Abstract
The mineralocorticoid hormone, aldosterone, is known to play a role in sodium homeostasis. We serendipitously found, however, highly significant association between single-nucleotide polymorphisms in the aldosterone synthase gene and plasma glucose levels in a large population of Chinese and Japanese origin. Two polymorphisms—one in the putative promoter (T-344C) and another resulting in a lysine/arginine substitution at amino acid 173, which are in complete linkage disequilibrium in this population—were associated with fasting plasma glucose levels (P = 0.000017) and those 60 (P = 0.017) and 120 (P = 0.0019) min after an oral glucose challenge. A C/T variant in intron 1, between these polymorphisms, was not associated with glucose levels. Arg-173 and -344C homozygotes were most likely to be diabetic [odds ratio 2.51; 95% confidence interval (C.I.) 1.39–3.92; P = 0.0015] and have impaired fasting glucose levels (odds ratio 3.53; 95% C.I. 2.02–5.5; P = 0.0000036). These results suggest a new role for aldosterone in glucose homeostasis.
Hypertension affects 15–50% of the adult population and is one of the major risk factors for stroke, myocardial infarction, and renal disease. Despite intense effort, our understanding of heritable factors in the etiology of hypertension is rather poor. Twin, adoption, and epidemiologic studies indicate that variation in blood pressure (BP) is genetically determined to some extent (1–3). Insulin resistance, defined as the sluggish uptake of glucose by peripheral tissue in the presence of normal or elevated levels of insulin, is frequently associated with hypertension (4). One of the goals of the Stanford, Asia, and Pacific Program for Hypertension and Insulin Resistance (SAPPHIRe) study is to identify predisposing genes for essential hypertension and insulin resistance.
To maximize our chances of finding susceptibility genes for these diseases, we took several steps. First, in an attempt to reduce genetic heterogeneity, we focused on a relatively homogeneous population of Chinese and Japanese descent. Second, we sampled subjects from tails of the systolic or diastolic BP distributions—the affected hypertensives were recruited from the upper 20% of the distribution, whereas the unaffected “low-normotensive” individuals had BP readings in the lower 30% of the distribution (5). Third, because recent theoretical work suggests that association studies might be adequately powerful to detect susceptibility loci that have a modest effect on the phenotype (6), we began candidate gene studies by testing specific variants in genes that might, for biological reasons, be expected to be involved in BP regulation or glucose homeostasis for association with hypertension or insulin resistance.
The mineralocorticoid hormone, aldosterone, by determining the amount of sodium and water reabsorbed by the kidney, plays a key role in regulating BP (7). Indeed, mutations in the aldosterone synthase gene or in the receptor for the hormone can cause Mendelian forms of hypertension and hypotension (8). For this reason, we evaluated single-nucleotide polymorphisms (SNPs) in the aldosterone synthase gene for association with hypertension. We unexpectedly found, however, that genetic variation in this gene is associated with plasma glucose levels and diabetes status in this population.
Materials and Methods
Subjects.
Individuals were recruited as part of the Stanford, Asia, and Pacific Program for Hypertension and Insulin Resistance study, and details of the study population have been published (9). Briefly, hypertensive subjects of Chinese and Japanese origin were recruited in the San Francisco Bay area, Hawaii, and Taiwan. Hypertension was defined as BP in the upper 20% of the systolic or diastolic BP distributions. In our population, these thresholds translated into the following values: systolic BP (SBP) ≥160 mm Hg or diastolic BP (DBP) ≥95 mm Hg, or taking 2 medications for high BP (stage II hypertension). Alternatively, the subjects had uncontrolled hypertension, i.e., took 1 medication for high BP and had either SBP ≥140 or DBP ≥90 mm Hg. Comparably hypertensive siblings of the proband, as well as low-normotensive siblings, were also recruited. “Low-normal” BP was defined as SBP ≤115 mm Hg and DBP ≤76 for males under 45 years of age. For males over 45 years of age, SBP ≤122 and DBP ≤78 were used. For females younger than 45 years of age, low-normal BP was defined as SBP ≤107 and DBP ≤70 mm Hg. For those females over 45 years of age, the cutoffs were SBP ≤118 and DBP ≤75. Individuals with chronic illnesses like diabetes, heart, liver, or kidney diseases and cancer were excluded from the study. However, individuals diagnosed with diabetes as a result of tests done by us were not excluded from the study.
In this study, 1,368 individuals were evaluated. The distribution of family sizes was 178 singletons, and 252, 117, 48, 19, and 8 with 2, 3, 4, 5, and 6 siblings, respectively. Glucose and insulin levels were measured after an overnight fast and 60 and 120 min after ingesting 75 gm of glucose. Total cholesterol, triglycerides, and high-density lipoprotein levels were measured after an overnight fast.
This study was approved by Institutional Review Boards at all participating sites, and all subjects gave written informed consent.
Genotyping.
