Abstract
Long non-coding RNAs (lncRNAs) have been increasingly studied during the past decade. This led to an immense number of annotated transcripts, out of which many were linked to a diverse range of biological mechanisms and diseases. Due to the variety of their regulatory potential, they are seen as an important link in understanding complex epigenetic mechanisms. Prominent examples of lncRNAs in the cardiovascular system are ANRIL, Braveheart, MALAT1 and HOTAIR which have been excessively studied. But despite the impressive number of described transcripts, only a few examples are characterized functionally. One way to do this is to identify accessible structural domains in the RNA secondary structure which have the ability to bind to DNA, RNA or proteins. Through recent improvements in computational as well as experimental methods, this exploration of secondary structure became not only more efficient than traditional methods like crystallization, but also feasible to investigate whole genome RNA structures. The purpose of this review is to highlight the recent advances in secondary structure probing methods and how these can be applied in order to investigate the functional roles of lncRNAs in the cardiovascular system.
Keywords: Long non-coding RNA, Cardiovascular system, Secondary structure
1. Introduction
For a long time, protein coding RNAs and their products were the focus of genetic research. Then slowly a group of small non-coding RNAs, including miRNAs, siRNAs, piRNAs, snRNAs and snoRNAs became a new trend in describing functional mechanisms in gene regulation. However, not until the discovery that the majority of genetic information is transcribed into a new group of lncRNAs [1], the functional potential of non-coding regions has been fully recognized and hence has become more and more predominant in research. LncRNAs are a special group of non-coding RNA with a length of over 200 nucleotides which can function through a variety of binding partners, including RNA binding proteins (RBPs), DNA and other RNAs. The reason they have been overlooked for so long is that their expression level is usually lower compared to protein coding genes and are highly celltype-specific [2]. Nonetheless it has already been shown that different groups of lncRNAs have a significant impact on major biological processes including metabolism [3], differentiation [4] and development [5]. Also there is a steadily growing number of lncRNA transcripts linked to the cardiovascular system [6]. This can be of great interest because a significant amount of genetic variation identified in cardiovascular diseases through genome-wide association studies is found in non-coding regions [7]. The greatest challenge in lncRNA research is that although the number of described lncRNAs is rapidly increasing, the molecular mechanisms are still largely unknown. Since lncRNAs lack the ability to code for a protein, they need to affect their targets through their structure. Therefore, the focus recently shifted to investigating their structure for functional hints. The structure of RNA molecules can be categorized in different depths. The primary structure is simply the sequence of nucleotides. Secondary structure predicts the structure based solely on base pairing due to hydrogen bonds within the lncRNA without taking the three-dimensional structure into account which would be the tertiary structure. These structures then can give hints on the formation of secondary or tertiary complexes with proteins or other genomic sequences and therefore affects gene regulation. For a long time, unreliable in silico predictions or difficult and laborious crystallization experiments were the only methods for investigating the secondary structure, but recent advances in structure probing led to a few very well described examples of secondary structure so far. Especially the combination of experimentally and computationally predicted structures seemed to produce the most reliable results and will help to understand the impact of these molecules in diverse complex cardiovascular diseases. This review will give an overview of the different developed methods as well as pointing out how they have improved the understanding of lncRNA function in the cardiovascular context so far.
2. Experimental RNA structure probing
During the last decade many different probing techniques evolved to explore the accurate secondary structures of RNA molecules. They can be categorized into two different groups, namely the enzymatic and the chemical probing techniques which are both combined with high-throughput sequencing and followed by bioinformatic analysis. The enzymatic approach involves nucleases that are able to bind to either paired or unpaired bases in a RNA structure and digest the according region. Consequently, the remaining RNA fragments can be sequenced and analyzed. The secondary structure is then derived from the characteristic pattern of the sequenced fragments. Chemical probing methods are based on several different chemical reagents that bind to unpaired nucleotides. This leads to interruption during the reverse transcription (RT) process, and the corresponding structure information can be retrieved after sequencing of the shortened reads. Previously, it was only possible to probe a single RNA structure at a time, but due to the recent combination of probing techniques with next generation sequencing, it is now feasible to investigate multiple targets or even a whole transcriptome in vivo or in vitro at once. Although all those approaches have the same goal in common, it is important to choose the correct approach to address each problem, due to certain variations in their protocol and experimental limitations (see Table 1). In the following section, the most prominent methods for structure probing will be discussed.
Table 1.
