Abstract
Expanded polyglutamine tracts are responsible for at least eight fatal neurodegenerative diseases. In mouse models, proteins with expanded polyglutamine cause transcriptional dysregulation before onset of symptoms, suggesting that this dysregulation may be an early event in polyglutamine pathogenesis. Transcriptional dysregulation and cellular toxicity may be due to interaction between expanded polyglutamine and the histone acetyltransferase CREB-binding protein. To determine whether polyglutamine-mediated transcriptional dysregulation occurs in yeast, we expressed polyglutamine tracts in Saccharomyces cerevisiae. Gene expression profiles were determined for strains expressing either a cytoplasmic or nuclear protein with 23 or 75 glutamines, and these profiles were compared to existing profiles of mutant yeast strains. Transcriptional induction of genes encoding chaperones and heat-shock factors was caused by expression of expanded polyglutamine in either the nucleus or cytoplasm. Transcriptional repression was most prominent in yeast expressing nuclear expanded polyglutamine and was similar to profiles of yeast strains deleted for components of the histone acetyltransferase complex Spt/Ada/Gcn5 acetyltransferase (SAGA). The promoter from one affected gene (PHO84) was repressed by expanded polyglutamine in a reporter gene assay, and this effect was mitigated by the histone deacetylase inhibitor, Trichostatin A. Consistent with an effect on SAGA, nuclear expanded polyglutamine enhanced the toxicity of a deletion in the SAGA component SPT3. Thus, an early component of polyglutamine toxicity, transcriptional dysregulation, is conserved in yeast and is pharmacologically antagonized by a histone deacetylase inhibitor. These results suggest a therapeutic approach for treatment of polyglutamine diseases and provide the potential for yeast-based screens for agents that reverse polyglutamine toxicity.
Polyglutamine diseases comprise a group of fatal hereditary movement disorders that share a number of clinical and molecular features. These maladies, which include Huntington's disease (HD), Kennedy's disease, and the spinocerebellar ataxias (SCAs), are caused by expansion in the number of glutamine-encoding CAG triplet repeats in specific genes. Clinical features usually include a late-onset progressive loss of motor and cognitive function associated with neuronal cell degeneration in the central nervous system (1, 2). At present, there are no effective treatments for these diseases.
Molecular genetic studies of the polyglutamine diseases indicate that mutant proteins containing expanded polyglutamine tracts acquire toxic properties involved in the etiology of the diseases. Perutz and coworkers (3) proposed that expanded polyglutamine tracts could alter the solubility and folding properties of the proteins in which they were embedded. Experiments in vitro have demonstrated that polyglutamine tracts can form insoluble aggregates with properties similar to amyloid fibrils. Furthermore, insoluble polyglutamine-containing cellular inclusions result when expanded forms of disease-causing proteins are expressed in vivo. Experiments with transgenic mice have shown that expression of polyglutamine-containing proteins in the central nervous system is sufficient to cause loss of motor function and premature death, which is associated with the presence of polyglutamine-containing inclusions in brain tissue (2). Inclusions have also been observed in brain tissue from patients with HD and other polyglutamine diseases (4, 5). In cultured animal cells, expression of expanded polyglutamine tracts leads to formation of inclusions and cellular toxicity (6–8). Cellular toxicity has also been observed in transgenic Drosophila melanogaster and Caenorhabditis elegans (9–12). Expanded polyglutamine tracts themselves are sufficient to cause neurotoxic phenotypes in transgenic mice and Drosophila, indicating that a particular protein context is not required for toxicity (13–15).
These observations indicate that cellular components and processes sensitive to polyglutamine toxicity insult are cell autonomous and conserved across divergent metazoan taxa. However, the key cellular targets of polyglutamine-mediated toxicity have not been firmly established. An area of increasing interest is the interaction of expanded polyglutamine with components of the transcriptional machinery (16). Polyglutamine-mediated effects on transcription have been reported in transgenic mice in a mouse model of SCA1, where expression of expanded ataxin-1 altered gene expression in a manner dependent on repeat-length and nuclear localization (17). In an HD mouse model, altered expression of many genes was detected by DNA microarray analysis (18).