Genotyping was performed by using the TaqMan assay as described (10). The probes and primers used in the TaqMan assay were as follows. For the T-344C SNP, the probes were CCAAGGCTCCCTC and CCAAGGCCCCCTC (SNP underlined), and the primers were TTGCAATGAACTAAATCTGTGGTATAAAA (forward) and GAGTAAAATGGATGGGGACTTTATCTTAT (reverse). For the intron 1 SNP, the probes were AGGGACATGACCCCGTCCAGCAG and AGGGACATGACCCTGTCCAGCAGG, and the primers were ACACTTTGGATTGGGACTGCA (forward) and CTTCTCCACATCCTCCGGC (reverse). For the Lys-173/Arg SNP, the probes were CCAGGCCCTGAAGAAGAAGGTGCTG and CCAGGCCCTGAGGAAGAAGGTGCT, and the primers were GATGCAGTGGCCAGGGACT (forward) and GCTGGACGTCCAGGGTCA (reverse).
Statistical Analysis.
Glucose values were transformed to approximate normality and then used in the statistical analyses. Fasting glucose values were raised to the power −1.4 and multiplied by a constant (−100,000). Glucose values 60 [oral glucose tolerance test (OGTT) 60] and 120 (OGTT120) minutes after an oral glucose tolerance test were natural log-transformed. To control for additional sources of variation in glucose values, all three transformed values were regressed on body mass index (BMI), age, gender, ethnicity, and geographical origin of subjects. Residuals from the regression were initially used in ANOVA and multivariate ANOVA to examine main effects of aldosterone synthase genotypes. These analyses, however, did not allow for residual correlation of phenotypes in sibships or the fact that sibling genotypes are not independent. Therefore, significant results were followed by variance-component analysis, as implemented in the program solar (11). This program estimates the proportion of variance explained by a covariate (in this case, the aldosterone synthase genotype) in the presence of background polygenic variance, thus allowing for familial correlation in glucose values. The program also takes into account the nonindependence of sibling genotypes. The heritabilities for fasting and OGTT120 glucose were 49% and 54%, respectively, and highly significant (P < 0.0000001). solar assumes multivariate normality under the null hypothesis of no association; and, although we transformed the data to approximate univariate normality, it is difficult to test this multivariate assumption. For this reason, we did analyses parallel to those from solar. The variance in residual fasting glucose contributed by the Lys-173/Arg or T-344C SNPs in the aldosterone synthase gene was estimated by using a “method of moments” approach, which accounts for familial correlation in trait values but does not make assumptions regarding the distribution of trait values. Details of this method are presented in supporting information, which is published on the PNAS web site, www.pnas.org. The inferences from these analyses were indistinguishable from those obtained from solar. Discrete traits like diabetes and impaired fasting glucose were analyzed by using a liability-threshold model as implemented in solar. Because we used siblings in the analysis, we could not rely on standard methods to estimate the significance of the odds ratios. Instead, we estimated odds ratios and their 95% confidence intervals (C.I.s) by using a percentile bootstrap method (12). For each trait, we drew 1,000 independent samples (with replacement) from the overall sample, each such sample being the same size as the overall sample. An odds ratio was then calculated for each bootstrap sample, and the overall mean of these 1,000 samples was taken as the bootstrap estimate of the odds ratio. To estimate C.I.s, these 1,000 odds ratios were ranked, and the 25th and 975th values were used as the 95% C.I.s.
To estimate linkage disequilibrium among SNPs, one individual was chosen at random from each of the families studied, and pairwise haplotype frequencies were estimated from these 479 individuals by using the expectation-maximization algorithm as implemented in the eh program (ftp://linkage.rockefeller.edu/software/eh).l Note that the number of individuals used in this analysis is less than the number of families noted above because not all individuals were typed for the intronic SNP. Linkage disequilibrium values (D′) were calculated as described (13).
Results
We typed 1,368 subjects of Chinese and Japanese origin (details of study population are given in Table 1) for three SNPs in the aldosterone synthase gene. The three SNPs were a T/C transition at position -344 in the putative promoter (T-344C; ref. 14); a C/T transition in intron 1 (recently identified in our laboratory) 353 bp downstream of the 3′ end of exon 1; and an A/G variant in exon 3, which results in a lysine/arginine amino acid substitution at residue 173 (Lys-173/Arg; ref. 15). None of the SNPs was associated with hypertension in our study population, however (data not shown).
Table 1.
Study population
| Strata | Chinese | Japanese |
|---|---|---|
| Gender (male/female) | 498/530 | 144/196 |
| Hypertensive (% medicated)* | 718 (86.5) | 300 (87.3) |
| Age† | 49.8 ± 8.3 | 55.2 ± 8.1 |
| Body mass index,† kg/m2 | 25.4 ± 3.4 | 26.6 ± 3.8 |
Percent taking antihypertensive medication.
Mean ± SD.
