Abstract
Measuring total cell-free DNA (cfDNA) or cancer-specific mutations herein has presented as new tools in aiding the treatment of cancer patients. Studies show that total cfDNA bears prognostic value in metastatic colorectal cancer (mCRC) and that measuring cancer-specific mutations could supplement biopsies. However, limited information is available on the performance of different methods. Blood samples from 28 patients with mCRC and known KRAS mutation status were included. cfDNA was extracted and quantified with droplet digital polymerase chain reaction (ddPCR) measuring Beta-2 Microglobulin. KRAS mutation detection was performed using ddPCR (Bio-Rad) and next-generation sequencing (NGS, Ion Torrent PGM). Comparing KRAS mutation status in plasma and tissue revealed concordance rates of 79% and 89% for NGS and ddPCR. Strong correlation between the methods was observed. Most KRAS mutations were also detectable in 10-fold diluted samples using the ddPCR. We find that for detection of KRAS mutations in ctDNA ddPCR was superior to NGS both in analysis success rate and concordance to tissue. We further present results indicating that lower amount of plasma may be used for detection of KRAS mutations in mCRC.
Introduction
Circulating cell-free DNA (cfDNA) present in blood and other body fluids has initiated a new era in cancer diagnosis and treatment. Apoptotic and necrotic cells from the entire body is the primary source of cfDNA (reviewed by Thierry et al. [1]), and total levels of cfDNA are found to be increased in various cancers [2]. Circulating tumor DNA (ctDNA) is the fraction of cfDNA originating from tumor cells, and ctDNA analyses enable detection of tumor-specific mutations in cfDNA. ctDNA has been estimated to contribute to 0.01%-93% of the total cfDNA [3], [4], and ctDNA has been suggested to reflect tumor burden [5], [6], [7], [8], [9]. Therefore, ctDNA analysis has been proposed as a noninvasive strategy for gaining insights into the tumor’s mutational profile.
Of particular interest is detection of KRAS, BRAF, and NRAS mutations in metastatic colorectal (mCRC) patients, as these are associated with intrinsic resistance to the anti-EGFR antibody treatments offered to this patient group (reviewed by Misale et al. [10]). Concordance rates of 80%-96% between KRAS mutations identified in ctDNA analyses and biopsies have been observed [6], [11], [12], [13], [14], [15], [16], [17], [18]. Next-generation sequencing (NGS) and polymerase chain reaction (PCR)–based techniques for ctDNA analysis are continuously being refined to accommodate the variety of mutations and the low frequencies. Several different approaches have been investigated to accommodate the increasing request for a clinically applicable method for ctDNA analysis [19], [20], but information on their performance on clinical material remains insufficient.
The aim of the present study was to evaluate and compare methods used for measuring KRAS mutations in ctDNA from mCRC patients. Further, in a clinical setting, sample material can be sparse. Therefore, the present study also undertook the aim of investigating the feasibility of analyzing ctDNA from smaller plasma volumes.
Materials and Methods
Patient Samples
Plasma samples originate from a Danish cohort of mCRC patients treated with irinotecan/cetuximab and everolimus according to a closed clinical trial (NCT01387880). Primary data are to be published elsewhere. The KRAS tissue genotype was recovered from patient files, but no other clinical data were used. KRAS tissue-genotype analysis was performed in a routine setting using Therascreen DXS KRAS mutation kit covering codon 12 and 13 (Qiagen, Hilden, Germany). Blood samples were centrifuged at 2300g for 15 minutes at 4°C. Plasma was collected and stored at −80°C until further use.
