Chini et al., 2007; Fonseca et al., 2009; Fernández-Calvo et al., 2011; Boter et al., 2015). Moreover, the COI1-independent LR inhibition dominated at higher concentrations, despite ASA1 induction and the accumulation of IAA both in wild type and coi1-16s (Supplemental Fig. S4). Conversely, a low concentration of JA has been reported to stimulate LR formation and to inhibit primary root elongation in a COI1-dependent manner (Sun et al., 2009, 2011; Chen et al., 2011). Small stimulation of COI1-dependent LR formation and inhibition of primary root elongation upon treatment with a lower concentration of either (−)-JA, (+)-JA, (±)-methyl-JA, or (±)-JA was also observed (Fig. 1; Supplemental Fig. S3). These results suggest that JA modulates LR formation via regulatory pathways transduced both by COI1-dependent and COI1-independent signaling. The positive/negative effect on LR formation depends on the concentration of JA.
Stabilization of Aux/IAAs by JAs and (+)-CFA
There are many known mutations in domain II of the Aux/IAA protein that confer improved functional activity and increase the stability of the protein, such as iaa1/axr5 (Park et al., 2002b; Yang et al., 2004), iaa3/shy2 (Tian and Reed, 1999), iaa6/shy1 (Reed, 2001), iaa7/axr2 (Nagpal et al., 2000), iaa12/bdl (Hamann et al., 2002), iaa14/slr (Fukaki et al., 2002), iaa17/axr3 (Rouse et al., 1998), iaa18/crane (Reed, 2001; Uehara et al., 2008; Ploense et al., 2009), iaa19/msg2 (Tatematsu et al., 2004), and iaa28 (Rogg et al., 2001). These mutants exhibit various abnormal phenotypes involved in auxin-mediated growth and development, as well as altered gene expression in response to auxin. The phenotypes are distinct in each Aux/IAA mutant, although some persistent similarities suggest that each Aux/IAA has both similar and unique roles in the auxin response. The phenotypes of preferential LR inhibition by JAs and (+)-CFA was most similar to those of iaa3/shy2, iaa14/slr, iaa18/crane, and iaa28 gain-of-function mutants (Figs. 1 and 8, A–D). The DII-VENUS reporter line was designed on IAA28 (Brunoud et al., 2012), and the stabilization of DII-VENUS by JAs and (+)-CFA would at least explain similar phenotypes in the inhibition of LR formation observed in both iaa28 gain-of-function mutants, and JA- and (+)-CFA-treated plants (Figs. 6 and 8E).
During LR formation, degradation of IAA14 and IAA28 activates the transcriptional activity of ARF5, 6, 7, 8, and 19 (De Rybel et al., 2010). Of these ARFs, ARF7 and ARF19 directly activate LBD16 and LBD29, leading to the induction of PUCHI and LR primordium development (Hirota et al., 2007; Okushima et al., 2007; Lavenus et al., 2015). LBD29 and PUCHI were found to be suppressed by JA treatments (Fig. 3), suggesting that JAs might also stabilize IAA14.
Relationships between JAs or (+)-CFA and TIR1/AFB Receptors
The degradation of Aux/IAAs by ubiquitination is initiated by auxin perception at the TIR1/AFB receptors (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). Previous phylogenetic studies revealed that TIR1/AFBs can be classified into three clades, represented by TIR1, AFB2, and AFB5 in Arabidopsis. The functionality of these receptors was found to overlap, but a functional difference was clearly demonstrated in planta; the tir1afb2 double mutant is deficient in a variety of auxin-regulated growth processes, including hypocotyl elongation and LR formation, and the afb5 mutant was tolerant to the piclolinate family of auxinic herbicides (Walsh et al., 2006; Parry et al., 2009; Prigge et al., 2016). Compared with wild type, afb5-5 showed resistance to the reductions in LR formation by JAs and (+)-CFA treatments, but tir1afb2-3 did not (Figs. 7 and 8, F–K). On the other hand, suppression of auxin-induced DR5::GUS and accumulation of DII-VENUS were observed after treatments of JA and (+)-CFA in wild-type plants (Figs. 5 and 6). Both TIR1 and AFB5 seem to be involved in the inhibition of Aux/IAA degradation by JAs and (+)-CFA, but it seems that AFB5 might have a dominant role, leading to reduced Aux/IAA breakdown and inhibition of LR formation.
