Abstract
Background:
Epidemiological evidence suggests a link between pesticide exposure and the development of metabolic diseases. However, most experimental studies have evaluated the metabolic effects of pesticides using individual molecules, often at nonrelevant doses or in combination with other risk factors such as high-fat diets.
Objectives:
We aimed to evaluate, in mice, the metabolic consequences of chronic dietary exposure to a pesticide mixture at nontoxic doses, relevant to consumers’ risk assessment.
Methods:
A mixture of six pesticides commonly used in France, i.e., boscalid, captan, chlorpyrifos, thiofanate, thiacloprid, and ziram, was incorporated in a standard chow at doses exposing mice to the tolerable daily intake (TDI) of each pesticide. Wild-type (WT) and constitutive androstane receptor–deficient () male and female mice were exposed for 52 wk. We assessed metabolic parameters [body weight (BW), food and water consumption, glucose tolerance, urinary metabolome] throughout the experiment. At the end of the experiment, we evaluated liver metabolism (histology, transcriptomics, metabolomics, lipidomics) and pesticide detoxification using liquid chromatography–mass spectrometry (LC-MS).
Results:
Compared to those fed control chow, WT male mice fed pesticide chow had greater BW gain and more adiposity. Moreover, these WT males fed pesticide chow exhibited characteristics of hepatic steatosis and glucose intolerance, which were not observed in those fed control chow. WT exposed female mice exhibited fasting hyperglycemia, higher reduced glutathione (GSH):oxidized glutathione (GSSG) liver ratio and perturbations of gut microbiota–related urinary metabolites compared to WT mice fed control chow. When we performed these experiments on mice, pesticide-exposed males did not exhibit BW gain or changes in glucose metabolism compared to the males fed control chow. Moreover, females fed pesticide chow exhibited pesticide toxicity with higher BWs and mortality rate compared to the females fed control chow.
Conclusions:
To our knowledge, we are the first to demonstrate a sexually dimorphic obesogenic and diabetogenic effect of chronic dietary exposure to a common mixture of pesticides at TDI levels, and to provide evidence for a partial role for CAR in an in vivo mouse model. This raises questions about the relevance of TDI for individual pesticides when present in a mixture. https://doi.org/10.1289/EHP2877
Introduction
The rates of metabolic disorders, including obesity and its complications, such as type 2 diabetes (T2D) and nonalcoholic fatty liver disease (NAFLD), have increased dramatically over the past three decades, especially in developed countries. Genetic factors and changes in lifestyle are key factors in the development of metabolic syndrome. Exposure to environmental contaminants is also suspected to be part of the etiological factors. Experimental and epidemiological studies suggest that some environmental pollutants, such as endocrine-disrupting chemicals (EDCs), may contribute to the onset and progression of obesity, diabetes, and NAFLD by interfering with various aspects of metabolism (Hong et al. 2012). Diethylstilbestrol, bisphenol A, phthalates, or tributyltin, which cause perturbations in endogenous hormonal regulation involved in weight homeostasis, have recently been reviewed as possible risk factors for obesity (De Long and Holloway 2017; Newbold et al. 2009). Moreover, EDCs are known to affect liver function and, therefore, may contribute to increasing the prevalence of NAFLD worldwide (Polyzos et al. 2012). Hepatic metabolic homeostasis is tightly controlled by various transcription factors sensing hormonal, nutritional, and environmental cues. Among transcription factors, nuclear receptors such as constitutive androstane receptor (CAR) (Qatanani and Moore 2005) and peroxisome proliferator–activated receptors (PPARs) (Desvergne et al. 2006) modulate the expression of genes involved in xenobiotic and energy homeostasis (Evans and Mangelsdorf 2014). Nuclear receptor transcriptional activity is regulated upon binding of small lipophilic ligands that include hormone and metabolites as well as endogenous compounds and xenobiotics.
Pesticides represent an increasingly widespread environmental contamination. Much epidemiological evidence has linked pesticides in water sources, groundwater, and/or foodstuffs with obesity, diabetes, insulin resistance (Casals-Casas and Desvergne 2011; Grün and Blumberg 2009), and NAFLD (Al-Eryani et al. 2015; Arciello et al. 2013; Wahlang et al. 2013). In a human cohort, the weight and body mass index of the offspring significantly correlated with the prenatal maternal levels of dichlorodiphenyldichloroethylene (DDE), a metabolite of the organochlorine (OC) pesticide dichlorodiphenyltrichloroethane (DDT) (Karmaus et al. 2009). A recent systematic review and meta-analysis clearly supports the hypothesis that environmental or occupational exposure to OC pesticides increases the risk of T2D (Evangelou et al. 2016). Similarly, in vivo and in vitro experimental studies support the notion that nonpersistent pesticides (e.g., pyrethroids, organophosphorus, neonicotinoids, dithiocarbamates, carbamates, triazines) are potential contributors to general or hepatic metabolic changes (Bhaskar and Mohanty 2014; Jin et al. 2014; Kim et al. 2013; Seidler and Slotkin 2011; Wang et al. 2014) and obesity (De Long and Holloway 2017; Newbold et al. 2009). However, epidemiological studies have mainly focused on OC pesticides, and evidence on other pesticide classes is limited. Most experimental studies assess the effect of individual compounds, using doses that are often far from the environmental levels to which the general population is exposed. Consumers are mainly exposed through the food chain to mixtures of pesticides at low doses (EFSA 2015) throughout their lifetime, and the consequences of such a chronic exposure on metabolic homeostasis have not been well described. Moreover, the metabolic impact of pesticides has often been observed in combination with other risk factors, such as high-fat diets (Adigun et al. 2010b; Howell et al. 2015; Lassiter et al. 2010; Maqbool et al. 2016) and/or after a relatively short exposure period, never exceeding a few months (Mostafalou and Abdollahi 2017). Finally, animals were often exposed to pesticides via oral gavage, an exposure route that is not representative of consumer exposure, which mainly occurs through food intake.
In the present study, we assessed the metabolic consequences of chronic dietary exposure to a cocktail of pesticides in mice. We selected six pesticides among those used for the treatment of apple orchards in the south of France. Therefore, the pesticide cocktail used may be commonly found in apples in the European Union, as recently described in an European Food Safety Authority (EFSA) report (EFSA 2015). Pesticides were incorporated at nontoxic doses in a standard chow and mice exposed to the tolerable daily intake (TDI) for each of these six individual pesticides. We followed the body weight (BW) and metabolic parameters of male and female mice using both the wild-type (WT) and CAR-deficient () strains over a period of 52 wk, determining, to our knowledge, for the first time, the in vivo effect of chronic exposure to a mixture of pesticides at very low doses.
Methods
Chemicals
High-purity analytical standards of boscalid, captan, chlorpyrifos (99.2%), thiacloprid, thiophanate, and ziram were purchased from Sigma-Aldrich. Information on the toxicity of pesticides was obtained from Agritox (http://www.agritox.anses.fr/).
Methanol was liquid chromatography–mass spectrometry (LC-MS) grade and purchased from Fisher Scientific (Thermo Fisher Scientific). Ultrapure water from the Milli-Q® system (Millipore) was used for mobile phases.
Pesticide Chow
Ziram, thiophanate, captan, chlorpyrifos, boscalid, and thiachlopride belong to various chemical families and are either fungicides or insecticides. Mice were exposed at the TDI of each pesticide as defined in BW per day; the TDIs were those defined by the EFSA until 2014 for humans by the Joint Food and Agriculture Organization of the United Nations/World Health Organization Meeting on Pesticide Residues and extrapolated for mice on the basis of mean BW. To calculate the quantity of pesticide needed to incorporate in rodent pellets to achieve TDI, we assumed a BW of and a daily food intake of for all mice. The properties and quantities of each pesticide incorporated in the mouse diet are described in Table 1. Pesticides were dissolved individually in a 9:1 volume/volume (v/v) mixture of methanol:acetone and then mixed together. The solution was dispersed over the vitamin powder [vitaminic mixture 200, Scientific Animal Food Engineering (SAFE)] and then homogenized in a rotavapor (Laborota 4000™; BUCHI Switzerland) for 30 min at 45°C to evaporate the solvents and at room temperature for 50 min. The vitamin powder enriched with pesticides was sent to the Animal Feed Preparation Unit [UPAE; National Institute of Agricultural Research (INRA)] and incorporated into the pellets. Control feed was prepared as described above with the 9:1 methanol:acetone mixture alone. Pesticides present in the diet were quantified by Eurofins using gas chromatography–tandem mass spectrometry and liquid chromatography–tandem mass spectrometry, and the final concentrations were confirmed, except for ziram, which could not be detected at such low concentration (Table 1). Analysis from Eurofins laboratory confirmed that control feed contained none of the pesticides being studied.
Table 1.
Chemical families, functions, and tolerable daily intake (TDI) ( ) of each pesticide and the expected and measured pesticides concentrations (determined level) ( food) in the animal pellets.
| Pesticide name | Chemical family | Function | TDI () | Expected quantity ( food)a | Determined level ( food) |
|---|---|---|---|---|---|
| Ziram | Dithiocarbamate | Fungicide | 0.006 | 36 | NDb |
| Chlorpyrifos | Organophosphorus | Insecticide | 0.01 | 60 | 47 |
| Thiacloprid | Neonicotinoid | Insecticide | 0.01 | 60 | 56 |
| Boscalid | Carboxamide | Fungicide | 0.04 | 240 | 240 |
| Thiofanate | Benzimidazole | Fungicide | 0.08 | 480 | 205 |
| Captan | Dicarboximide | Fungicide | 0.1 | 600 | 165 |
Note: BW, Body weight; ND, not determined; TDI, tolerable daily intake (http://www.agritox.anses.fr/).
Expected quantity refers to the incorporated quantities of pesticides in mice pellets.
Ziram was not present at a detectible level ().
Animal Experiment
The experimental work on animals was conducted in the Toxalim Unit, INRA, in a conventional laboratory animal room following the European Union guidelines for laboratory animal use and care, and was approved by an independent ethics committee (authorization number 05,185.02). The animals were treated humanely with due consideration to the alleviation of distress and discomfort and housed in polycarbonate cages (Charles River Type S, ). Eight-week-old female and male C57BL/6J mice were purchased from Janvier Laboratories and acclimatized for 4 wk. CAR-deficient mice (backcrossed on the C57BL/6J background) were engineered in Wei et al. laboratory (Figure S1A-B) and are bred for 10 y in our animal room. Figure S1C confirms that mice were deficient in CAR compared the WT C57BL/6J mice (Wei et al. 2000). The C57Bl/6J mice served as WT strain for comparison with the mice on the same genetic background (C57Bl/6J). Both strains were treated in the same manner and experiments conducted concurrently to allow direct comparison of results. After the initial acclimatization period, all mice were maintained on standard chow for 4 wk (weeks through 0), during which time we monitored BW weekly. At 16 wk of age (week 0), diets were switched to either a pesticide-enriched or -free diet, which was continued for a total of 52 wk to mimic a 30-y exposure in humans (Flurkey et al. 2007). Mice were randomly housed five or four per cage at their receipt (72 WT mice) or at their transfer from the breeding house into the animal room (72 mice) and allowed ad libitum access to food and water with a 12-h light/dark cycle (). Eight groups of 18 animals each were used in this study: WT males exposed, WT males nonexposed, WT females exposed, WT females nonexposed, males exposed, males nonexposed, females exposed, and females nonexposed. Mice were randomly distributed into unexposed and exposed groups. In this experiment, we used a standard synthetic diet from Animal Feed Preparation Unit (UPAE, INRA). This diet is composed of 63% carbohydrate, 5% fat, 22% protein, 2% cellulose, 1% vitamins (Safe 200, SAFE) and 7% minerals (Safe 205b, Safe France).
During the 6th, 26th, and 48th weeks of exposure, the mice were placed in individual metabolic cages (Tecniplast France) to obtain 24-h urine samples to use for metabolomics and mass spectrometry. The samples were stored at until analysis. After 52 wk of exposure, 4 to 5 mice from each experimental group were daily randomly sacrificed (in total, 16 to 20 animals from the four experimental groups were sacrificed per day) by cervical dislocation at Zeitgeber time 17 (i.e., in the fed state) at which hepatic activities are optimal (Lu et al. 2013). Blood was collected from the submandibular vein with a lancet into lithium heparin–coated tubes (BD Microtainer®; BD). Plasma was prepared by centrifugation (; 10 min; 4°C) and stored at to be used for enzyme-linked immunosorbent assay (ELISA), biochemical analysis, and metabolomics. Liver samples were collected, weighed, snap-frozen in liquid nitrogen, and kept at for further analyses (liver neutral lipid analyses, lipidomics, microarray, and metabolomics). Subcutaneous and epidydimal white adipose tissue (WAT) were collected and weighed. Liver samples () were fixed in formaldehyde (4%) for 24 h and embedded in paraffin for histology.
