CHLOROPLAST RIBOSOME ASSOCIATED plays a role in the biogenesis and/or stability of the chloroplast ribosome, especially under stress when ribosomal activity is compromised.
Abstract
Chloroplast ribosomes, which originated from cyanobacteria, comprise a large subunit (50S) and a small subunit (30S) containing ribosomal RNAs (rRNAs) and various ribosomal proteins. Genes for many chloroplast ribosomal proteins, as well as proteins with auxiliary roles in ribosome biogenesis or functioning, reside in the nucleus. Here, we identified Arabidopsis (Arabidopsis thaliana) CHLOROPLAST RIBOSOME ASSOCIATED (CRASS), a member of the latter class of proteins, based on the tight coexpression of its mRNA with transcripts for nucleus-encoded chloroplast ribosomal proteins. CRASS was acquired during the evolution of embryophytes and is localized to the chloroplast stroma. Loss of CRASS results in minor defects in development, photosynthetic efficiency, and chloroplast translation activity under controlled growth conditions, but these phenotypes are greatly exacerbated under stress conditions induced by the translational inhibitors lincomycin and chloramphenicol or by cold treatment. The CRASS protein comigrates with chloroplast ribosomal particles and coimmunoprecipitates with the 16S rRNA and several chloroplast ribosomal proteins, particularly the plastid ribosomal proteins of the 30S subunit (PRPS1 and PRPS5). The association of CRASS with PRPS1 and PRPS5 is independent of rRNA and is not detectable in yeast two-hybrid experiments, implying that either CRASS interacts indirectly with PRPS1 and PRPS5 via another component of the small ribosomal subunit or that it recognizes structural features of the multiprotein/rRNA particle. CRASS plays a role in the biogenesis and/or stability of the chloroplast ribosome that becomes critical under certain stressful conditions when ribosomal activity is compromised.
Chloroplasts are endosymbiotic organelles that are derived from a cyanobacterial ancestor. The chloroplast contains about 120 genes organized on a circular chromosome, and the majority of cyanobacterial genes have been transferred from the original symbiont to the nucleus or were lost in the course of evolution (Kleine et al., 2009). The chloroplast still hosts its own transcriptional and translational machineries for the synthesis of chloroplast-encoded proteins (Lyska et al., 2013). Two transcriptional systems coexist in chloroplasts. The bacterial-type plastid-encoded RNA polymerase works in concert with σ-factors that recognize specific promoters and are needed for chloroplast biogenesis (Woodson et al., 2013; Pfannschmidt et al., 2015). Additionally, the phage-type nucleus-encoded polymerase is required for the expression of a large number of chloroplast proteins and also is essential for plant survival (Hricová et al., 2006). Although the expression of some genes is controlled predominantly by either plastid-encoded RNA polymerase or nucleus-encoded polymerase, many genes possess promoters for both RNA polymerases, which adds to the complexity of plastid gene regulation (Börner et al., 2015).
In contrast to the case in bacteria, the regulation of chloroplast gene expression is mediated mainly by posttranscriptional processes, including RNA processing, splicing, editing, and RNA stabilization and translation, and requires a large number of nucleus-encoded proteins, predominantly RNA-binding factors (Manavski et al., 2018). For instance, members of the vast family of pentatricopeptide repeat proteins are essential for all aspects of RNA metabolism in the chloroplast. Ribonucleases are required for the maturation of ribosomal RNA (rRNA) and tRNA precursors, RNA processing, and mRNA degradation (Stoppel and Meurer, 2012). Likewise, DEAD-box RNA helicases are involved in intron splicing or rRNA processing because of their ability to alter RNA secondary structures (Nawaz and Kang, 2017).
Protein synthesis in the chloroplast is carried out by bacterial-type 70S ribosomes, each composed of a large subunit (50S) and a small subunit (30S). As in bacteria, three phases can be distinguished in the process. First, initiation factors associated with the 30S subunit recognize the start codon via Shine-Dalgarno sequences and/or local minima of mRNA secondary structure (Hirose and Sugiura, 2004; Scharff et al., 2011, 2017). Several initiation factors have been shown to be essential for chloroplast development (Miura et al., 2007; Zheng et al., 2016). Second, after the recruitment of the 50S subunit, elongation proceeds with the assistance of elongation factors (Albrecht et al., 2006; Ruppel and Hangarter, 2007; Liu et al., 2010). Finally, termination is triggered by the presence of the stop codon, involving peptide chain release factors (PrfA and PrfB) and the ribosomal recycling factor (Meurer et al., 2002; Motohashi et al., 2007).
In general, the structure of the plastid ribosome is highly conserved relative to its cyanobacterial counterpart (Ahmed et al., 2016, 2017; Bieri et al., 2017). In the chloroplast 30S subunit, one rRNA species (16S rRNA) is present, whereas three rRNAs (23S rRNA, 5S rRNA, and 4.5S rRNA) are found in the 50S subunit. Twenty-one plastid ribosomal proteins of the 30S subunit (PRPS proteins) and 31 plastid ribosomal proteins of the 50S subunit (PRPL proteins) descend from eubacterial orthologs. In addition, several plastid-specific ribosomal proteins (PSRPs) have been identified (Yamaguchi and Subramanian, 2000, 2003; Yamaguchi et al., 2000; Manuell et al., 2007; Sharma et al., 2007), of which five (PSRP2, PSRP3, PSRP4, PSRP5, and PSRP6) are found in stoichiometric amounts with the classical ribosomal proteins, indicating that they represent bona fide ribosomal proteins with a putative role in the light-dependent regulation of translation in photosynthetic organisms (for review, see Tiller and Bock, 2014). In fact, PSRP2, PSRP3, PSRP4, and PSRP5 have been localized in the three-dimensional structure of the chloroplast ribosome by cryoelectron microscopy (Ahmed et al., 2017; Bieri et al., 2017; Graf et al., 2017; Boerema et al., 2018). Hence, the acquisition of new components by the translational machinery may be related to the wider range of environmental fluctuations to which plants are exposed, and it may facilitate the regulation of translation in accordance with the specific needs of plant cells. Moreover, the sets of ribosomal proteins that are essential in eubacteria and plants have diverged, indicating a degree of flexibility that has emerged during the evolution of the green lineage (Tiller and Bock, 2014).
In this work, we describe the identification and characterization of CHLOROPLAST RIBOSOME ASSOCIATED (CRASS), a protein that is physically associated with the chloroplast ribosomal 30S subunit in Arabidopsis (Arabidopsis thaliana). The stromal protein CRASS is present in embryophytes but not in green algae, suggesting that its recruitment by the chloroplast ribosome may be a relatively recent event in the evolution of chloroplasts. CRASS is not essential for plant survival under controlled growth conditions, but it becomes limiting for chloroplast translational activity, photosynthesis, and growth under stressful conditions, such as the inhibition of chloroplast translation by lincomycin (LIN) or chloramphenicol (CAP) or the exposure of plants to low-temperature stress. Therefore, CRASS represents a chloroplast-associated protein that supports ribosomal activity under stressful conditions.
RESULTS
CRASS mRNA Is Coexpressed with Transcripts for Nucleus-Encoded Chloroplast Ribosomal Proteins
The expression of many nucleus-encoded chloroplast proteins is highly coregulated at the mRNA level in plants in order to ensure the coordinated activity of related processes within chloroplasts (Leister et al., 2011). In fact, two groups of genes (regulons), one for proteins linked to photosynthesis and one for proteins linked to plastid gene expression, are clearly recognizable (Biehl et al., 2005). A previous guilt-by-association approach based on the study of hitherto uncharacterized proteins encoded by genes assigned to the photosynthesis regulon resulted in the identification of a new component of photosynthetic cyclic electron flow (DalCorso et al., 2008).
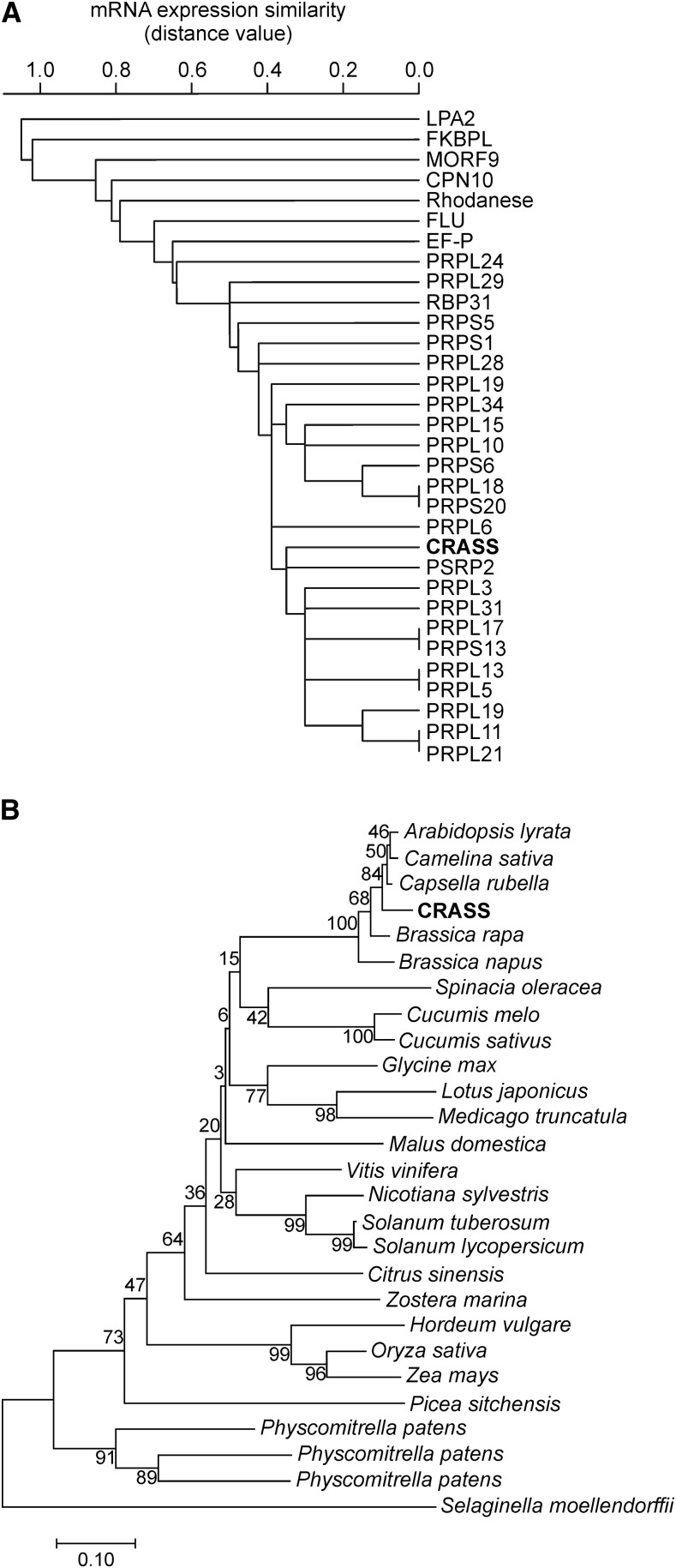
To identify novel factors involved in plastid translation, we performed coexpression analysis with the ATTED-II coexpression tool (atted.jp), using all Arabidopsis nuclear genes encoding plastid ribosomal proteins of the 30S and 50S subunits (PRPSs and PRPLs, respectively) as bait as well as the genes for plastid-specific ribosomal proteins (PSRPs; Supplemental Fig. S1). Besides the starting set of ribosomal proteins, other known factors involved in plastid RNA processing were found in the network, such as the 31-kD RNA-binding protein CP31A (Tillich et al., 2009) and the ribosome recycling factor (Wang et al., 2010). Interestingly, one protein of unknown function (AT5G14910) was found to be highly coexpressed with the bait transcripts and, therefore, was named CRASS. When CRASS was used in a reciprocal coexpression analysis as bait, employing hierarchical clustering with the single-linkage method provided by the HCluster tool (atted.jp), chloroplast ribosomal proteins were detected almost exclusively (Fig. 1A). From these results, we inferred that CRASS might be a new factor associated with protein synthesis in the chloroplast.