While DII-VENUS accumulated in the wild type after treatments of (+)-CFA, SPR assays revealed that (+)-CFA neither agonized nor antagonized binding of the AFB5-Aux/IAA complex. This suggests that (+)-CFA may not interact with the auxin-binding pocket in AFB5 (Figs. 6 and 8). The mutant of SGT1B, which is required for the degradation of Aux/IAA proteins, also showed resistance to picolinate auxins, as well as afb5 mutant (Walsh et al., 2006). The LR-preferential phenotype and the accumulation of DII-VENUS after treatments with JA and (+)-CFA suggest that their target might be proteins related to the Aux/IAAs degradation system (Figs. 1, 6, and 8).
Structural Requirements for JA-Mediated Inhibition of LR Formation
In our study, (−)-JA, (+)-JA, and (+)-CFA induced inhibition of LR formation. In Arabidopsis, (+)-7-iso-JA (with the 7S configuration) was biosynthesized and converted into (+)-7-iso- JA-l-Ile (Supplemental Fig. S8; Fonseca et al., 2009). However, (+)-7-iso-JA is easily epimerized to thermodynamically stable (−)-JA to give a mixture of 95% (−)-JA and 5% (+)-7-iso-JA (Mueller and Brodschelm, 1994). The decrease in LR number after treatment with (+)-7S-JA was more pronounced than that of (−)-7R-JA (Fig. 1, E and G). Considering the strong bioactivity of (+)-CFA, which is conformationally fixed in the 7S configuration, the S configuration at the C-7 position of JA seems likely to contribute to bioactivity in COI1-independent LR formation (Fig. 8).
CONCLUSION
Both enantiomers of JA and (+)-CFA have been confirmed to inhibit LR formation by Aux/IAA stabilization, independent of COI1 signaling. The study has also identified (+)-CFA as a chemical, specialized, and conditional LR auxin inhibitor, showing similar LR phenotypes to those observed in several aux/iaa mutants. This inhibitor could prove a useful tool for a deeper understanding of the molecular basis of LR formation.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Chemical Treatment
Arabidopsis (Arabidopsis thaliana) accession Col-0 was used as the wild type in all experiments. Seeds of P35S-JAZ1:GUS reporter line (Meesters et al., 2014), DR5::GUS (Ulmasov et al., 1997), DII-VENUS (Brunoud et al., 2012), PPUCHI-GUS (Hirota et al., 2007), and PLBD29-GUS (Okushima et al., 2007) were obtained as previously described. Seeds of coi1-1 and coi1-30 mutants were provided by Dr. R. Solano and Dr. A. Chini (National Center for Biotechnology, Madrid, Spain) and those of coi1-16s by Dr. H. Ohta (Tokyo Institute of Technology; Chen et al., 2013). The lines tir1 afb2-3 and afb5-5 were gifts from Dr. Mark Estelle (University of California at San Diego, CA). Sterilized seeds were sown on half-length Murashige-Skoog (1/2 MS) medium (2% [w/v] Suc, pH 5.8) hardened with 0.4% (w/v) gellan gum (Wako) and then vernalized for 3 d at 4°C. Seedlings were grown at 22°C under 12 h light (100–118 μmol·m−2·s−1; cool-white fluorescent light)/12 h dark in a BIOTRON LPH-240SP growth chamber (Nippon Medical and Chemical Instruments). For jasmonate quantification and expression analysis of JA responsive genes, seedlings were grown on 48-well plates containing 1/2 MS liquid medium (0.5% [w/v] Suc, pH 5.8) in the same chamber. The number of lateral roots and the primary root length were determined using a dissecting microscope and ImageJ software (https://imagej.nih.gov/ij/). Chemicals were stored as stock solutions, then directly added to the 50°C–60°C medium after autoclaving. For preparation of stock solutions, JA and (–)-12-OH-JA were dissolved in water and then filter sterilized (0.22 μm). Ethanol was used as the solvent for MeJA, JA-l-Ile, and CFA, and DMSO was used for jarin-1 (Meesters et al., 2014), JA-Trp, IAA, PCIB (Oono et al., 2003), and NPA.