We examined pairwise linkage disequilibrium among these three SNPs (Table 2). Consistent with a previous report (16), the T-334C and Lys-173/Arg SNPs were in complete linkage disequilibrium with each other (D′ = 1, χ21df = 832.5) and are referred to as a single SNP, Lys-173/Arg, in the following discussion. The SNP in intron 1, which lies 939 bp downstream from T-344C and 2,119 bp upstream of Lys-173/Arg, was in weak linkage disequilibrium (D′ = 0.37, χ21df = 70.4) with either SNP.
Table 2.
Aldosterone synthase genotype frequencies
| SNP
|
Lys-173/Arg
|
Intron 1 SNP
|
||||
|---|---|---|---|---|---|---|
| T-344C | Lys/Lys | Lys/Arg | Arg/Arg | C/C | C/T | T/T |
| T/T | 715 | 0 | 0 | 380 | 259 | 52 |
| T/C | 0 | 546 | 0 | 423 | 107 | 0 |
| C/C | 0 | 0 | 107 | 106 | 0 | 0 |
Because insulin resistance or syndrome X is frequently associated with hypertension (4), we also evaluated quantitative phenotypes like plasma glucose, insulin, and lipid levels for association with aldosterone synthase genotypes. As shown in Table 3, neither the Lys-173/Arg SNP nor the intron 1 SNP were associated with lipid or insulin levels.
Table 3.
Aldosterone synthase SNPs and intermediate phenotypes
| Trait | Lys173Arg
|
Intron 1 SNP
|
||||||
|---|---|---|---|---|---|---|---|---|
| Lys/Lys | Lys/Arg | Arg/Arg | P value | C/C | C/T | T/T | P value | |
| Cholesterol | 195.2 (38.7) | 193.7 (38.2) | 195.7 (35.6) | 0.8 | 194.8 (38.5) | 194.1 (38.2) | 195.6 (35.8) | 0.9 |
| HDL | 43.5 (11.8) | 44.8 (12.9) | 45.4 (12.9) | 0.1 | 43.7 (11.8) | 44.9 (12.8) | 45.2 (12.9) | 0.6 |
| Triglycerides | 146.3 (92.7) | 143.7 (94.1) | 133.3 (69.7) | 0.4 | 145.7 (92.7) | 145 (94.8) | 133.6 (70) | 0.7 |
| Fasting insulin | 7.5 (4.7) | 7.7 (5.2) | 7.6 (5.2) | 0.8 | 7.6 (4.8) | 7.8 (5.2) | 7.7 (5.5) | 0.4 |
| OGTT60 insulin | 73.3 (53.1) | 75.4 (54.3) | 70.4 (55.2) | 0.7 | 73.4 (53.5) | 76.3 (54.6) | 70.5 (55.5) | 0.8 |
| OGTT120 insulin | 67.4 (59.2) | 66.8 (60.0) | 62.6 (49.6) | 0.8 | 67.9 (59.7) | 67.1 (60.2) | 63 (49.7) | 0.8 |
Mean and SD for each genotype are given. The P value is derived from a one-way ANOVA with body mass index (BMI), age, gender, ethnicity, and geographical origin as covariates. Cholesterol, high-density lipoproteins (HDL), and triglycerides are expressed in mg/dl. Insulin values, fasting, and 60 and 120 min after an OGTT are expressed in μl units/ml.
In contrast, the Lys-173/Arg SNP showed highly significant association with fasting plasma glucose levels (P = 0.0000067) and those 60 (P = 0.0089) and 120 (P = 0.0005) minutes after an oral glucose load. Because all three glucose levels were correlated, we also analyzed the data by using multivariate ANOVA, with similar results. Taken together, all three glucose levels were highly significantly associated with Lys-173/Arg genotypes (P < 0.0001).
These analyses, however, did not take into account residual correlation in glucose levels within sibships or the fact that sibling genotypes were not independent. Both these factors would tend to reduce the significance of the association between Lys-173/Arg genotypes and glucose levels. For this reason, we analyzed the data by using a variance-components model that allowed for residual correlations in phenotype and nonindependence of sibling genotypes (see Methods). These results are presented in Table 4. Consistent with the above analysis, the Lys-173/Arg SNP showed highly significant association with fasting glucose levels, although, as expected, the P value was slightly less significant (P = 0.000017). Mean fasting glucose values for arginine homozygotes were higher than those for lysine/arginine or lysine/lysine genotypes; the difference in mean values between heterozygotes and lysine homozygotes, however, was not significant. These results suggest a model in which the arginine allele is recessive. The aldosterone synthase gene seemed to have a substantial impact on glucose levels because the difference in mean residual values between arginine homozygotes (15.99) and the other genotypes (−1.01 and −1.8) was approximately half an SD. This SNP explained ≈1.7% of the phenotypic variance and ≈3.5% of the genetic variance in fasting glucose levels.
Table 4.
