Abstract
LncRNAs appear to play a considerable role in tumourigenesis through regulating key processes in cancer cells such as proliferative signalling, replicative immortality, invasion and metastasis, evasion of growth suppressors, induction of angiogenesis and resistance to apoptosis. LncRNAs have been reported to play a role in prostate cancer, particularly in regulating the androgen receptor signalling pathway. In this review article, we summarise the role of 34 lncRNAs in prostate cancer with a particular focus on their role in the androgen receptor signalling pathway and the epithelial to mesenchymal transition pathway.
1. Introduction
Prostate cancer (PCa) is one of the most common forms of cancer and is the third leading cause of cancer-related deaths in men in the USA [1]. PCa is usually hormone-dependent at diagnosis highlighting the importance of androgen signalling in this disease. While the majority of patients with PCa are treated successfully with surgery and/or radiotherapy, metastatic disease develops in a significant proportion of patients. Androgen deprivation therapy (ADT) is initially effective in these patients but, invariably, resistance emerges. This form of PCa, known as castration resistant PCa (CRPC), is currently incurable.
Sustained androgen receptor (AR) signalling is a key feature of CRPC [2]. The AR is a ligand-activated transcription factor normally responsive to the androgens testosterone and dihydrotestosterone. However, CRPC progresses despite low/absent levels of circulating androgens. Mechanisms for sustained AR signalling despite low levels of circulating androgens include AR overexpression, aberrant activation of AR transcription and the development of AR variants [3,4].
It has now become apparent that the vast majority of human RNA transcripts are non-coding. Approximately 70–90% of the human genome is transcribed into RNA but only roughly 2% of the genome encodes for protein [5]. This large group of non-coding RNAs are broadly divided into two categories: small non-coding RNAs, <200 nucleotides long, and long non-coding RNAs (lncRNAs), >200 nucleotides long. While much work has focused on the function of small non-coding RNAs, we are only beginning to understand the role of lncRNAs in the regulation of gene expression.
Over 100,000 lncRNAs have been identified in the human genome to date [6]. They exhibit both nuclear and cytoplasmic localisation and are involved in the regulation of key cellular processes including transcription, translation, cell cycle regulation, cellular differentiation and nuclear-cytoplasmic trafficking [7,8]. The mechanisms through which this regulation is achieved are incompletely understood but include transcriptional interference, recruitment of chromatin remodelling complexes to specific gene loci, and by acting as competing endogenous RNAs [9].
This suggests a wide-ranging role for lncRNAs in gene regulation, so it is unsurprising that aberrant expression of lncRNAs appear to play a considerable role in human tumourigenesis [10,11]. LncRNAs appear to regulate key functions in cancer cells including sustained proliferative signalling, replicative immortality, invasion and metastasis, evasion of growth suppressors, induction of angiogenesis and resistance to apoptosis [12].
Although numerous lncRNAs have been implicated in prostate carcinogenesis [13], in many incidences, their functional role remains unknown or partially understood. The aim of this review is to collate the information in the literature related to lncRNAs in PCa, present a short summary on the most relevant lncRNAs and then discuss how these lncRNAs affect critical pathways in PCa pathogenesis.
2. LncRNAs associated with prostate cancer
2.1. CCAT2 (Colon Cancer-Associated Transcript 2)
The CCAT2 gene is located on chromosome 8q24, a known PCa locus. A recent study [14] examined fresh tissue from 96 patients with PCa and found that expression levels of CCAT2 were significantly higher in PCa tissues compared to adjacent non-tumour tissue. Upregulation of CCAT2 was positively associated with the histological grade and tumour stage, and Kaplan-Meier survival analysis revealed that patients with high CCAT2 expression levels had poorer overall survival and progression-free survival than patients with low CCAT2 expression [14]. Multivariate analysis also showed that the status of CCAT2 expression was an independent prognostic indicator [14].
In vitro, expression levels of CCAT2 were also demonstrated to be higher in PCa cell lines compared to a normal prostate stromal cell line [14]. Knockdown of CCAT2 impaired cellular proliferation, migration and invasion. Furthermore, it decreased expression of N-cadherin and vimentin and increased expression of E-cadherin, suggesting that CCAT2 may play a role in regulating EMT [14].
2.2. CTBP1-AS (C-Terminal Binding Protein 1 Antisense)
CTBP1-AS is located in the anti-sense region of CTBP1, a gene that is reported to function as an AR corepressor [15]. In support of this, lower CTBP1 expression is associated with poor cancer-specific survival and PSA-free survival, and CTBP1 overexpression is associated with reduced cell proliferation and repression of androgen-regulated genes [15].
CTBP1-AS represses CTBP1 expression by interacting with PTB-associated splicing factor (PSF), which binds at the CTBP1 promoter to induce histone deacetylation [15]. CTBP1-AS promotes cell cycle progression and tumour cell proliferation, is frequently overexpressed in PCa and is significantly correlated with high Gleason scores and overexpression of the AR. Expression levels of CTBP1-AS increase during ADT. In addition, PSF and CTBP1-AS promote cell cycle progression by repressing SMAD3 and p53, two cell cycle inhibitors. CTBP1-AS also mimics AR signalling by inducing upregulation of AR-regulated genes [15].
2.3. DANCR (Differentiation Antagonising Non-protein Coding RNA)
A single study reported that DANCR is upregulated in PCa and elevated expression is associated with high-grade tumours [16]. It promotes invasion and migration of prostate cancer cells in vitro and represses TIMP2/3 expression by mediating the binding of EZH2 on the TIMP2/3 promoter. TIMP2/3 has been reported to prevent invasion and metastatic spread in PCa [17].
2.4. DRAIC (Downregulated RNA in Androgen Independent Cells)
A recent study reported that DRAIC exerts suppressive functions by preventing the migration and metastatic spread of PCa cells [18]. This study reported that low levels of DRAIC are associated with increased rates of biochemical recurrence in patients with localised PCa. The AR pathway represses the transcription of DRAIC while FOXA1 and NKX3.1 induce the transcription of DRAIC. During the progression from PCa to CRPC, the expression of FOXA1 and NKX3.1 decrease and the AR pathway is aberrantly activated resulting in decreased levels of DRAIC.
2.5. FALEC (Focally Amplified LncRNA in Epithelial Cancer)
Zhao et al., reported that FALEC expression is significantly increased in PCa tissue samples compared to adjacent normal tissue [19]. This study reported that knockdown of FALEC inhibited cell proliferation, migration and invasion in vitro. In addition, PCa cells subjected to a hypoxic environment showed increased expression of FALEC and HIF-1α. Knockdown of HIF1α caused a reduction in FALEC expression whereas overexpression conferred the opposite effect.
2.6. GAS5 (Growth Arrest Specific 5)
GAS5 is abundant in cells whose growth has arrested due to a lack of nutrients or growth factors [20]. Suppression of mTOR activity, such as occurs upon growth arrest, prevents degradation of GAS5 transcripts by the nonsense mediated decay (NMD) pathway leading to an accumulation of GAS5 transcripts [21]. GAS5 accumulation results in increased susceptibility to apoptosis by inhibiting the anti-apoptotic action of glucocorticoids [22]. GAS5 binds to the DNA-binding domain of the glucocorticoid receptor, thus competing with DNA glucocorticoid response elements for binding. Importantly, GAS5 can also sequester the androgen/AR complex and prevent it binding to target DNA sequences [22].
In CRPC, GAS5 is downregulated and it has been shown to increase apoptosis and decrease cell survival in vitro [23]. The use of mTOR inhibitors leads to an increase in GAS5 levels in androgen-dependent and androgen-sensitive PCa cell lines but not in androgen-independent cell lines [24].
A further study [25] reported that GAS5 inactivates the PI3K-AKT-mTOR signalling pathway in PCa by targeting miR-103. Overexpression of GAS5 was shown to significantly decelerate PCa cell progression in vitro and tumour growth in vivo through inactivating the PI3K-AKT-mTOR signalling pathway.
In terms of cell cycle, GAS5 is also reported to induce the expression of P27Kip1, which regulates the progression at the G1 to S phase transition [26]. GAS5 interacts with E2F1, a transcription factor, and enhances the binding of E2F1 to the P27Kip1 promoter.
