Abstract
We used multilocus sequence typing (MLST) to analyze the diversity of natural isolates of Saccharomyces cerevisiae, the most important microorganism in alcoholic fermentation. Six loci, ADP1, RPN2, GLN4, ACC1, MET4, and NUP116, in S. cerevisiae genome were selected as MLST markers. To investigate genetic diversity within S. cerevisiae, 42 S. cerevisiae isolated from natural sources in Korea as well as six S. cerevisiae obtained from Genbank and four industrial S. cerevisiae were examined using MLST. Twenty-six polymorphic sites were found in the six loci. Among them, ACC1 had the most genetic variation with eight polymorphic sites. MLST differentiated the 52 strains into three clades. Alcohol fermentation results revealed that S. cerevisiae in Clade III produced less alcohol than those in Clades I and II. These results suggested that MLST is a powerful tool to differentiate S. cerevisiae and can potentially be used to select S. cerevisiae suitable for industrial use.
Keywords: Saccharomyces cerevisiae, MLST, Housekeeping genes, Makgeolli
Introduction
Saccharomyces cerevisiae is a unicellular budding yeast that has been used to make bread, wine, beer, and various other fermented foods since ancient times. S. cerevisiae is ubiquitous in nature and frequently isolated from sugary foods and alcoholic beverages. This yeast is capable of fermentative and oxidative metabolism and can reversibly switch between these two metabolism types depending on environmental conditions. Under anaerobic conditions, this microorganism can ferment sugars and produce ethanol and carbon dioxide [1]. However, under aerobic conditions, it propagates rapidly by assimilating sugar, resulting in high cell yields. Because both characteristics are utilized industrially, it is very important to select suitable strains for production of breads, beers, wines, and nutritional yeast biomass [2].
Numerous DNA and protein-based methods have been developed to identify and classify various microorganisms such as bacteria and yeast [3]. Different S. cerevisiae wine strains have been discriminated by amplified fragment length polymorphism (AFLP) [4], multilocus sequence typing (MLST) [5], pulsed-field gel electrophoresis (PFGE) [6], random amplified polymorphic DNA (RAPD) [7], and restriction fragment length polymorphism (RFLP) analyses [8]. Recently, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has been shown to be a rapid and reliable tool for identification of S. cerevisiae strains [9].
MLST is a molecular biological tool that can be used to differentiate among isolates of microbial species. The procedure of MLST involves PCR amplification of about 450–500 bp internal fragments of multiple housekeeping genes, followed by determination of their sequences using an automated DNA sequencing tool. Final, multilocus sequence analysis is used to differentiate among microbial strains [10, 11]. Muñoz et al. [5] successfully employed MLST analysis for molecular discrimination at the strain level of Spanish wine yeast. This study used five housekeeping genes (ADP1, ACC1, RPN2, GLN4, and ALA1) for the MLST analysis and reported 10 polymorphic sites in the amplified fragments of these five genes.
Makgeolli is a popular traditional Korean alcoholic beverage made mainly from rice and Nuruk (a Korean fermentation starter). Because fine-filtering is avoided during its preparation, Makgeolli contains numerous nutritional compounds originating from rice, yeast, and even lactic acid bacteria [12]. Those nutritional compounds include vitamins, essential amino acids, organic acids, oligosaccharides, and dietary fiber [13]. Makgeolli has been demonstrated to have health-promoting biological activities including antioxidant [14], anti-hypertensive [15], anti-diabetic, [16] and anti-cancer [17] activities.
In Korea, various alcoholic beverages, including Makgeolli, are traditionally made in home-based facilities containing various natural microorganisms. Even though many microorganisms are involved in Makgeolli fermentation, S. cerevisiae strains are considered the main determinants of the quality of the final product and to contribute to the diversity of Korean traditional alcoholic beverages [18, 19]. Hence, it is very important to identify and classify natural S. cerevisiae strains for their industrial application [20]. Many studies have selected S. cerevisiae strains from natural sources, mainly Nuruk, based on the alcoholic fermentation characteristics of the strains. However, molecular typing of selected S. cerevisiae strains from natural sources has not been performed. Therefore, we selected and examined various MLST markers from the S. cerevisiae genome to evaluate genetic diversity among natural S. cerevisiae strains in Korea.
Materials and methods
Yeast strains
Yeast strains used in this work are listed in Table 1. These strains are from our laboratory collection that includes previous isolates from natural sources such as Nuruk and flowers. Information about reference S. cerevisiae strains (ref 1, ref 2, ref 3, ref 4, ref 5, and ref 6) was obtained from the literature [21–24] and the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/). We also analyzed four commercial S. cerevisiae strains (com 1, com 2, com 3, and com 4) used in the bakery and alcohol fermentation industries in Korea.
Table 1.