Figure 1.
Coexpression and phylogeny of CRASS. A, The coexpression of CRASS transcripts with mRNAs for other nucleus-encoded proteins was analyzed using hierarchical clustering with the single-linkage method provided by the HCluster tool (atted.jp). B, Phylogenetic tree derived from the full-length sequences of Arabidopsis CRASS and its orthologs from other species. The tree was rooted at midpoint using the neighbor-joining method in MEGA6. Bootstrap values (expressed as percentages of 2,000 replicates) are indicated at the branches. All accession numbers of the proteins reported here are listed in “Materials and Methods.”
CRASS Is Present in Embryophytes But Not in Algae
CRASS is encoded by a single gene in Arabidopsis (AT5G14910) and is annotated as a heavy metal transport protein, due to the presence of a putative heavy metal-associated (HMA) domain. In fact, CRASS proteins from Arabidopsis, grapevine (Vitis vinifera), and the model lycophyte Selaginella moellendorffii contain regions with homology to HMA proteins from bacteria (Supplemental Fig. S2). However, the essential Cys residues present in the catalytic core of the HMA domain are not conserved in CRASS proteins of the green lineage. Without these Cys residues, heavy metal transporters are unable to bind cations (Lutsenko et al., 1997). Therefore, the HMA-like domain present in CRASS proteins is likely to serve some function other than cation binding. The phylogenetic analysis of Arabidopsis CRASS and its homologs in other species revealed that orthologs can be found in other embryophyte species (Fig. 1B) but not in green algae or cyanobacteria.
CRASS Is a Nonessential Protein Localized in the Chloroplast Stroma
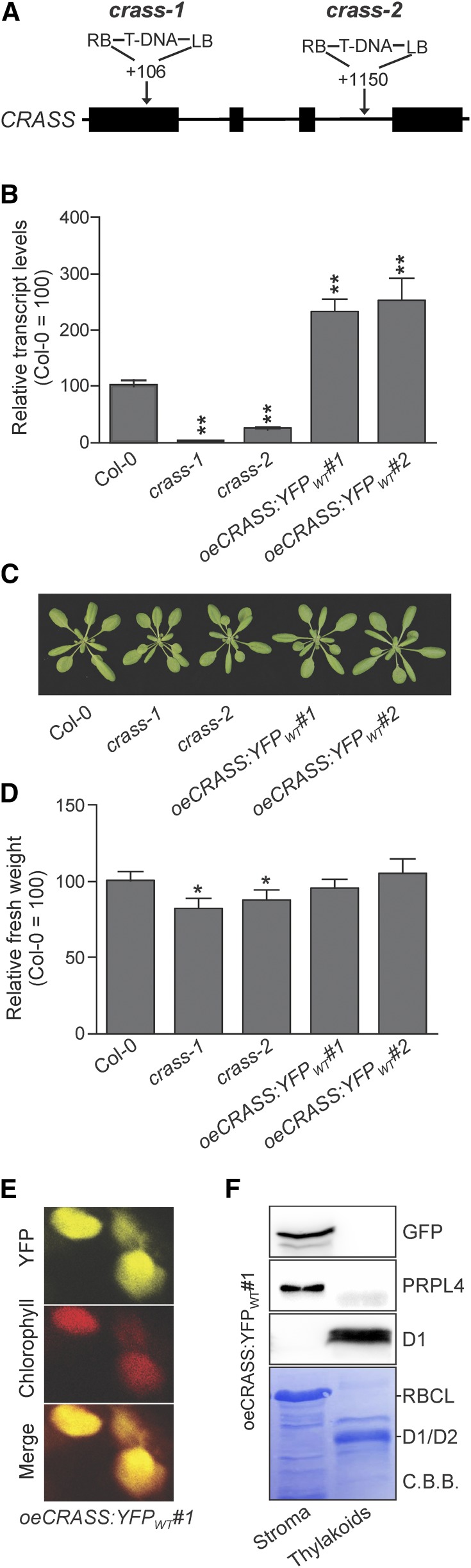
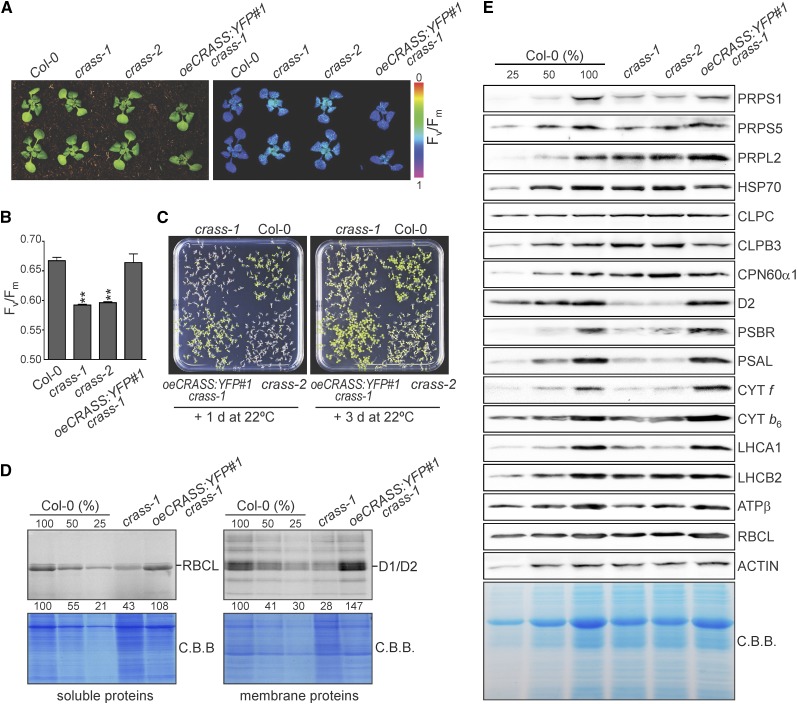
To investigate the biological function of CRASS in Arabidopsis, two independent T-DNA mutant lines, crass-1 (84-776) and crass-2 (72-131), were isolated from the Koncz T-DNA collection (Ríos et al., 2002), carrying T-DNA insertions in exon 1 and intron 3, respectively (Fig. 2A). Moreover, two independent transgenic Arabidopsis lines that overexpress a CRASS fusion protein with a YFP tag at its C terminus in the wild-type Columbia-0 (Col-0) background (oeCRASS:YFPWT#1 and oeCRASS:YFPWT#2) were generated to allow for fluorescence detection of the CRASS protein. While transcripts of CRASS were completely absent in crass-1 (a knockout allele) and reduced to about 20% of wild-type levels in crass-2 (a knockdown allele), both overexpressor lines oeCRASS:YFPWT#1 and oeCRASS:YFPWT#2 contained more than twice as many CRASS transcripts as wild-type plants (Fig. 2B). The overexpressor lines were visually indistinguishable from the wild type when grown under controlled greenhouse conditions, but the two crass mutants exhibited slight but statistically significant reductions in fresh weight (Fig. 2, C and D) and Fv/Fm (maximum quantum yield of PSII) values (Supplemental Fig. S3, A and B) relative to wild-type plants. However, the differences in Fv/Fm are subtle, and even after several biological replicates, other parameters of photosynthetic performance were practically unaltered under these conditions, as demonstrated by wild-type-like values for the effective quantum yield of PSII and nonphotochemical quenching, measured in light-response experiments using increasing light intensities (Supplemental Fig. S3, C and D). Hence, this is in contrast to other ribosomal proteins, whose absence results in lethality (Tiller and Bock, 2014). CRASS is not essential for plant survival, a feature it shares with several other ribosomal proteins and factors required for ribosome functioning (Fleischmann et al., 2011; Romani et al., 2012; Paieri et al., 2018).
Figure 2.
Mutants and subcellular localization of CRASS. A, Gene model of CRASS (AT5G14910). Exons are shown as black boxes and introns as lines. The positions of T-DNA insertions in the crass-1 and crass-2 mutants are indicated relative to the start codon. LB, Left border; RB, right border. B, Relative levels of CRASS transcripts were analyzed in the wild-type (Col-0), in crass mutants (crass-1 and crass-2), and in overexpressors of CRASS in the wild-type background (oeCRASS:YFPWT#1 and oeCRASS:YFPWT#2) using UBIQUITIN10 as a control (see “Materials and Methods”); levels in Col-0 = 100%. C, Representative images of 3-week-old plants from the same genotypes as in B. D, Relative fresh weight data for the same genotypes as in B and C (Col-0 = 100%). Relative values are means ± se from independent (n ≥ 6) experiments with at least 15 plants each, grown for 3 weeks under long-day conditions. E, Chloroplast localization of CRASS. Mesophyll cells of oeCRASS:YFPWT#1 plants were analyzed with a laser scanning confocal microscope. The overlap between the YFP signal and the chloroplast autofluorescence (Chlorophyll) indicates chloroplast localization. F, Subchloroplast localization of CRASS. Chloroplasts from oeCRASS:YFPWT#1 plants were isolated and separated into stroma and thylakoid fractions (see “Materials and Methods”). Immunoblot analysis was performed with an antibody directed against the YFP tag of the fusion protein, a PRPL4-specific antibody, and a D1-specific antibody (as a control for the thylakoid fraction). As an additional control, the gel was stained with Coomassie Brilliant Blue (C.B.B.) to detect D1 and D2 (thylakoid control) and RBCL (chloroplast stroma control). Student’s t test (*, P < 0.05 and **, P < 0.01) was used for statistical analysis.
Bioinformatic analyses with ChloroP and WoLF PSORT (suba.live) predicted an N-terminal chloroplast transit peptide in the CRASS sequence (Supplemental Fig. S2). Published proteomic studies previously detected the CRASS protein in chloroplast fractions (Friso et al., 2004; Zybailov et al., 2008), and confocal fluorescence microscopy of the oeCRASS:YFPWT#1 line confirmed the chloroplast localization of the fusion protein (Fig. 2E). Chloroplast isolation and fractionation into stroma and thylakoids under standard conditions that do not stabilize membrane-bound ribosomes revealed that CRASS is present exclusively in the chloroplast stroma, similar to the ribosomal protein PRPL4 (Fig. 2F). This localization is consistent with a putative role of CRASS in the translational apparatus of plastids. In fact, the abundance of CRASS reported in the literature is well within the range typical for bona fide chloroplast ribosomal proteins (see Supplemental Table S2 in Zybailov et al., 2008).
Absence of CRASS Does Not Affect Ribosomal Activity under Controlled Greenhouse Conditions
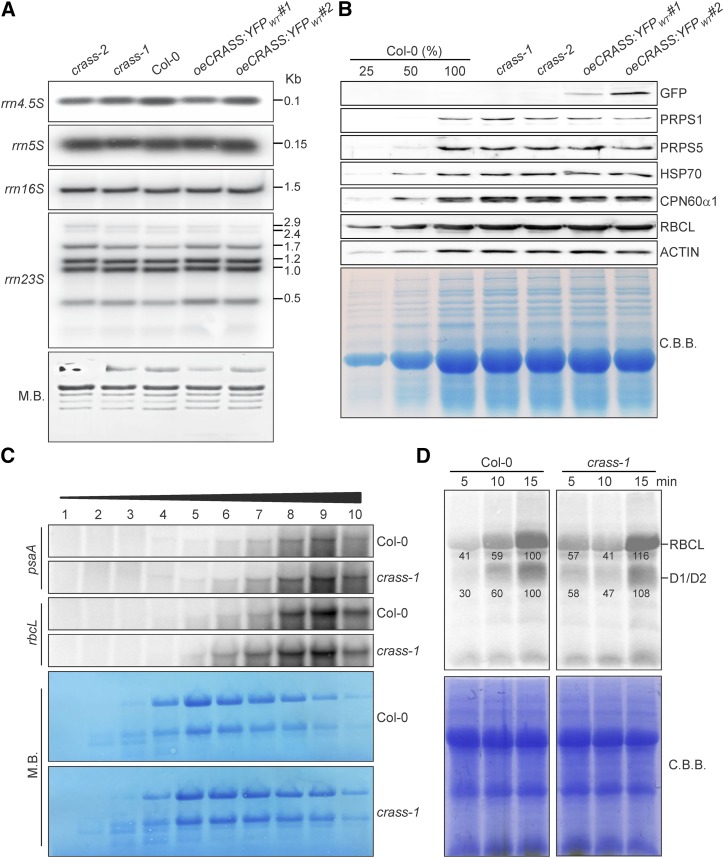
To explore the function of CRASS, we first quantified the levels of plastid rRNAs in crass mutants and oeCRASS:YFPWT lines by RNA gel-blot analysis. Neither the processing nor the abundance of rRNAs was altered in these lines (Fig. 3A). Moreover, steady-state amounts of chloroplast ribosomal proteins, chaperones, and the large subunit of Rubisco were not altered markedly in crass mutant or oeCRASS:YFPWT lines compared with wild-type plants (Fig. 3B; Supplemental Table S1).