2.7. HCG11 (HLA Complex Group 11)
HCG11 is an androgen-responsive lncRNA, downregulation of which is associated with a poor prognosis in PCa [27]. This study reported that expression of HCG11 was significantly lower in PCa tissue compared to non-tumour tissue.
2.8. HOTAIR (HOX Transcript Antisense RNA)
Zhang et al. reported that HOTAIR is repressed by androgens and is therefore upregulated in CRPC [28]. The study demonstrated that HOTAIR binds to the N-terminal domain of the AR, blocking the binding of E3 ubiquitin ligase MDM2 which interacts with the AR through the same domain. This prevents AR ubiquitination and degradation, and leads to increased AR gene expression. They also reported that HOTAIR overexpression can induce an AR-binding profile similar to that induced by androgens resulting in activation of the AR transcriptional programme in the absence of androgens. HOTAIR overexpression was found to increase PCa cell growth in vitro with knockdown of HOTAIR having the opposite effect. Levels of HOTAIR were also found to increase in LNCaP cell lines upon treatment with enzalutamide [28].
Another study reported that an increase in infiltrating mast cells enhanced PCa invasion via modulation of the interaction between HOTAIR and Polycomb Repressive Complex 2 (PRC2) [29]. PCa cells treated with ADT using casodex or enzalutamide were reported to recruit more mast cells.
Genistein, a soy isoflavone, has been reported to inhibit PCa cell growth in AR-negative cell lines through downregulation of HOTAIR [30]. This study also reported that HOTAIR is targeted by miR-34a and that knockdown of HOTAIR decreased PCa cell proliferation, migration and invasion and induced apoptosis and cell cycle arrest.
2.9. LINC01296 (Long Intergenic Non-protein-coding RNA 1296)
A single study reported that LINC01296 expression promotes PCa growth and metastasis by activating the PI3K-AKT-mTOR signalling pathway and promoting EMT [31]. Patients with higher LINC01296 expression displayed advanced clinical features and shorter biochemical recurrence-free survival time than those with lower LINC01296 expression. LINC01296 expression correlated with preoperative PSA level, lymph node metastasis, Gleason score and tumour stage. Multivariate analysis showed that LINC01296 expression was an independent predictor of biochemical recurrence-free survival.
2.10. LincRNA-p21 (Long Intragenic Non-coding RNA p21)
A single study reported that LincRNA-p21 suppresses the development of PCa [32]. LincRNA-p21 expression inhibited PCa cell proliferation and colony formation in vitro and reduced the rate of PCa cell population growth in vivo. It promoted apoptosis and induced expression of p53 responsive genes by regulating p53 binding to their promoters. In malignant prostate tissue, the expression of p53 downstream genes such as MDM2, Puma, Noxa and Bax, were reduced and correlated with reduced expression of lincRNA-p21. Low levels of LincRNA-p21 correlated with higher disease stage and predicted poor overall and disease-free survival.
2.11. LncRNA-ATB (Long non-coding RNA-activated by transforming growth factor β)
A single study reported that LncRNA-ATB expression was significantly upregulated in PCa tumour tissue compared to adjacent non-tumour tissue [33]. High levels of lncRNA-ATB correlated with high preoperative PSA level, pathological stage, high Gleason score, lymph node metastasis, angiolymphatic invasion and biochemical recurrence. LncRNA-ATB expression increased cell proliferation and increased expression levels of the cell cycle regulatory proteins cyclin E and cyclin D1 in vitro.
Knockdown of lncRNA-ATB increased expression levels of E-cadherin and ZO-1 and decreased expression levels of N-cadherin and Vimentin in vitro while overexpression had the opposite effect, suggesting a role in EMT. Upregulation of lncRNA-ATB also led to upregulation of ZEB1 and ZNF217, and to activation of the PI3K-AKT-mTOR and ERK signalling pathways, all of which have been implicated in EMT [33].
2.12. LOC283070
A single study reported that LOC283070 plays an important role in the transition of LNCaP cells from an androgen-dependent state to an androgen-independent state [34]. Overexpression of LOC283070 in LNCaP cells accelerated cell proliferation and migration in both androgen dependent and independent LNCaP cell lines. Overexpression of LOC283070 also promoted tumour growth in vivo in both normal and castrated mice. Overexpression of CAMK1D had similar effects to LOC283070 on the transition of LNCaP cells to an androgen-independent state while its knockdown fully abrogated the effect of LOC283070 overexpression suggesting a relationship between CAMK1D and LOC283070.
2.13. LOC400891
A single study reported that LOC400891 was significantly upregulated in PCa tissue compared to adjacent non-tumour tissue [35]. LOC400891 was also overexpressed in DU-145 and 22RV1 PCa cell lines compared to the normal human stromal cell line, WPMY-1. Patients with high LOC400891 expression had a significantly shorter biochemical recurrence-free survival time than those with low LOC400891 expression. In vitro experiments suggested that LOC400891 is involved in the promotion of PCa cell proliferation and metastasis and this may be mediated by modulation of the PI3K-AKT-mTOR pathway and PTEN gene. In addition, LOC400891 may function as an inducer of EMT.
2.14. LOC440040
A single study reported that LOC440040 was significantly upregulated in PCa tissue compared to non-tumour tissue [36]. LOC440040 expression level was also significantly increased in PC3 and 22RV1 cell lines but not in the DU-145 cell line when compared to the normal human stromal cell line, WPMY-1. Patients with high expression levels of LOC440040 had a significantly shorter biochemical recurrence-free survival time than those with low expression levels. LOC440040 promoted the proliferation of PCa cells in vitro, possibly mediated by the PI3K-AKT-mTOR pathway. In addition, LOC440040 may be associated with EMT.
2.15. MALAT-1 (Metastasis Associated Lung Adenocarcinoma Transcript 1)
Ren et al. reported that MALAT-1 is overexpressed in human PCa tissues and cell lines [37]. High MALAT-1 expression levels correlated with high Gleason score and tumour stage. Expression levels increased during the progression from hormone-sensitive to castration-resistant states. Suppression of MALAT-1 in vitro resulted in reduced cell growth, invasion, migration and colony formation, and increased rates of apoptosis and cell cycle arrest. Suppressing expression in vivo delayed tumour growth and reduced metastasis of PCa xenografts in castrated nude mice.
Another study reported that MALAT-1 binds to EZH2, a core subunit of PRC2 that is also frequently overexpressed in PCa, especially CRPC [38]. MALAT-1 enhanced EZH2-mediated PCa cell invasion and migration, and EZH2-mediated repression of PRC2-dependent target genes such as DAB2IP and BRACHYURY. MALAT-1 also enhanced the expression of EZH2 target genes that are independent of PRC2 such as CKS2 and TMEM48.
A recent study reported that MALAT-1 is required for enzalutamide induced AR-V7 production and may enhance splicing of AR-V7 through direct modulation of SF2 binding and activity [39]. MALAT-1 expression and SF2 activity were upregulated in enzalutamide resistant PCa cell lines, contributed to AR-V7 production and led to drug resistance.
MALAT-1 has also been reported to promote bone metastasis in PCa [40]. An in vitro co-culture model of PC3 cells and osteoblasts was used to identify PCa-bone microenvironment interactions. Factors secreted by PC3 cells resulted in the downregulation of Wnt inhibitor, Sost, in osteoblasts suggesting that Sost may play a role in facilitating PCa metastasis to bone. Downregulation of Sost led to a significant upregulation of MALAT-1 in PC3 cells.
2.16. MEG3 (Maternally Expressed 3)
A single study showed expression levels of MEG3 were significantly lower in PCa tissue compared to non-tumour tissue, although no significant relationship between MEG3 expression and clinical characteristics was identified [41]. MEG3 suppressed cell proliferation and induced apoptosis in vitro. It increased the proportion of cells in G0/G1 phase and decreased the proportion of cells in S phase. It induced Bax protein expression and activated caspase-3, both of which are associated with pro-apoptotic activity, and inhibited Bcl-2 and Cyclin D1 protein expression.
MEG3 has also been reported to act as a tumour suppressor by activating p53, causing impaired cell proliferation and promotion of apoptosis [42].
