Abstract
The heterochromatic state of the Drosophila Y chromosome has made the cloning and identification of Y-linked genes a challenging process. Here, we report application of a procedure to identify Y-linked gene fragments from the unmapped residue of the whole genome sequencing effort. Previously identified Y-linked genes appear in sequenced scaffolds as individual exons, apparently because many introns have become heterochromatic, growing to enormous size and becoming virtually unclonable. A TBLASTN search using all known proteins as query sequences, tested against a blastable database of the unmapped fragments, produced a number of matches consistent with this scenario. Reverse transcription–PCR and genetic methods were used to confirm those that are expressed, Y-linked genes. The five genes reported here include three protein phosphatases (Pp1-Y1, Pp1-Y2, and PPr-Y), an occludin-related gene (ORY), and a coiled-coils gene (CCY). This brings the total to nine protein-coding genes identified on the Drosophila Y chromosome. ORY and CCY may correspond, respectively, to the fertility factors ks-1 and ks-2, whereas the three protein phosphatases represent novel genes. There remains a strong functional coherence to male function among the genes on the Drosophila Y chromosome.
The Y chromosome of Drosophila has several unusual features that together have made the molecular identification of its genes difficult. In addition to ribosomal DNA and a few other multiple copy genes, it is known to contain six single-copy genes essential for male fertility (kl-1, kl-2, kl-3, kl-5, ks-1, and ks-2) (1, 2). X/0 Drosophila males are completely normal (except for the sterility), so the Y chromosome seems to have an unusual functional specialization, apparently containing only genes directly involved with male fertility (3, 4). This functional specialization is highlighted by the recent finding that kl-2 and kl-3 fertility factors encode dynein heavy chains (4), as previously found for kl-5 (5). Thus, three of the six known single-copy genes encode proteins belonging to the same gene family, a functional specialization unheard of in eukaryotic chromosomes. The structure of Y-linked genes is also peculiar. Several fertility factors produce lampbrush loop structures during spermatogenesis, a finding that first suggested that these genes are very large (6–8). This suggestion was confirmed by the painstaking study of Gatti and Pimpinelli (9), which produced a detailed cytogenetic map of the Y chromosome, and by Kurek et al. (10) and Reugels et al. (11), who showed that kl-5 contains gigantic heterochromatic introns.
These unusual features, plus the natural interest in an uncharted terrain of the Drosophila genome, led to extensive efforts to clone Y chromosome fertility factors in Drosophila. Despite these efforts, until recently only kl-5 had been identified at the molecular level (5). The slow progress was mainly caused by the heterochromatic state of the Y chromosome, which prevents or makes more difficult the use of powerful genetic tools such as recombination mapping, cytogenetic banding, P-element mutagenesis, and genome walking. Even the ultimate weapon, genome sequencing, did not immediately identify the Y genes, in part because heterochromatic sequences are not stable in the cloning vectors used in the whole genome shotgun (WGS). So the Drosophila Genome Project produced an essentially complete sequence of the 120-Mbp euchromatic portion of the genome, assembling it into the chromosome arms X, 2L, 2R, 3L, 3R, and 4, and produced also some 4 Mbp of sequence that did not fit in any of the chromosome arms (12). This unmapped residue was called “armU,” and it presumably corresponds to the small portion of the 60 Mbp of Drosophila heterochromatin (including Y-linked genes) that has unique, nonrepetitive sequence, immersed in a sea of satellite DNA and transposable elements.
We recently developed a method for the identification of putative genes in the unmapped portion of the Drosophila genome, and we successfully applied it to identify three Y genes (4). Two of the Y genes correspond to the fertility factors kl-2 and kl-3 and encode dynein heavy chains, whereas the third is a previously unknown gene (PRY). Here, we report the identification of five new Y genes, by using a generalization of this method. As was the case with the previously identified Y genes, these new genes have close homologs in the autosomes and presumably were acquired by translocation, rather than being present in the putative primitive X-Y pair. Most of them seem to have male-related functions, and three of them encode protein phosphatases. These results, coupled with the previous finding that three of the six fertility factors encode the molecular motor dynein, highlight the extreme functional coherence and unusual evolution of the Drosophila Y chromosome.
Materials and Methods
Drosophila Strains.
Kennison's X-Y translocation strains V24, E15, F12, W19, and V8 were kindly provided by D. L. Lindsley (University of California at San Diego, La Jolla), and strain W27 was obtained from the Bloomington Stock Center, which also provided the strains R(YL)/C(1;YS)1, y1 w1, and C(YS)2/C(1;YL)1, y1 v1 f1 bb− & C(1)DX, y1 f1/0. The iso-1 strain, used in the whole genome shotgun sequencing effort, was kindly provided by R. Hoskins from the Berkeley Drosophila Genome Project.
Test for Y Linkage and Mapping.