Chemicals
(±)-JA, prepared by hydrolysis of (±)-MeJA (Nihon-Zeon, Japan), was condensed with l-Trp-Me ester (Wako), followed by hydrolysis to yield the product as a yellow solid (Ueda et al., 2017). (−)-TA, (+)-TA, (+)-CFA, (−)-CFA, and Trp conjugate of (±)-JA were prepared as previously described (Nakamura et al., 2008, 2011). IAA (Wako), NPA (Tokyo Chemical Industry), and PCIB (Sigma-Aldrich) were purchased as fine chemicals and used as supplied.
Jasmonate Quantification
Jasmonates were extracted from plants and quantified by UPLC (Agilent)-TOFMS (Bruker) as described (Kurotani et al., 2015). In brief, 50–100 mg fresh weight samples with 100 ng/g JA-d6-Ile were homogenized in ethanol. After removing the debris by centrifugation, the supernatant was dried with N2 gas, then dissolved in water. The aqueous solution (recovery rate of JA-d6-Ile, 12.5%) was used for the following UPLC-TOF-MS measurement.
Anthocyanin Quantification
Anthocyanin was extracted from plants and quantified as described (Neff and Chory, 1998). Seedlings were snap-frozen with liquid N2, then homogenized in methanol acidified with 1% (w/v) HCl and allowed to stand overnight at 4°C in the dark for efficient extraction. Sterile water was added to the lysate to give 60% (w/w) methanol, followed by addition of equal amount of chloroform. After vigorous mixing and centrifugation, the aqueous layer was carefully collected, and absorbance measured at 530 nm and 657 nm using GeneQuant1300 (GE Healthcare).
IAA Quantification
IAAs were extracted from plants and quantified with a UPLC (Agilent)-TOFMS (Bruker) system as described (Barkawi et al., 2008; Kurotani et al., 2015). In brief, 10 to 50 mg fresh weight samples with 10 ng/g d5-IAA were homogenized in extraction buffer (65% [v/v] isopropanol and 35% [w/v] imidazole, pH 7.0). After removal of the debris by centrifugation, the supernatant was dried with N2 gas, then dissolved in 28% (v/v) aqueous acetonitrile containing 0.05% (w/v) acetic acid. The solution was used for the following UPLC-TOF-MS measurement.
Histochemical Staining of GUS
Transgenic plants expressing GUS were grown on 1/2 MS plates for 4 d, then transferred to plates containing the indicated compounds and grown for an additional 3 d. For histochemical staining, samples were immersed in GUS staining solution (40 mm Na-phosphate buffer [Wako] containing 1 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide [Wako], 3 mm K4[Fe(CN)6] [Wako], 0.5 mm K3[Fe(CN)6] [Wako] in 20% [v/v] methanol). After 30 min staining, samples were incubated in 70% [v/v] ethanol. Stemi 2000-C (Carl Zeiss), equipped with AxioCam ERc 5s (Carl Zeiss), was used to observe and photograph the samples, following the manufacturer’s instructions.
Quantitative RT-PCR
Immediately after sampling, seedlings were frozen with liquid N2. Extraction and purification of total RNA from plant samples were performed with RNeasy Plant Mini Kit (Qiagen), and the reverse transcription was achieved by ReverTra Ace (Toyobo). Quantitative RT-PCR was conducted with StepOnePlus real-time PCR system (Applied Biosystems) with primers (Supplemental Table S2). The sizes and sequences of the amplified fragments were confirmed by agarose gel electrophoresis and sequencing. AOS, ASA1, LBD29, and PUCHI were quantified with UBQ10.
Microarray Analysis
Total RNA was extracted from roots from 4-d-old seedlings using RNeasy Plant Mini Kit (QIAGEN). The microarray analyses, hybridization, data analyses, and data mining were carried out as previously described (Bashir et al., 2011). In brief, cDNAs were synthesized using 500 ng of total RNA and labeled with one color (Cy3) using a Quick Amp labeling kit (Agilent Technologies), followed by fragmentation and hybridization to the Arabidopsis Oligo 44K DNA microarray (Ver. 4.0; Agilent Technologies). Three biological replicates were performed for each treatment, making a total of nine hybridizations. All arrays were scanned with a microarray scanner (G2505B; Agilent Technologies) and analyzed using GeneSpring Ver. 11 (Agilent Technologies). The Student's t test (P value) was used as a parametric test and the Benjamini and Hochberg false-discovery rate (q value) procedure was used to control the certainty level. Genes with at least a 2-fold difference in their expression levels and a q value < 0.05 were considered to be differentially expressed.