In this study, we used DNA microarray analysis to examine the effects of polyglutamine expression on yeast transcript profiles. Our results indicate that expression of expanded polyglutamine in the yeast nucleus leads to significant defects in transcription. The patterns of transcript changes correlate with those seen in yeast carrying mutations in the conserved histone acetylase complex Spt/Ada/Gcn5 acetyltransferase (SAGA). These observations suggest that the polyglutamine-mediated effects on transcription seen in animal models may operate through effects on conserved transcriptional regulatory pathways and may represent a primary cellular defect in polyglutamine disease.
Materials and Methods
Plasmids and Strains.
The first 170 codons of human HD (with 23 and 75 CAG codons) were amplified by PCR. HD DNA fragments containing a precise in-frame deletion of codons 43–70 (in the polyproline tract adjacent to the polyglutamine tract) were cloned as galactose-inducible HD-green fluorescent protein (GFP) fusions in the centromere-based plasmid pTS544, a YCp50-derived plasmid driving GFP expression from the GAL1,10 promoter (provided by Tim Stearns, Stanford Univ., Stanford, CA). Epitope tagged versions were made by inserting the 3X-HA region of pGTEPI (provided by Bruce Futcher, State Univ. of New York, Stony Brook) between polyQ and GFP. Clones with a nuclear localization signal (NLS) were made by inserting oligonucleotides corresponding to the simian virus 40 NLS (MPKKKRKV) at the HD start. Constitutive expression clones were constructed in the centromere-based plasmid p416GPD, which carries the GPD1 promoter (19). The strain W303–1a (MATa ade2–1 trp1–1 can1–100 leu2–3,112 his3–11,15 ura3–1) was used for microscopy and gene expression experiments. Transcription factor mutant strains FY51 (spt3), FY963 (spt7), FY1258 (gal11), FY1294 (gcn5), and FY1658 (snf5) (20), were provided by Fred Winston.
Protein Expression and Microscopy.
Cells with GPD1 expression clones were grown in synthetic complete media−uracil. For galactose induction, cells were grown initially in 2% raffinose −uracil and diluted into media containing 2% raffinose + 2% galactose. For indirect immunofluorescence, cells were grown to mid-log and prepared as described (21). Cells were stained with rabbit anti-HA antibody (Santa Cruz Biotechnology) and followed by a rhodamine-conjugated goat-anti-rabbit antibody (Santa Cruz Biotechnology). Cells were visualized with a Zeiss Axiovert microscope by using a 63 × 1.4 NA objective.
Gene Expression Analysis.
Microarray construction and hybridization protocols were modified from those described (22). Yeast cells (100 ml) were grown to mid-log (OD600 0.7–0.9) in synthetic complete media−uracil + 2% glucose and lysed with hot acidic phenol. RNA was recovered from the aqueous fraction by ethanol precipitation. Poly(A)+ RNA was selected with Oligotex resin (Qiagen). cDNA labeling was as described (http://cmgm.stanford.edu/pbrown/protocols/aadUTPCouplingProcedure.htm). Two-color expression profiles were generated by using microarrays in which control and experimental cDNA targets were labeled with different fluors. After cohybridization to the chip, a fluorescent image of the microarray was collected at both emission wavelengths by using a GenePix 4000 fluorescent scanner (Axon Instruments, Foster City, CA). To compare expression data from different strains and replicates, the fold changes were transformed into a normalized standard score. The fold change (FC) was defined as the ratio of the median pixel intensity from the two dye channels. The normalized standard score for a gene was calculated as (log2 FC − x)/SD, where x is the mean log2FC and SD is the standard deviation of the log2FC distribution. This transforms the FC distribution of the experiment into a normalized distribution with unit standard deviation and mean of zero. Genes flagged by the analysis software (genepix 3.0, Axon Instruments) were removed from consideration before normalization.