The genomic location of each candidate scaffold (see below) was tested by performing PCR in male and virgin female DNA from the iso-1 and Oregon-R strains. Male-specific scaffolds (i.e., Y-linked) were then mapped to the regions of the Y chromosome with a set of male-fertile reciprocal X-Y translocations having one breakpoint in the proximal heterochromatin of the X and one in the Y chromosome (2). F1 progeny of pairs of these lines lack specific regions of the Y chromosome (e.g., kl-2− males). By performing PCR in males lacking each region of the Y chromosome, we were able to map unambiguously the tested scaffold. Y-deficiency males were obtained with the following crosses (females first): kl-5−, attached-X/0 × V24; kl-3−, V24 × W27; kl-2−, W27 × E15; kl-1−, E15 × F12; ks-1−, V8 × W19; ks-2−, attached-X/0 × V8. An independent mapping on YS and YL elements was made by crossing males from strains R(YL)/C(1;YS)1, y1 w1, and C(YS)2/C(1;YL)1, y1 v1 f1 bb− & C(1)DX, y1 f1/0, to Oregon-R females. This allowed recovery of flies carrying only R(YL), C(1;YS)1, C(YS)2, or C(1;YL).
Identification of Candidate Y-Linked Genes.
Y-linked genes were sought by two different approaches, described below.
tblastn Search of Y-Linked Genes in armU.
This approach was used in our previous paper (4), where a more detailed description may be found. Heterochromatic genes, and Y-linked ones in particular, are prone to have large introns composed of repetitive sequences that cannot be sequenced and assembled by WGS (nor by any available method). On the other hand, their exons and small intervening introns have a unique sequence and will appear at the end of WGS as small isolated scaffolds. These small pieces of genes usually remain undetected by first pass annotation procedures such as gene prediction programs. However, the coding sequence of the whole gene can be recovered if we have a good query sequence to use in a BLAST search. For known genes, we can use cDNA as a query in a BLASTN search, and for unknown genes, a related protein and TBLASTN. In both cases, the BLAST hits have a characteristic staggered pattern, resulting from the scattering (in different isolated scaffolds) of the exons belonging to the same gene (see Figs. 1, 3, and 4). To identify protein query sequences, we did a TBLASTN search of each of the roughly 500,000 protein sequences in the NCBI's nr database against a database of the unmapped Drosophila scaffolds (called “armU” in Celera's CD-ROM release of the Drosophila genome). This search was performed with StandAloneBlast, whose output was filtered by computer programs tailored for this purpose (to be published elsewhere). Proteins having hits in two or more armU scaffolds were then checked for the staggered pattern. For all such staggered candidates, we tested one or two of the involved armU scaffolds for Y linkage. Y-linked scaffolds were then mapped onto the Y chromosome regions by PCR against a set of Y deletion lines described above. Finally, we closed the gaps between scaffolds by performing reverse transcription–PCR (RT-PCR) with primers designed to the putative N and C termini. Note that this RT-PCR also tests the putative gene for expression. When necessary, rapid amplification of 3′ cDNA ends (3′-RACE) was used to complete the missing end of the genes.
Figure 1.
Y-linked protein phosphatases (Pp1-Y1 and Pp1-Y2 genes). (A) TBLASTN using a Drosophila PPP1 as the query sequence. (B and C) BLASTN searches using cDNA of Pp1-Y1 and Pp1-Y2, respectively, as query sequences. The numbers above the bars are the abridged accession nos. (AE003043 was abridged to 3043 and so on). The fragments labeled “RT-PCR” had no armU match and were sequenced de novo.
Figure 3.
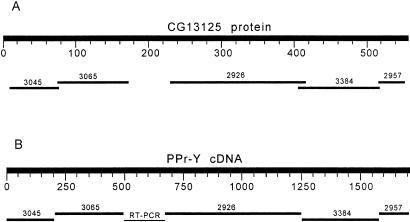
PPr-Y gene. (A) TBLASTN using CG13125 gene product as the query sequence. (B) BLASTN using the cDNA of the long splice variant of PPr-Y as the query sequence.
Figure 4.
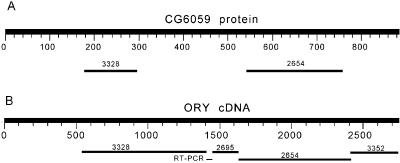
ORY gene. (A) TBLASTN using CG6059 gene product as the query sequence. (B) BLASTN using the cDNA of ORY as the query sequence.
Testis Expressed Sequence Tags (ESTs).
We used Drosophila testis EST sequences (ref. 13; accession nos. AI944400–AI947263) in a BLASTN search against the armU database and looked for perfect matches. The rationale was that a gene expressed in testis (present in the testis EST library) and heterochromatic (present in armU) has a good chance of being Y-linked, for testis cDNAs have revealed many Y-linked genes in humans (14). Promising candidates were tested for Y linkage and mapped as described above.
Molecular Biology Methods.
RNA and DNA extractions, PCR, and RT-PCR were performed by using standard protocols (see ref. 4 for details). 3′-RACE was performed with the 5′/3′-RACE kit (Roche Diagnostics) following the instructions of the manufacturer, using either testis or whole body total RNA (from Oregon-R males) as template. Primer sequences are available on request.
Results
The TBLASTN search of 500,000 proteins against the armU database yielded 18 candidates with a staggered pattern (i.e., at least two hits in nonoverlapping regions of the protein). For each one, we tested at least one scaffold for Y linkage and found that four genes are Y-linked. RT-PCR showed that all of these genes are expressed in adult males, allowing us to sequence the gaps and find the precise splice junctions.
