Abstract
Background
The identification of blood-based biomarkers specific to the diagnosis of amyotrophic lateral sclerosis (ALS) is an active field of academic and clinical research. While inheritance studies have advanced the field, a majority of patients do not have a known genetic link to the disease, making direct sequence-based genetic testing for ALS difficult. The ability to detect biofluid-based epigenetic changes in ALS would expand the relevance of using genomic information for disease diagnosis.
Methods
Assessing differences in chromosomal conformations (i.e. how they are positioned in 3-dimensions) represents one approach for assessing epigenetic changes. In this study, we used an industrial platform, EpiSwitch™, to compare the genomic architecture of healthy and diseased patient samples (blood and tissue) to discover a chromosomal conformation signature (CCS) with diagnostic potential in ALS. A three-step biomarker selection process yielded a distinct CCS for ALS, comprised of conformation changes in eight genomic loci and detectable in blood.
Findings
We applied the ALS CCS to determine a diagnosis for 74 unblinded patient samples and subsequently conducted a blinded diagnostic study of 16 samples. Sensitivity and specificity for ALS detection in the 74 unblinded patient samples were 83∙33% (CI 51∙59 to 97∙91%) and 76∙92% (46∙19 to 94∙96%), respectively. In the blinded cohort, sensitivity reached 87∙50% (CI 47∙35 to 99∙68%) and specificity was 75∙0% (34∙91 to 96∙81%).
Interpretations
The sensitivity and specificity values achieved using the ALS CCS identified and validated in this study provide an indication that the detection of chromosome conformation signatures is a promising approach to disease diagnosis and can potentially augment current strategies for diagnosing ALS.
Fund
This research was funded by Oxford BioDynamics and Innovate UK. Work in the Oxford MND Care and Research Centre is supported by grants from the Motor Neurone Disease Association and the Medical Research Council. Additional support was provided by the Northeast ALS Consortium (NEALS).
Research in context.
Evidence Before this Study
We searched primary research studies done in human ALS subjects in the last ten years and referenced in PubMed by use of the MeSH terms “(amyotrophic lateral sclerosis OR ALS) AND (biomarker OR marker) AND (diagnostic OR diagnosis) AND (blood OR PBMC OR plasma OR serum) AND (sensitivity OR specificity OR AUC)”. There were no language restrictions. We identified 16 studies where diagnostic biomarkers for ALS were determined in a non-invasive, clinically accessible biological sample and statistical power was reported. In these studies, eight used plasma as a biomarker source, five used serum and three studies used PBMCs or whole blood. The biomarkers ranged from mRNA, proteins/peptides, metabolites, metals, or combinations of several molecular modalities combined into a multi-marker panel. Neurofilament light (NfL) was the most extensively studied, and the sensitivity and specificity of the biomarkers in making a diagnosis of ALS ranged between 90 and 93% and 58–91%, respectively. Two studies showed no significant diagnostic power with the biomarkers under investigation.
Added Value of this Study
The current diagnosis of ALS remains mainly based on clinical parameters. However, the development of molecular tests can be used to exclude other diseases and help to confirm the diagnosis. Currently, there is a shortage of these molecular tests available to physicians. We were able to develop a highly discriminatory chromosome conformation signature that could accurately identify patients with ALS in an independent patient cohort. The test can be performed within a day on standard laboratory equipment, uses a readily accessible biofluid (blood) and requires and requires a minimal amount of volume.
Implications of all the Available Evidence
Our findings indicate that a simple and rapid blood-based test can distinguish patients with ALS from healthy controls with a high degree of sensitivity and can serve as a complementary tool in helping aid in disease diagnosis. These preliminary findings provide an initial proof of concept that the approach of looking at changes in genomic architecture provide a reliable readout of physiological changes associated with neurological disease. Additional studies that independently validate the biomarker panel identified here and assess the ability of the panel to discriminate ALS from other motor neuron diseases are required before application in pre-clinical and clinical settings.
