Abstract
Glycine is an inhibitory neurotransmitter in the brainstem and spinal cord. Glycine transporter 2 (GLYT2) is responsible for the uptake of extracellular glycine. GLYT2 is specifically expressed in glycinergic neurons and thus has been used as a marker of glycinergic neurons. Here, we generated GLYT2 promotor-driven Cre recombinase (Cre)-expressing mice (GLYT2-Cre knock-in mice) to develop a tool for manipulating gene expression in glycinergic neurons. Cre activity was examined by crossing the GLYT2-Cre knock-in mice with a Cre reporter mouse line, R26R, which express β-galactosidase (β-gal) in a Cre-dependent manner. X-gal staining of GLYT2-Cre/R26R double transgenic mouse brains and spinal cords revealed that the Cre activity was primarily distributed in the brainstem, cerebellum, and spinal cord. These areas are rich in glycinergic neurons. Furthermore, we performed immunohistochemistry for β-gal combined with in situ hybridization for GLYT2 in the GLYT2-Cre/R26R double transgenic mouse brains to determine whether Cre activity is specifically localized to glycinergic neurons. The β-gal protein and GLYT2 mRNAs were colocalized in the cerebellar Golgi cells, dorsal cochlear nucleus, gigantocellular reticular nucleus, spinal trigeminal nucleus, nucleus of the trapezoid body, and lateral lemniscus. More than 98% of the GLYT2 mRNA-expressing cells in these brain regions also expressed β-gal, whereas 90–98% of the β-gal-positive cells expressed the GLYT2 mRNAs. Thus, Cre activity is specifically localized to glycinergic neurons with high fidelity in the GLYT2-Cre knock-in mice. The GLYT2-Cre knock-in mouse line will be a useful tool for studying glycinergic neurons and neurotransmission.
Keywords: Glycine, Glycinergic neuron, GLYT2, Cre recombinase, Knock-in
Abbreviations:
- 4V
fourth ventricle
- 7
facial nucleus
- 7n
facial nerve
- 8n
vestibulocochlear nerve
- 12
hypoglossal nucleus
- Aq
aqueduct
- BAC
bacterial artificial chromosome
- β-gal
β-galactosidase
- Cre
Cre recombinase
- DAPI
4′,6-diamidino-2-phenylindole dihydrochloride
- DC
dorsal cochlear nucleus
- DIG
digoxigenin
- DTg
dorsal tegmental nucleus
- EGFP
enhanced green fluorescent protein
- GC
granule cell layer
- Gi
gigantocellular reticular nucleus
- GiA
gigantocellular reticular nucleus, alpha part
- GLYT
glycine transporter
- GLYT1
glycine transporter 1
- GLYT2
glycine transporter 2
- GM
gray matter
- Gr
granular layer
- IC
inferior colliculus
- icp
inferior cerebellar peduncle
- Int
interposed cerebellar nucleus
- IO
inferior olive
- Lat
lateral cerebellar nucleus
- MdD
medullary reticular nucleus, dorsal part
- MdV
medullary reticular nucleus, ventral part
- Mo
molecular layer
- MVe
medial vestibular nucleus
- neo
neomycin-resistant gene expression cassette
- Pa4
paratrochlear nucleus
- PBS
phosphate-buffered saline
- PC
Purkinje cell layer
- Pn
pontine nuclei
- PnC
pontine reticular nucleus, caudal part
- PnO
pontine reticular nucleus, oral part
- Pr
prepositus nucleus
- Pr5
principal sensory trigeminal nucleus
- py
pyramidal tract
- RMg
raphe magnus nucleus
- SC
superior colliculus
- SO
superior olive
- SP5
spinal trigeminal nucleus
- TBS
Tris-buffered saline
- TBST
TBS containing 0.1% Tween-20
- Tz
nucleus of the trapezoid body
- VC
ventral cochlear nucleus
- VLL
ventral nucleus of the lateral lemniscus
- WM
white matter
1. Introduction
Glycine is a major inhibitory neurotransmitter in the brainstem and spinal cord, and glycinergic neurotransmission participates in a variety of functions, such as sensory processing, motor rhythm generation, and control of the respiratory network (Legendre, 2001, Ezure et al., 2003, Yevenes and Zeilhofer, 2011). Glycine is released from presynaptic terminals in glycinergic neurons through a vesicular mechanism (Wojcik et al., 2006; Saito et al., 2010) and acts on postsynaptic glycine receptors (GlyRs) (Laube et al., 2002). GlyR activation leads to an influx of chloride to hyperpolarize and subsequently inhibit postsynaptic neurons (Aragón and López-Corcuera, 2003). Extracellular glycine is transported to the intracellular space by two types of glycine transporters (GLYTs), GLYT1 and GLYT2, which are encoded by two separate genes (Ebihara et al., 2004, Betz et al., 2006). Both GLYTs belong to a large family of Na+/Cl−-dependent transporter proteins (Nelson, 1998). GLYT1 and GLYT2 have different functions at glycinergic synapses. GLYT1 eliminates glycine from the synaptic cleft, whereas GLYT2 is essential for glycine uptake into the presynaptic terminal and the subsequent refilling of synaptic vesicles with glycine. Deficiencies of GLYT1 and GLYT2 cause the opposite effects on glycinergic neurotransmission. GLYT1 deficiency causes an increased extracellular glycine, which leads to a sustained activation of GlyRs. This effect generates symptoms found in human glycine encephalopathy. On the other hand, GLYT2 deficiency causes the decreased inhibitory postsynaptic currents via GlyRs and induces human hyperekplexia phenotype in vivo (Gomeza et al., 2003a, Gomeza et al., 2003b, Eulenburg et al., 2005, Rees et al., 2006, Carta et al., 2012). GLYT1 is predominantly expressed in glial cells of the brain, whereas GLYT2 is specifically expressed in glycinergic neurons and is a reliable marker of glycinergic neurons (Poyatos et al., 1997).