The testis EST search yielded five candidates, of which one proved to be Y-linked (testis EST AI946068/armU scaffold AE003014). Thus, we found five new Y genes, described below.
Protein Phosphatase 1 Catalytic Subunit, Y-Linked Gene 1 (Pp1-Y1).
The armU scaffolds AE002932, AE002921, and AE003043 showed strong similarity to catalytic subunits of protein phosphatases (Fig. 1A) and proved to be Y-linked. Given the overlap between AE002932 and the other two scaffolds, there must be at least two genes. The mapping confirmed this: AE002932 maps to the kl-5 region, whereas AE002921 and AE003043 produce a complex result (described below). RT-PCR using primers located in the putative coding region of AE002932 confirmed expression of Pp1-Y1.
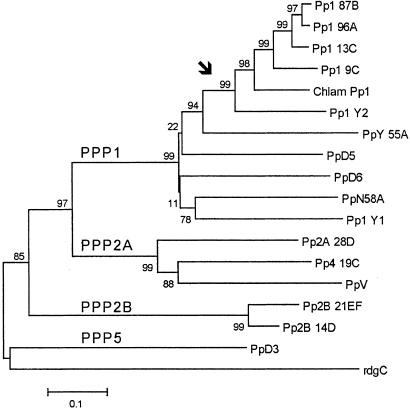
The coding region of the AE002932 scaffold presumably contains the complete N terminus of the protein and finishes abruptly at amino acid residue no. 199. We obtained the missing C terminus by 3′-RACE; this sequence was not included in the released Drosophila genome. A neighbor-joining analysis (15) of the complete protein sequence (306 residues) showed that Pp1-Y1 belongs to the PPP1 subfamily of serine–threonine protein phosphatases (Fig. 2), a group of enzymes involved in the control of a diversity of physiological processes, such as cellular divisions, flagellar motility, and muscle contraction (17). The closest Drosophila homolog seems to be the autosomal gene PpN58A (58% amino acid identity and 88% similarity; Fig. 2), which suggests that Pp1-Y1 originated from a duplication of PpN58A. Interestingly, this gene is expressed only in testis, apparently in the nucleus of undifferentiated germ cells (18). However, several other PPP1 gene family members may also be the parental gene (e.g., PpD5, with 57% amino acid identity, 84% similarity). We examined the intron–exon structure, searching for more clues on the parental gene of Pp1-Y1. There is no intron in the available genomic sequence of Pp1-Y1 (residues 1–199), but unfortunately the absence of an intron in this region is shared by most Drosophila PPP1 members (except for Pp1–96A and Pp1–9C; see below), so it is not informative.
Figure 2.
Phylogeny of the Drosophila serine–threonine protein phosphatase-catalytic subunits (neighbor-joining with Poisson correction and complete deletion; interior branch test for 1,000 replicates; ref. 15). The subfamilies are indicated in the figure. Chlam Pp1 is the Chlamydomonas protein phosphatase that controls flagellar beating (ref. 16; AAD38856). The bar indicates the number of amino acid substitutions per site.
Pp1-Y1 probably is not an essential gene, as the formal genetic data available indicate that the kl-5 region contains only one gene essential for male fertility (the β dynein kl-5).
Protein Phosphatase 1 Catalytic Subunit, Y-Linked Gene 2 (Pp1-Y2).
Both AE002921 and AE003043 produce PCR bands with all single-deletion Y lines (kl-5−, kl-3−, etc.), which indicates that the original Y chromosome used to construct them (BS Y y+) contains more than one copy of the gene. We determined the location of the copies by performing PCR with flies containing Y chromosomes deleted for several regions. In particular, females from the V24 strain carry the XDYP element and possess only the kl-5 region, whereas V8 females carry the YDXP element and possess only the ks-2 region (2). Both were PCR-positive for AE002921 and AE003043 scaffolds, and thus there must be one copy in kl-5 and one copy in ks-2. This duplication may be present on all Y chromosomes, or it may be an artifact induced during construction of BS Y y+. In support of the second hypothesis, the y+ marker was originally present in the long arm of the Y chromosome (YL) and somehow transferred to the short arm (YS; ref. 9). This exchange could have carried some YL sequences (including Pp1-Y2) with it. To resolve this ambiguity, we examined two pairs of independently obtained YL and YS elements (see Material and Methods), and we found that Pp1-Y2 scaffolds are present on R(YL) and C(1;YL) but not on C(1;YS)1 and C(YS)2. Thus, Pp1-Y2 is located in YL, most likely at its very tip.
RT-PCR produced a fragment of the expected size, whose identity was confirmed by sequencing. There is a gap of 45 bp between AE003043 and AE002921. Genomic PCR crossed this gap and showed that there is no intron in this region of the gene. Except for the gap, the AE003043 and AE002921 scaffolds contain the complete coding sequence of Pp1-Y2; in the expected positions (inferred from the alignment with other PPP1), there is a methionine start codon in AE003043 and a stop codon in AE002921.