1. Introduction
ALS is a fatal, degenerative neurologic disorder characterized by progressive muscle weakness and eventual paralysis. Disease presentation and rate of progression vary from patient-to-patient and while there are clinical tools to monitor disease progression after diagnosis (e.g. ALSFRS-R, FVC measures), no definitive clinically validated measure currently exists to diagnose the disease. Combined with the insidious nature of symptom onset, lack of recognition of symptoms and signs in non-specialists, and the need to perform multiple investigations to rule out conditions which can mimic ALS, lack of a diagnostic marker significantly contributes to diagnostic delay, which averages 1 year from symptom onset [1]. Given the rapidly progressing nature of ALS, this delay can have significant clinical and lifestyle impact on patients and limits recruitment of patients with early phase disease to clinical trials. With the potential for the approval of new therapies currently in different stages of clinical development over the coming years, there is a pressing need for a validated biomarker for the diagnosis of ALS.
While advances in genomic sequencing have provided new insights into the disease [2], gene sequencing is primarily used to assess familial risk and define biological subtypes in the 10% or so of patients carrying mutations once the clinical diagnosis of ALS has been established, and therefore does not in itself provide a way of improving early diagnosis [3]. Another approach that has been used to aid in diagnosis of other neurological disorders is the analysis of non-sequence based alterations in the genome (epigenetics) [4, 5]. This is particularly useful in understanding the pathogenesis of multifactorial neurodegenerative diseases like ALS, where epigenetics can expose an integrated view of cellular function and dysfunction via regulation of gene expression [6].
The exploration of patient-derived samples is an obvious choice in attempting to discover new tools for diagnosing ALS, as biofluid collection is increasingly being done in clinical settings to serve as a data source for potential biomarkers. In light of this, the Northeast ALS Consortium (NEALS) Biofluid Repository was established to provide researches and industry partners with biologic samples collected from the patients using standard operating procedures (SOPs) and linked to clinical information for use in research studies [7]. In this pilot study, samples from the NEALS Biofluid Repository were analyzed using EpiSwitch, a high-resolution technology to identify structural-functional epigenetic changes in genomic architecture associated with pathological phenotypes developed by Oxford BioDynamics [[8], [9], [10], [11], [12], [13]]. As multiple genomic regions contribute to phenotypic differences through changes in genomic architecture, this approach allows for the development of a chromosomal conformation signature (CCS) of alterations in genomic architecture between two states (disease vs. non-diseased, pre-treatment vs. post-treatment).
With the aim of identifying novel approaches to the diagnosis of ALS, this study was undertaken to discover and validate a CCS applicable to the diagnosis of ALS. After defining and refining a biomarker panel, we applied the signature to an independent sample cohort for validation. Last, the biological relevance in ALS of the top performing markers was assessed.
2. Methods
High-throughput screening of blood samples and a multi-step selection process was used to define a panel of epigenetic changes that could distinguish ALS patients from healthy patients.
2.1. Sample Collection
All samples banked and collected were done under IRB approved protocols (Oxford University samples: NRES Committee South West - Cornwall & Plymouth, reference 15/SW/0224). For the initial biomarker screening and development, the NEALS Biofluid Repository provided 50 ALS (Table 1) and 5 healthy control whole blood samples (Table 2). Oxford BioDynamics sample collection provided a further 37 controls (Table 2). In the validation stages of the project, Oxford University provided an independent sample cohort of eight ALS and eight spousal healthy control samples (Supplementary Table S1). Clinical characteristics for ALS patients were similar between the Discovery and Validation cohorts (Table 3). Whole blood samples were collected by peripheral venipucture using a 22-gauge large bore needle. Blood was collected directly into EDTA filled BD Vacutainer™ The tubes were immediately mixed by inversion and placed into a -80o C freezer. Samples were transported on dry ice and the frozen condition inspected on delivery.
Table 1.
NEALS Biofluid Repository samples used in this study.
