Abstract
Background and Purpose
Hematoma volume is an important determinant of clinical outcome in spontaneous intracerebral hemorrhage (ICH). We performed a genome-wide association study (GWAS) of hematoma volume with the aim of identifying novel biological pathways involved in the pathophysiology of primary brain injury in ICH.
Methods
We conducted a two-stage (discovery and replication) case-only genome-wide association study in ICH patients of European ancestry. We utilized the admission head computed tomography (CT) to calculate hematoma volume via semi-automated computer-assisted technique. After quality control and imputation, seven million genetic variants were available for association testing with ICH volume, which was carried out separately in lobar and non-lobar ICH cases using linear regression. Signals with p<5×10−8 were pursued in replication and tested for association with admission Glasgow Coma Scale (aGCS) and 3-month post-ICH dichotomized (0–2 vs 3–6) modified Rankin Scale (mRS) using ordinal and logistic regression, respectively.
Results
The discovery phase included 394 ICH cases (228 lobar and 166 non-lobar) and identified 2 susceptibility loci: a genomic region on 22q13 encompassing PARVB (top SNP rs9614326: beta 1.84, SE 0.32; p=4.4×10−8) for lobar ICH volume; and an intergenic region overlying numerous copy number variants on 17p12 (top SNP rs11655160: beta 0.95, SE 0.17; p=4.3×10−8) for non-lobar ICH volume. The replication included 240 ICH cases (71 lobar and 169 non-lobar) and corroborated the association for 17p12 (p=0.04; meta-analysis p=2.5×10−9; heterogeneity-p=0.16) but not for 22q13 (p=0.49). In multivariable analysis, rs11655160 was also associated with lower aGCS (OR 0.17; p=0.004) and increased risk of poor 3-month mRS (OR 1.94; p=0.045).
Conclusions
We identified 17p12 as a novel susceptibility risk locus for hematoma volume, clinical severity, and functional outcome in non-lobar ICH. Replication in other ethnicities and follow-up translational studies are needed to elucidate the mechanism mediating the observed association.
Keywords: Intracerebral Hemorrhage, Hemorrhagic stroke, Genome-Wide Association Study, Neuroimaging, ICH volume
INTRODUCTION
Hematoma volume is an established neuroimaging biomarker of primary injury in spontaneous intracerebral hemorrhage (ICH). It is a powerful predictor of clinical outcome1 and strongly correlates with markers of clinical and radiological severity, including admission Glasgow Coma Scale (aGCS), presence of contrast extravasation in computed tomography (CT) angiography (termed the ‘spot sign’), and hematoma expansion 2–4. Importantly, this predictive power applies to both lobar and non-lobar hemorrhages, making ICH volume a useful tool regardless of the specific biology underlying the brain bleed3,5. Beyond its predictive potential, ICH volume is easy to calculate and widely available in everyday clinical practice, as almost all ICH patients undergo a head CT upon admission to the hospital6. In combination, these characteristics make hematoma volume an ideal radiological marker to study the mechanisms of primary injury that determine severity and outcome in ICH.
Inherited genetic variation plays an important role in ICH. Heritability estimates indicate that 40% of the variation in ICH risk can be attributed to rare and common mutations7–9. Additionally, several monogenic diseases that follow a Mendelian pattern of inheritance manifest clinically through ICH, including familial cerebral amyloid angiopathy and COL4A1-related intracerebral hemorrhage10. Large association studies have also identified common mutations in APOE, PMF1 and COL4A1/COL4A210–12 that increase ICH risk with incomplete penetrance. Furthermore, mutations in genes related to blood pressure and lipid levels, two physiological pathways known to influence ICH occurrence, also modify the risk of ICH13–15.
Recent studies indicate that the role of genetic variation extends beyond risk to also influence clinical severity, radiological severity, and functional outcome in ICH. For lobar ICH, the APOE epsilon 2 allele associates with a 5 mL increase in mean ICH volume, resulting in a 50% increase in risk of poor outcome16. For deep ICH, the cumulative burden of hypertension-related mutations associates with a 2.7 mL increase in mean ICH volume, resulting in a 71% increase in risk of poor outcome14. Despite these findings, genome-wide association studies (GWAS) focused on ICH volume are lacking. Given the unique potential of this radiological marker and the valuable information provided by genome-wide scans, we conducted a GWAS of ICH volume, as measured on the first head CT scan upon admission to the hospital. Our hypothesis is that a substantial proportion of the variation observed in ICH volume can be attributed to genetic variation and that this genetic contribution translates into concrete genetic risk loci that influence not only the size of the hematoma, but also clinical severity.