A neighbor-joining analysis of the protein sequence (309 residues) showed that Pp1-Y2 belongs to the PPP1 subfamily (Fig. 2), being closely related to Pp1–96A, Pp1–87B, Pp1–9C, and Pp1–13C. Among them, only Pp1–87B and Pp1–13C are devoid of introns in the coding region (as Pp1-Y2). Sequence data suggest that Pp1-Y2 is more closely related to Pp1–87B (71% identity, 90% similarity; the corresponding values for Pp1–13C are 70% and 90%). Pp1–87B mutants have impaired chromatin condensation, neurogenesis, oogenesis, and adult behavior. As judged by its EST hits, Pp1–87B is expressed in embryo, ovary, head, larval-early pupae, and, interestingly, testis. Both sequence similarity and intron–exon structure suggest that Pp1–87B is the parental gene of Pp1-Y2, but the evidence is weak (all four PPP1 family members mentioned above are good candidates) because of the strong conservation of PPP1 and because intron–exon structure is not perfectly conserved between the Y-linked genes and their parent genes (e.g., Table 1).
Table 1.
Intron–exon structure of CG6059 and Occludin-related Y
Pp1-Y2 and Pp1-Y1 have 57% identity (85% similarity) at the amino acid level. The statistically significant node marked with an arrow in Fig. 2 does not include Pp1-Y1 and hence essentially rules out the possibility that they originated from a duplication after the translocation to the Y chromosome.
Protein Phosphatase 1 Regulatory Subunit (PPr-Y).
A TBLASTN search, using the autosomal gene CG13125 as the query, showed a clear staggered pattern with the armU scaffolds AE003045-AE003065-gap-AE002926-AE003384-AE002957 (Fig. 3A). RT-PCR using 3045-forward and 2957-reverse primers amplified a 1-kb fragment instead of the predicted 1.5 kb (predicted size is based on the CG13125 protein). Sequencing demonstrates that this RT-PCR product skips the AE002926 scaffold and that the gap (Fig. 3A) is real, corresponding to a whole exon missing in armU. As AE002926 is also Y linked, we thought that it may be present in a rarer splicing variant. We investigated this possibility with two RT-PCR reactions by using the following primers: (i) 3045-forward–2926-reverse, which produced a fragment with the expected size (1.1 kb). Sequencing confirmed the structure 3045–3065-gap-2926; and (ii) 2926-forward–2957-reverse, which produced a fragment of the expected size (900 bp). Sequencing confirmed the structure AE002926-AE003384-AE002957. Thus, it is likely that there are two splice variants of this gene: a rare one encompassing the scaffolds AE003045-AE003065-gap-AE002926-AE003384-AE002957 and a major variant, which skips the AE002926 sequence. There are methionine and stop codons at the expected positions in the AE003045 and AE002957 scaffolds, so the PPr-Y sequence is complete. It has 57% identity (75% similarity) with CG13125.
Sequencing of the RT-PCR products showed that the shared exons are identically spliced in both variants. However, omission of the AE002926 exon generates a stop codon in the junction gap AE003384. This region was sequenced with two different primers four times for each strand, so an error is unlikely. Thus, the common splice variant encodes a much shorter protein (223 residues), corresponding to the exons encoded by AE003045, AE003065, and the gap, whereas the rarer form encodes a protein of 569 residues (nearly the same size of CG13125, 557 residues).
BLASTP showed that PPr-Y is similar to the Sds22p gene of Saccharomyces cerevisiae (NP_012728) and other regulatory subunits of protein phosphatases of the PPP1 subfamily. These proteins associate with the catalytic PPP1 subunits and regulate their activity by targeting them to a specific intracellular location, determining the substrate specificity and modulating the enzymatic activity (19). A search for protein domains with CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) did not yield a single hit for the whole PPr-Y protein, whereas InterPro (http://www.ebi.ac.uk/interpro/scan.html) found five leucine-rich repeats, of which four are present in the short PPr-Y. Thus, PPr-Y belongs to the leucine-rich repeat superfamily. The consensus of leucine-rich repeats from PPr-Y (LxxLxNLxxLxLxxNxIExIEN) shows that it is a member of the Sds22-like subfamily (20, 21), a group that includes the Sds22p gene of S. cerevisiae (which is involved in cell cycle control; ref. 19) and the dynein light chain 1 of Chlamydomonas (AAD41040), a flagellar component thought to be involved in the control of beating (21). The similarity of PPr-Y with Sds22p and dynein light chain 1 (36% and 35% identity in the aligned region, respectively) is restricted to the residues encoded by the AE003065 exon and the gap (≈130 residues), whereas the remainder of the protein does not show significant homology to any known protein. The significance of the alternative splicing of PPr-Y remains to be determined. The shorter splice variant probably is functional because the premature stop codon is located exactly after the Sds22p homologous region of PPr-Y. On the other hand, the conservation of the C-terminal exons (AE002926, AE003384, AE002957) between PPr-Y and CG13125 strongly suggest that they too are functional.
The AE003045 scaffold maps to the kl-2 region, whereas AE002957 maps to kl-1. This shows that PPr-Y is not essential for male fertility because the gene is interrupted in the fertile E15 Kennison line (2).
Occludin-Related Y (ORY).
