Abstract
Sulforaphane (SFN), a natural compound derived from cruciferous vegetables, has been proved to possess potent anti-cancer activity. SMYD3 is a histone methyltransferase which is closely related to the proliferation and migration of cancer cells. This study showed that SFN could dose-dependently induce cell cycle arrest, stimulate apoptosis, and inhibit proliferation and migration of gastric carcinoma cells. Accompanied with these anti-cancer effects, SMYD3 and its downstream genes, myosin regulatory light chain 9, and cysteine-rich angiogenic inducer 61, was downregulated by SFN. Furthermore, overexpression of SMYD3 via transfection could abolish the effects of SFN, suggesting that SMYD3 might be an important mediator of SFN. To the best of our knowledge, this is the first report describing the role of SMYD3 in the anti-cancer of SFN. These findings might throw light on the development of novel anti-cancer drugs and functional food using SFN-rich cruciferous vegetables.
Keywords: Sulforaphane, SET and MYND domain containing 3, Cell cycle, Apoptosis, Migration
Introduction
Cancer is a group of diseases characterized by the growth of abnormal cells beyond their usual boundaries and the invasion to other parts of body. However, up to now, there are still many aspects of the mechanisms underlying cancer pathogenesis remain to be elucidated, and the study of diagnosis and treatment of cancer is still a long way to go.
Accumulating evidence showed that high consumption of cruciferous vegetables, such as broccoli, cabbage and cauliflower, could prevent the development of cancer cells, and sulforaphane (SFN), a dietary isothiocyanate, might be the most critical ingredient for the anti-cancer effects of cruciferous vegetables [1]. SFN is a product of glucoraphanin hydrolysis by myrosinase [2]. Previous studies showed that SFN could inhibit proliferation and migration of many types of cancer cells by modulating several cancer-related cell signaling, such as the reactive oxygen species-dependent pathway and ERK1/2 pathway [3–6].
Methylation of histone tails plays a pivotal role in the regulation of a wide range of biological processes. SET and MYND domain containing 3 (SMYD3) is an important histone methyltransferase which was discovered in 2004 [7]. SMYD3 could modulate the chromatin architecture via its methyltransferase activity, and then interact with RNA polymerase II and regulate the transcription of downstream genes by binding on the cis-acting element CCCTCC or GGAGGG in the promoter. Overexpression of SMYD3 has been confirmed in hepatocellular, colorectal, cervical and breast cancer [8–11]. In addition, our recent study showed that SMYD3 activation might also be an important characteristic in gastric cancer [12–14]. Furthermore, previous studies have shown that SMYD3 can specifically catalyze di/tri-methylation of H3K4 and activate the transcription of human telomerase reverse transcriptase (hTERT) and matrix metalloproteinase-9 (MMP-9) [7, 15–17], interestingly, SFN could also inhibit the expression and activity of hTERT and MMP-9 in cancer cells, and its effect was correlated with di-methylation of H3K4 [18–21]. In addition, several literatures have shown that SFN could induce cell cycle arrest and inhibit proliferation and migration of breast cancer cells, liver cancer cells and colon cancer cells [22–24], in which SMYD3 has been confirmed to be overexpressed and play essential roles [7, 8]. However, so far, the relationship between SMYD3 and SFN is absolutely unknown. To address these issues, in this study, the roles of SMYD3 in the anti-cancer effects of SFN on human gastric cancer cells were investigated using methods of MTT, quantitative real-time PCR, flow cytometry and wound scratch assay.
Materials and methods
Cell culture and transient transfection
Human gastric carcinoma cell line MGC803 and AGS were cultured in RPMI-1640 medium (Gibco) containing 10% fetal bovine serum (FBS, PAA), penicillin (100 U/mL) and streptomycin (100 U/mL) at 37 °C in humified air with 5% CO2. The plasmid pcDNA3.1-SMYD3 and siRNA targeted to SMYD3 (5′-CCCAGTATCTCTTTGCTCAATCAC-3′/5′-TTACGGGTGTTGAAGGT-3′) were transfected into cells with turbofect reagent (Thermo, USA) according to the manufacturer’s instructions. After the transfection, cells were treated with SFN (Yuanye, China) in different concentrations for 24–48 h.
Cell viability assay
Cell viability was determined by 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Solarbio). Briefly, cells were cultured in 96-well plates and treated with different concentrations of SFN for 24–48 h. Approximately 10 μL of the 5 mg/mL MTT solution was added to each well and the plates were then incubated again for 4 h at 37 °C. Subsequently, the media were removed, and 200 μL of dimethylsulfoxide (DMSO, Solarbio) was added to each well. Absorbance at 570 nm was detected using microplate reader (Synergy4, Biotek, USA), with 630 nm as a reference wavelength. All samples were tested in six replicates, and the experiments were repeated at least thrice.
Cell cycle analysis
Cell cycle were analyzed by flow cytometry. In brief, the cells were cultured in six-well plates and treated with different concentrations of SFN for 24 h. After the treatment, cells were collected, washed with cold PBS and fixed with 70% ethanol at − 20 °C overnight. Afterward, cells were incubated with 500 μL PBS containing RNAase (100 μg/mL), propidium iodide (PI) (50 μg/mL), 0.2% Trion X-100 for 30 min at 4 °C in the dark. Finally, the cells were analyzed by an Accuri C6 flow cytometer (Accuri, Ann Arbor, MI).