Summary of the most broadly applied secondary structure probing methods. Different techniques may fit best for different applications.
| Technique | Type | Pro | Contra | Reagent |
|---|---|---|---|---|
| PARS [10] | enzymatic | digests paired/unpaired bases; suited for riboswitches; transcriptome | poly(A) selected; over-digestion of RNA; in vitro | ssRNase S1 dsRNase V1 |
| Fraq-Seq [13] | enzymatic | P1 is thermal stable; transcriptome | control needed; for smaller RNA molecules; poly(A) selection; in vitro | ssRNAase P1 |
| DMS-Seq [14,16] | chemical | in vitro/in vivo; DMS cell permeable; transcriptome in vivo | poly(A) selection; needs control sample; RBPs influence; in vivo | DMS |
| SHAPE [18] | chemical | single nucleotide resolution; multiple RNA profiling | in vitro; needs control sample | IM7, NMIA, NAI-N3, FAI |
| IcSHAPE [22] | chemical | in vivo; single nucleotide resolution; transcriptome | influenced by RBP binding | IM7, NMIA, NAI-N3, FAI |
2.1. Enzymatic probing techniques
Enzymatic probing of RNA structures already dates back several decades [8,9], but only coupling them with high-throughput sequencing makes them widely applicable and superior to nuclear magnetic resonance (NMR) spectroscopy and crystallization methods. Especially since lncRNAs are in general too long and unstable for those methods. Recent efforts showed promising results and improvements in exploring structures from single RNAs and even transcriptomes.
2.1.1. PARS
Parallel analysis of RNA structure (PARS) [10] involves two enzymes which are structure-specific ribonucleases. One sample is treated with the RNase V1 which targets double stranded and another is treated with the RNase S1 which targets single stranded regions of ribonuleic acids. After sequencing both samples, the so-called PARS score is calculated for each RNA on the nucleotide level. It is described as the -ratio of fragments found in both samples. Therefore the PARS score gives an impression of the degree of structured RNA along the bases. With this method it is feasible to determine the secondary structure of many different RNAs at the same time. A previous study used PARS to characterize the transcriptome-wide coding and non-coding RNA secondary structures of a family trio [11]. Their findings suggested that around 15% of all single nucleotide variations observed can alter the secondary structure. Since this method targets both paired and unpaired bases, a high score in both enzyme treated regions might hint at a RNA structure with multiple possible states. These RNAs are called Riboswitches and they are an important regulatory mechanism in gene regulation by altering their structure when binding to small molecules [12].
2.1.2. Frag-Seq
Just like PARS, Fragmentation Sequencing (Frag-Seq) [13] specializes in determining the secondary structure of many RNAs simultaneously. It creates fragments by cleaving predominantly single-stranded nucleic acids with the nuclease P1. Subsequently, sequencing the remaining reads can give information about the cutting scores, the -ratio of expression between treated and control samples. This method was used to probe the whole mouse nuclear transcriptome [13] and correlated well with few available known structures. This method works best on stem-loop structures, but performs less well on small interior loops which it often fails to capture [13]. Therefore this method is less favorable if a high resolution of the structure is desired. Instead of reflecting details, it is best suited for a global view of the structure. Also the enzyme P1 is relatively spacious compared to other RNases which could lead to a falsely low cutting score. This means that this method is not equipped for very long RNA structures with multiple or diverse long-range tertiary interactions, where the enzyme is unable to access the bases.
2.2. Chemical probing techniques
Chemical probing techniques are a relatively recent invention to probe the secondary structure of RNA molecules. Their main advantage is that they can achieve better resolution due to the reagent's smaller size that can cover more nucleotides. Also some reagents can penetrate cells and probe RNAs in vivo in realistic conditions.
2.2.1. DMS
Dimethyl sulfate-Seq (DMS-Seq) [14] was one of the first chemical probing techniques. By adding the DMS reagent to the RNA of interest, it alters the methylation of unpaired adenine and cytosine bases. Those changes subsequently lead to the interruption of replication during RT. Sequencing these shortened fragments and comparing them to the untreated control allows for the calculation of pairing probabilities per base. Unlike previous methods, this application can be realized in vitro as well as in vivo due to the ability of DMS to rapidly penetrate the cell and recent studies showed that there can be a significant difference in the structure in both states [15]. Rouskin et al. treated yeast and mammalian cells with DMS in vivo, followed by sequencing [16]. The resulting structures largely agreed with known structures, but seemed to be less structured and more dynamic in vivo than in vitro. Another genome-wide study on arabidopsis thaliana found that in general in vitro structures were more similar to in silico predicted structures, than the in vivo determined ones [17]. This highlights the fact, that not only the sequence plays a role in forming secondary structure, but also the environment and interaction partners like RNA binding proteins (RBPs) should be taken into consideration. This would be an important aspect to consider when planning in vivo experiments.