The lacZ construct driven by the PHO84 promoter was derived from pRB003, a centromere-based plasmid driving GFP expression from a 600-bp PHO84 promoter (provided by Rusty Howson and Erin O'Shea, Univ. of California, San Francisco). The GFP ORF was replaced with the lacZ gene. β-Galactosidase assays were performed on mid-log cultures by using a yeast (β-galactosidase assay kit (Pierce). To create the ACT1-lacZ reporter, the 618-bp intragenic region immediately upstream from the start codon of ACT1 replaced the PHO84 promoter in the PHO84-lacZ reporter plasmid. Trichostatin A (TSA) experiments were done by growing the yeast cultures overnight in the presence of 20 μM TSA (Upstate Biotechnology, Lake Placid, NY) and 1% DMSO (Fisher). Cells were then diluted in TSA media and grown to log phase before the β-galactosidase assay.
Results
Aggregation of Polyglutamine in Yeast Is Affected by Nuclear Localization.
The presence of intranuclear polyglutamine inclusions in transgenic models of polyglutamine disease, and in brain tissue of polyglutamine disease patients, has generated interest into the nature and consequences of cytoplasmic vs. nuclear polyglutamine localization. In a mammalian cell culture model, nuclear localization of an expanded huntingtin fragment enhances its toxicity (23). Length-dependent cytoplasmic aggregation of polyglutamine without cellular toxicity has been reported in yeast (24, 25).
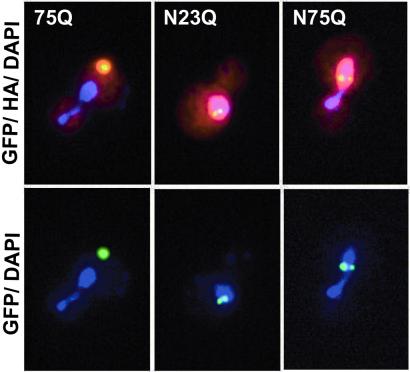
To examine the effects of nuclear localization in yeast, we fused the simian virus 40 NLS to residue 1 of the 23Q and 75Q-GFP fusions. In contrast to the 23Q fusion, which showed diffuse staining throughout the cell (data not shown), the NLS-23Q fusion localized to discrete foci in the yeast nucleus (Fig. 1 Center). An identical pattern of localization was also observed with the NLS-75Q fusion (Fig. 1 Right). These data indicate that the nuclear environment, or transport into the nucleus, can facilitate the aggregation of nonexpanded polyglutamine repeats. Nuclear aggregation of nonexpanded ataxin-3 expressed in tissue culture cells also has been observed (26).
Figure 1.
Effect of nuclear localization on polyglutamine aggregation. The simian virus 40 NLS was fused to the N terminus of the 23Q- and 75Q-GFP fusions, and an HA epitope tag was inserted between the polyglutamine and GFP. Cells were induced for 6 h, fixed, and stained with a rhodamine conjugated anti-HA antibody and 4′,6-diamidino-2-phenylindole. Yellow features indicate colocalization of HA (red) and GFP (green); purple indicates localization of DNA. (Left) Cytoplasmic localization of a 75Q inclusion in a budding cell. (Center) NLS-23Q-GFP present in two discrete nuclear inclusions. (Right) Nuclear inclusions observed with NLS-75Q-GFP expression.
Expanded Nuclear Polyglutamine Causes Altered Transcription in Yeast.
To determine whether the ability of expanded polyglutamine to interfere with transcription was a conserved cell-autonomous phenomenon, we grew yeast cells constitutively expressing either vector, 23Q-GFP, 75Q-GFP, NLS-23Q-GFP, or NLS-75Q-GFP in parallel, and poly(A)+ RNA was prepared, fluorescently labeled, and hybridized to yeast ORF DNA arrays (22). RNA from cells expressing a form of polyglutamine was cohybridized with RNA prepared from the vector-containing yeast. Experiments were done in duplicate with dye orientations reversed.