Genetically modified mice have become a powerful tool in the analysis of neural networks. The Cre/loxP system, in which the Cre recombinase (Cre) transgene activates a reporter gene by inducing recombination at loxP sites, is a widely used approach. In addition, this system facilitates cell-specific gene expression and inactivation in the mouse (Wouterlood et al., 2014). We generated a GLYT2-Cre knock-in mouse line using knock-in techniques to develop a tool for the manipulation of gene expression in glycinergic neurons and the inactivation of the endogenous GLYT2 gene. GLYT2-Cre transgenic mouse lines have been developed using bacterial artificial chromosomes (BACs), which contains more than 100 kb DNA sequences (Ishihara et al., 2010, Foster et al., 2015, Rahman et al., 2015). BAC-based transgenesis can result in ectopic expression due to the lack of all cis-regulatory elements required for proper expression and random integration into the genome (Harris et al., 2014). We generated a GLYT2-Cre knock-in mouse line expressing Cre under the control of the endogenous GLYT2 promoter using the knock-in strategy to overcome this problem. The knock-in strategy uses homologous recombination in ES cells to insert the Cre into the endogenous gene locus. The knock-in strategy generally captures endogenous gene expression patterns better than BAC transgenic strategies (Harris et al., 2014).
In the present study, we report the generation and characterization of a GLYT2-Cre knock-in mouse line. Cre activity is efficient and restricted to glycinergic neurons in the GLYT2-Cre knock-in mice. The GLYT2-Cre knock-in mouse line is very useful for studies of glycinergic neurons and neurotransmission.
2. Materials and methods
2.1. Mice
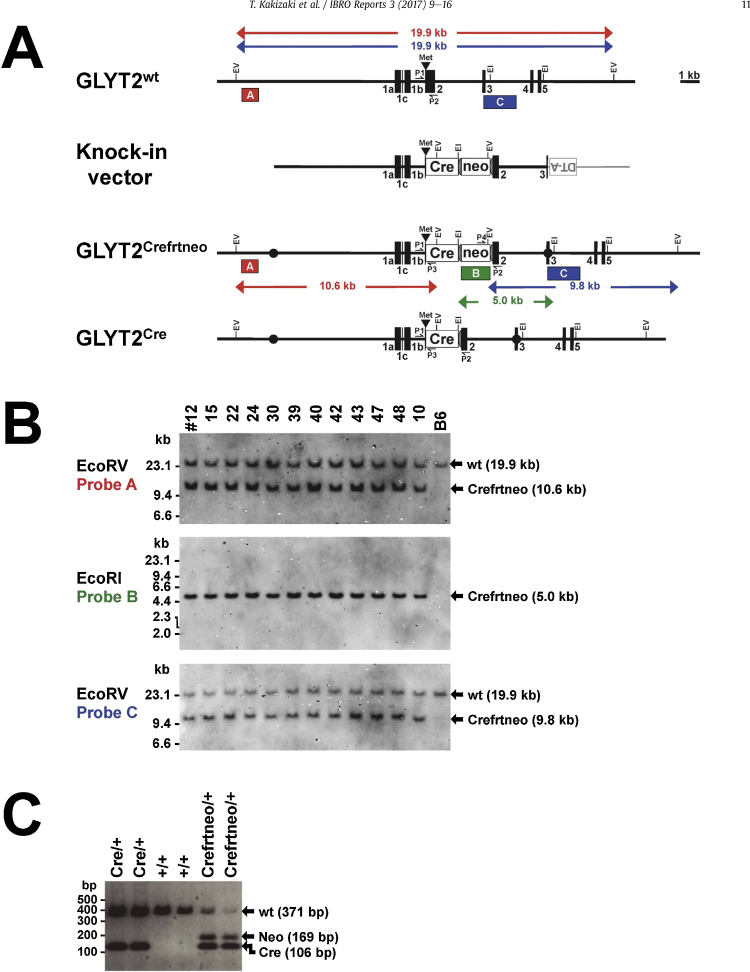
Two C57BL/6 mouse genomic BAC clones, RP23-83C16 and RP23-155A11 (BACPAC Resources, Oakland, CA, USA), containing the GLYT2 gene were purchased and used to construct the targeting vector. The knock-in vector contained an 8.1 kb fragment from the SalI site (in the 5′-flanking region) to the translation initiation site (in exon 2) of the GLYT2 gene, a Cre cDNA, a polyadenylation signal (pA) from the human granulocyte-colony stimulating factor gene, a phosphoglycerate kinase (PGK) promoter-driven neomycin resistance gene (neo)-pA cassette (PGK-neo-pA cassette) flanked by two frt sites (Takeuchi et al., 2002), a 2.9 kb AflII (in exon 2)-NcoI (in intron 2) fragment of the GLYT2 gene, and a MC1 promoter-driven diphtheria toxin A-fragment gene. A 440 bp KpnI (in intron 1b of the GLYT2 gene)-AgeI (in the Cre gene) fragment was synthesized by two-step PCR to fuse the GLYT2 gene (360 bp) to the Cre gene (80 bp) at their translation initiation sites. The Cre gene was isolated from the Cre-MC vector (Akashi et al., 2009). The PGK-neo-pA cassette and the MC1 promoter-driven diphtheria toxin A-fragment gene were isolated from the DT-NeoMC vector (Akashi et al., 2009).
The knock-in vector was linearized with SalI and electroporated into the RENKA mouse C57BL/6 ES cell line (Mishina and Sakimura, 2007). G418-resistant ES clones were isolated and screened using Southern blotting to detect homologous recombination. Genomic DNAs prepared from the ES cell colonies were digested with EcoRV and EcoRI and were hybridized with the 5′ external probe (Probe A), the 3′ external probe (Probe C), and internal probe (Probe B) (see Fig. 1). These DNA probes were labeled with digoxigenin using the DIG DNA Labeling Mix, 10x conc. (Roche, Basel, Switzerland). The recombinant ES cells were microinjected into eight cell-stage embryos of the ICR mouse strain to obtain chimeric mice.