2.17. NEAT1 (Nuclear Enriched Abundant Transcript 1)
Oestrogen receptor alpha (ERα) is expressed in PCa and may comprise an important AR-independent mechanism of signalling in CRPC [43]. ERα regulates the expression of TMPRSS2-ERG fusion gene and induces the transcription of NEAT1. A single study reported that NEAT1 and ERα levels are significantly higher in CRPC compared to PCa, and that NEAT1 levels increase upon treatment with tamoxifen, bicalutamide or enzalutamide [43]. NEAT1 promoted cellular proliferation and invasion both in vitro and in vivo, and NEAT1 levels were positively associated with biochemical recurrence and metastasis. PCa cells expressing high levels of NEAT1 were resistant to ADT or AR antagonists.
2.18. PCA3 (Prostate Cancer Antigen 3)
PCA3 expression is highly specific for PCa and therefore has been used as a clinical biomarker [44]. PCA3 appears to be involved in EMT, as knockdown of PCA3 leads to upregulation of epithelial markers E-cadherin, claudin-3 and CK18 and downregulation of vimentin [45]. PCA3 knockdown also inhibited AR signalling, cell growth and viability [45]. It has been reported to regulate the expression of genes involved in angiogenesis, signal transduction and apoptosis and to regulate the tumour suppressor, PRUNE2 [46].
The transcription factor Snail is reported to activate the expression of PCA3 by binding to its promoter region [47]. PCA3, in turn, negatively regulates the expression of miR-1261 through competitive sponging [47]. PRKD3 is negatively regulated by miR-1261 meaning that increased PCA3 can result in high expression of PRKD3, which can promote invasion and metastasis of PCa.
Another study reported that PCA3 knockdown reduced the expression of AR-related genes PSA and PCGEM1, and sensitised PCa cells to enzalutamide-induced loss of cell growth [48].
2.19. PCAT1 (Prostate Cancer Associated Transcript 1)
Double-stranded DNA breaks (DSBs) are repaired by several mechanisms including homologous recombination (HR) [49]. BRCA1/2 mutations lead to defective HR and increased DSBs [49]. PCAT1 expression has been reported to cause repression of BRCA2 which leads to an impairment in HR which, in turn, imparts a high sensitivity to PARP1 inhibitors [50].
PCAT1 has also been reported to increase cell proliferation by enhancing the expression of cMyc protein [51]. PCAT1 post-transcriptionally upregulates cMyc and interferes with the downregulation of MYC by miR-34a. Conversely, targeting of PCAT1 by another microRNA, miR-3667-3p, reverses its effect on cMyc [51]. A further study reported that PCAT1 upregulates FSCN1 by acting as a miR-145-5p sponge resulting in increased PCa cell proliferation, migration, invasion and suppressed apoptosis [52].
2.20. PCAT5 (Prostate Cancer Associated Transcript 5)
A single study reported that knockdown of PCAT5 resulted in enhanced apoptosis and reduced cell proliferation and invasion of PCa cells in vitro [53]. PCAT5 was reported to be exclusively overexpressed in ERG-positive PCa and CRPC tissue compared to non-tumour prostate tissue. Knockdown of ERG resulted in a reduction of PCAT5 levels.
2.21. PCAT14 (Prostate Cancer Associated Transcript 14)
A study reported that PCAT14 is regulated by the AR and overexpression of PCAT14 suppresses the invasive capabilities of PCa cells [54]. PCAT14 expression was associated with favourable outcomes in biochemical progression-free survival, metastasis-free survival and PCa-specific survival. Another study analysed radical prostatectomy microarray and clinical data from 910 patients and found PCAT14 to be the most prevalent aberrantly expressed lncRNA [55]. Lower PCAT14 expression was significantly associated with unfavourable outcomes in distant metastasis-free survival, PCa specific survival and overall survival.
2.22. PCAT18 (Prostate Cancer Associated Transcript 18)
A single study reported PCAT18 was upregulated in PCa tissue compared to non-tumour tissue and was highly expressed in CRPC [56]. AR activation dramatically upregulated PCAT18 expression in vitro and in vivo, and knockdown of PCAT18 significantly inhibited PCa cell proliferation, migration and invasion.
2.23. PCAT29 (Prostate Cancer Associated Transcript 29)
It has been reported that PCAT29 functions as a tumour suppressor and is repressed by androgen signalling [57]. AR binds to the PCAT29 promoter and suppresses its transcription [57]. In this study [57], knockdown of PCAT29 increased cell proliferation and the migration of PCa cells in vitro whereas overexpression conferred the opposite effect. In patient specimens, low PCAT29 expression correlated with significantly higher rates of biochemical recurrence.
Another study [58] reported IL-6, a cytokine which is elevated in prostate tumours [59], reduced the expression of PCAT29 in both DU145 and LNCaP cell lines. IL-6 activates STAT3 which induces miR-21. This interaction leads to the repression of PCAT29. Inhibition of miR-21 was shown to increase PCAT29 expression [58].
2.24. PCGEM1 (Prostate Cancer Gene Expression Marker 1)
Yang et al. reported that PCGEM1 activates the AR in vitro and that PCGEM1 acts in association with PRNCR1 to physically bind to the AR [60]. Both PCGEM1 and PRNCR1 were highly upregulated in CRPC cell line models and were reported to activate the AR in the absence of androgen.
However, these findings were disputed by Prensner et al., who reported that PCGEM1 and PRNCR1 were not implicated in CRPC or AR signalling [61]. A further study by Parolia et al. reported that PCGEM1 was highly expressed in PCa while PRNCR1 showed scant expression. PCGEM1 was implicated in early stage PCa but not in metastatic PCa, was significantly downregulated in metastatic tumours relative to primary tumours, and significantly repressed in patients exhibiting poor clinical outcomes [62]. They also reported that in vivo stimulation of the AR led to upregulation of PCGEM1 (Refer to section 3.1.1).
Zhang et al. reported that ADT induced PCGEM1 and that this expression was associated with the expression of AR-V7 [63]. ADT led to an upregulation of PCGEM1, which relocated to the nucleus, and enhanced interaction with the splicing factors U2AF65 and hnRNP A1. The interaction with U2AF65, an enhancer of AR-V7, led to increased activity of the splicing factor to AR pre-mRNA. Interaction with hnRNP A1, a negative regulator of AR-V7, reduced the affinity of hnRNP A1 to AR pre-mRNA.
Another study by Ho et al. reported that PCGEM1 is regulated by p54/nrb, a multifunctional nuclear protein involved in a variety of nuclear processes [64]. PCGEM1 was upregulated through the action of p54/nrb in response to androgen deprivation which also led to upregulation of AR-V7.
Hung et al. reported that PCGEM1 regulates tumour metabolism by enhancing activation of cMyc and the AR [65]. PCGEM1 directly bound cMyc and facilitated the recruitment of cMyc to target sites.
A further study reported that downregulation of PCGEM1 increased the expression of miR-145, a miRNA that appears to function as a tumour suppressor [66]. In addition, overexpression of miR-145 decreased PCGEM1 expression as well as inhibiting cell proliferation, migration and invasion, and also induced early apoptosis. It was suggested that miR-145 may regulate PCGEM1 expression by directly binding to target sites within the PCGEM1 sequence.
2.25. PlncRNA-1
A study reported that PlncRNA-1 is overexpressed in PCa and knockdown of PlncRNA-1 leads to enhanced apoptosis and reduced cell proliferation [67]. Knockdown also led to decreased expression of AR and NKX3-1, a downstream target of AR. Knockdown of AR caused decreased levels of PlncRNA-1.
PlncRNA-1 appears to protect the AR from miRNA-mediated suppression [68]. Experiments showed that PlncRNA-1 acts as a sponge for certain AR targeting miRNAs such as miR-34c and miR-297.
PlncRNA-1 is also reported to regulate both the cell cycle and EMT through the TGF-β1 pathway [69], and to regulate CyclinD1 levels through the Her-2 pathway [70]. Downregulation of PlncRNA-1 was associated with a decrease in Her-2 protein expression and an increase in apoptosis.