Expanded polyglutamine induced a number of genes involved in protein-folding and chaperone function (Fig. 2). Of 35 genes induced in the NLS-75Q yeast, 14 are involved in protein folding. Fourteen of these 35 genes are common to the set of genes induced in the 75Q strain and nine of these 14 are involved in protein folding. These data indicate that yeast constitutively expressing expanded polyglutamine mount a mild heat-shock response. Polyglutamine-mediated induction of heat-shock factors has been observed in C. elegans, and chaperone overexpression suppresses polyglutamine toxicity in a number of Drosophila models (12, 14, 27, 28). Interestingly, of the chaperone genes, HSP104 was induced to the greatest degree in both N75Q and NLS-N75Q strains. HSP104 is required for prion propagation and is also required for cytoplasmic aggregation of expanded polyglutamine in yeast (24). The gene induced to the greatest degree by expanded polyglutamine is BTN2, a gene involved in cellular pH homeostasis. Like HSP104, the induction of BTN2 is not sensitive to cytoplasmic vs. nuclear localization of expanded polyglutamine. BTN2 is one of five genes whose expression is elevated in yeast deleted for YCH3 (BTN1), a gene with similarity to the human CLN3 gene (29). Mutations in CLN3 are responsible for Batten's disease, a progressive neurodegenerative disorder affecting children.
Figure 2.
Genes in the NLS-75Q strain whose expression was repressed or induced by at least two SDs from the mean (of normalized average fold change) in duplicate experiments are shown. Lists are sorted by the N75 data, and changes observed in other strains are indicated (dark blue/red: >4 SD; medium blue/red: 3–4 SD; pale blue/red: 2–3 SD). Fold changes of ≥1.5 and ≤0.67 are shown.
The gene expression changes of greatest magnitude were genes repressed by nuclear expanded polylgutamine. Of the 25 genes repressed by at least a factor of 0.66 times, 10 encode small molecule transporters and 10 encode proteins involved in phosphate metabolism (Fig. 2). PHO84 mRNA, which consistently showed the greatest degree of repression, encodes a high-affinity phosphate transporter gene that is required for growth under conditions of low phosphate (30, 31). Yeast used for the microarray experiments were grown in standard synthetic media with phosphate. Because PHO84 is repressed under these conditions, the effects of polyglutamine are on the basal activity of this promoter. However, cells expressing NLS-75Q showed no obvious growth defect in media containing low phosphate (30 mg/liter) (not shown).
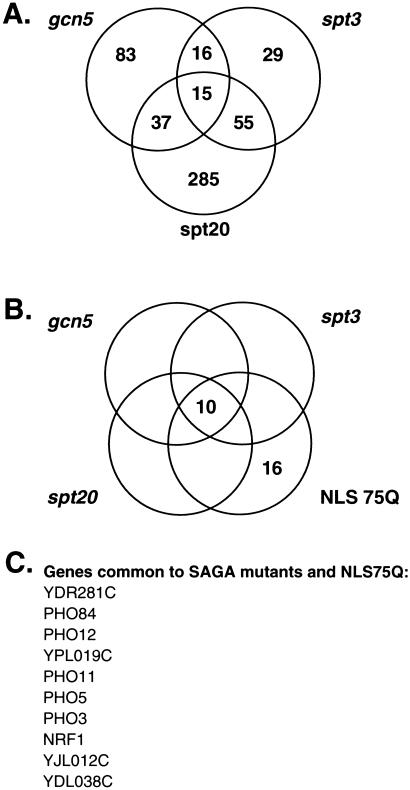
Comparison of the data from the NLS75Q-expressing strain with gene expression profiles from numerous mutant yeast strains revealed significant overlap with several strains that were deficient in SAGA complex proteins (database: http://chipdb.wi.mit.edu/chipdb_public/index.html). Of genes repressed in yeast containing mutations in components of the histone acetyltransferase complex SAGA, 15 are repressed in common among the SAGA mutants gcn5, spt3, and spt20 (32) (Fig. 3A). Of these core 15 genes, 10 are also repressed in the NLS-75Q expressing yeast (Fig. 3 B and C). Only one gene, PHO84, is similarly repressed in the strains expressing other forms of polyglutamine, and this repression is observed only in yeast expressing NLS-23Q. In contrast to gene induction, which appears primarily responsive to polyglutamine length, repression appears sensitive to both polyglutamine length and nuclear localization (Fig. 2).