Two subcohorts were used for the current study. Cohort 1 included 28 plasma samples from patients with known KRAS mutations in tissue (mutation in codon 12 or 13). Cohort 2 included 16 plasma samples containing at least 2.2 ml plasma and randomly chosen among the available samples. Each sample was divided into a 2 ml and a 200 μl portion, and extraction was performed on these separately. Samples were used to study how extraction of cfDNA from two different volumes affects the total cfDNA measurements. cfDNA was extracted from plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) from a range of 1.5-2 ml plasma for cohort 2 (see Supplementary Table 1) and from either 2 ml or 200 μl for cohort 2 following manufacturer’s protocol. Samples were eluted in 100 μl (for 1.5-2 ml plasma samples) or 50 μl (for 200 μl plasma samples) elution buffer (supplied with the kit). cfDNA was frozen at −80°C until further analysis.
After extraction of cfDNA, a 10-fold dilution was made from each sample in cohort 1. Original and diluted samples were used for evaluating how dilution of samples affected the total cfDNA measurements and how the Bio-Rad ddPCR assays performed.
Droplet Digital PCR
Droplet digital PCRs (ddPCRs) were performed using 2X ddPCR Supermix for Probes (no UTP, Bio-Rad), relevant assay (Bio-Rad), and 5 μl cfDNA in a total reaction volume of 20 μl following manufacturer’s recommendations. ddPCRs were performed using the QX200 Droplet Digital PCR System (Bio-Rad). Data analyses were performed as recommended by the manufacturer using the QuantaSoft Software version 1.7.4.
Total cfDNA was quantified using an assay targeting beta-2-microglobuline (B2M). Lymphocyte contamination of total cfDNA was detected using an immunoglobulin gene specific assay (PBC) as previously described [21]. The assays were multiplexed on the ddPCR platform. Information on assays for B2M and PBC quantification can be found in Supplementary Table 1.
The Bio-Rad KRAS PrimePCR ddPCR Mutation assays (Bio-Rad) were used for ddPCR (see Supplementary Table 2). The limit of detection for the assays was determined as recommended by Milbury and colleagues [22].
ddPCR analyses was performed in triplicates. Data from triplicates were merged in the QuantaSoft software and were used for further analysis. Erroneous wells were not included in the merge.
Next-Generation Sequencing
Libraries were prepared using the Oncomine Solid Tumor DNA kit on 1.1-10 ng of cfDNA following manufacturer’s instructions (Thermo Fisher Scientific). Amplicon sizes may be found in Supplementary Table 2. Sample concentrations were estimated from the B2M measurements. NGS was performed using the Ion Chef Instrument and Ion Personal Genome Machine (PGM) System (Thermo Fisher Scientific). Sequencing was performed using Ion 316 v2 BC chips with eight cfDNA samples per chip.
Primary data processing was performed using the Torrent Suite Software (version 5.0.4) on a Torrent Server and the Ion AmpliSeq Colon and Lung Cancer panel v2 template (Thermo Fisher Scientific). Variant calling was performed using the Ion Reporter Software (version 5.0) and the AmpliSeq CHPv2 peripheral/CTC/CF DNA single sample workflow (Thermo Fisher Scientific). Reference and hotspot BED files were replaced with those supplied with the kit. Default settings were used. Sequencing was considered successful if the mean sequencing depth was ≥2000. If this criterion could not be met, the sample was disqualified. Called variants were only accepted if AF ≥1%. For visualization and manual inspection of variants, the Integrative Genomics Viewer (Broad Institute) was used [23].
The median number of mapped reads per sample was 408,401.5 (range: 233,435-1,414,524), mean depth was 4066.5 (range: 2332-13,438), median uniformity was 100% (range: 94.31-100), and median reads on target were 94.5% (range: 90.3-98.8).
Statistics
All statistics were performed using Stata 13 (StataCorp 2013). Datasets were tested for normality. If data were not normally distributed, log-transformation (using the natural logarithm) was performed to achieve normality, and graphs were produced with log-transformed data. In the KRAS mutation analyses, the value 0 was given to wild-type samples. For these datasets, the value 1 was added to all results to enable log-transformation.
For comparing the data produced in this study, the guidelines for comparing methods by Giavarina were used [24]. To test correlation between measurements, linear regression was performed. To test agreement between methods/measurements, Bland-Altman plots and limits of agreement (95% prediction intervals) were used. Results from log-transformed data were back-transformed to achieve meaningful median ratios.
