Abstract
Myeloperoxidase (MPO) promoter single nucleotide polymorphisms (SNPs) rs2243828 (−764T>C) and rs2333227 (G-463A) program malignant phenotypes by regulating MPO transcriptional activity. In this study, we enrolled a total of 1,175 controls and 1,078 colorectal cancer (CRC) patients with comprehensive clinical and survival information to assess whether these SNPs could affect the susceptibility and development of CRC. The MPO rs2333227 TT genotype significantly increased the risk of CRC and decreased the overall survival time of patients. CRC cells with the rs2333227 TT genotype exhibited enhanced proliferation, migration, and invasion capacity in vitro and in vivo. Mechanistically, we found that MPO SNP rs2333227 C to T mutation altered the binding affinity of the transcription factors AP-2α to the rs2333227 mutation region, sequentially enhancing expression levels of MPO and activating further IL23A-MMP9 axis-mediated oncogenic signaling. Taken together, our findings indicate that MPO SNP rs2333227 serves as a marker of enhanced risk for development of CRC.
Keywords: colorectal cancer, MPO, survival, AP-2α, polymorphism
Introduction
Globally, colorectal cancer (CRC) is the third most-common form of cancer and the second-leading cause of cancer-related death in the western world, with a lifetime risk in the United States of approximately 7%(1). Several factors are routinely used as prognostic markers for predictive prognosis including depth of tumor invasion, nodal status and distant metastasis(2). The 5-year relative survival rate ranges from 90% in CRC patients with stage І to 10% in CRC patients with stage IV(3). Other environmental factors thought to influence CRC prognosis include lifestyle (4) and the tumor immunologic microenvironment(5).
Myeloperoxidase (MPO) is a lysosomal enzyme that is present in neutrophils, monocytes and tissue macrophages. It protects against microbial infections (6) by generating various free radicals and reactive oxidants. Published data provides evidence of high circulating levels of MPO in patients with heart failure or coronary artery diseases(7–9). The single nucleotide polymorphisms (SNPs) rs2333227 (G-463A) and rs2243828 (T-764C), located in the 5' upstream region of the MPO gene, are functional SNPs, as reported in the Carotene and Retinol Efficacy Trial (CARET) cohort (10) and other studies on rectal cancer (11). It is well known that the MPO rs2333227 polymorphism is associated with increased risk for disease, including breast cancer (12), prostate cancer (13,14) and peripheral arterial disease (15). However, an association between MPO variants and CRC risk has yet to be reported.
In the current study, we investigated the association between MPO gene polymorphisms and the risk of CRC in a Chinese population. Experiments were performed to assess the role of SNPs rs2333227 in promoting CRC.
Materials and methods
Study subjects
This case-control study included 1,277 patients with CRC and 1,175 cancer-free controls. Newly diagnosed and histologically confirmed CRC patients were recruited between January 2007 and October 2011 the Jiangsu province (China). Controls were randomly selected from patients attending the same hospital for physical examination. The recruited patients had no history of previous cancer in other organs, or preoperative chemotherapy or radiotherapy, and the controls were genetically unrelated to the CRC patients. The pathological stage of CRC was assessed using the Sixth Edition of the American Joint Committee on Cancer (AJCC) Cancer Staging Manual. The time of death or the last follow-up (in June 2016) of each patient was confirmed by a trained clinical specialist. The median survival time (MST) was 75.0 months and the maximum follow-up time was 112.7 months. Of the recruited patients, 1,078 patients who completed the follow-up were enrolled in the study, with a response rate of 84.4%. Written informed consent was obtained from each enrolled subject before recruitment. The study was conducted in accordance with Declaration of Helsinki and approved by the Institutional Review Board of Southeast University.
DNA extraction and genotype analysis
Due to lack of blood samples, the genomic DNA of the CRC patients was extracted from paraffin-embedded tissues using the EZNA Tissue DNA Kit (Omega Bio-Tek, GA, USA). For the controls, the genomic DNA was extracted from venous blood using the RelaxGene Blood DNA kit (Tiangen Biotech, Beijing, China). The selected polymorphisms were genotyped using TaqMan allelic discrimination assays with the Quant Studio 6 Flex system (Applied Biosystems, CA, USA). The loading wells without DNA were used as negative controls. Ten percent of the samples were randomly selected for confirmation and the results were 100% consistent with previous genotyping results.
Tissue microarray construction and Immunohistochemistry (IHC)
The colorectal cancer TMAs were created by the National Engineering Center for Biochip (Shanghai, China). Duplicate 1.0 mm diameter cores of tissue from each sample were punched from paraffin tumor blocks and adjacent colorectal tissues. The immunohistochemistry (IHC) assay procedure used has been previously described (16). In brief, the specimens were incubated overnight with rabbit polyclonal anti-MPO antibody (BOSTER, Wuhan, China) and anti-MMP9 antibody (Abcam, Cambridge, UK) at 4°C. Each specimen was stained with 3, 3′-diaminobenzidine (DAB; Zhongshan Biotech, Beijing, China). The evaluation of MPO and MMP9 expression were blindly conducted by two independent pathologists. The specimens were categorized using a semi-quantitative immunoreactivity score(17). Briefly, the intensity of immunostaining was deemed as 0–3 (0, negative; 1, weak; 2, moderate; 3,strong, and the percentage of immunoreactive cells were regarded as 1 (0%–25%), 2 (26%–50%), 3 (51%–75%), and 4 (76%–100%). The immunoreactivity score (IRS) was calculated by the intensity multiply the percentage. The optimal cutoff value for the MPO and MMP9 IRS was obtained by the receiver operator characteristic (ROC) analysis. Briefly, the area under the curve (AUC) at different cutoff values of MPO IRS for survival time from 1 to 9 years were calculated, respectively. Under this condition, samples with IRS 0–4 and IRS 6–12 were defined as low and high expression for MPO, respectively. While, samples with IRS 0–6 and IRS 8–12 were defined as low and high expression for MMP9, respectively
Cell culture
The human colorectal cancer cell lines (SW620 and SW480) were purchased from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China) in 2016 and maintained in Dulbecco’s modified Eagle’s medium (DMEM, HyClone) at 37°C under 5% CO2. The medium was supplemented with 10% fetal bovine serum (FBS) (Sigma, St. Louis, MO, USA), 100U/ml penicillin and 100µg/ml streptomycin (HyClone, Logan, UT, USA). The cells were periodically tested and validated to be free of mycoplasma. These two cell lines were tested by short-tandem repeat analysis and used within 6 months. The cells were grown for no more than 25 passages for any expreiaments.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) experiments were performed using the ChIP-ITTM magnetic chromatin immunoprecipitation kit (Cat. No. 53008, Active Motif) according to the manufacturer’s instructions. Briefly, samples were fixed in formaldehyde for 10 min at room temperature. Then, samples were neutralized by treated with glycine stop-fix solution for 5 min at room temperature. The isolated nuclei and chromatin were incubed with antibody against AP-2α, or control IgG at 4 overnight. After sequential ChIP, quantitative PCR was performed to determine the precipitated genomic DNA using the Quant Studio 6 Flex system (Applied Biosystems, Life Technologies, USA), followed by human MPO rs2333227 (G-463A) promoter primers identifying the AP-2α binding sequence: forward: 5'- GCCTCTAGCCACATCATCAA −3'; reverse: 5'- AATTTAGCACTACCAGCCCA-3'.