Glucose Tolerance Test
All experiments were performed on conscious mice. For the oral glucose tolerance test at weeks 36 and 48 or the intraperitoneal glucose tolerance test at week 16, mice were fasted for 6 h and received an oral ( BW) or intraperitoneal ( BW) glucose load. Blood glucose was measured at the tail vein using an Accu-Check® Performa glucometer (Roche Diagnostics) 15 min prior to and at 0, 15, 30, 45, 60, 90, and 120 min after the glucose load.
Plasma Analysis
Plasma insulin was assayed after 52 wk of pesticide exposure using the ultrasensitive mouse insulin ELISA kit (EMD Millipore) following the manufacturer's instructions. Aspartate transaminase, alanine transaminase (ALT), total cholesterol, low-density lipoprotein (LDL) cholesterol, high-density lipoprotein (HDL) cholesterol, free fatty acids (FFAs), and triglycerides (TGs) were determined using a Pentra 400 biochemical analyzer (HORIBA, Montpellier, France; Anexplo facility, Toulouse, France).
Liver Neutral Lipid Analysis
Hepatic lipid contents were determined at the end of the experiment as described elsewhere (Bligh and Dyer 1959). Briefly, following homogenization of tissue samples in 2:1 (v/v) methanol/ethylene glycol-bis(-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA; ), lipids corresponding to an equivalent of of tissue were extracted according to Bligh and Dyer in chloroform:methanol:water (2.5:2.5:2.1, v/v/v) in the presence of the internal standards glyceryltrinonadecanoate, stigmasterol, and cholesteryl heptadecanoate (Sigma). TGs, free cholesterol, and cholesterol esters were analyzed by gas chromatography on a Focus Thermo Electron system using a Zebron-1 Phenomenex (Phenomenex Zebron-1, England) fused-silica capillary column [; internal diameter (i.d.); film thickness]. The oven temperature was programmed from 200 to 350°C at a rate of , and the carrier gas was hydrogen (0.5 bar). The injector and detector were at 315°C and 345°C, respectively.
Liver Fatty Acid Analysis
To measure all hepatic fatty acid methyl ester (FAME) molecular species, lipids that corresponded to an equivalent of of liver were extracted in the presence of the internal standard glyceryl triheptadecanoate (). The lipid extract was transmethylated with boron trifluoride (BF3) in methanol (14% solution; Sigma) and heptane for 60 min at 80°C and evaporated to dryness. The FAMEs were extracted with heptane/water (2:1). The organic phase was evaporated to dryness and dissolved in ethyl acetate. A sample () of total FAME was analyzed with gas–liquid chromatography [Clarus 600 system (PerkinElmer), with FAMEWAX fused silica capillary columns (Restek), i.d., film thickness]. Oven temperature was programmed to increase from 110°C to 220°C at a rate of , and the carrier gas was hydrogen [7.25 pounds per square inch (psi)]. Injector and detector temperatures were 225°C and 245°C, respectively.
Liver Phospholipid and Sphingolipid Analysis
The liquid chromatography solvent acetonitrile was high-performance liquid chromatography (HPLC) grade and purchased from Acros Organics. Ammonium formate () was supplied by Sigma-Aldrich. Synthetic lipid standards [ceramide (Cer) d18:1/18:0, Cer d18:1/15:0, phosphatidylethanolamine (PE) 12:0/12:0, PE 16:0/16:0, phosphatidlcholine (PC) 13:0/13:0, PC 16:0/16:0, sphingomyelin (SM) d18:1/18:0, SM d18:1/12:0] were purchased from Avanti Polar Lipids.
Lipids were extracted from the liver () as described by Bligh and Dyer (1959) in dichloromethane/methanol (2% acetic acid)/water (2.5:2.5:2 v/v/v). Internal standards were added (Cer d18:1/15:0, ; PE 12:0/12:0, ; PC 13:0/13:0, ; SM d18:1/12:0, ; PI 16:0/17:0, ; PS 12:0/12:0, ). The solution was centrifuged at for 3 min. The organic phase was collected and dried under azote, then dissolved in MeOH. Sample solutions were analyzed with an Agilent 1290 UPLC system coupled to a G6460 triple quadripole spectrometer (Agilent, version 2.1). MassHunter software was used for data acquisition and analysis. A Kinetex® HILIC column (Phenomenex; ; ) was used for LC separations. The column temperature was maintained at 40°C. The mobile phase, A, was Acetonitrile; and B was ammonium formate in water at pH 3.2. The gradient was as follows: from 10% to 30% B in 10 min, 100% B from 10 to 12 min, and then back to 10% B at 13 min for 1 min to re-equilibrate prior to the next injection. The flow rate of the mobile phase was , and the injection volume was . An electrospray source was employed in positive (for Cer, PE, PC, and SM analysis) or negative ion mode (for PI and PS analysis). The collision gas was nitrogen. Needle voltage was set at . Several scan modes were used. First, to obtain the naturally different masses of different species, we analyzed cell lipid extracts with a precursor ion scan at , , and for PC/SM, PI, and Cer, respectively. We performed a neutral loss scan at and for PE and PS, respectively. The collision energy optimums for Cer, PE, PC, SM, PI, and PS were 25 electrovolt (eV), 20 eV, 30 eV, 25 eV, 45 eV, and 22 eV, respectively. Then the corresponding Single Reaction Monitoring (SRM) transitions were used to quantify different phospholipid species for each class. Two Multiple Reaction Monitoring (MRM) acquisitions were necessary, due to important differences between phospholipid classes. Data were treated with QqQ Quantitative (version B.05.00; Agilent) and Qualitative analysis software (version B.04.00; Agilent).
Histology
Paraformaldehyde-fixed, paraffin-embedded liver tissue was sliced into sections, deparaffinized, rehydrated, and then stained with hematoxylin and eosin for histopathological analysis. The staining was visualized with a Leica microscope DM4000 B equipped with a Leica DFC450 C camera (Leica Microsystems). The 72 slides (one slide per animal, 18 slides per groups, and four groups: WT males and females fed control or pesticide chow) were full-blind analyzed by two individuals (C.L. and L.G.P.), and observations were approved by an histopathologist (T.A.S.). The score of steatosis was calculated according to the Kleiner method (Kleiner et al. 2005) and was the sum of three scores using the entire slice field: (a) the first score was defined according to the surface covered by lipid droplets (i.e., a surface of lipid droplet of the entire slice correspond to a score 0; a surface of lipid droplet of the entire slice correspond to a score 3); (b) for the second score, a predominant azonal distribution patter of lipid droplets was graded with the highest value (score 3); and (c) the third score quantified the presence (score 1) or absence (score 0) of microvesicles.
Gene Expression Studies
RNAs were extracted from liver samples collected at the end of the exposure in WT and females and males fed control or pesticide chow using the method (Chomczynski and Sacchi 1987) with TRI Reagent® (Sigma-Aldrich). RNAs were quantified using nanodrop (NanoDrop™ 1000; Thermo Scientific). Two micrograms of total RNA were reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems™). The SYBR Green (Applied Biosystems, California) assay primers are presented in Table S1. Amplification was performed using an ABI Prism 7,300 Real-Time PCR System (Applied Biosystems™). Quantitative real-time polymerase chain reaction (qPCR) data were normalized to TATA-box-binding protein mRNA levels and analyzed with LinRegPCR (version 2015.3; Jan Ruijter) to get mean efficiency (NO), which is calculated as follows: with Eff mean: mean PCR efficiency and CQ: quantification cycle.
Microarray Gene Expression Studies
Gene expression profiles were obtained for six liver samples per group at the GeT‐TRiX facility (GénoToul, Génopole Toulouse Midi-Pyrénées, France) using Sureprint G3 Mouse GE v2 microarrays (8x60K; design 074,809; Agilent) following the manufacturer's instructions. For each sample, Cyanine-3 (Cy3)–labeled complementary RNA (cRNA) was prepared from of total RNA using the One-Color Quick Amp Labeling kit (Agilent) according to the manufacturer's instructions, followed by Agencourt RNAClean XP (Beckman Coulter Inc.). Dye incorporation and cRNA yield were checked using a Dropsense™ 96 ultraviolet/visible (VIS) droplet reader (Trinean). A total of of Cy3-labeled cRNA was hybridized on the microarray slides following the manufacturer’s instructions. Immediately after washing, the slides were scanned on an Agilent G2505C Microarray Scanner using Agilent Scan Control A.8.5.1 software and the fluorescence signal extracted using Agilent Feature Extraction software (version 10.10.1.1) with default parameters. Microarray data and experimental details are available in National Center for Biotechnology Information's Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO Series accession number GSE (GSE101405; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101405).
Microarray data were analyzed using R (version 3.1.2; R Development Core Team) Bioconductor packages (version 3.0) as described in GEO accession GSE 101,405. Raw data (median signal intensity) were filtered, transformed, and normalized using the quantile method (Bolstad et al. 2003). A model was fitted using the limma lmFit function (Smyth 2004). Pair-wise comparisons between biological conditions were applied using specific contrasts. A correction for multiple testing was applied using the Benjamini-Hochberg procedure (Klipper-Aurbach et al. 1995) for false discovery rate. Hierarchical clustering was applied to the samples and the differentially expressed probes using 1-Pearson correlation coefficient as distance and Ward’s criterion for agglomeration. The clustering results are illustrated as a heatmap of expression signals. Functional annotation clustering of GO Biological Process were performed using DAVID Bioinformatics Resources 6.7 (Huang et al. 2009a, 2009b).
Proton Nuclear Magnetic Resonance () Based Metabolomics
Urine, plasma, and liver polar extracts were prepared for NMR analysis as described previously (Beckonert et al. 2007). All spectra were obtained on a Bruker DRX-600-Avance NMR spectrometer (Bruker) using the AXIOM metabolomics platform (MetaToul) operating at for resonance frequency using an inverse detection cryoprobe attached to a cryoplatform (the preamplifier cooling unit). The spectra were acquired at 300 K using a standard one-dimensional noesypr1D pulse sequence with water presaturation and a total spin-echo delay () of . A total of 128 transients were collected into 64,000 data points using a spectral width of , a relaxation delay of 2.5 s, and an acquisition time of 2.28 s. , , and Heteronuclear Single Quantum Coherence (HSQC) were obtained for each biological matrix on one representative sample for metabolite identification.
Data were analyzed by applying an exponential window function with a 0.3-Hz line broadening prior to Fourier transformation. The resultant spectra were phased, baseline corrected, and calibrated to trimethylsilylpropanoic acid (TSP) ( ) manually using Mnova NMR (version 9.0; Mestrelab Research S.L.). The spectra were subsequently imported into MatLab (R2014a; MathsWorks, Inc.). All data were analyzed using full-resolution spectra. The region containing the water resonance ( ) was removed, and the spectra were normalized to the probabilistic quotient (Dieterle et al. 2006) and aligned using a previously published function (Veselkov et al. 2009).
Data were mean-centered and scaled using the unit variance scaling prior to analysis with orthogonal projection on latent structure-discriminant analysis (O-PLS-DA). data were used as independent variables (X matrix) and regressed against a dummy matrix (Y matrix) indicating the class of samples (mice fed control or pesticide chow) (Trygg and Wold 2002). The O-PLS–derived model was evaluated for goodness of prediction ( value) using -fold cross-validation, where n depends on the sample size. The parameters of the final models are indicated in the figure legends. To identify metabolites responsible for discrimination between the groups, the O-PLS-DA correlation coefficients () were calculated for each variable and back-scaled into a spectral domain so that the shapes of the NMR spectra and the signs of the coefficients were preserved (Cloarec et al. 2005b, 2005a). The weights of the variables were color-coded according to the square of the O-PLS-DA correlation coefficients. Correlation coefficients extracted from significant models were filtered so that only significant correlations above the threshold defined by Pearson’s critical correlation coefficient (; ; for per group) were considered significant. For illustration purposes, the area under the curve of several signals of interest was integrated and significance tested with the t-test.
NMR data are freely available at MetaboLights (http://www.ebi.ac.uk/metabolights/) under the accession number MTBLS602.