Figure 3.
CRASS is not required for chloroplast translational activity under nonstressed conditions. A, RNA gel-blot analysis of 10-μg samples of total RNA from 3-week-old wild-type (Col-0), mutant (crass-1 and crass-2), and overexpressor (oeCRASS:YFPWT#1 and oeCRASS:YFPWT#2) lines with probes specific for the four plastid rRNAs (rrn23, rrn16, rrn5, and rrn4.5). The sizes of the transcripts are given in kb on the right. M.B., Methylene Blue. B, The steady-state levels of representative chloroplast proteins were analyzed by immunoblotting of samples isolated from 3-week-old plants. Total protein extracts from the same genotypes as in A were examined (loading equal amounts of proteins), together with a dilution series of the Col-0 sample as indicated. Representative images of immunoblots probed with antibodies specific for the indicated proteins are shown. Quantification of the results of five biological replicates (by ImageJ) relative to Col-0 plants is provided in Supplemental Table S1. C, CRASS is not required for efficient polysome loading. RNA gel-blot analysis is shown for psaA and rbcL transcripts in polysome fractions 1 to 10 collected after Suc-gradient centrifugation of wild-type (Col-0) and crass-1 extracts (see “Materials and Methods”). rRNA was stained with Methylene Blue. D, Pulse-labeling analysis of D1/D2 synthesis. Leaves isolated from plants at the six-leaf rosette stage were pulse labeled with [35S]Met under low-light illumination (20 μmol photons m−2 s−1) for 5, 10, and 15 min in the presence of cycloheximide to inhibit cytosolic protein synthesis. Total leaf proteins were then isolated, fractionated by SDS-PAGE, and detected by autoradiography. A portion of the SDS-polyacrylamide gel corresponding to the RBCL region was stained with Coomassie Brilliant Blue (C.B.B.) and served as an internal standard for loading normalization. Quantification of signals (by ImageJ) relative to Col-0 at the 15-min time point (=100%) is provided below each relevant band.
To investigate the impact of CRASS on translation efficiency, CRASS loss-of-function lines and overexpressors, together with Col-0 plants as the wild-type control, were subjected to polysome loading and in vivo labeling experiments. First, we chose to examine psaA and rbcL mRNAs, coding for the PSI subunit A and the large subunit of Rubisco, respectively, which are loaded efficiently with ribosomes and, hence, migrate deeply into the Suc gradient. The profiles of psaA and rbcL RNAs obtained for extracts from crass and wild-type plants grown under controlled greenhouse conditions were indistinguishable, peaking in fractions 8 and 9, thus indicating that their polysome densities were unchanged by the mutation (Fig. 3C). Similarly, when extracts of the same genotypes were evaluated by in vivo labeling of newly synthesized chloroplast proteins (see “Materials and Methods”), the rate of incorporation of [35S]Met in the crass mutant was very similar to that of wild-type control leaves (Fig. 3D).
Taken together, these results suggest that the absence of CRASS does not substantially alter plastid translational activity in plants grown under normal greenhouse conditions.
Absence of CRASS Enhances the Effects of the Translation Inhibitors LIN and CAP
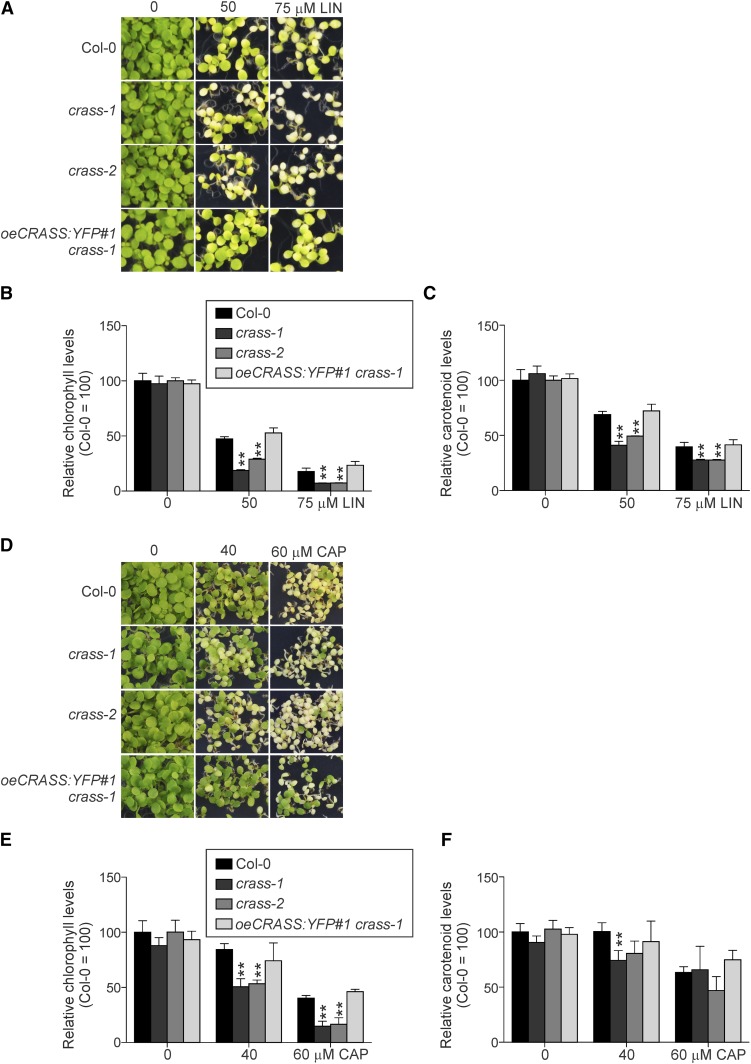
As the lack of CRASS under controlled growth conditions had only minor effects in planta, we next studied the impact of its loss on ribosomal activity under stressful conditions. LIN and CAP specifically inhibit the elongation of the nascent polypeptide chain during chloroplast protein synthesis (Sohmen et al., 2009; Liao et al., 2016). Therefore, crass mutant plants and wild-type controls were grown on plates in the presence of different LIN or CAP concentrations under sterile conditions. Ten days after germination, crass mutants clearly exhibited hypersensitivity to the inhibitors, manifested as enhanced loss of leaf coloration relative to the wild-type control (Fig. 4, A and D). Quantification of chlorophyll and carotenoid levels showed that both LIN- and CAP-treated crass seedlings accumulated significantly less of these pigments than did wild-type seedlings (Fig. 4, B, C, E and F). Furthermore, overexpression of CRASS-YFP in the crass-1 background (oeCRASS:YFP crass-1) suppressed the LIN- or CAP-induced phenotype of the crass-1 mutant (Fig. 4), implying that the recombinant CRASS-YFP protein is functional and able to fully compensate for the mutant’s hypersensitivity to the antibiotics.
Figure 4.
Absence of CRASS enhances the sensitivity of plants to the inhibition of chloroplast translation by LIN or CAP. A and D, Representative images of 10-d-old wild-type (Col-0), mutant (crass-1 and crass-2), and oeCRASS:YFP#1 crass-1 overexpressor plants germinated on Murashige and Skoog (MS) medium containing the indicated concentrations of LIN (A) or CAP (D). B, C, E, and F, Quantification of total chlorophyll (B and E) and carotenoid (C and F) levels (see “Materials and Methods”) demonstrates the differences between wild-type or oeCRASS:YFP#1 crass-1 plants compared with mutant plants grown in the presence of LIN or CAP. Relative data are shown (wild-type plants grown in the absence of LIN/CAP = 100%), and means ± se values (n = 4) are provided. Student’s t test (**, P < 0.01) was used for statistical analysis.
These results demonstrate that CRASS has a positive effect on chloroplast translation under conditions that limit the activity of the chloroplast ribosome, as LIN or CAP treatments are known to do.
CRASS Is Necessary for Tolerance to Cold Stress
Many mutants affected in plastid ribosomal activity are more sensitive to cold treatments than wild-type plants (Rogalski et al., 2008; Fleischmann et al., 2011; Wang et al., 2016; Zhang et al., 2016; Paieri et al., 2018). Experiments with plants grown for 2 weeks at 22°C and then transferred for 5 weeks to 4°C showed that both crass mutants were more susceptible to this treatment than wild-type plants or overexpressors of CRASS1:YFP in the crass-1 background (oeCRASS:YFP#1 crass-1; Fig. 5A). The Fv/Fm was reduced significantly in the crass mutants relative to wild-type and oeCRASS:YFP#1 crass-1 lines (Fig. 5B). Even more pronounced phenotypes were obtained by germinating seeds directly at 4°C for 6 weeks followed by a few days at 22°C. This growth regime resulted in crass plants that were paler and smaller than the other genotypes (Fig. 5C). After 3 d of recovery, the chloroplast translation efficiency was clearly reduced in crass-1 mutant plants compared with the other genotypes, as demonstrated by in vivo labeling experiments (see “Materials and Methods”; Fig. 5D). In fact, after 15 min of light exposure, the incorporation of [35S]Met into the plastid proteins RBCL and the PSII core proteins D1/D2 in the crass-1 mutant was reduced to 43% and 28%, respectively, relative to wild-type plants. Immunoblot analysis showed that, after 3 d of recovery, the levels of the plastid ribosomal proteins PRPS1, PRPS5, and PRPSL2 were reduced to 63%, 70%, and 92% of wild-type amounts, respectively, in the crass-1 mutants compared with wild-type and oeCRASS:YFP#1 crass-1 lines (Fig. 5E; Supplemental Table S2). This could be explained by the reduced stability of plastid ribosomes, impaired ribosome biogenesis, or decreased translation rate under stress conditions in the absence of CRASS. While the levels of nucleus-encoded chloroplast chaperones (HSP70, CLPC, CLPB3, and CPN60α1) were unchanged, the amounts of chloroplast-encoded proteins (D2, CYT f, CYT b6, ATPβ, and RBCL) were reduced strongly in the absence of CRASS, most likely as a result of reduced plastid translation efficiency. Lower levels of nucleus-encoded photosynthetic proteins (PSBR, PSAL, LHCA1, and LHCB2) are likely to reflect secondary effects, due to either reduced accumulation of plastid-encoded proteins in the corresponding multiprotein complexes or retrograde signaling that down-regulates the expression of the corresponding nuclear genes. The analysis of samples directly after 6 weeks at 4°C (without recovery time at 22°C) showed an even more pronounced reduction in PRPS1 levels (Supplemental Fig. S4A). However, besides a mild reduction in the levels of rrn16S transcripts, there were no differences in the processing and amounts of 23S and 16S rRNAs in crass-1 compared with wild-type plants at the end of the cold period (Supplemental Fig. S4B).
Figure 5.