RNA extraction and real-time RT-PCR analysis
Total RNA was isolated with Trizol reagent (Invitrogen, USA) from frozen tissues and colorectal cancer cells. Quantitative PCR reactions were performed using SYBR Green real-time PCR kits (Toyobo, Osaka, Japan). Primers were 5’-GACAAATACCGCACCATC-3’ (forward) and 5’-AAGCCGTCCTCATACTCC-3’ (reverse) for MPO; 5’-ATCCGCAAAGACCTGT-3’ and 5’-GGGTGTAACGCAACTAAG-3’ for β-actin.
Western blot analysis
Total proteins were extracted from CRC tumor tissues with radioimmunoprecipitation assay buffer (RIPA) lysis buffer. Immunoblot assays were used to detect the MPO protein level by using primary antibodies human MPO (1:1000 dilution, BOSTER Biological Technology, Wuhan) and β-actin (1:10000 dilution; sigma, USA). Semi-quantitative analysis of MPO protein levels was performed with Image Lab 3.0 software (BIO-RAD, USA).
CRISPR/Cas9-mediated genome engineering
The CRISPR/Cas9 expression vectors expressing Cas9, puromycin resistant gene and sgRNA were constructed (Supplementary Fig.S1A) based on a pX330 vector. The sequences of sgRNA targeting the rs2333227 mutation region are shown in Fig S1A. One day before the transfection, the cells were seeded in 6-well plates (2×105 cells/well). CRC cells were co-transfected with single-stranded DNA donor construct (0.1 ug/ml) and CRISPR/Cas9 expression plasmids (1ug/ml) using Lipofectamine 2000 according to the manufacturer’s protocol (Invitrogen, CA, USA). After 24 hr, the medium was replaced with complete medium (2ml) containing puromycin (0.6µg/ml) (VWR Pty Ltd, Brisbane, Australia), then every 2 days until day 14. Surviving cells were plated in a 96 well plate (10 cells/ well) for single-cell-derived colonies. Cells were then harvested for DNA sequencing (rs2333227, Fig. S1B).
Analysis of CRC cell colony formation, migration and invasion
For colony formation ability assays, 500 cells were seeded in 100-mm culture dishes. We added Dulbecco’s modified Eagle’s medium (DMEM) (Hyclone) containing 10% FBS (Sigma), and placed the dishes at 37°C with 5% CO2 in an incubator, allowing the colonies to grow for 10 days. Cell colonies were then stained and imaged according to our previous study (18). Transwell (Corning, New York, USA) assays were performed to detect the effect of different MPO rs2333227 CRC cell genotypes on CRC cell migration and invasion. Cells (5×104) were added to transwell chamber with a membrane pore size of 8 µm (Corning, New York, USA) in triplicate. The cells (5×104) were added to the 8-µm pore insert in serum-free medium. The pore inserts were coated with Matrigel (Cell Biolabs, USA). As a chemoattractant, DMEM containing 10% FBS was added to the lower chambers. After 24 hours, cells were fixed and stained with crystal violet (1%) in PBS/ethanol (85%/15%). Cell migration was determined in an analogous fashion, but in the absence of Matrigel in the transwell chambers.
In vivo nude mice flank tumor experiments
All animal studies were performed with the approval of the Committee on Animal Care and Use of Southeast University. A total of 36 Nude mice (6 mice for each group) were injected subcutaneously with 5×106 luciferase-expressing stably transformed CRC cells (SW620 and SW480) with CC, CT, and TT genotype, respectively. Two weeks later, the nude mice were euthanized and tumor, lung and liver tissues were removed, and luciferase activity was determined with a luminometer (Sirius, Berthold Detection Systems, Germany).
ELISA
The MPO and IL-23A levels of the CRC patients’ serum and CRC cell culture supernatant were detected by enzyme-linked immunosorbent assay (ELISA)(Abcam, Cambridge) according to the manufacturer’s instructions.
Statistical analysis
The genotype frequencies of the controls were analyzed for deviations from the Hardy–Weinberg equilibrium (HWE) using the goodness-of-fit χ2 test. T-test (for continuous variables) and χ2 test (for categorical variables) were used to evaluate the differences in demographic characteristics between the CRC patients and controls and the genotype results of the SNPs. The association between different genotypes and CRC risk were estimated using the logistic regression models with adjustment for age, Gender, smoking and drinking status. The Kaplan-Meier method and log rank test were used to analyze the overall survival in different subgroups classified by demographic characteristics, genotype, and clinical information. Univariate or multivariate Cox regression analysis was used to analyze hazard ratios (HRs) and 95% CIs, adjusted for age, sex, drinking or smoking status, tumor site (colon or rectum), grade (low or intermediate/high differentiated) and clinical stage (I to IV). Stepwise Cox regression analysis was conducted to assess predictive factors of CRC survival, with a significance level of P < 0.05 for entering and P > 0.10 for removing the variables adjusted in Cox regression analysis. The mediation model was used to explore whether the association between MPO rs2333227 C>T polymorphism and CRC patients’ survival was mediated by MPO protein level. Briefly, the first assumption of the mediation model was applied to evaluate the association between rs2333227 CT/TT genotypes and CRC patients’ survival. The second assumption of the mediation model was applied to assess the association between rs2333227 CT/TT genotypes and MPO protein levels. For the third assumption, the Cox regression analysis was performed to test the association between levels of MPO protein and CRC patients’ survival. The statistical significance criterion was set with a two-sided P < 0.05. All the statistical analyses were performed using SAS version 9.1.3 (SAS Institute, Cary, NC).