Fig. 1.
Generation of the GLYT2-Cre knock-in mouse line. (A) Schematic representations of the wild-type allele (GLYT2+), knock-in vector, targeted allele (GLYT2Crefrtneo) and modified targeted allele after deletion of the PGK-neo-p(A) cassette (neo) (GLYT2Cre). The black numbered boxes represent exons. The black triangle and the gray semicircles represent translation initiation sites and FRT sequences, respectively. The white boxes represent the Cre recombinase gene followed by polyA signal (Cre) and the neo cassette, respectively. The gray box and the line represent the MC1 promoter-driven diphtheria toxin A-fragment gene (DT-A) and the plasmid vector, respectively. The black circles represent the 5′ and 3′ termini of the homologous recombination region in the knock-in targeting vector. The red, green and blue boxes represent the positions of probes A, B and C, respectively. The expected restriction fragment lengths (in kb) for Southern blot analyses are shown. The small black arrows represent the positions of primers P1, P2, P3 and P4 for the PCR analysis. EI, EcoRI; EV, EcoRV. (B) Southern blot analysis of genomic DNA extracted from neomycin-resistant ES cell clones. B6 indicates the negative control. All twelve analyzed clones were correctly targeted. (C) PCR analysis of the progeny of GLYT2Crefrtneo/+ mice crossed with FLP mice. GLYT2-Cre knock-in (GLYT2Cre/+) mice and wild-type (GLYT2+/+) mice were obtained. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The chimeric mice were crossed with C57BL/6 mice, and mice with germline transmission of the recombined allele were then crossed with FLP66 transgenic mice (Takeuchi et al., 2002) to delete the PGK-neo-pA cassette. GLYT2-Cre knock-in mice were crossed with R26R mice to detect Cre activity (Soriano, 1999), and GLYT2-Cre/R26R double transgenic (GLYT2Cre/+; R26RlacZ/+) mice (Yang et al., 2003) were used for histological analyses. GLYT2+/+; R26RlacZ/+ or GLYT2+/+; R26R+/+ mice were used as control mice.
All animal procedures were conducted in accordance with the guidelines of the NIH and were reviewed and approved by the Animal Care and Experimentation Committee of Gunma University, Showa Campus (Maebashi, Japan). Every effort was made to minimize the number of animals used and their suffering.
2.2. Genotyping of mutant mice
Genotypes of the GLYT2-Cre knock-in mice were determined by PCR using the following oligonucleotides: primer P1 (5′- GGATTGCAGTGCTCCCAAGG-3′), primer P2 (5′- ATCACTCGTTGCATCGACCG-3′), primer P3 (5′-ATCCCTCGATCGAGTGGATC-3′), and primer P4 (5′- GAATCTTGCAGTGCAGAGCG-3′). Primers P1, P2, P3, and P4 correspond to intron 1b and exon 2 of the GLYT2 gene, Cre gene, and PGK-neo-pA cassette, respectively. Primers P1 and P2 amplified a 371 bp fragment specific to the wild-type allele. Primers P1 and P3 amplified a 106 bp fragment specific to the GLYT2-Cre allele and independent of the PGK-neo-pA cassette. Primers P2 and P4 amplified a 169 bp fragment specific to the GLYT2-Cre knock-in allele.
Genotypes of the R26R mice were determined by PCR using the following oligonucleotides: primer R1295 (5′-GCGAAGAGTTTGTCCTCAACC-3′), primer R523 (5′-GGAGCGGGAGAAATGGATATG-3′), and primer R26F2 (5′- AAAGTCGCTCTGAGTTGTTAT-3′) (Rijnkels and Rosen, 2001). Primer R1295 corresponds to the reporter transgene, and primers R523 and R26F2 correspond to the wild-type ROSA26 locus. Primers R523 and R26F2 amplified a 650 bp fragment specific to the wild-type allele. Primers R1295 and R26F2 amplified a 340 bp fragment specific to the mutant allele.
2.3. X-gal staining
Adult control and GLYT2-Cre/R26R double transgenic mice were decapitated under ether anesthesia, and whole brains were removed. Sagittal brain slices were obtained by cutting the brains into 300 μm thick sections using a microslicer (D.S.K. Super Microslicer Zero 1, Dosaka EM, Kyoto, Japan). The brain slices were fixed with 2% formaldehyde at RT for 15 min. Adult control and mutant mice were transcardially perfused with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) under ether anesthesia to prepare coronal spinal cord slices. The spinal cords were removed and embedded in Tissue-Tek OCT compound (Sakura Finetek Japan, Tokyo, Japan). The frozen spinal cords were cut into 100 μm thick sections using a cryostat (CM 1510 S, Leica, Nussloch, Germany). These fixed brain and spinal cord slices were washed with PBS and then subjected to X-gal staining (X-Gal Staining Kit, Genlantis, San Diego, USA).
2.4. Combination of fluorescent in situ hybridization and immunohistochemistry
The 1000-bp cDNA fragment corresponding to nucleotides 1443–2442 of mouse GLYT2 (GenBank accession number AB118159.1) was amplified by PCR and subcloned into iSiP2 vector to generate the cRNA probe for GLYT2. The plasmids were linearized with the appropriate restriction enzymes and used as templates (2 μg) to synthesize anti-sense or sense cRNA probes for GLYT2 in an in vitro transcription reaction with digoxigenin-11-uridine 5′-triphosphate (DIG RNA labeling kit, Roche Diagnostics, Indianapolis, IN).