2.2.2. SHAPE-Seq
Selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) [18] is a novel form of RNA secondary structure probing. It is just like DMS-Seq dependent on a reagent that can bind to unbound bases of the RNA molecule. The main steps are to modify the folded RNA with chemical probes like 1-methyl-7-nitroisatoic anhydride (1M7) and run RT. This will be interrupted where the probe was previously bound and produce according fragments which can be sequenced and analyzed [18]. Compared to a non-modified control, SHAPE-seq is then able to assess single RNA secondary structures by assigning an agent reaction probability to each nucleotide. The significant advantage compared to DMS techniques is the ability to bind to any of the four bases, leading to a higher resolution of the secondary structure. This can be extremely beneficial in point mutation studies like investigating RiboSNitches [19]. RiboSNitches are genetic variants which have the ability to significantly alter the RNA structure and therefore also affect the RNA's functionality as a regulator. Additionally, SHAPE-Seq does not filter for poly(A) RNA. This feature is most indispensable for investigating non-coding RNAs, since many of them do not have a poly(A) tail. It was shown in a study [18] that the average precision of pure in silico folding algorithms raises from 61.4% up to 88.9% by taking SHAPE-Seq data into account. Another study focused on the structure of Escherichia coli 16S rRNA and was able to reproduce the known structure by incorporating SHAPE-Seq data into the RNAstructure [20] prediction algorithm with an accuracy of 97% [21]. There is also an in vivo version of SHAPE-Seq, the so-called icSHAPE-Seq [22] which compares in vivo to in vitro folded RNAs. Changes in their reactivities could identify new protein-binding motifs, since the binding makes the RNA inaccessible to the modification reagent. An important part to consider when performing this probing technique is to choose the right one among several chemical probes during preparation. After establishing the 1M7 reagent, several other substitutes such as NMIA, NAI-N3 and FAI followed. A study [23] found that depending of the experiment setup, the results might differ significantly depending on the probe. It was suggested that for example the 1M7 reagent might have high temporal specificity, but performs worse for in vivo compared to other reagents.
2.2.3. Shotgun secondary structure
In general, lncRNAs can be quite long and for each base only the general pairing probability can be assessed. This leads to the problem that using conventional methods there is no option to differentiate between close and long-range base pairing. The longer the RNA is, the more possibilities exist and predictions can become less reliable. Therefore another addition to these methods is the shotgun secondary structure (3S) technique [24] which tries to tackle this issue. The whole intact RNA structure profile is compared to overlapping fragments of the same RNA which preferably are shorter than 100 nucleotides. If the profile is similar, then this region can be assumed to be modular, i.e. independent from influence of far distance regions. The structure possibilities then can be reduced and the predicted structure becomes far more reliable. It is important to keep in mind that each method is not powerful enough yet to predict structure fully on its own, but is able to improve in silico predictions immensely by serving as additional constraints. Therefore the choice of computational algorithms are still vital to the overall secondary structure reconstruction process.
3. Computational structure prediction
Since it was not feasible to experimentally determine the secondary structure of a large amount of long sequences, the in silico prediction was for a long time the only realistic source for studying RNA structure. Common approaches for investigating the secondary structure are comparative methods. They compare evolutionarily conserved regions and identify structural elements. But they require a relatively large input of conserved regions in different species which is a problem for lncRNAs, that are predominantly poorly conserved [25]. Therefore, the only alternative for computational prediction is using thermodynamics. A widely applied approach in this case is RNAfold [26]. This includes an implementation of the Zuker algorithm [27] which calculates the minimal free energy (MFE) of a structure to any given RNA sequence. Every RNA molecule tries to adjust its structure to their surrounding conditions so it has a stable structure. This can be achieved by reducing the molecule's Gibbs free energy (G) in a structure. When G is maximally reduced, the structure of the molecule found its stable equilibrium state [28] and is called the MFE structure. The drawback of this method is that a single RNA can exist in different structures, where the MFE structure might not even be the most functionally important one [29]. With growing length of the RNA molecule, the complexity of the prediction increases as well which can lead to incorrect predictions. Instead of calculating the mere MFE structure of a given sequence, another approach is sFold [30]. This technique is considering the whole Boltzmann ensemble of secondary structures from which it statistically samples suboptimal structures. Following, the ensemble centroid structure, which is the center of all sampled structures, is chosen as a representative. Compared to several known structures, it was found that the ensemble centroid structure could improve prediction accuracy significantly [31].