Mass Spectrometry
Urine from seven to eight mice per group was screened for pesticides and their metabolites using liquid chromatography coupled to high-resolution mass spectrometry (HPLC-HRMS). The chromatographic system consisted of a RSLC3000 UHPLC (Thermo Scientific) operating with an Hypersil Gold C18 column (; ) (Thermo Scientific). A gradient program of (A) (95:5:0.1, v/v/v) and (B) (1,000:1, v/v) was used at 40°C and a flow rate of as follows: 0 min, 0% B; from 30 to 35 min, 100% B; and from 36 to 45 min, 0% B. A volume of of each sample diluted twice with mobile phase A injected. The mass spectrometer was an LTQ Orbitrap XL™ (Thermo Scientific) fitted with an electrospray ionization source (ESI) operating in the negative mode. ESI parameters were set as follows: spray voltage, ; sheath gas flow rate (), 55 arbitrary units (a.u.); auxiliary gas flow rate (), 10 a.u.; capillary temperature, 300°C; capillary voltage, ; and tube lens offset, . High-resolution mass spectra were acquired between m/z 80 and 800 at a resolution of 30,000. The mass spectrometer was calibrated using the Thermo Scientific calibration solution in agreement with their protocol. Tandem mass spectrometry (MS-MS) spectra were obtained using the collision-induced dissociation mode of the ion trap analyzer at low resolution and a normalized collision energy of 25%.
Data were extracted from mass spectrometry acquisition files using xcms software, version 3.5 (Scripps Research Institute, La Jolla, CA) (Smith et al. 2006) with centwave algorithm (Tautenhahn et al. 2008). To avoid the effect of urine dilution, variation among sample data was normalized using probabilistic quotient normalization (Dieterle et al. 2006). In the resulting normalized data, univariate analysis of variance was used to study the effect of sex on the different molecule intensities. All these data analysis steps were performed using workflow4metabolomics, a collaborative portal dedicated to metabolomics data processing (Giacomoni et al. 2015).
HPLC-HRMS data were analyzed using a methodology previously developed for exposomics studies (Jamin et al. 2014). From the six studied pesticides, a list of suspected metabolites was compiled from previously described metabolites of these pesticides, as well as all other putative phase II metabolites (Table S2). Thus, the signals of 16 compounds were monitored for the metabolism of boscalid (), 13 for thiachlopride (), 30 for captan (), 9 for thiophanate (), 4 for chlorpyrifos (), and 9 for ziram (9 metabolites). However, standards of these metabolites are not commercially available. In this study, a direct comparison with nonexposed animals was used as a validation strategy. From data extracted with xcms, 16 features matched the theoretical m/z of monitored compounds with a mass precision . MS-MS experiments were performed to confirm the identity of suspected compounds. Conjugation to glucuronic acid was confirmed by detection of the characteristic loss of 176 unified atomic mass (u), the conjugation to sulphate confirmed by the loss of 80 u, and the conjugation to mercapturic acid confirmed by the loss of 129 u. Concerning captan metabolites, MS-MS spectra of the ion detected at m/z 150.0564, corresponding to the in-source fragmentation of sulfate and glucuronic conjugates of tetrahydrophtalimide (THPI), showed a loss of 43 u corresponding to neutral isocyanic acid CONH, validating the identification of these metabolites. Finally, seven features were confirmed by MS-MS experiments, corresponding to five metabolites: two metabolites of captan {[M-H]- and [(M-SO3)-H]- of THPI conjugated to sulphate and [(M-GlcAc)-H]- of THPI conjugated to glucuronic acid}, one metabolite of chlorpyrifos {[M-H]- of 3,5,6-trichloro-2-pyridinol}, and two metabolites of boscalid {[M-H]- and [(M-GlcAc)-H]- of 2-chloro-N-(4′-chloro-5-hydroxybiphenyl-2-yl) nicotinamide conjugated to glucuronic acid and the [M-H]- of the mercapturic conjugation of boscalid}. Structures are available in Figure S2A-B. HRMS and MS-MS data are freely available at MetaboLights (http://www.ebi.ac.uk/metabolights/) under the accession number MTBLS596.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism for Windows (version 4.00; GraphPad Software). One-way or two-way analysis of variance (ANOVA) was performed, followed by appropriate posthoc tests (Dunn’s or Bonferroni) when differences were found statistically significant. When only two groups were compared, the student’s t-test was used; was considered significant.
Results
Body Weight and White Adipose Tissue Gain in Wild-Type Mice Exposed to Pesticide Chow Compared to Those Fed Control Chow
Pesticide levels were assessed in pellets before the beginning of the experiment (Table 1), and we confirmed that pesticides were present in the diet at levels significantly below the expected quantities. We calculated the actual pesticide levels to which mice were exposed after accounting for actual animal BW and food intake during the exposure (Figure S3A). We confirmed that males and females were exposed to similar levels of pesticides and that these levels were below the TDI for each pesticide (Figure S3B).
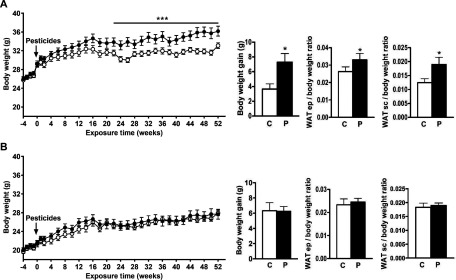
Beginning at 20 wk postexposure and through the end of the exposure period, male WT mice fed the pesticide mixture gained significantly more weight than those fed the control chow (Figure 1A). After 52 wk of exposure, the BW gain in exposed male mice was twice that of the male mice fed control chow ( vs. , respectively). Epididymal and subcutaneous WAT weights were significantly higher in exposed WT male mice than the WT males fed control chow group (subcutaneous WAT: vs. ; epididymal WAT: vs. ; Figure 1A). As shown in Figure S4, WT male and female fed control chow mice had distinct food and water consumption patterns, and specifically female mice fed control chow consumed more food and less water over the period evaluated. Food intake was not different in WT male or female mice fed pesticide chow compared to those fed control chow (Figure S4). WT female mice fed pesticide chow consumed more water than those fed the control chow. In contrast, WT female mice did not exhibit any significant changes in BW or fat mass upon exposure to pesticides (Figure 1B). No cage effect in BW was observed during the experience in WT male fed control or pesticide chow (Figure S5A) and in WT female fed control or pesticide chow (Figure S5B).
Figure 1.
Body weight and white adipose tissue (WAT) weight in male (A) and female (B) wild-type mice. The line graphs show body weight (BW) for pesticide-exposed mice (black circles) and mice fed control chow (white circles) from 4 wk prior to exposure through 52 wk. The bar graphs show the gains in BW and epididymal (ep) and subcutaneous (sc) white adipose tissue (WAT) for pesticide-exposed mice (P) and mice fed control chow (C) after 52 wk. Data are presented as error of the mean. * compared to mice fed control chow as determined by a student’s t-test. *** compared to mice fed control chow as determined by analysis of variance (ANOVA). mice per group.
Glucose Tolerance and Fasting Glucose Levels in Mice Fed Control or Pesticide Chow
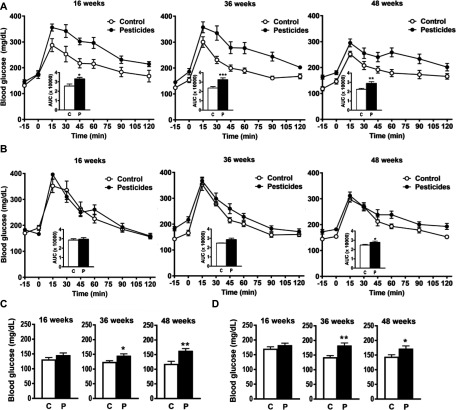
We evaluated the fasting blood glucose and glucose tolerance of WT male and female mice fed control or pesticide chow (Figure 2). At 16, 36, and 48 wk of exposure, WT male mice fed pesticide chow exhibited significantly higher blood glucose following a glucose load compared to those fed control chow (Figure 2A). In addition, at 36 and 48 wk of exposure, WT male mice fed pesticide chow had significantly higher fasting blood glucose ( and , respectively) compared to those fed control chow ( and , respectively) (Figure 2C). WT female mice fed pesticide chow also exhibited significantly higher fasting blood glucose at 36 and 48 wk (Figure 2D) and significantly higher blood glucose after a glucose load at 48 wk (Figure 2B) ( and at 36 and 48 wk, respectively) compared to those fed control chow ( and at 36 and 48 wk, respectively). After 16, 36, and 48 wk of exposure, WT female mice exhibited significantly higher fasting blood glucose than WT male mice (Table S3). This difference was observed in both mice that were fed pesticide chow and those fed control chow. No significant differences in insulin levels were observed in WT males (Figure S6A) or females (Figure S6B) fed pesticide chow compared to those fed control chow. Further biochemical analyses of serum revealed significantly higher ALT activity in WT males fed pesticide chow than in those fed control chow (Figure 3A).
Figure 2.
Fasting blood glucose in male (A and C) and female (B and D) mice after administration of an intraperitoneal [ body weight (BW); 16 wk] or an oral ( BW; 36 and 48 wk) glucose load. (A and B) Line graphs show blood glucose 15 min before and up to 120 min after administration of glucose load. Inset shows the area under the curve for mice fed control chow (C) and those fed pesticide chow (P). (C and D) Bar graphs show blood glucose before administration of glucose load and after a 6-h fasting period in mice fed control chow (C) and in those fed pesticide chow (P). Results are presented as error of the mean. * ** ***. p-Values represent the difference between mice fed pesticide chow and those fed control chow as determined using a student’s t-test. mice per group.
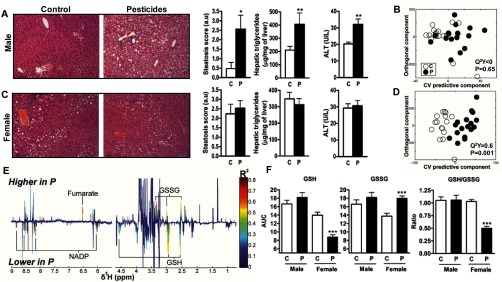
Figure 3.
Evaluation of hepatic steatosis and oxidative stress in male and female mice fed either control chow (C) or pesticide chow (P) for 52 wk. (A and C) Histopathology, steatosis score, hepatic triglycerides (TGs), and alanine transaminase (ALT) levels in male (A) and female (C) mice. Scale bar represents . (B and D) orthogonal projection on latent structure-discriminant analysis (O-PLS-DA) score plots derived from the liver extract spectra of male (B) and female (D) mice. represents the goodness of fit for the PLS-DA models, and p-values were derived using 1,000 permutations of the Y matrix. (E) Coefficient plots related to the O-PLS-DA models discriminating between wild-type (WT) control and WT exposed females. Figure shows the discriminant metabolites that are higher or lower in WT fed pesticide chow (P) compared to WT fed control chow. Metabolites are color-coded according to their correlation coefficient, red indicating a very strong positive correlation (). The direction of the metabolite indicates the group with which it is positively associated as labeled on the diagram. (F) Area under the curve of the spectra was integrated for the glutathione signals (GSSG, oxidized form, multiplet at ; GSH, reduced form, multiplet at ). Data are presented as the error of the mean. * ***. p-Values represent the difference between mice fed pesticide chow and those fed control chow as determined using a student’s t-test. mice per group.
Effect of Eating Pesticide Chow on Liver Metabolism
We further investigated pesticide-induced metabolic perturbations in the liver. Hematoxylin and eosin staining revealed that the livers from pesticide-exposed male mice presented hepatocellular vacuolizations localized predominantly in the centrilobular zone, characteristic of an emerging steatosis (Figure 3A). The steatosis score, calculated using the Kleiner method as described in the “Material and Methods” section (Kleiner et al. 2005), was in WT males fed control chow and in exposed WT male mice. Importantly, exposed male mice had significantly higher levels of hepatic TGs than unexposed male mice ( vs. liver) (Figure 3A). This difference in hepatic TGs between exposed and unexposed males (Figure 3A) correlated with weak but significant differences in serum FFAs and TG levels (Figure S7A) between the two groups without significant impact on the plasma metabolome (Figure S8A). Plasma analysis revealed no significant differences in total or HDL and LDL cholesterol (Figure S7B) between male mice fed pesticide and those fed control chow.
By contrast, females fed the pesticide chow did not exhibit observable differences in liver histology, with steatosis scores of compared to for mice fed control chow (Figure 3C). Moreover, female mice fed pesticide chow did not exhibit higher hepatic TGs or ALT than those fed control chow. Plasma analysis revealed no significant differences in FFA or TG (Figure S7C) or total or HDL cholesterol (Figure S7D) between female mice fed pesticide and those fed control chow, and showed no significant differences in plasma metabolomics (Figure S8B). Exposed females exhibited a slight decrease of LDL cholesterol compared to animals fed control chow (Figure S7D).