Figure 3.
Expanded nuclear polyglutamine induces transcriptional changes that parallel those in SAGA complex mutants. (A) An existing database (http://web.wi.mit.edu/young/TFIID_SAGA/) was used to identify genes that were decreased in gcn5, spt3, and spt20 mutants. Genes that were decreased in one or more strains are enumerated in the Venn diagram. (B) Of the 26 genes that were decreased in the strain expressing NLS75Q, 10 were also decreased in all three SAGA complex mutants. (C) List of the 10 genes common to SAGA complex mutants and NLS75Q.
Expanded Nuclear Polyglutamine Enhances Toxicity of an spt3 Deletion.
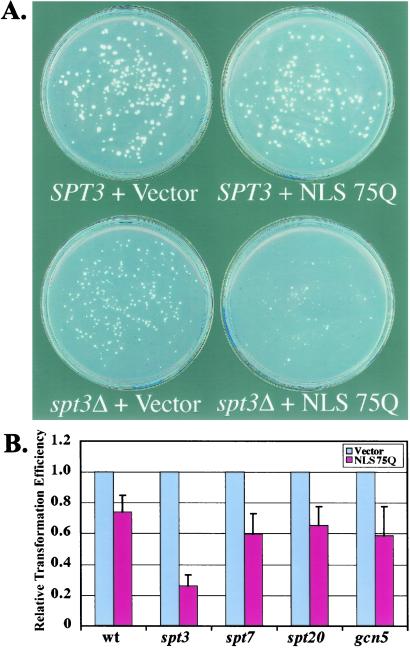
Because the transcript profile of the NLS75Q strain indicated a possible SAGA defect, we tested for possible genetic interactions between polyglutamine and mutations in genes encoding components of the SAGA complex, or in genes encoding proteins that interact with SAGA. We transformed strains carrying deletions in SAGA components with equal amounts of either vector DNA, DNA encoding a nonexpanded polyglutamine tract (23Q), or DNA encoding an expanded nuclear polyglutamine tract (NLS-75Q). The strains were deleted for GCN5, SPT3, or SPT7 (SAGA components), GAL11 (a component of the RNA polymerase II mediator complex) or SNF5 (a component of the SWI/SNF complex). The latter two genes (which contain glutamine tracts) were examined because of their genetic interaction with SAGA (20).
Toxicity was scored by comparing transformation efficiencies of polyglutamine constructs to each other and to vector. Significant toxicity was observed when nuclear expanded polyglutamine was expressed in a strain deleted for the SAGA component SPT3. Fig. 4B shows transformation plates comparing a wild-type to spt3 strain transformed either with vector or NLS-75Q construct. The spt3 strain alone has a slow growth phenotype and lower transformation efficiency than the wild-type. However, expression of NLS-75Q led to an ≈75% reduction in the total number of colonies and a dramatic decrease in average colony size (Fig. 4B). These effects were not seen with 23Q expression (not shown). These results are consistent with NLS-75Q interfering with an SPT3-related SAGA function.
Figure 4.
Enhanced toxicity of spt3 deletion by expression of nuclear expanded polyglutamine. (A) Wild-type or spt3 cells were transformed with equivalent amounts of vector plasmid or plasmid expressing simian virus 40 NLS-tagged expanded polyglutamine (75Q) from the constitutive GPD1 promoter. (B) Comparison of transformation efficiencies of an NLS-75Q plasmid into yeast strains deleted for SAGA components. Bars show the average and SE for three independent transformations (values are relative to transformation with vector).
Expanded Polyglutamine Repression of the PHO84 Promoter Is Mitigated by TSA.