Sections of freshly frozen brains from the third postnatal week from control and double heterozygous mice were generated on a cryostat at a thickness of 20 μm. The sections were fixed with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) for 10 min, washed in phosphate-buffered saline (PBS) containing 2 mg/ml glycine, and acetylated with 0.25% acetic anhydride in 0.1 M triethanolamine for 10 min. After dehydration through a graded ethanol series, the sections were prehybridized for 30 min in a buffer consisting of 50% formamide, 4x SSC, 0.02% Ficoll, 0.02% polyvinylpyrrolidone, 0.02% bovine serum albumin, 1 mM EDTA, 0.1% sodium N-lauroyl sarcosinate, and 200 μg/ml tRNA. Hybridization was performed overnight at 65 °C in prehybridization buffer supplemented with 10% dextran sulfate, 100 mM dithiothreitol, and heat-denatured cRNA probes. On the following day, the sections were washed twice with 5x SSC containing 50% formamide for 30 min, and three times with 2x SSC containing 50% formamide for 30 min at 65 °C.
For the immunodetection of the GLYT2 hybridization signals, the sections were incubated with 0.5% DIG blocking reagent (Roche) in Tris-buffered saline (TBS: 100 mM Tris•HCl [pH 7.5] and 150 mM NaCl) for 30 min, followed by 0.5% TSA blocking reagent in TBS containing 0.1% Tween-20 (TBST) for 30 min. The sections were subsequently incubated with a sheep anti-digoxigenin antibody conjugated to polymerized horseradish peroxidase (1:500, anti-digoxigenin-POD, Fab fragments, Roche) in TBST overnight. After extensive washes with TBST, the sections were subjected to tyramide signal amplification with Cy3-tyramide according to the manufacturer's instructions (TSA Plus Cy3 system, Perkin-Elmer, Waltham, MA).
For immunodetection of β-galactosidase (β-gal), the sections were subsequently rinsed with PBS, blocked with 5% normal donkey serum in PBS for 30 min, and incubated with a rabbit polyclonal anti-β-galactosidase antibody (5 Prime → 3 Prime, Boulder, CO) overnight at room temperature. Immunoreactions were visualized by incubating the sections with an AlexaFluor 594-conjugated anti-rabbit IgG secondary antibody (Invitrogen, Eugene, OR). Sections were subjected to nuclear staining with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) and mounted in Fluormount (Diagnostic Biosystems Inc., Pleasanton, CA).
For the colocalization analysis, fluorescent images (512 × 512 pixels) were captured in various brain regions as single optical slices with a Plan Apochromat objective lens × 20/0.8 using a confocal laser scanning microscope (LSM 710, Carl Zeiss, Jena Germany). Colocalization was evaluated by a visual inspection with the aid of the ZEN2012 software (Carl Zeiss).
3. Results
3.1. Generation of GLYT2-Cre knock-in mice
The mouse GLYT2 gene encodes three splice variants, GLYT2a, GLYT2b and GLYT2c, which differ in their 5′ regions (Ebihara et al., 2004). GLYT2b and GLYT2c share the same translation initiation codon (ATG), and it is located in exon 2 of the GLYT2 gene. The translation initiation codon of GLYT2a is located in exon 1a of the GLYT2 gene. GLYT2b and GLYT2c produce proteins with identical amino acid sequences; on the other hand, the N-terminus of the GLYT2a protein is eight amino acids longer than the GLYT2b and GLYT2c proteins. We constructed a targeting vector that placed the Cre gene and the polyadenylation signal of the human granulocyte-colony stimulating factor gene into exon 2 of the GLYT2 gene, in-frame with the first ATG codon (Fig. 1A). This strategy places the Cre expression cassette under the control of the endogenous GLYT2a, GLYT2b and GLYT2c promoters.
Among the 147 neomycin-resistant ES clones obtained, 40 correctly targeted clones were identified by Southern blot analysis (Fig. 1B). Fourteen chimeric mice were obtained from twelve targeted ES clones. Germline transmission was confirmed in three chimeric mice derived from the two recombinant targeted ES clones. The germline chimeras were mated to C57BL/6 mice to generate heterozygous mice carrying one Crefrtneo allele (GLYT2Crefrtneo/+). The GLYT2Crefrtneo/+ mice were crossed with Flp66 transgenic mice (Takeuchi et al., 2002), and the male offspring were mated to C57BL/6 mice to eliminate the PGK-neo cassette from the genome through Flp/frt-mediated excision (Fig. 1C). The resulting GLYT2Cre/+ mice are hereafter referred to as GLYT2-Cre knock-in mice.
3.2. Localization of Cre activity in the GLYT2-Cre knock-in mice
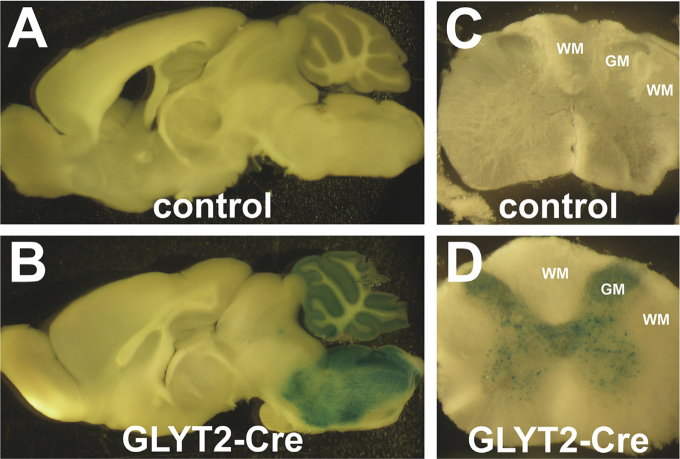
We crossed the GLYT2-Cre knock-in mice with R26R mice, a reporter line used to monitor Cre activity (Soriano, 1999), and produced GLYT2-Cre/R26R double transgenic mice to examine the spatial localization of GLYT2-Cre activity. The R26R mice express β-gal in cells in which the Cre-dependent excision of the floxed stop codon occurs (Soriano, 1999). Cre recombination was visualized using X-gal staining. Many X-gal-positive cells were present in the brainstem, cerebellum, and spinal cord gray matter of the double transgenic mice, but not in the control mice (Fig. 2). Glycinergic neurons are largely restricted to these brain regions (Zeilhofer et al., 2005). Therefore, Cre is likely expressed in glycinergic neurons in the GLYT2-Cre knock-in mice.