Aldosterone synthase SNPs and plasma glucose levels
| Glucose trait | Lys173/Arg
|
Intron 1 SNP
|
||||||
|---|---|---|---|---|---|---|---|---|
| Lys/Lys | Lys/Arg | Arg/Arg | P value | C/C | C/T | T/T | P value | |
| Fasting | ||||||||
| Raw | 94.1 (17.7) | 92.6 (14.2) | 102 (27.4) | 94 (17.2) | 94.7 (19.4) | 90.6 (12.4) | ||
| Residual | −1.01 (37.5) | −1.8 (30.6) | 15.98 (32.9) | 0.000017 | 0.21 (35.1) | −1.37 (34.4) | 8.09 (39.8) | 0.2 |
| OGTT60 | ||||||||
| Raw | 177.3 (47.6) | 173.3 (47.3) | 188.8 (59.1) | 177.7 (49.3) | 174.9 (47.6) | 171.3 (43) | ||
| Residual | 0.006 (0.26) | −0.02 (0.26) | 0.067 (0.29) | 0.017 | 0.003 (0.26) | 0.004 (0.26) | −0.05 (0.24) | 0.4 |
| OGTT120 | ||||||||
| Raw | 143.3 (49.8) | 140 (48.2) | 158.1 (65) | 143.9 (52.1) | 139.9 (48.7) | 146.5 (43.9) | ||
| Residual | 0.01 (0.32) | −0.03 (0.31) | 0.1 (0.35) | 0.0019 | 0.005 (0.32) | −0.005 (0.31) | 0.01 (0.32) | 0.9 |
Mean and SD for each genotype are given. Residuals were obtained after regressing glucose values on body mass index (BMI), age, gender, ethnicity, and geographical origin. For the sake of clarity, mean values in the untransformed scale (Raw, in mg/dl) are also given. The P values were obtained from variance-component analysis, allowing for familial correlations in glucose levels, using the program solar.
There was a similar trend for glucose levels after an OGTT. Arginine homozygotes had higher glucose levels compared with individuals with other genotypes, 60 (OGTT60) and 120 min (OGTT120) after ingesting glucose, although the effect was more pronounced for the 120-min levels (Table 4). The Lys-173/Arg SNP contributed ≈0.7% of the phenotypic and ≈1.3% of the genetic variance in glucose levels 2 h after an OGTT. Taken together, these observations suggest that the aldosterone synthase gene contributes more to variation in base-line glucose levels than to the response to glucose loading. In contrast to these results, the SNP in intron 1 showed no association with either fasting or OGTT glucose levels (Table 4).
We could envision two confounding factors that might result in spurious association between aldosterone synthase and glucose levels: population admixture and treatment for hypertension. To determine whether admixture resulted in the association, we stratified the population based on ethnicity and site of recruitment. As shown in Table 5, arginine homozygotes had consistently higher mean fasting glucose levels in both Chinese and Japanese populations. This difference persisted across sites; arginine homozygotes recruited in the San Francisco Bay area, Hawaii, or Taiwan had higher mean fasting glucose values. Finally, we examined sibships that were segregating the arginine allele. Even in such sibships, arginine homozygotes had higher mean glucose values than their siblings with lysine/lysine or lysine/arginine genotypes. These observations make it unlikely that the association between Lys-173/Arg and glucose levels is merely the result of population stratification.
Table 5.
Aldosterone synthase genotypes and fasting glucose levels by ethnicity, geographical origin, medication status, and within siblingships
| Strata | Genotype
|
||
|---|---|---|---|
| Lys/Lys (N) | Lys/Arg (N) | Arg/Arg (N) | |
| Ethnicity | |||
| Chinese | 91.8 ± 16 (553) | 90.7 ± 12.1 (401) | 100.8 ± 30.6 (74) |
| Japanese | 101.7 ± 20.9 (162) | 97.7 ± 17.9 (145) | 104.8 ± 20.9 (33) |
| Site* | |||
| SF Bay | 95.2 ± 15.0 (89) | 92.1 ± 9.6 (42) | 102.7 ± 23 (10) |
| Hawaii | 101.3 ± 23 (170) | 97.6 ± 17.6 (158) | 104.3 ± 17.9 (34) |
| TSGH | 94.8 ± 16.6 (135) | 90.8 ± 10.0 (76) | 104 ± 11.4 (15) |
| NTU | 91.5 ± 14.0 (97) | 92.9 ± 14.1 (72) | 106.6 ± 58.2 (17) |
| VGH | 88.7 ± 13.7 (224) | 89.2 ± 12.2 (198) | 95.9 ± 14.6 (31) |
| Medication† | |||
| UL | 88.8 ± 11.0 (182) | 87.9 ± 9.4 (140) | 92.7 ± 12.2 (28) |
| UH | 92.3 ± 14.0 (74) | 93.2 ± 13.1 (50) | 127.0 ± 70.5 (11)‡ |
| MH | 96.4 ± 19.8 (459) | 94.3 ± 15.5 (356) | 101.8 ± 15.7 (68) |
| Sibships | |||
| Families with Arg/Arg subjects | 95.9 ± 9.6 (22) | 93.7 ± 10.7 (69) | 102.6 ± 31.5 (65) |
| Families without Arg/Arg subjects | 93.9 ± 17.8 (693) | 92.4 ± 14.7 (477) | |
Mean ± SD for fasting glucose level (mg/dl) are given.
Geographical origin of subjects. SF Bay, San Francisco Bay area; TSGH, Tri-Services General Hospital, Taipei, Taiwan; NTU, National Taiwan University, Taipei, Taiwan; VGH, Veterans General Hospital, Taipei/Taichung, Taiwan.
UL, unmedicated low-normotensive; UH, unmedicated hypertensive; MH, hypertensive subjects taking antihypertensive medication.
The median for this group is 107.0 mg/dl.
Certain antihypertensive drugs, like diuretics and β-blockers, can affect glucose levels (17–19). If the association between aldosterone synthase and glucose levels was the result of an interaction between antihypertensive medication and genotype, then one would expect that the effect would be restricted to medicated individuals. This is not the case, however. Even among unmedicated subjects, arginine homozygotes had higher mean glucose values compared with the other genotypes (Table 5).
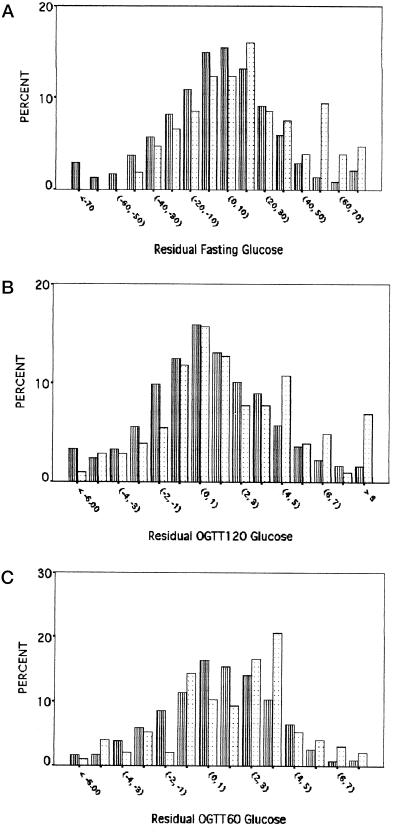
The observed statistical significance could arise in two ways. First, the entire distribution of glucose values among the arginine homozygotes might be displaced from those of the other two genotypes. Alternatively, arginine homozygotes might have a preponderance of high-glucose values. To distinguish between these possibilities, we compared the distribution of residual glucose values for arginine homozygotes to those for the other two genotypes (because the mean values suggested a recessive arginine model, we combined the lysine homozygotes and heterozygotes). As shown in the Fig. 1A, the distribution of residual fasting glucose values for the arginine homozygotes, compared with the other genotypes, was shifted slightly toward higher values, with a paucity of arginine homozygotes at the lower end of the glucose spectrum. Remarkably, the arginine/arginine distribution seemed to be bimodal—a major peak between values 10 and 20 and a second minor peak between values 50 and 60. The distribution of glucose values 2 h after an OGTT was similar (Fig. 1B), with a slight decrease in the frequency of arginine homozygotes at low glucose values and a sharp increase in the frequency of arginine homozygotes between values 4 and 5. There was a similar trend for glucose values 1 h after an OGTT (Fig. 1C), although the pattern was not so consistent. Thus, although there was a shift toward the right in the distribution of residual fasting and 2-h OGTT glucose values for the arginine homozygotes, it seemed that the largest difference in distributions occurred in the right tail, i.e., past the definition of impaired glucose levels.
Figure 1.
Distributions of residual fasting (A), 2-h OGTT (OGTT120, B), and 1-h OGTT (OGTT60, C) glucose values for lysine/lysine + lysine/arginine (striped bars) and arginine/arginine (dotted bars) genotypes.
Consistent with these observations, the Lys-173/Arg SNP was associated with diabetes status. Although we intentionally excluded diabetics from the study, there were 159 subjects whose diabetes was uncovered as a result of our tests. These individuals had 2-h OGTT values that met the World Health Organization threshold (≥200 mg/dl) for diabetes (20, 21). The frequency of arginine homozygotes in this group was 15% (Table 6). In contrast, the frequency of arginine homozygotes in the nondiabetic group was only 7% (odds ratio 2.51; 95% C.I. 1.39–3.92, P = 0.0015). When we considered individuals with impaired fasting glucose levels (≥110 mg/dl) as affected (20–22), the difference in genotype frequencies was even more striking: 6% of the normal individuals, and 19% of the affected subjects, were homozygous for the arginine allele (odds ratio 3.53; 95% C.I. 2.02–5.5, P = 0.0000036).
Table 6.