DII-VENUS Analysis
Six-day-old DII-VENUS seedlings were incubated with indicated compounds for 3 h. For jarin-1 treatment, seedlings were treated with jarin-1 for 1 h before the application of indicated compounds. Half-strength MS liquid medium (0.5% [w/v] Suc, pH 5.8) was used as an incubation buffer. After the treatment, roots were cut off and mounted on slide grasses with 1:1 phosphate-buffered saline:glycerol solution. Photographic and fluorescent images of DII-VENUS seedlings were taken with an LSM-710 laser scanning confocal microscope (Carl Zeiss), following the manufacturer’s instructions. Acquisition parameters were as follows: master gain was always set for 726 with a digital gain of 1, a digital offset of 3, excitation at 488 nm (2%), and emission at 490 to 555 nm.
Microscopy
Photographic and fluorescent images were taken with LSM-710 laser scanning confocal microscope (Carl Zeiss). For histochemical analysis using transgenic plants carrying GUS, a Stemi 2000-C equipped with AxioCam ERc 5s (Carl Zeiss) was used to observe and photograph the samples, following the manufacturer’s instructions.
Surface Plasmon Resonance
Surface plasmon resonance assays were done as described (Lee et al., 2014). Jasmonates were dissolved as stocks at 10 mm in DMSO and diluted to 50 µm for the assay with controls run in the equivalent DMSO concentration. Auxin-like activity and antiauxin activity were compared in each run against signals for IAA, with auxin-like activity recorded if the test compound acted to assemble the respective coreceptor complex (acting as “molecular glue”), and antiauxin activity recorded as a reduction in IAA-supported coreceptor complex. In the antiauxin assay, IAA was present at 5 µm, the test compound at 50 µm.
Statistical Analysis
The data were analyzed with one-way ANOVA followed by SNK post-hoc test among all means. Statistical analyses were conducted using CoStat Version 6.400 (CoHort Software).
Accession Numbers
COI1, AT2G39940; JAZ1, AT1G19180; TIR1, AT3G62980; AFB2, AT3G26810; AFB5, AT5G49980; Aux/IAA7, AT3G23050; Aux/IAA28, AT5G25890; AOS, AT5G42650; ASA1, AT5G05730; LBD29, AT3G58190; and PUCHI, AT5G18560.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Effects of (+)-JA on wild type.
Supplemental Figure S2. Effects of (−)-JA or (+)-JA on coi1-1 null mutant.
Supplemental Figure S3. Effects of (±)-MeJA or (±)-JA treatment on LR number and primary root length.
Supplemental Figure S4. Expressions of ASA1 and IAA concentration treated with JAs.
Supplemental Figure S5. Effects of known bioactive JA metabolites on LR number and the primary root length on coi1-1 null mutant.
Supplemental Figure S6. Effects by the cotreatment of JAs and IAA on LR number and the primary root length on coi1-1 null mutant.
Supplemental Figure S7. DII-VENUS fluorescence at the root tip.
Supplemental Figure S8. Structures of JAs and CFA.
Supplemental Figure S9. AFB5 antiauxin activity sensorgram.
Supplemental Table S1. Summary of the microarray analysis.
Supplemental Table S2. Primers for quantitative RT-PCR.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dr. R. Solano and Dr. A. Chini (National Center for Biotechnology), Dr. H. Ohta (Tokyo Institute of Technology), and Dr. M. Estelle (University of California at San Diego) for providing mutants and reporter line seeds. We also thank Dr. T. Kakimoto (Osaka University), Dr. T. Usui (Tsukuba University), Dr. T. Chinen (Tsukuba University), Dr. T. Hashimoto (Nara Institute of Science and Technology), Dr. H. Takahashi, Dr. S. Tamura, Dr. Y. Takaoka, Dr. T. Oikawa, Mr. K. Nakatani, and Mr. H. Kaneko (Tohoku University) for discussions and help. We thank laboratory members for supporting the research.