Results
ctDNA Analysis
All cfDNA samples from cohort 1 were sequenced, and the analysis was feasible for 86% of the samples (24 of 28). Among the 24 sequenced samples, there was a 79% (19 of 24) concordance between tissue and cfDNA (see Table 1). For further information on the NGS analysis, see Supplementary Table 4. For samples where the tissue genotype was not confirmed, BAM files were visualized using Integrative Genomics Viewer, and the mutation was identified in three additional samples (10, 11, 12). Common for these was low variant frequency. For sample 26, neither of the mutations detected in tumor were found by NGS ctDNA analysis; instead, a p.Gly12Ala mutation was detected.
Table 1.
CtDNA Analysis
| Sample ID | Tissue Genotype | NGS |
ddPCR-2 ml |
ddPCR-200 μl |
||
|---|---|---|---|---|---|---|
| AF (%) | AF (%) | Copies/ml | AF (%) | Copies/ml | ||
| 1 | p.Gly12Val | 19.08 | 22.80 | 4114.00 | 22.10 | 4620.00 |
| 2 | p.Gly12Ala | 37.93 | 40.40 | 15708.00 | 39.80 | 14300.00 |
| 3 | p.Gly13Asp | 5.51 | 12.00 | 607.20 | 12.20 | 715.00 |
| 4 | p.Gly13Asp | 19.32 | 27.20 | 1848.00 | 26.50 | 1969.00 |
| 5 | p.Gly12Val | 1.75 | 2.40 | 180.40 | 1.60 | 132.00 |
| 6 | p.Gly12Asp | 14.24 | 26.80 | 5104.00 | 25.80 | 5573.33 |
| 7 | p.Gly12Val | 39.70 | 60.50 | 12430.00 | 60.60 | 9460.00 |
| 8 | p.Gly12Ser | 1.99 | 6.00 | 280.50 | 7.90 | 357.50 |
| 9 | p.Gly13Asp | 7.84 | 16.80 | 1848.00 | 15.00 | 1826.00 |
| 10 | p.Gly12Ser | 0.00 | 0.70 | 30.80 | 0.00 | 0.00 |
| 11 | p.Gly12Asp | 0.00 | 1.13 | 112.20 | 0.00 | 0.00 |
| 12 | p.Gly13Asp | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 13 | p.Gly13Asp | 23.38 | 20.60 | 926.32 | 16.00 | 648.42 |
| 14 | p.Gly12Ala | 19.16 | 26.60 | 2917.89 | 25.60 | 2929.47 |
| 15 | p.Gly12Val | 31.11 | 44.10 | 7526.32 | 43.30 | 7410.53 |
| 16 | p.Gly12Val | 2.09 | 2.20 | 259.11 | 1.40 | 158.89 |
| 17 | p.Gly13Aspa | 7.55 | NA | NA | ||
| 18 | p.Gly12Asp | NA | 2.60 | 92.40 | 6.20 | 198.00 |
| 19 | p.Gly12Val | NA | 15.20 | 2002.00 | 13.20 | 1628.00 |
| 20 | p.Gly13Asp | 20.91 | 31.30 | 11088.00 | 30.40 | 11550.00 |
| 21 | p.Gly12Asp | 33.36 | 54.81 | 255200.00 | 54.40 | 242115.80 |
| 22 | p.Gly12Val | 32.27 | 36.80 | 35288.00 | 35.90 | 33770.00 |
| 23 | p.Gly12Asp | NA | 14.90 | 1650.00 | 13.10 | 1353.00 |
| 24 | p.Gly12Val | 3.56 | 8.30 | 682.00 | 7.80 | 759.00 |
| 25 | p.Gly12Val | 58.15 | 66.45 | 170720.00 | NA | |
| 26 | p.Gly12Ser | 0.00 | 0.00 | 0.00 | NA | |
| p.Gly12Asp | 0.00 | 0.00 | 0.00 | NA | ||
| p.Gly12Alab | 31.68 | 35.70 | 40304.00 | 35.60 | 39930.00 | |
| 27 | p.Gly13Asp | NA | 13.10 | 875.60 | 11.10 | 957.00 |
| 28 | p.Gly12Asp | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
Abbreviations: AF, allele frequency; ctDNA, circulating tumor DNA; ddPCR, droplet digital PCR; NA, not available; NGS, next-generation sequencing.