Aldosterone synthase genotypes and diabetes
| Trait | Genotype
|
Odds ratio‡ (95% C.I.), P value§ | |
|---|---|---|---|
| Lys/Lys + Lys/Arg N (%) | Arg/Arg N (%) | ||
| Diabetes* | |||
| Normal | 1,051 (93.0) | 79 (7.0) | |
| Diabetes | 135 (84.9) | 24 (15.1) | 2.51 (1.39–3.92), 0.0015 |
| Impaired fasting glucose† | |||
| Normal | 1,138 (93.6) | 78 (6.4) | |
| Impaired | 123 (80.9) | 29 (19.1) | 3.53 (2.02–5.5), 0.0000036 |
Diabetes is defined as OGTT120 ≥ 200 mg/dl.
Impaired fasting glucose is defined as fasting glucose levels ≥ 110 mg/dl.
Odds ratios and their 95% C.I. are bootstrap estimates.
P values were obtained from a liability-threshold model, allowing for familial correlations in diabetes and impaired fasting glucose. Heritabilities for diabetes and impaired fasting glucose were 65% and 55%, respectively.
Discussion
Because the T-344C and Lys-173/Arg SNPs are in complete linkage disequilibrium with each other in the populations studied here, we cannot determine which SNP is causative. Deletion of Lys-173 causes pathologically low levels of aldosterone (hypoaldosteronism; ref. 23), and there was a small but statistically insignificant difference in the activity of aldosterone synthase bearing lysine or arginine residues at position 173 in in vitro studies (14, 24). The T-344C SNP is in a region that is conserved among several mammalian species and is bound in vitro by the steroidogenic transcription factor SF-1. Deletion of this region, however, did not affect transcription of the aldosterone synthase gene in in vitro experiments (25). This SNP was associated also with plasma and urinary aldosterone levels (26–30). Linkage disequilibrium between the T-344C and Lys-173/Arg SNPs, however, was not examined in these studies, therefore raising the possibility that, in fact, the Lys-173/Arg SNP (or another undetected polymorphism in this gene) was causatively associated with differences in aldosterone levels in these studies.
It might, at first sight, appear puzzling that the T-344C and Lys-173/Arg SNPs are in complete linkage disequilibrium with each other and were associated with plasma glucose levels, but an SNP in intron 1 between these two SNPs was only in weak linkage disequilibrium with either SNP and showed no association with glucose levels. One explanation for these observations is that there were two common ancestral haplotypes, T-344/Intron 1-C/Lys-173 and C-344/Intron 1-C/Arg-173, and the T allele of the intronic SNP arose on the former haplotype, resulting in the third less common T-344/Intron 1-T/Lys-173 haplotype.
How could nucleotide variation in the aldosterone synthase gene contribute to variation in glucose levels and susceptibility to diabetes? One possibility is that the SNPs might affect the activity/expression of the enzyme, which in turn would affect aldosterone levels. Differences in aldosterone levels might then contribute to variation in the expression/activity of glucose transporters or sodium/glucose cotransporters, thereby affecting plasma glucose levels. Alternatively, changes in aldosterone levels might affect plasma potassium levels, which might directly or indirectly, by hindering the action of insulin, contribute to hyperglycemia. Indeed, diabetes is associated with both hyperkalemia and hypoaldosteronism (31, 32), and glucose infusions decreased plasma aldosterone (33, 34) and potassium levels (34, 35) in nondiabetic individuals. Furthermore, there is evidence that aldosterone can affect insulin receptor mRNA levels, at least in cultured cells (36), and aldosterone levels have been correlated with insulin resistance (37). Finally, aldosterone induces the expression of the serum glucocorticoid-inducible kinase gene, whose expression level also responds to changes in glucose and insulin levels (38, 39), suggesting yet another potential connection between aldosterone and glucose homeostasis. Alternatively, it is possible that arginine at residue 173 affects the activity of aldosterone synthase such that the rate of conversion of an intermediate to aldosterone is reduced. This defect might result in a slight accumulation of an intermediate like corticosterone, a minor glucocorticoid. This mechanism would be consistent with the recessive effect of the arginine allele on glucose levels.
It is also conceivable that, in fact, the adjacent and highly homologous gene 11-β-hydroxylase, which is involved in glucocorticoid synthesis, determines glucose levels and susceptibility to diabetes, and the SNPs we investigated are merely in linkage disequilibrium with variants in that gene. Additional studies of nucleotide variation and haplotype structure in this region of chromosome 8 will shed light on this issue. Preliminary studies, however, showed no association between variation in the 11-β-hydroxylase gene and glucose levels in our population (K.R., unpublished observations).
In the present context, it is noteworthy that the rat homolog of aldosterone synthase is located in a region on rat chromosome 7 that is highly significantly linked to plasma glucose levels in the Otsuka Long-Evans Tokushima Fatty rat, a model for type II diabetes (40). Thus, the aldosterone synthase gene might also contribute to variation in glucose levels in this rat model.
In summary, our results, showing highly significant association between genetic variation in the aldosterone synthase gene and plasma glucose levels, suggest a new role for aldosterone in glucose homeostasis. Furthermore, that two flanking SNPs showed highly significant association, but that an SNP in the middle fewer than 3,000 bp from these SNPs showed no association with phenotype, suggests caution in the use of linkage disequilibrium for mapping complex traits.
Supplementary Material
Acknowledgments
We thank patients for participating in this study. We also thank Stephen Mockrin and Susan Old of the National Heart, Lung, and Blood Institute; and other members of the Stanford, Asia, and Pacific Program for Hypertension and Insulin Resistance project for their help. We thank Ken Livak and Mike Lucero of Perkin–Elmer (Applied Biosystems Division) for help with the TaqMan genotyping assay. K.R. thanks Winnie Koo for pointing out the Decode study. This work was supported by National Institutes of Health Grant U01 HL54527-0151 from the National Heart, Lung, and Blood Institute.
Abbreviations
- BP
blood pressure
- SBP
systolic BP
- DBP
diastolic BP
- SNP
single-nucleotide polymorphism
- OGTT
oral glucose tolerance test
- C.I.
confidence interval
Footnotes
Xie, X. & Ott, J. (1993) Am. J. Hum. Genet. 53, Suppl., 1107 (abstr.).
References
- 1.Christian J C. In: Children's Blood Pressure. Flier L J, Lauer R M, editors. Columbus, OH: Ross Laboratories; 1985. pp. 51–55. [Google Scholar]
- 2.Longini I M, Higgins M W, Hinton P C, Moll P C, Keller J B. Am J Epidemiol. 1984;120:131–144. doi: 10.1093/oxfordjournals.aje.a113862. [DOI] [PubMed] [Google Scholar]
- 3.Biron P, Mongeau J G, Bertrand D. Can Med Assoc J. 1976;115:773–774. [PMC free article] [PubMed] [Google Scholar]
- 4.Reaven G M, Lithell H, Landsberg L. N Engl J Med. 1996;334:374–381. doi: 10.1056/NEJM199602083340607. [DOI] [PubMed] [Google Scholar]
- 5.Risch N, Zhang H. Science. 1995;268:1584–1589. doi: 10.1126/science.7777857. [DOI] [PubMed] [Google Scholar]
- 6.Risch N, Merikangas K. Science. 1996;13:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 7.Vinson G P, Whitehouse B, Hinson J. The Adrenal Cortex. Englewood Cliffs, NJ: Prentice–Hall; 1992. [Google Scholar]
- 8.Lifton R P. Science. 1996;272:676–680. doi: 10.1126/science.272.5262.676. [DOI] [PubMed] [Google Scholar]
- 9.Ranade K, Hsuing A C, Wu K D, Chang M S, Chen Y T, Hebert J, Chen Y I, Olshen R, Curb D, Dzau V, et al. Am J Hypertens. 2000;13:704–709. doi: 10.1016/s0895-7061(00)00238-7. [DOI] [PubMed] [Google Scholar]
- 10.Ranade K, Chang M S, Ting C T, Pei D, Hsiao C F, Olivier M, Pesich R, Hebert J, Chen Y D, Dzau V J, et al. Genome Res. 2001;11:1262–1268. doi: 10.1101/gr.157801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Almasy L, Blangero J. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Efron B, Tibshirani R J. An Introduction to the Bootstrap. New York: Chapman & Hall; 1993. [Google Scholar]
- 13.Lewontin R C. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White P C, Slutsker L. Endocr Res. 1995;21:437–442. doi: 10.3109/07435809509030459. [DOI] [PubMed] [Google Scholar]
- 15.Fardella C E, Rodriguez H, Montero J, Zhang G, Vignolo P, Rojas A, Villarroel L, Miller W L. J Clin Endocrinol Metab. 1996;81:4347–4351. doi: 10.1210/jcem.81.12.8954040. [DOI] [PubMed] [Google Scholar]
- 16.Komiya I, Yamada T, Takara M, Asawa T, Shimabukuro M, Nishimori T, Takasu N. Hypertension. 2000;35:699–703. doi: 10.1161/01.hyp.35.3.699. [DOI] [PubMed] [Google Scholar]
- 17.Greenberg A. Am J Med Sci. 2000;319:10–24. [PubMed] [Google Scholar]
- 18.Suter P M, Vetter W. J Hypertens. 1995;13,Suppl.:S11–S17. doi: 10.1097/00004872-199512002-00003. [DOI] [PubMed] [Google Scholar]