3.1. Combining computational and probing methods
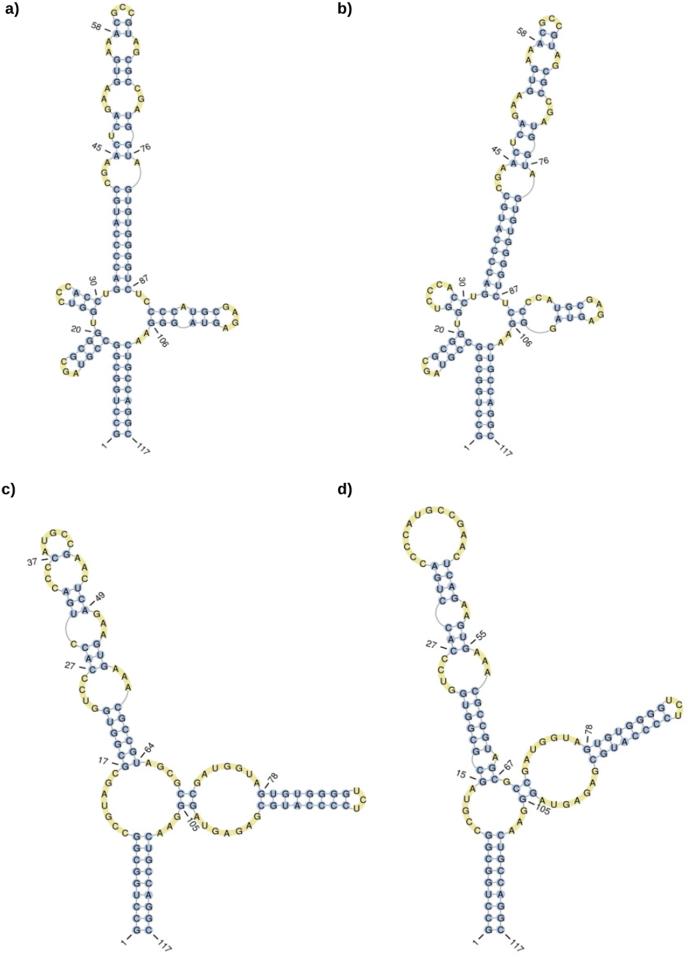
Despite recent efforts to improve prediction methods, it was clear that further information was needed than just the sequence in order to improve structure prediction. Comparative experiments showed also that structure probing methods alone might often not generate enough evidence to conclude on the correct structure [32]. Based on a handful of known RNA structures, the sensitivity and positive prediction value of the prediction algorithm improved from 61.9% and 55.3% to 83.3% and 79.5% respectively when used together with probing data. Therefore a combined method is highly preferred which can additionally deal with noise introduced data. In Fig. 1, an exemplary comparison between the computational, chemically probed and crystallized structures can be seen and the combination with SHAPE-Seq is the closest to the crystallization structure which is assumed to be realistic. A popular method, which can combine computational prediction with structural probing data, is RNAstructure [20]. It can combine different methods like ensemble centroid and MFE structure prediction with different experimental data from e.g. SHAPE-seq or NMR experiments and use them as constraints during prediction in order to improve accuracy [20]. Seqfold [33] is another method to incorporate experimental data into classical computational-based prediction methods by looking at a group of possible formations. It samples structures from the Boltzmann-weighted ensemble and clusters them. Simultaneously it assigns, based on experimental probing data, for each base its structural preference. The final predicted structure is then the centroid in that cluster which shows the most consensus with the structure preference profile. During comparative studies, the authors were able to show that the combination improved the prediction significantly and outperformed the other MFE methods [34]. The SeqFold method can be applied with standard structure probing data from SHAPE, PARS and Frag-Seq experiments. Recognizing the recent advances in structural probing techniques for investigating lncRNAs functionally was an important step and already lead to some very promising results. While lncRNAs are an emerging and essential part in regulatory mechanisms of various fields, their functionality is still widely unknown due to a lack of protein products. In the following section, some examples of successfully probed lncRNA structures in the cardiovascular system will be introduced. Although the examples are still very scarce, they highlight the positive prospect of this approach.
Fig. 1.
a) MFE based structure prediction by RNAfold [26] b) Centroid based structure prediction by RNAfold [26] c) Crystallization structure (PDB ID: 4V7T) d) SHAPE-Seq based predicted structure (RMDB ID: 5SRRNA_SHP_0001) of the E. coli 5S rRNA. Visualization with PseudoViewer [35].