| Study | Sample ID | Basic Annotation | Sample Type | Gender | Ethnic Category | Race | Age at Diagnosis | Disease Duration (Month from Symptom Onset to Sample Collection) | ALSFRS Total Score | ALS Type |
|---|---|---|---|---|---|---|---|---|---|---|
| Skin biopsy | 701001 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 67 | 34.20 | 38 | Sporadic |
| 701002 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 42 | 22.51 | 24 | Sporadic | |
| 701024 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 67 | 15.9 | 42 | Sporadic | |
| 701026 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 35 | 31.11 | 41 | Sporadic | |
| 701028 | ALS | Whole blood | Male | Non-Hispanic or Latino | Black | 48 | 14.06 | 43 | Sporadic | |
| 701032 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 57 | 9.66 | 40 | Familial | |
| 701035 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 63 | 39.26 | 29 | Sporadic | |
| 701037 | ALS | Whole Blood | Female | Non-Hispanic or Latino | White | 49 | 15.28 | 40 | Sporadic | |
| 701040 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 39 | 19.52 | 43 | Sporadic | |
| 701043 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 50 | 7.92 | 29 | Sporadic | |
| 701054 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 44 | 16.99 | 39 | Sporadic | |
| 701060 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 46 | 3.02 | 42 | Sporadic | |
| 701080 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 66 | 12.65 | 36 | Sporadic | |
| 701021 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 64 | 13.96 | 17 | Sporadic | |
| ALS Sample repository | 701001 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 57 | 13.57 | N/A | Sporadic |
| 701026 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 47 | 0 | N/A | Sporadic | |
| 701027 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 55 | 0.4 | N/A | Familial | |
| 701038 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 73 | 0.92 | N/A | Sporadic | |
| 701042 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 56 | 8.76 | N/A | Sporadic | |
| 701043 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 59 | 10.61 | N/A | Sporadic | |
| 701044 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 58 | 1.5* | N/A | Sporadic | |
| 701045 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 47 | 5.9 | N/A | Sporadic | |
| 701048 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 62 | 14 | N/A | Sporadic | |
| 701049 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 47 | 11 | N/A | Sporadic | |
| 701051 | ALS | Whole Blood | Male | Non-Hispanic or Latino | White | 65 | 26.23 | N/A | Sporadic | |
| 701052 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 46 | 63.26 | N/A | Sporadic | |
| 701055 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 55 | 2.1 | N/A | Sporadic | |
| 701056 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 53 | 0.43* | N/A | Sporadic | |
| 701064 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 67 | 0.74* | N/A | Sporadic | |
| 701066 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 41 | 9 | N/A | Sporadic | |
| 701069 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 51 | 0.48 | N/A | Familial | |
| 701082 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 52 | 11.36* | N/A | Sporadic | |
| 701083 | ALS | Whole blood | Male | Hispanic, Latino | White | 55 | 50.26* | N/A | Sporadic | |
| 701084 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 51 | 1.17 | N/A | Sporadic | |
| 701085 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 47 | 38.67* | N/A | Familial | |
| 701101 | ALS | Whole blood | Male | Non-Hispanic or Latino | Black | 43 | 43.73* | N/A | Sporadic | |
| 701107 | ALS | Whole blood | Female | Non-Hispanic or Latino | Unknown/Not reported | 65 | 2.83 | N/A | Familial | |
| 701108 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 70 | 0 | N/A | Familiar | |
| 701116 | ALS | Whole blood | Male | Unknown/Not reported | Unknown/Not reported | 68 | 3.89* | N/A | Sporadic | |
| 701122 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 58 | 0.45 | N/A | Sporadic | |
| 701132 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 61 | 2.86* | N/A | Sporadic | |
| 701136 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 53 | 18.86 | N/A | Sporadic | |
| 701139 | ALS | Whole blood | Female | Non-Hispanic or Latino | White | 47 | 7.03 | N/A | Familial | |
| 701142 | ALS | Whole blood | Male | Unknown/Not reported | Unknown/Not reported | 31 | 0 | N/A | Sporadic | |
| 701148 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 44 | 3.26 | N/A | Sporadic | |
| 701154 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | N/A | N/A | N/A | Disease Control | |
| 701158 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 52 | 29.48 | N/A | Sporadic | |
| 701161 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 55 | 8.5 | N/A | Sporadic | |
| 701163 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 46 | 4.54* | N/A | Sporadic | |
| 701185 | ALS | Whole blood | Male | Non-Hispanic or Latino | White | 43 | 108–120* | N/A | Sporadic | |
| 701151 | Healthy control | Whole blood | Female | Non-Hispanic or Latino | White | N/A | N/A | N/A | Healthy control | |
| 701143 | Healthy control | Whole blood | Male | Non-Hispanic or Latino | White | N/A | N/A | N/A | Healthy control | |
| 701174 | Healthy control | Whole blood | Female | Non-Hispanic or Latino | White | N/A | N/A | N/A | Healthy control | |
| 701172 | Healthy control | Whole blood | Male | Non-Hispanic or Latino | White | N/A | N/A | N/A | Healthy control | |
| 701141 | Healthy control | Whole blood | Female | Non-Hispanic or Latino | Asian | N/A | N/A | N/A | Healthy control |
Table 2.