Results
Subject characteristics
The detailed information on the subjects have been showed previously(19). Briefly, the age and sex distribution of CRC patients and controls were similar. More smokers and drinkers were observed in CRC patients than the controls (P = 0.0116 and 0.0347 for smoking and drinking status, respectively).
Association between MPO polymorphism and risk of CRC
The genotype distribution of MPO rs2243828 and rs2333227 among the controls was consistent with HWE (P = 0.1924 for rs2243828, P = 0.5097 for rs2333227). As shown in Table 1, frequencies of MPO rs2243828 genotypes were not significantly different between the CRC patients and controls (P = 0.3981). However, the frequency distributions of rs2333227 genotypes were significantly different between CRC patients and controls (P = 0.0066). When the rs2333227 CC genotype was used as a reference, the heterozygous CT and TT genotypes imparted increased risk of CRC (adjusted OR = 1.27, 95% CI = 1.05–1.53 for the CT genotype; 2.00, 1.12–3.56 for the TT genotype), and the increased risk did not change substantially in a dominant genetic model (adjusted OR = 1.31, 95% CI = 1.09–1.58).
Table 1.
Association between the MPO polymorphisms and risk of CRC
| SNPs | genotype | Cases | Controls | P | Adjusted OR(95%CI)a |
||
|---|---|---|---|---|---|---|---|
|
| |||||||
| n | % | n | % | ||||
| rs2243828 | AA | 714 | 66.2 | 787 | 67 | 0.3981 | 1.00(ref) |
| AG | 308 | 28.6 | 341 | 29 | 0.99(0.82–1.19) | ||
| GG | 56 | 5.2 | 47 | 4 | 1.33(0.89–1.99) | ||
| HWE | 0.0034 | 0.1924 | |||||
| P trend | 0.4212 | ||||||
| rs2333227 | CC | 746 | 69.2 | 875 | 74.5 | 0.0066 | 1.00(ref) |
| CT | 300 | 27.8 | 281 | 23.9 | 1.27(1.05–1.53) | ||
| TT | 32 | 3 | 19 | 1.6 | 2.00(1.12–3.56) | ||
| CC | 746 | 69.2 | 875 | 74.5 | 0.0054 | 1.00(ref) | |
| CT/TT | 332 | 30.8 | 300 | 25.5 | 1.31(1.09–1.58) | ||
| T allele | 0.1688 | 0.1357 | 0.002 | ||||
| HWE | 0.7824 | 0.5097 | |||||
| P trend | 0.002 | ||||||
Adjusted for age, gender, smoking and drinking status.
The stratification analysis revealed that rs2333227 CT/TT imparted a higher risk for CRC, especially in subgroups of age (>56 years), female, colon cancer, and TNM stages (III-IV) (adjusted OR = 1.42, 95% CI = 1.09–1.85 for age >56 years; 1.53, 1.15–2.04 for female; 1.64, 1.30–2.08 for patients with colon cancer; 1.44, 1.11–1.87 for patients with TNM stage III tumors; 2.49, 1.70–3.66 for patients with TNM stage IV tumors), compared with CRC cancer patients with rs2333227 CC genotype(Table S1).
Association between rs2333227 and CRC survival
As shown in Table 2 and Fig. 1 A–B, Kaplan–Meier survival curves were used to assess the association between CRC survival and rs2333227 genotype. In the overall model, the MST of CRC patients with MPO rs2333227 CC genotype was 79.5 months, and 59.4 and 58.4 months for the CT and TT genotypes, respectively (log-rank P = 0.0026). Compared with CRC patients with rs2333227 CC genotype, those with rs2333227 CT/TT had a shortened survival time in the dominant model (MST = 79.5 and 59.3 months, respectively, log-rank P = 0.0006). Although previous studies noted that SNP rs2243828 was positively associated with other cancers (8,16), we found that the SNP rs2243828 does not contribute to the clinical occurrence of CRC.
Table 2.
Association between MPO rs2333227 polymorphisms and overall survival of the CRC patients.
| Genotype | Patients (n) | Deaths (n) | MST | Log-rank P | Adjusted HRa (95% CI) |
|---|---|---|---|---|---|
| CC | 746 | 427 | 79.5 | 0.0026 | 1.00(ref) |
| CT | 300 | 194 | 59.4 | 1.18(0.99–1.40) | |
| TT | 32 | 23 | 58.4 | 1.28(0.84–1.96) | |
| CT/TT | 332 | 217 | 59.3 | 0.0006 | 1.19(1.01–1.40) |
Adjusted for age, gender, smoke and drink status, location, grade, TNM.
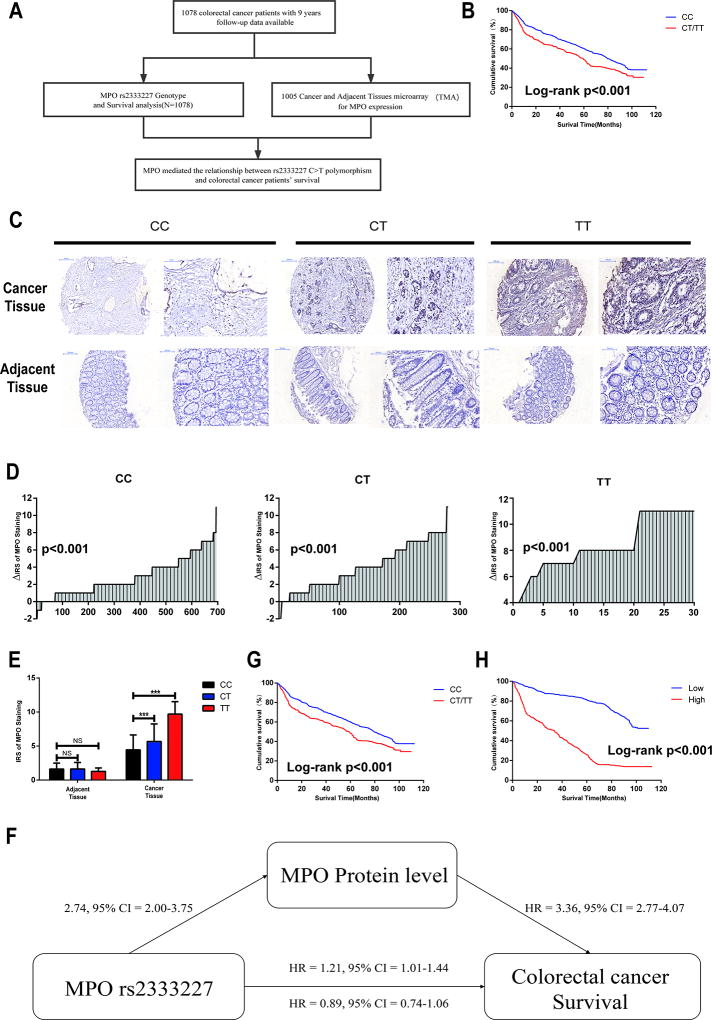
Fig 1. MPO mediated the relationship between rs2333227 C>T polymorphism and colorectal cancer patients’ survival.