Fig. 2.
Cre-mediated recombination in the GLYT2-Cre knock-in mice. X-gal staining of parasagittal brain sections (A, B) and coronal spinal cord sections (C, D) from the control (A, C) and GLYT2-Cre/R26R double transgenic mice (B, D). WM, white matter; GM, gray matter.
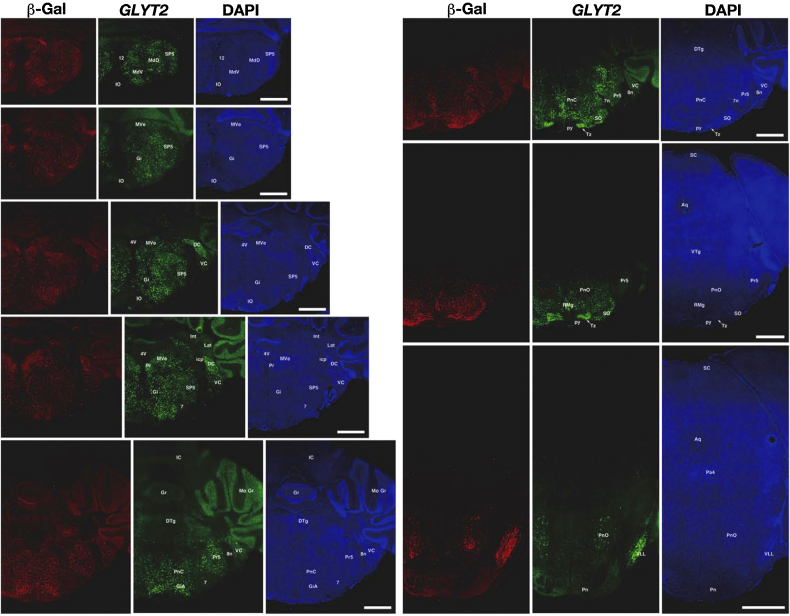
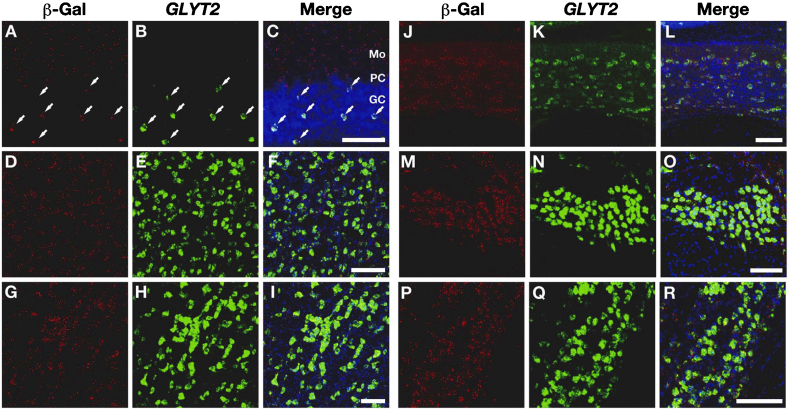
Next, we performed immunohistochemistry for β-gal combined with in situ hybridization for GLYT2 in the GLYT2-Cre/R26R double transgenic mouse brain to evaluate the specific cell type in which Cre recombination occurred. GLYT2 mRNAs were widely expressed in the brainstem and cerebellar cortex, and the distribution pattern was similar to β-gal (Fig. 3). Among these regions, the dorsal cochlear nucleus, nucleus of the trapezoid body, and ventral nucleus of lateral lemniscus expressed particularly high levels of the GLYT2 mRNA (Fig. 3) (Zeilhofer et al., 2005). We investigated the colocalization of β-gal and GLYT2 in six brain regions, including the three areas mentioned above. GLYT2 mRNAs were localized in the soma, and it was easy to identify the positive neurons. However, immuno-positive signals for β-gal were not uniformly distributed in the cells but were scattered as small patches (Fig. 4). More than 98% of the GLYT2 mRNA-expressing cells were positive for β-gal in these brain regions, whereas 90–98% of the β-gal-positive cells expressed GLYT2 mRNAs (Table 1). One possible explanation for the difference is that β-gal immuno-positive signals were distributed in the proximal neuropil in addition to the soma, whereas GLYT2 mRNAs were restricted to the soma. In summary, Cre activity was undetectable in GLYT2-negative cells, with the exception of the interneurons in the molecular layer of the cerebellar cortex, and almost all glycinergic neurons expressed Cre in the GLYT2-Cre knock-in mice, indicating that the Cre activity is specifically localized to the glycinergic neurons in the GLYT2-Cre knock-in mice.
Fig. 3.