Footnotes
This work was supported in part by the Ministry of Education, Culture, Sports, Science, and Technology through a Grant-in-Aid for Scientific Research (no. 23102012 to M.U.) on Innovative Areas “Chemical Biology of Natural Products (no. 2301),” by the Japan Society for the Promotion of Science (JSPS; no. 17H00885 to M.U.), by a JSPS Grant-in-Aid for Scientific Research (no. 26282207 to M.U.), by a SUNBOR grant (to Y.I.), and by a Botanical Research Grant from the New Technology Development Foundation (to Y.I.).
References
- Asamitsu Y, Nakamura Y, Ueda M, Kuwahara S, Kiyota H (2006) Synthesis and odor description of both enantiomers of methyl 4,5-didehydrojasmonate, a component of jasmin absolute. Chem Biodivers 3: 654–659 [DOI] [PubMed] [Google Scholar]
- Barkawi LS, Tam Y-Y, Tillman JA, Pederson B, Calio J, Al-Amier H, Emerick M, Normanly J, Cohen JD (2008) A high-throughput method for the quantitative analysis of indole-3-acetic acid and other auxins from plant tissue. Anal Biochem 372: 177–188 [DOI] [PubMed] [Google Scholar]
- Bashir K, Ishimaru Y, Shimo H, Nagasaka S, Fujimoto M, Takanashi H, Tsutsumi N, An G, Nakanishi H, Nishizawa NK (2011) The rice mitochondrial iron transporter is essential for plant growth. Nat Commun 2: 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao RP, Eklöf J, Ljung K, Marchant A, Bennett M, Sandberg G (2002) Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J 29: 325–332 [DOI] [PubMed] [Google Scholar]
- Biswas KK, Ooura C, Higuchi K, Miyazaki Y, Van Nguyen V, Rahman A, Uchimiya H, Kiyosue T, Koshiba T, Tanaka A, et al. (2007) Genetic characterization of mutants resistant to the antiauxin p-chlorophenoxyisobutyric acid reveals that AAR3, a gene encoding a DCN1-like protein, regulates responses to the synthetic auxin 2,4-dichlorophenoxyacetic acid in Arabidopsis roots. Plant Physiol 145: 773–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M, Golz JF, Giménez-Ibañez S, Fernandez-Barbero G, Franco-Zorrilla JM, Solano R (2015) FILAMENTOUS FLOWER is a direct target of JAZ3 and modulates responses to jasmonate. Plant Cell 27: 3160–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, et al. (2012) A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482: 103–106 [DOI] [PubMed] [Google Scholar]
- Calderon-Villalobos LI, Tan X, Zheng N, Estelle M (2010) Auxin perception—structural insights. Cold Spring Harb Perspect Biol 2: a005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inzé D, Sandberg G, Casero PJ, et al. (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B (2012) The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sonobe K, Ogawa N, Masuda S, Nagatani A, Kobayashi Y, Ohta H (2013) Inhibition of arabidopsis hypocotyl elongation by jasmonates is enhanced under red light in phytochrome B dependent manner. J Plant Res 126: 161–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun J, Zhai Q, Zhou W, Qi L, Xu L, Wang B, Chen R, Jiang H, Qi J, et al. (2011) The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Hilson P, Sedbrook J, Rosen E, Caspar T, Masson PH (1998) The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc Natl Acad Sci USA 95: 15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chung HS, Howe GA (2009) A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Audenaert D, Beeckman T, Kepinski S (2009) The past, present, and future of chemical biology in auxin research. ACS Chem Biol 4: 987–998 [DOI] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, et al. (2010) A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol 20: 1697–1706 [DOI] [PubMed] [Google Scholar]
- Devoto A, Nieto-Rostro M, Xie D, Ellis C, Harmston R, Patrick E, Davis J, Sherratt L, Coleman M, Turner JG (2002) COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J 32: 457–466 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egoshi S, Takaoka Y, Saito H, Nukadzuka Y, Hayashi K, Ishimaru Y, Yamakoshi H, Dodo K, Sodeoka M, Ueda M (2016) Dual function of coronatine as a bacterial virulence factor against plants: possible COI1-JAZ-independent role. RSC Advances 6: 19404–19412 [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter A. (1996) The first new phytologist symposium—molecular approaches to the study of plant-microbe symbioses. New Phytol 133: 1–2 [Google Scholar]
- Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R (2009) (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Guranowski A, Miersch O, Staswick PE, Suza W, Wasternack C (2007) Substrate specificity and products of side-reactions catalyzed by jasmonate:amino acid synthetase (JAR1). FEBS Lett 581: 815–820 [DOI] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G (2002) The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16: 1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Neve J, Hirose M, Kuboki A, Shimada Y, Kepinski S, Nozaki H (2012) Rational design of an auxin antagonist of the SCF(TIR1) auxin receptor complex. ACS Chem Biol 7: 590–598 [DOI] [PubMed] [Google Scholar]
- He W, Brumos J, Li H, Ji Y, Ke M, Gong X, Zeng Q, Li W, Zhang X, An F, et al. (2011) A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota A, Kato T, Fukaki H, Aida M, Tasaka M (2007) The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis. Plant Cell 19: 2156–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kurotani K, Hayashi K, Hatanaka S, Toda Y, Ogawa D, Ichikawa H, Ishimaru Y, Tashita R, Suzuki T, Ueda M, et al. (2015) Elevated levels of CYP94 family gene expression alleviate the jasmonate response and enhance salt tolerance in rice. Plant Cell Physiol 56: 779–789 [DOI] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Guyomarc’h S, Hill K, Lucas M, Voß U, Kenobi K, Wilson MH, Farcot E, Hagen G, et al. (2015) Inference of the Arabidopsis lateral root gene regulatory network suggests a bifurcation mechanism that defines primordia flanking and central zones. Plant Cell 27: 1368–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sundaram S, Armitage L, Evans JP, Hawkes T, Kepinski S, Ferro N, Napier RM (2014) Defining binding efficiency and specificity of auxins for SCF(TIR1/AFB)-Aux/IAA co-receptor complex formation. ACS Chem Biol 9: 673–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM (2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. (1995) Root architecture and plant productivity. Plant Physiol 109: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW (1994) The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol 106: 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW (1996) Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8: 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meesters C, Mönig T, Oeljeklaus J, Krahn D, Westfall CS, Hause B, Jez JM, Kaiser M, Kombrink E (2014) A chemical inhibitor of jasmonate signaling targets JAR1 in Arabidopsis thaliana. Nat Chem Biol 10: 830–836 [DOI] [PubMed] [Google Scholar]
- Mueller MJ, Brodschelm W (1994) Quantification of jasmonic acid by capillary gas chromatography-negative chemical ionization-mass spectrometry. Anal Biochem 218: 425–435 [DOI] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Miyatake R, Inomata S, Ueda M (2008) Synthesis and bioactivity of potassium beta-D-glucopyranosyl 12-hydroxy jasmonate and related compounds. Biosci Biotechnol Biochem 72: 2867–2876 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Mithöfer A, Kombrink E, Boland W, Hamamoto S, Uozumi N, Tohma K, Ueda M (2011) 12-hydroxyjasmonic acid glucoside is a COI1-JAZ-independent activator of leaf-closing movement in Samanea saman. Plant Physiol 155: 1226–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narukawa-Nara M, Nakamura A, Kikuzato K, Kakei Y, Sato A, Mitani Y, Yamasaki-Kokudo Y, Ishii T, Hayashi K, Asami T, et al. (2016) Aminooxy-naphthylpropionic acid and its derivatives are inhibitors of auxin biosynthesis targeting l-tryptophan aminotransferase: structure-activity relationships. Plant J 87: 245–257 [DOI] [PubMed] [Google Scholar]
- Neff MM, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Hayashi K, Suzuki H, Gyohda A, Takaoka C, Sakaguchi Y, Matsumoto S, Kasahara H, Sakai T, Kato J, et al. (2014) Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J 77: 352–366 [DOI] [PubMed] [Google Scholar]
- Niu Y, Figueroa P, Browse J (2011) Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono Y, Ooura C, Rahman A, Aspuria ET, Hayashi K, Tanaka A, Uchimiya H (2003) p-Chlorophenoxyisobutyric acid impairs auxin response in Arabidopsis root. Plant Physiol 133: 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A (2001) IAA17/AXR3: biochemical insight into an auxin mutant phenotype. Plant Cell 13: 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-H, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R (2002a) A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Park J-Y, Kim H-J, Kim J (2002b) Mutation in domain II of IAA1 confers diverse auxin-related phenotypes and represses auxin-activated expression of Aux/IAA genes in steroid regulator-inducible system. Plant J 32: 669–683 [DOI] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M (2009) Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci USA 106: 22540–22545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Goossens A (2011) The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23: 3089–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts RJ, Cernac A, Estelle M (1998) Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J 16: 553–560 [DOI] [PubMed] [Google Scholar]