CRASS is required for cold stress tolerance. A, Representative images and chlorophyll fluorescence images of the wild type (Col-0), crass-1 and crass-2 mutants, and an oeCRASS:YFP#1 crass-1 overexpresser grown on soil for 2 weeks at 22°C, followed by 5 weeks at 4°C. The color scale indicates Fv/Fm signal intensities. B, Quantification of Fv/Fm values (means ± se) from A, demonstrating that both crass-1 and crass-2 mutants display reduced efficiency of PSII after the cold treatment (Student’s t test: **, P < 0.01). C, Representative images of wild-type (Col-0) and crass-1 and crass-2 mutant plants grown on MS plates for 6 weeks at 4°C, followed by 1 or 3 d at 22°C. D, Analysis of the translation rate. Seedlings exposed to cold treatment (as in C) and allowed to recover for 3 d were incubated with [35S]Met under low-level illumination (20 μmol photons m−2 s−1) for 15 min in the presence of cycloheximide (to inhibit cytosolic protein synthesis), and total protein extracts were subjected to SDS-PAGE and autoradiography. The stained (Coomassie Brilliant Blue [C.B.B.]) gels loaded with soluble and membrane proteins and the corresponding autoradiographs are shown. E, Total protein extracts were analyzed by immunoblotting (loading equal amounts of proteins), using material obtained from cold-treated plants (after 3 d of recovery) grown on MS plates. Representative images of immunoblot analyses with specific antibodies are shown. Quantification of the results of five biological replicates (by ImageJ) relative to Col-0 plants is provided in Supplemental Table S2.
Together, these results suggest that, like mutants defective in plastid ribosomal activity or ribosome biogenesis, the CRASS knockout mutant shows a higher sensitivity to cold stress. This phenotype, in turn, is associated with reduced plastid translational capacity.
Chloroplast 16S rRNA Specifically Coimmunoprecipitates with CRASS
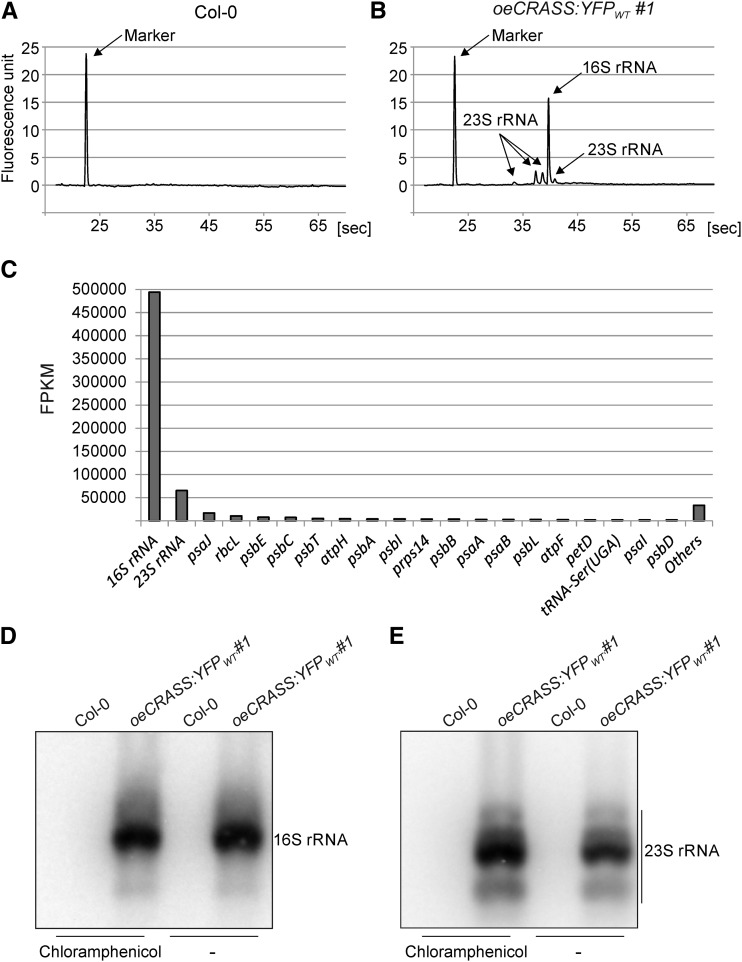
To further analyze whether CRASS interacts with components of plastid ribosomes, we first performed RNA coimmunoprecipitation experiments combined with RNA deep sequencing (RIP-seq). Experiments were carried out with samples from oeCRASS:YFPWT#1 plants and wild-type plants (Col-0) as the negative control using an antibody specific for GFP. As expected, no RNA was detected in the pellet fraction obtained from the control sample (Fig. 6, A, D, and E; Supplemental Fig. S5), but experiments with the oeCRASS:YFPWT#1 line resulted in quantities of immunoprecipitated RNA sufficient for RIP-seq (see “Materials and Methods”). Interestingly, the major RNA found in association with CRASS in vivo in the two biological replicates was the chloroplast 16S rRNA, and the chloroplast 23S rRNA was present in lesser amounts (Fig. 6, B and C; Supplemental Table S3). RNA gel-blot experiments confirmed the specific enrichment of chloroplast 16S rRNA in coimmunoprecipitates when compared with 23S rRNA (Fig. 6, D and E). Only the coimmunoprecipitation of CRASS with the 23S rRNA was increased by the addition of CAP, which stabilizes elongating ribosomes (Fig. 6E).
Figure 6.
Chloroplast 16S rRNA coimmunoprecipitates with CRASS. RNA species associated with CRASS were identified by immunoprecipitation of the fusion protein with a GFP-specific antibody, followed by RIP-seq. Plants overexpressing CRASS-YFP in the wild-type background (oeCRASS:YFPWT#1) were employed, as well as wild-type (Col-0) plants as a control. A and B, Analysis of the RNA from samples obtained from the wild type (A) and the oeCRASS:YFPWT#1 line (B) by capillary electrophoresis using Bioanalyzer (Agilent). Two replicates were analyzed (Supplemental Fig. S5). In the wild-type sample, no RNA was detected in the pelleted fraction; therefore, the sample was not subjected to RIP-seq analysis. In the oeCRASS:YFPWT#1 sample, RNA species of the size of the plastid 16S and 23S rRNAs were detected. C, Analysis of the oeCRASS:YFPWT#1 samples by RNA sequencing (RNA-seq). Average values (fragments per kilobase of exon per million reads mapped [FPKM] for the 20 most abundant immunoprecipitated RNAs from two independent RIP-seq experiments are shown (Supplemental Table S3). The remaining 153 genes with fewer than 1,500 FPKM are combined into the Others column. D, RNA gel-blot analysis of immunoprecipitated material of the wild-type negative control and the oeCRASS:YFPWT#1 line using a probe against the plastid 16S rRNA. The 16S rRNA coimmunoprecipitates with CRASS both in the presence and absence of CAP, which stabilizes elongating ribosomes. In contrast, there was no signal in the wild-type control. E, RNA gel-blot analysis using a probe against the plastid 23S rRNA. The coimmunoprecipitation of the 23S rRNA is increased by the addition of CAP.
This result provides the first indication that CRASS is not only functionally associated with chloroplast ribosomes but that it might interact physically with the 30S ribosomal subunit.
CRASS Is Associated with the Chloroplast 30S Ribosomal Subunit
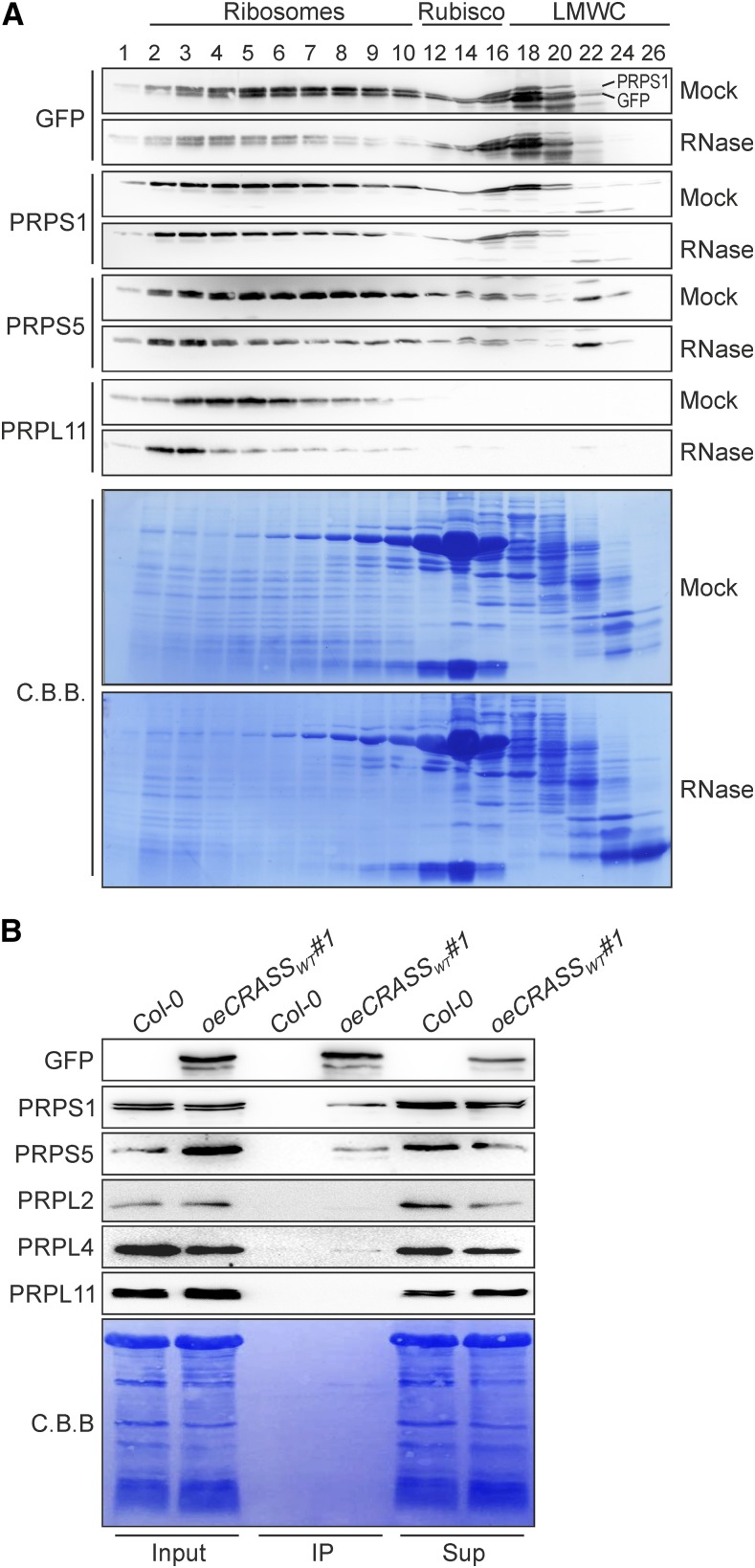
To further investigate whether CRASS associates with ribosomes in vivo, size-exclusion chromatography (SEC) of chloroplast stroma extracts from plants expressing YFP-tagged CRASS (oeCRASS:YFPWT#1) was carried out to visualize the different assembly states of ribosomal subunits. Immunodetection of CRASS-YFP revealed that it was present in the same fraction (1-22) as PRPS1 and PRPS5, indicating that CRASS indeed comigrates with the 30S ribosomal subunit (Fig. 7A). Moreover, gentle RNase treatment of stroma extracts prior to SEC analysis decreased the signal intensities of PRPS1, PRPS5, and PRPL11 in those fractions (Fig. 7A) that were described previously to contain RNase-sensitive, preassembled ribosomal particles (Meurer et al., 2017). This trend also was observed for CRASS proteins, suggesting that CRASS is associated with both mature and immature ribosomal particles.
Figure 7.
CRASS is associated with chloroplast 30S subcomplexes. A, Size distribution of CRASS complexes. SEC analysis was performed as described in “Materials and Methods” with native stroma extracts from 3-week-old oeCRASS:YFPWT#1 lines with and without RNase treatment. Precipitated fractions were fractionated on SDS gels, and blots were stained with Coomassie Brilliant Blue (C.B.B.). Complexes are indicated above and were deduced from previously published data (Olinares et al., 2010). The proteins detected by specific antibodies are shown on the left. Membranes were reprobed, and the top band visible on the gel showing the results obtained with the GFP-specific antibody derives from PRPS1 signals from a previous immunodecoration. The curvature of signals in fractions 12 to 16 is caused by the large amounts of Rubisco present (see also C.B.B. gels). LMWC, Low-molecular-weight complexes. B, Coimmunoprecipitation experiments were performed using 3-week-old plants overexpressing CRASS-YFP (oeCRASS:YFPWT#1) and wild-type (Col-0) plants as a control. Input, supernatant (Sup), and fractions immunoprecipitated (IP) with the antibody against the YFP tag were electrophoresed, blotted, and probed with antibodies against PRPS1, PRPS5, PRPL2, PRPL4, and PRPL11. The immunoblot shows the interaction of CRASS with the PRPS1 and PSPS5 proteins.