Healthy control blood samples provided by Oxford BioDynamics and the NEALS Consortium used in this study.
| Sample type | Participant no | Date sample collected | Baseline diagnosis |
|---|---|---|---|
| Healthy control | 10088 | 11/13/2013 | NFG |
| 10917 | 9/20/2013 | NFG | |
| 14376 | 9/11/2013 | NFG | |
| 16345 | 12/2/2013 | NFG | |
| 16391 | 11/13/2013 | NFG | |
| 16771 | 11/18/2013 | NFG | |
| 17139 | 9/20/2013 | NFG | |
| 17152 | 12/6/2013 | NFG | |
| 17238 | 10/29/2013 | NFG | |
| 17265 | 10/24/2013 | NFG | |
| 17280 | 11/8/2013 | NFG | |
| 17328 | 11/15/2013 | NFG | |
| 17410 | 12/19/2013 | NFG | |
| 17411 | 1/17/2014 | NFG | |
| 17414 | 1/2/2014 | NFG | |
| 17415 | 1/10/2014 | NFG | |
| 17416 | 2/18/2014 | NFG | |
| 17417 | 2/4/2014 | NFG | |
| 17419 | 1/20/2014 | NFG | |
| 17422 | 1/6/2014 | NFG | |
| 17426 | 12/19/2013 | NFG | |
| 17427 | 1/15/2014 | NFG | |
| 17432 | 2/4/2014 | NFG | |
| 17433 | 1/10/2014 | NFG | |
| 17434 | 1/2/2014 | NFG | |
| 17445 | 1/2/2014 | NFG | |
| 17446 | 1/10/2014 | NFG | |
| 17447 | 1/10/2014 | NFG | |
| 17448 | 1/17/2014 | NFG | |
| 17449 | 1/27/2014 | NFG | |
| 17451 | 1/29/2014 | NFG | |
| 17452 | 2/4/2014 | NFG | |
| 17454 | 1/16/2014 | NFG | |
| 17456 | 1/21/2014 | NFG | |
| 17457 | 1/31/2014 | NFG | |
| 17459 | 2/10/2014 | NFG | |
| 17467 | 2/10/2014 | NFG | |
| 17480 | 2/11/2014 | NFG | |
| 17495 | 2/17/2014 | NFG | |
| 17508 | 2/19/2014 | NFG | |
| 17669 | 2/24/2014 | NFG | |
| 701141 | 12/5/2014 | NEALS respository control | |
| 701143 | 12/5/2014 | NEALS respository control | |
| 701151 | 12/5/2014 | NEALS respository control | |
| 701172 | 12/5/2014 | NEALS respository control | |
| 701174 | 12/5/2014 | NEALS respository control |
Abbreviations. NFG: Normal Fasting Glucose.
Table 3.
ALS clinical characteristics of the Discovery and Validation cohorts.
| Discovery cohort (N = 50) | Validation cohort (N = 8) | |
|---|---|---|
| Gender | ||
| Male (N, (%)) | 32 (64) | 5 (63) |
| Female (N, (%)) | 18 (36) | 3 (37) |
| Ethnicity | ||
| Non-Hispanic or Latino | 47 | N/A |
| Hispanic, Latino | 1 | N/A |
| Unknown/not reported | 2 | N/A |
| Race | ||
| White (N, (%)) | 45 (90) | N/A |
| Black (N, (%)) | 2 (4) | N/A |
| Unknown/not reported (N, (%)) | 3 (6) | N/A |
| ALS type | ||
| Sporadic (N, (%)) | 42 (84) | 6 (75) |
| Familial (N, (%)) | 8 (16) | 2 (25) |
| Age at diagnosis (Average, (SD)) | 53.4 (9.7) | 54.9 (11.9) |
| Disease duration (Average, (SD)) | 15.6 (20.4) | 11.0 (7.9) |
| ALSFRS-R (Average, (SD)) | 35.9 (8.1)⁎ | 36.4 (7.7) |
N/A = Not Available.
ALSFRS-R scores were available for 14 of the 50 patients in the Discovery cohort.