2.26. POTEF-AS1 (Prostate Ovary Testis Expressed Family member Antisense 1)
A single study reported that the expression of POTEF-AS1 was regulated by the AR in vitro [71]. It promoted cell growth, inhibited apoptosis and repressed genes related to the Toll-like receptor signalling pathway including TLR3, CXCL10 and TNFSF10.
2.27. PRNCR1 (Prostate Cancer Associated Non-coding RNA 1)
A study reported that PRNCR1 expression was upregulated in PCa cells and knockdown of PRNCR1 reduced the viability of PCa cells and the activity of the AR [72]. PRNCR1 was also reported to associate with PCGEM1 and physically bind to the AR [60]. However, this result has been disputed and as two studies have failed to show upregulation of PRNCR1 in PCa [61,62] (Refer to Section 2.24).
2.28. PVT1 (Plasmacytoma Variant Translocation 1)
PVT1 is overexpressed in PCa and is reported to be involved in the regulation of tumour growth [73,74]. PVT1 can regulate PCa cell viability and apoptosis in association with miR-146a [75]. MiR-146a was downregulated and negatively correlated with PVT1 levels in PCa. PVT1 regulated miR-146a expression by inducing the methylation of CpG Island in its promoter. Another study reported that PVT1 knockdown significantly upregulated the expression of cleaved caspase-3 and cleaved caspase-9 and downregulated the expression of cMyc in PCa [74].
2.29. SChLAP1 (Second Chromosome Locus Associated with Prostate 1)
Multiple studies have reported that SChLAP1 is overexpressed in PCa, especially CRPC, and is predictive for poor patient outcomes [[76], [77], [78], [79]]. It interacts with the SWI/SNF complex, impairing its ability to regulate gene expression, resulting in the promotion of cell invasion and metastasis [78]. SChLAP1 appears to counteract the tumour-suppressive effects of the SWI/SNF complex by directly disrupting SNF5, a core subunit of the SWI/SNF complex [80]. It has also been reported that SChLAP1 acts via targeting miR-198 and promoting the MAPK1 pathway [81]. SChLAP1 acts as a negative regulator of miR-198.
2.30. SNHG1 (Small Nucleolar RNA Host Gene 1)
A single study reported that SNHG1 was upregulated in PCa and negatively regulates miR-199a-3p by acting as a competing endogenous RNA [82]. This resulted in enhanced CDK7 expression and promotion of PCa cell proliferation and cell cycle progression.
2.31. SOCS2-AS1 (Suppressor of cytokine signalling 2 antisense)
A single study reported that SOCS2-AS1, an androgen-induced lncRNA, promoted castration resistant and androgen-dependent cell growth, and enhanced the migration potential of PCa cells in vitro [83]. The authors reported that SOCS2-AS1 regulates genes involved in cellular proliferation, apoptosis and the cell cycle such as TNFSF10, FOXM1 and CENPF. They also suggested that high AR signalling in CRPC may lead to overexpression of SOCS-AS1.
2.32. TRPM2-AS (Transient Receptor Potential Cation Channel Subfamily M Member 2 Antisense)
A single study reported that TRPM2-AS is overexpressed in PCa and is associated with a poor prognosis [84]. As well as regulating the TRPM2 gene, TRPM2-AS regulates the expression of a number of genes involved in cell survival and the cell cycle such as the oncogenes FYN and AKT1 [85].
2.33. UCA1 (Urothelial Cancer Associated 1)
Studies have reported that UCA1 is abnormally upregulated in PCa tissue and is associated with poor clinical outcomes [86,87]. It appears to regulate cell growth, migration and invasion, and inhibit apoptosis [86,87]. It is also associated with drug resistance in various cancers, including PCa [88]. In one study, expression levels of KLF4 in PCa tissue correlated with UCA1 levels [86] and knockdown of UCA1 decreased KLF4 expression. KLF4 plays a key role in cancer cellular differentiation and proliferation [89]. Knockdown of UCA1 also resulted in the downregulation of KRT6 and KRT13.
UCA1 has been reported to act as a competitive endogenous RNA and downregulates the expression of miR-204 [87]. In one study, the authors reported that this resulted in upregulation of the ATF2 gene as miR-204 causes the degradation of ATF2 [87]. Another study reported that miR-204 negatively modulates Sirt1 expression and that UCA1 overexpression results in increased Sirt1 expression [90]. Upregulation of Sirt1 is associated with enhanced tumour cell proliferation, invasion and migration and the promotion of EMT.
2.34. ZEB1-AS1 (Zinc Finger E-box Binding Protein 1)
ZEB1-AS1 has been reported to promote the proliferation and migration of PCa cells and also promote ZEB1 expression by binding and recruiting histone methyltransferase MLL1 to the promoter region of ZEB1 [91]. ZEB1 promotes many tumour biological behaviours including EMT.
ZEB1-AS1 also indirectly regulates BMI1, a regulator of prostate stem cells [92]. BMI1 is associated with metastatic disease and poor clinical outcomes [93]. BMI1 is directly repressed by miR200c, which is itself directly inhibited by ZEB1 and ZEB1-AS1 [91]. Therefore, upregulation of ZEB1-AS1 results in the upregulation of BMI1 via the repression of miR-200c.
3. Involvement of lncRNAs in specific pathways in PCa
Certain pathways are important in the pathogenesis of PCa, the most important of which is the pathway regulated by the AR. In addition, the EMT pathway is an important pathway in the pathogenesis of metastatic PCa. Below, we summarise the roles of lncRNAs currently identified as playing roles in these pathways.
3.1. Androgen/AR Pathway
3.1.1. Positive regulation of the AR
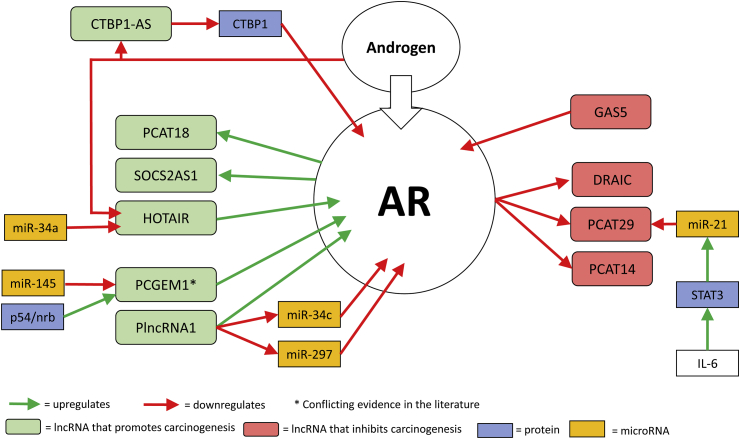
Upregulation of the AR is a feature of CRPC and may be one mechanism facilitating AR signalling despite absent/low levels of endogenous androgens. Certain lncRNAs (CTBP1-AS and HOTAIR) are upregulated after ADT suggesting that they are negatively regulated by androgens (Fig. 1). The upregulation of these lncRNAs result in upregulation of the AR.
Fig. 1.
lncRNAs that have been shown to interact with the Androgen Receptor (AR).
CTBP1-AS upregulation results in downregulation of CTBP1. CTBP1 negatively regulates the AR by inhibiting androgen-mediated demethylation [15]. Therefore, downregulation of CTBP1 by CTBP1-AS results in upregulation of the AR (Fig. 1).
HOTAIR binds to the N-terminal domain of the AR (Fig. 1), blocking the binding of E3 ubiquitin ligase MDM2, which interacts with the AR through the same domain [28]. This prevents AR ubiquitination and degradation. HOTAIR overexpression can also induce an AR-binding profile similar to that induced by androgen thus resulting in activation of the AR transcriptional programme in the absence of androgen.
There are conflicting reports regarding PCGEM1 and whether it is upregulated after ADT and if it interacts with the AR. Yang et al. reported that PCGEM1 and PRNCR1 bind to the AR and enhance AR-mediated gene activation programmes [60]. PCGEM1 was reported to be dependent on PRNCR1 for binding. Both PCGEM1 and PRNCR1 were highly upregulated in CRPC cell line models and were reported to activate the AR in the absence of androgen. Knockdown of either PCGEM1 or PRNCR1 significantly inhibited in vivo tumour growth in a xenograft mouse model, and ADT also resulted in PCGEM1 relocalisation from the cytoplasm to the nucleus.