4. LncRNAs in the cardiovascular system
4.1. Braveheart
Braveheart was characterized by Klattenhoff et al. [36] while identifying embryonic stem cell (ESC) differentiation-specific genes in mice. They were able to show by RIP experiments that Braveheart interacts with the SUZ12 component of the PRC2 complex. This complex can suppress expression of its target genes by altering the epigenetic chromatin modifications, but needs to be guided to its destination first. LncRNAs were previously shown to also function as mediators for chromatin remodelling complexes, such as Myheart [37], Chaer [38], MANTIS [39], Carmen [40], Fendrr [41] and ANRIL [42]. Further, it was shown that loss of Braveheart affects the expression of its target genes significantly, which are known to be important for cardiovascular lineage commitment like MesP1, GATA4, HAND1, HAND2, NKX2.5 and TBX5. Later, the highly modulated secondary structure was determined by a combination of SHAPE, DMS and 3S probing experiments [43]. As a result, Braveheart was found to be very modular and in particular its 5′ asymmetric G-righ internal loop (AGIL) motif was investigated. This motif was later shown to interact with even more binding proteins including CNBP/ZNF9 and HNRNPF. Loss of this motif through CRISPR/Cas9 was followed by a decrease in beating cardiomyocytes which developed from the mutant ESCs. This could only be reversed by reducing CNBP as well. This led to the assumption that the interaction between Braveheart and CNBP is essential for cardiomyocyte differentiation.
4.2. SRA
The case of SRA is special, because its gene expresses not only protein coding transcripts, but also the lncRNA SRA. The difference is only based on alternative splicing influencing the first intron of the gene. Previously associated to breast cancer [44], it is increasingly linked to adipogenesis [45], steroidogenesis [46] and myogenesis [47] as well. SRA can enhance MyoD activity, a major transcription factor in cardiac muscle differentiation [48]. Knockout experiments in zebrafish were able to show reduced cardiac contractility predominantly in ventricular heart chambers [49]. Since the SRA lncRNA can also interact with the SRA protein through the RNA recognition motif (RRM), it can even control this enhancing effect during differentiation [48]. Other binding partners include the protein complexes TrxG and PRC2 which can influence gene expression by altering histone modifications [50]. In 2012, SRA was the first lncRNA that had its secondary structure probed by chemical and enzymatic methods by Novikova et al. [51]. The resulting structure consists mainly of four domains with a range of several loop and helical structures. To see how the different probing methods accord with each other or even might complement each other, they probed the structure with several different experimental methods. The results showed that the chemical probing methods predominantly agree with each other, but the enzymatic characterization was lacking concordance sometimes. This could be explained by the relatively spacious enzyme, which might not be able to bind to the RNA in regions that are tightly structured. Through comparative studies, certain substructures were found to be highly conserved across vertebrates [51], leading to the assumption of functional significance of these structures. Consequently which of the conserved domains are responsible for binding its target proteins still needs to be addressed by further analysis.
4.3. HOTAIR
HOTAIR (HOX transcript antisense intergenic RNA) is a roughly 2.2 kb long lncRNA that acts on the HoxD locus in trans by binding to several chromatin modifying proteins like PRC2 [52] and LSD1 [53]. Although HOTAIR was mainly studied in cancer [54,55], there were several studies conducted which hint at its importance in heart development and disease. Among other lncRNAs, HOTAIR was found to be differentially expressed in heart failure patients as well as in the mouse model of cardiac hypertrophy obtained by transverse aortic constriction (TAC) [56]. Also it was found to play a significant role in cardiomyocytes from sepsis mice [57]. Functional and in vivo studies were able to show the regulatory potential of HOTAIR on p65 phosphorylation and NF-κB which in turn leads to myocardial dysfunction by increased TNF-α production. The structure of the HOTAIR transcript was determined by Somarowthu et al. by different probing methods including SHAPE-Seq and DMS. This study resulted in four highly structured modules with more than 90% agreement between the different probing methods [58]. Two of those modules were also found to be in concordance with previously investigated protein binding sites and phylogenetic comparison analysis showed significant evolutionary conservation of those sites.