This mutation was revealed as a c.38_39delGCinsAT and not the expected c.38G>A by NGS. The mutation was not investigated by ddPCR.
The p.GlyAla was not detected by the tissue genotyping, but exclusively in the ctDNA analysis.
The p.Gly13Asp mutation in sample 17 was revealed as a c.38_39delGCinsAT and not the expected c.38G>A by NGS. Unfortunately, this was not detected in time to allow a change in ddPCR assay, and ddPCR results are therefore unavailable. For the investigated samples, the ddPCR confirmed 89% (24 of 27) of the tissue genotypes (Table 1). The ddPCR analysis confirmed the p.Gly12Ala mutation in sample 26. Further information on the ddPCR analyses can be found in Supplementary Table 4.
Comparison of KRAS Mutation Analyses on ctDNA
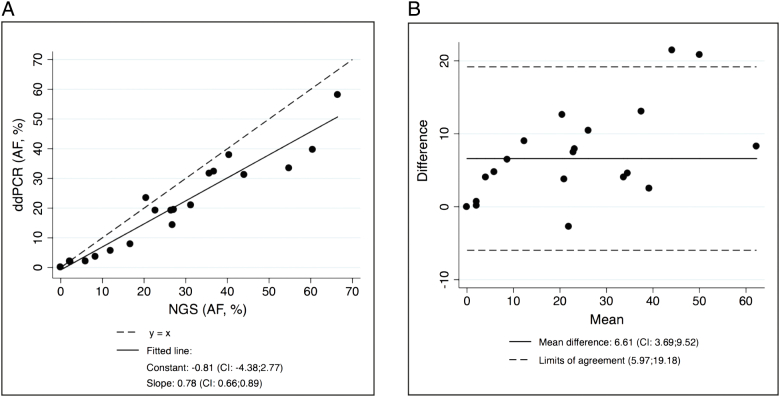
Among the 23 samples where both NGS and ddPCR results were available, there was a 91% concordance between the methods (21 of 23, see Table 1). The results obtained from NGS and ddPCR analyses were compared. Three ddPCR analyses were performed on sample 26, and all analyses were included in the comparison. Only samples where methods agreed in either identifying or not identifying the mutation were included (n=23). Good correlation between AFs obtained from the two analyses was obtained (R2 values of 0.91, see Figure 1A). The Bland-Altman analysis presented a mean difference of 6.61 [95% confidence interval (CI): 0.3.69-9.52] and limits of agreement of 5.97-19.18. The 95% prediction interval predicted that measured AFs in NGS versus ddPCR could vary, but the AFs did correlate to those of the NGS analysis.
Figure 1.
Comparison of the NGS and ddPCR ctDNA analyses. (A) Linear regression from the comparison of NGS and ddPCR AFs (R2 = 0.91). Results related to the regression are presented below the graph. (B) Bland-Altman plot of the differences (NGS (AF) − ddPCR (AF)).
KRAS Mutation Detection in Small-Volume Samples
KRAS mutation detection was investigated in the diluted samples from cohort 1 to determine if small-volume samples can be used for this purpose. A total of 26 of 28 samples had available material for dilution. We found a 92% concordance with the analysis performed in the original samples (24 of 26, Table 1).