However, Prensner et al. subsequently reported that PCGEM1 and PRNCR1 were not implicated in CRPC or AR signalling [61]. They assessed the expression levels of PCGEM1 and PRNCR1 in 171 human prostatic tissues using RNA sequencing data aggregated from four independent studies of PCa. They reported that PCGEM1 but not PRNCR1 was associated with PCa. They stated that neither PCGEM1 nor PRNCR1 were associated with poor patient outcomes after examining a cohort of 235 high-risk PCa patients with long-term outcome data. They were also unable to observe PCGEM1 or PRNCR1 interacting with the AR or acting as a component of AR signalling.
This issue was further investigated by Parolia et al. who noted that the published data on the relationship between the AR and PCGEM1/PRNCR1 had been derived from in vitro experiments, using androgen-sensitive LNCaP cells [62]. As the AR has been reported to induce varied AR transcriptional programmes in patient tumour tissue compared to PCa cell lines [94], they used Living Tumour Laboratory (LTL) patient-derived xenograft PCa models with the intention of more accurately mirroring the pathogenesis of PCa. They analysed RNA sequencing data from two distinct androgen-dependent models and reported that PCGEM1 was considerably expressed in PCa, while PRNCR1 showed scant expression in keeping with the findings of Prensner et al. [61]. PCGEM1 was reported to be implicated in early stage PCa but not in metastatic PCa, was significantly downregulated in metastatic tumours relative to primary tumours and significantly repressed in patients exhibiting poor clinical outcomes [62]. They also observed a significant decrease in PCGEM1 expression and reduced AR activity post-castration, contradicting findings made by Yang et al. [60]. However, in contrast to Prensner et al. [61], they found in vivo stimulation of the AR led to upregulation of PCGEM1 [62]. AR activation did not alter the subcellular localisation of PCGEM1, and it was evenly distributed between the nucleus and the cytoplasm.
Two subsequent studies by Zhang et al. [63]and Ho et al. [63,64] found that PCGEM1 was upregulated in response to ADT in vitro which is supportive of the original conclusions by Yang et al. [60]. Zhang et al. [63] reported that suppression of PCGEM1 significantly reduced tumour growth and tumour weight in castrated mice, and also resulted in subcellular redistribution of PCGEM1 from the cytoplasm to the nucleus. Ho et al. [64]also identified p54/nrb, a ubiquitously expressed nuclear protein, as a positive regulator of PCGEM1 levels.
Another mechanism of lncRNAs regulating the AR is through interactions with miRNAs. PlncRNA-1 appears to protect the AR from miRNA-mediated suppression [68] by acting as a sponge for certain AR-targeting microRNAs such as miR-34c and miR-297. HOTAIR was reported to be targeted by miR-34a [30] and PCGEM1 by miR-145 [66].
3.1.2. Negative regulation of the AR
GAS5 negatively regulates the AR by binding to the DNA-binding domain of the AR preventing it from binding to target DNA sequences [22].
3.1.3. LncRNAs regulated by the androgen receptor
AR activation is reported to upregulate lncRNAs PCAT18 and SOCS-AS1 resulting in enhanced PCa cell proliferation, migration and invasion [56,83]. In vitro analysis suggested that the AR indirectly activates PCAT18 expression. Microarray data revealed 402 genes positively and significantly associated with PCAT18 expression. This expression signature was strongly associated with AR activation. SOCS2-AS1 is induced by androgen and may modulate AR activity by altering the epigenetic control for AR target genes.
AR activation is reported to repress the transcription of tumour suppressors DRAIC and PCAT29 and also repress PCAT14, a lncRNA associated with favourable clinical outcomes in PCa [18,54,56]. PCAT29 is also reported to be indirectly repressed by IL-6, a cytokine which is elevated in prostate tumours [58]. IL-6 activates STAT3 which induces miR-21 and this interaction leads to the repression of PCAT29.
3.1.4. AR splice variants
MALAT-1 is reported to induce AR-V7 production in vitro and may enhance splicing of AR-V7 through direct modulation of SF2 binding and activity [39].
Zhang et al. reported that after ADT, PCGEM1 relocated from the cytoplasm to nuclear speckles where it interacted with U2AF65, promoting its interaction with AR pre-mRNA and promoting AR-V7 production [63]. PCGEM1 also interacted with hnRNPA1, a repressor of AR-V7, causing a significant decrease in the ability of hnRNPA1 to interact with AR pre-mRNA. As mentioned previously, there are conflicting reports in the literature regarding PCGEM1 and some studies did not show subnuclear relocalisation of PCGEM1 after ADT or promotion of AR-V7 production [61,62].
3.2. EMT
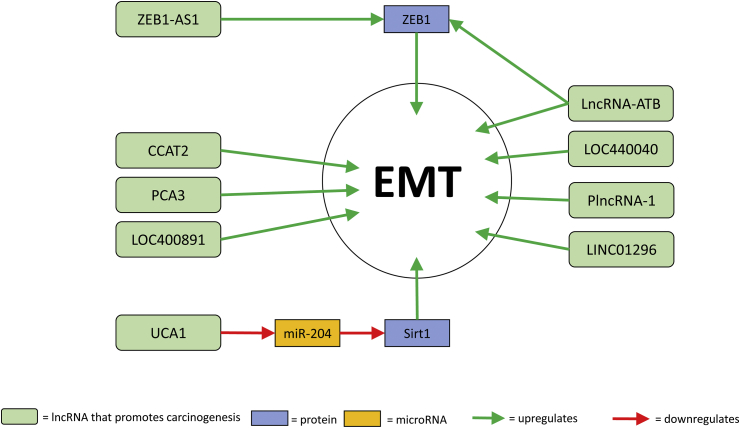
EMT is an important step in metastasis, during which non-motile polarised epithelial cells convert into individual, motile mesenchymal cells [95]. EMT has also been associated with resistance to various anti-cancer therapies [96]. In PCa, numerous lncRNAs are associated with the promotion of EMT (Fig. 2).
Fig. 2.
lncRNA associated with EMT and Prostate Cancer.
Many of these, such as CCAT2, LOC400891, LOC440040, PCA3 and LINC01296 have been associated with increased/decreased expression of N-cadherin, vimentin, β-catenin, Twist, Snail, claudin-3 and E-cadherin [14,31,35,36,45]. PlncRNA-1 induces EMT through the TGF-β1 pathway [69]. ZEB1-AS1 promotes ZEB1 expression by binding and recruiting histone methyltransferase MLL1 to the promoter region of ZEB1 [91]. ZEB1 is known to promote EMT [97,98]. LncRNA-ATB also upregulates ZEB1 as well as modulating expression levels of E-cadherin, ZO-1, N-cadherin and vimentin [33]. UCA1 promotes EMT through the targeting of miR-204 [90]. MiR-204 negatively modulates Sirt1 expression so UCA1 overexpression results in increased Sirt1 expression. Upregulation of Sirt1 is associated with the promotion of EMT.
4. Discussion
The study of lncRNAs has emerged as a burgeoning area of cancer research and, accordingly, there has been a surge in publications in this area in recent years. LncRNAs appear to regulate key functions in cancer cells including sustained proliferative signalling, replicative immortality, invasion and metastasis, evasion of growth suppressors, induction of angiogenesis and resistance to apoptosis [12]. However, with over 100,000 lncRNAs identified to date, the mechanisms of action of most lncRNAs remains unknown.
In this report, we have reviewed the literature and provided an up to date summary of the lncRNAs reported to be involved in prostate carcinogenesis. We have reviewed 34 specific lncRNAs and found that lncRNAs in PCa exert their actions through various mechanisms including, but not limited to, regulation of (i) AR expression pathway, (ii) EMT, (ii) miRNAs and (iv) PI3K/AKT/mTOR pathway. We have focused on lncRNAs involved in the regulation of the AR expression pathway as sustained AR activity is known to be a critical component in prostate carcinogenesis, even after ADT [99].
The clinical use of lncRNAs has, so far, been restricted to their use as biomarkers. Their potential as therapeutic targets to halt disease progression has yet to be realised. In addition, as in any nascent research field, the number of large scale studies validating original findings are small and there are conflicting findings in the literature with regards to the role of some lncRNAs. Many of the summaries provided in this article are based on single reports, often involving predominantly or solely in vitro experiments. Further large scale in vivo studies are required to validate many of the findings mentioned in this article.