List of S. cerevisiae strains analyzed by MLST
| Isolate no. | Date isolated | Source | |
|---|---|---|---|
| Geographical location | Sample | ||
| 95 | 2016 | Korea | Nuruk |
| 97 | 2016 | Korea | Nuruk |
| 98 | 2016 | Korea | Nuruk |
| 99 | 2016 | Korea | Nuruk |
| 100 | 2016 | Korea | Nuruk |
| 101 | 2016 | Korea | Nuruk |
| 102 | 2016 | Korea | Nuruk |
| 103 | 2016 | Korea | Nuruk |
| 104 | 2016 | Korea | Nuruk |
| 105 | 2016 | Korea | Nuruk |
| 106 | 2016 | Korea | Nuruk |
| 107 | 2016 | Korea | Nuruk |
| W | 2016 | Korea | Nuruk |
| 22S | 2016 | Korea | Nuruk |
| 3-1 | 2017 | Korea | Nuruk |
| S1-1 | 2017 | Korea | Cornus officinalis |
| S1-2 | 2017 | Korea | Cornus officinalis |
| S1-5 | 2017 | Korea | Cornus officinalis |
| S2-6 | 2017 | Korea | Cornus officinalis |
| S2-7 | 2017 | Korea | Cornus officinalis |
| S2-8 | 2017 | Korea | Cornus officinalis |
| S2-9 | 2017 | Korea | Cornus officinalis |
| S2-10 | 2017 | Korea | Cornus officinalis |
| S3-1 | 2017 | Korea | Cornus officinalis |
| S3-3 | 2017 | Korea | Cornus officinalis |
| S3-4 | 2017 | Korea | Cornus officinalis |
| S3-5 | 2017 | Korea | Cornus officinalis |
| 3O1 | 2017 | Korea | Prunus serrulata |
| 3O2 | 2017 | Korea | Prunus serrulata |
| 3O3 | 2017 | Korea | Prunus serrulata |
| 3O4 | 2017 | Korea | Prunus serrulata |
| 3O5 | 2017 | Korea | Prunus serrulata |
| 3X1 | 2017 | Korea | Prunus serrulata |
| 3X2 | 2017 | Korea | Prunus serrulata |
| 3X3 | 2017 | Korea | Prunus serrulata |
| 3X4 | 2017 | Korea | Prunus serrulata |
| 3X5 | 2017 | Korea | Prunus serrulata |
| 5O1 | 2017 | Korea | Prunus serrulata |
| 5O2 | 2017 | Korea | Prunus serrulata |
| 5O3 | 2017 | Korea | Prunus serrulata |
| 5O4 | 2017 | Korea | Prunus serrulata |
| 5O5 | 2017 | Korea | Prunus serrulata |
| Ref 1a | USA | Wine barrel | |
| Ref 2a | Japan | Sake | |
| Ref 3a | Chile | Wine | |
| Ref 4a | South Africa | White wine | |
| Ref 5a | Singapore | Bakery | |
| Ref 6a | Laboratory strain [21] | ||
| Com 1b | Korea | Beer | |
| Com 2b | Korea | Bakery | |
| Com 3b | Korea | Bakery | |
| Com 4b | Korea | Beer | |
aReference strains isolated from various countries. Genetic information for these strains was obtained from Genbank
bCommercial strains used for industrial-scale baking and alcohol fermentation in Korea
Genomic DNA extraction
Genomic DNA extraction was performed as described by Tavanti et al. [25] with slight modifications. Yeast cells were grown overnight at 30 °C in 10 mL YPD medium containing 1% yeast extract, 2% peptone, and 2% dextrose (Difco, Detroit, MI, USA). Yeast cells were centrifuged at 13,000 rpm for 5 min and the supernatant was discarded. Liquid nitrogen was used to break the yeast cell walls. Cells were then resuspended in 500 μl lysis buffer (200 mM Tris–HCl pH 8.5, 250 mM NaCl, 25 mM EDTA, and 0.5% SDS) and 500 μl phenol/chloroform/iso-amyl alcohol (25:24:1) was added. Cell lysate were pelleted at 13,000 rpm for 30 min in a microcentrifuge. Sodium acetate (3 M) and isopropyl alcohol were added to the supernatant at 1/10 and 6/10 of the supernatant volume, respectively. Following centrifugation at 13,000 rpm for 5 min, the supernatant was discarded and the pellet was washed with 1 mL ethanol. The pellet was then completely dried and 30 μl of TE-RNase [100 mM Tris–HCl (pH 8), 10 mM EDTA, and 20 μg of RNase/mL] was added to resuspend the genomic DNA.