2.2. Application of EpiSwitch and the Stepwise Biomarker Discovery Process
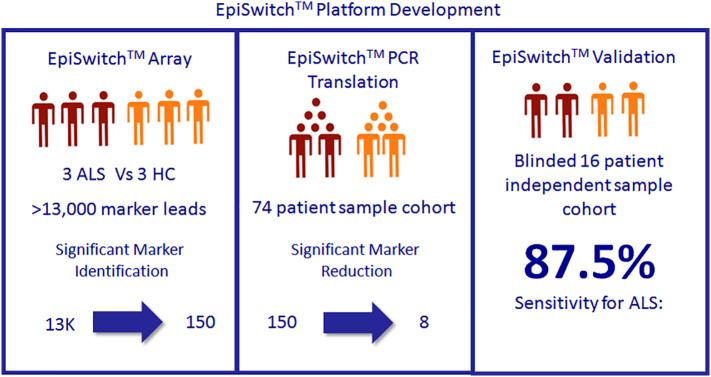
EpiSwitch is a high throughput technology platform that pairs high resolution chromosome conformational capture results with regression analysis and machine learning to develop disease classifications [8]. Screening and selection of statistically significant differences in conditional and stable profiles of genome architecture associated with samples from patients suffering from a disease, in comparison to healthy control samples, serves as a way to select epigenetic biomarkers that can diagnose and stratify pathological conditions [[8], [9], [10], [11], [12], [13]]. In this study, EpiSwitch was used on blood samples in a three-step process to identify, evaluate, and validate statistically-significant differences in chromosomal conformations between ALS patients and healthy controls (Fig. 1).
Fig. 1.
Three-step biomarker discovery workflow. Starting with an initial pool of over 13,000 markers, a series of statistical comparisons between ALS and healthy controls samples refined the final ALS chromosome conformation signature panel into a set of 8 markers that could diagnose ALS patients in a blinded, independent cohort with 87.5% sensitivity.
An initial customized CGH Agilent microarray (8x60k) was designed to test technical and biological repeats for 13,880 potential chromosome conformations across 308 genetic loci. With the focus on the development of a non-invasive blood based diagnostic test, we first concentrated our attention on potential chromosome conformations in genomic loci specific to ALS's immuno-footprint. A literature search was conducted to identify loci that would be used on the microarray. In an earlier study, we compared and reported unique and common aspects of the ALS immuno-footprint to other Autoimmune Conditions such as Systemic Lupus Erythematosus (SLE), Ulcerative Colitis (UC), Rheumatoid Arthritis (RA), Multiple Sclerosis (MS) and Relapse-Remitting MS (MSRR) [14]. The genetic loci selected for the array were genes primarily involved in immunodeficiency, adaptive and innate immune systems, and cytokine signaling (Table 4).
Table 4.
List of immune-related genomic loci tested in the initial ALS array.
| Gene name | Array probe count | Gene name | Array probe count | Gene name | Array probe count |
|---|---|---|---|---|---|
| VCAM1 | 10 | IGHV3-23 | 13 | CD40 | 7 |
| RAP1A | 332 | IGHV1-46 | 2 | CXADR | 8 |
| NRAS | 12 | TRAV19 | 5 | IFNAR2 | 24 |
| CD160 | 24 | TRAC | 12 | IFNAR1 | 4 |
| FCGR1A | 23 | ADCY4 | 10 | IFNGR2 | 90 |
| RFX5 | 24 | LGALS3 | 7 | DONSON | 5 |
| THEM4 | 88 | RASGRP1 | 9 | ICOSLG | 12 |
| IL6R | 138 | B2M | 30 | ICOSLG;AIRE | 3 |
| FCER1A | 2 | CIITA | 4 | ITGB2 | 14 |
| FCER1G | 24 | PRKCB | 415 | VPREB1 | 1 |
| FCGR2A | 19 | PDPK1 | 51 | IGLV7-43 | 8 |
| FCGR3A;FCGR2A | 4 | IL21R | 56 | IGLC3;IGLC7;IGLC2;IGLC1;IGLC6 | 2 |
| FCGR3A | 42 | CD19 | 10 | BCR | 100 |
| FCGR2B;FCGR3A | 125 | LAT | 22 | IGLL1 | 10 |
| CD247 | 254 | ITGAL | 23 | SEC14L3 | 12 |
| SELL | 52 | ITGAM | 54 | CSF2RB | 15 |
| DNM3 | 1121 | ADCY9 | 132 | IL2RB | 4 |
| PTPRC | 154 | CDH1 | 154 | MKL1 | 183 |
| CHI3L1 | 38 | PLCG2 | 221 | TNFRSF13C | 4 |
| C4BPB | 9 | GINS2 | 27 | IRAK2 | 38 |
| C4BPB;C4BPA | 10 | TNFRSF13B | 7 | CBLB | 51 |
| C4BPA | 35 | CPD | 61 | CD96 | 159 |
| CD55 | 33 | AP2B1 | 44 | CD200 | 26 |
| CR2 | 15 | ERBB2 | 6 | CD200R1 | 9 |
| CR1;CR2 | 6 | PLD2 | 10 | CD80 | 21 |
| CR1 | 205 | C1QBP | 36 | CD86 | 11 |
| CD46 | 11 | MRC2 | 51 | ADCY5 | 153 |
| CD34 | 70 | CD300LB | 5 | ITGB5 | 63 |
| DDOST | 11 | CD300E | 10 | NCK1 | 95 |
| TLR5 | 53 | GRB2 | 270 | TRPC1 | 21 |
| LYPLA2 | 30 | GUCY2D | 13 | PLD1 | 201 |
| AKT3 | 437 | SIGLEC15 | 2 | AP2M1 | 13 |
| CLIC4 | 479 | MALT1 | 32 | IL1RAP | 29 |
| IFI6 | 116 | CD226 | 15 | IL5RA | 11 |
| PTAFR | 49 | ICAM1 | 9 | MYD88 | 8 |
| ATPIF1;PTAFR | 14 | ICAM1;ICAM4 | 1 | CCR2 | 2 |
| ATPIF1 | 24 | DNM2 | 235 | DAPP1 | 26 |
| LCK | 158 | PRKCSH | 31 | NFKB1 | 27 |
| BCL10 | 18 | ACP5 | 23 | TLR2 | 5 |
| PIK3CD | 191 | JAK3 | 21 | CD38 | 24 |
| CHUK | 15 | RFXANK | 33 | TLR10 | 2 |
| NFKB2 | 5 | HCST | 3 | TLR1;TLR10 | 3 |
| PPAPDC1A | 19 | TYROBP;HCST | 5 | TLR1 | 4 |
| FGFR2 | 95 | TYROBP | 12 | TLR1;TLR6 | 3 |
| MRC1 | 23 | MATK | 27 | KLB | 27 |
| ITGB1 | 29 | AKT2 | 34 | PDGFRA | 69 |
| IL2RA | 7 | AKT2;PLD3 | 6 | KIT | 32 |
| PRKCQ | 164 | PLD3 | 23 | TICAM2 | 12 |
| FAS | 16 | CD79A | 6 | CD14 | 55 |
| BLNK | 35 | MADCAM1 | 3 | PDGFRB | 18 |
| PIK3AP1 | 210 | PVR | 8 | CD74 | 6 |
| AP2A2 | 20 | PVRL2 | 43 | FGFR4 | 28 |
| NCAM1 | 43 | PTGIR | 42 | PRLR | 41 |
| AMICA1 | 16 | AP2S1 | 10 | IL7R | 14 |
| CD3E | 13 | AP2A1 | 22 | GHR | 114 |
| CD3G | 3 | SIGLEC16 | 10 | CCNO | 7 |
| CD3D | 2 | SIGLEC14 | 3 | IL6ST | 24 |
| CD3G;CD3D | 1 | LILRB2 | 2 | CD180 | 1 |
| CXCR5 | 13 | LILRB1 | 10 | PIK3R1 | 14 |
| CBL | 41 | LILRB4 | 13 | ADCY2 | 364 |
| CRTAM | 11 | KIR2DL4;KIR3DL1;KIR2DL3 | 13 | FYN | 332 |
| TIRAP | 2 | KIR2DL4;KIR3DL1 | 11 | IFNGR1 | 15 |
| CD81 | 3 | KIR3DL1 | 4 | TAB2 | 150 |
| IFITM1;IFITM2 | 4 | KIR2DL1;KIR2DL4;KIR3DL1;KIR2DL3 | 5 | ULBP1 | 14 |
| IFITM3 | 6 | KIR2DS4;KIR3DL1 | 1 | ULBP3 | 34 |
| CD59 | 7 | KIR2DS4;KIR3DL1;KIR3DL2 | 2 | CCR6 | 11 |
| CD44 | 24 | C3 | 28 | MICB | 28 |
| ART1 | 1 | VAV1;C3 | 3 | CFB | 2 |
| RAG1;TRAF6 | 8 | VAV1 | 49 | C4A | 3 |
| RAG1 | 46 | IL1R2 | 5 | C4B | 1 |
| RAG2;RAG1 | 10 | IL1R1 | 56 | AGER | 7 |
| RAG2 | 8 | IL1RN | 9 | TAP2;TAP1 | 3 |
| SIGIRR | 1 | RAPGEF4 | 195 | TAP1 | 4 |
| HRAS | 14 | ITGA4 | 1 | TREM2 | 5 |
| MS4A2 | 12 | ITGAV | 1 | TREM1 | 9 |
| SCYL1 | 9 | CD28 | 2 | NCR2 | 5 |
| PANX1 | 29 | ICOS | 10 | LY86 | 54 |
| KLRD1 | 53 | IRS1 | 6 | MAP3K7 | 2 |
| KLRK1 | 3 | PDCD1 | 2 | EPHB4 | 30 |
| UNG | 18 | ADCY3 | 30 | CLEC5A | 7 |
| P2RX7 | 53 | RASGRP3 | 41 | CARD11 | 50 |
| ORAI1 | 22 | SOS1 | 177 | ADCY1 | 30 |
| KRAS | 5 | ACTR2 | 46 | EGFR | 209 |
| RAPGEF3 | 15 | CD8A | 9 | LAT2 | 9 |
| ADCY6 | 7 | CD8A;CD8B | 14 | CD36 | 127 |
| ITGB7 | 6 | CD8B | 22 | ADCY8 | 121 |
| GLYCAM1 | 6 | IGKV1-5 | 10 | PPAPDC1B | 26 |
| ERBB3 | 2 | IGKV3-7 | 7 | FGFR1 | 7 |
| CD4 | 57 | IGKV3-11 | 3 | IKBKB | 18 |
| RAP1B | 33 | IGKV3-15 | 10 | LYN | 179 |
| FRS2 | 63 | IGKV3-20 | 3 | PAG1 | 73 |
| AICDA | 3 | IGKV2-30 | 8 | TLR4 | 10 |
| KLRG1 | 15 | IGKV1D-16 | 8 | ANGPTL2 | 4 |
| IRS2 | 1 | MAL | 3 | DNM1 | 11 |
| ARHGEF7 | 94 | ADAM17 | 17 | SH3GL2 | 101 |
| KL | 23 | ZAP70 | 10 | CLTA | 110 |
| RFXAP | 26 | SIRPB1 | 4 | CD274 | 1 |
| DLEU2 | 20 | GINS1 | 39 | PDCD1LG2 | 1 |
| AKT1 | 18 | TRIB3 | 21 | SYK | 25 |
| CD40LG | 3 | PLCG1 | 12 | BTK | 8 |
| IL3RA;CSF2RA | 3 | ADA | 9 | CSF2RA | 25 |
| IL3RA | 15 | IKBKG | 2 | TAB3 | 35 |
| IRAK1 | 5 | SH3KBP1 | 291 | Total | 13,880 |
A comparative microarray analysis was conducted using samples from individual ALS patients from the NEALS Biofluid Repository and pooled healthy control samples. Array readouts were analyzed with linear regression modeling to select the 153 chromosomal interactions with the ability to best discriminate ALS from controls (Table 5).
Table 5.
Top 153 markers produced from the second array.