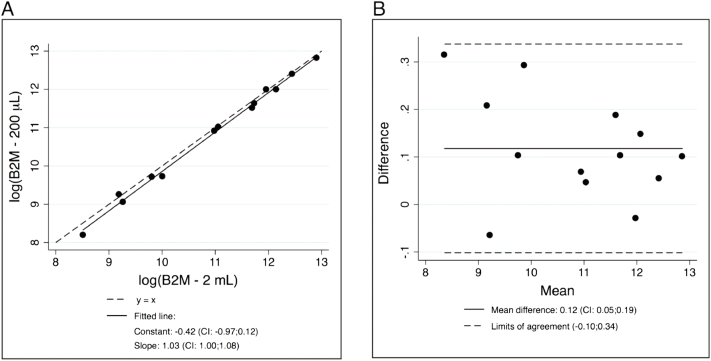
Prior to analysis the diluted samples, we tested how extraction of cfDNA from two different plasma volumes influenced total cfDNA measurement. The B2M-PBC multiplex ddPCR analysis was performed on the samples from cohort 2. Further information can be found in Supplementary Table 5. Three samples were PBC positive, and PBC exceeded 0.1% of the total cfDNA (see Supplementary Table 5); therefore, these were excluded from further analysis. Comparison of the original and diluted samples resulted in a median ratio (B2M 2 ml/B2M 200 μl) of 1.15 (95% CI: 1.08-1.22), and limits of agreement were 0.92-1.43, corresponding to a median underestimation of 15% (95% CI: 8-23) of B2M in 200 μl samples as compared to 2 ml samples (Figure 2).
Figure 2.
Comparison of log-transformed B2M measurements from cohort 2. Constant and slope for the regression and mean difference and limits of agreements are found in the figure. (A) Linear regression on measurements from cohort 2. The regression resulted in an R2 value of 0.994. (B) Bland-Altman plot of the differences (log(B2M 2 ml) − log(B2M 200 μl)).
Discussion
In the present study, we investigated two different methods, NGS and ddPCR, for investigating KRAS mutations in ctDNA. We further investigated the possibility using ddPCR for KRAS detection in small-volume samples.
We find that when considering the tissue as gold standard, concordance rates of 79% and 89% for NGS and ddPCR were found. The concordance rates are similar to those found by others [6], [11], [12], [13], [14], [15], [16], [17], [18]. The deviation between tissue and plasma analyses may be explained by various causes. First, the tissue biopsies were taken at the time of cancer diagnosis, while the plasma samples were collected after at least three lines of palliative chemotherapy. Hence, the mutations found in the tissue biopsy may originate from a clone that has been eliminated by the following treatments [25], [26]. Also, new mutations may have arisen during the treatment, explaining our findings in sample 26, where the tissue genotype was not confirmed,but a new KRAS mutation was securely detected. Secondly, the NGS analysis only securely detects variants present in ≥1% of cfDNA, while the ddPCR assays have limits of detection ranging between 0.1% and 0.3% (Supplementary Table 2). Studies suggest that the ctDNA fractions are low and highly variable (reviewed by Siravegna et al. [27]), challenging the detection limit of especially the NGS panel used here.
When comparing the results obtained from the ctDNA analyses, we find that ddPCR is superior in identifying the KRAS mutations in ctDNA when considering the analysis success rates (NGS: 86%; ddPCR: 100%) and the concordance to tissue genotype. This is in line with other studies [13], [28]. When comparing the AFs obtained from the two methods, we find good correlation between them, with a tendency of higher AFs obtained by ddPCR. If a quantitative measure of ctDNA is needed, this finding is relevant since the results may depend on the method used. The best methods for investigating ctDNA and cfDNA are under continuous evaluation (recently reviewed by Sacher et al. [29]). If the mutational tissue status is known and the purpose is to investigate the presence of this mutation in cfDNA, our results suggest that ddPCR is the best method because of the higher success rate, lower time demands, and lower detection limits and costs as compared to the NGS method used here. However, in situations where tissue mutational status is unknown or cannot be recovered, NGS analysis could be preferable for screening the samples. Also, NGS would reveal more information about the constitution of the ctDNA since several genes are often investigated.