Extensive lipid profiling revealed distinct hepatic lipid profiles for WT male and female mice that were fed control and pesticide chow for 52 wk (Figure S9A). Cluster 5 showed lipid species specifically down-regulated in WT males exposed to pesticides. Exposure to pesticide mixture led in WT males to slight but significant changes in the relative PS32:0, PS34:0, and PS36:1 abundance (Figure S9B). Cluster 4 showed lipid species up-regulated in WT males and females exposed to pesticides. It is noteworthy that pesticide exposure led in both WT males and females to increased abundance of SM18:1/16:0, PC30:0, and PC32:0 (Figure S9C). Lipid species down-regulated in WT males and females exposed to pesticides (cluster 2) were C18:2n-6, Cer d18:1/26:1, and SM18:1/24:1 (Figure S9D). The relative TG51, TG53, PI36:0 abundance (Cluster 1) was up-regulated in WT males exposed to pesticides, while exposed WT females exhibited higher PI36:0 (Figure S9E).
Metabolomics
We used metabolomics of aqueous liver extracts to evaluate the metabolic profiles of WT male (Figure 3B) and female (Figure 3D) mice fed control or pesticide chow for 52 wk. Whereas there was no significant difference detected in male mice (Figure 3B), we observed a clear discrimination between the metabolic profiles of female mice fed control and pesticide chow (see also partially assigned 600 MHz 1D NMR spectra of aqueous liver extract in Figure S10A and and assignments for identified metabolites in Table S4). Analysis of the discriminant metabolites revealed higher levels of fumarate and oxidized glutathione (GSSG) and lower levels of nicotinamide adenine dinucleotide phosphate and reduced glutathione (GSH) in the livers of pesticide-exposed females (Figures 3E and 3F). Thus, the GSH/GSSG ratio was significantly lower in the livers of exposed females (Figure 3F).
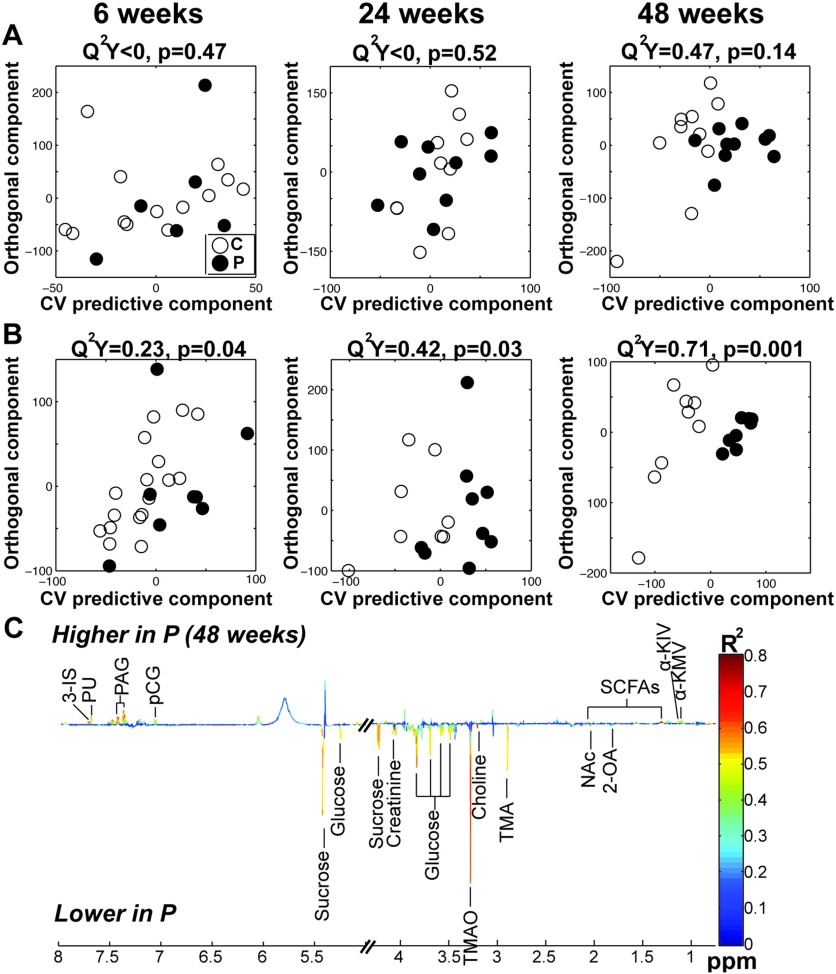
metabolomics of urinary samples was performed after 6 wk, 24 wk, and 48 wk of exposure (Figure 4, Figure S10B, and Table S5). We did not observe any significant difference in the endogenous urinary metabolic profiles of exposed and nonexposed males (Figure 4A). In contrast, and confirming our previous metabolomics observations in liver extracts, pesticide exposure induced significant urinary metabolic changes in females. After only 6 wk of exposure, an O-PLS-DA statistical model was fitted that could significantly discriminate exposed females from their nonexposed counterparts (parameters of the O-PLS-DA model: ; ; Figure 4B). The metabolic profiles were differentiated mainly by the level of 2-ketoadipic acid (2-KAA), the identification of which was confirmed by additional two dimensional sequences and spike in NMR experiments (Figures S11A and B). This metabolite was undetectable in urine samples from male mice (both those fed control and those fed pesticide chow), whereas females mice (both those fed control and those fed pesticide chow) exhibited high levels of 2-KAA at the beginning of the experiment (e.g., week 6; Figure S12) . This metabolite decreased gradually upon pesticide exposure, with a 10-fold difference in 2-KAA levels between exposed and nonexposed female mice at 48 wk (Figure S12). At the end of the experiment, the metabolic profiles of WT females fed control and pesticide chow were distinct (parameters of the PLS-DA model: ; ; Figure 4C). WT-exposed females clearly exhibited higher urinary levels of branched-chain keto acids (, , and ) and lower levels of sucrose, glucose, and creatinine than WT females fed control chow (Table S5). We also observed in WT exposed females lower choline levels and its main metabolites, trimethylamine and trimethylamine oxide, which are both derived from dietary choline fermentation by commensal bacteria than in WT females fed control chow (Craciun and Balskus 2012). Other metabolites levels in the urine of pesticide-treated WT females, including the indole derivative 3-indoxyl sulphate and phenyl derivatives phenylacetylglycine and p-cresol glucuronide, which have been demonstrated to be derived from host gut microbiota co-metabolism, were higher than in WT female fed control chow (Wikoff et al. 2009). These gut microbiota–related metabolites were significantly different between WT females fed pesticide and WT females fed control chow after only 48 wk of pesticide exposure.
Figure 4.
Orthogonal projection on latent structure-discriminant analysis (O-PLS-DA) score plots derived from the spectra of urine from male (A) and female (B) mice fed control or pesticide chow for 6, 24, and 48 wk and related coefficient plots (C) Coefficient plots related to the O-PLS-DA models discriminating between wild-type (WT) control and WT exposed females. Figure shows the discriminant metabolites that are higher or lower in WT fed pesticide chow (P) compared to WT fed control chow. Metabolites are color-coded according to their correlation coefficient, red indicating a very strong positive correlation (). The direction of the metabolite indicates the group with which it is positively associated as labeled on the diagram. Note: 2-OA, 2-oxoadipate; 3-IS, 3-indoxyl sulphate; NAc, N-acetyl groups, PAG, phenylacetylglycine; pCG, p-cresol glucuronide; PU, pseudouridine; SCFAs: short-chain fatty acids; TMA, trimethylamine; TMAO, trimethylamine oxide; , ; , ; : .
Nuclear Receptor Activation in Female and Male Mice
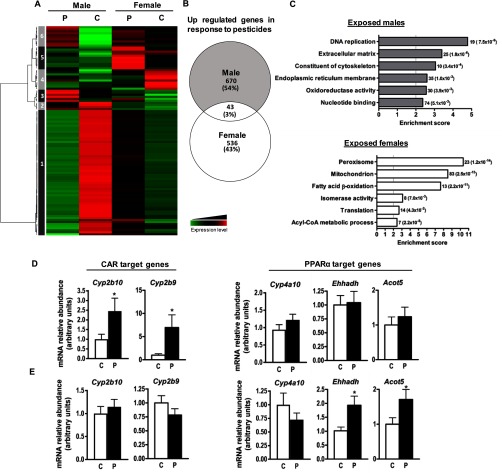
To further investigate the mechanisms involved in the sexually dimorphic response to pesticides, we examined the hepatic transcriptome through microarray analysis of six samples per group. Differentially expressed genes were classified using hierarchical clustering, which illustrated distinct gene regulation in response to pesticides between the two sexes (Figure 5A). One hundred and two probes were selected as differentially regulated in response to pesticide exposure in males and females ( and ). Six clusters of genes were identified (Figure 5A); clusters 1 and 4 clearly highlight genes down-regulated upon pesticide exposure in WT males and females, respectively. Clusters 5 in WT females and clusters 3 and 6 in WT males show genes whose expression was up-regulated in response to pesticides. The Venn diagram in Figure 5B represents the numbers of hepatic genes specifically upregulated in response to pesticides in male and female mice selected with a p-value in order to get a larger regulated gene sampling. A total of 536 genes (43%) were significantly up-regulated in females and 670 (54%) in males upon pesticide exposure, among which only 43 were common between the two sexes (Figure 5B). For the down-regulated genes, 511 (36.4%) were significantly altered in females, 853 (60.8%) significantly altered in males, and 38 were common (Figure S13).
Figure 5.
Hepatic gene expression profile in male and female mice fed control (C) or pesticide (P) chow for 52 wk. One hundred and two probes were selected as differentially regulated in response to pesticide exposure in males and females ( and ). per group. (A) Heat map showing averaged gene expression values per condition, with hierarchical clustering obtained from individual expression values using Pearson correlation coefficient as distance and the Ward’s criterion for agglomeration. Red and green indicate values above and below the mean averaged centered and scaled expression values (z-score), respectively. Black indicates values close to the mean. According to the probe clustering (dendrogram, left panel), six probe clusters showed specific gene expression profiles. (B) Venn diagram representing the numbers of hepatic genes specifically upregulated in response to pesticides in male and female mice (). (C) Gene ontology pathway analysis of the 670 and 536 significantly induced genes in male and female mice, respectively. Histograms show the enrichment score for each identified pathway. Gene number and the corresponding p-value are indicated to the right of the histograms. The relative gene expression of Cyp2b9, Cyp2b10, Cyp4a10, Ehhadh, and Acot5 was confirmed by RT-qPCR in male (D) and female (E) mice ( per group). Data represent error of the mean. * compared to mice fed control chow as determined using a student’s t-test.
Pathway enrichment analysis identified the top biological functions associated with the genes identified as up-regulated in response to pesticide exposure for WT male and female mice (Figure 5C). In WT pesticide-exposed male mice, we observed increased expression of genes linked to DNA replication (19 genes), extracellular matrix (25 genes), cytoskeleton (10 genes), endoplasmic reticulum membrane (35 genes), oxidoreductase activity (30 genes), and nucleotide binding (74 genes) compared to WT males fed control chow (Figure 5C). Among the genes involved in the oxidoreductase activity pathway, Cyp2b9 and Cyp2b10 exhibited the largest differences in mRNA expression between WT male mice fed control chow and those fed pesticide chow (3.4-fold and 2.5-fold increases, respectively) (Figure 5D).
In WT female mice, the genes up-regulated by pesticide exposure were associated primarily with the peroxisome (23 genes), mitochondrion (83 genes), fatty acid oxidation and acyl-coA metabolic processes (20 genes), isomerase activity (8 genes), and translation processes (14 genes) (Figure 5C). The upregulation of Ehhadh and Acot5, two genes involved in fatty acid oxidation, was confirmed by Quantitative Reverse Transcription - Polymerase Chain Reaction (RT-qPCR) of the 18 liver samples (Figure 5E). Additional comparison of our regulated genes upon exposure to pesticide with previously published lists of target genes show that 7.6% of genes regulated in response to pesticides in females are well-described target genes of (Figure S14A). Analysis of the expression profile for the 41 hepatic genes identified in our study and reported as related to –dependent pathways revealed that pesticide exposure changed several gene expressions, mainly in WT females (Figure S14B).
Evaluation of the Pesticide-Detoxifying Capacities of Male and Female Mice
Next, we focused the analysis of the hepatic microarray on genes involved in xenobiotic metabolism in males compared to females. Analysis of 82 genes present in the microarray and involved in this pathway revealed that pesticide exposure significantly affected 14 xenobiotic metabolism enzymes in WT males compared to only three in WT females (Figure 6A). These data show higher xenobiotic metabolic activity in the livers of exposed WT males compared to WT females.
Figure 6.