METHODS
Study design and participating cohorts
This work adheres to the Transparency and Openness Promotion Guidelines. The full set of genome-wide association results of this study will be available through the Cerebrovascular Disease Knowledge Portal (http://www.cerebrovascularportal.org)17. The individual level data of this study are available through two different publicly available resources: the American Heart Association Precision Medicine Platform (https://precision.heart.org/) and the database of Genotypes and Phenotypes (https://www.ncbi.nlm.nih.gov/gap). We utilized a case-only study design that included discovery and replication stages. The qualifying event for all cohorts was the occurrence of a non-traumatic, spontaneous ICH. ICH cases included in the discovery phase were subjects of European ancestry enrolled in the Genetics of Cerebral Hemorrhage with Anticoagulation (GOCHA) study, a multicenter study conducted in the US13. ICH cases included in the replication phase were subjects of European ancestry enrolled in the Hospital del Mar Intracerebral Hemorrhage study (Barcelona, Spain)18, Vall d’Hebron Hospital ICH study (Barcelona, Spain)19 and Lund Stroke Register study (Lund, Sweden)20. Due to their limited sample sizes, data from the three European studies (ESs) utilized for replication were pooled for genomic quality control, imputation, and association testing. Included studies had approval from the local institutional review board or ethics committee at each participating institution. Informed consent was either obtained from patients or their legally authorized representatives.
Cases
Subjects were enrolled in participating studies according to predefined criteria. Included subjects were aged >55 years (GOCHA) or >18 years (ESs) and had a diagnosis of spontaneous, non-traumatic ICH. Spontaneous ICH was defined as a new and acute neurological deficit with compatible neuroimaging findings showing the presence of intraparenchymal bleeding. Patients with hemorrhage resulting from anticoagulation, trauma, conversion of an ischemic infarct, rupture of a vascular malformation or aneurysm, or brain tumor were excluded.
Ascertainment of ICH volume
For all subjects, ICH volume in the first available CT upon admission was centrally read at the Massachusetts General Hospital using Analyze Direct 11.0 software, a semi-automated computer-assisted technique with excellent inter-rater reliability (intraventricular bleeding was not included in volume calculations)21. In accordance with the known biology of the disease, ICH cases were classified as lobar or non-lobar based on the location of the bleed within the brain. Hematomas originating in the cerebral cortex or cortical-subcortical junction were classified as lobar, whereas hematomas originating in the thalamus, internal capsule, basal ganglia, deep periventricular white matter, cerebellum, or brainstem were classified as non-lobar. Bleeds involving both deep and lobar territories were defined as mixed ICH, and these individuals were not eligible for inclusion in our analysis.
Clinical data
Demographics, clinical variables and past medical history were collected through retrospective review of hospital charts. We identified comorbidities, medications, laboratory values and aGCS22. The modified Rankin Scale (mRS)23 was assessed through follow-up telephone calls 3 months after the index ICH14,24.
Ascertainment of genotypes
Genotyping
Cases were genotyped using Illumina HumanHap610-Quad (San Diego, CA), as part of a previous published study11. Standardized pre-specified quality control procedures were implemented separately for the discovery and replication datasets25. These filters excluded single nucleotide polymorphisms (SNPs) with genotype call rate <0.95, significant differential missingness across bins of ICH volume, deviation from Hardy-Weinberg equilibrium (p<1×10−6), or minor allele frequency (MAF) <0.01. At the sample level, quality control excluded individuals with a genotype call rate of <95%, who had inconsistency between self-reported and genotypic sex, who had an inferred first- or second-degree relative in the sample identified based on pairwise allele sharing estimates (estimated genome proportion shared identical by descent; π >0.1875), and who had extreme genome-wide heterozygosity f statistics.
Population stratification
Following quality control procedures, principal-component analysis26 was implemented to account for population structure.
Imputation
Imputation was performed separately in the discovery and replication datasets using IMPUTE2 V2.227 and 1000 Genomes28 integrated reference panels (Phase I interim release in NCBI build 37). Post-imputation filters excluded imputed SNPs with MAF <0.01, IMPUTE2 information score <0.7, confidence score <0.9 and missing estimates in association testing for 1 or more studies.
Statistical analysis
Descriptive statistics
Discrete data are presented as count (percentages [%]) and continuous data as mean (standard deviation[SD]) or median (interquartile range [IQR]) as appropriate.
Association testing for risk loci discovery
Genome-wide association analyses were computed separately for lobar ICH and non-lobar ICH volume. We chose this approach given the known differences in biology across locations29, which were confirmed by a previous GWAS of ICH risk11. Following standard practices in the field, ICH volume was natural log-transformed to approximate normality30–32. Analyses were computed separately for the discovery and replication phases via linear regression, assuming additive genetic effects (1-degree-of-freedom additive trend test) and adjusting for age, sex, and the first four principal components. Following standard practices in the field, these covariates were added to the regression models to improve model fitting and enhance statistical precision, not to adjust for confounding. Quantile-quantile plots were utilized to assess systematic inflation in association results resulting from population stratification or other systematic causes of bias. A Bonferroni-corrected significance threshold of 5×10−8, adjusting for 1 million independent genetic loci across the human genome, was used to declare statistical significance.
Meta-analysis
Results from the discovery and replication stages were pooled in meta-analysis using an inverse-variance weighed, fixed-effects approach. Heterogeneity across effects estimates was evaluated using I2 values.
Association testing to evaluate clinical significance
We used ordinal multivariable logistic regression to test for association between the identified genetic risk loci and aGCS, as well as multivariable logistic regression to test for association between these same loci and a dichotomized version (0–2 vs 3–6) of 3-month post bleed modified Rankin Scale (mRS), as previously done33,34. All these models were adjusted for multiple confounders (age, sex, hypertension, diabetes, antiplatelet use, Principal Components 1 to 4.).