- 19.Ramsay L E, Yeo W W, Jackson P R. J Cardiovasc Pharmacol. 1992;20:S49–S53. [PubMed] [Google Scholar]
- 20.Alberti K G, Zimmet P Z. Diabetes Med. 1998;15:539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 21.DECODE Study Group; European Diabetes Epidemiology Group. Lancet. 1999;354:617–621. [PubMed] [Google Scholar]
- 22.Gabir M M, Hanson R L, Dabelea D, Imperatore G, Roumain J, Bennett P H, Knowler W C. Diabetes Care. 2000;23:1108–1112. doi: 10.2337/diacare.23.8.1108. [DOI] [PubMed] [Google Scholar]
- 23.Peter M, Nikischin W, Heinz-Erian P, Fussenegger W, Kapelari K, Sippell W G. Horm Res. 1998;50:222–225. doi: 10.1159/000023278. [DOI] [PubMed] [Google Scholar]
- 24.Portrat-Doyen S, Tourniaire J, Richard O, Mulatero P, Aupetit-Faisant B, Curnow K M, Pascoe L, Morel Y. J Clin Endocrinol Metab. 1998;83:4156–4161. doi: 10.1210/jcem.83.11.5258. [DOI] [PubMed] [Google Scholar]
- 25.Clyne C D, Zhang Y, Slutsker L, Mathis J M, White P C, Rainey W E. Mol Endocrinol. 1997;11:638–649. doi: 10.1210/mend.11.5.9920. [DOI] [PubMed] [Google Scholar]
- 26.Hautanena A, Lankinen L, Kupari M, Janne O A, Adlercreutz H, Nikkila H, White P C. J Intern Med. 1998;244:11–18. doi: 10.1046/j.1365-2796.1998.00308.x. [DOI] [PubMed] [Google Scholar]
- 27.Pojoga L, Gautier S, Blanc H, Guyene T T, Poirier O, Cambien F, Benetos A. Am J Hypertens. 1998;11:856–860. doi: 10.1016/s0895-7061(98)00048-x. [DOI] [PubMed] [Google Scholar]
- 28.Brand E, Chatelain N, Mulatero P, Fery I, Curnow K, Jeunemaitre X, Corvol P, Pascoe L, Soubrier F. Hypertension. 1998;32:198–204. doi: 10.1161/01.hyp.32.2.198. [DOI] [PubMed] [Google Scholar]
- 29.Paillard F, Chansel D, Brand E, Benetos A, Thomas F, Czekalski S, Ardaillou R, Soubrier F. Hypertension. 1999;34:423–429. doi: 10.1161/01.hyp.34.3.423. [DOI] [PubMed] [Google Scholar]
- 30.Davies E, Holloway C D, Ingram M C, Inglis G C, Friel E C, Morrison C, Anderson N H, Fraser R, Connell J M. Hypertension. 1999;33:703–707. doi: 10.1161/01.hyp.33.2.703. [DOI] [PubMed] [Google Scholar]
- 31.Perez G O, Lespier L, Knowles R, Oster J R, Vaamonde C A. Arch Intern Med. 1977;137:1018–1022. [PubMed] [Google Scholar]
- 32.Cox M, Sterns R H, Singer I. N Engl J Med. 1978;299:525–532. doi: 10.1056/NEJM197809072991007. [DOI] [PubMed] [Google Scholar]
- 33.Hochberg Z, Dickerman Z, Kaufman H, Laron Z. Horm Res. 1980;12:16–21. doi: 10.1159/000179101. [DOI] [PubMed] [Google Scholar]
- 34.Rosenstock J, Loizou S A, Brajkovich I E, Mashiter K, Joplin G F. Diabetologia. 1982;22:184–187. doi: 10.1007/BF00283750. [DOI] [PubMed] [Google Scholar]
- 35.Ferrannini E, Galvan A Q, Santoro D, Natali A. J Hypertens. 1992;10,Suppl.:S5–S10. doi: 10.1097/00004872-199204001-00002. [DOI] [PubMed] [Google Scholar]
- 36.Campion J, Maestro B, Mata F, Davila N, Carranza M C, Calle C. J Steroid Biochem Mol Biol. 1999;70:211–218. doi: 10.1016/s0960-0760(99)00117-x. [DOI] [PubMed] [Google Scholar]
- 37.Goodfriend T L, Egan B M, Kelley D E. Prostaglandins Leukotrienes Essent Fatty Acids. 1999;60:401–405. doi: 10.1016/s0952-3278(99)80020-9. [DOI] [PubMed] [Google Scholar]
- 38.Lang F, Klingel K, Wagner C A, Stegen C, Warntges S, Friedrich B, Lanzendorfer M, Melzig J, Moschen I, Steuer S, et al. Proc Natl Acad Sci. 2000;97:8157–8162. doi: 10.1073/pnas.97.14.8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Barbry P, Maiyar A C, Rozansky D J, Bhargava A, Leong M, Firestone G L, Pearce D. Am J Physiol Renal Physiol. 2001;280:F303–F313. doi: 10.1152/ajprenal.2001.280.2.F303. [DOI] [PubMed] [Google Scholar]
- 40.Wei S, Wei K, Moralejo D H, Ogino T, Koike G, Jacob H J, Sugiura K, Sasaki Y, Yamada T, Matsumoto K. Mamm Genome. 1999;10:249–258. doi: 10.1007/s003359900982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.