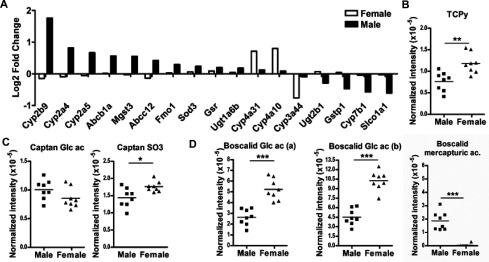
Expression profiles of hepatic genes and high-performance liquid chromatography (HPLC)–high-resolution mass spectrometry (HRMS) features of pesticide metabolites identified in 24-h urine samples from exposed male and female mice fed pesticide chow for 48 or 52 wk, respectively. (A) Expression profiles of hepatic genes involved in detoxifying pathways in females and males in response to pesticides after 52 wk. A list of 14 and 3 probes was selected as differentially regulated in response to pesticide exposure in males and females, respectively (; per group). (B–D) Normalized intensities of (B) one feature of one metabolite of chlorpyrifos: [deprotonated molecular ion (M-H)] of 3,5,6-trichloro-2-pyridinol (TCPy); (C) two features of two metabolites of captan: [M-H] of tetrahydrophthalimide (THPI) conjugated to sulphate (captan S03) and [(M-GlcAc)-H]- of THPI conjugated to glucuronic acid (captan Glc ac); and (D) three features of three metabolites of boscalid: [M-H]- of 2-chloro-N-(4′-chloro-5-hydroxybiphenyl-2-yl)nicotinamide conjugated to glucuronic acid [Boscalid Glc ac (a)], [(M-GlcAc)-H]- of 2-chloro-N-(4′-chloro-5-hydroxybiphenyl-2-yl) nicotinamide conjugated to glucuronic acid [Boscalid Glc ac (b)], and the [M-H]- feature of the mercapturic conjugation of boscalid metabolite (Boscalid mercapturic ac) in male and female mice fed pesticide chow for 48 wk. *, **, ***. p-Values represent differences between male and female mice fed pesticide chow for 52 wk. mice per group.
Thus, we used an untargeted LC-MS–based exposomics approach (Jamin et al. 2014) to compare the urinary end products of pesticide metabolism after 52 wk of exposure (Figures 6B–D). Among the six assayed pesticides, only metabolites of boscalid, captan, and chlorpyrifos were clearly identified (Figure S2). Pesticides were confirmed to be absent in controls. Only one suspected metabolite of thiachlopride and two metabolites of thiophanate were detected according to their exact mass (oxazole derivate of thiachlopride, hydroxylated and oxidized derivates of thiophanate), but could not be confirmed by MS-MS due to their weak signals. Three suspected metabolites of ziram were detected (dithiocarbamic acid conjugated or not to sulfate or glucuronic acid), but these nonspecific structures were also detected in urine of nonexposed mice. Therefore, metabolites of thiachlopride, thiophanate, and ziram were not further considered for the exposomic data analysis. The metabolism of chlorpyrifos, boscalid, and captan differed between WT male and female mice fed pesticide chow for 52 wk (Figures 6B–D). Exposed WT females exhibited higher levels than exposed WT males of 3,5,6-trichloro-2-pyridinol (TCPy), a metabolite resulting from either the dearylation of chlorpyrifos oxon or its oxidation by Cyp3a11 (Croom et al. 2010). WT Females also presented higher levels of sulfated captan and boscalid–glucuronide metabolites than exposed WT males. Therefore, despite less transcriptional regulation of hepatic xenobiotic metabolizing enzymes in the liver, WT females appeared to efficiently metabolize these three pesticides, suggesting that the metabolism of pesticides did not only occur in the liver. Interestingly, the mercapturic acid metabolite of boscalid was only found in WT males and was almost undetectable in WT females.
Role of Constitutive Androstane Receptor in Pesticide-Induced Metabolic Perturbations
We investigated the involvement of a key player in the transcriptional regulation of xenobiotic metabolizing enzymes, the nuclear receptor CAR. We utilized a constitutive CAR knockout mouse model to evaluate the role of CAR in pesticide-induced metabolic perturbations. We compared male and female mice fed pesticide chow for 52 wk with male and female mice fed control chow for 52 wk (18 mice per group). Male mice fed pesticide chow did not have significantly different BWs than those fed control chow (Figure 7A). Moreover, male mice fed control chow and those fed pesticide chow exhibited similar blood glucose levels following a glucose bolus at 16, 36, and 48 wk (Figure S15A). In the liver of males, pesticide exposure decreased the expression of Cyp2b9 and did not alter the expression of Cyp2b10 (Figure 8C). It is noteworthy that pesticide exposure significantly decreased the prototypical target gene (Cyp4a10) in the male mice, which was not observed in WT male mice (Figure 5D). Finally, in male mice, knock out of CAR did not result in significant differences in the urinary levels of pesticide metabolites (Figure 8A). analysis of plasma samples showed no significant differences in serum metabolomics between male mice fed control chow and those fed pesticide chow (Figure S8C).
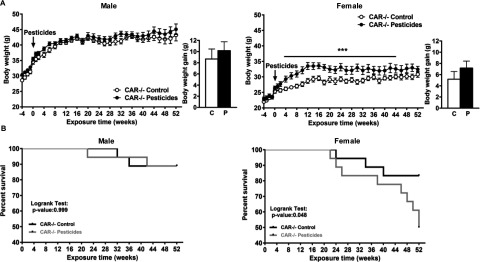
Figure 7.
Body weight (A) and survival (B) of male and female constitutive androstane receptor mice fed control (C) or pesticide (P) chow for 52 wk. (A) The line graphs show body weight (BW) for pesticide-exposed mice (black circles) and mice fed control chow (white circles) from 4 wk prior to exposure through 52 wk. The bar graphs show the gains in BW for pesticide-exposed mice after 52 wk. (B) Kaplan-Meier survival curves.
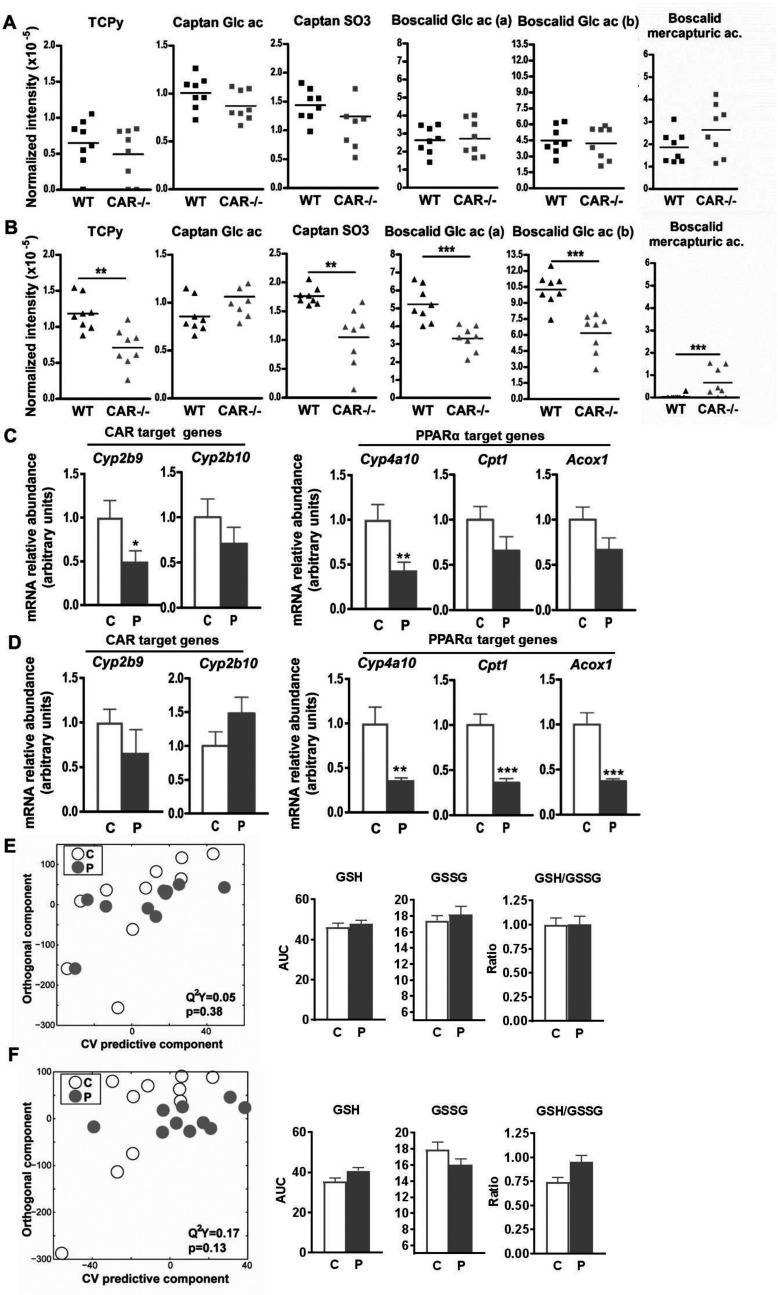
Figure 8.
(A and B) Levels of high-performance liquid chromatography (HPLC)–high-resolution mass spectrometry (HRMS) features of pesticide metabolites identified in urine samples from male (A) and female (B) wild-type (WT) and constitutive androstane receptor mice fed pesticide chow for 52 wk. **; *** compared to WT as determined using a student’s t-test. (C and D) Relative hepatic expression of CAR and target genes as determined by RT-qPCR in male (C) and female (D) mice fed control (C) or pesticide chow (P) for 52 wk. (E and F) Orthogonal projection on latent structure-discriminant analysis (O-PLS-DA) score plots derived from the liver extract –based spectra of male (E) and female (F) mice fed control or pesticide chow for 52 wk. represents the goodness of fit for the PLS-DA models, and p-values were derived using 1,000 permutations of the Y matrix. Area under the curve of the spectra was integrated for the glutathione signals (GSSG, oxidized form, multiplet at ; GSH, reduced form, multiplet at ). Data are presented as error of the mean. *; **; ***. p-Values represent differences between mice fed control chow and those fed pesticide chow as determined using a student’s t-test. Note: C, control; P, pesticide mixtures. mice per group.
In contrast, female mice fed pesticide chow had significantly larger BWs than mice fed control chow beginning at 4 wk and lasting through 48 wk (Figure 7A). At week 12, the weight gain of exposed females was compared to in unexposed females. This significant difference in BW was maintained over 40 wk (Figure 7A). However, the BW of mice fed control chow gradually increased over the duration of the experiment and was not significantly different from that of mice fed pesticide chow from week 44 through week 52. As observed in male mice fed pesticide chow, female mice did not exhibit significantly higher serum glucose levels (either fasting or after a glucose bolus), as was observed for WT animals fed pesticide chow (Figure S15B). Moreover, pesticide exposure led to a high death rate in females (Figure 7B). After 52 wk of pesticide exposure, only 50% of female mice survived, whereas the survival rate was nearly 88.8% in unexposed females. In female WT mice, the survival rate was similar between mice fed control chow and those fed pesticide chow (Figure S16). Knockout of CAR in female mice modified the levels of almost all measured urinary pesticide metabolites (Figure 8B); females had lower levels of chlorpyrifos, captan (captan SO3) and boscalid metabolites compared to their WT counterparts, and higher levels of boscalid mercapturic acid. analysis of plasma samples showed no significant differences in plasma metabolomics between female mice fed control chow and those fed pesticide chow (Figure S8D).
In contrast to WT female mice, for which we identified several genes as up-regulated in mice fed pesticide chow compared to those fed control chow (Figure 5E), we detected significantly lower mRNA expression levels of genes in female mice fed pesticide chow (Cyp4a10, Acox1, and Cpt1; Figure 8D). Finally, metabolic profiling of liver extracts of male mice did not show any significant difference between animals fed control chow and those fed pesticide chow (Figure 8E). Similarly, we observed no significant difference in the metabolic profiles of liver extracts for female mice fed control chow compared to those fed pesticide chow (Figure 8F), in contrast to the results in WT mice (Figure 3D).
Discussion
Metabolic diseases have multifactorial origins, often resulting from a combination of sedentary lifestyles and high-energy diets. The involvement of endocrine-disruptive environmental contaminants in the onset of diabetes and obesity is of growing interest. Various epidemiological (Evangelou et al. 2016), animal (Grün and Blumberg 2009), and mechanistic data (Grün and Blumberg 2009; Kim et al. 2013) support the hypothesis that exposure to several classes of pesticides, including organophosphorus and OCs, may be considered a risk factor for obesity and diabetes. However, in previous animal studies, metabolic disruptions were observed with specific individual compounds, such as DDT, DDE, dieldrin, glyphosate, mancozeb, imidacloprid, or chlorpyrifos (Androutsopoulos et al. 2013; Bhaskar and Mohanty 2014; De Long and Holloway 2017), and often at doses that are too high to be relevant for human exposure (Wang et al. 2014). Moreover, most studies investigate the obesogenic effect of pesticides in a context of high-fat diets (Adigun et al. 2010b; Howell et al. 2015; Lasram et al. 2014; Lassiter et al. 2010; Maqbool et al. 2016) or using a genetically induced model of obesity (Ob/Ob mice) (Mulligan et al. 2017; Peris-Sampedro et al. 2015). In our study, we aimed to use a pesticide exposure protocol relevant to consumers’ exposure. A mixture of six pesticides was incorporated into a standard rodent diet to mimic the exposure of consumers (EFSA), which occurs mainly as a mixture of several chemicals through food intake. We chose to assess a reference dose, the TDI, an estimated amount of a substance in food or drinking water that can be consumed over a lifetime without presenting an appreciable risk to health (EFSA).