Next, we studied whether CRASS interacts directly with chloroplast ribosomes. To this end, we subjected oeCRASS:YFPWT#1 plants to a coimmunoprecipitation assay employing an antibody directed against the YFP tag. The coimmunoprecipitates from two independent experiments and from a wild-type control sample were then analyzed by liquid chromatography-tandem mass spectrometry. A large number of proteins were identified as putative interactors of CRASS (Supplemental Table S4). Among the peptides with the highest scores, the most abundant group clearly derives from chloroplast ribosomal proteins. In fact, the best 10 candidates belonged to the plastid ribosomal group of protein components of both 50S and 30S subunits (Table 1). Although we cannot completely exclude the possibility that CRASS interacts with other chloroplast proteins, we found no obvious candidates for CRASS interaction partners other than ribosomal proteins (Supplemental Table S4).
Table 1. Plastid ribosomal proteins coimmunoprecipitated with CRASS.
Proteins from the oeCRASS:YFPWT#1 line were immunoprecipitated with antibodies recognizing YFP and fractionated by SDS-PAGE. Gel fractions were analyzed by liquid chromatography-tandem mass spectrometry with an electrospray ionization ion trap instrument to identify CRASS-associated proteins. To check for unspecific contaminants, the output was compared with a similar immunoprecipate from wild-type (Col-0) control plants (Supplemental Table S4) as described in “Materials and Methods.” Two independent biological replicates (Exp 1 and Exp 2) were analyzed, and the best 10 candidates based on their scores from these two biological replicates (Supplemental Table S4) are shown. Note that because ribosomal proteins belong to the Arg/Lys-rich class of proteins and often are quite small, a high protein coverage after trypsination cannot be expected. Therefore, proteins with low coverage but high peptide scores also were considered.
| Arabidopsis Genome Initiative Code | Identifier | Molecular Mass | Score, Exp 1 | Score, Exp 2 | Coverage, Exp 1 | Coverage, Exp 2 | Most Abundant Peptide |
|---|---|---|---|---|---|---|---|
| kD | % | ||||||
| AT5G14910 | CRASS | 19.1 | 3,023 | 2,303 | 27.5 | 27.5 | ALQDIDGVSNLK |
| ATCG00770 | PRPS8 | 15.5 | 560 | 492 | 33.6 | 43.3 | DTIADIITSIR |
| AT1G07320 | PRPL4 | 30.5 | 541 | 223 | 25.5 | 20.6 | TLNLFDILNADK |
| AT2G33800 | PRPS5 | 32.7 | 426 | 300 | 10.9 | 4.6 | IVLEMAGVENALGK |
| AT5G30510 | PRPS1 | 45.3 | 410 | 466 | 26.4 | 34.1 | GGLVALVEGLR |
| AT1G05190 | PRPL6 | 24.7 | 409 | 296 | 31.8 | 31.8 | GPLGELALTYPR |
| AT1G74970 | PRPS9 | 22.6 | 379 | 188 | 16.3 | 16.3 | EYLQGNPLWLQYVK |
| AT3G44890 | PRPL9 | 22.2 | 331 | 289 | 17.8 | 17.8 | LIFGSVTAQDLVDIIK |
| AT5G14320 | PRPS13 | 19.4 | 305 | 243 | 17.8 | 19.5 | DMAEEELIILR |
| AT2G43030 | PRPL3 | 29.3 | 295 | 155 | 13.7 | 8.1 | TLATDGYDAVQIGYR |
| AT1G32990 | PRPL11 | 23.3 | 209 | 254 | 16.2 | 25.2 | AGYIIPVEITVFDDK |
To further study the interactions of CRASS with plastid ribosomal proteins in vivo, yeast two-hybrid (Y2H) analyses and immunoblot analyses of coimmunoprecipitates were performed. The Y2H experiments failed to demonstrate direct interactions of CRASS with any of the ribosomal proteins in the selected set (Supplemental Fig. S6), indicating that CRASS might not recognize the individual proteins tested but perhaps binds to another ribosomal protein or interacts with more complex structures that are accessible on ribosomal particles. Examination of CRASS-YFP coimmunoprecipitates by immunoblot analysis clearly confirmed the presence of PRPS1 and PRPS5 (Fig. 7B). Furthermore, similar experiments employing specific antibodies in the presence of RNase A showed that the interaction of CRASS with PRPS1 and PRPS5 is independent of the presence of chloroplast RNAs (Supplemental Fig. S7).
Taken together, our results suggest that CRASS forms a complex with the chloroplast ribosomal 30S subunit. The formation of this complex is based primarily on protein-protein interactions, since CRASS does not require RNA to interact with PRPS1- or PRPS5-containing complexes. The interaction between CRASS and the 30S ribosomal subunits presumably involves the recognition by CRASS of additional, as yet unknown, proteins or of a composite feature of the 30S structure rather than the binding of individual proteins, as indicated by the negative Y2H results.
DISCUSSION
CRASS Is Associated with Plastid Ribosomes
Various protein components of the plastid ribosome have been functionally characterized (Pesaresi et al., 2001; Rogalski et al., 2006, 2008; Fleischmann et al., 2011; Romani et al., 2012; Tiller and Bock, 2014). Additional proteins transiently associate with plastid ribosomes during the different steps of protein synthesis (Ramakrishnan, 2002; Tiller and Bock, 2014), and the inactivation of auxiliary proteins such as initiation factors (Miura et al., 2007; Zheng et al., 2016), elongation factors (Albrecht et al., 2006; Ruppel and Hangarter, 2007), or ribosome recycling factors (Motohashi et al., 2007) can have severe phenotypic effects due to impaired translation, ranging from marked decreases in pigments and photosynthetic protein levels to lethality. In addition, a large number of RNA processing factors, including pentatricopeptide repeat/tetratricopeptide repeat proteins and DEAD-box RNA helicases, participate in the biogenesis of plastid rRNAs (Stern et al., 2010). Their functional and physical associations with ribosomes (Meurer et al., 2017; Paieri et al., 2018) justify the inclusion of such plastid rRNA maturation factors in the family of auxiliary proteins involved in the assembly of plastid ribosomes.
As their name implies, most PSRPs lack eubacterial homologs (Tiller et al., 2012), and their functions are thought to be related to the regulation of translation (Manuell et al., 2007; Sharma et al., 2007; Tiller et al., 2012) and may possibly require the binding and stabilizing of specific regions of chloroplast rRNAs (Bieri et al., 2017). Based on their mutant phenotypes, PSRPs have been classified into two types (Tiller et al., 2012): (1) PSRPs with a structural role in the ribosome, which are bona fide subunits of the plastid ribosome; and (2) nonessential PSRPs, which, when mutated, have no effect on ribosome levels or function under normal greenhouse conditions. Our results indicate that CRASS strongly resembles a nonessential PSRP. Like most PSRPs, CRASS represents an embryophyte invention (Fig. 1A), its transcriptional expression profile is very similar to that of nucleus-encoded ribosomal proteins (Fig. 1B; Supplemental Fig. S1), and it is associated physically with plastid ribosomes (Figs. 6 and 7; Table 1). Moreover, the abundance of CRASS reported in the literature is well within the range typical for chloroplast ribosomal proteins (see Supplemental Table S2 in Zybailov et al., 2008). Because crass mutants do not show any marked phenotype under normal greenhouse conditions (Figs. 2 and 3; Supplemental Fig. S3), the CRASS protein qualifies as a nonessential PSRP-like protein rather than as a PSRP with an indispensable structural role.
CRASS Becomes Critical for Translation Only under Certain Conditions
In accordance with their mild growth phenotype, rRNA processing, levels of photosynthetic proteins, and polysome densities were unchanged in crass mutants grown under controlled greenhouse conditions (Fig. 3). Specific treatments were required to uncover conditions in which the presence of CRASS becomes critical for efficient translation in chloroplasts. Thus, crass-1 plants were found to be more sensitive to treatments with the translation inhibitors LIN and CAP than wild-type plants (Fig. 4) and to exposure to low temperatures (Fig. 5), and in both cases, translation rates were lower in the mutants than in wild-type plants. The photosynthetic phenotype is more pronounced in young leaves (Fig. 5A), indicating that the impaired translation in crass-1 plants under cold stress affects the biogenesis of the photosystems more than repair. Abnormal sensitivity to antibiotics and cold is well known in mutants defective in proteins involved in plastid translation. For instance, loss of the 23S-4.5S maturation factor RH50 leads to erythromycin sensitivity and reduced chloroplast protein synthesis upon cold exposure (Paieri et al., 2018). Moreover, mutants for plastid ribosomal subunits (like PRPS5, PRPS6, and PRPL33) or the elongation factor SVR3 display reduced tolerance to cold stress (Rogalski et al., 2008; Liu et al., 2010; Zhang et al., 2016; Wang et al., 2017). Furthermore, the cytosolic pre-18S rRNA processing factor RH7 is involved in cold tolerance (Huang et al., 2016), providing a general link between temperature on the one hand and translation or ribosome biogenesis on the other. Low temperatures might stabilize certain secondary RNA structures, and the above-mentioned proteins might be required for unfolding and/or refolding of rRNAs under these conditions (Herschlag, 1995; Jones and Inouye, 1996), such that, in the absence of these proteins, the processing and function of plastid rRNAs is impaired.
What Is the Molecular Function of CRASS?
After the exposure of crass mutants to cold stress, the abundance of the ribosomal proteins PRPS1 and PRPS5 was reduced, which probably explains the accompanying perturbation in chloroplast translation (Fig. 5; Supplemental Table S2). The high degree of enrichment of 16S rRNA in RIP-seq experiments suggests that CRASS may interact in particular with the small ribosomal 30S subunit (Fig. 6; Supplemental Table S3). However, in contrast to many mutants for known RNA-interacting proteins (Nishimura et al., 2010; Fristedt et al., 2014; Kleinknecht et al., 2014; Liu et al., 2015; Wu et al., 2016), the absence of CRASS failed to alter the accumulation of its putative target rRNA, the 16S rRNA, under standard conditions (Fig. 3A). Moreover, CRASS lacks a classical RNA-interacting domain, prompting us to speculate that it interacts primarily with proteins and that coimmunoprecipitation of rRNA is due to its association with ribosomal proteins or associated factors rather than with the rRNA itself. In fact, the overwhelming majority of proteins detected by mass spectrometry in coimmunoprecipitates of CRASS belong to the ribosomal protein family (Table 1; Supplemental Table S4). Interestingly, only direct interactions between CRASS and primary components of the small ribosomal subunit (PRPS1 and PRPS5) could be confirmed by immunoblot analysis of coimmunoprecipitates (Fig. 7B), and these interactions also occur in the absence of rRNAs (Supplemental Fig. S7). CRASS comigrates with ribosomal particles of the small subunit (Fig. 7A), suggesting that CRASS interacts with proteins of the 30S subunit either directly or via another factor. In consequence, the coimmunoprecipitation of 16S rRNA is most probably a secondary effect of pulling down the native complex.
CONCLUSION
Since CRASS is present in immature and fully assembled ribosomes, it may act as an auxiliary ribosomal factor, like the nonessential type of PSRPs (Tiller et al., 2012; Tiller and Bock, 2014). Indeed, CRASS shares many characteristics with nonessential PSRPs. Its mutant phenotype, mRNA expression profile, association with plastid ribosomes, noncyanobacterial origin, and abundance all are strikingly reminiscent of plastid ribosomal proteins. Nevertheless, in contrast to PSRPs, CRASS has not been identified directly in plastid ribosomal preparations (Yamaguchi et al., 2000; Yamaguchi and Subramanian, 2003), implying that CRASS is only weakly associated with ribosomes and is easily dissociated from them.