Annotation
Variants identified at genetic risk loci were evaluated for gene expression in cis using publicly available resources. SNPs with p<1×10−5 were assessed in four publicly available eQTL databases: SCAN–SNP and CNV Annotation Database, the NCBI GTEx (Genotype-Tissue Expression) eQTL Browser,35 the Pritchard laboratory UChicago eQTL browser,36 and mRNA by SNP Browser v1.0.137. The positions of SNPs in ICH volume-associated loci were overlapped with DNase I hotspot regions from the Encyclopedia of DNA Elements (ENCODE) Project that mark generalized chromatin accessibility, mapped for each of 125 diverse cell lines and tissues.38 In addition, ICH volume-associated SNPs were analyzed for overlap with ENCODE data, including transcription factor motifs, using RegulomeDB.39
Copy number variant analysis
Because one of the identified loci overlaid multiple copy number variants, PennCNV was used to ascertain this type of variant in a 62 Mb region that began at the start of the CNV farthest away from the locus of interest in one direction and finished at the end of the CNV that was farthest away from this same locus in the opposite direction.
Statistical analyses were completed using PLINK40, SAS 9.3 and PennCNV41.
RESULTS
After excluding subjects based on principal components analysis (n=43) and quality control procedures (n=16), 394 ICH cases (mean age 73 [SD 10]; females 47%) were included in the discovery stage (Table 1). Lobar ICH cases comprised 58% (n=228) of the discovery cohort. The median ICH volumes, as measured in the first available CT scan upon admission to the hospital, were 50 mL (IQR 43 mL) and 27 mL (IQR 34 mL) for lobar and non-lobar hemorrhages, respectively. After preimputation quality-control procedures, imputation to 1000 Genomes reference panels, and postimputation quality-control filters, a total of 7,567,631 and 7,569,876 SNPs were available for association testing in the discovery sample for lobar and non-lobar ICH, respectively. Quantile-quantile plots showed no significant inflation in association results (Supplemental Figure I, II).
Table 1.
Baseline characteristics.
| Covariate | Discovery | Replication | ||||
|---|---|---|---|---|---|---|
| GOCHA Study | ESs | |||||
| All ICH | Non-lobar ICH | Lobar ICH | All ICH | Non-lobar ICH | Lobar ICH | |
| Total, n (%) | 394 (100) | 166 (42) | 228 (58) | 240 (100) | 169 (70) | 71 (30) |
| Age, mean (±SD) | 73 (10) | 72 (10) | 74 (11) | 74 (12) | 74 (11) | 74 (13) |
| Female, n (%) | 194 (47) | 81 (49) | 112 (48) | 102 (43) | 66 (39) | 36 (51) |
| Baseline ICH volume, mean (±SD) | 39 (41) | 27 (34) | 50 (43) | 40 (62) | 28 (38) | 68 (92) |
| Baseline ICH volume, median (IQR) | 23 (51) | 13 (27) | 36 (64) | 16 (43) | 12 (31) | 26 (76) |
| Imputed SNPs, n | 7,365,864 | 7,274,346 | 7,274,921 | 2 | 2 | 2 |
ESs: European studies; ICH: intracerebral hemorrhage; SD: standard deviation; IQR: interquartile range; SNP: single nucleotide polymorphism
Genome-wide association results
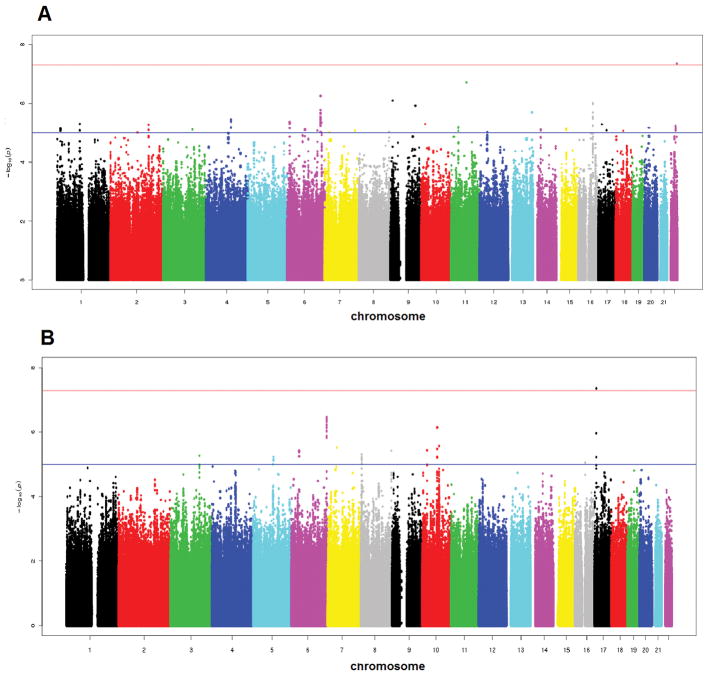
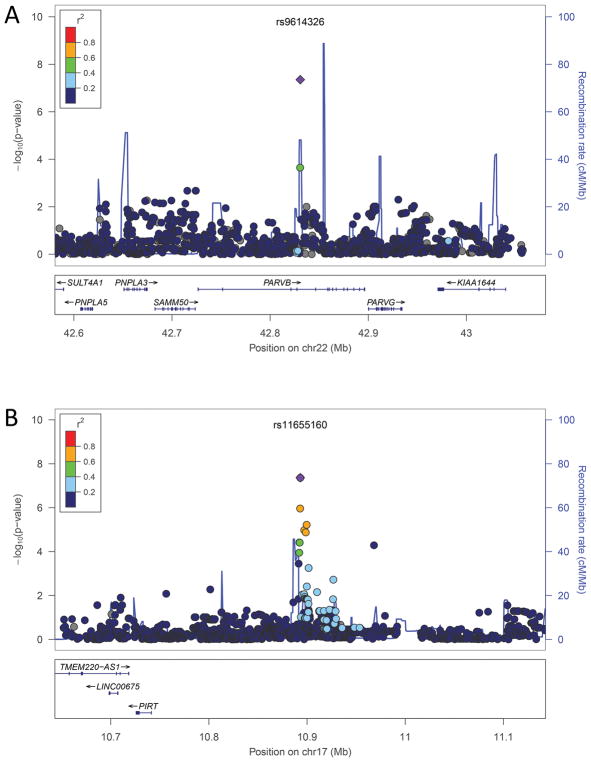