Obesogenic Impact of the Pesticide Mixture in Wild-Type Males
We observed that chronic dietary exposure of WT male mice to a mixture of pesticides at levels that are below the TDI resulted in a greater BW gain and adiposity than in WT males fed control chow. This suggests that adipose tissue might be a possible target of pesticides in male mice, as observed previously with several compounds (Chapados et al. 2012). The proadipogenic properties of pesticides has been correlated with their capacity to induce oxidative stress in 3T3-L1 adipocytes in vitro (Shen et al. 2017) and in mice (Armstrong et al. 2013). Importantly, adipose , a major player in WAT homeostasis, has been reported to be activated by several environmental pollutants, including mono-(2-ethylhexyl)phtalate in COS7, C2C12, and 3T3L1 cell lines (Feige et al. 2007) and triflumizole in mouse 3T3-L1 preadipocytes and human multipotent mesenchymal stromal stem cells (Li et al. 2012).
Hepatic lipid accumulation has also been reported in mice upon exposure to perfluoroalkyl acids (Das et al. 2017), perfluorooctanoic acid (Hui et al. 2017), and cadmium (Zhang et al. 2015), and this raises the question whether the hepatic alterations observed in our study are specific to pesticide exposure. In our study, pesticide exposure led not only to steatosis glucose intolerance and elevated blood glucose, but also resulted in greater WAT mass and BW and more circulating FFAs in WT male mice compared to unexposed WT male mice. Previous studies showed that steatosis induced in mice by perfluoroalkyl acids (Das et al. 2017) and perfluorooctanoic acid (Yan et al. 2015) correlates with an increased expression of lipogenic genes in the liver. In the present work, we did not detect significant differences in the mRNA expression of hepatic genes involved in de novo fatty acid synthesis between mice fed control and pesticide chow. It is therefore possible that TGs accumulating in the liver of WT male mice exposed to pesticides originate from adipose tissue lipolysis rather than from hepatic de novo synthesis. Therefore, the molecular mechanism leading to steatosis in our model might be distinct from those described in other studies. However, whether hepatic steatosis is a generic adaptive response of this organ to chronic exposure to pollutants remains an open question. These results demonstrate that chronic low-dose pesticide exposure leads to sex-specific hepatic metabolic disturbances; WT male mice present with significantly increased hepatic TGs, whereas WT females present with metabolic changes related to oxidative stress.
Male WT mice fed pesticide chow exhibited higher mRNA expression of genes involved in hepatic detoxifying pathways compared to those fed control chow, suggesting that pesticide metabolism occurs mainly in the liver. This metabolism was independent of CAR, as male mice exhibited the similar urinary pesticide metabolite profiles as WT males. Liver CAR activation may be linked to the observed increased expression of hepatic genes involved in DNA replication, extracellular matrix, and cytoskeleton constituents, intracellular events that could predispose an individual to hepatocellular adenoma or carcinomas (Huang et al. 2005). The organophosphates, pyrethroids, triazole, and carbamate pesticides have been shown to activate CAR in vitro (Baldwin and Roling 2009; Tamura et al. 2013) or in vivo (Tamura et al. 2013). Thus, it is likely that, at least in our pesticide cocktail, chlorpyrifos (OP) and/or thiophanate (carbamate) may be involved in the activation of CAR in male mice, though one cannot rule out the interaction of the other compounds with CAR.
Diabetogenic Effect of Pesticide Exposure in Females
In female WT mice, there were not significant differences in BW or WAT mass between WT mice fed control chow and those fed pesticide chow. Moreover, female WT mice fed pesticide chow did not experience hepatic steatosis as determined by histology and hepatic TG quantification. However, transcriptomics and metabolomics profiling revealed a significant impact of pesticides in female livers, including a marked oxidative status and activation of signaling pathways, a transcription factor crucial for liver fatty acid degradation [Montagner et al. 2016; Régnier et al. (In press)]. Therefore, pesticide-induced activation of is in good agreement with a reduced hepatic lipid accumulation in the livers of female mice. is also involved in controlling the expression of antioxidant defense enzymes (Anderson et al. 2004), such as those leading to the conjugation of GSH, which could explain at least part of the drop in reduced glutathione liver content in exposed female mice. also leads to the production of free radicals, which could also induce significant oxidative stress (Pessayre 2007). Therefore, we can hypothesize that some of the pesticides or their metabolites may interact with in WT females, leading to increased hepatic lipid catabolism. Previous studies demonstrated that some pesticides can activate . Pyrethrin and diclofop-methyl, for example, has been shown to induce transcriptional activities in in vitro reported gene assays using CV-1 monkey kidney cells and in mice (Takeuchi et al. 2006). The pesticides methiocarb and carbaryl exhibited agonist activities in in vitro reporter gene assays using COS1 cells (Abass and Pelkonen 2013; Fujino et al. 2016). The pesticide adjuvant Toximul® (Stepan Company) was reported to modulate hepatic metabolism through the activation of (Upham et al. 2007). However, no data are yet available regarding a possible interaction of one of the six pesticides of our study with this nuclear receptor.
Blood metabolomic assay on samples from WT exposed and nonexposed male and female mice displayed similar metabolite profiles. However, urinary metabolic profiling using identified 2-KAA as an early biomarker that discriminates pesticide-treated WT females from WT females fed control chow and that may be linked to the observed increase in blood glucose in WT female exposed to pesticides. There was a trend of lower 2-KAA levels in pesticide-exposed WT females after only 6 wk of exposure and a significant difference after 36 and 48 wk of pesticide exposure. A product of lysine and tryptophan degradation, 2-KAA is further degraded in the mitochondria via the DHTKD1 enzyme, and 2-aminoadipic and 2-KAA aciduria have been linked to mutations in DHTKD1 (Hagen et al. 2015). Wang et al. (2013) identified 2-aminoadipic acid, the immediate precursor of 2-KAA, as a predictor of diabetes development in normoglycemic individuals in observational study in human and demonstrated in vivo in mice that it modulates glucose homeostasis, whereas in vitro studies in murine and human islets support an effect of 2-aminoadipic acid on insulin secretion (Wang et al. 2013). Using 40 strains of a well-characterized murine genetic reference population and a multilayered omics approach, Wu et al. (2014) identified Dhtkd1 as a primary regulator of plasmatic 2-aminoadipic acid, explaining variance in fasted glucose and diabetes status. Changes in 2-aminoadipate levels were highly correlated with changes in the Homeostatic Model Assessment (HOMA) index and therefore with insulin sensitivity. Finally, in a cohort of 835 Caucasian adults in the CoLaus study, including 43 diabetics and 792 nondiabetics males and females, 2-aminoadipic acid levels were also lower in the urine of diabetic patients compared to healthy patients (Wu et al. 2014). In this context, our findings suggest that 2-KAA may be an early biomarker of altered glucose homeostasis in WT female mice, and point out the pancreas as a possible early target of pesticides (Kamath and Rajini 2007).
We also observed significant differences in concentration of gut microbiota–related metabolites in the urine of WT female mice fed pesticide chow compared to those fed control chow. We found lower levels of metabolites from the methylamine pathway; trimethylamine is derived from dietary choline fermentation by commensal bacteria and metabolized into trimethylamine oxide in the liver (Craciun and Balskus 2012). We also observed higher levels of 3-indoxylsulfate, a metabolite derived from the conversion of dietary tryptophan to indole by enteric bacteria and further conversion of indole in the liver (Wikoff et al. 2009). Finally, the phenyl derivatives p-cresol glucuronide and phenylacetylglycine, which were also lower in the urine of pesticide-treated WT females, are well-described host gut microbiota cometabolites (Wikoff et al. 2009). The gut microbiota possesses extensive xenobiotic metabolizing capacities (Claus et al. 2016) and may be involved in pesticide metabolism in females, thereby modifying their toxicity. A recent study demonstrated that the gut microbiota is involved in organophosphate metabolism, thereby inducing hepatic gluconeogenesis and glucose intolerance (Velmurugan et al. 2017). Our results suggest that pesticide treatment impacted the gut microbiota of WT female mice. However, these perturbations were observed only after 48 wk of pesticide exposure, after metabolic perturbations (such as hyperglycemia) were set up, which could imply that perturbations in the gut microbiota are a consequence of the pesticide-induced metabolic disorders and not the cause. Complementary experiments would be necessary to fully understand the role of the gut microbiota in the diabetogenic impact of our pesticide mixture in females.
The Role of Constitutive Androstane Receptor in the Dimorphic Response to Pesticide Exposure
One of the main findings of our experiment was that the response to pesticide exposure was sexually dimorphic. This sexual dimorphism appeared to be related to CAR activity. Compared to WT male mice fed control chow, those fed pesticide chow exhibited higher expression levels of xenobiotic metabolizing enzymes in the liver, particularly those reflecting increased CAR activity (Cyp2b9 and Cyp2b10) (Maglich et al. 2002). As expected, this difference in the expression CAR-dependent xenobiotic metabolizing enzymes was not seen with males fed control or pesticide chow. Moreover, pesticide-exposed males did not exhibit greater BW gains or changes in glucose metabolism compared to those fed control chow. As urinary profiles of pesticide metabolites were similar between and WT males, we hypothesize that CAR was likely not involved in pesticide metabolism in males and that CAR-dependent changes in pesticide metabolism cannot account for the observed protective effects in males upon pesticide exposure.
Unlike males, CAR activity and other xenobiotic detoxifying enzymes were not induced in livers from WT females upon pesticide exposure, therefore suggesting extrahepatic pesticide metabolism. However, the urinary pesticide metabolite profile of female mice was different from that of their WT counterparts, suggesting that CAR played a role in pesticide detoxification in females. females fed pesticide chow also exhibited larger BWs throughout most of the experiment and exhibited higher mortality than those fed control chow. Reduced metabolic capacities in female mice may lead to accumulation of the parent compounds and may be more toxic than their metabolites or may accumulate in vital organs, explaining the high rate of mortality. Given our observation that gut microbiota–related metabolites are changed in urine of pesticide-exposed WT females and the fact that CAR is highly expressed in the intestine (Bookout et al. 2006), we speculate that CAR-dependent metabolism occurs in the gut. We additionally found no considerable differences in GSH:GSSG ratios between mice fed control and pesticide chow, suggesting no significant increase in oxidative stress. This may be attributed to the lack of activation, as determined by the similar pattern of expression of its prototypical targets genes in mice fed control and pesticide chow. Moreover, activation of has been shown to be involved in the control of the expression of antioxidant defense enzymes, such as those leading to the conjugation of GSH (Anderson et al. 2004). More experiments would be necessary to confirm these hypothesis. Moreover, the findings of altered target genes in the mice after pesticide exposure may reflect cross-talks between these two nuclear receptors as previously reported by others (Anderson et al. 2004; Rosen et al. 2017). The dimorphic effect of pesticides may also be linked to estrogenic protection. A growing body of evidence demonstrates a protective role of estrogenic signaling in the development of metabolic syndrome (Matic et al. 2013; Riant et al. 2009; Zhu et al. 2013, 2014). Thus, we postulate that in our model, females may be protected from pesticide-induced obesity and hepatic fat accumulation through estrogenic signaling.
A Cocktail Effect of Pesticides
We observed significant metabolic dysregulation in animals fed a pesticide mixture at doses at which each individual pesticide is supposed to not exert any health impact. The observed disturbances may be linked to a cocktail effect resulting from dose addition or a synergistic interaction between two or more compounds. Interactions between the pesticides in the mixture may occur at various levels or on various targets and/or cell signaling pathways (Delfosse et al. 2015; Rizzati et al. 2016). Organophosphates are well known endocrine–metabolic disruptors (Adigun et al. 2010a) acting on liver adenylate cyclase, oxidative status, and proinflammatory markers (Adigun et al. 2010a; Lasram et al. 2015). Neonicotinoides have been shown to disrupt the thyroid (Bhaskar and Mohanty 2014). Carbamate may alter liver glucose, fat, and oxidative balance (Wang et al. 2014), and dicarboximide (captan) has been shown to reduce liver reduced glutathione (Della Morte et al. 1994). Comparison of the metabolic effects of the pesticide mixture vs. those of each of the single pesticides requires further investigations. Such comparisons are needed to understand whether a synergistic deregulation of metabolic pathways or an additive effect of pesticides on the induction of oxidative stress could explain the observed metabolic disruptions.