It is tempting to speculate that CRASS might play a regulatory or stabilizing role in stressful conditions, such as upon cold treatment. Future biochemical studies will be required to determine the precise nature of its association with the ribosome or its assembly as well as additional auxiliary factors involved in the interaction with proteins from the small ribosomal subunit. To identify further auxiliary proteins of plastid ribosomes such as CRASS that interact only transiently with the plastid ribosome, our guilt-by-association strategy can be extended for the identification of additional candidate proteins that might be present in the periphery of the previously described plastid ribosome coexpression regulon (Supplemental Fig. S1).
MATERIALS AND METHODS
Coexpression Analysis
The coexpression database for plants, ATTED-II (atted.jp), is based on both microarray and RNA-seq data (Obayashi et al., 2018). For guilt-by-association approaches, the NetworkDrawer tool from ATTED-II was used in Cytoscape mode to analyze genes coregulated with genes for ribosomal proteins. Additionally, the HCluster tool with the single linkage method was employed to perform hierarchical clustering using CRASS as bait.
Phylogenetic Analysis
CRASS orthologs were identified using BLAST (blast.ncbi.nlm.nih.gov). Sequences were aligned with MUSCLE (www.ebi.ac.uk/Tools/msa/muscle) and BioEdit Sequence Alignment Editor version 7.0.5 (www.mbio.ncsu.edu/BioEdit/bioedit.html). The phylogenetic tree rooted at midpoint was constructed using the neighbor-joining method in MEGA6 (megasoftware.net). The evolutionary distances were computed using the Poisson correction method, and the bootstrap test was performed with 2,000 replications.
Confocal Microscopy
Transgenic 7-d-old plants were analyzed for YFP fluorescence by confocal laser-scanning microscopy using a Leica TCS SP2 device. Samples were excited at 514 nm, and fluorescence was detected in the range 550 to 600 nm for YFP and 600 to 700 nm for chlorophyll emission.
Plant Material, Propagation, and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) crass-1 (line 84-776) and crass-2 (line 72-131) mutants in the Col-0 background were obtained from the Czaba Koncz collection (Max Planck Institute for Plant Breeding Research; Ríos et al., 2002). To generate the oeCRASS:YFP-overexpressing lines in the wild-type background, the coding sequence of CRASS (AT5G14910) was PCR amplified using gene-specific primers (Supplemental Table S5), and the gel-purified PCR product was used for BP and LR clonase reactions (Invitrogen). The resulting product was cloned in frame into the Gateway binary vector pGWB641 containing a 35S promoter and YFP (Invitrogen). Agrobacterium tumefaciens-mediated transformation was performed by floral dipping densely sown Col-0 wild-type plants in a solution of transformed A. tumefaciens (strain GV3101). After seed set, transgenic plants were selected on the basis of their resistance to BASTA, propagated, and genotyped. The oeCRASS:YFP crass-1 plants were generated by crossing oeCRASS:YFPWT with crass-1 plants and selecting lines expressing oeCRASS:YFP in the homozygous crass-1 background in the F2 generation.
After stratification for 3 d in darkness at 4°C, wild-type (Col-0) and transgenic plants were grown on soil or MS agar plates supplemented with 1% (w/v) Suc. Plants were grown under controlled conditions in growth chambers at 22°C under long-day (16 h of light/8 h of dark, 100 μmol photons m−2 s−1) or short-day (8 h of light/16 h of dark, 100 μmol photons m−2 s−1) conditions. At 21 d after germination, plants were harvested for weight measurements or transferred to liquid nitrogen for protein extraction. For coimmunoprecipitation experiments, plants were grown in soil on a 12-h-light/12-h-dark cycle (100 μmol photons m−2 s−1) for 2 weeks. For cold stress experiments, adult plants were grown in soil under long-day conditions for 2 weeks at 22°C and then moved to a 4°C chamber (Percival Scientific LED 41HL2) equipped with white and red LEDs set at 18% intensity (equivalent to 100 μmol photons m−2 s−1) for 5 weeks. Alternatively, plants were germinated and grown on plates for 6 weeks in the same chamber and under the same conditions. Seedlings were otherwise grown for 10 d on plates supplemented with the indicated concentrations of LIN or CAP.
Chlorophyll Fluorescence Measurements
In vivo room temperature chlorophyll a fluorescence of leaves was analyzed using an Imaging PAM chlorophyll fluorometer equipped with the computer-operated PAM control unit IMAG-MAXI (Walz) as described previously (Zagari et al., 2017). The minimal fluorescence (F0) was measured after acclimation for at least 30 min in the dark. For cold tolerance experiments, dark acclimation was carried out at 4°C. To determine the maximum fluorescence (Fm), a pulse (0.8 s) of saturating white light (5,000 μmol photon m−1 s−1) was applied. The ratio (Fm − F0)/Fm was calculated as Fv/Fm, the maximum quantum yield of PSII. False-color images representing Fv/Fm levels in wild-type and mutant leaves were produced by the Imaging PAM device, and representative images were selected. The effective quantum yield of PSII [(Fm′ – Fs)/Fm′] and the level of nonphotochemical quenching [(Fm – Fm′)/Fm′)] were monitored at increasing light intensities and plotted as light-response curves.
Nucleic Acid Analysis
Arabidopsis genomic DNA was isolated by a phenol- and chloroform-free method (Edwards et al., 1991). The crass-1 and crass-2 T-DNA insertion-junction sites were recovered by PCR using combinations of insertion- and gene-specific primers (Supplemental Table S5) and subsequent sequencing of the PCR products.
RNA extraction, electrophoresis, transfer, and probe labeling were performed as described (Manavski et al., 2015). Blots were routinely stripped and reprobed. For primer information, see Supplemental Table S5. To ensure equal loading, quantification of RNA was performed on a Nanodrop (Thermo Scientific) device.
Polysome loading experiments were performed as described previously (Barkan, 1993). The psaA probe was amplified with specific primers, whereas an 80-mer oligonucleotide was used as a probe for rbcL (Supplemental Table S5). Labeling was performed as described (Manavski et al., 2015).
The RNA samples used for RNA-seq were analyzed by capillary electrophoresis using a 2100 Bioanalyzer and the RNA 6000 Nano Kit (Agilent) according to the manufacturer’s instructions.
For reverse transcription-quantitative PCR, 1-μg aliquots of RNA were reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative PCR experiments were performed in a final volume of 15 μL containing 7.5 μL of iQ SYBR Green Supermix solution (Bio-Rad), cDNA derived from 15 ng of input RNA, and sense and antisense primers (0.5 μm). A standard thermal profile (95°C for 5 min, 40 cycles of 95°C for 10 s, 55°C for 30 s, and 72°C for 20 s) was employed in an iQ5 real-time PCR detection system (Bio-Rad). Relative CRASS transcript levels were determined by analysis of the threshold cycles with the iQ5 software (Bio-Rad), and UBIQUITIN10 was used as the internal control (Supplemental Table S5).
In Vivo Translation Assay
In vivo labeling of newly synthesized chloroplast protein with [35S]Met was performed as described previously (Meurer et al., 2017). Labeling was performed for 15 min in ambient light. Soluble and insoluble fractions were prepared as described previously (Torabi et al., 2014), and proteins were loaded onto 12% SDS-polyacrylamide gels according to the calculated total counts (100% corresponds to 100,000 cpm for insoluble proteins and 1,000,000 cpm for soluble proteins). Gels were stained for 1 h with Roti-Blue quick (Carl Roth) and dried.
RNA Sequencing, Mapping, and Slot-Blot Analysis
RNA was isolated from the coimmunoprecipitates (see below) obtained from two replicates of the oeCRASS:YFPWT#1 line and two replicates of the wild type (Col-0) and analyzed with Bioanalyzer (Agilent). Specific RNA peaks were detected only in the samples from the oeCRASS:YFPWT#1 line, and the samples were subjected to strand-specific transcriptome sequencing without enrichment for mRNA (i.e. without poly[T] oligonucleotides) and without rRNA depletion (Beijing Genomics Institute). The RNA was fragmented to lengths of 160 to 180 nucleotides and reverse transcribed, dATP was then added, the fragments were size selected by gel electrophoresis, and the selected fragments were amplified by PCR. The sequencing was done using a paired-end 100-nucleotide protocol on an Illumina HiSeq 4000. Paired-end reads were mapped to the TAIR10 Arabidopsis genome (version 31) using STAR aligner 2.5.0 (Dobin et al., 2013) with the following options: -alignIntronMax 5000 - outFilterMismatchNmax 4 - outSAMmultNmax 1 - outMultimapperOrder Random. Next, the bam file was loaded into R and reads were counted with the summarizeOverlaps function from the GenomicAlignments package (Lawrence et al., 2013). Genes with at least 10 mapped reads were used for further analysis. Fragments per kilobase per million mapped fragment values were calculated using the fpkm function from the DEseq2 package (Love et al., 2014).
Slot-blot experiments were performed as described (Manavski et al., 2015). Primers for PCR probes are listed in Supplemental Table S5.
Protein Isolation and Immunoblot Analyses
Protein analyses were performed as described previously (Pulido et al., 2013). Briefly, total plant protein extracts were obtained from 50 mg of 21-d-old fresh tissue by grinding samples in liquid nitrogen. The powder was resuspended in 100 μL of ice-cold TKMES homogenization buffer (100 mm Tricine-KOH, pH 7.5, 10 mm KCl, 1 mm MgCl2, 1 mm EDTA, and 10% [w/v] Suc) supplemented with 0.2% (v/v) Triton X-100, 1 mm DTT, and 20 μL mL−1 protease inhibitor cocktail (Roche). The resuspended sample was centrifuged at 2,300g for 10 min at 4°C, and the supernatant was recovered for a second centrifugation. The supernatant protein concentration was determined using the Bio-Rad protein assay. After SDS-PAGE, the proteins were electrotransferred to Hybond-P polyvinylidene difluoride membranes (GE Healthcare). After protein transfer was complete, membranes were incubated overnight at 4°C with the appropriate specific primary antibody, diluted 1:5,000 to detect YFP, PRPS1, PRPS5, PRPL2, PRPL4, PRPL11, CLPC, CLPB3, CPN60α1, D2, PSBR, PSAL, CYT f, CYT b6, and ACTIN and 1:10,000 for HSP70, LHCA1, LHCB2, RBCL, and ATPβ. With the exception of α-GFP (Life Technologies) and α-PRPL11 (Meurer et al., 2017), all antibodies were purchased from Agrisera. Incubation with the horseradish peroxidase-conjugated secondary antibody (diluted 1:10,000) was performed for 1 h at room temperature. The detection of immunoreactive bands was performed using the ECL Plus reagent (GE Healthcare). Chemiluminescent signals were visualized using a ChemiDoc MP analyzer (Bio-Rad).
Pigment Analysis
Chlorophyll quantification was performed as described previously (Lichtenthaler and Wellburn, 1983). Briefly, pigments were extracted by shaking 50-mg (fresh weight) samples with 1 mL of 80% (v/v) ice-cold acetone in the dark at 4°C for 30 min. After centrifugation (10,000g, 10 min, 4°C), A663, A647, and A470 were recorded with a spectrophotometer (Ultrospec3100; Amersham Biosciences) and pigment levels were calculated according to the following equations: chlorophyll a = 12.25 A663 – 2.79 A647; chlorophyll b = 21.50 A647 – 5.10 A663; chlorophyll tot = 7.15 A663 + 18.71 A647; carotenoids = (1,000 A470 – 1.82 Cla – 85.02 Clb)/198.
Chloroplast Fractionation
Chloroplasts were isolated from 3-week-old CRASS-GFP plants as described previously (Stoppel et al., 2012). Chloroplasts were suspended in lysis buffer (30 mm HEPES, pH 8, 10 mm magnesium acetate, 60 mm potassium acetate, and protease inhibitor cocktail [Roche]) and passed 20 times through a 0.5-mm needle. After centrifugation (45,000g for 30 min at 4°C), the supernatant (stroma) was placed on ice, while the pellet (thylakoids) was washed twice with washing buffer (30 mm HEPES, pH 8, 100 mm sorbitol, and 10 mm MgCl2) and centrifuged at 5,000g for 5 min at 4°C. Thirty micrograms of stroma extract and thylakoid proteins corresponding to 5 µg of chlorophyll were separated via SDS-PAGE.