(A) Schematic of the study. (B) MPO rs2333227 genotypes and CRC survival in a dominant genetic model in all patients. (C) Protein expression of MPO obtained by IHC assay in human CRC and adjacent tissues TMA. (D) The distribution of MPO protein expression of human CRC and adjacent tissues TMA in patients carrying three rs2333227 genotypes. P values were calculated with the Wilcoxon test. (E) MPO rs2333227 genotypes and protein expression of MPO in human CRC and adjacent tissues TMA. ***P < 0.001, NS means no significance. (F) MPO rs2333227 genotypes and CRC survival in a dominant genetic model in those who was used to analysis MPO expression (Log-rank P <0.001). (G) The relationship between MPO protein expression and CRC patients’ survival (Log-rank P <0.001). KM analysis was calculated with 1005 CRC patients (H) Schematic of mediation model for rs2333227 C>T polymorphism, MPO protein level, and CRC patients’ survival.
In the stratification analysis (Table S2), we found that the CT/TT genotypes were significantly associated with a decreased survival time in the subgroup of age >56 years, female gender, never drink and smoke (adjusted HR = 1.33, 95% CI = 1.05–1.67; adjusted HR = 1.61, 95% CI = 1.24–2.10, adjusted HR = 1.24, 95% CI = 1.01–1.53; adjusted HR = 1.25, 95% CI = 1.01–1.54,respectively). We also found a similar result in the subgroups of Intermediate/High grade and TNM stage III (adjusted HR = 1.25, 95% CI = 1.02–1.54; adjusted HR = 1.29, 95% CI = 1.01–1.65, respectively).
We then performed a Cox stepwise regression analysis to evaluate the effects of selected demographic, clinical information and the rs2333227 variant on colorectal cancer survival. As a result, Four variables (age, drinking status, TNM stage, and rs2333227 dominant model) were included in the model (P < 0.0001 for age, P < 0.0001 for drinking status, P < 0.0001 for TNM stage, and P = 0.013 for rs2333227 dominant model) (Table S3).
MPO rs2333227 genotype and MPO expression in CRC
To further determine whether the MPO rs2333227 polymorphism can affect its protein expression, tissue microarray construction (TMAs) from each genotype were collected for IHC analysis. IHC assay revealed that MPO protein expression levels were up-regulated in the cancer tissue of CRC patients as well as patients with the MPO rs2333227 CT/TT genotype (Figs. 1C–E).
Next, we used the mediation model to explore whether MPO mediated the relationship between rs2333227 C>T polymorphism and CRC patients’ survival. We found that the levels of MPO protein were significantly up-regulated in the patients carrying the CT/TT genotypes compared with those with the CC genotype (by the logistic regression analysis) (adjusted OR = 2.74, 95% CI = 2.00–3.75) (Figs. 1 E and F), thus supporting the second assumption of the mediation model. Moreover, the first assumption of the mediation model showed the CT/TT genotypes significantly decreased the CRC patients’ survival time when compared with those with the CC genotype (adjusted HR = 1.21, 95% CI = 1.01–1.44) (Fig. 1F and G). Further, Cox regression analysis indicated that high MPO expressions was associated with shorter survival time (adjusted HR = 3.36, 95% CI = 2.77–4.07) (Figs. 1F and H), confirming the third assumption of the mediation model. Moreover, the relationship between rs2333227 C>T polymorphism and CRC patients’ survival became insignificant once the MPO was included in the model (adjusted HR = 0.89, 95% CI = 0.74–1.06) (Figs. 1 F).
MPO rs2333227 T genotype recruited AP-2α to bind on the −463 region
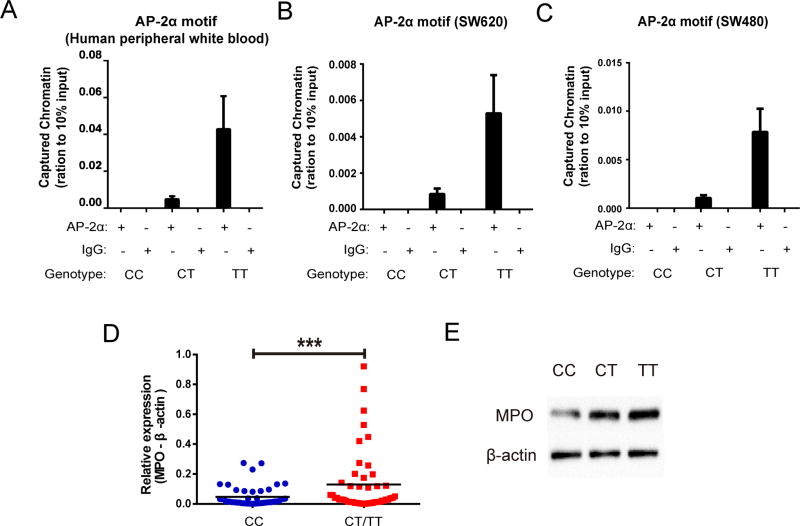
According to the JASPAR CORE database and Gene Regulation, the MPO rs2333227 SNP site, located at the −463 promoter, is contained in the AP-2α binding motif. Accordingly, we hypothesized that the MPO rs2333227 variant might affect the binding affinity of AP-2α to the MPO rs2333227 mutation region. Thus, the efficacy binding affinity of AP-2α to the −463 site was assessed by ChIP analysis using CRC Human peripheral white blood. Results showed that the rs2333227 CT/TT genotype promoted AP-2α binding to the MPO promoter, but the CC genotype did not in CRC Human peripheral white blood (Fig. 2A). Similar results were also confirmed in SW620 and SW480 cells with the indicated genotypes (Figs. 2B–C; three separate colonies with rs2333227 CT/TT genotype). To further determine whether the MPO rs2333227 polymorphism can affect MPO transcriptional activity, the MPO mRNA level of CRC patients was determined. Results showed that individuals with mutant CT/TT genotypes at the rs2333227 had significantly higher MPO levels than those with the CC genotype (Fig. 2D). The MPO protein expression levels of CRC patients with the CT/TT genotype were significantly higher than those with the CC genotype (Fig. 2E). Moreover, the serum MPO levels of CRC patients with the CT/TT genotype were statistically significantly higher than those with the CC genotype (Table.S4). Thus, we posit that higher AP-2α binding efficiency is driving increased MPO protein expression in the rs2333227 genotypes.