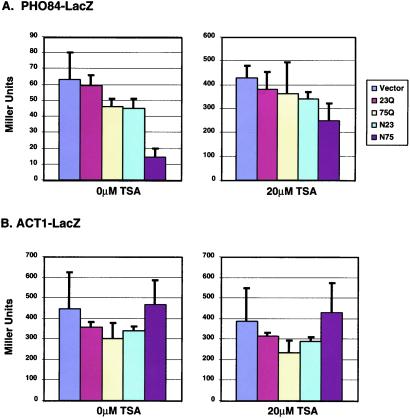
To further explore the role of histone acetylation in the NLS-75Q-mediated transcription defect, we tested whether repression of PHO84 was responsive to the histone deacetylase inhibitor TSA. In yeast, TSA inhibits the histone deacetylases Rpd3 and Hda1 (33). Using a PHO84-lacZ reporter, we observed a fivefold difference in β-galactosidase activity between cells expressing nuclear expanded polyglutamine (14 units) and cells with vector (63 units) (Fig. 5A Left). This reporter gene assay correlates well with the array data. Cells expressing other forms of polyglutamine showed no significant changes in PHO84-lacZ activity (Fig. 5A Left). Equivalent cells treated with 20 μM TSA for 20 h showed significantly increased levels of PHO84-lacZ activity (Fig. 5A Right), indicating that histone deacetylation plays a role in the down-regulation of PHO84 promoter basal activity. In the presence of TSA, the repressive effect of nuclear expanded polyglutamine was greatly reduced; cells with NLS-75Q produced 250 units and cells with vector, 427 units (less than a twofold difference). The ACT1 promoter was unresponsive to either polyglutamine expression or TSA (Fig. 5B).
Figure 5.
Effect of polyglutamine and TSA on expression of the PHO84 and ACT1 promoters (A and B). LacZ activity was measured in cells co-expressing different forms of polyglutamine and vector alone. Graphs on the right of each panel show LacZ activity in cells grown in the presence of 20 μM TSA for 20 h. Bars show the average and SEM for three independent transformants.
Discussion
A growing body of evidence indicates that transcriptional dysregulation resulting from effects on histone acetylation may be a primary pathogenic process in polyglutamine disease. Expanded polyglutamine-containing proteins have been shown to interact with transcription factors and co-activators (e.g., CREB-binding protein (CBP), p53, mSin3, N-CoR, and TAFII130) and to interfere directly with transcription (34–37). A genetic screen for enhancers and suppressors of SCA1 toxicity in Drosophila identified the transcriptional cofactors Sin3a, Rpd3, dCtBP, and dSir2, all known to regulate gene expression through histone acetylation (28). Loss of CBP function appears to play a direct role in polyglutamine pathogenesis. CBP is present in polyglutamine inclusions in human tissues from spinal and bulbar muscular atrophy (SBMA), HD, and dentatorubral–pallidoluysian atrophy (DRPLA) patients, and cellular toxicity associated with expanded polyglutamine expression is suppressed by overexpression of CBP (38, 39). These data suggest that polyglutamine-mediated loss of function in a critical transcription factor (CBP) may be a primary defect driving pathogenesis in a diverse group of polyglutamine diseases. Of transcription factors shown to interact with polyglutamine disease proteins, Sin3, Rpd3 and Sir2 have yeast orthologs, and all (like human CBP) are involved in regulating histone acetylation. CBP shares homology with Gcn5 and Spt7, components of the SAGA complex. CBP also interacts with the histone acetylase P/CAF, a human homolog of Gcn5 (40). Recent evidence indicates that decreased levels of acetylated histones are associated with polyglutamine toxicity in cell culture models and that toxicity in cell culture and a Drosophila model can be reversed with histone deacetylase inhibitors (A. McCampbell and K. Fischbeck, personal communication; ref. 41). Furthermore, polyglutamine can interact directly with the histone acetyltransferase domains of CBP, p300, and P/CAF and inhibit their enzymatic activity (41).
The homology of CBP and P/CAF to yeast SAGA components, together with the patterns of transcriptional changes, indicate that primary cellular events observed in polyglutamine disease pathogenesis are conserved between yeast and metazoan cells. The mitigating effect of histone deacetylase inhibitors in yeast, cell culture, and animal models of polyglutamine toxicity suggests that these may be a class of compounds useful in the treatment of polyglutamine disease. These observations indicate that yeast may provide a useful system for screening and analysis of chemical compounds that target this part of the polyglutamine toxicity pathway. The data also further demonstrate the value of yeast in modeling the molecular pathology of complex human diseases whose key defects are rooted in conserved cell autonomous pathways.