Coimmunoprecipitation Analysis
Stroma extracts and coimmunoprecipitates were prepared as described (Paieri et al., 2018), except for the RNA-coimmunoprecipitated GFP antibodies (A-6455; Thermo Fisher Scientific) and 500 µg of stroma extracts for each coimmunoprecipitation was used. For RNase treatment, stroma extracts were incubated with 0.2 µg of RNaseA (Qiagen)/1 µg of stroma at room temperature for 15 min prior to coimmunoprecipitation. Washing, RNA isolation, and slot-blot analysis were conducted as described (Meurer et al., 2017). For mass spectrometry analyses, the eluates were fractionated on an SDS-PAGE gel (12% (w/v) polyacrylamide) and stained with Colloidal Coomassie for mass spectrometry analyses.
The extractions for the immunoprecipitations for RNA-seq and mass spectrometry were done with polysome extraction buffer (Barkan, 1993). No translation inhibitors were added except in one control experiment (Fig. 6, D and E), in which 100 µg mL−1 CAP was used.
Mass Spectrometry
In-gel tryptic digestion was done as described previously (Shevchenko et al., 2006), and peptides were resolubilized in 2.5% (v/v) acetonitrile and 0.5% (v/v) trifluoroacetic acid. Desalting was done in the nano RSLC Ultimate 3000 system (Dionex) on a 500-mm Acclaim PepMap C18 column (particle size, 3 µm, with nano viper fingertight fittings), and peptides were separated on a 150-mm Acclaim PepMap C18 (particle size, 2 µm) using a linear acetonitrile gradient made up of solvent A (0.1% (v/v) formic acid in water) and solvent B (0.1% (v/v) formic acid in 90% (v/v) acetonitrile and 10% water). A gradient from 2% B to 45% B was first applied over a period of 30 min. For eluting and washing the column, B was then increased to 90% over 1 min and maintained for a further 5 min. A 15-min reequilibration step followed. The connected ion-trap AmZon ETD instrument (Bruker) measured the peptides with the factory proteomics AutoMS/MS CID method (capillary voltage 1,300, temperature 180°C, mode Ultrascan for parental masses, Xtreme Scan for fragmented masses with Smart fragmentation on, top4 fragmentation, dynamic exclusion 0.2 min). A total of 5,000 compound spectra with a TIC intensity higher than 10,000 were converted by the DataAnalysis software (Bruker) to mgf files and searched against the TAIR10 peptide database including contaminants with the Mascot Daemon 2.5.1. An error of 0.5 D was allowed for the parental mass and the fragmented masses. Carbamidomethylation was set as a fixed modification and oxidation as a variable one. Peptides were taken as identified with a score above 21. Proteins were taken as identified with two peptides for one protein or one reproducible peptide between the experiments with a score above 60. As significance threshold of P < 0.01 was used.
Y2H Analysis
The coding sequences of CRASS, PRPS5, and PRPS8 (excluding the transit peptides) were cloned into pGKBT7 (CRASS and GUN1) and pGADT7 (PRPS1, PRPS5, PRPS8, and PRPL24) vectors (Clontech). The constructs for PRPS1, PRPL24, and GUN1 were described previously (Tadini et al., 2016). Interactions in yeast were analyzed as described earlier (DalCorso et al., 2008).
SEC and Suc-Gradient Fractionation
Chloroplasts were isolated from 2-week-old plants as described previously (Stoppel et al., 2012). Chloroplasts were lysed in extraction buffer (10 mm HEPES-KOH, pH 8, 5 mm MgCl2, and protease inhibitor cocktail [Roche]) by passing the suspension 20 times through a 0.5-mm needle. Membranes were pelleted by centrifugation at 45,000g for 30 min at 4°C.
SEC analysis of stroma fractions from CRASS-YFP-overexpressing lines (oeCRASS:YFPWT#1) was performed as described (Meurer et al., 2017). Aliquots (3 mg) of stroma were treated with 300 mg of RNase A (Qiagen) for 1 h on ice and centrifuged at 16,000g for 10 min at 4°C to pellet precipitates prior to SEC.
Accession Numbers
The following genes were used to create the gene network in Supplemental Figure S1: PRPS1 (AT5G30510), PRPS5 (AT2G33800), PRPS6 (AT1G64510), PRPS7 (AT5G30510), PRPS9 (AT1G74970), PRPS10 (AT3G13120), PRPS13 (AT5G14320), PRPS17 (AT1G79850), PRPS20 (AT3G15190), PRPS21 (AT3G27160), PRPL1 (AT3G63490), PRPL3 (AT2G43030), PRPL4 (AT1G07320), PRPL5 (AT4G01310), PRPL6 (AT1G05190), PRPL9 (AT3G44890), PRPL10 (AT5G13510), PRPL11 (AT1G32990), PRPL13 (AT1G78630), PRPL15 (AT3G25920), PRPL17 (AT3G54210), PRPL18 (AT1G48350), PRPL19 (AT5G47190 and AT4G17560), PRPL21 (AT1G35680), PRPL24 (AT5G54600), PRPL27 (AT5G40950), PRPL28 (AT2G33450), PRPL29 (AT5G65220), PRPL31 (AT1G75350), PRPL34 (AT1G29070), PRPL35 (AT2G24090), PSRP2 (AT3G52150), PSRP3 (AT1G68590), PSRP4 (AT2G38140), PSRP5 (AT3G56910), and PSRP6 (AT5G17870).
Arabidopsis genes found to be coregulated with CRASS in Figure 1A have the following accession numbers: LPA2 (AT5G51545), FKBPL (AT3G60370), MORF9 (AT1G11430), CPN10 (AT2G44650), Rhodanese (AT3G08920), FLU (AT3G14110), EF-P (AT3G08740), PRPL24 (AT5G546000), PRPL29 (AT5G65220), RBP31 (AT4G24770), PRPS5 (AT2G33800), PRPS1 (AT5G30510), PRPL28 (AT2G33450), PRPL19 (AT5G47190), PRPL34 (AT1G29070), PRPL15 (AT3G25920) PRPL10 (AT5G13510), PRPS6 (AT1G64510), PRPL18 (AT1G48350), PRPS20 (AT3G15190), PRPL6 (AT1G05190), PSRP2 (AT3G52150), PRPL3 (AT2G43030), PRPL31 (AT1G75350), PRPL17 (AT3G54210), PRPS13 (AT5G14320), PRPL13 (AT1G78630), PRPL5 (AT4G01310), PRPL19 (AT4G17560), PRPL11 (AT1G32990), and PRPL21 (AT1G35680).
CRASS proteins from Figure 1B have the following accession numbers: AT5G14910, CRASS Arabidopsis; XP_006286392, Capsella rubella; XP_002871636, Arabidopsis lyrata; XP_010453619, Camelina sativa; CDX85578, Brassica napus; XP_009131426, Brassica rapa; XP_008341487, Malus domestica; KDO72385, Citrus sinensis; XP_002275276, Vitis vinifera; KNA24776, Spinacia oleracea; XP_008443474, Cucumis melo; XP_004147445, Cucumis sativus; XP_006338529, Solanum tuberosum; KMZ72790, Zostera marina; AFK34041, Lotus japonicus; XP_004232276, Solanum lycopersicum; XP_009786795, Nicotiana sylvestris; KRH18591, Glycine max; XP_013450869, Medicago truncatula; EEE63302, Oryza sativa; NP_001143959, Zea mays; BAJ98599, Hordeum vulgare; ABK23791, Picea sitchensis; XP_001772064, Physcomitrella patens; XP_002968622, Selaginella moellendorffii; XP_001758023, P. patens; and XP_001763071, P. patens.
Sequences used in the sequence alignment of CRASS with HMA proteins (Supplemental Fig. S2) have the following accession numbers: AT5G14910, CRASS Arabidopsis; XP_002275276, V. vinifera; XP_002968622, S. moellendorffii; WP_003720172, Listeria ivanovii; WP_014093194, L. ivanovii; WP_051872593, Chryseobacterium haifense; WP_059344219, Elizabethkingia genomosp. 2; WP_007292244, Delta proteobacterium MLMS-1; and ANC24349, Streptococcus pyogenes.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The plastid ribosome coexpression regulon.
Supplemental Figure S2. Sequence alignment of CRASS and bacterial HMA proteins.
Supplemental Figure S3. Under standard growth conditions, altered CRASS levels have only minor effects on photosynthesis.
Supplemental Figure S4. Analysis of the ribosomal protein and rRNA content at the end of the cold period.
Supplemental Figure S5. RNA content of coimmunoprecipitates of CRASS-YFP.
Supplemental Figure S6. Y2H analysis of interactions between CRASS and plastid ribosomal proteins.
Supplemental Figure S7. RNA dependence of CRASS-PRPS1 interaction.
Supplemental Table S1. Quantification of immunoblot signals (nonstressed conditions), including visualization as histogram.
Supplemental Table S2. Quantification of immunoblot signals (cold stress), including visualization as histogram.
Supplemental Table S3. RIP-seq data set obtained with tagged CRASS.
Supplemental Table S4. Mass spectrometry data set obtained by coimmunoprecipitation of tagged CRASS protein.
Supplemental Table S5. List of primers used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Paul Hardy for critical reading of the article.
Footnotes
This work was funded by grants from the European Union Marie Curie IIF program (ChloroQuality, grant agreement no. 656822) to P.P., the Fondazione Edmund Mach (CRI-FEM) to N.Z., the Deutsche Forschungsgemeinschaft (TRR 175, projects A03 and C05) to J.M. and D.L., and the Polish National Science Centre (SONATA12, UMO-2016/23/D/NZ3/02491) to P.G.
Articles can be viewed without a subscription.