Fig 2. AP-2α promoted tumor MPO transcription by binding to the rs2333227 T genotype in CRC patients.
(A–C) The binding affinity between AP-2α and the indicated MPO rs2333227 genotypes were determined by ChIP assay in CRC Human peripheral white blood (A), SW620 cells (B) and SW480 cells (C). (D) MPO mRNA levels in CRC tissues expressing CC or CT/TT genotype were detected by qPCR analysis. (E) MPO protein levels in CRC tissues expressing CC, CT or TT genotype were detected by immunobloting. ***P < 0.001.
MPO rs2333227 TT genotype promotes MPO expression and CRC cell proliferation, invasion and migration
The single nucleotide polymorphisms (SNPs) rs2333227 (G-463A) and rs2243828 (T-764C), located in the 5’ upstream region of MPO gene, are functional SNPs, as reported in various diseases (10,11).
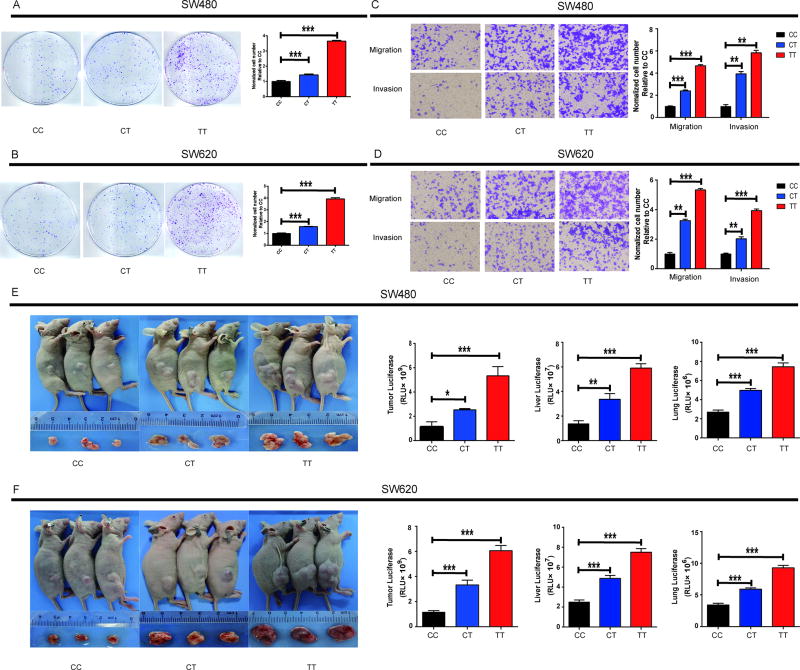
We hypothesized that the MPO mutant rs2333227 genotype might alter CRC cell invasion and migration. Our results indicated that colony formation in both SW620 and SW480 cells were markedly enhanced by the rs2333227 CT/TT genotype compared with the CC genotype (Figs. 3A–B). The MPO rs2333227 TT genotype significantly promoted the migration and invasion of SW480 and SW620 CRC cell lines (Figs. 3C–D).
Fig 3. MPO rs2333227 T genotype promotes in vitro and in vivo cell proliferation, migration and invasion of human CRC cell lines.
(A,B) Representation colony formation images and colony formation counting are shown. (A: SW620, and B: SW480). (C,D) Transwell assay performed to determine the migration and invasion of cell lines SW620 and SW480(C: SW620, and D: SW480). (E,G) Flank tumor, liver and lung metastatic burden in nude mice (n = 6 for each group) injected with CRC cell lines with rs2333227 CC,CT and TT genotypes, measured by luciferase activity. ***P < 0.001, *P < 0.05, **P < 0.01.
Next, we injected the CRC cell lines (SW480 and SW620 with rs2333227 CC, CT, and TT genotype) into the dorsal flanks of nude mice. The results showed that the growth rate of xenograft CRC cells was significantly higher with the rs2333227 CT/TT genotype than CC genotype (Figs. 3E–F).