Localization of Cre activity in the GLYT2-Cre knock-in mice. Combined immunohistochemistry for β-gal (red) and in situ hybridization for GLYT2 (green) in coronal sections of the brainstem from the GLYT2-Cre/R26R double transgenic mouse. Images in the right columns show the same sections counter-stained with DAPI (blue) as a reference. 4V, fourth ventricle; 7, facial nucleus; 7n, facial nerve; 8n, vestibulocochlear nerve; 12, hypoglossal nucleus; Aq, aqueduct; DC, dorsal cochlear nucleus; DTg, dorsal tegmental nucleus; Gi, gigantocellular reticular nucleus; GiA, gigantocellular reticular nucleus, alpha part; Gr, granular layer; IC, inferior colliculus; icp, inferior cerebellar peduncle; Int, interposed cerebellar nucleus; IO, inferior olive; Lat, lateral cerebellar nucleus; MdD, medullary reticular nucleus, dorsal part; MdV, medullary reticular nucleus, ventral part; Mo, molecular layer; MVe, medial vestibular nucleus; Pa4, paratrochlear nucleus; Pn, pontine nuclei; PnC, pontine reticular nucleus, caudal part; PnO, pontine reticular nucleus, oral part Pr, prepositus nucleus; Pr5, principal sensory trigeminal nucleus; py, pyramidal tract; RMg, raphe magnus nucleus; SC, superior colliculus; SO, superior olive; SP5, spinal trigeminal nucleus; Tz, nucleus of the trapezoid body; VC, ventral cochlear nucleus; VLL, ventral nucleus of the lateral lemniscus. Scale bars, 1 mm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Cell type specific localization of Cre activity in the GLYT2-Cre knock-in mice. Combined immunohistochemistry for β-gal (A, D, G, J, M, P) and in situ hybridization for GLYT2 (B, E, H, K, N, Q) in the cerebellar cortex (A–C) and various brainstem nuclei, including the spinal trigeminal nucleus (D–F), gigantocellular reticular nucleus (G–I), dorsal cochlear nucleus (J–L), nucleus of the trapezoid body (M–O), and ventral nucleus of lateral lemniscus (P–R), of the GLYT2-Cre/R26R double transgenic mice. Arrows in A-C indicate the coexpression of the β-galactosidase protein and GLYT2 mRNA in Golgi cells. GC, granule cell layer; Mo, molecular layer; PC, Purkinje cell layer. Scale bars, 100 μm.
Table 1.
Colocalization of neurons expressing GLYT2 mRNAs and β-galactosidase in various brain regions.
| Brain region | % Colocalization |
|
|---|---|---|
| β-gal(+) neurons in GLYT2 mRNA-expressing neurons | GLYT2 mRNA-expressing neurons in β-gal(+) neurons | |
| Cerebellar Golgi cells | 98.4% (n = 316) | 93.7% (n = 332) |
| Spinal trigeminal nucleus | 99.9% (n = 775) | 96.0% (n = 806) |
| Gigantocellular reticular nucleus | 100% (n = 466) | 97.1% (n = 480) |
| Dorsal cochlear nucleus | 100% (n = 637) | 97.1% (n = 656) |
| Nucleus of the trapezoid body | 100% (n = 437) | 98.4% (n = 444) |
| Ventral nucleus of the lateral lemniscus | 98.9% (n = 182) | 90.5% (n = 199) |
Fluorescent images were acquired with a confocal laser scanning microscope, and the number of labeled neurons was counted by visual inspection.
4. Discussion
We generated a novel GLYT2-Cre knock-in mouse line and showed that the Cre activity was highly efficient and restricted to glycinergic neurons. The GLYT2-Cre knock-in mice will enable us to specifically manipulate gene expression in glycinergic neurons. The GLYT2-Cre knock-in mice will allow us to develop mice with a deletion of the floxed genes of interest or overexpression of genes using a floxed-stop-transgene strategy specifically in glycinergic neurons, enabling studies of the cellular properties of glycinergic neurons and their functions in the neural network (Gavériaux-Ruff and Kieffer, 2007). Targeting of fluorescent proteins such as EGFP and tdTomato will facilitate the electrophysiological and anatomical characterization of glycinergic neurons (Wouterlood et al., 2014). The GLYT2-Cre knock-in mice will be used in combination with viral vectors carrying floxed opsin genes such as channelrhodopsin-2 and halorhodopsin to enable the genetically driven activation and inactivation of glycinergic neurons (Pupe and Wallén-Mackenzie, 2015, Zhao et al., 2015).
Some transgenic mouse lines expressing enhanced green fluorescent protein (EGFP) or Cre under control of the GLYT2 promoter were generated using a BAC system and have contributed to research on glycinergic neurons (Zeilhofer et al., 2005, Ishihara et al., 2010, Foster et al., 2015). On the other hand, our GLYT2-Cre mouse line was generated using knock-in system. Transgene expression is theoretically controlled by all cis-regulatory elements in knock-in mice, providing a greater ability to recapitulate the endogenous gene expression pattern than BAC transgenic mice (Harris et al., 2014). In fact, nearly 100% of the GLYT2 mRNA-expressing cells were positive for β-gal in our GLYT2-Cre knock-in mouse line (Table 1). We identified β-gal-positive interneurons in the molecular layer of the cerebellum in adult GLYT2-Cre/R26R double transgenic mice. These observations in the knock-in mice are similar to the observations in the GLYT2-Cre BAC transgenic mice (Ishihara et al., 2010). Furthermore, EGFP was transiently expressed in molecular interneurons in the cerebellum of the GLYT2-EGFP BAC transgenic mice (Simat et al., 2007). Therefore, the recombination appears to reflect the transient GLYT2-Cre expression in an early developmental period.
Our GLYT2-Cre knock-in mouse line was generated using a gene targeting method. This strategy allowed us to inactivate the GLYT2 gene. Therefore, homozygous GLYT2-Cre knock-in (GLYT2Cre/Cre) mice will also be used as GLYT2 knockout mice. Similar to the conventional GLYT2−/− mice (Gomeza et al., 2003b), GLYT2Cre/Cre mice died at approximately postnatal day 14. Researchers have not been able to easily identify the GLYT2-deficient glycinergic neurons with a reduced glycine content (Latal et al., 2010). The expression of the fluorescent protein in glycinergic neurons will allow us to characterize the GLYT2-deficient neurons using the GLYT2Cre/Cre mice that also express a floxed fluorescent protein gene. However, analogous to the conventional GLYT2+/− mice (Gomeza et al., 2003b), GLYT2Cre/+ mice were viable and fertile. It has been reported that glycine transport activities in P2 membrane fractions prepared from CNS were not significantly different between heterozygous GlyT2-deficient and wild-type mice (Gomeza et al., 2003b). Furthermore, motor functions of the GLYT2Cre/+ mice were not different from those of control wild-type mice (Nishiyama et al., 2013). These results imply that the heterozygous deletion of the GLYT2 gene does not make a drastic impact on glycinergic neurotransmission. Despite the lack of one GLYT2 allele, the GLYT2Cre/+ mice are a versatile tool for unraveling the properties of glycinergic neurons and the mechanisms of glycinergic neurotransmission.