5. Conclusion
LncRNAs are involved in the regulation of key processes in prostate carcinogenesis, particularly the AR expression pathway and EMT pathway. However, the extent of the role that lncRNAs play in prostate carcinogenesis has yet to be fully elucidated. This lack of knowledge demands that we exercise caution when interpreting results, particularly when based on single studies. That being said, the study of lncRNAs is currently an exciting area of cancer research precisely because there is much that is unknown. Unlocking the role of lncRNAs may provide crucial insights into the pathogenesis of PCa and we hope that this knowledge can be transferred into clinical benefits for patients through the use of better biomarkers to detect disease progression, to identify patients who will respond to targeted therapies and to act as targets for future therapies that may halt the progression of this prevalent disease.
A summary of known interactions/associations of various lncRNAs (and miRNAs) discussed in the text that are linked to the Androgen Receptor.
A summary of lncRNAs which have been linked to EMT in prostate cancer, some of which have been directly linked to the AR, and which have been discussed in more detail in the text.
References
- 1.Siegel R.L., Miller K.D., Jemal A., Statistics Cancer. CA cancer. J. Clim. 2017;67(1):7–30. doi: 10.3322/caac.21387. 2017. [DOI] [PubMed] [Google Scholar]
- 2.Chen C.D., Welsbie D.S., Tran C., Baek S.H., Chen R., Vessella R. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 2004;10(1):33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 3.Waltering K.K., Helenius M.A., Sahu B., Manni V., Linja M.J., Janne O.A. Increased expression of androgen receptor sensitizes prostate cancer cells to low levels of androgens. Cancer Res. 2009;69(20):8141–8149. doi: 10.1158/0008-5472.CAN-09-0919. [DOI] [PubMed] [Google Scholar]
- 4.Antonarakis E.S., Lu C., Wang H., Luber B., Nakazawa M., Roeser J.C. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014;371(11):1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kung J.T., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma L., Li A., Zou D., Xu X., Xia L., Yu J. LncRNAWiki: harnessing community knowledge in collaborative curation of human long non-coding RNAs. Nucleic Acids Res. 2015;43(Database issue):D187–D192. doi: 10.1093/nar/gku1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J.T. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 9.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Gibb E.A., Brown C.J., Lam W.L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prensner J.R., Chinnaiyan A.M. The emergence of lncRNAs in cancer biology. Canc. Discov. 2011;1(5):391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutschner T., Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh A.L., Tuzova A.V., Bolton E.M., Lynch T.H., Perry A.S. Long noncoding RNAs and prostate carcinogenesis: the missing 'linc'? Trends Mol. Med. 2014;20(8):428–436. doi: 10.1016/j.molmed.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Zheng J., Zhao S., He X., Zheng Z., Bai W., Duan Y. The up-regulation of long non-coding RNA CCAT2 indicates a poor prognosis for prostate cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2016;480(4):508–514. doi: 10.1016/j.bbrc.2016.08.120. [DOI] [PubMed] [Google Scholar]
- 15.Takayama K., Horie-Inoue K., Katayama S., Suzuki T., Tsutsumi S., Ikeda K. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013;32(12):1665–1680. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia J., Li F., Tang X.S., Xu S., Gao Y., Shi Q. Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget. 2016;7(25):37868–37881. doi: 10.18632/oncotarget.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X., Du W.W., Li H., Liu F., Khorshidi A., Rutnam Z.J. Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res. 2013;41(21):9688–9704. doi: 10.1093/nar/gkt680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakurai K., Reon B.J., Anaya J., Dutta A. The lncRNA DRAIC/PCAT29 locus constitutes a tumor-suppressive nexus. Mol. Canc. Res. 2015;13(5):828–838. doi: 10.1158/1541-7786.MCR-15-0016-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao R., Sun F., Bei X., Wang X., Zhu Y., Jiang C. Upregulation of the long non-coding RNA FALEC promotes proliferation and migration of prostate cancer cell lines and predicts prognosis of PCa patients. Prostate. 2017;77(10):1107–1117. doi: 10.1002/pros.23367. [DOI] [PubMed] [Google Scholar]
- 20.Schneider C., King R.M., Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54(6):787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 21.Pickard M.R., Williams G.T. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: implications for chemotherapy. Breast Canc. Res. Treat. 2014;145(2):359–370. doi: 10.1007/s10549-014-2974-y. [DOI] [PubMed] [Google Scholar]
- 22.Kino T., Hurt D.E., Ichijo T., Nader N., Chrousos G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3(107):ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pickard M.R., Mourtada-Maarabouni M., Williams G.T. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim. Biophys. Acta. 2013;1832(10):1613–1623. doi: 10.1016/j.bbadis.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Yacqub-Usman K., Pickard M.R., Williams G.T. Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. Prostate. 2015;75(7):693–705. doi: 10.1002/pros.22952. [DOI] [PubMed] [Google Scholar]
- 25.Xue D., Zhou C., Lu H., Xu R., Xu X., He X. LncRNA GAS5 inhibits proliferation and progression of prostate cancer by targeting miR-103 through AKT/mTOR signaling pathway. Tumor Biol. 2016;37(12):16187–16197. doi: 10.1007/s13277-016-5429-8. [DOI] [PubMed] [Google Scholar]
- 26.Luo G., Liu D., Huang C., Wang M., Xiao X., Zeng F. LncRNA GAS5 inhibits cellular proliferation by targeting P27Kip1. Mol. Canc. Res. 2017;15(7):789–799. doi: 10.1158/1541-7786.MCR-16-0331. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y., Zhang P., Wan X., Su X., Kong Z., Zhai Q. Downregulation of long non-coding RNA HCG11 predicts a poor prognosis in prostate cancer. Biomed. Pharmacother. 2016;83:936–941. doi: 10.1016/j.biopha.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 28.Zhang A., Zhao J.C., Kim J., Fong K.W., Yang Y.A., Chakravarti D. LncRNA HOTAIR enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate cancer. Cell Rep. 2015;13(1):209–221. doi: 10.1016/j.celrep.2015.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L., Dang Q., Xie H., Yang Z., He D., Liang L. Infiltrating mast cells enhance prostate cancer invasion via altering LncRNA-HOTAIR/PRC2-androgen receptor (AR)-MMP9 signals and increased stem/progenitor cell population. Oncotarget. 2015;6(16):14179–14190. doi: 10.18632/oncotarget.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiyomaru T., Yamamura S., Fukuhara S., Yoshino H., Kinoshita T., Majid S. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PLoS One. 2013;8(8):e70372. doi: 10.1371/journal.pone.0070372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J., Cheng G., Zhang C., Zheng Y., Xu H., Yang H. Long noncoding RNA LINC01296 is associated with poor prognosis in prostate cancer and promotes cancer-cell proliferation and metastasis. OncoTargets Ther. 2017;10:1843–1852. doi: 10.2147/OTT.S129928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Xu Y., Wang X., Jiang C., Han S., Dong K. LincRNA-p21 suppresses development of human prostate cancer through inhibition of PKM2. Cell Prolif. 2017;50(6) doi: 10.1111/cpr.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu S., Yi X.M., Tang C.P., Ge J.P., Zhang Z.Y., Zhou W.Q. Long non-coding RNA ATB promotes growth and epithelial-mesenchymal transition and predicts poor prognosis in human prostate carcinoma. Oncol. Rep. 2016;36(1):10–22. doi: 10.3892/or.2016.