IL-23A and MMP9 mediated the relationship between MPO and development of colorectal cancer
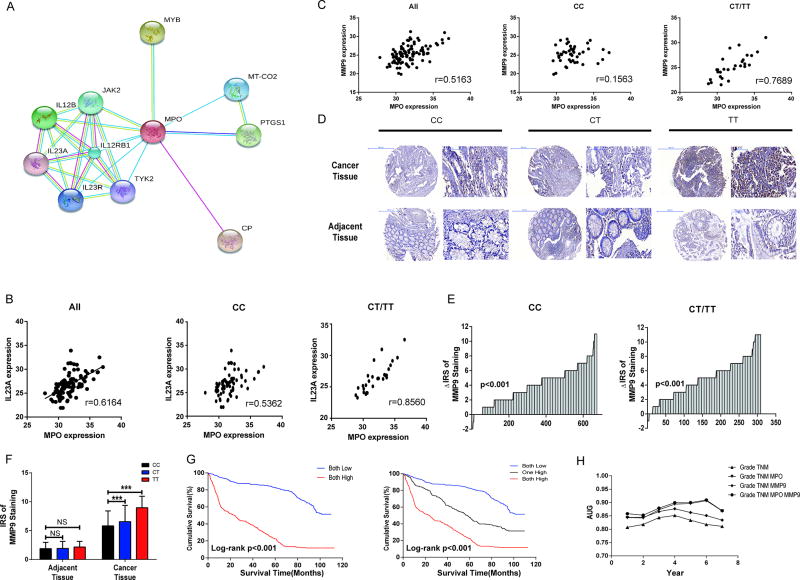
To further explore the potential mechanism underlying the biological effects of MPO in colorectal cancer development, we constructed a protein interaction network for genes regulated by MPO using STRING version 10.0 (search tool for the retrieval of interacting genes) (Fig.4 A). The correlations between mRNA levels of MPO and key components in the predicted network were validated in CRC tissues (Fig.4 B and Fig S2 A – I). Among all the components, IL23A transcriptional levels exhibited the strongest correlation with MPO, especially in patients carrying with CT/TT genotype(r=0.8560 for CT/TT, and r=0.5362 for CC, respectively, Fig.4 B). Given that IL23 up-regulates the matrix metalloproteinase 9 (MMP9) expression (20), we next evaluated the mRNA and protein levels of MMP9 in CRC patients. We found a significantly positive correlation between MPO and MMP9 mRNA expression levels (r=0.5163, P <0.001, Fig.4 C). Moreover, the correlation in patients with CT/TT genotypes was stronger than with the CC genotype(r=0.7689, and r=0.1563, respectively), corroborating the CT/TT genotypes might be a higher risk factors for CRC metastasis (Fig.4 C). Similar to our finding in MPO, MMP9 were up-regulated in CRC patients’ cancer tissues and patients carrying the MPO rs2333227 CT/TT genotypes (Fig.4 D–F.). Furthermore, CRC cells carrying the CT/TT genotypes also showed increased mRNA levels of MPO, IL-23A and MMP9 compared with the CC genotype (Fig.S3 A – F.). The trend for increased MPO and IL-23 protein levels was also consistent with the mRNA levels in the cell culture supernatants assayed by ELISA (Fig.S3 G – J.). KM analysis showed that both high expression of those two proteins showed worsening outcome for survival compared with the other groups (one high or both low) (Fig.4 G.). Furthermore, multivariate Cox regression analysis indicated that high MPO and MMP9 expressions were independent prognostic markers for CRC (Table 3). We then performed a time-dependent ROC analysis to evaluate the prognostic efficacy of MPO and MMP9 expressions on colorectal cancer patients. We found that the combination of the clinical variables, MPO and MMP9 risk score contributed more than either one of these alone (Fig.4 H.).
Fig 4. IL-23A and MMP9 mediated the relationship between MPO and development of colorectal cancer.
(A) Protein interaction network for genes regulated by MPO analyzed by STRING (version 10.0). (B) Correlation analysis between mRNA expression levels of MPO and IL-23A by qRT-PCR. (C) Correlation analysis between mRNA expression levels of MPO and MMP9 by qRT-PCR. (D) Protein expression of MMP9 obtained by IHC assay in human CRC and adjacent tissues TMA. (E) MPO protein expressions of human CRC and adjacent tissues TMA in patients carrying three rs2333227 genotypes were determined. P values were calculated with the Wilcoxon test. (F) MPO rs2333227 genotypes and protein expression of MMP9 in human CRC and adjacent tissues TMA. ***P < 0.001, NS means no significance. (G) The relationship between MPO and MMP9 protein expression and CRC patients’ survival (Log-rank P <0.001). KM analysis was calculated in a sample of 915 CRC patients. (H) Time-dependent ROC analysis for the clinical risk score (TNM, Grade), MPO, MMP9 or the combined MPO, MMP9 and the clinical risk score (TNM, Grade).
Discussion
In the present study, we explored the role of promoter MPO SNPs in susceptibility and development of CRC. By conducting a hospital based case-control study, we found that subjects carrying rs2333227 CT/TT genotypes had a significantly higher risk for CRC and poorer survival outcomes when compared to the CC genotype. With in vivo and in vitro assays, we demonstrated that transcription factor AP-2α presented stronger binding affinity with rs2333227 CT/TT genotype than CC genotype, thus upregulating MPO expressions, and ultimately promoting CRC proliferation and metastasis. Therefore, our results verified the critical role of MPO functional variant (rs2333227) in CRC.
MPO catalyzes oxidants generation, thus contributing to the risk of cancer(21). It has been reported that MPO rs2333227 T allele alters the susceptibility of breast(12) and prostate cancer(14). However, few studies were paid attention to the role of MPO rs2333227 in CRC. In our cohort, we demonstrated that rs2333227 TT genotype was a risk factor for CRC and associated with worse clinical features, suggesting that different clinical features of colorectal cancer might be regulated by different molecular mechanisms. Interestingly, we found that age, drinking status, TNM stage and rs2333227 dominant model were independent factors affecting CRC in our stepwise Cox regression analysis. Taken together, our results have verified the oncogenic role of rs2333227 TT genotype in CRC.
Transcription factor AP-2α promotes the expressions of target genes (such as DEK(22)) through directly binding to the promoter region. By conducting the ChIP assay, we demonstrated that AP-2α preferred to binding with MPO rs2333227 CT/TT genotype. As CRC tissues, serum and cells carrying CT/TT genotype exhibited much higher MPO protein levels, we speculated that AP-2α was responsible for rs2333227 CT/TT genotype induced higher transcriptional activity. For few reports were focused on AP-2α/rs2333227 T allele axis, further researches are still needed to confirm our findings.
It have been reported that MPO plays an important role in cellular processes, such as DNA damage, oxidative stress (23) and innate immune response (24). Its major function is to mediate hypochlorous acid (HOCl) generation (25). However, HOCl and other oxidants derived by MPO are considered important mediators of oxidative damage to cellular biomolecules (26) in that they may damage the structure of host tissues (27). Moreover, HOCl can decrease the adhesiveness of extracellular matrix proteins and affect endothelial function (11), destabilizing the tissue environment surrounding endothelial cells (28). This suggests that over-expressed MPO in human tumors may be associated with enhanced cell invasion and migration. Moreover, we found that MPO protein expression levels were up-regulated in the cancer tissue of CRC patients, which corroborating that MPO could be an oncogene in CRC.