Cell apoptosis analysis
Cells were treated with different concentrations of SFN for 24 h. After the treatment, cells were collected, washed with cold PBS and then resuspended in the 200 μL 1 × Binding buffer. Afterward, cells were incubated with 5 μL Annexin V and 5 μL PI for 5 min at room temperature in the dark. Finally, the cells were analyzed by an Accuri C6 flow cytometer (Accuri, Ann Arbor, MI).
Quantitative real-time PCR analysis
Total RNA from cells was extracted using TRIzol and then reverse transcribed into cDNA. The mRNA level of SMYD1, SMYD2, SMYD3, myosin regulatory light chain 9 (MYL9), cysteine-rich angiogenic inducer 61 (CYR61) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were quantified by quantitative real-time RT-PCR analysis using specific primers. The sequences of primers were as follows (forward/reverse sequences): SMYD1 (5′-TTCCGCAGTGGTTTTTG-3′/5′-CGCCAGCCTGATGTTCT-3′), SMYD2 (5′-ATGGGTGTCTGCTTGTA-3′/5′-CTGCGGCTTTGTGTTCC-3′), SMYD3 (5′-CCCAGTATCTCTTTGCTCAATCAC-3′/5′-TTACGGGTGTTGAAGGT-3′), MYL9 (5′-CACCCACCAGAAGCCAAGAT-3′/5′-TGCCCTCCAGGTATTCGTCT-3′), CYR61 (5′-AGCAGCGTTTCCCTTCTAC-3′/5′-TGAGTCCCATCACCCACA-3′) and GAPDH (5′-ATTCAACGGCACAGTCAAGG-3′/5′-GCAGAAGGGGCGGAGATGA-3′). The conditions of PCR were as follows: 5 min at 95 °C followed by 30 cycles of incubation at 95 °C for 30 s, then 54–56 °C for 30 s, and 72 °C for 30 s. Finally 72 °C for 10 min. PCR products were electrophoretically separated in 2% agarose gels and the densities of the bands were analyzed with Quantity One software (v4.31; Bio Rad Laboratories, Hercules, CA, USA). The quantitative real-time RT-PCR was carried out in triplicate using the SYBR Green PCR Master Mix (Bio-Rad) on a StepOne™ Real-Time PCR Detection System (Applied Biosystems), according to the manufacturer’s instructions. Relative quantification was performed using the ddCT method. Data were shown as fold change after being normalized by GAPDH.
Western blot analysis
Cells were harvested and lysed in RadioImmunoprecipitation Assay (RIPA) buffer with protease inhibitors. Proteins were separated on 10% sodium dodecyl sulfate (SDS)—polyacrylamide gel and transferred to Polyvinylidene Fluoride (PVDF) membranes. After being incubate with appropriate primary antibodies against human SMYD3 (rabbit anti human monoclonal antibody; dilution, 1:1000; catalog NO. ab187149; Abcam, USA), CYR61 (rabbit anti-human Polyclonal antibody; dilution, 1:1000; catalog NO. ab24448; Bioss, Beijing, China), MYL9 (rabbit anti-human Polyclonal antibody; dilution, 1:2000; catalog NO. ab191393; Bioss, Beijing, China) and GAPDH (mouse anti human; dilution, 1:5000; catalog NO. UM4002; Utibody, Beijing, China) overnight at 4 °C, the membrane was incubated with IR DyeTM-800 conjugated anti-rabbit or anti-mouse secondary antibodies for 30 min at room temperature. Specific proteins were visualized by Odyssey™ Infrared Imaging System (Gene Company, Li-Cor, USA). The expression of GAPDH (Santa Cruz NO. sc-25778) was detected as an internal control to show equal loading of the protein samples.
Cell migration assay
Cell migration ability was determined by the wound scratch assay. The cells grown in a 6-well plate to confluence were scraped with a sterile 10 μL pipette tip to create a cell-free area and then washed 3 times with phosphate-buffered saline (PBS) to remove cell debris. Subsequently, cells were treated with different concentrations of SFN for 0, 24, 48 and 72 h. The scratches were examined with microscope.
Statistical analysis
All experimental data were presented as the mean ± standard deviations. Data were analyzed using Graphpad. prism.v5.0 software (GraphPad, San Diego, CA, USA). Comparisons among groups were performed with Student’s t test. Values of P < 0.05 were considered statistically significant.
Results and discussion
SFN dose-dependently inhibited proliferation and induced cell cycle arrest and apoptosis of gastric cells
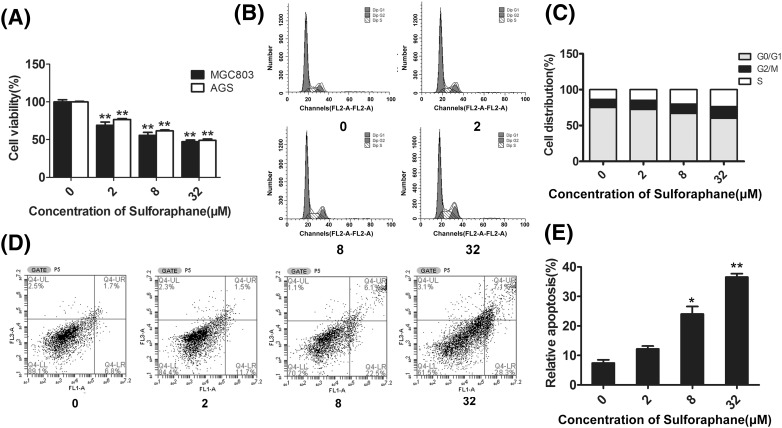
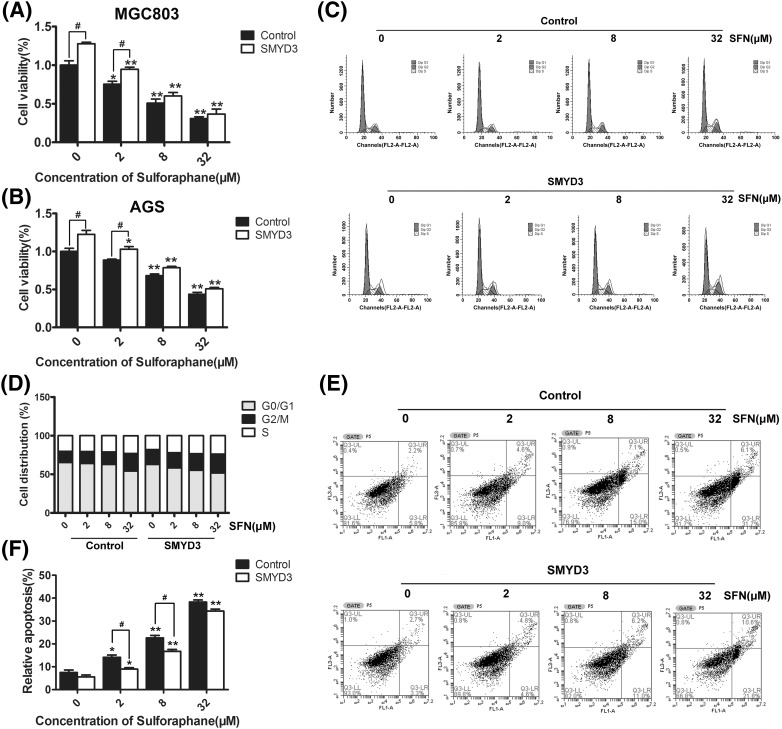
In order to detect the anti-tumor effect of SFN on gastric cells, firstly, MTT assay was performed. As shown in Fig. 1(A), SFN could significantly inhibit the proliferation of either MGC803 or AGS gastric cancer cells in a dose-dependent manner (P < 0.01). Furthermore, the effects of SFN on cell-cycle distribution and apoptosis were also examined using the flow cytometry methods. As shown in Fig. 1(B, C), with the increase of SFN dosage, the percentage of cells in G0/G1 phase was markedly reduced (32 μΜ, P < 0.05), while cells in S (8 μΜ, P < 0.05; 32 μΜ, P < 0.01) phase and G2/M (32 μΜ, P < 0.05) phase were accumulated, suggesting that SFN could halt gastric cancer cells at G2/M phase. Besides, Fig. 1(D, E) showed that SFN could dose-dependently increase the proportion of apoptosis cells. Kokotou and his colleague have reported that the content of sulforaphane varied between 72 ± 9 and 304 ± 2 mg per 100 g of fresh broccoli [25]. By calculation, about 0.37–1.58 mg broccoli could provide 32 μM SFN in our cell assay. Furthermore, previous study also showed that dietary broccoli could lessen development of fatty liver and liver cancer in mice [26]. Therefore, in our opinion, it might not be difficult for human to get expect chemopreventive effects by ingesting broccoli.
Fig. 1.
Effects of SFN on proliferation, cell-cycle and apoptosis of gastric cancer cells. (A) MGC803 or AGS cells were treated with 0, 2, 8 and 32 μM of SFN for 48 h, and the viability of cells was detected by MTT assay. In addition, the distribution of cell-cycle phases (B, C) and apoptosis of cells (D, E) being treated with 0, 2, 8, and 32 μM of SFN for 48 h were detected with flow cytometry analysis. *P < 0.05, **P < 0.01 compared with the corresponding group without SFN treatment (0 μM)
SFN suppressed SMYD3 via both transcriptional and post-transcriptional regulation
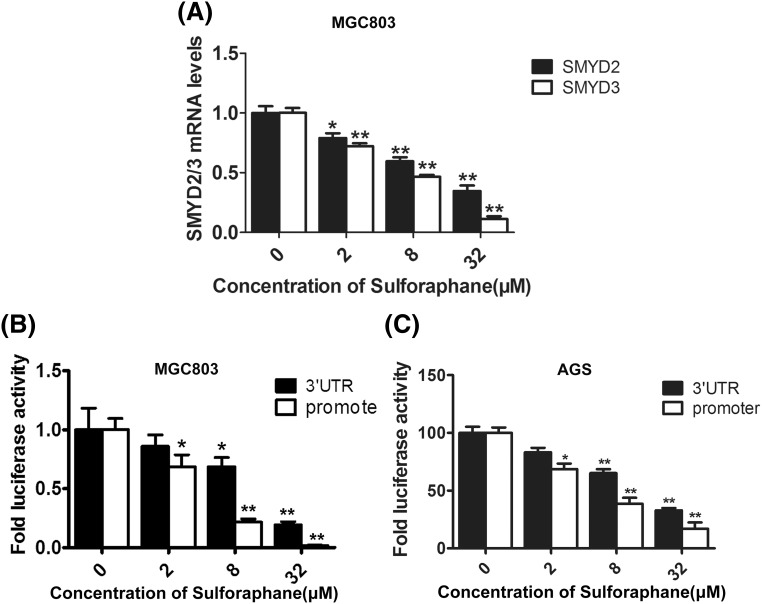
With the purpose of verifying the association between SFN and SMYD3, firstly, the quantitative real-time RT-PCR was performed to detect the effect of SFN on the transcriptional level of endogenous SMYD3 and another two important members of SMYD family, SMYD1 and SMYD2. The results showed that SFN significantly inhibited the mRNA level of SMYD3 and SMYD2 in a dose-dependent manner [Fig. 2(A)], whereas SMYD1 could not be detected in the gastric cancer cells (data not shown). These data were coincident with the previous reports that SMYD2 contributes to malignant outcome in gastric cancer, while SMYD1 is restrictedly expressed in skeletal muscle and heart tissues [27, 28]. Furthermore, as the suppressing effect of SFN on SMYD3 was higher than SMYD2, we then continued choosing SMYD3 but not SMYD2 in the further investigation.
Fig. 2.
SFN suppressed SMYD3 via both transcriptional and post-transcriptional regulation. (A) MGC803 cells were treated with 0, 2, 8 and 32 μM of SFN for 48 h, and then the mRNA level of SMYD2 and SMYD3 was detected by qRT-PCR analysis. GAPDH was used as an internal control. (B, C) The effects of SFN on the activity of 3′UTR (black columns) and promoter (white columns) of SMYD3 were analyzed by luciferase reporter assay in MGC803 and AGS cells. *P < 0.05, **P < 0.01 compared with the corresponding group without SFN treatment (0 μM)
What’s more, the results of luciferase reporter assay further showed that SFN could markedly inhibit the activity of both promoter and 3′UTR of SMYD3 in either MGC803 or AGS cells [Fig. 2(B, C)]. Previous study has reported that SMYD3 could be suppressed by miR-124 through binding 3′UTR, and miR-124 was also found to be an important downstream miRNAs affected by SFN [29, 30]. Taken together, these findings indicated that SFN might suppress SMYD3 via both transcriptional and post-transcriptional regulation.
CYR61 and MYL9, two downstream genes related to SMYD3, were also downregulated by SFN
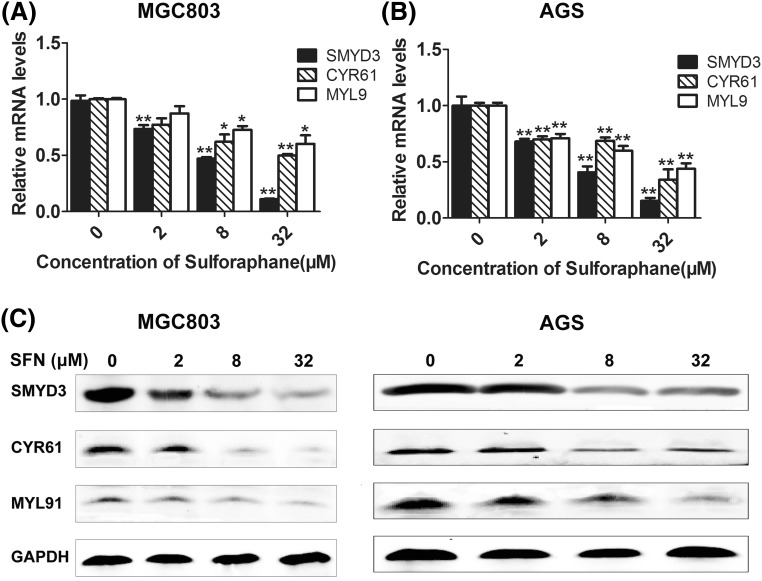
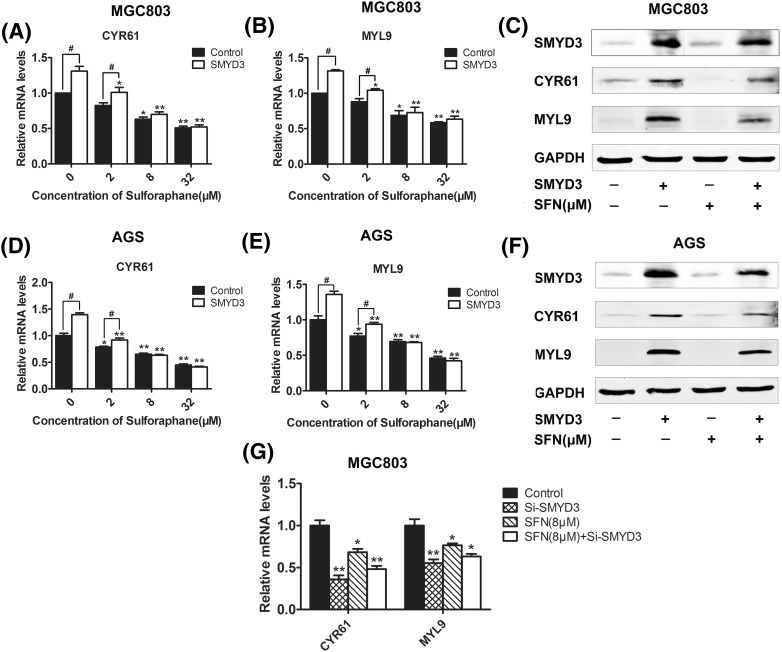
To further confirm the regulation effect of SFN on SMYD3, we also examined the expression level of CYR61 and MYL9, two downstream cancer-related genes regulated by the SMYD3-associated signal pathway [31–33]. CYR61 is a member of CCN family which is involved in diverse biological processes, such as regulation of cell adhesion, migration, proliferation, differentiation and survival [34, 35]. MYL9 is one of the four light chains of myosin, the important component of the cytoskeletal structure involved in cell movement, deformation process, and cell migration [36, 37]. Therefore, they have been regards as important markers of cancer. Here, the results of real-time RT-PCR and western blot showed that SFN dose-dependently inhibited the expression of both CYR61 and MYL9 in either MGC803 or AGS cells (Fig. 3). Furthermore, overexpression of SMYD3 could upregulate CYR61 and MYL9 and abolished the inhibitory effect of SNF on these two downstream genes [Fig. 4(A–F)]. Hung and his colleagues found that SFN could suppress RhoA signal pathway [38], while our previous study demonstrated that SMYD3 could promote MRTF-A, a critical mediator of RhoA signal pathway, and stimulate migration of breast cancer cells [31, 39]. Taken together, our present study provided the first evidence that SMYD3 and its downstream genes, CYR61 and MYL9, could be inhibited by SFN. However, interestingly, there seemed no difference in the expression of CYR61 and MYL9 between control and SMYD3-overexpressed groups when the cells were treated with high dose of SFN, and what’s more, when the endogenous expression of SMYD3 was knocked down, although both SFN (8 μM) and SMYD3-specific siRNAs could down-regulate CYR61 and MYL9, synergistic effects were not observed, these data implied that there might still be some other mediators also involved in the regulation of SFN on CYR61 and MYL9.
Fig. 3.
Effects of SFN on the transcriptional level of CYR61 and MYL9. MGC803 or AGS cells were treated with 0, 2, 8 and 32 μM of SFN for 48 h, and then the mRNA level of SMYD3, CYR61 and MYL9 was detected by qRT-PCR analysis (A, B). The protein level was further evaluated by western blot (C). GAPDH was used as an internal control. *P < 0.05, **P < 0.01 compared with the corresponding group without SFN treatment (0 μM)
Fig. 4.
Combining effects of SFN and SMYD3 on the regulation of CYR61 and MYL9. After being transfected with SMYD3 overexpression plasmid or its specific siRNAs, MGC803 or AGS cells were treated with 0, 2, 8 and 32 μM of SFN for 48 h, and then the mRNA level of MYL9 and CYR61 was detected by qRT-PCR analysis (A, B, D, E, G). The protein level was evaluated by western blot (C, F). GAPDH was used as an internal control. *P < 0.05, **P < 0.01 compared with the corresponding group without SFN treatment (0 μM). #P < 0.05 compared with the corresponding group transfected with empty plasmids (control)
Overexpression of SMYD3 repressed the anti-cancer effects of SFN
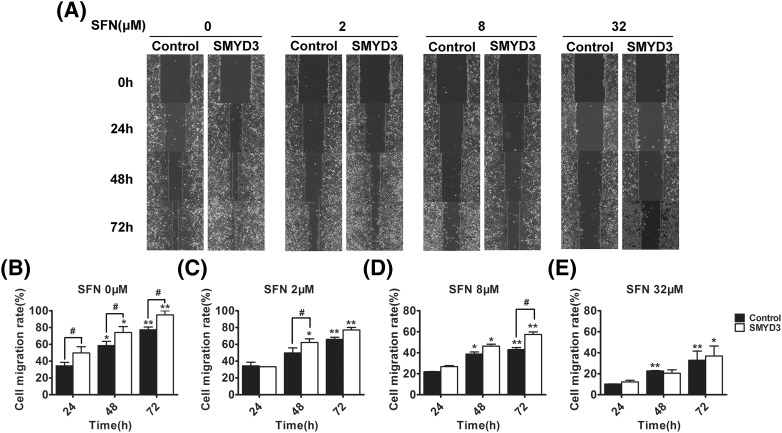
Finally, to elucidate the influence of SMYD3 on the anti-cancer efficiency of SFN, the gastric cancer cells were transfected with SMYD3-expressing plasmids, and then the effects of SFN on cell proliferation, cell cycle and apoptosis were detected again. As shown in Fig. 5, SFN could dose-dependently inhibit cell proliferation (groups of either 8 μM or 32 μM vs control group: P < 0.01; group of 2 μM in MGC803 cells vs control group: P < 0.05), induce cell cycle arrest (group of 32 μΜ vs control group: P < 0.05) and apoptosis (groups of either 8 μM or 32 μM vs control group: P < 0.01; group of 2 μM vs control group: P < 0.05), what’s more, although overexpression of SMYD3 did not influence the cell cycle arrest effect of SFN, it could repress the anti-proliferation and apoptosis-stimulating effects of SFN (especially at the dosage 2, 8, P < 0.05). In addition, since SMYD3, CYR61 and MYL9 play essential role in the migration of cells, the combined effects of SFN and SMYD3 on migration ability of gastric cancer cells were also investigated by the wound scratch assay. As showed in Fig. 6, the migration rate of SFN-treated cells was significantly lower than that of cells without SFN treatment, and overexpression of SMYD3 could partially repress this migration inhibitory effect of SFN.
Fig. 5.
Effects of SMYD3 on the proliferation-inhibitory, cell-cycle-arrest and apoptosis-stimulating effects of SFN. MGC803 or AGS cells were transfected with SMYD3 and then treated with different concentration of SFN for 48 h. The viability of cells was qualified by MTT assay (A, B), and the cell-cycle distribution (C, D) and apoptosis (E, F) were examined using the flow cytometry analysis. *P < 0.05, **P < 0.01 compared with the corresponding group without SFN treatment (0 μM). #P < 0.05 compared with the corresponding group transfected with empty plasmids (control)
Fig. 6.
Overexpression of SMYD3 repressed the inhibitory effect of SFN on cell migration ability. MGC803 gastric cancer cells were treated with SFN and/or transfected with SMYD3, and then scratch-wounded using sterile pipette tips (T = 0 h). The space between the wound fronts was photographed (A), measured and cell migration rate was calculated (B–E). *P < 0.05, **P < 0.01 compared with the corresponding group with SFN treatment for 24 h (0 μM). #P < 0.05 compared with the corresponding group transfected with empty plasmids (control)
In conclusion, SMYD3 might be an important mediator in the anti-cancer effects of SFN. To our best knowledge, this is the first report that SMYD3-associated pathway is implicated in the effects of SFN. These findings would help to better develop the novel chemotherapeutic agents derived from SFN and functional food using broccoli or other SFN-rich cruciferous vegetables.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Nos. 31470816; 31300642), the College Students’ Innovation and Entrepreneurship Training Program of Tianjin (No. 201510057057) and the Young Teachers’ Innovation Fund of Tianjin University of Science and Technology (No. 2016LG06).
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Qing-Qing Dong and Qiu-Tong Wang: Co-first authors.
References
- 1.Bayat Mokhtari R, Baluch N, Homayouni TS, Morgatskaya Z, Kumar S, Kazemi P, Yeger H. The role of Sulforaphane in cancer chemoprevention and health benefits: a mini-review. J Cell Commun Signal. Epub ahead of print (2017). [DOI] [PMC free article] [PubMed]
- 2.Houghton CA, Fassett RG, Coombes JS. Sulforaphane: translational research from laboratory bench to clinic. Nutr Rev. 2013;71:709–726. doi: 10.1111/nure.12060. [DOI] [PubMed] [Google Scholar]
- 3.Juengel E, Maxeiner S, Rutz J, Justin S, Roos F, Khoder W, Tsaur I, Nelson K, Bechstein WO, Haferkamp A, Blaheta RA. Sulforaphane inhibits proliferation and invasive activity of everolimus-resistant kidney cancer cells in vitro. Oncotarget. 2016;7:85208–85219. doi: 10.18632/oncotarget.13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, Tian Z, Yang Q, Li H, Guan H, Shi B, Hou P, Ji M. Sulforaphane inhibits thyroid cancer cell growth and invasiveness through the reactive oxygen species-dependent pathway. Oncotarget. 2015;6:25917–25931. doi: 10.18632/oncotarget.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mondal A, Biswas R, Rhee YH, Kim J, Ahn JC. Sulforaphene promotes Bax/Bcl2, MAPK-dependent human gastric cancer AGS cells apoptosis and inhibits migration via EGFR, p-ERK1/2 down-regulation. Gen Physiol Biophys. 2016;35:25–34. doi: 10.4149/gpb_2015033. [DOI] [PubMed] [Google Scholar]
- 6.Xu C, Shen G, Yuan X, Kim JH, Gopalkrishnan A, Keum YS, Nair S, Kong AN. ERK and JNK signaling pathways are involved in the regulation of activator protein 1 and cell death elicited by three isothiocyanates in human prostate cancer PC-3 cells. Carcinogenesis. 2006;27:437–445. doi: 10.1093/carcin/bgi251. [DOI] [PubMed] [Google Scholar]
- 7.Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 8.Hamamoto R, Silva FP, Tsuge M, Nishidate T, Katagiri T, Nakamura Y, Furukawa Y. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113–118. doi: 10.1111/j.1349-7006.2006.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang SZ, Luo XG, Shen J, Zou JN, Lu YH, Xi T. Knockdown of SMYD3 by RNA interference inhibits cervical carcinoma cell growth and invasion in vitro. BMB Rep. 2008;41:294–299. doi: 10.5483/BMBRep.2008.41.4.294. [DOI] [PubMed] [Google Scholar]
- 10.Medjkane S, Cock-Rada A, Weitzman JB. Role of the SMYD3 histone methyltransferase in tumorigenesis: local or global effects? Cell Cycle. 2012;11:1865. doi: 10.4161/cc.20415. [DOI] [PubMed] [Google Scholar]
- 11.Sarris ME, Moulos P, Haroniti A, Giakountis A, Talianidis I. Smyd3 Is a Transcriptional Potentiator of Multiple Cancer-Promoting Genes and Required for Liver and Colon Cancer Development. Cancer Cell. 2016;29:354–366. doi: 10.1016/j.ccell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Luo X, Deng J, Pan Y, Zhang L, Liang H. SMYD3 overexpression was a risk factor in the biological behavior and prognosis of gastric carcinoma. Tumour Biol. (2014). [DOI] [PubMed]
- 13.Liu Y, Deng J, Luo X, Pan Y, Zhang L, Zhang R, Liang H. Overexpression of SMYD3 was associated with increased STAT3 activation in gastric cancer. Med Oncol. 2015;32:404. doi: 10.1007/s12032-014-0404-y. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Liu H, Luo X, Deng J, Pan Y, Liang H. Overexpression of SMYD3 and matrix metalloproteinase-9 are associated with poor prognosis of patients with gastric cancer. Tumour Biol. 2015;36:4377–4386. doi: 10.1007/s13277-015-3077-z. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Wang QT, Liu YP, Dong QQ, Hu HJ, Miao Z, Li S, Liu Y, Zhou H, Zhang TC, Ma WJ, Luo XG. ATM Signaling Pathway Is Implicated in the SMYD3-mediated Proliferation and Migration of Gastric Cancer Cells. Journal of Gastric Cancer. 2017;17:e33. doi: 10.5230/jgc.2017.17.e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cock-Rada AM, Medjkane S, Janski N, Yousfi N, Perichon M, Chaussepied M, Chluba J, Langsley G, Weitzman JB. SMYD3 promotes cancer invasion by epigenetic upregulation of the metalloproteinase MMP-9. Cancer Res. 2012;72:810–820. doi: 10.1158/0008-5472.CAN-11-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu C, Fang X, Ge Z, Jalink M, Kyo S, Bjorkholm M, Gruber A, Sjoberg J, Xu D. The telomerase reverse transcriptase (hTERT) gene is a direct target of the histone methyltransferase SMYD3. Cancer Res. 2007;67:2626–2631. doi: 10.1158/0008-5472.CAN-06-4126. [DOI] [PubMed] [Google Scholar]
- 18.Rose P, Huang Q, Ong CN, Whiteman M. Broccoli and watercress suppress matrix metalloproteinase-9 activity and invasiveness of human MDA-MB-231 breast cancer cells. Toxicol Appl Pharmacol. 2005;209:105–113. doi: 10.1016/j.taap.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Mao L, Wang HD, Wang XL, Qiao L, Yin HX. Sulforaphane attenuates matrix metalloproteinase-9 expression following spinal cord injury in mice. Ann Clin Lab Sci. 2010;40:354–360. [PubMed] [Google Scholar]
- 20.Abbas A, Hall JA, Patterson WL, 3rd, Ho E, Hsu A, Al-Mulla F, Georgel PT. Sulforaphane modulates telomerase activity via epigenetic regulation in prostate cancer cell lines. Biochem Cell Biol. 2016;94:71–81. doi: 10.1139/bcb-2015-0038. [DOI] [PubMed] [Google Scholar]
- 21.Annabi B, Rojas-Sutterlin S, Laroche M, Lachambre MP, Moumdjian R, Beliveau R. The diet-derived sulforaphane inhibits matrix metalloproteinase-9-activated human brain microvascular endothelial cell migration and tubulogenesis. Mol Nutr Food Res. 2008;52:692–700. doi: 10.1002/mnfr.200700434. [DOI] [PubMed] [Google Scholar]
- 22.Lewinska A, Adamczyk-Grochala J, Deregowska A, Wnuk M. Sulforaphane-Induced Cell Cycle Arrest and Senescence are accompanied by DNA Hypomethylation and Changes in microRNA Profile in Breast Cancer Cells. Theranostics. 2017;7:3461–3477. doi: 10.7150/thno.20657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou X, Qu Z, Fang Y, Shi X, Ji Y. Endoplasmic reticulum stress mediates sulforaphane-induced apoptosis of HepG2 human hepatocellular carcinoma cells. Mol Med Rep. 2017;15:331–338. doi: 10.3892/mmr.2016.6016. [DOI] [PubMed] [Google Scholar]
- 24.Liu KC, Shih TY, Kuo CL, Ma YS, Yang JL, Wu PP, Huang YP, Lai KC, Chung JG. Sulforaphane Induces Cell Death Through G2/M Phase Arrest andTriggers Apoptosis in HCT 116 Human Colon Cancer Cells. Am J Chin Med. 2016;44(6):1289–1310. doi: 10.1142/S0192415X16500725. [DOI] [PubMed] [Google Scholar]
- 25.Kokotou MG, Revelou PK, Pappas C, Constantinou-Kokotou V. High resolution mass spectrometry studies of sulforaphane and indole-3-carbinol in broccoli. Food Chem. 2017;237:566–573. doi: 10.1016/j.foodchem.2017.05.139. [DOI] [PubMed] [Google Scholar]
- 26.Chen YJ, Wallig MA, Jeffery EH. Dietary Broccoli Lessens Development of Fatty Liver and Liver Cancer in Mice Given Diethylnitrosamine and Fed a Western or Control Diet. J Nutr. 2016;146:542–550. doi: 10.3945/jn.115.228148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komatsu S, Ichikawa D, Hirajima S, Nagata H, Nishimura Y, Kawaguchi T, Miyamae M, Okajima W, Ohashi T, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Tsuda H, Imoto I, Inazawa J, Otsuji E. Overexpression of SMYD2 contributes to malignant outcome in gastric cancer. Br J Cancer. 2015;112:357–364. doi: 10.1038/bjc.2014.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagandla H, Lopez S, Yu W. Defective myogenesis in the absence of the muscle-specific lysine methyltransferase SMYD1. Dev Biol. 2016;410:86–97. doi: 10.1016/j.ydbio.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Zeng B, Li Z, Chen R, Guo N, Zhou J, Zhou Q, Lin Q, Cheng D, Liao Q, Zheng L, Gong Y. Epigenetic regulation of miR-124 by hepatitis C virus core protein promotes migration and invasion of intrahepatic cholangiocarcinoma cells by targeting SMYD3. FEBS Lett. 2012;586:3271–3278. doi: 10.1016/j.febslet.2012.06.049. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Li Y, Dai Y, Liu Q, Ning S, Liu J, Shen Z, Zhu D, Jiang F, Zhang J, Li Z. Sulforaphane improves chemotherapy efficacy by targeting cancer stem cell-like properties via the miR-124/IL-6R/STAT3 axis. Sci Rep. 2016;6:36796. doi: 10.1038/srep36796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo XG, Zhang CL, Zhao WW, Liu ZP, Liu L, Mu A, Guo S, Wang N, Zhou H, Zhang TC. Histone methyltransferase SMYD3 promotes MRTF-A-mediated transactivation of MYL9 and migration of MCF-7 breast cancer cells. Cancer Lett. 2014;344:129–137. doi: 10.1016/j.canlet.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 32.Zhang C, Luo X, Liu L, Guo S, Zhao W, Mu A, Liu Z, Wang N, Zhou H, Zhang T. Myocardin-related transcription factor A is up-regulated by 17beta-estradiol and promotes migration of MCF-7 breast cancer cells via transactivation of MYL9 and CYR61. Acta Biochim Biophys Sin (Shanghai). 2013;45:921–927. doi: 10.1093/abbs/gmt104. [DOI] [PubMed] [Google Scholar]
- 33.Gilles L, Bluteau D, Boukour S, Chang Y, Zhang Y, Robert T, Dessen P, Debili N, Bernard OA, Vainchenker W, Raslova H. MAL/SRF complex is involved in platelet formation and megakaryocyte migration by regulating MYL9 (MLC2) and MMP9. Blood. 2009;114:4221–4232. doi: 10.1182/blood-2009-03-209932. [DOI] [PubMed] [Google Scholar]
- 34.Wei J, Yu G, Shao G, Sun A, Chen M, Yang W, Lin Q. CYR61 (CCN1) is a metastatic biomarker of gastric cardia adenocarcinoma. Oncotarget. 2016;7:31067–33178. doi: 10.18632/oncotarget.8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeger H, Perbal B. CCN family of proteins: critical modulators of the tumor cell microenvironment. J Cell Commun Signal. 2016;10:229–240. doi: 10.1007/s12079-016-0346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang YQ, Han ZD, Liang YX, Lin ZY, Ling XH, Fu X, Cai C, Bi XC, Dai QS, Chen JH, He HC, Chen YR, Jiang FN, Zhong WD. Decreased expression of myosin light chain MYL9 in stroma predicts malignant progression and poor biochemical recurrence-free survival in prostate cancer. Med Oncol. 2014;31:820. doi: 10.1007/s12032-013-0820-4. [DOI] [PubMed] [Google Scholar]
- 37.Park I, Han C, Jin S, Lee B, Choi H, Kwon JT, Kim D, Kim J, Lifirsu E, Park WJ, Park ZY, Kim DH, Cho C. Myosin regulatory light chains are required to maintain the stability of myosin II and cellular integrity. Biochem J. 2011;434:171–180. doi: 10.1042/BJ20101473. [DOI] [PubMed] [Google Scholar]
- 38.Hung CN, Huang HP, Wang CJ, Liu KL, Lii CK. Sulforaphane inhibits TNF-alpha-induced adhesion molecule expression through the Rho A/ROCK/NF-kappaB signaling pathway. J Med Food. 2014;17:1095–1102. doi: 10.1089/jmf.2013.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol. 2009;11:257–268. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]