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L., Lin Y., Meng H., Liu C., Xue J., Zhang Q. Long non-coding RNA LOC283070 mediates the transition of LNCaP cells into androgen-independent cells possibly via CAMK1D. Am. J. Transl. Res. 2016;8(12):5219–5234. [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J., Cheng G., Li X., Pan Y., Qin C., Yang H. Overexpression of long non-coding RNA LOC400891 promotes tumor progression and poor prognosis in prostate cancer. Tumor Biol. 2016;37(7):9603–9613. doi: 10.1007/s13277-016-4847-y. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C., Liu C., Wu J., Zheng Y., Xu H., Cheng G. Upregulation of long noncoding RNA LOC440040 promotes tumor progression and predicts poor prognosis in patients with prostate cancer. OncoTargets Ther. 2017;10:4945–4954. doi: 10.2147/OTT.S138354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren S., Liu Y., Xu W., Sun Y., Lu J., Wang F. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J. Urol. 2013;190(6):2278–2287. doi: 10.1016/j.juro.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Wang D., Ding L., Wang L., Zhao Y., Sun Z., Karnes R.J. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget. 2015;6(38):41045–41055. doi: 10.18632/oncotarget.5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang R., Sun Y., Li L., Niu Y., Lin W., Lin C. Preclinical study using Malat1 small interfering RNA or androgen receptor splicing variant 7 degradation enhancer ASC-J9(R) to suppress enzalutamide-resistant prostate cancer progression. Eur. Urol. 2017;72(5):835–844. doi: 10.1016/j.eururo.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sebastian A., Hum N.R., Hudson B.D., Loots G.G. Cancer-osteoblast interaction reduces Sost expression in osteoblasts and up-regulates lncRNA MALAT1 in prostate cancer. Microarrays (Basel) 2015;4(4):503–519. doi: 10.3390/microarrays4040503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo G., Wang M., Wu X., Tao D., Xiao X., Wang L. Long non-coding RNA MEG3 inhibits cell proliferation and induces apoptosis in prostate cancer. Cell. Physiol. Biochem. 2015;37(6):2209–2220. doi: 10.1159/000438577. [DOI] [PubMed] [Google Scholar]
- 42.Zhou Y., Zhong Y., Wang Y., Zhang X., Batista D.L., Gejman R. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007;282(34):24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 43.Chakravarty D., Sboner A., Nair S.S., Giannopoulou E., Li R., Hennig S. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schalken J.A., Hessels D., Verhaegh G. New targets for therapy in prostate cancer: differential display code 3 (DD3(PCA3)), a highly prostate cancer-specific gene. Urology. 2003;62(5 Suppl 1):34–43. doi: 10.1016/s0090-4295(03)00759-3. [DOI] [PubMed] [Google Scholar]
- 45.Lemos A.E., Ferreira L.B., Batoreu N.M., de Freitas P.P., Bonamino M.H., Gimba E.R. PCA3 long noncoding RNA modulates the expression of key cancer-related genes in LNCaP prostate cancer cells. Tumor Biol. 2016;37(8):11339–11348. doi: 10.1007/s13277-016-5012-3. [DOI] [PubMed] [Google Scholar]
- 46.Salameh A., Lee A.K., Cardo-Vila M., Nunes D.N., Efstathiou E., Staquicini F.I. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc. Natl. Acad. Sci. U. S. A. 2015;112(27):8403–8408. doi: 10.1073/pnas.1507882112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He J.H., Li B.X., Han Z.P., Zou M.X., Wang L., Lv Y.B. Snail-activated long non-coding RNA PCA3 up-regulates PRKD3 expression by miR-1261 sponging, thereby promotes invasion and migration of prostate cancer cells. Tumor Biol. 2016;37(12):16163–16176. doi: 10.1007/s13277-016-5450-y. [DOI] [PubMed] [Google Scholar]
- 48.Ozgur E., Celik A.I., Darendeliler E., Gezer U. PCA3 silencing sensitizes prostate cancer cells to enzalutamide-mediated androgen receptor blockade. Anticancer Res. 2017;37(7):3631–3637. doi: 10.21873/anticanres.11733. [DOI] [PubMed] [Google Scholar]
- 49.Roy R., Chun J., Powell S.N. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Canc. 2011;12(1):68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prensner J.R., Chen W., Iyer M.K., Cao Q., Ma T., Han S. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74(6):1651–1660. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prensner J.R., Chen W., Han S., Iyer M.K., Cao Q., Kothari V. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16(11):900–908. doi: 10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu W., Chang J., Du X., Hou J. Long non-coding RNA PCAT-1 contributes to tumorigenesis by regulating FSCN1 via miR-145-5p in prostate cancer. Biomed. Pharmacother. 2017;95:1112–1118. doi: 10.1016/j.biopha.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 53.Ylipaa A., Kivinummi K., Kohvakka A., Annala M., Latonen L., Scaravilli M. Transcriptome sequencing reveals PCAT5 as a novel ERG-regulated long noncoding RNA in prostate cancer. Cancer Res. 2015;75(19):4026–4031. doi: 10.1158/0008-5472.CAN-15-0217. [DOI] [PubMed] [Google Scholar]
- 54.Shukla S., Zhang X., Niknafs Y.S., Xiao L., Mehra R., Cieslik M. Identification and validation of PCAT14 as prognostic biomarker in prostate cancer. Neoplasia. 2016;18(8):489–499. doi: 10.1016/j.neo.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White N.M., Zhao S.G., Zhang J., Rozycki E.B., Dang H.X., McFadden S.D. Multi-institutional analysis shows that low PCAT-14 expression associates with poor outcomes in prostate cancer. Eur. Urol. 2017;71(2):257–266. doi: 10.1016/j.eururo.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 56.Crea F., Watahiki A., Quagliata L., Xue H., Pikor L., Parolia A. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5(3):764–774. doi: 10.18632/oncotarget.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malik R., Patel L., Prensner J.R., Shi Y., Iyer M.K., Subramaniyan S. The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Mol. Canc. Res. 2014;12(8):1081–1087. doi: 10.1158/1541-7786.MCR-14-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Al Aameri R.F.H., Sheth S., Alanisi E.M.A., Borse V., Mukherjea D., Rybak L.P. Tonic suppression of PCAT29 by the IL-6 signaling pathway in prostate cancer: reversal by resveratrol. PLoS One. 2017;12(5):e0177198. doi: 10.1371/journal.pone.0177198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lou W., Ni Z., Dyer K., Tweardy D.J., Gao A.C. Interleukin-6 induces prostate cancer cell growth accompanied by activation of stat3 signaling pathway. Prostate. 2000;42(3):239–242. doi: 10.1002/(sici)1097-0045(20000215)42:3<239::aid-pros10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 60.Yang L., Lin C., Jin C., Yang J.C., Tanasa B., Li W. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500(7464):598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prensner J.R., Sahu A., Iyer M.K., Malik R., Chandler B., Asangani I.A. The IncRNAs PCGEM1 and PRNCR1 are not implicated in castration resistant prostate cancer. Oncotarget. 2014;5(6):1434–1438. doi: 10.18632/oncotarget.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parolia A., Crea F., Xue H., Wang Y., Mo F., Ramnarine V.R. The long non-coding RNA PCGEM1 is regulated by androgen receptor activity in vivo. Mol. Cancer. 2015;14:46. doi: 10.1186/s12943-015-0314-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Z., Zhou N., Huang J., Ho T.T., Zhu Z., Qiu Z. Regulation of androgen receptor splice variant AR3 by PCGEM1. Oncotarget. 2016;7(13):15481–15491. doi: 10.18632/oncotarget.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho T.T., Huang J., Zhou N., Zhang Z., Koirala P., Zhou X. Regulation of PCGEM1 by p54/nrb in prostate cancer. Sci. Rep. 2016;6:34529. doi: 10.1038/srep34529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hung C.L., Wang L.Y., Yu Y.L., Chen H.W., Srivastava S., Petrovics G. A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl. Acad. Sci. U. S. A. 2014;111(52):18697–18702. doi: 10.1073/pnas.1415669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He J.H., Zhang J.Z., Han Z.P., Wang L., Lv Y.B., Li Y.G. Reciprocal regulation of PCGEM1 and miR-145 promote proliferation of LNCaP prostate cancer cells. J. Exp. Clin. Canc. Res. 2014;33:72. doi: 10.1186/s13046-014-0072-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cui Z., Ren S., Lu J., Wang F., Xu W., Sun Y. The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor. Urol. Oncol. 