To further ascertain the downregulators of MPO that drives aggressive phenotype, protein interaction network were constructed(29,30). Among the 10 candidate, IL-23A showed the strongest correlation with MPO levels, especially in patients carrying TT genotype. Interestingly, Lan et al. showed that IL-23A was up-regulated in the CRC tissues(31). Previous study have showed that IL-23A promotes tumor incidence and growth by targeting MMP9(20). In our study, we again demonstrated that MMP9 was not only the downregulator of IL-23A, but also synergistically predicted CRC patients’ prognosis with MPO. Taken together, our results indicated that MPO/IL-23A/MMP9 axis might promote CRC progression and metastasis.
In summary, our results revealed that functional rs2333227 TT genotype upregulated MPO expressions in CRC, thus activating IL-23A/MMP9 signaling medicated tumor growth and development. However, further studies are still required to confirm our findings.
Supplementary Material
Table 3.
Multivariate Cox regression analysis of MPO and MMP9 expression and selected clinical pathological variables in colorectal cancer survival
| Variables | MPO a
|
MMP9 a
|
Combined ab
|
Combined ac
|
||||
|---|---|---|---|---|---|---|---|---|
| Adjusted HR(95%CI) | P | Adjusted HR(95%CI) | P | Adjusted HR(95%CI) | P | Adjusted HR(95%CI) | P | |
| Age(>56 vs. ≤56) | 1.41(1.18–1.67) | 0.0001 | 1.47(1.24–1.75) | <.0001 | 1.39(1.17–1.65) | 0.0002 | 1.52(1.24–1.85) | <.0001 |
| Gender (Females vs. Males) | 0.86(0.71–1.03) | 0.0975 | 0.82(0.68–0.98) | 0.0284 | 0.80(0.67–0.96) | 0.019 | 0.89(0.72–1.10) | 0.2861 |
| Smoke (Ever vs. Never) | 1.04(0.83–1.30) | 0.7538 | 0.94(0.76–1.18) | 0.5959 | 0.97(0.78–1.21) | 0.7728 | 1.11(0.85–1.45) | 0.4503 |
| Drink(Ever vs. Never) | 1.16(0.91–1.46) | 0.2312 | 1.19(0.94–1.50) | 0.1410 | 1.17(0.93–1.47) | 0.1819 | 1.05(0.79–1.39) | 0.7327 |
| Location (Rectum vs. Colon) | 0.89(0.75–1.06) | 0.2060 | 0.90(0.76–1.07) | 0.2307 | 0.86(0.72–1.03) | 0.0935 | 0.95(0.78–1.17) | 0.6402 |
| Grade(Intermediate/High vs. low) | 0.95(0.80–1.14) | 0.5813 | 0.93(0.78–1.12) | 0.4515 | 0.98(0.82–1.17) | 0.832 | 0.94(0.76–1.16) | 0.5652 |
| TNM(3/4 vs. 1/2) | 5.17(4.24–6.29) | <.0001 | 6.10(5.04–7.39) | <.0001 | 5.63(4.64–6.84) | <.0001 | 5.71(4.55–7.18) | <.0001 |
| Protein (High vs. Low) | 3.48(2.89–4.21) | <.0001 | 2.23(1.88–2.64) | <.0001 | 2.91(2.40–3.53) | <.0001 | 4.04(3.25–5.03) | <.0001 |
Multivariate Cox regression analysis were performed with 915 subjects because of loss of sample during MPO and MMP9 antigen retrieval
Multivariate Cox regression analysis were calculated with follows proteins expression status (One High vs. Both Low)
Multivariate Cox regression analysis were calculated with follows proteins expression status (Both High vs. Both Low)
Acknowledgments
This work was financially supported by National Natural Science Foundation of China (81472938, and 91643109 to R. Chen, 81703261 to J. Xu), the National Key Research and Development Program of China(2017YFC0211600, and 2017YFC0211603 to R. Chen), the Natural Science Foundation of Jiangsu Province (BK20151418 to X. Li, BK20171060 to J. Xu), the Fund of the Distinguished Professor of Jiangsu Province to R. Chen, the Fund of the Distinguished Talents of Jiangsu Province to R. Chen (BK20150021), the Six talent peaks project in Jiangsu Province to R. Chen( 2016-WSN-002), the Natural Science Foundation of Jiangsu Higher Education Institutions to J. Xu (17KJB330002), the Fund of the Post-graduate Innovative Talents to Q. Meng(KYZZ16_0137) and the Fundamental Research Funds for the Central Universities to R. Chen and X. Li, the National Institute of Environmental health Sciences ( NIEHS) (R01 ES10563, R01 ES07331 and R01 ES020852 to M. Aschner).
Footnotes
Disclosure of Potential Conflicts of Interests
The authors declare no conflict of interest.