Lastly, 92% of the investigated mutations could be recovered in a 10-fold dilution of the plasma samples compared to the full-volume samples. Since the ddPCR reaction is expected to be linear, this may not be surprising. In the present study, extraction of DNA from two different volumes was not feasible in the cohort of patients with known tissue genotype. Instead, we tested extraction on a different cohort where plasma was sufficient. We found that in the extraction of cfDNA from 200 μl plasma, the yield was 15% lower than extraction from 2 ml plasma. This could have influenced our results in the KRAS mutation analysis, and the recovery would most likely have been lower (see Supplementary Table 4). For complete elucidation of the extraction efficiency for different input volumes, a larger study is needed.
Despite this, our results indicate that for mCRC low-volume samples may often be sufficient for detection of KRAS mutations. This may also be relevant in other solid cancer with high ctDNA shedding.
Conclusion
In this study, we show that the NGS and ddPCR methods investigated have high concordance to tumor genotype (79% and 89%). When comparing the methods, we find that ddPCR is superior to NGS. And lastly, we find that in cases with sparse material from mCRC patients, smaller plasma volumes may often be sufficient for KRAS mutation detection by ddPCR.
Acknowledgement
The authors wish to acknowledge Birgit Westh Mortensen for exceptional technical assistance. We also thank members of Professor Boe Sandahl Sorensen’s group and Rikke Hjortebjerg for contributing to the statistical discussion.
The authors have no conflicts of interest. The study was supported by the Novo Nordic Foundation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2018.07.013.
Appendix A. Supplementary data
Supplementary tables
References
- 1.Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35:347–376. doi: 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 3.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA, Goodman SN, David KA, Juhl H. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jahr S, Hentze H, Englisch S, Hardt D, Fachelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 5.Winther-Larsen A, Demuth C, Fledelius J, Madsen AT, Hjorthaug K, Meldgaard P, Sorensen BS. Correlation between circulating mutant DNA and metabolic tumour burden in advanced non–small cell lung cancer patients. Br J Cancer. 2017;117:704–709. doi: 10.1038/bjc.2017.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spindler KL, Pallisgaard N, Andersen RF, Brandslund I, Jakobsen A. Circulating free DNA as biomarker and source for mutation detection in metastatic colorectal cancer. PLoS One. 2015;10 doi: 10.1371/journal.pone.0108247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tissot C, Toffart AC, Villar S, Souquet PJ, Merle P, Moro-Sibilot D, Pérol M, Zavadil J, Brambilla C, Olivier M. Circulating free DNA concentration is an independent prognostic biomarker in lung cancer. Eur Respir J. 2015;46:1773–1780. doi: 10.1183/13993003.00676-2015. [DOI] [PubMed] [Google Scholar]
- 8.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thierry AR, Mouliere F, Gongora C, Ollier J, Robert B, Ychou M, Rio MD, Molina F. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 2010;38:6159–6175. doi: 10.1093/nar/gkq421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to anti-EGFR therapy in colorectal cancer: from heterogeneity to convergent evolution. Cancer Discov. 2014;4:1269–1280. doi: 10.1158/2159-8290.CD-14-0462. [DOI] [PubMed] [Google Scholar]
- 11.Thierry AR, Mouliere F, El Messaoudi S, Mollevi C, Lopez-Crapez E, Rolet F, Gillet B, Gongora C, Dechelotte P, Robert B. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med. 2014;20:430–435. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 12.Thierry AR, El Messaoudi S, Mollevi C, Raoul JL, Guimbaud R, Pezet D, Artru P, Assenat E, Borg C, Mathonnet M. Clinical utility of circulating DNA analysis for rapid detection of actionable mutations to select metastatic colorectal patients for anti-EGFR treatment. Ann Oncol. 2017;28:2149–2159. doi: 10.1093/annonc/mdx330. [DOI] [PubMed] [Google Scholar]