5. Discussion
Since the emergence of lncRNAs as a major regulatory mechanism in development and disease, the list of cardiac-related lncRNAs and the pressure to investigate them functionally keeps growing. Since lncRNAs do not encode for proteins, but instead can bind to various partners including proteins, DNA and other RNAs, their structure is the key in exploring their functionality. Recent research of Alu motifs in the lncRNAs MANTIS [39] and ANRIL [42] highlighted the role of specific patterns identifying their function. In ANRIL this motif is located in a central loop-like structure in the RNA transcript as well as in promoter regions of its trans-targeted genes, suggesting a RNA-chromatin interaction. Therefore exploring RNA structures could identify similar mechanisms. Improvements in computational and experimental techniques, and especially the combination of both, have shown an astonishing improvement in structure prediction. This led to predictions which already are very close to the real structure. It is important to keep in mind though, that each technique has certain advantages and flaws which need to be considered before planning an experiment, but since the determination of lncRNA structure is a relatively new field, there is still a lot of research being done. Also after probing the structures of lncRNAs, follow-up experiments are still essential to fully understand the importance of certain domains, but in the end more and more exciting examples of probed RNAs will emerge which will significantly improve our understanding of the cardiovascular system and disease.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the fund Innovative Medical Research of the University of Münster Medical School (RÜ121510).
References
- 1.Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S. The landscape of long noncoding rnas in the human transcriptome. Nat. Genet. 2015;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. The gencode v7 catalog of human long noncoding rnas: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kornfeld JW, Brüning JC. Regulation of metabolism by long, non-coding RNAs. Front. Genet. 2014;5:57. doi: 10.3389/fgene.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding rnas. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding rnas: insights into functions. Nat. Rev. Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 6.Ounzain S., Burdet F., Ibberson M., Pedrazzini T. Discovery and functional characterization of cardiovascular long noncoding rnas. J. Mol. Cell. Cardiol. 2015;89:17–26. doi: 10.1016/j.yjmcc.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 7.Zhang F., Lupski J.R. Non-coding genetic variants in human disease. Hum. Mol. Genet. 2015;24(R1):R102–R110. doi: 10.1093/hmg/ddv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knapp G. [16] enzymatic approaches to probing of rna secondary and tertiary structure. Meth. Enzymol. 1989;180:192–212. doi: 10.1016/0076-6879(89)80102-8. [DOI] [PubMed] [Google Scholar]
- 9.Stern S., Moazed D., Noller H.F. [33] structural analysis of rna using chemical and enzymatic probing monitored by primer extension. Meth. Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 10.Wan Y., Qu K., Ouyang Z., Chang H.Y. Genome-wide mapping of rna structure using nuclease digestion and high-throughput sequencing. Nat. Protoc. 2013;8(5):849–869. doi: 10.1038/nprot.2013.045. [DOI] [PubMed] [Google Scholar]
- 11.Wan Y., Qu K., Zhang Q.C., Flynn R.A., Manor O., Ouyang Z., Zhang J., Spitale R.C., Snyder M.P., Segal E. Landscape and variation of rna secondary structure across the human transcriptome. Nature. 2014;505(7485):706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henkin T.M. Riboswitch rnas: using rna to sense cellular metabolism. Gene Dev. 2008;22(24):3383–3390. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Underwood J.G., Uzilov A.V., Katzman S., Onodera C.S., Mainzer J.E., Mathews D.H., Lowe T.M., Salama S.R., Haussler D. Fragseq: transcriptome-wide rna structure probing using high-throughput sequencing. Nat. Meth. 2010;7(12):995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tijerina P., Mohr S., Russell R. Dms footprinting of structured rnas and rna–protein complexes. Nat. Protoc. 2007;2(10):2608–2623. doi: 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyrrell J., McGinnis J.L., Weeks K.M., Pielak G.J. The cellular environment stabilizes adenine riboswitch rna structure. Biochemistry. 2013;52(48):8777–8785. doi: 10.1021/bi401207q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rouskin S., Zubradt M., Washietl S., Kellis M., Weissman J.S. Genome-wide probing of rna structure reveals active unfolding of mrna structures in vivo. Nature. 2014;505(7485):701. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding Y., Tang Y., Kwok C.K., Zhang Y., Bevilacqua P.C., Assmann S.M. In vivo genome-wide profiling of rna secondary structure reveals novel regulatory features. Nature. 2014;505(7485):696. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 18.Loughrey D., Watters K.E., Settle A.H., Lucks J.B. Shape-seq 2.0: systematic optimization and extension of high-throughput chemical probing of rna secondary structure with next generation sequencing. Nucleic Acids Res. 2014;42(21) doi: 10.1093/nar/gku909. 000–000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solem A.C., Halvorsen M., Ramos S.B., Laederach A. The potential of the ribosnitch in personalized medicine. Wiley Interdiscipl. Rev. RNA. 2015;6(5):517–532. doi: 10.1002/wrna.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reuter J.S., Mathews D.H. Rnastructure: software for rna secondary structure prediction and analysis. BMC Bioinf. 2010;11(1):129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deigan K.E., Li T.W., Mathews D.H., Weeks K.M. Accurate shape-directed rna structure determination. Proc. Natl. Acad. Sci. 2009;106(1):97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flynn R.A., Zhang Q.C., Spitale R.C., Lee B., Mumbach M.R., Chang H.Y. Transcriptome-wide interrogation of rna secondary structure in living cells with icshape. Nat. Protoc. 2016;11(2):273–290. doi: 10.1038/nprot.2016.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee B., Flynn R.A., Kadina A., Guo J.K., Kool E.T., Chang H.Y. Comparison of shape reagents for mapping rna structures inside living cells. RNA. 2017;23(2):169–174. doi: 10.1261/rna.058784.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novikova I.V., Dharap A., Hennelly S.P., Sanbonmatsu K.Y. 3s: shotgun secondary structure determination of long non-coding rnas. Methods. 2013;63(2):170–177. doi: 10.1016/j.ymeth.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 25.Rivas E., Clements J., Eddy S.R. Lack of evidence for conserved secondary structure in long noncoding rnas. Nat. Meth. 2017;14(1):45. doi: 10.1038/nmeth.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorenz R., Bernhart S.H., Zu Siederdissen C.H., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. Viennarna package 2.0. Algorithm Mol. Biol. 2011;6(1):1. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuker M., Stiegler P. Optimal computer folding of large rna sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H., Zheng H., Wang C., Lu X., Zhao X., Li X. Insight into hotair structural features and functions as landing pads for transcription regulation proteins. Biochem. Biophys. Res. Commun. 2017;485(3):679–685. doi: 10.1016/j.bbrc.2017.02.100. [DOI] [PubMed] [Google Scholar]
- 29.Washietl S., Will S., Hendrix D.A., Goff L.A., Rinn J.L., Berger B., Kellis M. Computational analysis of noncoding rnas. Wiley Interdiscipl. Rev. RNA. 2012;3(6):759–778. doi: 10.1002/wrna.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding Y., Lawrence C.E. A statistical sampling algorithm for rna secondary structure prediction. Nucleic Acids Res. 2003;31(24):7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Y., Chan C.Y., Lawrence C.E. Rna secondary structure prediction by centroids in a boltzmann weighted ensemble. RNA. 2005;11(8):1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kladwang W., VanLang C.C., Cordero P., Das R. Understanding the errors of shape-directed rna structure modeling. Biochemistry. 2011;50(37):8049–8056. doi: 10.1021/bi200524n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouyang Z., Snyder M.P., Chang H.Y. Seqfold: genome-scale reconstruction of rna secondary structure integrating high-throughput sequencing data. Genome Res. 2013;23(2):377–387. doi: 10.1101/gr.138545.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quarrier S., Martin J.S., Davis-Neulander L., Beauregard A., Laederach A. Evaluation of the information content of rna structure mapping data for secondary structure prediction. RNA. 2010;16(6):1108–1117. doi: 10.1261/rna.1988510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byun Y., Han K. Pseudoviewer: web application and web service for visualizing rna pseudoknots and secondary structures. Nucleic Acids Res. 2006;34(suppl_2):W416–W422. doi: 10.1093/nar/gkl210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L., Ding H., Butty V.L., Torrey L., Haas S. Braveheart, a long noncoding rna required for cardiovascular lineage commitment. Cell. 2013;152(3):570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han P., Li W., Lin C.-H., Yang J., Shang C., Nurnberg S.T., Jin K.K., Xu W., Lin C.-Y., Lin C.-J. A long noncoding rna protects the heart from pathological hypertrophy. Nature. 2014;514(7520):102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z., Zhang X.-J., Ji Y.-X., Zhang P., Deng K.-Q., Gong J., Ren S., Wang X., Chen I., Wang H. The long noncoding rna chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 2016;22(10):1131–1139. doi: 10.1038/nm.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leisegang M.S., Fork C., Josipovic I., Richter F., Preussner J., Hu J., Miller M.J., Epah J.N., Hofmann P., Günther S. Long noncoding rna mantis facilitates endothelial angiogenic function. Circulation. 