References
- 1.Quan B, Qi X, Yu Z, Jiang Y, Liao M, Wang G, et al. Pathway analysis of genome-wide association study and transcriptome data highlights new biological pathways in colorectal cancer. Molecular genetics and genomics : MGG. 2015;290:603–10. doi: 10.1007/s00438-014-0945-y. [DOI] [PubMed] [Google Scholar]
- 2.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9:489–99. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 3.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 4.Haydon AM, Macinnis RJ, English DR, Giles GG. Effect of physical activity and body size on survival after diagnosis with colorectal cancer. Gut. 2006;55:62–7. doi: 10.1136/gut.2005.068189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 6.Arnhold J, Flemmig J. Human myeloperoxidase in innate and acquired immunity. Archives of biochemistry and biophysics. 2010;500:92–106. doi: 10.1016/j.abb.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 7.Baldus S, Heeschen C, Meinertz T, Zeiher AM, Eiserich JP, Munzel T, et al. Myeloperoxidase serum levels predict risk in patients with acute coronary syndromes. Circulation. 2003;108:1440–5. doi: 10.1161/01.CIR.0000090690.67322.51. [DOI] [PubMed] [Google Scholar]
- 8.Karakas M, Koenig W. Myeloperoxidase production by macrophage and risk of atherosclerosis. Current atherosclerosis reports. 2012;14:277–83. doi: 10.1007/s11883-012-0242-3. [DOI] [PubMed] [Google Scholar]
- 9.Reichlin T, Socrates T, Egli P, Potocki M, Breidthardt T, Arenja N, et al. Use of myeloperoxidase for risk stratification in acute heart failure. Clinical chemistry. 2010;56:944–51. doi: 10.1373/clinchem.2009.142257. [DOI] [PubMed] [Google Scholar]
- 10.Choi JY, Neuhouser ML, Barnett MJ, Hong CC, Kristal AR, Thornquist MD, et al. Iron intake, oxidative stress-related genes (MnSOD and MPO) and prostate cancer risk in CARET cohort. Carcinogenesis. 2008;29:964–70. doi: 10.1093/carcin/bgn056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slattery ML, Lundgreen A, Welbourn B, Wolff RK, Corcoran C. Oxidative balance and colon and rectal cancer: interaction of lifestyle factors and genes. Mutation research. 2012;734:30–40. doi: 10.1016/j.mrfmmm.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He C, Tamimi RM, Hankinson SE, Hunter DJ, Han J. A prospective study of genetic polymorphism in MPO, antioxidant status, and breast cancer risk. Breast cancer research and treatment. 2009;113:585–94. doi: 10.1007/s10549-008-9962-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng TY, King IB, Barnett MJ, Ambrosone CB, Thornquist MD, Goodman GE, et al. Serum phospholipid fatty acids, genetic variation in myeloperoxidase, and prostate cancer risk in heavy smokers: a gene-nutrient interaction in the carotene and retinol efficacy trial. American journal of epidemiology. 2013;177:1106–17. doi: 10.1093/aje/kws356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng TY, Barnett MJ, Kristal AR, Ambrosone CB, King IB, Thornquist MD, et al. Genetic variation in myeloperoxidase modifies the association of serum alpha-tocopherol with aggressive prostate cancer among current smokers. The Journal of nutrition. 2011;141:1731–7. doi: 10.3945/jn.111.141713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haslacher H, Perkmann T, Gruenewald J, Exner M, Endler G, Scheichenberger V, et al. Plasma myeloperoxidase level and peripheral arterial disease. European journal of clinical investigation. 2012;42:463–9. doi: 10.1111/j.1365-2362.2011.02601.x. [DOI] [PubMed] [Google Scholar]
- 16.Wu DM, Zhu HX, Zhao QH, Zhang ZZ, Wang SZ, Wang ML, et al. Genetic variations in the SMAD4 gene and gastric cancer susceptibility. World journal of gastroenterology. 2010;16:5635–41. doi: 10.3748/wjg.v16.i44.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Wu X, Chen Y, Zhang J, Ding J, Zhou Y, et al. Prognostic and predictive role of JWA and XRCC1 expressions in gastric cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:2987–96. doi: 10.1158/1078-0432.CCR-11-2863. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Lv Y, Gao N, Sun H, Lu R, Yang H, et al. microRNA-802/Rnd3 pathway imposes on carcinogenesis and metastasis of fine particulate matter exposure. Oncotarget. 2016;7:35026–43. doi: 10.18632/oncotarget.9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu S, Meng Q, Zhang C, Sun H, Lu R, Gao N, et al. DR4 mediates the progression, invasion, metastasis and survival of colorectal cancer through the Sp1/NF1 switch axis on genomic locus. International journal of cancer Journal international du cancer. 2018 doi: 10.1002/ijc.31318. [DOI] [PubMed] [Google Scholar]
- 20.Langowski JL, Zhang X, Wu L, Mattson JD, Chen T, Smith K, et al. IL-23 promotes tumour incidence and growth. Nature. 2006;442:461–5. doi: 10.1038/nature04808. [DOI] [PubMed] [Google Scholar]
- 21.Winterbourn CC, Kettle AJ. Biomarkers of myeloperoxidase-derived hypochlorous acid. Free radical biology & medicine. 2000;29:403–9. doi: 10.1016/s0891-5849(00)00204-5. [DOI] [PubMed] [Google Scholar]
- 22.Qiao MX, Li C, Zhang AQ, Hou LL, Yang J, Hu HG. Regulation of DEK expression by AP-2alpha and methylation level of DEK promoter in hepatocellular carcinoma. Oncology reports. 2016;36:2382–90. doi: 10.3892/or.2016.4984. [DOI] [PubMed] [Google Scholar]
- 23.Klebanoff SJ. Myeloperoxidase: friend and foe. Journal of leukocyte biology. 2005;77:598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- 24.Mayadas TN, Cullere X, Lowell CA. The multifaceted functions of neutrophils. Annual review of pathology. 2014;9:181–218. doi: 10.1146/annurev-pathol-020712-164023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hickey MJ. MPO and neutrophils: a magnetic attraction. Blood. 2011;117:1103–4. doi: 10.1182/blood-2010-11-317479. [DOI] [PubMed] [Google Scholar]
- 26.Himmelfarb J, McMenamin ME, Loseto G, Heinecke JW. Myeloperoxidase-catalyzed 3-chlorotyrosine formation in dialysis patients. Free radical biology & medicine. 2001;31:1163–9. doi: 10.1016/s0891-5849(01)00697-9. [DOI] [PubMed] [Google Scholar]
- 27.Hansson M, Olsson I, Nauseef WM. Biosynthesis, processing, and sorting of human myeloperoxidase. Archives of biochemistry and biophysics. 2006;445:214–24. doi: 10.1016/j.abb.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Fu X, Kassim SY, Parks WC, Heinecke JW. Hypochlorous acid generated by myeloperoxidase modifies adjacent tryptophan and glycine residues in the catalytic domain of matrix metalloproteinase-7 (matrilysin): an oxidative mechanism for restraining proteolytic activity during inflammation. The Journal of biological chemistry. 2003;278:28403–9. doi: 10.1074/jbc.M304739200. [DOI] [PubMed] [Google Scholar]
- 29.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic acids research. 2017;45:D362–D8. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang DS, Zhang L, Han K, Deng S, Yang K, Zhang H. Prediction of protein-protein interactions based on protein-protein correlation using least squares regression. Current protein & peptide science. 2014;15:553–60. doi: 10.2174/1389203715666140724084019. [DOI] [PubMed] [Google Scholar]
- 31.Lan F, Zhang L, Wu J, Zhang J, Zhang S, Li K, et al. IL-23/IL-23R: potential mediator of intestinal tumor progression from adenomatous polyps to colorectal carcinoma. International journal of colorectal disease. 2011;26:1511–8. doi: 10.1007/s00384-011-1232-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.