Amplification of 5.8S-ITS region and identification
Amplification was performed using an ASTEC PC-320 thermal cycler (Astec Inc., Fukuoka, Japan). The ITS1 forward primer and ITS4 reverse primer were used to amplify the conserved 5.8S-ITS region [26]. Amplifications were performed in 30 μl reaction mixtures containing DNA polymerase (TaKaRa Taq™, 5 U/μl), 10X PCR buffer, dNTPs (2.5 mM each), forward primer (0.2–1.0 μM, final conc.), reverse primer (0.2–1.0 μM, final conc.), template (< 500 ng), and sterilized distilled water. After denaturation at 94 °C for 5 min, 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 1 min were performed. This was followed by a final extension step of 5 min at 72 °C. The presence of amplified product was monitored on agarose gels. Sequencing of the amplified fragments was performed by Macrogen (Seoul, Korea). Isolates were identified using LaserGene (http://www.dnastar.com) and BLAST software (https://www.ncbi.nlm.nih.gov/BLAST/).
MLST analysis
Even though 10 genes were examined for the MLST analysis, only six genes were selected for the final analysis. The six genes encoded the following proteins: acetyl-CoA carboxylase (ACC1), ATP-dependent permease (ADP1), glutamine tRNA synthetase (GLN4), leucine-zipper transcriptional activator (MET4), the FG-nucleoporin component of central core of the nuclear pore complex (NUP116), and a subunit of the 26S proteasome (RPN2) [5, 27]. Internal fragments of each gene were amplified by PCR from genomic DNA of the sample S. cerevisiae strains. Primer sequences used for MLST analysis in this study are listed in Table 2. PCR was implemented in reaction volumes of 25 μl containing DNA polymerase (TaKaRa Ex Taq™, 5 U/μl), 10X Ex Taq buffer, dNTPs (2.5 mM each), forward primer (0.2–1.0 μM, final conc.), reverse primer (0.2–1.0 μM, final conc.), template (< 500 ng), and sterilized distilled water. One cycle of denaturation for 10 min at 95 °C was followed by 30 cycles of denaturation at 95 °C for 1 min, annealing at various temperatures for 30 s, and elongation at 72 °C for 1 min, and then a final post-extension step of 10 min at 72 °C [27].
Table 2.
Primer sequences used for MLST analysis of S. cerevisiae strains
| ORF | Gene | Primers | Sequence 5′ → 3′ | Amplicon size (bp) |
|---|---|---|---|---|
| YNR016R | ACC1 | ACC1 F | GCAAGAGAAATTTTGATTCAAGG | 492 |
| ACC1 R | TTCATCAACATCATCTAAATG | |||
| YOR168W | GLN4 | GLN4 F | GAGATTGTCAAGAATAAAAAGGT | 489 |
| GLN4 R | GTCTCTCTCATCCTTTGGACC | |||
| YCR011C | ADP1 | ADP1 F | GAGCCTTCTATGAATGATTTG | 585 |
| ADP1 R | TTGATCGACGAACCCGATTAT | |||
| YIL075C | RPN2 | RPN2 F | TTTATGCACGCTGGTACTAC | 450 |
| RPN2 R | GAGACCCATACCTAATGCAG | |||
| YNL103W | MET4 | MET4 F | CGAGGATAAGCCGAGCAA | 395 |
| MET4 R | GCGCATCCACTCCATTGT | |||
| YMR047C | NUP116 | NUP116 F | AAGCAACTGTCACCAACACG | 501 |
| NUP116 R | CTTCCCCATCGTTCTTTGAG |
PCR fragments obtained for each locus were sequenced and compared with each other and/or the reference S. cerevisiae strains using Lagergene software (http://www.dnastar.com) and Clustal omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) [28]. Heterozygosity was defined as the occurrence of a mixed peak. Different sequences at each locus were assigned an arbitrary number. Concatenation of sequences from the six markers was performed and edited sequences were saved in FASTA file format using the MLSTest program (https://mlstest.codeplex.com/) [29]. A neighbor-joining (NJ) phylogenetic tree was generated using MEGA 6 software (www.megasoftware.net/). Confidence in nodes was determined using the bootstrap procedure with 1000 randomizations. A neighbor-joining tree shows the relationships among strains, and the branch lengths of the tree are proportional to the evolutionary distances among strains [30]. Because NJ trees are unrooted, common ancestry cannot be inferred.
Makgeolli brewing
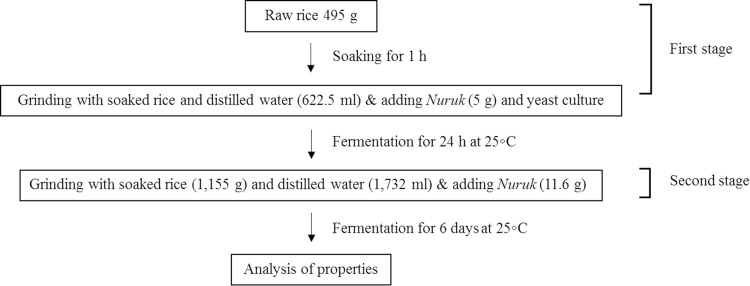
The process used to brew Makgeolli is illustrated in Fig. 1. Raw rice was washed and soaked in water for 1 h. After soaking, the rice was drained for 30 min and ground in water using a blender. The ground rice was added to a bottle along with Nuruk (Kooksoondang Brewery CO., Ltd, Korea), yeast, and distilled water followed by fermentation at 25 °C for 24 h. After fermentation, more ground rice and distilled water were added to the first brew followed by fermentation at 25 °C for 6 days.
Fig. 1.
Process used to brew Makgeolli
Alcohol content analysis
The alcohol content of Makgeolli was measured using a method described previously with slight modifications [31]. Briefly, 100 mL of sample was distilled using a distiller until around 80 mL was collected. The collected sample was adjusted to 100 mL with distilled water. The alcohol content (%) was measured using a density meter (DMA 4500, Anton-Paar, Ashland, VA, USA).
Alcohol production capacity of Saccharomyces cerevisiae strains
To measure alcohol production capacity, 100 mL of GY medium (20% glucose, and 3% yeast extract) was placed in a flask, and Saccharomyces cerevisiae was inoculated at a 0.1% concentration followed by incubation without shaking at 30 °C for 7 days. Culture medium was centrifuged at 10,000 rpm for 10 min and the supernatant was used to measure the alcohol content using the method described above [31].
Results and discussion
Selection of housekeeping genes for MLST analysis
The main objective of this research was to examine the suitability of MLST to differentiate 42 S. cerevisiae strains from natural sources as well as four Korean commercial strains (com 1, com 2, com 3, and com 4). In addition, the genomic diversity of those strains was compared with each other and those isolated from other countries (ref 1, ref 2, ref 3, ref 4, ref 5, and ref 6).
The first step in MLST analysis is the selection of adequate housekeeping genes. Previously, there were only two published reports in which MLST was applied to study the diversity of S. cerevisiae wine yeasts [5, 27]. Ayoub et al. [27] adopted seven loci (ATF1, MET4, RPN2, NUP116, STE50, YBL081W, and IntAY) to analyze the biodiversity of indigenous S. cerevisiae wine yeasts from Lebanon whereas Munoz et al. [5] used six different loci (ACC1, ADP1, GLN4, MET4, NUP116, and RPN2) to discriminate Spanish wine yeast strains at the strain level. Among those 12 loci, YBL081W and IntAY were not housekeeping genes [27]. YBL081W was the hypothetical ORF while IntAY was an intergenic locus between ORFs APP1 and YPT53). Therefore, we examined 10 loci (ACC1, ADP1, ALA1, ATF1, GLN4, MET4, NUP116, RPN2, STE50 and VPS13) to assess the degree of genetic variability in S. cerevisiae strains isolated from Nuruk and flowers in Korea.
Munoz et al. [5] examined 14 nuclear genes as potential MLST target genes, but found only a few single nucleotide polymorphisms (SNPs) (0–3) in the subset of strains analyzed depending on the particular locus. Therefore, they used six loci (ACC1, ADP1, ALA1, GLN4, RPN2, and VPS13) previously used to characterize clinical isolates of Candida albicans for MLST analysis and found 10 polymorphic sites and 13 different genotypes based on these five loci.
When we examined 10 loci, the sequences of the ALA1 and ATF1 fragments were conserved among all tested S. cerevisiae strains (data not shown), indicating that these genes had low discriminatory power. Therefore, ALA1 and ATF1 were eliminated from the list of target housekeeping genes for MLST analysis. MET4 and NUP116 genes were chosen because these genes have shown high discriminatory power in MLST analysis of wine yeast strains [27]. STE50 and VPS 13 were also considered target genes, but were excluded because they also displayed relatively low discriminatory power (data not shown). As a conclusion, six of the 10 loci (ACC1, ADP1, GLN4, MET4, NUP116, and RPN2) were finally selected. The PCR primers used for MLST analysis are listed in Table 2.
Effectiveness of MLST analysis and genetic diversity
Sequencing results for the 42 S. cerevisiae strains for the six loci revealed 26 polymorphic sites among 2617 bp, which corresponds to a sequence diversity of 0.99%. The proportion of polymorphic sites per locus varied between 0.74% (GLN4) and 1.73% (ACC1) with an average of 1.09%. All mutations were base substitutions. Approximately 23% (6 of 26) of the sites were heterozygous. ACC1 contained eight polymorphic sites that resulted into a classification of the 42 S. cerevisiae strains into five genotypes, while the MET4 gene yielded four genotypes among the 42 strains based on three polymorphic sites (Table 3). These results indicated that MLST can be used to type S. cerevisiae strains.
Table 3.
Polymorphic sites in the fragments of the six housekeeping genes tested
| GLN4 | 1 | 1 | 4 | NUP116 | 2 | 2 | 2 | 2 | 2 | 2 |
| 3 | 9 | 5 | 3 | 3 | 4 | 4 | 4 | 6 | ||
| 6 | 5 | 0 | 3 | 8 | 2 | 4 | 5 | 0 | ||
| 5 | 2 | 2 | 2 | 4 | 4 | |||||
| Genotype 1 (38) | C | G | T | Genotype 1 (36) | A | A | G | G | C | C |
| Genotype 2 (1) | G | G | T | Genotype 2 (3) | G | G | C | T | A | T |
| Genotype 3 (3) | C | A | C | Genotype 3 (3) | R | A | G | G | C | C |
| MET4 | 6 | 7 | 7 | ADP1 | 9 | 1 | 1 | |||
| 3 | 1 | 5 | 6 | 0 | 1 | |||||
| 0 | 6 | 9 | 9 | 5 | 0 | |||||
| 0 | 9 | |||||||||
| Genotype 1 (29) | T | G | Y | Genotype 1 (34) | G | G | G | |||
| Genotype 2 (10) | T | G | T | Genotype 2 (1) | R | R | G | |||
| Genotype 3 (1) | C | T | T | Genotype 3 (4) | A | R | G | |||
| Genotype 4 (2) | C | K | T | Genotype 4 (3) | G | A | A |
| ACC1 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | RPN2 | 3 | 3 | 3 |
| 3 | 6 | 6 | 6 | 6 | 7 | 7 | 7 | 0 | 1 | 1 | ||
| 6 | 0 | 4 | 6 | 6 | 0 | 4 | 5 | 1 | 0 | 4 | ||
| 3 | 3 | 1 | 3 | 9 | 2 | 4 | 4 | 8 | 4 | 7 | ||
| Genotype 1 (33) | C | T | A | C | C | A | A | T | Genotype 1 (39) | T | A | C |
| Genotype 2 (5) | C | C | T | T | C | A | A | C | Genotype 2 (3) | C | C | T |
| Genotype 3 (1) | C | T | W | C | C | A | A | C | ||||
| Genotype 4 (2) | T | C | A | C | T | G | G | T | ||||
| Genotype 5 (1) | T | T | A | C | C | A | A | T |
The number of the polymorphic sites (vertical format) is the order from the first nucleotide of each housekeeping gene. All polymorphic sequences are shown. (Y = T or C, K = G or T, R = G or A, and W = A or T)
The major advantage of MLST is that simple PCR and sequencing of several house-keeping genes can generate sequence data that can be easily compared to data in a database; thus, it is fast and unambiguous. This technique has been successfully employed to discriminate among clinically important bacterial strains such as Neisseria meningitides, Streptococcus pneumoniae, Staphylococcus aureus, Campylobacter jejuni, and Listeria monocytogenes [10, 32–35]. Recently, Wu et al. [36] used MLST to study the epidemiology and evolution of C. albicans by typing 62 C. albicans isolates. MLST has also been shown to be a useful tool for typing S. cerevisiae isolated from wine [5, 27]. Until now, however, it has not been employed to study the biodiversity of wild-type S. cerevisiae found in Korea.
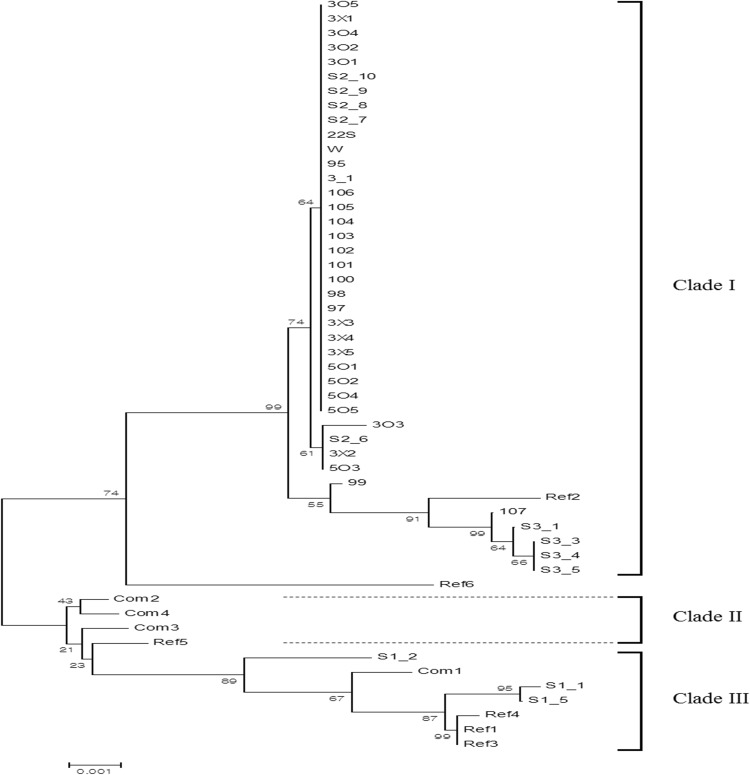
MLST analysis using six housekeeping genes allowed construction of a reliable phylogenetic tree (Fig. 2). To verify the effectiveness of MLST, the sequence information of six loci (ACC1, ADP1, GLN4, MET4, NUP116, and RPN2) from ten more S. cerevisiae strains including six reference and four commercial strains (Table 1) were included in the phylogenetic tree.
Fig. 2.
Unrooted neighbor-joining tree of the genetic relationships among 52 S. cerevisiae strains with branch lengths proportional to the p-distance. Numbers within the tree indicate the bootstrap values for nodes
Genetic information for four wine S. cerevisiae strains (ref 1–ref 4 in Table 1) from various countries (USA, Japan, Chile, and South Africa), a baking yeast (ref 5, Singapore), and a laboratory strain (ref 6, USA) were obtained from Genbank. The sequence information of six loci (ACC1, ADP1, ALA1, GLN4, MET4, NUP112, and RPN2) of these strains were extracted from their whole genome sequences. As shown in Fig. 2, three of the reference strains (ref 1, ref 3, and ref 4, all of which are wine-making strains) were clustered together. However, ref 2, a strain used in sake fermentation, which is different from typical wine fermentation but close to Makgeolli fermentation, was closely related to the S. cerevisiae strains isolated in Korea. In addition, ref 6, which is the most common laboratory strain used in academia, and ref 5, which is a baker’s yeast from Singapore, were isolated from other strains.
Furthermore, we examined four commercial S. cerevisiae strains (com 1–com 4 in Table 1), used in the baking and alcohol fermentation industries in Korea. They all clustered together except com 1. In fact, all baking yeasts including com 2, com 3, and ref 5 were closely grouped together in the MLST analysis (Fig. 2). This suggests that there is little variation among commercial S. cerevisiae strains. Also, it is obvious that the wild S. cerevisiae strains isolated in Korea are somewhat different from commercial S. cerevisiae strains isolated in other countries. It was interesting that two commercial S. cerevisiae (com 1 and com 4) used in beer fermentation in Korea were not tightly linked together. Especially, com 4 was grouped with baker’s yeast, suggesting that it was originally used in baking industry.
A total of 42 S. cerevisiae strains isolated from natural sources, such as Nuruk and flowers, were broadly classified into three clades based on MLST analysis and their applications as shown Fig. 2. Clade I contained 39 of the 42 S. cerevisiae strains (93%). No S. cerevisiae strains isolated from natural sources were members of Clade II, while Clade III contained the remaining three isolated S. cerevisiae strains (7%). None of the clades showed any correlation with the original sample sources from which the strains were isolated.
Alcohol fermentation characteristics of selected S. cerevisiae strains
To investigate if there is any difference between S. cerevisiae strains belong to each Clade groups in MLST analysis, Makgeolli fermentation characteristics of each S. cerevisiae strains were examined. In fact, there was not much difference in Makgeolli fermentation characteristics in Clade I and Clade II groups by showing typical Makgeolli fermentation. However, S. cerevisiae strains belonged to Clade III showed rather different properties in Makgeolli fermentation. The final alcohol contents of S. cerevisiae strains belonged to Clade III (14.7–17.5%) was lower than other S. cerevisiae strains in Clade I (19.7–20.7%) and Clade II (18.5%) groups (Table 4).
Table 4.
Alcohol fermentation characteristics of representative Saccharomyces cerevisiae strains isolated from natural sources
| Number | Alcohol content of Makgeolli (%) | Alcohol production capacity (%) | Clade |
|---|---|---|---|
| S2-6 | 20.7 | 11.0 | Clade I |
| S2-7 | 20.5 | 10.6 | |
| S2-8 | 20.5 | 11.0 | |
| S2-9 | 20.6 | 10.5 | |
| S2-10 | 19.9 | 10.4 | |
| 3O1 | 20.7 | 11.1 | |
| 3O3 | 19.7 | 11.4 | |
| 3X1 | 20.2 | 11.2 | |
| 3X2 | 20.2 | 10.9 | |
| 5O1 | 20.0 | 10.6 | |
| 5O3 | 20.0 | 11.4 | |
| Com2 | 18.5 | 11.2 | Clade II |
| S1-1 | 14.7 | 9.5 | Clade III |
| S1-2 | 16.0 | 9.2 | |
| S1-5 | 15.4 | 9.5 | |
| Com1 | 17.5 | 9.7 |
The ability of the S. cerevisiae strains to produce alcohol when the GY medium was supplemented with 20% glucose was examined. While there was little difference between strains from Clades I and II, there was distinct difference between strains in these clades and the alcohol production capacity of S. cerevisiae strains in Clade III. S. cerevisiae strains in Clade III produced less alcohol (9.2–9.7%) than strains in Clades I and II (10.4–11.6%) (Table 4). These results indicate that classification of S. cerevisiae based on MLST analysis is somewhat related to the alcohol fermentation properties of the S. cerevisiae strains.
In conclusion, MLST analysis was performed to differentiate among S. cerevisiae strains isolated from natural sources. MLST analysis can be used to assess genetic variation. Clustering of allele profiles differentiated the 52 S. cerevisiae strains examined into three groups (Clade I, Clade II, and Clade III). Among those groups, strains belonging to Clade III exhibited lower alcohol fermentation capacity than strains belonging to Clades I and II. This result indicates that MLST analysis could be a highly reliable method to study the genetic diversity of S. cerevisiae strains and a useful tool to classify S. cerevisiae, an important baking and brewing yeast.
Acknowledgements
This work was supported by a grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR201629202).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Dalawai N, Krupa K, Nadkarni S, Bharani S, Harinikumar K. Screening of Efficient Ethanol Tolerant Yeast Strain for Production of Ethanol. Int. J. Pure App. Biosci. 2017;5:744–752. doi: 10.18782/2320-7051.2587. [DOI] [Google Scholar]
- 2.Novak J, Basarova G, Teixeira J, Vicente A. Monitoring of brewing yeast propagation under aerobic and anaerobic conditions employing flow cytometry. J. I. Brewing. 2007;113:249–255. doi: 10.1002/j.2050-0416.2007.tb00284.x. [DOI] [Google Scholar]
- 3.Maslow JN, Mulligan ME, Arbeit RD. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin. Infect. Dis. 1993;17:153–162. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- 4.Esteve-Zarzoso B, Hierro N, Mas A, Guillamón JM. A new simplified AFLP method for wine yeast strain typing. LWT-Food Sci. Technol. 2010;43:1480–1484. doi: 10.1016/j.lwt.2010.05.016. [DOI] [Google Scholar]
- 5.Muñoz R, Gómez A, Robles V, Rodríguez P, Cebollero E, Tabera L, Carrascosa AV, Gonzalez R. Multilocus sequence typing of oenological Saccharomyces cerevisiae strains. Food Microbiol. 2009;26:841–846. doi: 10.1016/j.fm.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Bidenne C, Blondin B, Dequin S, Vezinhet F. Analysis of the chromosomal DNA polymorphism of wine strains of Saccharomyces cerevisiae. Curr. Genet. 1992;22:1–7. doi: 10.1007/BF00351734. [DOI] [PubMed] [Google Scholar]
- 7.Grando MS, Ubeda J, Briones A. RAPD analysis of wine Saccharomyces cerevisiae strains differentiated by pulsed field gel electrophoresis. Biotechnol. Tech. 1994;8:557–560. doi: 10.1007/BF00152145. [DOI] [Google Scholar]
- 8.Masneuf I, Aigle M, Dubourdieu D. Development of a polymerase chain reaction/restriction fragment length polymorphism method for Saccharomyces cerevisiae and Saccharomyces bayanus identification in enology. FEMS Microbiol. Lett. 1996;138:239–244. doi: 10.1111/j.1574-6968.1996.tb08164.x. [DOI] [PubMed] [Google Scholar]
- 9.Guillamón JM, Barrio E. Genetic polymorphism in wine yeasts: mechanisms and methods for its detection. Front. Microbiol. 2017;8:1–20. doi: 10.3389/fmicb.2017.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan CB, Diggle MA, Clarke SC. Multilocus sequence typing. Mol. Biotechnol. 2005;29:245. doi: 10.1385/MB:29:3:245. [DOI] [PubMed] [Google Scholar]
- 12.Lee S, Yoo M, Shin D. The identification and quantification of biogenic amines in Korean turbid rice wine, Makgeolli by HPLC with mass spectrometry detection. LWT-Food Sci. Technol. 2015;62:350–356. doi: 10.1016/j.lwt.2015.01.016. [DOI] [Google Scholar]
- 13.Jung H, Lee S-J, Lim JH, Kim BK, Park KJ. Chemical and sensory profiles of Makgeolli, Korean commercial rice wine, from descriptive, chemical, and volatile compound analyses. Food Chem. 2014;152:624–632. doi: 10.1016/j.foodchem.2013.11.127. [DOI] [PubMed] [Google Scholar]
- 14.Jeong J-W, Nam P-W, Lee S-J, Lee K-G. Antioxidant activities of Korean rice wine concentrates. J. Agric. Food Chem. 2011;59:7039–7044. doi: 10.1021/jf200901j. [DOI] [PubMed] [Google Scholar]
- 15.Kang M-G, Kim J-H, Ahn B-H, Lee J-S. Characterization of new antihypertensive angiotensin I-converting enzyme inhibitory peptides from Korean traditional rice wine. J. Microbiol. Biotechnol. 2012;22:339–342. doi: 10.4014/jmb.1109.09015. [DOI] [PubMed] [Google Scholar]
- 16.Choi J-S, Seo HJ, Lee Y-R, Kwon S-J, Moon SH, Park S-M, Sohn JH. Characteristics and in vitro anti-diabetic properties of the Korean rice wine, makgeolli fermented with Laminaria japonica. Prev. Nutr. Food Sci. 2014;19:98. doi: 10.3746/pnf.2014.19.2.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin M-O, Kang D-Y, Kim M-H, Bae S-J. Effect of growth inhibition and quinone reductase activity stimulation of Makgeoly fractions in various cancer cells. J. Korean Soc. Food Sci. Nutr. 2008;37:288–293. doi: 10.3746/jkfn.2008.37.3.288. [DOI] [Google Scholar]
- 18.Jeon H, Yu JC, Kim G, Kong H-S. Quality characteristics of Takju by yeast strain type. Korean J Food Nutr. 2014;27:971–978. doi: 10.9799/ksfan.2014.27.5.971. [DOI] [Google Scholar]
- 19.Baek SY, Lee YJ, Kim M-D, Yi J-H, Mun J-Y, Yeo S-H. Characterization of Ethanol Fermentation with Wild Type Yeast Strains. Microbiol. Biotechnol. Lett. 2015;43:227–235. doi: 10.4014/mbl.1507.07002. [DOI] [Google Scholar]
- 20.Kang SH, Kim HR, Kim JH, Ahn BH, Kim TW, Lee J-E. Identification of wild yeast strains and analysis of their β-glucan and glutathione levels for use in Makgeolli brewing. Mycobiology. 2014;42:361–367. doi: 10.5941/MYCO.2014.42.4.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mortimer RK, Johnston JR. Genealogy of principal strains of the yeast genetic stock center. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carter DM, Liti G, Moses AM, Parts L, James SA, Davey RP, Roberts IN, Blomberg A, Warringer J, Burt A. Population genomics of domestic and wild yeasts. Nature. 2008;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akao T, Yashiro I, Hosoyama A, Kitagaki H, Horikawa H, Watanabe D, Akada R, Ando Y, Harashima S, Inoue T. Whole-genome sequencing of sake yeast Saccharomyces cerevisiae Kyokai no. 7. DNA Res. 2011;18:423–434. doi: 10.1093/dnares/dsr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song G, Dickins BJ, Demeter J, Engel S, Dunn B, Cherry JM. AGAPE (Automated Genome Analysis PipelinE) for pan-genome analysis of Saccharomyces cerevisiae. PLoS One. 2015;10:e0120671. doi: 10.1371/journal.pone.0120671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tavanti A, Davidson AD, Johnson EM, Maiden MC, Shaw DJ, Gow NA, Odds FC. Multilocus sequence typing for differentiation of strains of Candida tropicalis. J. Clin. Microbiol. 2005;43:5593–5600. doi: 10.1128/JCM.43.11.5593-5600.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao G, Liu Z, Hyde K, Lui X, Yu Z. Whole rDNA analysis reveals novel and endophytic fungi in Bletilla ochracea (Orchidaceae) Fungal Divers. 2008;33:101–122. [Google Scholar]
- 27.Ayoub MJ, Legras JL, Saliba R, Gaillardin C. Application of multi locus sequence typing to the analysis of the biodiversity of indigenous Saccharomyces cerevisiae wine yeasts from Lebanon. J. Appl. Microbiol. 2006;100:699–711. doi: 10.1111/j.1365-2672.2006.02817.x. [DOI] [PubMed] [Google Scholar]
- 28.Pais FS-M, de Cássia Ruy P, Oliveira G, Coimbra RS. Assessing the efficiency of multiple sequence alignment programs. Algorithms Mol. Biol. 2014;9:144. doi: 10.1186/1748-7188-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomasini N, Lauthier JJ, Llewellyn MS, Diosque P. MLSTest: Novel software for multi-locus sequence data analysis in eukaryotic organisms. Infect. Genet. Evol. 2013;20:188–196. doi: 10.1016/j.meegid.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Woo K-S, Ko J-Y, Song S-B, Lee J-S, Oh B-G, Kang J-R, Nam M-H, Ryu I-S, Jeong H-S, Seo M-C. Physicochemical characteristics of Korean traditional wines prepared by addition of sorghum (Sorghum bicolor L. Moench) using different Nuruks. J. Korean Soc. Food Sci. Nutr. 2010;39:548–553. doi: 10.3746/jkfn.2010.39.4.548. [DOI] [Google Scholar]
- 32.Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 33.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Nati. Acad. Sci. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dingle K, Colles F, Wareing D, Ure R, Fox A, Bolton F, Bootsma H, Willems R, Urwin R, Maiden M. Multilocus Sequence Typing System for Campylobacter jejuni. J. Clin. Microbiol. 2001;39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salcedo C, Arreaza L, Alcala B, De La Fuente L, Vazquez J. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 2003;41:757–762. doi: 10.1128/JCM.41.2.757-762.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu K, Luo T, Li L, Zhang Q, Zhu J, Gao Q, Chen M, Zhu M. Multilocus sequence typing of pathogenic Candida albicans isolates collected from a teaching hospital in Shanghai, China: a molecular epidemiology study. PloS one. 2015;10:e0125245. doi: 10.1371/journal.pone.0125245. [DOI] [PMC free article] [PubMed] [Google Scholar]