Conclusions
We demonstrated that a pesticide cocktail to which consumers may be exposed through food intake, induced at a nontoxic dose (TDI level as defined for human exposure but adjusted to the BW of mice) in a mouse model, caused metabolic disruption consistent with diabetic status. Moreover, the sexual dimorphic impact of this pesticide cocktail may be governed by distinct xenobiotic metabolic capacities and distinct nuclear receptor activation. Our results thus question the relevance of TDI levels for individual pesticides when present in a mixture.
Supplemental Material
Acknowledgments
C.L. is the recipient of a PhD grant from Région Occitanie, INRA (Département Santé Animale), and EI-Purpan. This work was supported by grant from Région Occitanie (subvention number 14054410) and Fonds Européens de DEvelopment Regional (FEDER; No. 12055324). T.I.M. is supported by ANR and region Occitanie. We thank all of the members of the EZOP staff, particularly C. Bétoulières and C. Sommer, for their careful and outstanding help from the start of this project. We thank A. Dequesnes, F. Capilla, and L. Monbrun from Anexplo Genotoul MetaToul for their excellent work on plasma biochemistry and tissue histology. All MS and NMR experiments were performed on the instruments of the MetaToul platform, partner of the national infrastructure of metabolomics and fluxomics: MetaboHUB (MetaboHUB-ANR-11-INBS-0010). We thank L. Peyriga and E. Cahoreau from INSA GenoToul MetaToul for their technical support with the NMR analysis. We thank X. Blanc from INRA UPAE (Jouy-en-Josas) for his technical support for the pellets preparation. We thank S. Claus from Reading University for help in metabolite identification. We thank Professor D. Moore (Baylor College of Medicine, Houston, Texas, United States) for providing us with the CAR-deficient mice.
References
- Abass K, Pelkonen O. 2013. The inhibition of major human hepatic cytochrome P450 enzymes by 18 pesticides: comparison of the N-in-one and single substrate approaches. Toxicol In Vitr 27(5):1584–1588, PMID: 22634058, 10.1016/j.tiv.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Adigun AA, Ryde IT, Seidler FJ, Slotkin TA. 2010a. Organophosphate exposure during a critical developmental stage reprograms adenylyl cyclase signaling in PC12 cells. Brain Res 1329:36–44, PMID: 20298678, 10.1016/j.brainres.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adigun AA, Wrench N, Levin ED, Seidler FJ, Slotkin TA. 2010b. Neonatal parathion exposure and interactions with a high-fat diet in adulthood: adenylyl cyclase-mediated cell signaling in heart, liver and cerebellum. Brain Res Bull 81(6):605–612, PMID: 20074626, 10.1016/j.brainresbull.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Eryani L, Wahlang B, Falkner KC, Guardiola JJ, Clair HB, Prough RA, et al. 2015. Identification of environmental chemicals associated with the development of toxicant-associated fatty liver disease in rodents. Toxicol Pathol 43(4):482–497, PMID: 25326588, 10.1177/0192623314549960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SP, Howroyd P, Liu J, Qian X, Bahnemann R, Swanson C, et al. 2004. The transcriptional response to a peroxisome proliferator-activated receptor alpha agonist includes increased expression of proteome maintenance genes. J Biol Chem 279(50):52390–52398, PMID: 15375163, 10.1074/jbc.M409347200. [DOI] [PubMed] [Google Scholar]
- Androutsopoulos VP, Hernandez AF, Liesivuori J, Tsatsakis AM. 2013. A mechanistic overview of health associated effects of low levels of organochlorine and organophosphorous pesticides. Toxicology 307:89–94, PMID: 23041710, 10.1016/j.tox.2012.09.011. [DOI] [PubMed] [Google Scholar]
- Arciello M, Gori M, Maggio R, Barbaro B, Tarocchi M, Galli A, et al. 2013. Environmental pollution: a tangible risk for NAFLD pathogenesis. Int J Mol Sci 14(11):22052–22066, PMID: 24213605, 10.3390/ijms141122052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong LE, Driscoll MV, Donepudi AC, Xu J, Baker A, Aleksunes LM, et al. 2013. Effects of developmental deltamethrin exposure on white adipose tissue gene expression. J Biochem Mol Toxicol 27(2):165–171, PMID: 23401056, 10.1002/jbt.21477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin WS, Roling JA. 2009. A Concentration addition model for the activation of the constitutive androstane receptor by xenobiotic mixtures. Toxicol Sci 107(1):93–105, PMID: 18832183, 10.1093/toxsci/kfn206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, et al. 2007. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc 2(11):2692–2703, PMID: 18007604, 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- Bhaskar R, Mohanty B. 2014. Pesticides in mixture disrupt metabolic regulation: in silico and in vivo analysis of cumulative toxicity of mancozeb and imidacloprid on body weight of mice. Gen Comp Endocrinol 205:226–234, PMID: 24530807, 10.1016/j.ygcen.2014.02.007. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37(8):911–917, PMID: 13671378, 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19(2):185–193, PMID: 12538238, 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. 2006. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126(4):789–799, PMID: 16923397, 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casals-Casas C, Desvergne B. 2011. Endocrine disruptors: from endocrine to metabolic disruption. Annu Rev Physiol 73:135–162, PMID: 21054169, 10.1146/annurev-physiol-012110-142200. [DOI] [PubMed] [Google Scholar]
- Chapados NA, Casimiro C, Robidoux MA, Haman F, Batal M, Blais JM, et al. 2012. Increased proliferative effect of organochlorine compounds on human preadipocytes. Mol Cell Biochem 365(1–2):275–278, PMID: 22350817, 10.1007/s11010-012-1268-0. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162(1):156–159, PMID: 2440339, 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- Claus SP, Guillou H, Ellero-Simatos S. 2016. The gut microbiota: a major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes 2:16003, PMID: 28721242, 10.1038/npjbiofilms.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloarec O, Dumas M-E, Craig A, Barton RH, Trygg J, Hudson J, et al. 2005a. Statistical total correlation spectroscopy: an exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Anal Chem 77(5):1282–1289, PMID: 15732908, 10.1021/ac048630x. [DOI] [PubMed] [Google Scholar]
- Cloarec O, Dumas ME, Trygg J, Craig A, Barton RH, Lindon JC, et al. 2005b. Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in 1H NMR spectroscopic metabonomic studies. Anal Chem 77(2):517–526, PMID: 15649048, 10.1021/ac048803i. [DOI] [PubMed] [Google Scholar]
- Craciun S, Balskus EP. 2012. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci USA 109(52):21307–21312, PMID: 23151509, 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croom EL, Wallace AD, Hodgson E. 2010. Human variation in CYP-specific chlorpyrifos metabolism. Toxicology 276(3):184–191, PMID: 20709133, 10.1016/j.tox.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Das KP, Wood CR, Lin MT, Starkov AA, Lau C, Wallace KB, et al. 2017. Perfluoroalkyl acids-induced liver steatosis: effects on genes controlling lipid homeostasis. Toxicology 378:37–52, PMID: 28049043, 10.1016/j.tox.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Long NE, Holloway AC. 2017. Early-life chemical exposures and risk of metabolic syndrome. Diabetes Metab Syndr Obes 10:101–109, PMID: 28367067, 10.2147/DMSO.S95296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfosse V, Dendele B, Huet T, Grimaldi M, Boulahtouf A, Gerbal-Chaloin S, et al. 2015. Synergistic activation of human pregnane X receptor by binary cocktails of pharmaceutical and environmental compounds. Nat Commun 6:8089, PMID: 26333997, 10.1038/ncomms9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Morte R, Villani GR, Di Martino E, Squillacioti C, De Marco L, Vuotto P, et al. 1994. Glutathione depletion induced in rat liver fractions by seven pesticides. Boll Soc Ital Biol Sper 70(8–9):185–192, PMID: 7893475. [PubMed] [Google Scholar]
- Desvergne B, Michalik L, Wahli W. 2006. Transcriptional regulation of metabolism. Physiol Rev 86(2):465–514, PMID: 16601267, 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- Dieterle F, Ross A, Schlotterbeck G, Senn H. 2006. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem 78(13):4281–4290, PMID: 16808434, 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. 2002. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30(1):207–210, PMID: 11752295, 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority). 2015. The 2013 European Union report on pesticide residues in food. EFSA J 13(3):4038, 10.2903/j.efsa.2015.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelou E, Ntritsos G, Chondrogiorgi M, Kavvoura FK, Hernández AF, Ntzani EE, et al. 2016. Exposure to pesticides and diabetes: a systematic review and meta-analysis. Environ Int 91:60–68, PMID: 26909814, 10.1016/j.envint.2016.02.013. [DOI] [PubMed] [Google Scholar]
- Evans RM, Mangelsdorf DJ. 2014. Nuclear receptors, RXR, and the big bang. Cell 157(1):255–266, PMID: 24679540, 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige JN, Gelman L, Rossi D, Zoete V, Métivier R, Tudor C, et al. 2007. The endocrine disruptor monoethyl-hexyl-phthalate is a selective peroxisome proliferator-activated receptor gamma modulator that promotes adipogenesis. J Biol Chem 282(26):19152–19166, PMID: 17468099, 10.1074/jbc.M702724200. [DOI] [PubMed] [Google Scholar]
- Flurkey K, Currer J, Harrison D. 2007. “Mouse models in aging research.” In: Faculty Research 2000– 2009. https://mouseion.jax.org/stfb2000_2009/1685. [Google Scholar]
- Fujino C, Tamura Y, Tange S, Nakajima H, Sanoh S, Watanabe Y, et al. 2016. Metabolism of methiocarb and carbaryl by rat and human livers and plasma, and effect on their PXR, CAR and PPARα activities. J Toxicol Sci 41(5):677–691, PMID: 27665777, 10.2131/jts.41.677. [DOI] [PubMed] [Google Scholar]
- Giacomoni F, Le Corguillé G, Monsoor M, Landi M, Pericard P, Pétéra M, et al. 2015. Workflow4Metabolomics: a collaborative research infrastructure for computational metabolomics. Bioinformatics 31(9):1493–1495, PMID: 25527831, 10.1093/bioinformatics/btu813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün F, Blumberg B. 2009. Endocrine disrupters as obesogens. Mol Cell Endocrinol 304(1–2):19–29, PMID: 19433244, 10.1016/j.mce.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen J, te Brinke H, Wanders RJ, Knegt AC, Oussoren E, Hoogeboom AJ, et al. 2015. Genetic basis of alpha-aminoadipic and alpha-ketoadipic aciduria. J Inherit Metab Dis 38(5):873–879, PMID: 25860818, 10.1007/s10545-015-9841-9. [DOI] [PubMed] [Google Scholar]
- Hong NS, Kim KS, Lee IK, Lind PM, Lind L, Jacobs DR, et al. 2012. The association between obesity and mortality in the elderly differs by serum concentrations of persistent organic pollutants: a possible explanation for the obesity paradox. Int J Obes (Lond) 36(9):1170–1175, PMID: 21946706, 10.1038/ijo.2011.187. [DOI] [PubMed] [Google Scholar]
- Howell GE III, Mulligan C, Meek E, Chambers JE. 2015. Effect of chronic p, p′-dichlorodiphenyldichloroethylene (DDE) exposure on high fat diet-induced alterations in glucose and lipid metabolism in male C57BL/6H mice. Toxicology 328:112–122, PMID: 25541407, 10.1016/j.tox.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37(1):1–13, PMID: 19033363, 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57, PMID: 19131956, 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang W, Zhang J, Washington M, Liu J, Parant JM, Lozano G, et al. 2005. Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol Endocrinol 19(6):1646–1653, PMID: 15831521, 10.1210/me.2004-0520. [DOI] [PubMed] [Google Scholar]
- Hui Z, Li R, Chen L. 2017. The impact of exposure to environmental contaminant on hepatocellular lipid metabolism. Gene 622:67–71, PMID: 28431976, 10.1016/j.gene.2017.04.024. [DOI] [PubMed] [Google Scholar]
- Jamin EL, Bonvallot N, Tremblay-Franco M, Cravedi JP, Chevrier C, Cordier S, et al. 2014. Untargeted profiling of pesticide metabolites by LC-HRMS: an exposomics tool for human exposure evaluation. Anal Bioanal Chem 406(4):1149–1161, PMID: 23892877, 10.1007/s00216-013-7136-2. [DOI] [PubMed] [Google Scholar]