Acknowledgments
We thank Marcy MacDonald for providing human HD constructs, Peter Young and John Barnett for assistance with ChipDB, and Jeff Delrow for helpful discussion. DNA arrays were prepared by Jeff Delrow of the Fred Hutchinson Cancer Research Center DNA Array Facility. Alex McCampbell suggested the TSA experiment. R.E.H. is supported by the Milton Wexler Postdoctoral Fellowship of the Hereditary Disease Foundation. R.E.H, A.D.S., and J.M.O are supported by the Cure HD Initiative of the Hereditary Disease Foundation. S.F. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- CBP
CREB-binding protein
- SAGA
Spt/Ada/Gcn5 acetyltransferase
- TSA
Trichostatin A
- NLS
nuclear localization signal
- HD
Huntington's disease
- GFP
green fluorescent protein
- FC
fold change
References
- 1.Ross C A, Wood J D, Schilling G, Peters M F, Nucifora F C, Jr, Cooper J K, Sharp A H, Margolis R L, Borchelt D R. Philos Trans R Soc London B. 1999;354:1005–1011. doi: 10.1098/rstb.1999.0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zoghbi H Y, Orr H T. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 3.Perutz M F, Johnson T, Suzuki M, Finch J T. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 5.Paulson H L. Am J Hum Genet. 1999;64:339–345. doi: 10.1086/302269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paulson H L, Perez M K, Trottier Y, Trojanowski J Q, Subramony S H, Das S S, Vig P, Mandel J L, Fischbeck K H, Pittman R N. Neuron. 1997;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 7.Cooper J K, Schilling G, Peters M F, Herring W J, Sharp A H, Kaminsky Z, Masone J, Khan F A, Delanoy M, Borchelt D R, et al. Hum Mol Genet. 1998;7:783–790. doi: 10.1093/hmg/7.5.783. [DOI] [PubMed] [Google Scholar]
- 8.Hackam A S, Singaraja R, Wellington C L, Metzler M, McCutcheon K, Zhang T, Kalchman M, Hayden M R. J Cell Biol. 1998;141:1097–1105. doi: 10.1083/jcb.141.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warrick J M, Paulson H L, Gray-Board G L, Bui Q T, Fischbeck K H, Pittman R N, Bonini N M. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 10.Jackson G R, Salecker I, Dong X, Yao X, Arnheim N, Faber P W, MacDonald M E, Zipursky S L. Neuron. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 11.Faber P W, Alter J R, MacDonald M E, Hart A C. Proc Natl Acad Sci USA. 1999;96:179–184. doi: 10.1073/pnas.96.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satyal S H, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer J M, Morimoto R I. Proc Natl Acad Sci USA. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. . (First Published May 16, 2000; 10.1073/pnas.100107297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marsh J L, Walker H, Theisen H, Zhu Y Z, Fielder T, Purcell J, Thompson L M. Hum Mol Genet. 2000;9:13–25. doi: 10.1093/hmg/9.1.13. [DOI] [PubMed] [Google Scholar]
- 14.Kazemi-Esfarjani P, Benzer S. Science. 2000;287:1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- 15.Ordway J M, Tallaksen-Greene S, Gutekunst C A, Bernstein E M, Cearley J A, Wiener H W, Dure L S t, Lindsey R, Hersch S M, Jope R S, et al. Cell. 1997;91:753–763. doi: 10.1016/s0092-8674(00)80464-x. [DOI] [PubMed] [Google Scholar]
- 16.Cha J J. Trends Neurosci. 2000;23:387–392. doi: 10.1016/s0166-2236(00)01609-x. [DOI] [PubMed] [Google Scholar]
- 17.Lin X, Antalffy B, Kang D, Orr H T, Zoghbi H Y. Nat Neurosci. 2000;3:157–163. doi: 10.1038/72101. [DOI] [PubMed] [Google Scholar]