We identified two genome-wide significant susceptibility loci for ICH volume, one for lobar and one for non-lobar ICH (Table 2). In lobar ICH, several SNPs in a genomic region of 22q13 encompassing PARVB were significantly associated with ICH volume (Figure 1A, 2A) (supplemental Table I). For rs9614326, the strongest correlated SNP, each additional C allele was associated with a mean decrease of 6.3 mL in ICH volume (tested allele frequency 3%, beta 1.58, SE 0.29; p=4.5×10−8). In non-lobar ICH, several SNPs in an intergenic region of 17p12 were significantly associated with ICH volume (Figure 1B). For the strongest associated SNP, rs11655160, each additional A allele was associated with a mean decrease of 2.6 mL in ICH volume (tested allele frequency 24%, beta 0.95, SE 0.17; p=4.3×10−8).
Table 2.
Effect size of significant GWAS SNPs for ICH volume
| Genotyping Information | Discovery (GOCHA Study) | Replication (ESs) | Discovery + Replication Meta - Analysis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Phenotype | CHR | A1 | A2 | FRQ A1 | Beta | SE | P | Beta | SE | P | Beta | SE | P | P-het |
| rs11655160 | Non-lobar ICH | 17 | G | A | 76% | 0.95 | 0.17 | 4.33E-08 | 0.52 | 0.25 | 0.036 | 0.82 | 0.14 | 2.5×E-9 | 0.16 |
| rs9614326 | Lobar ICH | 22 | A | C | 97% | 1.84 | 0.32 | 4.45E-08 | 0.45 | 0.66 | 0.491 | 1.58 | 0.29 | 5.8×E-8 | 0.06 |
ESs: European studies; ICH: intracerebral hemorrhage; SE: standard error; A1: tested allele; A2: reference allele; SNP: single nucleotide polymorphism; CHR: chromosome: P-het: study heterogeneity; FRQ: allele frequency
Figure 1.
Genome-wide Association results of autosomal SNPs in with lobar ICH volume (A) and non-lobar ICH volume (B)
The plots show –log10-transformed p values for genotyped and imputed SNPs with respect to their physical positions. The threshold for association at genome-wide significance (p=5×10−8) is shown by the upper line, and the lower line corresponds to p=5×10−5
Figure 2.
Zoom Plots of the regional association results.
(A) Chromosomal region 22q13.3 in lobar ICH. (B) Chromosomal region 17p12 in non-lobar ICH. The index-associated SNP is labeled in violet.
Replication of SNPs associated with ICH volume
The replication stage confirmed the association between rs11655160 in 17p12 and ICH volume in non-lobar ICH. This stage comprised 240 ICH cases (mean age 74 [SD 12]; females 43%), of which 71 (30%) corresponded to lobar ICH. Median ICH volume was 68 mL (IQR 92 mL) and 28 mL (IQR 38 mL) for lobar and non-lobar ICH, respectively, in the replication sample (Table 1). The pre-imputation quality control procedures, imputation pipeline, and post-imputation quality control filters were similar to those utilized in the discovery phase. For 17p12 in non-lobar ICH, each additional A allele of rs11655160 was associated with a 1.7 mL decrease in mean ICH volume (tested allele frequency 26%, beta 0.52, SE 0.25; p=0.036). In a meta-analysis across the replication and discovery stages, the same rs11655160 allele was associated with a 6.6 mL decrease in mean ICH volume (beta 0.82, SE 0.14; p=2.5×10−9), with no significant heterogeneity detected across effect estimates (heterogeneity p=0.16). In lobar ICH, no significant association was found between 22q13 and ICH volume (tested SNP rs9614326: beta 0.4, SE 0.66; p=0.49) (Table 2).