2013;31(7):1117–1123. doi: 10.1016/j.urolonc.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 68.Fang Z., Xu C., Li Y., Cai X., Ren S., Liu H. A feed-forward regulatory loop between androgen receptor and PlncRNA-1 promotes prostate cancer progression. Canc. Lett. 2016;374(1):62–74. doi: 10.1016/j.canlet.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 69.Jin Y., Cui Z., Li X., Jin X., Peng J. Upregulation of long non-coding RNA PlncRNA-1 promotes proliferation and induces epithelial-mesenchymal transition in prostate cancer. Oncotarget. 2017;8(16):26090–26099. doi: 10.18632/oncotarget.15318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Q., Cui Z.L., Wang Q., Jin X.B., Zhao Y., Wang M.W. PlncRNA-1 induces apoptosis through the Her-2 pathway in prostate cancer cells. Asian J. Androl. 2017;19(4):453–457. doi: 10.4103/1008-682X.178849. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Misawa A., Takayama K.I., Fujimura T., Homma Y., Suzuki Y., Inoue S. Androgen-induced lncRNA POTEF-AS1 regulates apoptosis-related pathway to facilitate cell survival in prostate cancer cells. Canc. Sci. 2017;108(3):373–379. doi: 10.1111/cas.13151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chung S., Nakagawa H., Uemura M., Piao L., Ashikawa K., Hosono N. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Canc. Sci. 2011;102(1):245–252. doi: 10.1111/j.1349-7006.2010.01737.x. [DOI] [PubMed] [Google Scholar]
- 73.Meyer K.B., Maia A.T., O'Reilly M., Ghoussaini M., Prathalingam R., Porter-Gill P. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS Genet. 2011;7(7):e1002165. doi: 10.1371/journal.pgen.1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang J., Li C., Mudd A., Gu X. LncRNA PVT1 predicts prognosis and regulates tumor growth in prostate cancer. Biosci. Biotechnol. Biochem. 2017:1–6. doi: 10.1080/09168451.2017.1387048. [DOI] [PubMed] [Google Scholar]
- 75.Liu H.T., Fang L., Cheng Y.X., Sun Q. LncRNA PVT1 regulates prostate cancer cell growth by inducing the methylation of miR-146a. Canc. Med. 2016;5(12):3512–3519. doi: 10.1002/cam4.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mehra R., Shi Y., Udager A.M., Prensner J.R., Sahu A., Iyer M.K. A novel RNA in situ hybridization assay for the long noncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia. 2014;16(12):1121–1127. doi: 10.1016/j.neo.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mehra R., Udager A.M., Ahearn T.U., Cao X., Feng F.Y., Loda M. Overexpression of the long non-coding RNA SChLAP1 independently predicts lethal prostate cancer. Eur. Urol. 2016;70(4):549–552. doi: 10.1016/j.eururo.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prensner J.R., Iyer M.K., Sahu A., Asangani I.A., Cao Q., Patel L. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 2013;45(11):1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prensner J.R., Zhao S., Erho N., Schipper M., Iyer M.K., Dhanasekaran S.M. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014;15(13):1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee R.S., Roberts C.W. Linking the SWI/SNF complex to prostate cancer. Nat. Genet. 2013;45(11):1268–1269. doi: 10.1038/ng.2805. [DOI] [PubMed] [Google Scholar]
- 81.Li Y., Luo H., Xiao N., Duan J., Wang Z., Wang S. Long noncoding RNA SChLAP1 accelerates the proliferation and metastasis of prostate cancer via targeting miR-198 and promoting the MAPK1 pathway. Oncol. Res. 2018;26(1):131–143. doi: 10.3727/096504017X14944585873631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li J., Zhang Z., Xiong L., Guo C., Jiang T., Zeng L. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem. Biophys. Res. Commun. 2017;487(1):146–152. doi: 10.1016/j.bbrc.2017.03.169. [DOI] [PubMed] [Google Scholar]
- 83.Misawa A., Takayama K., Urano T., Inoue S. Androgen-induced long noncoding RNA (lncRNA) SOCS2-AS1 promotes cell growth and inhibits apoptosis in prostate cancer cells. J. Biol. Chem. 2016;291(34):17861–17880. doi: 10.1074/jbc.M116.718536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Orfanelli U., Jachetti E., Chiacchiera F., Grioni M., Brambilla P., Briganti A. Antisense transcription at the TRPM2 locus as a novel prognostic marker and therapeutic target in prostate cancer. Oncogene. 2015;34(16):2094–2102. doi: 10.1038/onc.2014.144. [DOI] [PubMed] [Google Scholar]
- 85.Lavorgna G., Chiacchiera F., Briganti A., Montorsi F., Pasini D., Salonia A. Expression-profiling of apoptosis induced by ablation of the long ncRNA TRPM2-AS in prostate cancer cell. Genom. Data. 2015;3:4–5. doi: 10.1016/j.gdata.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Na X.Y., Liu Z.Y., Ren P.P., Yu R., Shang X.S. Long non-coding RNA UCA1 contributes to the progression of prostate cancer and regulates proliferation through KLF4-KRT6/13 signaling pathway. Int. J. Clin. Exp. Med. 2015;8(8):12609–12616. [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang S., Dong X., Ji T., Chen G., Shan L. Long non-coding RNA UCA1 promotes cell progression by acting as a competing endogenous RNA of ATF2 in prostate cancer. Am. J. Transl. Res. 2017;9(2):366–375. [PMC free article] [PubMed] [Google Scholar]
- 88.Wang H., Guan Z., He K., Qian J., Cao J., Teng L. LncRNA UCA1 in anti-cancer drug resistance. Oncotarget. 2017;8(38):64638–64650. doi: 10.18632/oncotarget.18344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee H.Y., Ahn J.B., Rha S.Y., Chung H.C., Park K.H., Kim T.S. High KLF4 level in normal tissue predicts poor survival in colorectal cancer patients. World J. Surg. Oncol. 2014;12:232. doi: 10.1186/1477-7819-12-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang X., Yang B., Ma B. The UCA1/miR-204/Sirt1 axis modulates docetaxel sensitivity of prostate cancer cells. Canc. Chemother. Pharmacol. 2016;78(5):1025–1031. doi: 10.1007/s00280-016-3158-8. [DOI] [PubMed] [Google Scholar]
- 91.Su W., Xu M., Chen X., Chen N., Gong J., Nie L. Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol. Cancer. 2017;16(1):142. doi: 10.1186/s12943-017-0711-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lukacs R.U., Memarzadeh S., Wu H., Witte O.N. Bmi-1 is a crucial regulator of prostate stem cell self-renewal and malignant transformation. Cell Stem Cell. 2010;7(6):682–693. doi: 10.1016/j.stem.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van Leenders G.J., Dukers D., Hessels D., van den Kieboom S.W., Hulsbergen C.A., Witjes J.A. Polycomb-group oncogenes EZH2, BMI1, and RING1 are overexpressed in prostate cancer with adverse pathologic and clinical features. Eur. Urol. 2007;52(2):455–463. doi: 10.1016/j.eururo.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 94.Sharma N.L., Massie C.E., Ramos-Montoya A., Zecchini V., Scott H.E., Lamb A.D. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Canc. Cell. 2013;23(1):35–47. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 95.Savagner P. Leaving the neighborhood: molecular mechanisms involved during epithelial-mesenchymal transition. Bioessays. 2001;23(10):912–923. doi: 10.1002/bies.1132. [DOI] [PubMed] [Google Scholar]
- 96.Voulgari A., Pintzas A. Epithelial-mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim. Biophys. Acta. 2009;1796(2):75–90. doi: 10.1016/j.bbcan.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 97.Dave N., Guaita-Esteruelas S., Gutarra S., Frias A., Beltran M., Peiro S. Functional cooperation between Snail1 and twist in the regulation of ZEB1 expression during epithelial to mesenchymal transition. J. Biol. Chem. 2011;286(14):12024–12032. doi: 10.1074/jbc.M110.168625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wels C., Joshi S., Koefinger P., Bergler H., Schaider H. Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial-mesenchymal transition-like phenotype in melanoma. J. Invest. Dermatol. 2011;131(9):1877–1885. doi: 10.1038/jid.2011.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Balk S.P., Knudsen K.E. AR, the cell cycle, and prostate cancer. Nucl. Recept. Signal. 2008;6:e001. doi: 10.1621/nrs.06001. [DOI] [PMC free article] [PubMed] [Google Scholar]