5. Conclusions
We have generated a novel GLYT2-Cre knock-in mouse line and have shown that Cre activity is specifically localized in glycinergic neurons. The GLYT2-Cre knock-in mouse line will be a useful tool for manipulating gene expression in glycinergic neurons.
Acknowledgements
The authors thank Dr. R Kaneko for the gift of the iSip2 vector; and Mss. T Honma, K Harada, A Morita, and Y Shimoda for providing technical and secretarial assistance. We thank the staff at the Department of Genetic and Behavioral Neuroscience and Bioresource Center, Gunma University Graduate School of Medicine for their critical comments and technical assistance. This study was supported by Grants-in-Aid for Scientific Research (23115503, 26290002, 15H01415 and 15H05872 to Y.Y.), a Grant-in-Aid for Scientific Research on Innovative Areas (Comprehensive Brain Science Network) (to Y.Y.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, a grant from the Co-operative Study Program of the National Institute for Physiological Sciences, Japan (to Y.Y.), and a grant from the Takeda Science Foundation (to Y.Y.).
References
- Akashi K., Kakizaki T., Kamiya H., Fukaya M., Yamasaki M., Abe M., Natsume R., Watanabe M., Sakimura K. NMDA receptor GluN2B (GluR epsilon 2/NR2B) subunit is crucial for channel function, postsynaptic macromolecular organization, and actin cytoskeleton at hippocampal CA3 synapses. J. Neurosci. 2009;29:10869–10882. doi: 10.1523/JNEUROSCI.5531-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón C., López-Corcuera B. Structure, function and regulation of glycine neurotransporters. Eur. J. Pharmacol. 2003;31:249–262. doi: 10.1016/j.ejphar.2003.08.074. [DOI] [PubMed] [Google Scholar]
- Betz H., Gomeza J., Armsen W., Scholze P., Eulenburg V. Glycine transporters: essential regulators of synaptic transmission. Biochem. Soc. Trans. 2006;34:55–58. doi: 10.1042/BST0340055. [DOI] [PubMed] [Google Scholar]
- Carta E., Chung S.K., James V.M., Robinson A., Gill J.L., Remy N., Vanbellinghen J.F., Drew C.J., Cagdas S., Cameron D., Cowan F.M., Del Toro M., Graham G.E., Manzur A.Y., Masri A., Rivera S., Scalais E., Shiang R., Sinclair K., Stuart C.A., Tijssen M.A., Wise G., Zuberi S.M., Harvey K., Pearce B.R., Topf M., Thomas R.H., Supplisson S., Rees M.I., Harvey R.J. Mutations in the GlyT2 gene (SLC6A5) are a second major cause of startle disease. J. Biol. Chem. 2012;287:28975–28985. doi: 10.1074/jbc.M112.372094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara S., Yamamoto T., Obata K., Yanagawa Y. Gene structure and alternative splicing of the mouse glycine transporter type-2. Biochem. Biophys. Res. Commun. 2004;317:857–864. doi: 10.1016/j.bbrc.2004.03.125. [DOI] [PubMed] [Google Scholar]
- Eulenburg V., Armsen W., Betz H., Gomeza J. Glycine transporters: essential regulators of neurotransmission. Trends Biochem. Sci. 2005;30:325–333. doi: 10.1016/j.tibs.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Ezure K., Tanaka I., Kondo M. Glycine is used as a transmitter by decrementing expiratory neurons of the ventrolateral medulla in the rat. J. Neurosci. 2003;23:8941–8948. doi: 10.1523/JNEUROSCI.23-26-08941.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster E., Wildner H., Tudeau L., Haueter S., Ralvenius W.T., Jegen M., Johannssen H., Hösli L., Haenraets K., Ghanem A., Conzelmann K.K., Bösl M., Zeilhofer H.U. Targeted ablation, silencing, and activation establish glycinergic dorsal horn neurons as key components of a spinal gate for pain and itch. Neuron. 2015;85:1289–1304. doi: 10.1016/j.neuron.2015.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavériaux-Ruff C., Kieffer B.L. Conditional gene targeting in the mouse nervous system: insights into brain function and diseases. Pharmacol. Ther. 2007;113:619–634. doi: 10.1016/j.pharmthera.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Gomeza J., Hülsmann S., Ohno K., Eulenburg V., Szöke K., Richter D., Betz H. Inactivation of the glycine transporter 1 gene discloses vital role of glial glycine uptake in glycinergic inhibition. Neuron. 2003;40:785–796. doi: 10.1016/s0896-6273(03)00672-x. [DOI] [PubMed] [Google Scholar]
- Gomeza J., Ohno K., Hülsmann S., Armsen W., Eulenburg V., Richter D.W., Laube B., Betz H. Deletion of the mouse glycine transporter 2 results in a hyperekplexia phenotype and postnatal lethality. Neuron. 2003;40:797–806. doi: 10.1016/s0896-6273(03)00673-1. [DOI] [PubMed] [Google Scholar]
- Harris J.A., Hirokawa K.E., Sorensen S.A., Gu H., Mills M., Ng L.L., Bohn P., Mortrud M., Ouellette B., Kidney J., Smith K.A., Dang C., Sunkin S., Bernard A., Oh S.W., Madisen L., Zeng H. Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front. Neural Circuits. 2014;8:76. doi: 10.3389/fncir.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Armsen W., Papadopoulos T., Betz H., Eulenburg V. Generation of a mouse line expressing Cre recombinase in glycinergic interneurons. Genesis. 2010;48:437–445. doi: 10.1002/dvg.20640. [DOI] [PubMed] [Google Scholar]