The armU scaffolds AE003328 and AE002654 seem to encode a protein similar to the autosomal gene CG6059 (Fig. 4A). RT-PCR using 3328-forward and 2654-reverse primers produced a product with the expected size, confirming that both scaffolds are part of the same gene. We sequenced this RT-PCR product and found the structure AE003328-gap-AE002695-AE002654. Scaffold AE002695 also belongs to armU, and its homology with CG6059 was not detected by our initial TBLASTN search. The same is true of AE003352, which was detected by 3′-RACE and encodes the C terminus exon. The 49-bp gap between AE003328 and AE002695 was caused by incompleteness of the AE003328 scaffold. Besides the presumably very large introns between the armU scaffolds, there is a short intron (55 bp) in AE003328 and another one (54 bp) in AE002654. The positions of the second and third introns are identical in relation to CG6059 (Table 1). Similarity between the AE003328 scaffold and the CG6059 protein starts at residue 180, which is close to the exon 1–exon 2 boundary of CG6059. Thus, it seems that the N terminus exon of ORY is missing, but we identified most of the gene (CG6059 has 884 residues, and the available sequence of ORY has 734). The similarity in intron–exon structure (Table 1) and sequence (31% identity, 54% similarity) strongly suggests that ORY originated from CG6059.
Regarding the function of ORY, both AE003328 and AE002654 map to the ks-1 region, so this gene may correspond to the ks-1 fertility factor. Most of the sequence of ORY and CG6059 is occupied by coiled-coils motifs (detected with the COILS program; http://www.ch.embnet.org/software/COILS_form.html; ref. 22). This explains their BLASTP hits with widely different proteins known to contain coiled-coils motifs, such as myosin heavy chain, early endosome antigen 1, etc., and implies that these hits do not indicate orthology. Coiled-coils constitute a ubiquitous protein-folding motif responsible for protein–protein associations (22). BLAST detected a very significant a similarity (42% identity, 66% similarity) of ORY and CG6059 to a fragment of a human protein (CAC16042), and to a lesser extent to sea-urchin ESTs (BG787439 and BG787478, both from mesenchymal cells) and to mouse ESTs (AA607756 and AI594587, both from skin); in these cases, similarity goes beyond the coiled-coils regions. Progress in the identification of these proteins may shed light on the function of ORY. Hardy et al. (23) described the cytological defects of males deficient for each Y chromosome region (kl-1, kl-2, etc.). This work might provide an alternative way to ascertain the function of ORY (say, if ks-1− males have defective endosomes). Unfortunately, these authors concluded that the syndromes described for deficiencies of the ks-1 region “are the indirect consequences of earlier lesions, which we have failed to identify.” Finally, a possible hint regarding the function of ORY is given by CDD, which indicated a weak similarity between CG6059 and occludin/ELL domain. Occludin is a membrane protein essential for male fertility in mice (24). The expression pattern of occludin and CG6059 are also similar: the mammalian gene is known to be highly expressed in the testis, kidney, lung, liver, and brain, whereas CG6059 is expressed, probably at a high level, in testis and head (a BLASTN search of CG6059 mRNA against Drosophila EST gave 23 matches, all in head and testis libraries). Thus, we tentatively suggest that CG6059 is a homolog of occludin and that ORY has a similar function.
Coiled-Coils Y (CCY).
The only gene identified with the scan of testis ESTs, armU scaffold AE003014, has a nearly perfect match with the testis ESTs AI946068 and BE978207. AE003014 contains a short fragment of the gene; 3′-RACE yielded additional 2.2 kb of sequence (including the stop codon), all of which are missing in armU. The N terminus is still missing. PCR mapping places AE003014 in the ks-2 region, and thus CCY may correspond to the ks-2 fertility factor. Unfortunately, the phenotype of ks-2− males (“misalignment of the axoneme with respect to the furrow separating the two halves of the nebenkern”; ref. 23) is not very informative.
CDD and InterPro did not detect any domain in the available sequence of CCY (939 residues), whereas BLAST detected similarity with coiled-coils proteins in the same region (residues 80–250) where the COILS program (22) found coiled-coils. In contrast to all other Y-linked genes, no clear Drosophila homolog could be found among described proteins, with all hits being weak and restricted to the ubiquitous coiled-coils motif. It is possible that the CCY parental gene might have escaped annotation, but a TBLASTN search of CCY against the entire Drosophila genome again failed to detect a convincing candidate. Thus, CCY diverged too much from its parental gene, or the parental gene degenerated after the duplication and no longer exists in the Drosophila genome, or the parental gene lies in a gap of sequence, for even the euchromatic portion of the genome is not yet completely seamless (12). Full sequencing of CCY may shed light on its origin and function; its singularity makes it especially interesting.
Discussion
In this article, we describe five new genes in the Y chromosome of Drosophila melanogaster, raising the number of identified single-copy genes on this chromosome to nine. Among the four previously identified genes (kl-2, kl-3, kl-5, and PRY), three encode dynein heavy chains (4, 5), whereas among the five new genes, three encode serine–threonine protein phosphatases. Our new results reinforce the notion that the gene content of the Drosophila Y shows unusual functional specialization. The precise function of these new genes remains to be determined; knowledge of their molecular identity is the first step toward this goal. With the completion of the first draft sequence of the human genome, and ongoing sequencing projects of mouse and other mammals, there is a void of computational methods tailored to detect heterochromatic genes in WGS projects. The staggered TBLASTN method we developed is a step in this direction, and the Drosophila armU sequences provide an ideal pilot experiment.