- 13.Beije N, Helmijr JC, Weerts MJA, Beaufort CM, Wiggin M, Marziali A, Verhoef C, Sleijfer S, Jansen MPHM, Martens JWM. Somatic mutation detection using various targeted detection assays in paired samples of circulating tumor DNA, primary tumor and metastases from patients undergoing resection of colorectal liver metastases. Mol Oncol. 2016;10:1575–1584. doi: 10.1016/j.molonc.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tie J, Kinde I, Wang Y, Wong HL, Roebert J, Christie M, Tacey M, Wong R, Singh M, Karapetis CS. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015;26:1715–1722. doi: 10.1093/annonc/mdv177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sefrioui D, Mauger F, Leclere L, Beaussire L, Di Fiore F, Deleuze JF, Sarafan-Vasseur N, Tost J. Comparison of the quantification of KRAS mutations by digital PCR and E-ice-COLD-PCR in circulating-cell-free DNA from metastatic colorectal cancer patients. Clin Chim Acta. 2017;465:1–4. doi: 10.1016/j.cca.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Danese E, Minicozzi AM, Benati M, Montagnana M, Paviati E, Salvagno GL, Lima-Oliveira G, Gusella M, Pasini F, Lippi G. Comparison of genetic and epigenetic alterations of primary tumors and matched plasma samples in patients with colorectal cancer. PLoS One. 2015;10 doi: 10.1371/journal.pone.0126417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spindler KL, Pallisgaard N, Appelt AL, Andersen RF, Schou JV, Nielsen D, Pfeiffer P, Yilmaz M, Johansen JS, Hoegdall EV. Clinical utility of KRAS status in circulating plasma DNA compared to archival tumour tissue from patients with metastatic colorectal cancer treated with anti-epidermal growth factor receptor therapy. Eur J Cancer. 2015;51:2678–2685. doi: 10.1016/j.ejca.2015.06.118. [DOI] [PubMed] [Google Scholar]
- 19.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, Stehr H, Liu CL, Bratman SV, Say C. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34:547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pallisgaard N, Spindler KL, Andersen RF, Brandslund I, Jakobsen A. Controls to validate plasma samples for cell free DNA quantification. Clin Chim Acta. 2015;446:141–146. doi: 10.1016/j.cca.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 22.Milbury CA, Zhong Q, Lin J, Williams M, Olson J, Link DR, Hutchison B. Determining lower limits of detection of digital PCR assays for cancer-related gene mutations. Biomol Detect Quantif. 2014;1:8–22. doi: 10.1016/j.bdq.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giavarina D. Understanding Bland Altman analysis. Biochem Med (Zagreb) 2015;25:141–151. doi: 10.11613/BM.2015.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Del Re M, Tiseo M, Bordi P, D'Incecco A, Camerini A, Petrini I, Lucchesi M, Inno A, Spada D, Vasile E. Contribution of KRAS mutations and c.2369C > T (p.T790M) EGFR to acquired resistance to EGFR-TKIs in EGFR mutant NSCLC: a study on circulating tumor DNA. Oncotarget. 2017;8:13611–13619. doi: 10.18632/oncotarget.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imamura F, Uchida J, Kukita Y, Kumagai T, Nishino K, Inoue T, Kimura M, Oba S, Kato K. Monitoring of treatment responses and clonal evolution of tumor cells by circulating tumor DNA of heterogeneous mutant EGFR genes in lung cancer. Lung Cancer. 2016;94:68–73. doi: 10.1016/j.lungcan.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 27.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 28.Takai E, Totoki Y, Nakamura H, Morizane C, Nara S, Hama N, Suzuki M, Furukawa E, Kato M, Hayashi H. Clinical utility of circulating tumor DNA for molecular assessment in pancreatic cancer. Sci Rep. 2015;5:18425. doi: 10.1038/srep18425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sacher AG, Komatsubara KM, Oxnard GR. Application of plasma genotyping technologies in non–small cell lung cancer: a practical review. J Thorac Oncol. 2017;12:1344–1356. doi: 10.1016/j.jtho.2017.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables