Abstract
GC–MS coupled with multivariate statistical analysis was performed to understand metabolites difference between pileus and stipe of Sparassis crispa (cauliflower mushroom). Metabolic changes of S. crispa after fermentation by different microorganisms were also investigated. PCA score plot showed a clear separation between pileus and stipe of S. crispa regardless of fermentation. However, OPLS-DA score plot showed clear separation among fermented S. crispa samples according to microbial strain used, indicating that both pileus and stipe fermented with the same strain showed similar pattern of metabolites. Fructose, lactic acid, citric acid, malic acid, and phosphoric acid were metabolites that contributed to the discrimination of fermented S. crispa samples. Results of this study provide novel insights into intrinsic characteristics of stipe of S. crispa which is cheaper than pileus as ingredient for alternative functional food.
Keywords: Metabolomics, Sparassis crispa, Stipe, GC–MS, Fermentation
Introduction
Sparassis crispa (SC) is a species of fungus, commonly called Ggotsongyi (meaning a blossom) in Korean and cauliflower mushroom in English. SC is well known for its taste and functionalities due to β-glucan [1]. It has been reported that SC possesses anti-thrombotic [2], anti-tumor [3], and immune-modulating activities [4]. It also has anti-angiogenic and anti-metastatic effects [5].
Physically, the lobes of SC are flat and curly with color of cream to pale yellow. Fruiting bodies of SC are approximately 5–20 cm tall and 6–30 mm in diameter [6]. SC consists of pileus and stipe that account for approximate 65–70 and 30–35% of the mushroom on fresh weight basis. In most mushroom processing, stipes of SC (lower part of mushroom) are not fully utilized. They are treated as waste. However, our previous study has confirmed that the stipe of SC has great potential as ingredient for functional food through component and immunoregulatory analysis [7]. For example, the mortality rate of the eels fed supplemented diets of SC stipe was decreased significantly [8].
Fermentation has been employed for a long time to improve the functionality, texture, shelf life, and flavor of food products [9]. Many metabolic changes occur during fermentation, thus affecting product properties such as bioactivity and digestibility [10]. Recently, fermentation has been applied to increase bioactive compounds in mushroom [11–13]. Through fermentation, various functions of mushroom can be improved. For example, fermentation has been applied to decrease the concentration of nitrite in mushroom (Pleurotus spp.) [11]. Yang et al. [14] have reported that fermentation of mushroom by lactic acid bacteria can increase its antioxidant activity. Our previous study has also found that fermented SC extracts possess antioxidant and immunological activities [15].
Since β-glucan is a major active component of SC, most studies have only dealt with the contents of β-glucan in SC without determining its full metabolites profiles. However, fermented SC contains many metabolites, including byproducts of microorganism during fermentation. To fully understand the effect of fermentation, a powerful analytical approach is needed to investigate metabolites of fermented SC.
NMR spectroscopy, GC–MS, and LC–MS are commonly used for identification and quantification of metabolome [16]. Among these techniques, GC–MS is one of the most widely used techniques for metabolomics study, allowing for identification of more than 300 compounds [17]. Metabolomics approaches have been applied to study food fermentation such as fruit [18], vegetables [19, 20], cereals [21, 22], and legumes [23]. They are also used to unravel unknown metabolites in food fermentation. However, little is known about the dynamics of metabolites during mushroom fermentation or the relationship between microbial strain and metabolic changes during mushroom fermentation.
Therefore, this study was performed to determine metabolites differences of pileus and stipe of SC and to investigate metabolic changes after SC fermentation according to the inoculation of different microbial strains by using GC–MS based metabolomics study.
Materials and methods
Preparation of S. crispa
Fresh S. crispa (SC) mushrooms were collected from Baegasan Cauliflower Mushroom (Hwasun, Jeollanamdo, Korea). SC was separated into pileus and stipe manually. These separated pilei and stipes of SC were hot-air-dried at 40 °C for 2 d and subsequently powdered using a blender. Next, pileus or stipe powder was dissolved in distilled water (20 times in volume) and extracted by autoclaving at 121 °C for 1 h. These extracts of SC pilei and stipes were then air-cooled to room temperature for fermentation.
Microbial strains and fermentation conditions
Lactobacillus plantarum KCCM 11322 was obtained from Korean Culture Center of Microorganisms (KCCM, Seoul, Korea). Leuconostoc mesenteroides KCTC 3718 and Bacillus subtilis KCTC 2023 were purchased from Korean Collection for Type Cultures (KCTC, Daejeon, Korea). One commercial Saccharomyces cerevisiae yeast strain (K1-V1116, Lalvin, Montreal, Canada) was also used for fermentation.
Before inoculation, strains were pre-cultured at 37 °C for 48 h in de Man Rogosa Sharpe broth (Difco, Sparks, MD, USA) for L. plantarum and L. mesenteroides, nutrient broth (Difco, Sparks, MD, USA) for B. subtilis, and yeast malt broth (Difco, Sparks, MD, USA) for S. cerevisiae to obtain a final cell count of more than 107 CFU/mL. One percent (v/v) of each starter culture was inoculated into each SC extract for fermentation. Fermentation was carried out in triplicates for each microbial strain at 30 °C for 48 h. Samples before and after fermentation were taken for GC–MS analysis. The samples were then centrifuged at 14,000 rpm for 10 min at 4 °C, and 100 μL of the supernatant was freeze dried for derivatization.
GC–MS methods for metabolites profiling
Sample derivatization protocol and GC–MS analysis conditions were the same as described in our previous study [24], with slight modifications. Briefly, after lyophilization of samples (100 μL), 100 μL of o-methoxyamine hydrochloride in pyridine solution (15 mg/mL) was added to freeze-dried sample residue. After vortex mixing each sample for 10 min, the sample was incubated at 25 °C for 16 h for oximation. Analytes were then trimethylsilylated using 100 μL of BSTFA (N,O-bis-(trimethylsilyl)-trifluoroacetamide) derivatization agent containing 1% TMCS (trimethylchlorosilane) as a catalyst at 70 °C for 1 h. Samples were then cooled at 25 °C in the dark for 1 h. Next, 600 µL of methyl stearate in heptane (10 ppm) was added as an internal standard. After centrifuging samples at 13,000 rpm for 15 min, the supernatant was subjected to GC–MS analysis.
QP-2020 gas Chromatography Mass Spectrometry (Shimadzu, Kyoto, Japan) was used for metabolites analysis. Separation was achieved using a Rtx-5MS capillary column (length, 30 m; diameter, 0.25 mm; film thickness, 0.25 μm). GC–MS temperatures were as follows: injector, 250 °C; column, 280 °C with an increment of 10 °C/min; transfer line, 280 °C; ion source, 230 °C; and quadrupole temperature, 150 °C. The oven temperature was maintained at 60 °C for 1 min, increased to 280 °C at 10 °C/min, and then held at 280 °C for 10 min. Ionization was achieved with electron beam at 70 eV. The mass spectrometer was programmed under electron impact (EI) in a full scan mode at m/z 50–550 with a scanning rate of a 2 scans/s.
Data processing and multivariate analysis
GC–MS files were analyzed with XCMS web software (https://xcmsonline.scripps.edu) for data pretreatment procedure, such as noise removal, baseline correction, alignment, and extraction of characteristic ions. A default Centwave method for GC Single Quadruple was selected for peak detection and alignment with the following parameters: signal/noise threshold, 2; mzdiff, 0.1; integration methods, 1; prefilter peaks, 3; prefilter intensity, 100; mzwid, 0.25; minfrac, 0.5; and bandwidth, 3. Identification of metabolites was made by comparing their mass spectra with NIST 14.0. Peak intensities of identified compounds were normalized by internal standard (methyl stearate) before multivariate statistical analysis.
GC–MS pre-processing data files were imported into SIMCA-P version 14.0 software package (Umetrics, Umea, Sweden) for multivariate analysis. GC–MS data were processed with principal component analysis (PCA) and orthogonal projections to latent structures discriminant analysis (OPLS-DA). A permutation test of two hundred iterations with cross-validation step was performed to avoid model overfitting. Discriminating variables were selected out according to high variable importance in projection value and low p value (p < 0.05).
Statistical analysis
SPSS version 22.0 statistical package (SPSS Inc., Chicago, IL, USA) was used for all data analyses, including analysis of variance (ANOVA) and Duncan multiple-range test. The intensity of individual metabolite identified by GC–MS was subjected to statistical comparison.
Results and discussion
Analysis of S. crispa metabolome by GC–MS
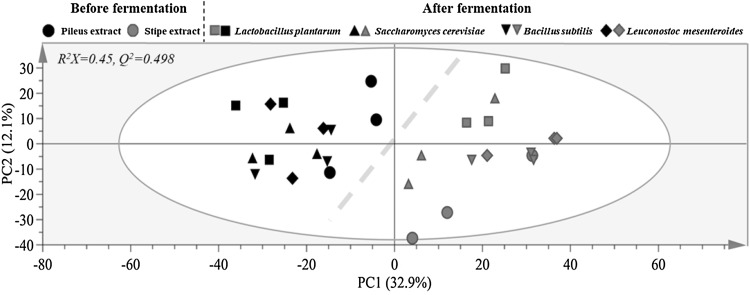
In order to understand metabolic differences of SC according to mushroom parts and fermentation strains, metabolites of SC were analyzed using GC–MS. After peak alignment and exclusion of ion features, 1769 ion features were imported for PCA analysis. PCA score plot derived from GC–MS profiles of all SC samples is shown in Fig. 1. There was a clear separation between pileus and stipe of SC, with cumulative R2X and Q2 values of 0.450 and 0.498, respectively, when two components were calculated. These results indicated that metabolites of SC were different according to mushroom parts. After fermentation, difference in metabolites between pileus and stipe of SC was successfully captured by PC1 regardless of microorganisms used for fermentation. This suggests that metabolic difference was affected by mushroom parts more than by fermentation microorganisms.
Fig. 1.
Principal component analysis (PCA) score plot derived from GC–MS data of pileus and stipe of Sparassis crispa before and after fermentation with different microbial strains, illustrating clear metabolic differences between pileus and stipe of S. crispa. Symbols with black and red color after fermentation denote fermented pileus and stipe of S. crispa, respectively. (Color figure online)
Metabolites differences between pileus and stipe extracts of S. crispa
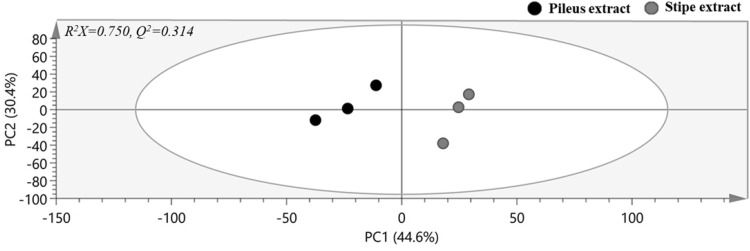
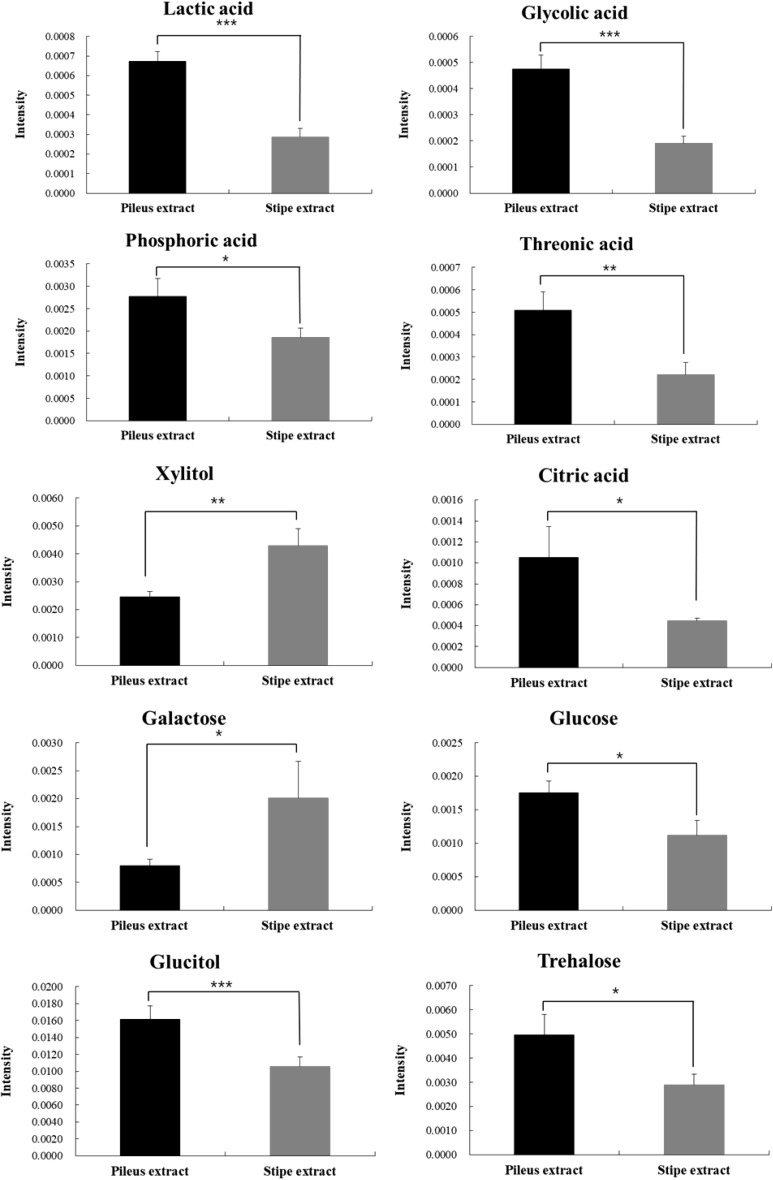
PCA score plot derived from GC–MS profiles of pileus and stipe extracts of SC is shown in Fig. 2. Quality parameters of the generated PCA model revealed that the model had a high fitting (R2X = 0.750) but a low predicting (Q2 = 0.314) quality. To maximize separation, OPLS-DA was also applied in the same datasets (Data not shown). To identify which metabolites were responsible for the separation, VIP statistics in OPLS-DA model was used. According to criteria of VIP > 1.0 and p < 0.05, a total of 10 metabolites were identified as contributing factors to metabolic differences between the two extracts (Fig. 3). Theses ten metabolites were identified with the help of fragmentation patterns of GC–MS experimental data in NIST 14 database. Higher levels of glucose, trehalose, glucitol, lactic acid, phosphoric acid, citric acid, threonic acid, and glycolic acid were observed in pileus extract of SC whereas levels of xylitol and galactose were higher in stipe extract of SC.
Fig. 2.
PCA score plot built on GC–MS data of pileus and stipe extracts of Sparassis crispa before fermentation
Fig. 3.
Metabolites that contribute to the difference between pileus and stipe extracts of Sparassis crispa before fermentation. Data are given as mean ± standard deviations (n = 3). *p < 0.05, **p < 0.01, and ***p < 0.001
Metabolites changes after S. crispa fermentation
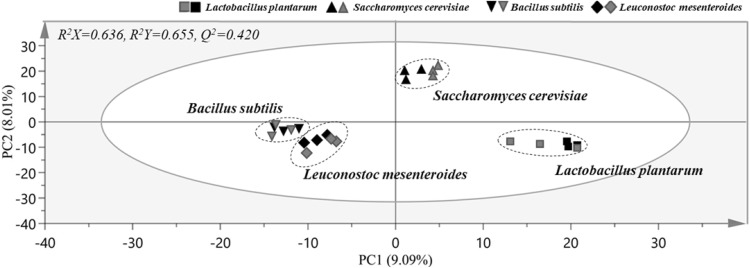
Metabolic changes after fermentation of pileus or stipe of SC were then analyzed. Four microbial food cultures commonly used in food production (generally recognized as safe, GRAS) were selected for SC fermentation. No significant differentiations were observed between SC samples according to different starter cultures in the PCA model with a poor predictive capability as indicated by a Q2 value of 0.420 (Data not shown). To maximize the separation, OPLS-DA score plot was made in the same datasets to remove non-correlated variations (Fig. 4). The OPLS-DA score plot showed clear separation among fermented SCs according to microbial strains used, revealing good fitness and predictability (R2X, 0.636; R2Y, 0.655, and Q2, 0.420). Cross validation was performed using a permutation test that was repeated 200 times. This test showed that Q2 and R2 values were higher than those in the OPLS-DA model, demonstrating both high predictive capability and high goodness of fit for the OPLS-DA model.
Fig. 4.
Orthogonal projections to latent structures discriminant analysis (OPLS-DA) score plot derived from GC–MS data of pileus and stipe of Sparassis crispa fermented with different microbial strains, showing clear metabolic differences among groups fermented with identical strains. The OPLS-DA model was generated with two predictive components and three orthogonal components. The OPLS-DA model was validated by permutation test with 200 random permutations. Symbols with black and red color denote fermented pileus and stipe of S. crispa, respectively. (Color figure online)
Interestingly, different SC fermentation behaviors were observed for each microbial strain, which is in agreement with the results of fermented ginseng with different starter cultures [24]. Samples fermented with identical strains were closely located in the OPLS-DA score plot, indicating that metabolites of these samples were more similar than those of samples fermented with other strains. These results also suggested that metabolites of fermented SC were different depending on strains used. The closely located SC samples fermented with B. subtilis and L. mesenteroides in the OPLS-DA score plot indicated that metabolites patterns of these two groups were more similar than those of other samples. Likewise, SC samples fermented with L. plantarum were located on the right side of PC 1, indicating that their metabolites patterns were more different than those of other strains. However, these results do not mean that the metabolites of fermented SC are affected by the strain rather than the part of mushroom. These results suggest that metabolites between pileus and stipe of SC fermented with identical starter culture might become similar to each other, although metabolites of pileus and stipe of SC were very different before fermentation.
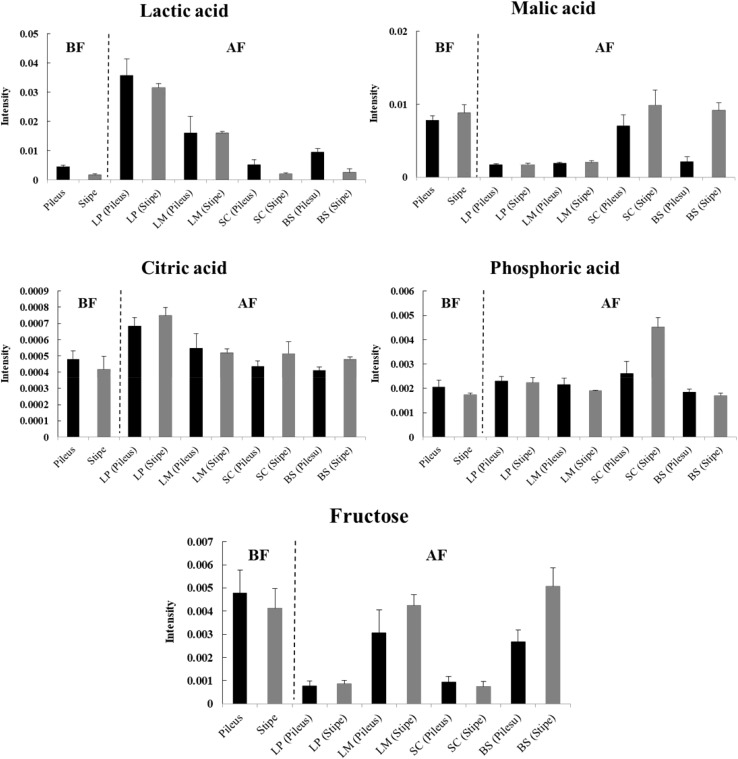
According to the criterion of VIP > 1.5, a total of five metabolites were identified as factors contributing to the discrimination among groups in the OPLS-DA model. Quantitative differences in identified metabolites after fermentation are shown in Fig. 5. Concentrations of lactic acid were higher in fermented SC samples with lactic acid bacteria (L. plantarum and L. mesenteroides). On the other hand, malic acid concentrations were the lowest in these two groups. Increased lactic acid contents together with decreased malic acid contents in fermented SC samples indicate that malolactic fermentation occurred, as lactic acid bacteria metabolize malic acid into lactic acid [25]. The content of citric acid was the highest in samples fermented with L. plantarum. Levels of malic acid in SC fermented with S. cerevisiae were the highest, indicating active malate metabolism by S. cerevisiae such as malic acid production from glucose during fermentation via fumarate or oxaloacetate pathway [26]. Dramatic decrease of fructose was observed in SC samples fermented with L. plantarum and S. cerevisiae, suggesting that these strains might have higher consumption of fructose during fermentation. Generally, microorganisms have higher preference for glucose than for fructose during fermentation. For example, fructose becomes the main sugar present during later stages of wine fermentation [27]. The significant decrease of fructose level in fermented SC samples suggests that there was an active fermentation.
Fig. 5.
Relative comparison of major metabolites (VIP > 1.5) obtained from pileus and stipe extracts of Sparassis crispa before and after fermentation with different microbial strains. Data are given as mean ± standard deviations (n = 3). BF, before fermentation; AF, after fermentation; LP, Lactobacilus plantarum; LM, Leuconostoc mesenteroides; SB, Saccharomyces cerevisiae; BS, Bacillus subtilis
Although SC is expensive, it is very popular among consumers due to its known functionality. Generally, the stipe of mushroom is composted, burned, blended into soil, or discarded after harvesting the fruiting body [28]. Fresh mushrooms are highly perishable to decay due to high moisture content and microorganisms [29]. Therefore, it is necessary to apply fermentation technology to increase the storage period of these mushrooms [11].
Medicinal value of SC is mainly attributable to its abundant content of β-glucan [30]. Our previous study has shown that the stipe of SC has higher β-glucan content than its pileus, although there is no significant difference in general component analysis between the two [7]. In the present study, different metabolites patterns were observed between pileus and stipe extracts of SC. However, pileus and stipe of SC fermented with the same strain of microorganism showed similar pattern of metabolites. These results suggest that different metabolites can be produced during fermentation according to microbial strains used. Similar profiles of metabolites of fermented pileus and stipe of SC may be related to similar taste and functionality. These metabolomics study could provide better understanding of nutritional and healthy potential of SC stipes as alternative ingredients for functional food.
Acknowledgements
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Carvalho JCT, Perazzo FF, Machado L, Bereau D. Biologic activity and biotechnological development of natural products. BioMed Res. Int. Article ID 971745 (2013) [DOI] [PMC free article] [PubMed]
- 2.Choi JH, Lee HJ, Kim S. Purification and antithrombotic activity of wulfase, a fibrinolytic enzyme from the fruit bodies of the edible and medicinal mushroom Sparassis crispa Wulf. ex. Fr. Appl. Biochem. Microbiol. 2016;52:608–614. doi: 10.1134/S000368381606003X. [DOI] [Google Scholar]
- 3.Ohno N, Harada T, Masuzawa S, Miura NN, Adachi Y, Nakajima M, Yadomae T. Antitumor activity and hematopoietic response of a b-glucan extracted from an edible and medicinal mushroom Sparassis crispa Wulf.: Fr.(Aphyllophoromycetideae) Int. J. Med. Mushrooms. 2002;4:13–26. doi: 10.1615/IntJMedMushr.v4.i3.40. [DOI] [Google Scholar]
- 4.Harada T, Kawaminami H, Miura NN, Adachi Y, Nakajima M, Yadomae T, Ohno N. Comparison of the Immunomodulating Activities of 1, 3-β-glucan Fractions from the Culinary-Medicinal Mushroom Sparassis crispa Wulf.: Fr.(Aphyllophoromycetideae) Int. J. Med. Mushrooms. 2006;8:231–244. doi: 10.1615/IntJMedMushr.v8.i3.50. [DOI] [Google Scholar]
- 5.Yamamoto K, Kimura T, Sugitachi A, Matsuura N. Anti-angiogenic and anti-metastatic effects of β-1, 3-D-glucan purified from Hanabiratake. Sparassis crispa. Biol. Pharm. Bull. 2009;32:259–263. doi: 10.1248/bpb.32.259. [DOI] [PubMed] [Google Scholar]
- 6.Chandrasekaran G, Kim GJ, Shin HJ. Purification and characterisation of an alkaliphilic esterase from a culinary medicinal mushroom. Sparassis crispa. Food Chem. 2011;124:1376–1381. doi: 10.1016/j.foodchem.2010.07.094. [DOI] [Google Scholar]
- 7.Seo SH, Park SE, Moon YS, Lee YM, Na CS, Son HS. Component analysis and immuno-stimulating activity of Sparassis crispa stipe. Korean J. Food Sci. Technol. 2016;48:515–520. doi: 10.9721/KJFST.2016.48.5.515. [DOI] [Google Scholar]
- 8.Kim EJ, Seo SH, Park SE, Kang MS, Son HS. Effects of fermented Sparassis crispa stipe extract supplemented diet on the immune responses of Philippines eel, Anguilla bicolor. J. Korean Soc. Food Sci. Nutr. 2017;46:1151–1157. [Google Scholar]
- 9.Hill D, Sugrue I, Arendt E, Hill C, Stanton C, Ross RP. Recent advances in microbial fermentation for dairy and health. F1000Res 6: 751 (2017). [DOI] [PMC free article] [PubMed]
- 10.Zhang Z, Lv G, Pan H, Fan L, Soccol CR, Pandey A. Production of powerful antioxidant supplements via solid-state fermentation of wheat (Triticum aestivum Linn.) by cordyceps militaris. Food Technol. Biotechnol. 2012;50:32–39. [Google Scholar]
- 11.Liu Y, Xie XX, Ibrahim SA, Khaskheli SG, Yang H, Wang YF, Huang W. Characterization of Lactobacillus pentosus as a starter culture for the fermentation of edible oyster mushrooms (Pleurotus spp.). LWT-Food. Sci. Technol. 2016;68:21–26. [Google Scholar]
- 12.Zhang D, Zhang Y, Liu B, Jiang Y, Zhou Q, Wang J, Wang H, Xie J, Kuang Q. Effect of replacing fish meal with fermented mushroom bran hydrolysate on the growth, digestive enzyme activity, and antioxidant capacity of allogynogenetic crucian carp (Carassius auratus gibelio) Turk. J. Fish. Aquat. Sci. 2017;17:1039–1048. doi: 10.4194/1303-2712-v17_5_20. [DOI] [Google Scholar]
- 13.Jabłońska-Ryś E, Sławińska A, Szwajgier D. Effect of lactic acid fermentation on antioxidant properties and phenolic acid contents of oyster (Pleurotus ostreatus) and chanterelle (Cantharellus cibarius) mushrooms. Food Sci. Biotechnol. 2016;25:439–444. doi: 10.1007/s10068-016-0060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang HS, Choi YJ, Oh HH, Moon JS, Jung HK, Kim KJ, Choi BS, Lee JW, Huh CK. Antioxidative activity of mushroom water extracts fermented by lactic acid bacteria. J. Korean Soc. Food Sci. Nutr. 2014;43:80–85. doi: 10.3746/jkfn.2014.43.1.080. [DOI] [Google Scholar]
- 15.Park SE, Seo SH, Moon YS, Lee YM, Na CS, Son HS. Antioxidant and immunological activities of Sparassis crispa fermented with Meyerozyma guilliermondii FM. J. Korean Soc. Food Sci. Nutr. 2016;45:1398–1405. doi: 10.3746/jkfn.2016.45.10.1398. [DOI] [Google Scholar]
- 16.More T, RoyChoudhury S, Gollapalli K, Patel SK, Gowda H, Chaudhury K, Rapole S. Metabolomics and its integration with systems biology: PSI 2014 conference panel discussion report. J. Proteomics. 2015;127:73–79. doi: 10.1016/j.jprot.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 17.Jonsson P, Gullberg J, Nordström A, Kusano M, Kowalczyk M, Sjöström M, Moritz T. A strategy for identifying differences in large series of metabolomic samples analyzed by GC/MS. Anal. Chem. 2004;76:1738–1745. doi: 10.1021/ac0352427. [DOI] [PubMed] [Google Scholar]
- 18.Khakimov B, Mongi RJ, Sørensen KM, Ndabikunze BK, Chove BE, Engelsen SB. A comprehensive and comparative GC–MS metabolomics study of non-volatiles in Tanzanian grown mango, pineapple, jackfruit, baobab and tamarind fruits. Food Chem. 2016;213:691–699. doi: 10.1016/j.foodchem.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Park SE, Yoo SA, Seo SH, Lee KI, Na CS, Son HS. GC–MS based metabolomics approach of Kimchi for the understanding of Lactobacillus plantarum fermentation characteristics. LWT-Food Sci. Technol. 2016;68:313–321. doi: 10.1016/j.lwt.2015.12.046. [DOI] [Google Scholar]
- 20.Jang GJ, Kim DW, Gu EJ, Song SH, Lee JI, Lee SB, Kim JH, Ham KS, Kim HJ. GC/MS-based metabolomic analysis of the radish water Kimchi, Dongchimi, with different salts. Food Sci. Biotechnol. 2015;24:1967–1972. doi: 10.1007/s10068-015-0259-9. [DOI] [Google Scholar]
- 21.Seo SH, Park SE, Yoo SA, Lee KI, Na CS, Son HS. Metabolite profiling of Makgeolli for the understanding of yeast fermentation characteristics during fermentation and aging. Process Biochem. 2016;51:1363–1373. doi: 10.1016/j.procbio.2016.08.005. [DOI] [Google Scholar]
- 22.Ponnusamy K, Lee S, Lee CH. Time-dependent correlation of the microbial community and the metabolomics of traditional barley nuruk starter fermentation. Biosci. Biotechnol. Biochem. 2013;77:683–690. doi: 10.1271/bbb.120665. [DOI] [PubMed] [Google Scholar]
- 23.Benkeblia N, Shinano T, Osaki M. Metabolite profiling and assessment of metabolome compartmentation of soybean leaves using non-aqueous fractionation and GC–MS analysis. Metabolomics. 2007;3:297–305. doi: 10.1007/s11306-007-0078-y. [DOI] [Google Scholar]
- 24.Park SE, Seo SH, Lee KI, Na CS, Son HS. Metabolite profiling of fermented ginseng extracts by gas chromatography mass spectrometry. J. Ginseng Res. 2018;42:57–67. doi: 10.1016/j.jgr.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avenoza A, Busto JH, Canal N, Peregrina JM. Time course of the evolution of malic and lactic acids in the alcoholic and malolactic fermentation of grape must by quantitative 1H NMR (qHNMR) spectroscopy. J. Agric. Food Chem. 2006;54:4715–4720. doi: 10.1021/jf060778p. [DOI] [PubMed] [Google Scholar]
- 26.Zelle RM, de Hulster E, van Winden WA, de Waard P, Dijkema C, Winkler AA, Geertman JM, van Dijken JP, Pronk JT, van Maris AJ. Malic acid production by Saccharomyces cerevisiae: engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export. Appl. Environ. Microbiol. 2008;74:2766–2777. doi: 10.1128/AEM.02591-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guillaume C, Delobel P, Sablayrolles JM, Blondin B. Molecular basis of fructose utilization by the wine yeast Saccharomyces cerevisiae: a mutated HXT3 allele enhances fructose fermentation. Appl. Environ. Microbiol. 2007;73:2432–2439. doi: 10.1128/AEM.02269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phan CW, Sabaratnam V. Potential uses of spent mushroom substrate and its associated lignocellulosic enzymes. Appl. Microbiol. Biotechnol. 2012;96:863–873. doi: 10.1007/s00253-012-4446-9. [DOI] [PubMed] [Google Scholar]
- 29.Burton K, Noble R. The influence of flush number, bruising and storage temperature on mushroom quality. Postharvest Biol. Technol. 1993;3:39–47. doi: 10.1016/0925-5214(93)90025-X. [DOI] [Google Scholar]
- 30.Kimura T. Natural products and biological activity of the pharmacologically active cauliflower mushroom Sparassis crispa. Biomed Res. Int. Article ID 982317 (2013) [DOI] [PMC free article] [PubMed]