Clinical impact of ICH volume-associated loci in the discovery sample
The identified genetic risk locus at 17p12 was associated with both admission clinical status and functional outcome in the discovery dataset (Table 3). On admission to the hospital, median aGCS was 7 (IQR 7–15). Ninety days after the ICH, 132 (40%) patients had died, 147 (26%) had an mRS of 0–2, and 109 (33.6%) had a mRS of 3–6. After adjustment for multiple potential confounders, each additional G allele of rs11655160 was associated with better clinical status on admission, as measured by the aGCS (beta 0.17, SE 0.60; p=0.004), and better functional outcome, as measured by a dichotomized version of the 3-month post ICH mRS (OR 1.94, SE 0.33; p=0.045).
Table 3.
Ordinal and logistic analysis for clinical G allele and clinical variables.
| SNP and allele | Admission GCS* | 3 months post ICH mRS 3–6* | ||
|---|---|---|---|---|
| OR (SE) | p | OR (SE) | p | |
| rs11655160_G | 0.17 (0.60) | 0.004 | 1.94 (0.33) | 0.045 |
Adjusted for age (continuously coded), sex, hypertension, diabetes, antiplatelet use, Principal Components 1 to 4.
Abbreviations: GCS = Glasgow Coma Scale; mRS = modified Rankin Scale; OR = odds ratio; SE = standard error; SNP= single nucleotide polymorphism.
Locus mapping and regional functional annotation
The susceptibility risk locus for non-lobar ICH volume identified in 17p12 encompasses a narrow 32kb intergenic region (Figure 2B). This locus has no reported associations with other traits. A total of 20 SNPs within this locus achieved p<5×10−5 (supplemental Table II). Two of these variants were directly genotyped in the Illumina 660 BeadChip utilized for genotyping. 17p12 does not contain coding genes or DNAse I hypersensitivity sites. rs11655160 is greatly enriched for copy number variation, overlapping 154 different CNVs (Supplemental Figure III and Supplemental Table III) that cover a 62 megabase region around rs11655160. These CNVs included duplications, insertions, inversions, intrachromosomal breakpoints and short tandem repeats of different sizes, from single base pair alterations to large chromosomal abnormalities spanning. Neither the presence of any CNV at this locus, nor the total sum of CNVs called by PennCNV, were associated with ICH volume, although power and genotyping considerations limit the conclusiveness of this analysis.
DISCUSSION
In this GWAS of hematoma volume in spontaneous, non-traumatic ICH, we evaluated genetic, radiological and clinical data on 634 well-phenotyped cases of European ancestry across discovery and independent replication phases. We identified two novel genome-wide significant susceptibility risk loci for ICH volume: 17p12 for non-lobar ICH and 22q13 for lobar ICH. The association at 17p12 was replicated in an independent dataset of ICH patients without evidence of effect heterogeneity across the discovery and replication phases. Further supporting our findings, this same locus correlated with admission clinical status, as measured by the aGCS, and functional outcome three months after the stroke, as evaluated by the mRS. While the 17p12 locus corresponds to an intergenic region, it overlays numerous CNVs of various sizes and types.
Our study provides new evidence regarding the role of common genetic variation in hematoma formation in spontaneous ICH. Beyond well-known Mendelian conditions that manifest with ICH, recent candidate gene and GWAS studies identified APOE, COL4A1/COL4A2, SLC22A44 and PMF1 as susceptibility loci for ICH risk10,42,43. The identification of these susceptibility loci for risk hinted that genetic variation could also influence ICH volume, as converging lines of evidence indicate that biological processes related to risk also influence volume in ICH. One possible explanation for this phenomenon is the “cascade hypothesis,” which posits that the volume of hematoma is influenced by the severity of the small vessel disease that initially caused the hemorrhage44. According to this model, the final size of the hematoma depends on the additional bleeding contributed by the rupture of previously diseased small vessels located in the periphery of the artery from which the ICH originated. This line of thinking was supported by candidate gene studies that showed that APOE variants related to risk also associated with an increase in the mean volume of the ICH43.
The present study identified a genome-wide significant locus for ICH volume at 17p12. In line with results yielded by prior GWAS of ICH risk, this susceptibility locus was location-specific, exerting its effect on non-lobar hemorrhages only. Importantly, the top SNP within this locus, rs11655160, was also associated with clinical status on admission and functional outcome 3 months after the bleed. The identified locus mapped to an intergenic region that has not been associated with stroke or other phenotypes by previous genome-wide studies. The most important finding of our functional characterization efforts is that the locus of interest is greatly affected by copy number variation, as shown by the 154 CNVs identified in this genomic region. Neither single-CNV nor aggregate-CNV analyses yielded significant associations between copy number variation and ICH volume; however, these findings are greatly limited by our sample size. Despite this negative preliminary analysis, the number and variety of CNVs found at this locus highlight a possible mechanism mediating the observed association.
To further understand the interpretability of our results, we hypothesize that the signal found may be tagging the pathophysiological process of the small vessel disease. To explore this possibility, given the close relation between white matter hyperintensity and cerebral small vessel burden, we looked at the effect of the top SNP rs11655160 on this trait. Three main GWAS have been performed to identify genetic associations with this neuroimaging marker45–47. Among the top SNPs found associated with white matter hyperintensities, none mapped on the 17p12 region.
A second genome-wide significant locus, located at 22q13, was identified for lobar ICH. This locus encompasses PARVB, a gene that codes for parvin beta, a member of the parvin family of actin-binding proteins, which plays an important role in cytoskeleton organization and cell adhesion48. These proteins are associated with focal contacts and contain calponin homology domains that bind to actin filaments. Unfortunately, we were not able to replicate the association between 22q13 and ICH volume in lobar hemorrhages.
Our study has a numer of limitations that are worthwhile mentioning. Patients with massive brain hemorrhages may have died before reaching the hospital, or immediately after admission to the emergency department, and were thus excluded from the present analysis. Second, some misclassification between lobar and non-lobar ICH could have occurred, although, the reliability of the utilized classification approach has proven to be excellent in prior studies49. Our data do not include MRI. Although MRI are less used in the acute setting of stroke, they can provide additional information and a better classification of the underlying pathology compared to an analysis CT based (cerebral amyloid angiopathy vs small vessel disease burden). Possible directions for future research efforts may involve a better classification of ICH based also on features coming from advanced neuroimaging data. Finally, our sample size is small for GWAS studies and mostly composed of individuals of European ancestry. This raises the possibility of false positive associations and precludes a complete fine mapping of the LD structure at the locus. Larger sample sizes will be crucial to further validate our findings and provide trans-ethnic replications.
SUMMARY
In summary, we report the first genome-wide association study of hematoma volume in spontaneous, non-traumatic ICH. We identified a novel susceptibility locus for non-lobar ICH (chromosomal region 17p12) that influences radiological severity, clinical severity, and functional outcome in ICH that occurs in deep regions of the brain. Our results indicate that the influence of common genetic variation in ICH extends beyond risk to also modify the severity and outcome of this condition. While the specific biological mechanism underlying the observed association remains to be elucidated, the presence of numerous CNVs co-localizing with the identified locus suggests that this specific type of mutation could be responsible for the observed changes in ICH volume. Ongoing genetic studies with larger sample sizes will likely answer this question.
Supplementary Material
Acknowledgments
Funding
This work was supported by the NIH National Institute of Neurological Disorders and Stroke (K23NS086873, U01NS069763, and R01NS093870). Project support for the GLGC through C.J.W. and S.K. was provided by the NIH National Heart, Lung, and Blood Institute (R01HL127564). Lund Stroke Register has been supported by the Swedish Heart and Lung Foundation, Skåne University Hospital, Region Skåne, the Freemasons Lodge of Instruction EOS in Lund, King Gustaf V and Queen Victoria’s Foundation, Lund University, the Swedish Stroke Association. Studies from Spain have been supported by Spain’s Ministry of Health (Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III FEDER, RD12/0042/0020). Dr. Anderson was supported by the American Brain Foundation, and received support from the Massachusetts General Hospital Institute for Heart, Vascular, and Stroke Care. GJF is supported by a Yale Pepper Scholar Award (P30AG021342) and the Neurocritical Care Society Research Fellowship.
Footnotes
Author Disclosures
Dr. Lindgren has consulted for Bayer, Astra Zeneca, Boehringer Ingelheim, BMS Pfizer, Reneuron. Dr. Lindgren has consulted for ApoPharma. Dr Rosand has consulted for Boehringer Ingelheim.
Conflicts of interest
None.
References
- 1.Broderick J, Brott T, Duldner J, Tomsick T, Huster G. Volume of Intracerebral Hemorrhage A Powerful and Easy-to-Use Predictor of 30-Day Mortality. Stroke. 1993;24:987–93. doi: 10.1161/01.str.24.7.987. [DOI] [PubMed] [Google Scholar]
- 2.Balami JS, Buchan AM. Complications of intracerebral haemorrhage. Lancet Neurol. 2012;11:101–118. doi: 10.1016/S1474-4422(11)70264-2. [DOI] [PubMed] [Google Scholar]
- 3.Falcone GJ, Biffi A, Brouwers HB, Anderson CD, Battey TWK, Ayres AM, et al. Predictors of hematoma volume in deep and lobar supratentorial intracerebral hemorrhage. JAMA Neurol [Internet] 2013;70:988–94. doi: 10.1001/jamaneurol.2013.98. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3808840&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romero JM, Heit JJ, Delgado Almandoz JE, Goldstein JN, Lu J, Halpern E, et al. Spot sign score predicts rapid bleeding in spontaneous intracerebral hemorrhage. Emerg Radiol. 2012;19:195–202. doi: 10.1007/s10140-012-1020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radmanesh F, Falcone GJ, Anderson CD, Battey TWK, Ayres AM, Vashkevich A, et al. Risk factors for computed tomography angiography spot sign in deep and lobar intracerebral hemorrhage are shared. Stroke. 2014;45:1833–1835. doi: 10.1161/STROKEAHA.114.005276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmerman RD, Maldjian JA, Brun NC, Horvath B, Skolnick BE. Radiologic estimation of hematoma volume in intracerebral hemorrhage trial by CT scan. Am J Neuroradiol. 2006;27:666–670. [PMC free article] [PubMed] [Google Scholar]
- 7.Rost NS, Greenberg SM, Rosand J. The genetic architecture of intracerebral hemorrhage. Stroke. 2008;39:2166–2173. doi: 10.1161/STROKEAHA.107.501650. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter A, Singh I, Gandhi C, Prestigiacomo C. Genetic risk factors for spontaneus intracerebral haemorrhage. Nat Rev. 2016;12:40–49. doi: 10.1038/nrneurol.2015.226. [DOI] [PubMed] [Google Scholar]
- 9.Falcone GJ, Rosand J. Genetic Determinants of Risk, Severity, and Outcome in Intracerebral Hemorrhage. Semin Neurol. 2016;36:298–305. doi: 10.1055/s-0036-1582134. [DOI] [PubMed] [Google Scholar]
- 10.Rannikmäe K, Davies G, Thomson PA, Bevan S, Devan WJ, Falcone GJ, et al. Common variation in COL4A1/COL4A2 is associated with sporadic cerebral small vessel disease. Neurology. 2015;84:918–926. doi: 10.1212/WNL.0000000000001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woo D, Falcone GJ, Devan WJ, Brown WM, Biffi A, Howard TD, et al. Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. Am J Hum Genet. 2014;94:511–521. doi: 10.1016/j.ajhg.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biffi A, Anderson CD, Falcone GJ, Kissela B, Norrving B, Tirschwell DL, et al. Novel insights into the genetics of intracerebral hemorrhage. Stroke. 2013:44. doi: 10.1161/STROKEAHA.113.001912. [DOI] [PubMed] [Google Scholar]
- 13.Anderson CD, Falcone GJ, Phuah CL, Radmanesh F, Brouwers HB, Battey TWK, et al. Genetic variants in CETP increase risk of intracerebral hemorrhage. Ann Neurol. 2016;80:730–740. doi: 10.1002/ana.24780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falcone G, Alessandro B, William DJ, Bart BH, Anderson C, Valerie V, et al. Burden of Blood Pressure-Related Alleles is Associated with Larger Hematoma Volume and Worse Outcome in Intracerebral Hemorrhage. Stroke. 2013;44:321–326. doi: 10.1161/STROKEAHA.112.675181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phuah C-L, Raffeld MR, Ayres AM, Gurol ME, Viswanathan A, Greenberg SM, et al. APOE polymorphisms influence longitudinal lipid trends preceding intracerebral hemorrhage. Neurol Genet [Internet] 2016;2:e81. doi: 10.1212/NXG.0000000000000081. Available from: http://ng.neurology.org/lookup/doi/10.1212/NXG.0000000000000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biffi A, Anderson CD, Jagiella JM, Schmidt H, Kissela B, Hansen BM, et al. APOE Genotype Predicts Extent of Bleeding and Outcome in Lobar Intracerebral Hemorrhage. Lancet Neurol. 2011;10:702–709. doi: 10.1016/S1474-4422(11)70148-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crawford KM, Gallego-Fabrega C, Kourkoulis C, Miyares L, Marini S, Flannick J, et al. Cerebrovascular Disease Knowledge Portal: An Open-Access Data Resource to Accelerate Genomic Discoveries in Stroke. Stroke. 2018:49. doi: 10.1161/STROKEAHA.117.018922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomis M, Ois A, Rodríguez-Campello A, Cuadrado-Godia E, Jiménez-Conde J, Subirana I, et al. Outcome of intracerebral haemorrhage patients pre-treated with statins. Eur J Neurol. 2010;17:443–448. doi: 10.1111/j.1468-1331.2009.02838.x. [DOI] [PubMed] [Google Scholar]
- 19.Domingues-Montanari S, Hernandez-Guillamon M, Fernandez-Cadenas I, Mendioroz M, Boada M, Munuera J, et al. ACE variants and risk of intracerebral hemorrhage recurrence in amyloid angiopathy. Neurobiol Aging [Internet] 2011;32:551e13–551.e22. doi: 10.1016/j.neurobiolaging.2010.01.019. Available from: [DOI] [PubMed] [Google Scholar]
- 20.Hallström B, Jönsson AC, Nerbrand C, Norrving B, Lindgren A. Stroke incidence and survival in the beginning of the 21st century in Southern Sweden: Comparisons with the late 20th century and projections into the future. Stroke. 2008;39:10–15. doi: 10.1161/STROKEAHA.107.491779. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez-Luna D, Boyko M, Subramaniam S, Klourfeld E, Jo P, Diederichs BJ, et al. Magnitude of hematoma volume measurement error in intracerebral hemorrhage. Stroke. 2016;47:1124–1126. doi: 10.1161/STROKEAHA.115.012170. [DOI] [PubMed] [Google Scholar]
- 22.Teasdale G, Jennett B. ASSESSMENT OF COMA AND IMPAIRED CONSCIOUSNESS. A Practical Scale. Lancet. 1974;304:81–84. doi: 10.1016/s0140-6736(74)91639-0. [DOI] [PubMed] [Google Scholar]
- 23.Bonita R, Beaglehole R. Modification of Rankin Scale: Recovery of motor function after stroke. Stroke [Internet] 1988;19:1497–500. doi: 10.1161/01.str.19.12.1497. Available from: http://www.ncbi.nlm.nih.gov/pubmed/3201508. [DOI] [PubMed] [Google Scholar]
- 24.Janssen PM, Visser NA, Dorhout Mees SM, Klijn CJM, Algra A, Rinkel GJE. Comparison of telephone and face-to-face assessment of the modified Rankin Scale. Cerebrovasc Dis. 2010;29:137–139. doi: 10.1159/000262309. [DOI] [PubMed] [Google Scholar]