2017;136:65–79. doi: 10.1161/CIRCULATIONAHA.116.026991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ounzain S., Micheletti R., Arnan C., Plaisance I., Cecchi D., Schroen B., Reverter F., Alexanian M., Gonzales C., Ng S.Y. Carmen, a human super enhancer-associated long noncoding rna controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell. Cardiol. 2015;89:98–112. doi: 10.1016/j.yjmcc.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 41.Grote P., Wittler L., Hendrix D., Koch F., Währisch S., Beisaw A., Macura K., Bläss G., Kellis M., Werber M. The tissue-specific lncrna fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013;24(2):206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holdt L.M., Hoffmann S., Sass K., Langenberger D., Scholz M., Krohn K., Finstermeier K., Stahringer A., Wilfert W., Beutner F. Alu elements in anril non-coding rna at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9(7) doi: 10.1371/journal.pgen.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue Z., Hennelly S., Doyle B., Gulati A.A., Novikova I.V., Sanbonmatsu K.Y., Boyer L.A. A g-rich motif in the lncrna braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage. Mol. Cell. 2016;64(1):37–50. doi: 10.1016/j.molcel.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hube F., Guo J., Chooniedass-Kothari S., Cooper C., Hamedani M.K., Dibrov A.A., Blanchard A.A., Wang X., Deng G., Myal Y. Alternative splicing of the first intron of the steroid receptor rna activator (sra) participates in the generation of coding and noncoding rna isoforms in breast cancer cell lines. DNA Cell Biol. 2006;25(7):418–428. doi: 10.1089/dna.2006.25.418. [DOI] [PubMed] [Google Scholar]
- 45.Xu B., Gerin I., Miao H., Vu-Phan D., Johnson C.N., Xu R., Chen X.-W., Cawthorn W.P., MacDougald O.A., Koenig R.J. Multiple roles for the non-coding rna sra in regulation of adipogenesis and insulin sensitivity. PLoS One. 2010;5(12) doi: 10.1371/journal.pone.0014199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu B., Yang W.-H., Gerin I., Hu C.-D., Hammer G.D., Koenig R.J. Dax-1 and steroid receptor rna activator (sra) function as transcriptional coactivators for steroidogenic factor 1 in steroidogenesis. Mol. Cell Biol. 2009;29(7):1719–1734. doi: 10.1128/MCB.01010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caretti G., Schiltz R.L., Dilworth F.J., Di Padova M., Zhao P., Ogryzko V., Fuller-Pace F.V., Hoffman E.P., Tapscott S.J., Sartorelli V. The rna helicases p68/p72 and the noncoding rna sra are coregulators of myod and skeletal muscle differentiation. Dev. Cell. 2006;11(4):547–560. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 48.Hubé F., Velasco G., Rollin J., Furling D., Francastel C. Steroid receptor rna activator protein binds to and counteracts sra rna-mediated activation of myod and muscle differentiation. Nucleic Acids Res. 2010;39(2):513–525. doi: 10.1093/nar/gkq833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friedrichs F., Zugck C., Rauch G.-J., Ivandic B., Weichenhan D., Müller-Bardorff M., Meder B., El Mokhtari N.E., Regitz-Zagrosek V., Hetzer R. Hbegf, sra1, and ik: three cosegregating genes as determinants of cardiomyopathy. Genome Res. 2009;19(3):395–403. doi: 10.1101/gr.076653.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wongtrakoongate P., Riddick G., Fucharoen S., Felsenfeld G. Association of the long non-coding rna steroid receptor rna activator (sra) with trxg and prc2 complexes. PLoS Genet. 2015;11(10) doi: 10.1371/journal.pgen.1005615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novikova I.V., Hennelly S.P., Sanbonmatsu K.Y. Structural architecture of the human long non-coding rna, steroid receptor rna activator. Nucleic Acids Res. 2012;40(11):5034–5051. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E. Functional demarcation of active and silent chromatin domains in human hox loci by noncoding rnas. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsai M.-C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding rna as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.-C., Hung T., Argani P., Rinn J.L. Long noncoding rna hotair reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T., Tanaka F., Shibata K., Suzuki A., Komune S. Long noncoding rna hotair regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Canc. Res. 2011;71(20):6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 56.Greco S., Zaccagnini G., Perfetti A., Fuschi P., Valaperta R., Voellenkle C., Castelvecchio S., Gaetano C., Finato N., Beltrami A.P. Long noncoding rna dysregulation in ischemic heart failure. J. Transl. Med. 2016;14(1):183. doi: 10.1186/s12967-016-0926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu H., Liu J., Li W., Liu G., Li Z. Lncrna-hotair promotes tnf-α production in cardiomyocytes of lps-induced sepsis mice by activating nf-κb pathway. Biochem. Biophys. Res. Commun. 2016;471(1):240–246. doi: 10.1016/j.bbrc.2016.01.117. [DOI] [PubMed] [Google Scholar]
- 58.Somarowthu S., Legiewicz M., Chillón I., Marcia M., Liu F., Pyle A.M. Hotair forms an intricate and modular secondary structure. Mol. Cell. 2015;58(2):353–361. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]