- 18.Luthi-Carter R, Strand A, Peters N L, Solano S M, Hollingsworth Z R, Menon A S, Frey A S, Spektor B S, Penney E B, Schilling G, et al. Hum Mol Genet. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- 19.Mumberg D, Muller R, Funk M. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 20.Roberts S M, Winston F. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pringle J R, Adams A E, Drubin D G, Haarer B K. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 22.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 23.Peters M F, Nucifora F C, Jr, Kushi J, Seaman H C, Cooper J K, Herring W J, Dawson V L, Dawson T M, Ross C A. Mol Cell Neurosci. 1999;14:121–128. doi: 10.1006/mcne.1999.0773. [DOI] [PubMed] [Google Scholar]
- 24.Krobitsch S, Lindquist S. Proc Natl Acad Sci USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muchowski P J, Schaffar G, Sittler A, Wanker E E, Hayer-Hartl M K, Hartl F U. Proc Natl Acad Sci USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez M K, Paulson H L, Pendse S J, Saionz S J, Bonini N M, Pittman R N. J Cell Biol. 1998;143:1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warrick J M, Chan H Y, Gray-Board G L, Chai Y, Paulson H L, Bonini N M. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez-Funez P, Nino-Rosales M L, de Gouyon B, She W C, Luchak J M, Martinez P, Turiegano E, Benito J, Capovilla M, Skinner P J, et al. Nature (London) 2000;408:101–106. doi: 10.1038/35040584. [DOI] [PubMed] [Google Scholar]
- 29.Pearce D A, Ferea T, Nosel S A, Das B, Sherman F. Nat Genet. 1999;22:55–58. doi: 10.1038/8861. [DOI] [PubMed] [Google Scholar]
- 30.Persson B L, Petersson J, Fristedt U, Weinander R, Berhe A, Pattison J. Biochim Biophys Acta. 1999;1422:255–272. doi: 10.1016/s0304-4157(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 31.Lenburg M E, O'Shea E K. Trends Biochem Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- 32.Lee T I, Causton H C, Holstege F C, Shen W C, Hannett N, Jennings E G, Winston F, Green M R, Young R A. Nature (London) 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 33.Bernstein B E, Tong J K, Schreiber S L. Proc Natl Acad Sci USA. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. . (First Published November 28, 2000; 10.1073/pnas.250477697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazantsev A, Preisinger E, Dranovsky A, Goldgaber D, Housman D. Proc Natl Acad Sci USA. 1999;96:11404–11409. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steffan J S, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu Y Z, Gohler H, Wanker E E, Bates G P, Housman D E, Thompson L M. Proc Natl Acad Sci USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. . (First Published May 23, 2000; 10.1073/pnas.100110097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boutell J M, Thomas P, Neal J W, Weston V J, Duce J, Harper P S, Jones A L. Hum Mol Genet. 1999;8:1647–1655. doi: 10.1093/hmg/8.9.1647. [DOI] [PubMed] [Google Scholar]
- 37.Shimohata T, Nakajima T, Yamada M, Uchida C, Onodera O, Naruse S, Kimura T, Koide R, Nozaki K, Sano Y, et al. Nat Genet. 2000;26:29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- 38.McCampbell A, Taylor J P, Taye A A, Robitschek J, Li M, Walcott J, Merry D, Chai Y, Paulson H, Sobue, Fischbeck K H. Hum Mol Genet. 2000;9:2197–2202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
- 39.Nucifora F C, Jr, Sasaki M, Peters M F, Huang H, Cooper J K, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson V L, et al. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 40.Berger S L. Curr Opin Cell Biol. 1999;11:336–341. doi: 10.1016/S0955-0674(99)80046-5. [DOI] [PubMed] [Google Scholar]
- 41.Steffan, J., Bodai, L., Pallos, J., Poelman, M., McCampbell, A., Apostol, B. L., Kazantsev, A., Schmidt, E., Zhu, Y.-Z., Greenwald, M., et al. (2001) Nature (London), in press. [DOI] [PubMed]