Function of the Y Genes and Their Correspondence with the Fertility Factors.
Six fertility genes were detected on the Y chromosome by formal genetics (1, 2), whereas molecular studies identified nine single-copy genes (refs. 4 and 5 and this study). Knowledge of the correspondence between the genes defined by these two types of evidence is still incomplete. The best known cases are kl-2, kl-3, and kl-5, where the cytological, biochemical, and molecular data are coherent and clear: mutants for these genes lack a cellular structure (outer arm of the sperm tail axoneme) known to contain the motor protein dynein heavy chain (23); the mutants also lack proteins with the same molecular weight as dynein heavy chains (25); and finally, molecular studies detected ORFs encoding axonemal dynein heavy chains in the regions of the Y chromosome known to contain the fertility factors kl-2, kl-3, and kl-5 (4, 5). Furthermore, axonemal dynein heavy chains have only one function, the beating of cilia and flagella (26). This situation contrasts with the other Y-linked genes. ORY and CCY map, respectively, to the ks-1 and ks-2 regions and thus may correspond to these fertility factors. ORY may be related to occludin, whereas the function of CCY is entirely mysterious. Unfortunately, the ks-1 and ks-2 fertility factors do not have an informative mutant phenotype. Thus, the attribution of function to ORY and CCY is at best tentative. In the cases of Pp1-Y1, Pp1-Y2, and PPr-Y, mapping experiments revealed that they do not correspond to any fertility factor. We know their basic function: Pp1-Y1 and Pp1-Y2 encode catalytic subunits of serine–threonine protein phosphatases (PPP1 subfamily), and PPr-Y encodes a regulatory subunit. PPP1 phosphatases control a multitude of cellular processes such as flagellar beating, cell-cycle progression, and glycogen metabolism, and given the ambiguity of the sequence similarity evidence, all are possible functions of the Y-linked PPP1. It is quite possible that they are involved in the control of flagellar beating, for Pp1-Y1 and Pp1-Y2 are similar to the Chlamydomonas PPP1 that performs this function (ref. 16; Fig. 2), and PPr-Y is similar to a dynein light chain that is a component of the Chlamydomonas flagella (21). None of the three genes seems to be essential for male fertility, possibly because their function can be partially rescued by their autosomal parents. Intriguingly, the male germ-line of Caenorhabditis elegans was found to specifically express a large number of protein kinases and phosphatases (27). As occurs with the protein phosphatases, the PRY gene (4) could not be one of the fertility factors. Its parental gene (DS07721.6; AAF44887) contains an REJ domain, which is present in a protein of the sea urchin sperm (AAB08448). This protein recognizes the egg jelly and participates in egg–sperm fusion (28). Thus, it is possible that DS07721.6 and PRY are involved in egg–sperm recognition. Finally, none of the nine molecularly defined genes can correspond to kl-1, which probably remains to be found amidst the 631 scaffolds (4 Mb) that comprise armU.
How Many Y Genes Are There?
We made an exhaustive search with the TBLASTN method and tested all candidates that showed staggered hits. This method may fail to detect all Y-linked genes because it is likely that some Y genes are encompassed by a single armU scaffold, and as of yet, none of the 109 single-hit proteins (i.e., a protein that has significant similarity with only one armU scaffold) has been tested for Y linkage. CCY is an example of a Y-linked gene that was not detected by the TBLASTN method, and Pp1-Y1 was only detected because of Pp1-Y2 (Fig. 1A). Furthermore, we used a rather high stringency in TBLASTN (e = 0.001) to avoid false positives, and the armU database itself is incomplete. Regarding the testis EST approach, at the time we performed the searches there were ≈2,900 EST sequences, and now there are 28,000. So it is not unlikely that an additional 10 Y-linked genes remain to be found.
Drosophila Y Chromosome Evolution.
A striking feature of almost all single-copy Y-linked genes is that their closest homologs are autosomal, rather than X-linked (the exception is CCY, devoid of any clear homolog). PRY, ORY, and PPr-Y do not have even weakly similar genes on the X chromosome, whereas kl-2, kl-3, kl-5, Pp1-Y1, and Pp1-Y2 (which belong to large, well conserved gene families) do have similar genes on the X, but an even closer homolog on the autosomes. A second feature of these genes is their exceptional functional coherence. These two features strongly suggest that these genes were not present in the hypothetical chromosome pair that gave rise to the X and Y. Instead, the most likely explanation is that they were acquired from the autosomes and are retained because they confer a specific fitness advantage to their carriers (4). Thus, we can delineate the following scenario. Step 1: Mutant Y chromosomes carrying autosomal (or X-linked) genes sporadically arise through translocation, transposition, etc. Step 2: Most of these mutant Y chromosomes are lost or the translocated gene degenerates, whereas a few of them confer some advantage to their carriers, spread, and became fixed. The mst77F pseudogene (29) seems to be an example of a gene translocated to the Y that degenerated. Step 3: As the Y genes are duplicates of autosomal genes and are localized on a male-specific region of the genome, they are free to evolve and suffer natural selection only for male fitness. Thus, it is probable that the sequence divergence between the Y genes and their parent genes is at least partly adaptive and that the Y copies are “fine-tuned” for male fitness. Step 4: Shortly after their origin, the Y copies would not be essential, although some may have increased male fitness (as still seems to be the case of PPr-Y, Pp1-Y1, Pp1-Y2, and PRY). But at some point, some of these Y genes became essential for male physiology (as is the case of the fertility factors). (5) The cycle may start again with a new translocation; each cycle would contribute one gene for the present-day Y-linked genome.