References
- Ahmed T, Yin Z, Bhushan S (2016) Cryo-EM structure of the large subunit of the spinach chloroplast ribosome. Sci Rep 6: 35793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed T, Shi J, Bhushan S (2017) Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation. Nucleic Acids Res 45: 8581–8595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht V, Ingenfeld A, Apel K (2006) Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: the impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol Biol 60: 507–518 [DOI] [PubMed] [Google Scholar]
- Barkan A. (1993) Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biehl A, Richly E, Noutsos C, Salamini F, Leister D (2005) Analysis of 101 nuclear transcriptomes reveals 23 distinct regulons and their relationship to metabolism, chromosomal gene distribution and co-ordination of nuclear and plastid gene expression. Gene 344: 33–41 [DOI] [PubMed] [Google Scholar]
- Bieri P, Leibundgut M, Saurer M, Boehringer D, Ban N (2017) The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J 36: 475–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerema AP, Aibara S, Paul B, Tobiasson V, Kimanius D, Forsberg BO, Wallden K, Lindahl E, Amunts A (2018) Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Nat Plants 4: 212–217 [DOI] [PubMed] [Google Scholar]
- Börner T, Aleynikova AY, Zubo YO, Kusnetsov VV (2015) Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim Biophys Acta 1847: 761–769 [DOI] [PubMed] [Google Scholar]
- DalCorso G, Pesaresi P, Masiero S, Aseeva E, Schünemann D, Finazzi G, Joliot P, Barbato R, Leister D (2008) A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132: 273–285 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann TT, Scharff LB, Alkatib S, Hasdorf S, Schöttler MA, Bock R (2011) Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23: 3137–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier JB, Rudella A, Sun Q, Wijk KJ (2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16: 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Scharff LB, Clarke CA, Wang Q, Lin C, Merchant SS, Bock R (2014) RBF1, a plant homolog of the bacterial ribosome-binding factor RbfA, acts in processing of the chloroplast 16S ribosomal RNA. Plant Physiol 164: 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf M, Arenz S, Huter P, Dönhöfer A, Novácek J, Wilson DN (2017) Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions. Nucleic Acids Res 45: 2887–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D. (1995) RNA chaperones and the RNA folding problem. J Biol Chem 270: 20871–20874 [DOI] [PubMed] [Google Scholar]
- Hirose T, Sugiura M (2004) Functional Shine-Dalgarno-like sequences for translational initiation of chloroplast mRNAs. Plant Cell Physiol 45: 114–117 [DOI] [PubMed] [Google Scholar]
- Hricová A, Quesada V, Micol JL (2006) The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol 141: 942–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CK, Shen YL, Huang LF, Wu SJ, Yeh CH, Lu CA (2016) The DEAD-box RNA helicase AtRH7/PRH75 participates in pre-rRNA processing, plant development and cold tolerance in Arabidopsis. Plant Cell Physiol 57: 174–191 [DOI] [PubMed] [Google Scholar]
- Jones PG, Inouye M (1996) RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol Microbiol 21: 1207–1218 [DOI] [PubMed] [Google Scholar]
- Kleine T, Maier UG, Leister D (2009) DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol 60: 115–138 [DOI] [PubMed] [Google Scholar]
- Kleinknecht L, Wang F, Stübe R, Philippar K, Nickelsen J, Bohne AV (2014) RAP, the sole octotricopeptide repeat protein in Arabidopsis, is required for chloroplast 16S rRNA maturation. Plant Cell 26: 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ (2013) Software for computing and annotating genomic ranges. PLOS Comput Biol 9: e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D, Wang X, Haberer G, Mayer KF, Kleine T (2011) Intracompartmental and intercompartmental transcriptional networks coordinate the expression of genes for organellar functions. Plant Physiol 157: 386–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JC, Hsieh WY, Tseng CC, Hsieh MH (2016) Dysfunctional chloroplasts up-regulate the expression of mitochondrial genes in Arabidopsis seedlings. Photosynth Res 127: 151–159 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR (1983) Determination of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem Soc Trans 603: 591–592 [Google Scholar]
- Liu J, Zhou W, Liu G, Yang C, Sun Y, Wu W, Cao S, Wang C, Hai G, Wang Z, et al. (2015) The conserved endoribonuclease YbeY is required for chloroplast ribosomal RNA processing in Arabidopsis. Plant Physiol 168: 205–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Rodermel SR, Yu F (2010) A var2 leaf variegation suppressor locus, SUPPRESSOR OF VARIEGATION3, encodes a putative chloroplast translation elongation factor that is important for chloroplast development in the cold. BMC Plant Biol 10: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutsenko S, Petrukhin K, Cooper MJ, Gilliam CT, Kaplan JH (1997) N-terminal domains of human copper-transporting adenosine triphosphatases (the Wilson’s and Menkes disease proteins) bind copper selectively in vivo and in vitro with stoichiometry of one copper per metal-binding repeat. J Biol Chem 272: 18939–18944 [DOI] [PubMed] [Google Scholar]
- Lyska D, Meierhoff K, Westhoff P (2013) How to build functional thylakoid membranes: from plastid transcription to protein complex assembly. Planta 237: 413–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavski N, Torabi S, Lezhneva L, Arif MA, Frank W, Meurer J (2015) HIGH CHLOROPHYLL FLUORESCENCE145 binds to and stabilizes the psaA 5′ UTR via a newly defined repeat motif in embryophyta. Plant Cell 27: 2600–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavski N, Schmid LM, Meurer J (2018) RNA-stabilization factors in chloroplasts of vascular plants. Essays Biochem 62: 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuell AL, Quispe J, Mayfield SP (2007) Structure of the chloroplast ribosome: novel domains for translation regulation. PLoS Biol 5: e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J, Lezhneva L, Amann K, Gödel M, Bezhani S, Sherameti I, Oelmüller R (2002) A peptide chain release factor 2 affects the stability of UGA-containing transcripts in Arabidopsis chloroplasts. Plant Cell 14: 3255–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J, Schmid LM, Stoppel R, Leister D, Brachmann A, Manavski N (2017) PALE CRESS binds to plastid RNAs and facilitates the biogenesis of the 50S ribosomal subunit. Plant J 92: 400–413 [DOI] [PubMed] [Google Scholar]
- Miura E, Kato Y, Matsushima R, Albrecht V, Laalami S, Sakamoto W (2007) The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant Cell 19: 1313–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi R, Yamazaki T, Myouga F, Ito T, Ito K, Satou M, Kobayashi M, Nagata N, Yoshida S, Nagashima A, et al. (2007) Chloroplast ribosome release factor 1 (AtcpRF1) is essential for chloroplast development. Plant Mol Biol 64: 481–497 [DOI] [PubMed] [Google Scholar]
- Nawaz G, Kang H (2017) Chloroplast- or mitochondria-targeted DEAD-box RNA helicases play essential roles in organellar RNA metabolism and abiotic stress responses. Front Plant Sci 8: 871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Ashida H, Ogawa T, Yokota A (2010) A DEAD box protein is required for formation of a hidden break in Arabidopsis chloroplast 23S rRNA. Plant J 63: 766–777 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Aoki Y, Tadaka S, Kagaya Y, Kinoshita K (2018) ATTED-II in 2018: a plant coexpression database based on investigation of the statistical property of the Mutual Rank Index. Plant Cell Physiol 59: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares PD, Ponnala L, van Wijk KJ (2010) Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol Cell Proteomics 9: 1594–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paieri F, Tadini L, Manavski N, Kleine T, Ferrari R, Morandini PA, Pesaresi P, Meurer J, Leister D (2018) The DEAD-box RNA helicase RH50 is a 23S-4.5S rRNA maturation factor that functionally overlaps with the plastid signaling factor GUN1. Plant Physiol 176: 634–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Varotto C, Meurer J, Jahns P, Salamini F, Leister D (2001) Knock-out of the plastid ribosomal protein L11 in Arabidopsis: effects on mRNA translation and photosynthesis. Plant J 27: 179–189 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Blanvillain R, Merendino L, Courtois F, Chevalier F, Liebers M, Grübler B, Hommel E, Lerbs-Mache S (2015) Plastid RNA polymerases: orchestration of enzymes with different evolutionary origins controls chloroplast biogenesis during the plant life cycle. J Exp Bot 66: 6957–6973 [DOI] [PubMed] [Google Scholar]
- Pulido P, Toledo-Ortiz G, Phillips MA, Wright LP, Rodríguez-Concepción M (2013) Arabidopsis J-protein J20 delivers the first enzyme of the plastidial isoprenoid pathway to protein quality control. Plant Cell 25: 4183–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V. (2002) Ribosome structure and the mechanism of translation. Cell 108: 557–572 [DOI] [PubMed] [Google Scholar]
- Ríos G, Lossow A, Hertel B, Breuer F, Schaefer S, Broich M, Kleinow T, Jásik J, Winter J, Ferrando A, et al. (2002) Rapid identification of Arabidopsis insertion mutants by non-radioactive detection of T-DNA tagged genes. Plant J 32: 243–253 [DOI] [PubMed] [Google Scholar]
- Rogalski M, Ruf S, Bock R (2006) Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res 34: 4537–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski M, Schöttler MA, Thiele W, Schulze WX, Bock R (2008) Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell 20: 2221–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani I, Tadini L, Rossi F, Masiero S, Pribil M, Jahns P, Kater M, Leister D, Pesaresi P (2012) Versatile roles of Arabidopsis plastid ribosomal proteins in plant growth and development. Plant J 72: 922–934 [DOI] [PubMed] [Google Scholar]
- Ruppel NJ, Hangarter RP (2007) Mutations in a plastid-localized elongation factor G alter early stages of plastid development in Arabidopsis thaliana. BMC Plant Biol 7: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff LB, Childs L, Walther D, Bock R (2011) Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites. PLoS Genet 7: e1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff LB, Ehrnthaler M, Janowski M, Childs LH, Hasse C, Gremmels J, Ruf S, Zoschke R, Bock R (2017) Shine-Dalgarno sequences play an essential role in the translation of plastid mRNAs in tobacco. Plant Cell 29: 3085–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MR, Wilson DN, Datta PP, Barat C, Schluenzen F, Fucini P, Agrawal RK (2007) Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc Natl Acad Sci USA 104: 19315–19320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Sohmen D, Harms JM, Schlunzen F, Wilson DN (2009) SnapShot: antibiotic inhibition of protein synthesis I. Cell 138: 1248.e1. [DOI] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR (2010) Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Stoppel R, Meurer J (2012) The cutting crew: ribonucleases are key players in the control of plastid gene expression. J Exp Bot 63: 1663–1673 [DOI] [PubMed] [Google Scholar]
- Stoppel R, Manavski N, Schein A, Schuster G, Teubner M, Schmitz-Linneweber C, Meurer J (2012) RHON1 is a novel ribonucleic acid-binding protein that supports RNase E function in the Arabidopsis chloroplast. Nucleic Acids Res 40: 8593–8606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadini L, Pesaresi P, Kleine T, Rossi F, Guljamow A, Sommer F, Mühlhaus T, Schroda M, Masiero S, Pribil M, et al. (2016) GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol 170: 1817–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N, Bock R (2014) The translational apparatus of plastids and its role in plant development. Mol Plant 7: 1105–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N, Weingartner M, Thiele W, Maximova E, Schöttler MA, Bock R (2012) The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. Plant J 69: 302–316 [DOI] [PubMed] [Google Scholar]
- Tillich M, Hardel SL, Kupsch C, Armbruster U, Delannoy E, Gualberto JM, Lehwark P, Leister D, Small ID, Schmitz-Linneweber C (2009) Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc Natl Acad Sci USA 106: 6002–6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabi S, Umate P, Manavski N, Plöchinger M, Kleinknecht L, Bogireddi H, Herrmann RG, Wanner G, Schröder WP, Meurer J (2014) PsbN is required for assembly of the photosystem II reaction center in Nicotiana tabacum. Plant Cell 26: 1183–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ouyang M, Li Q, Zou M, Guo J, Ma J, Lu C, Zhang L (2010) The Arabidopsis chloroplast ribosome recycling factor is essential for embryogenesis and chloroplast biogenesis. Plant Mol Biol 74: 47–59 [DOI] [PubMed] [Google Scholar]
- Wang S, Bai G, Wang S, Yang L, Yang F, Wang Y, Zhu JK, Hua J (2016) Chloroplast RNA-binding protein RBD1 promotes chilling tolerance through 23S rRNA processing in Arabidopsis. PLoS Genet 12: e1006027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WJ, Zheng KL, Gong XD, Xu JL, Huang JR, Lin DZ, Dong YJ (2017) The rice TCD11 encoding plastid ribosomal protein S6 is essential for chloroplast development at low temperature. Plant Sci 259: 1–11 [DOI] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J (2013) Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J 73: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Liu S, Ruwe H, Zhang D, Melonek J, Zhu Y, Hu X, Gusewski S, Yin P, Small ID, et al. (2016) SOT1, a pentatricopeptide repeat protein with a small MutS-related domain, is required for correct processing of plastid 23S-4.5S rRNA precursors in Arabidopsis thaliana. Plant J 85: 607–621 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Subramanian AR (2000) The plastid ribosomal proteins: identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J Biol Chem 275: 28466–28482 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Subramanian AR (2003) Proteomic identification of all plastid-specific ribosomal proteins in higher plant chloroplast 30S ribosomal subunit. Eur J Biochem 270: 190–205 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, von Knoblauch K, Subramanian AR (2000) The plastid ribosomal proteins: identification of all the proteins in the 30 S subunit of an organelle ribosome (chloroplast). J Biol Chem 275: 28455–28465 [DOI] [PubMed] [Google Scholar]
- Zagari N, Sandoval-Ibañez O, Sandal N, Su J, Rodriguez-Concepcion M, Stougaard J, Pribil M, Leister D, Pulido P (2017) SNOWY COTYLEDON 2 promotes chloroplast development and has a role in leaf variegation in both Lotus japonicus and Arabidopsis thaliana. Mol Plant 10: 721–734 [DOI] [PubMed] [Google Scholar]
- Zhang J, Yuan H, Yang Y, Fish T, Lyi SM, Thannhauser TW, Zhang L, Li L (2016) Plastid ribosomal protein S5 is involved in photosynthesis, plant development, and cold stress tolerance in Arabidopsis. J Exp Bot 67: 2731–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Liu X, Liang S, Fu S, Qi Y, Zhao J, Shao J, An L, Yu F (2016) Chloroplast translation initiation factors regulate leaf variegation and development. Plant Physiol 172: 1117–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]