- 25.Anderson C, Pettersson F, Clarke G, Cardon L, Morris A, Zondervan K. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick Na, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet [Internet] 2006;38:904–909. doi: 10.1038/ng1847. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16862161%5Cnhttp://www.nature.com/doifinder/10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 27.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;6:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durbin RM, Altshuler DL, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, et al. A map of human genome variation from population-scale sequencing. Nature [Internet] 2010;467:1061–73. doi: 10.1038/nature09534%0A. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keep RF, Hua Y, Xi G. Intracerebral haemorrhage: mechanisms of injury and therapeutic targets. Lancet Neurol [Internet] 2012;11 doi: 10.1016/S1474-4422(12)70104-7. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3884550/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biffi A, Battey TWK, Ayres AM, Cortellini L, Schwab K, Gilson AJ, et al. Warfarin-related intraventricular hemorrhage: Imaging and outcome. Neurology. 2011;77:1840–1846. doi: 10.1212/WNL.0b013e3182377e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arima H, Wang JG, Huang Y, Heeley E, Skulina C, Parsons MW, et al. Significance of perihematomal edema in acute intracerebral hemorrhage: The INTERACT trial. Neurology. 2009;73:1963–1968. doi: 10.1212/WNL.0b013e3181c55ed3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu TY, Putaala J, Sharma G, Strbian D, Tatlisumak T, Davis SM, et al. Persistent hyperglycemia is associated with increased mortality after intracerebral hemorrhage. J Am Heart Assoc. 2017:6. doi: 10.1161/JAHA.117.005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roquer J, Rodriguez-Campello A, Jimenez-Conde J, Cuadrado-Godia E, Giralt- Steinhauer E, Vivanco Hidalgo RM, et al. Sex-related differences in primary intracerebral hemorrhage. Neurology. 2016;87:257–262. doi: 10.1212/WNL.0000000000002792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delcourt C, Sandset EC, Zheng D, Chen X, Salman RA, Robinson T, et al. Intracerebral hemorrhage location and outcome among INTERACT2 participants. Neurology. 2017;88:1408–1414. doi: 10.1212/WNL.0000000000003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De La Vega FM, Isaac HI, Scafe CR. A tool for selecting SNPs for association studies based on observed linkage disequilibrium patterns. Pac Symp Biocomput. 2006:487–498. [PubMed] [Google Scholar]
- 38.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SFA, et al. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woo D, Sauerbeck LR, Kissela BM, Khoury JC, Szaflarski JP, Gebel J, et al. Genetic and environmental risk factors for intracerebral hemorrhage: preliminary results of a population-based study. Stroke [Internet] 2002;33:1190–1195. doi: 10.1161/01.str.0000014774.88027.22. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=11988589&retmode=ref&cmd=prlinks%5Cnpapers2://publication/uuid/F4CCED2B-2029-4760-BD5C-E16B7C9F166E. [DOI] [PubMed] [Google Scholar]
- 43.Biffi A, Anderson CD, Jagiella JM, Schmidt H, Kissela B, Hansen BM, et al. APOE genotype and extent of bleeding and outcome in lobar intracerebral haemorrhage: A genetic association study. Lancet Neurol [Internet] 2011;10:702–709. doi: 10.1016/S1474-4422(11)70148-X. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlunk F, Greenberg SM. The Pathophysiology of Intracerebral Hemorrhage Formation and Expansion. Transl Stroke Res. 2015;6:257–263. doi: 10.1007/s12975-015-0410-1. [DOI] [PubMed] [Google Scholar]
- 45.Traylor M, Zhang CR, Adib-Samii P, Devan WJ, Parsons OE, Lanfranconi S, et al. Genome-wide meta-analysis of cerebral white matter hyperintensities in patients with stroke. Neurology. 2016;86:146–153. doi: 10.1212/WNL.0000000000002263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verhaaren BFJ, Debette S, Bis JC, Smith JA, Ikram MK, Adams HH, et al. Multiethnic Genome-Wide Association Study of Cerebral White Matter Hyperintensities on MRI. Circ Cardiovasc Genet. 2015;8:398–409. doi: 10.1161/CIRCGENETICS.114.000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Opherk C, Gonik M, Duering M, Malik R, Jouvent E, Hervé D, et al. Genome-wide genotyping demonstrates a polygenic risk score associated with white matter hyperintensity volume in CADASIL. Stroke. 2014;45:968–972. doi: 10.1161/STROKEAHA.113.004461. [DOI] [PubMed] [Google Scholar]
- 48.Sepulveda JL, Wu C. The parvins. Cell Mol Life Sci. 2006;63:25–35. doi: 10.1007/s00018-005-5355-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rannikmäe K, RW, Anderson C, Charidimou APC, Greenberg S, et al. Reliability of intracerebral hemorrhage classification systems: A systematic review. Int J Stroke. 2016;11:626–636. doi: 10.1177/1747493016641962. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.