- Ploense SE, Wu MF, Nagpal P, Reed JW (2009) A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 136: 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Greenham K, Zhang Y, Santner A, Castillejo C, Mutka AM, O’Malley RC, Ecker JR, Kunkel BN, Estelle M (2016) The Arabidopsis auxin receptor F-box proteins AFB4 and AFB5 are required for response to the synthetic auxin picloram. G3 (Bethesda) 6: 1383–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Bannigan A, Sulaman W, Pechter P, Blancaflor EB, Baskin TI (2007) Auxin, actin and growth of the Arabidopsis thaliana primary root. Plant J 50: 514–528 [DOI] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J (2001) Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Brady SR, Reed RC, Ante SJ, Muday GK (2000) Basipetal auxin transport is required for gravitropism in roots of Arabidopsis. Plant Physiol 122: 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW. (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6: 420–425 [DOI] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK (1998) Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol 118: 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot C, Zimmerli C, Farmer EE, Reymond P, Poirier Y (2008) Induction of the Arabidopsis PHO1;H10 gene by 12-oxo-phytodienoic acid but not jasmonic acid via a CORONATINE INSENSITIVE1-dependent pathway. Plant Physiol 147: 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B (2001) A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13: 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O (1998) Changes in auxin response from mutations in an AUX/IAA gene. Science 279: 1371–1373 [DOI] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M (2015) SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27: 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y, Hamada S, Matsuura H, Matsushige M, Satou C, Takahashi K, Masuta C, Ito H, Matsui H, Nabeta K (2009) Purification and cDNA cloning of a wound inducible glucosyltransferase active toward 12-hydroxy jasmonic acid. Phytochemistry 70: 370–379 [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeno K, Goda H, Ishii T, Ogura T, Tachikawa T, Sasaki E, Yoshida S, Fujioka S, Asami T, Shimada Y (2010) Auxin biosynthesis inhibitors, identified by a genomics-based approach, provide insights into auxin biosynthesis. Plant Cell Physiol 51: 524–536 [DOI] [PubMed] [Google Scholar]
- Staswick PE. (2009) The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol 150: 1310–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar P, Edwards KS, Rahman A, Delong A, Muday GK (2009) PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiol 150: 722–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Xu Y, Ye S, Jiang H, Chen Q, Liu F, Zhou W, Chen R, Li X, Tietz O, et al. (2009) Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21: 1495–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Chen Q, Qi L, Jiang H, Li S, Xu Y, Liu F, Zhou W, Pan J, Li X, et al. (2011) Jasmonate modulates endocytosis and plasma membrane accumulation of the Arabidopsis PIN2 protein. New Phytol 191: 360–375 [DOI] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Tian Q, Reed JW (1999) Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126: 711–721 [DOI] [PubMed] [Google Scholar]
- Ueda M, Egoshi S, Dodo K, Ishimaru Y, Yamakoshi H, Nakano T, Takaoka Y, Tsukiji S, Sodeoka M (2017) Noncanonical function of a small-molecular virulence factor coronatine against plant immunity: an in vivo Raman imaging approach. ACS Cent Sci 3: 462–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H (2008) Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol 49: 1025–1038 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh TA, Neal R, Merlo AO, Honma M, Hicks GR, Wolff K, Matsumura W, Davies JP (2006) Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol 142: 542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D (2002) The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, Cheng Z, Peng W, Luo H, Nan F, et al. (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lee S, So JH, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle M (2004) The IAA1 protein is encoded by AXR5 and is a substrate of SCF(TIR1). Plant J 40: 772–782 [DOI] [PubMed] [Google Scholar]