This scenario raises many questions and has two important consequences. Regarding the mechanism that originates the mutants (step 1), most of the Y genes have introns, some of which are conserved in relation to the parental gene (e.g., Table 1). Thus, at least some of the genes originated from genomic DNA rather than from reverse transcription of mRNA. The genic content of chromosome Y is far from being a random sample of the whole genome, so step 2 has an underlying logic and mechanism. For example, a female-specific gene inserted in the Y is expected to be lost because the inserted sequence will be at most neutral, and will degenerate even if fixed by drift. House-keeping genes would behave like a normal duplication and, in principle, could confer some advantage and be retained. However, examination of the nine Y-linked genes suggests that none of them has general, house-keeping functions. Instead, most, if not all, of the Y genes have a male-related function. A possible explanation for this pattern was proposed by Fisher, who noted that the Y chromosome is expected to accumulate male-related genes because male–female antagonistic effects of genes may hamper the evolution of male-related traits, unless they are located in a male-specific region of the genome (30, 31). In addition, Y-linked copies of the autosomal genes are hemizygous, and the spread of beneficial mutations cannot be retarded by recessive masking. Both factors may confer an advantage to the mutated Y chromosomes, and we do not know their relative importance. Release from dominance would occur also with a house-keeping gene translocated to the Y; the apparent absence of these genes in the Y chromosome suggests that escape from male–female antagonistic effects of genes is the main advantage of Y translocated genes.
Steps 3 and 4 dealt with the origin of male-relatedness of the Y genes, i.e., were their parents already male-related, or did the specialization evolve after transposition to the Y? Table 2 suggests that most of the parental genes were already male-related. That this path is not obligatory is shown by the X-linked Sdic gene of D. melanogaster, which arose from a duplication and fusion of two genes. Sdic encodes an axonemal dynein intermediate chain that is expressed in testis, but neither of the parent genes is male related; male-specific regulatory and structural motifs were created de novo (32).
Table 2.
Evidence for male-related function of the autosomal paralogs of the Y-linked genes
| Y gene | Parental gene
|
||
|---|---|---|---|
| Name | Location | Evidence for male-related function | |
| kl-2 | CG9068 | 2R | Axonemal dyneins are a component of sperm flagella |
| kl-3 | CG9492 | 3R | Idem |
| kl-5 | Dhc 93AB | 3R | Idem |
| PRY | DS07721.6 | 2L | Contain receptor of egg jelly (REJ) domain |
| ORY | CG6059 | 3R | Contain Occludin domain; testis expressed (EST) |
| CCY | None found | — | — |
| Pp1-Y1 | PpN58A | 2R | Testis-restricted expression (ref. 17) |
| Pp1-Y2 | ? Pp1-87B | 3R | Testis expressed (EST) |
| PPr-Y | CG13125 | 2L | Testis expressed (data not shown) |
There are two important consequences of the above scenario. First, the evolutionary history of the Y chromosome would be shaped by selective sweeps; the accretion of each new Y-linked gene is accompanied by fixation of a single Y chromosome (and there may be additional sweeps caused by the fine-tuning of the genes to male fitness). These selective sweeps span the entire chromosome (there is no recombination in the Y) and may help to explain the low nucleotide diversity of the Drosophila Y chromosome (33, 34). Second, the genic content of the Y chromosome would be fluid and may differ among Drosophilid species.
In contrast with the usual eukaryotic chromosomes, the Drosophila Y contains a coherent set of genes, being an assemblage of male-related genes collected from the whole genome. The molecular identification of the Y-linked genes is revealing the underlying logic of the process of gene recruitment. The presence of three axonemal motor proteins shows that sperm motility, possibly via sperm competition, is at premium for male fitness (4). The presence of three protein phosphatases is an intriguing finding whose meaning remains to be elucidated. Further progress in the identification of Y genes, coupled with analysis of DNA sequence variation and interspecific divergence, is bound to reveal much about the evolutionary forces at play on the Drosophila Y chromosome.
Acknowledgments
We thank D. L. Lindsley for sharing Drosophila stocks and advice, X. Huang for sending the computer programs NAP and GAP2, Jim Kennison for discussions and enthusiasm, C. A. M. Russo for help with phylogenetic methods, and two anonymous reviewers for valuable criticisms. This work was supported by fellowships from the Pew Latin American Fellows Program and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (to A.B.C.), Conselho Nacional de Desenvolvimento Cientifico e Technológico (to M.D.V.), and by grants from the National Institutes of Health and National Science Foundation (to A.G.C.).