- Latal A.T., Kremer T., Gomeza J., Eulenburg V., Hülsmann S. Development of synaptic inhibition in glycine transporter 2 deficient mice. Mol. Cell Neurosci. 2010;44:342–352. doi: 10.1016/j.mcn.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Laube B., Maksay G., Schemm R., Betz H. Modulation of glycine receptor function: a novel approach for therapeutic intervention at inhibitory synapses? Trends Pharmacol. Sci. 2002;23:519–527. doi: 10.1016/s0165-6147(02)02138-7. [DOI] [PubMed] [Google Scholar]
- Legendre P. The glycinergic inhibitory synapse. Cell Mol. Life Sci. 2001;58:760–793. doi: 10.1007/PL00000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina M., Sakimura K. Conditional gene targeting on the pure C57BL/6 genetic background. Neurosci. Res. 2007;58:105–112. doi: 10.1016/j.neures.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Nelson N. The family of Na+/Cl- neurotransmitter transporters. J. Neurochem. 1998;71:1785–1803. doi: 10.1046/j.1471-4159.1998.71051785.x. [DOI] [PubMed] [Google Scholar]
- Nishiyama A., Shimomura H., KanbaraKume N., Kakizaki T., Tanizawa T., Yanagawa Y., Arata A. The behavioral development in the GlyT2 deficient mice. J. Physiol. Sci. 2013;63:S145. [Google Scholar]
- Poyatos I., Ponce J., Aragón C., Giménez C., Zafra F. The glycine transporter GLYT2 is a reliable marker for glycine-immunoreactive neurons. Brain Res. Mol. Brain Res. 1997;49:63–70. doi: 10.1016/s0169-328x(97)00124-1. [DOI] [PubMed] [Google Scholar]
- Pupe S., Wallén-Mackenzie Å. Cre-driven optogenetics in the heterogeneous genetic panorama of the VTA. Trends Neurosci. 2015;38:375–386. doi: 10.1016/j.tins.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Rahman J., Besser S., Schnell C., Eulenburg V., Hirrlinger J., Wojcik S.M., Hülsmann S. Genetic ablation of VIAAT in glycinergic neurons causes a severe respiratory phenotype and perinatal death. Brain Struct. Funct. 2015;220:2835–2849. doi: 10.1007/s00429-014-0829-2. [DOI] [PubMed] [Google Scholar]
- Rees M.I., Harvey K., Pearce B.R., Chung S.K., Duguid I.C., Thomas P., Beatty S., Graham G.E., Armstrong L., Shiang R., Abbott K.J., Zuberi S.M., Stephenson J.B., Owen M.J., Tijssen M.A., van den Maagdenberg A.M., Smart T.G., Supplisson S., Harvey R.J. Mutations in the gene encoding GlyT2 (SLC6A5) define a presynaptic component of human startle disease. Nat. Genet. 2006;38:801–806. doi: 10.1038/ng1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijnkels M., Rosen J.M. Adenovirus-Cre-mediated recombination in mammary epithelial early progenitor cells. J. Cell Sci. 2001;114:3147–3153. doi: 10.1242/jcs.114.17.3147. [DOI] [PubMed] [Google Scholar]
- Saito K., Kakizaki T., Hayashi R., Nishimaru H., Furukawa T., Nakazato Y., Takamori S., Ebihara S., Uematsu M., Mishina M., Miyazaki J., Yokoyama M., Konishi S., Inoue K., Fukuda A., Fukumoto M., Nakamura K., Obata K., Yanagawa Y. The physiological roles of vesicular GABA transporter during embryonic development: a study using knockout mice. Mol. Brain. 2010;30:40. doi: 10.1186/1756-6606-3-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simat M., Ambrosetti L., Lardi-Studler B., Fritschy J.M. GABAergic synaptogenesis marks the onset of differentiation of basket and stellate cells in mouse cerebellum. Eur. J. Neurosci. 2007;26:2239–2256. doi: 10.1111/j.1460-9568.2007.05846.x. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Takeuchi T., Nomura T., Tsujita M., Suzuki M., Fuse T., Mori H., Mishina M. Flp recombinase transgenic mice of C57BL/6 strain for conditional gene targeting. Biochem. Biophys. Res. Commun. 2002;293:953–957. doi: 10.1016/S0006-291X(02)00321-2. [DOI] [PubMed] [Google Scholar]
- Wojcik S.M., Katsurabayashi S., Guillemin I., Friauf E., Rosenmund C., Brose N., Rhee J.S. A shared vesicular carrier allows synaptic corelease of GABA and glycine. Neuron. 2006;50:575–587. doi: 10.1016/j.neuron.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Wouterlood F.G., Bloem B., Mansvelder H.D., Luchicchi A., Deisseroth K. A fourth generation of neuroanatomical tracing techniques: exploiting the offspring of genetic engineering. J. Neurosci. Methods. 2014;30:331–348. doi: 10.1016/j.jneumeth.2014.07.021. [DOI] [PubMed] [Google Scholar]
- Yang Z., Ding K., Pan L., Deng M., Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev. Biol. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Yevenes G.E., Zeilhofer H.U. Allosteric modulation of glycine receptors. Br. J. Pharmacol. 2011;164:224–236. doi: 10.1111/j.1476-5381.2011.01471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilhofer H.U., Studler B., Arabadzisz D., Schweizer C., Ahmadi S., Layh B., Bösl M.R., Fritschy J.M. Glycinergic neurons expressing enhanced green fluorescent protein in bacterial artificial chromosome transgenic mice. J. Comp. Neurol. 2005;482:123–141. doi: 10.1002/cne.20349. [DOI] [PubMed] [Google Scholar]
- Zhao M., Alleva R., Ma H., Daniel A.G., Schwartz T.H. Optogenetic tools for modulating and probing the epileptic network. Epilepsy Res. 2015;116:15–26. doi: 10.1016/j.eplepsyres.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]