- Jin Y, Lin X, Miao W, Wu T, Shen H, Chen S, et al. 2014. Chronic exposure of mice to environmental endocrine-disrupting chemicals disturbs their energy metabolism. Toxicol Lett 225(3):392–400, PMID: 24440342, 10.1016/j.toxlet.2014.01.006. [DOI] [PubMed] [Google Scholar]
- Kamath V, Rajini P. 2007. Altered glucose homeostasis and oxidative impairment in pancreas of rats subjected to dimethoate intoxication. Toxicology 231(2-3):137–146, PMID: 17197067, 10.1016/j.tox.2006.11.072. [DOI] [PubMed] [Google Scholar]
- Karmaus W, Osuch JR, Eneli I, Mudd LM, Zhang J, Mikucki D, et al. 2009. Maternal levels of dichlorodiphenyl-dichloroethylene (DDE) may increase weight and body mass index in adult female offspring. Occup Environ Med 66(3):143–149, PMID: 19060027, 10.1136/oem.2008.041921. [DOI] [PubMed] [Google Scholar]
- Kim J, Park Y, Yoon KS, Clark JM, Park Y. 2013. Imidacloprid, a neonicotinoid insecticide, induces insulin resistance. J Toxicol Sci 38(5):655–660, PMID: 24025781, 10.2131/jts.38.655. [DOI] [PubMed] [Google Scholar]
- Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, et al. 2005. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 41(6):1313–1321, PMID: 15915461, 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- Klipper-Aurbach Y, Wasserman M, Braunspiegel-Weintrob N, Borstein D, Peleg S, Assa S, et al. 1995. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med Hypotheses 45(5):486–490, PMID: 8748093, 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
- Lasram MM, Dhouib IB, Bouzid K, Lamine AJ, Annabi A, Belhadjhmida N, et al. 2014. Association of inflammatory response and oxidative injury in the pathogenesis of liver steatosis and insulin resistance following subchronic exposure to malathion in rats. Environ Toxicol Pharmacol 38(2):542–553, PMID: 25180440, 10.1016/j.etap.2014.08.007. [DOI] [PubMed] [Google Scholar]
- Lasram MM, El-Golli N, Lamine AJ, Douib IB, Bouzid K, Annabi A, et al. 2015. Changes in glucose metabolism and reversion of genes expression in the liver of insulin-resistant rats exposed to malathion. The protective effects of N-acetylcysteine. Gen Comp Endocrinol 215:88–97, PMID: 25449180, 10.1016/j.ygcen.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Lassiter TL, Ryde IT, Levin ED, Seidler FJ, Slotkin TA. 2010. Neonatal exposure to parathion alters lipid metabolism in adulthood: interactions with dietary fat intake and implications for neurodevelopmental deficits. Brain Res Bull 81(1):85–91, PMID: 19615431, 10.1016/j.brainresbull.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Pham HT, Janesick AS, Blumberg B. 2012. Triflumizole is an obesogen in mice that acts through peroxisome proliferator activated receptor gamma (PPARγ). Environ Health Perspect 120(12):1720–1726, PMID: 23086663, 10.1289/ehp.1205383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YF, Jin T, Xu Y, Zhang D, Wu Q, Zhang YK, et al. 2013. Sex differences in the circadian variation of cytochrome p450 genes and corresponding nuclear receptors in mouse liver. Chronobiol Int 30(9):1135–1143, PMID: 23926955, 10.3109/07420528.2013.805762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. 2002. Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol 62(3):638–646, PMID: 12181440, 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- Maqbool F, Mostafalou S, Bahadar H, Abdollahi M. 2016. Review of endocrine disorders associated with environmental toxicants and possible involved mechanisms. Life Sci 145:265–273, PMID: 26497928, 10.1016/j.lfs.2015.10.022. [DOI] [PubMed] [Google Scholar]
- Matic M, Bryzgalova G, Gao H, Antonson P, Humire P, Omoto Y, et al. 2013. Estrogen signalling and the metabolic syndrome: targeting the hepatic estrogen receptor alpha action. PLoS One 8(2):e57458, PMID: 23451233, 10.1371/journal.pone.0057458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagner A, Polizzi A, Fouché E, Ducheix S, Lippi Y, Lasserre F, et al. 2016. Liver PPARα is crucial for whole-body fatty acid homeostasis and is protective against NAFLD. Gut 65(7):1202–1214, PMID: 26838599, 10.1136/gutjnl-2015-310798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafalou S, Abdollahi M. 2017. Pesticides: an update of human exposure and toxicity. Arch Toxicol 91(2):549–599, PMID: 27722929, 10.1007/s00204-016-1849-x. [DOI] [PubMed] [Google Scholar]
- Mulligan C, Kondakala S, Yang EJ, Stokes JV, Stewart JA, Kaplan BL, et al. 2017. Exposure to an environmentally relevant mixture of organochlorine compounds and polychlorinated biphenyls promotes hepatic steatosis in male Ob/Ob mice. Environ Toxicol 32(4):1399–1411, PMID: 27533883, 10.1002/tox.22334. [DOI] [PubMed] [Google Scholar]
- Newbold RR, Padilla-Banks E, Jefferson WN. 2009. Environmental estrogens and obesity. Mol Cell Endocrinol 304(1–2):84–89, PMID: 19433252, 10.1016/j.mce.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris-Sampedro F, Cabré M, Basaure P, Reverte I, Domingo JL, Teresa Colomina M. 2015. Adulthood dietary exposure to a common pesticide leads to an obese-like phenotype and a diabetic profile in apoE3 mice. Environ Res 142:169–176, PMID: 26162960, 10.1016/j.envres.2015.06.036. [DOI] [PubMed] [Google Scholar]
- Pessayre D. 2007. Role of mitochondria in non-alcoholic fatty liver disease. J Gastroenterol Hepatol 22 (suppl 1):S20–S27, PMID: 17567459, 10.1111/j.1440-1746.2006.04640.x. [DOI] [PubMed] [Google Scholar]
- Polyzos SA, Kountouras J, Zavos C, Deretzi G. 2012. Nonalcoholic fatty liver disease: multimodal treatment options for a pathogenetically multiple-hit disease. J Clin Gastroenterol 46(4):272–284, 10.1097/MCG.0b013e31824587e0. [DOI] [PubMed] [Google Scholar]
- Qatanani M, Moore DD. 2005. CAR, the continuously advancing receptor, in drug metabolism and disease. Curr Drug Metab 6(4):329–339, PMID: 16101572, 10.2174/1389200054633899. [DOI] [PubMed] [Google Scholar]
- Régnier M, Polizzi A, Lippi Y, Fouché E, Michel G, Lukowicz C, et al. in Press. Insights into the role of hepatocyte PPARα activity in response to fasting. Mol Cell Endocrinol, PMID: 28774777, 10.1016/j.mce.2017.07.035. [DOI] [PubMed] [Google Scholar]
- Riant E, Waget A, Cogo H, Arnal J-F, Burcelin R, Gourdy P. 2009. Estrogens protect against high-fat diet-induced insulin resistance and glucose intolerance in mice. Endocrinology 150(5):2109–2117, PMID: 19164473, 10.1210/en.2008-0971. [DOI] [PubMed] [Google Scholar]
- Rizzati V, Briand O, Guillou H, Gamet-Payrastre L. 2016. Effects of pesticide mixtures in human and animal models: an update of the recent literature. Chem Biol Interact 254:231–246, PMID: 27312199, 10.1016/j.cbi.2016.06.003. [DOI] [PubMed] [Google Scholar]
- Rosen MB, Das KP, Rooney J, Abbott B, Lau C, Corton JC. 2017. PPARα-independent transcriptional targets of perfluoroalkyl acids revealed by transcript profiling. Toxicology 387:95–107, PMID: 28558994, 10.1016/j.tox.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler FJ, Slotkin TA. 2011. Developmental neurotoxicity targeting hepatic and cardiac sympathetic innervation: effects of organophosphates are distinct from those of glucocorticoids. Brain Res Bull 85(3–4):225–230, PMID: 21453761, 10.1016/j.brainresbull.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P, Hsieh TH, Yue Y, Sun Q, Clark JM, Park Y. 2017. Deltamethrin increases the fat accumulation in 3T3-L1 adipocytes and Caenorhabditis elegans. Food Chem Toxicol 101:149–156, PMID: 28119079, 10.1016/j.fct.2017.01.015. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. 2006. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78(3):779–787, PMID: 16448051, 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Smyth GK. 2004. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat Appl Genet Mol Biol 3(1):1–25, PMID: 16646809, 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Takeuchi S, Matsuda T, Kobayashi S, Takahashi T, Kojima H. 2006. In vitro screening of 200 pesticides for agonistic activity via mouse peroxisome proliferator-activated receptor (PPAR)α and PPARγ and quantitative analysis of in vivo induction pathway. Toxicol Appl Pharmacol 217(3):235–244, PMID: 17084873, 10.1016/j.taap.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Tamura K, Inoue K, Takahashi M, Matsuo S, Irie K, Kodama Y, et al. 2013. Dose-response involvement of constitutive androstane receptor in mouse liver hypertrophy induced by triazole fungicides. Toxicol Lett 221(1):47–56, PMID: 23721867, 10.1016/j.toxlet.2013.05.011. [DOI] [PubMed] [Google Scholar]
- Tautenhahn R, Böttcher C, Neumann S. 2008. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 9:504, PMID: 19040729, 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trygg J, Wold S. 2002. Orthogonal projections to latent structures (O-PLS). J Chemom 16(3):119–128. 10.1002/cem.695. [DOI] [Google Scholar]
- Upham J, Acott PD, O'Regan P, Sinal CJ, Crocker JF, Geldenhuys L, et al. 2007. The pesticide adjuvant, Toximul™, alters hepatic metabolism through effects on downstream targets of PPARα. Biochim Biophys Acta 1772(9):1057–1064, PMID: 17643967, 10.1016/j.bbadis.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Velmurugan G, Ramprasath T, Swaminathan K, Mithieux G, Rajendhran J, Dhivakar M, et al. 2017. Gut microbial degradation of organophosphate insecticides-induces glucose intolerance via gluconeogenesis. Genome Biol 18(1):8, PMID: 28115022, 10.1186/s13059-016-1134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselkov KA, Lindon JC, Ebbels TM, Crockford D, Volynkin VV, Holmes E, et al. 2009. Recursive segment-wise peak alignment of biological 1H NMR spectra for improved metabolic biomarker recovery. Anal Chem 81(1):56–66, PMID: 19049366, 10.1021/ac8011544. [DOI] [PubMed] [Google Scholar]
- Wahlang B, Falkner KC, Gregory B, Ansert D, Young D, Conklin DJ, et al. 2013. Polychlorinated biphenyl 153 is a diet-dependent obesogen that worsens nonalcoholic fatty liver disease in male C57BL6/J mice. J Nutr Biochem 24(9):1587–1595, PMID: 23618531, 10.1016/j.jnutbio.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Ngo D, Psychogios N, Dejam A, Larson MG, Vasan RS, et al. 2013. 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest 123(10):4309–4317, PMID: 24091325, 10.1172/JCI64801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Wang HP, Xu MY, Liang YJ, Sun YJ, Yang L, et al. 2014. Combined subchronic toxicity of dichlorvos with malathion or pirimicarb in mice liver and serum: a metabonomic study. Food Chem Toxicol 70:222–230, PMID: 24907623, 10.1016/j.fct.2014.05.027. [DOI] [PubMed] [Google Scholar]
- Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. 2000. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature 407(6806):920–923, PMID: 11057673, 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. 2009. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA 106(10):3698–3703, PMID: 19234110, 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Williams EG, Dubuis S, Mottis A, Jovaisaite V, Houten SM, et al. 2014. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell 158(6):1415–1430, PMID: 25215496, 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Wang J, Dai J. 2015. Activation of sterol regulatory element-binding proteins in mice exposed to perfluorooctanoic acid for 28 days. Arch Toxicol 89(9):1569–1578, PMID: 25092180, 10.1007/s00204-014-1322-7. [DOI] [PubMed] [Google Scholar]
- Zhang S, Jin Y, Zeng Z, Liu Z, Fu Z. 2015. Subchronic exposure of mice to cadmium perturbs their hepatic energy metabolism and gut microbiome. Chem Res Toxicol 28(10):2000–2009, PMID: 26352046, 10.1021/acs.chemrestox.5b00237. [DOI] [PubMed] [Google Scholar]
- Zhu L, Brown WC, Cai Q, Krust A, Chambon P, McGuinness OP, et al. 2013. Estrogen treatment after ovariectomy protects against fatty liver and may improve pathway-selective insulin resistance. Diabetes 62(2):424–434, PMID: 22966069, 10.2337/db11-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Martinez MN, Emfinger CH, Palmisano BT, Stafford JM. 2014. Estrogen signaling prevents diet-induced hepatic insulin resistance in male mice with obesity. Am J Physiol Endocrinol Metab 306(10):E1188–E1197, PMID: 24691030, 10.1152/ajpendo.00579.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.