Abbreviations
- WGS
whole genome shotgun
- armU
unmapped scaffolds of the Drosophila Genome Project
- RT
reverse transcription
- 3′-RACE
3′-rapid amplification of cDNA ends
- EST
expressed sequence tag
Footnotes
References
- 1.Brosseau G E. Genetics. 1960;45:257–274. doi: 10.1093/genetics/45.3.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennison J A. Genetics. 1981;98:529–548. doi: 10.1093/genetics/98.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan T H. Science. 1910;32:120–122. doi: 10.1126/science.32.812.120. [DOI] [PubMed] [Google Scholar]
- 4.Carvalho A B, Lazzaro B P, Clark A G. Proc Natl Acad Sci USA. 2000;97:13239–13244. doi: 10.1073/pnas.230438397. . (First Published November 7, 2000; 10.1073/pnas.230438397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gepner J, Hays T S. Proc Natl Acad Sci USA. 1993;90:11132–11136. doi: 10.1073/pnas.90.23.11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer G F, Hess O, Beermann W. Chromosoma. 1961;12:676–716. doi: 10.1007/BF00328946. [DOI] [PubMed] [Google Scholar]
- 7.Hennig W. Proc Natl Acad Sci USA. 1993;90:10904–10906. doi: 10.1073/pnas.90.23.10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonaccorsi S, Pisano C, Puoti F, Gatti M. Genetics. 1988;120:1015–1034. doi: 10.1093/genetics/120.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gatti M, Pimpinelli S. Chromosoma. 1983;88:349–373. [Google Scholar]
- 10.Kurek R, Reugels A M, Lammermann U, Bünemann H. Genetica. 2000;109:113–123. doi: 10.1023/a:1026552604229. [DOI] [PubMed] [Google Scholar]
- 11.Reugels A M, Kurek R, Lammermann U, Bünemann H. Genetics. 2000;154:759–769. doi: 10.1093/genetics/154.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams M, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 13.Andrews J, Bouffard G G, Cheadle C, Lu J, Becker K G, Oliver B. Genome Res. 2000;10:2030–2043. doi: 10.1101/gr.10.12.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lahn B, Page D C. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S, Tamura K, Nei M. Comput Appl Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 16.Yang P, Fox L, Colbran R J, Sale WS. J Cell Sci. 2000;113:91–102. doi: 10.1242/jcs.113.1.91. [DOI] [PubMed] [Google Scholar]
- 17.Cohen P T. Trends Biochem Sci. 1997;22:245–251. doi: 10.1016/s0968-0004(97)01060-8. [DOI] [PubMed] [Google Scholar]
- 18.Armstrong C G, Dombradi V, Mann D J, Cohen P T. Biochim Biophys Acta. 1998;1399:234–238. doi: 10.1016/s0167-4781(98)00117-1. [DOI] [PubMed] [Google Scholar]
- 19.Hong G, Trumbly R J, Reimann E M, Schlender K K. Arch Biochem Biophys. 2000;376:288–298. doi: 10.1006/abbi.2000.1715. [DOI] [PubMed] [Google Scholar]
- 20.Kajava A V. J Mol Biol. 1998;277:519–527. doi: 10.1006/jmbi.1998.1643. [DOI] [PubMed] [Google Scholar]
- 21.Benashski S E, Patel-King R S, King S M. Biochemistry. 1999;38:7253–7264. doi: 10.1021/bi990466y. [DOI] [PubMed] [Google Scholar]
- 22.Lupas A. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 23.Hardy R W, Tokuyasu K T, Lindsley D L. Chromosoma. 1981;83:593–617. doi: 10.1007/BF00328522. [DOI] [PubMed] [Google Scholar]
- 24.Saitou M, Furuse M, Sasaki H, Schulzke J D, Fromm M, Takano H, Noda T, Tsukita S. Mol Biol Cell. 2000;11:4131–4142. doi: 10.1091/mbc.11.12.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldstein L S B, Hardy R W, Lindsley D L. Proc Natl Acad Sci USA. 1982;79:7405–7409. doi: 10.1073/pnas.79.23.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibbons I R. Cell Motil Cytoskeleton. 1995;32:136–144. doi: 10.1002/cm.970320214. [DOI] [PubMed] [Google Scholar]
- 27.Reinke V, Smith H E, Nance J, Wang J, Van Doren C, Begley R, Jones S J, Davis E B, Scherer S, Ward S, Kim S K. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- 28.Moy G W, Mendoza L M, Schulz J R, Swanson W J, Glabe C G, Vacquier V D. J Cell Biol. 1996;133:809–817. doi: 10.1083/jcb.133.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell S R H, Kaiser K. Genetics. 1993;134:293–308. doi: 10.1093/genetics/134.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher R A. Biol Rev. 1931;6:345–368. [Google Scholar]
- 31.Roldan E R S, Gomendio M. Trends Ecol Evol. 1999;14:58–62. doi: 10.1016/s0169-5347(98)01567-5. [DOI] [PubMed] [Google Scholar]
- 32.Nurminsky D I, Nurminskaya M V, De Aguiar D, Hartl D L. Nature (London) 1998;396:572–575. doi: 10.1038/25126. [DOI] [PubMed] [Google Scholar]
- 33.Zurovcova M, Eanes W F. Genetics. 1999;153:1709–1715. doi: 10.1093/genetics/153.4.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachtrog D, Charlesworth B. Curr Biol. 2000;10:1025–1031. doi: 10.1016/s0960-9822(00)00656-4. [DOI] [PubMed] [Google Scholar]