| Probe | GeneLocus | logFC | AveExpr | t | P·Value | adj.P·Val | B | FC | FC_1 | Binary |
|---|---|---|---|---|---|---|---|---|---|---|
| ACOXL_2_110997162_111004405_111109898_111117325_RF | ACOXL | 0.3298921 | 0.3298921 | 11.076483 | 1.09E-09 | 1.62E-07 | 12.487822 | 1.256919 | 1.2569193 | 1 |
| ACOXL_2_110704616_110714102_110875631_110879536_RF | ACOXL | 0.3570591 | 0.3570591 | 9.638804 | 1.03E-08 | 7.06E-07 | 10.263069 | 1.280812 | 1.2808123 | 1 |
| ADAMTS20_12_43377477_43383257_43480754_43482402_FR | ADAMTS20 | −0.246307 | −0.246307 | −9.267993 | 1.91E-08 | 1.10E-06 | 9.6485602 | 0.843052 | −1.186167 | −1 |
| ADAMTS20_12_43377477_43383257_43588948_43590670_FR | ADAMTS20 | −0.322419 | −0.322419 | −6.16799 | 6.55E-06 | 7.78E-05 | 3.7882954 | 0.799728 | −1.250425 | −1 |
| ALDH1A2_15_58325151_58334051_58485549_58488054_FR | ALDH1A2 | 0.345862 | 0.345862 | 13.477242 | 4.03E-11 | 2.85E-08 | 15.710641 | 1.27091 | 1.2709101 | 1 |
| ALDH1A2_15_58325151_58334051_58452555_58457099_FR | ALDH1A2 | 0.3633487 | 0.3633487 | 13.188649 | 5.84E-11 | 3.35E-08 | 15.352222 | 1.286408 | 1.2864084 | 1 |
| ALDH1A2_15_58325151_58334051_58538695_58540885_FF | ALDH1A2 | 0.3398615 | 0.3398615 | 7.222828 | 7.78E-07 | 1.61E-05 | 5.9304456 | 1.265635 | 1.2656351 | 1 |
| ANKRD29_18_23664553_23665914_23692491_23699757_RR | ANKRD29 | 0.2493527 | 0.2493527 | 8.6194055 | 5.83E-08 | 2.37E-06 | 8.5308864 | 1.188674 | 1.1886737 | 1 |
| ATXN7L1_7_105654123_105657510_105741521_105750599_FR | ATXN7L1 | 0.18956 | 0.18956 | 6.5975399 | 2.70E-06 | 3.98E-05 | 4.678643 | 1.140416 | 1.1404158 | 1 |
| BANK1_4_101510635_101519668_101619789_101624381_RF | BANK1 | −0.390246 | −0.390246 | −10.63806 | 2.11E-09 | 2.34E-07 | 11.83492 | 0.763 | −1.310616 | −1 |
| BANK1_4_101476784_101489895_101732279_101733831_FF | BANK1 | 0.3134993 | 0.3134993 | 10.813911 | 1.62E-09 | 2.03E-07 | 12.099391 | 1.242718 | 1.2427183 | 1 |
| BTBD11_12_107272037_107279968_107305640_107312061_FF | BTBD11 | 0.3569279 | 0.3569279 | 13.088383 | 6.65E-11 | 3.57E-08 | 15.225963 | 1.280696 | 1.2806959 | 1 |
| C2CD2_21_41868909_41872387_41979620_41986582_FR | C2CD2 | 0.3304577 | 0.3304577 | 9.7178208 | 9.05E-09 | 6.41E-07 | 10.391775 | 1.257412 | 1.2574123 | 1 |
| C6orf132_6_42104999_42110975_42124471_42126639_RR | C6orf132 | −0.151906 | −0.151906 | −10.15532 | 4.48E-09 | 3.96E-07 | 11.090489 | 0.900061 | −1.111036 | −1 |
| C6orf58_6_127480771_127483471_127600017_127604343_FR | C6orf58 | −0.206941 | −0.206941 | −7.65767 | 3.38E-07 | 8.58E-06 | 6.7690901 | 0.866372 | −1.154238 | −1 |
| CAMK1D_10_12516951_12526338_12633776_12637357_FF | CAMK1D | −0.363678 | −0.363678 | −8.185719 | 1.27E-07 | 4.23E-06 | 7.7521485 | 0.777181 | −1.286702 | −1 |
| CAPN9_1_230738572_230739927_230752057_230757333_RR | CAPN9 | −0.224589 | −0.224589 | −8.238209 | 1.15E-07 | 3.91E-06 | 7.8477638 | 0.855839 | −1.168444 | −1 |
| CCDC3_10_12924962_12934165_13046815_13049059_FF | CCDC3 | 0.2758308 | 0.2758308 | 9.9013068 | 6.72E-09 | 5.34E-07 | 10.687663 | 1.210691 | 1.2106911 | 1 |
| CD1A_1_158226961_158229550_158243613_158252050_FR | CD1A | 0.179006 | 0.179006 | 4.472859 | 0.000266 | 0.001399 | 0.0795021 | 1.132104 | 1.1321036 | 1 |
| CDH12_5_21870983_21872848_21898498_21914941_RF | CDH12 | 0.3146402 | 0.3146402 | 9.742694 | 8.69E-09 | 6.31E-07 | 10.432129 | 1.243701 | 1.2437014 | 1 |
| CDK14_7_90936378_90943000_91140859_91158297_FF | CDK14 | 0.3051704 | 0.3051704 | 5.623921 | 2.08E-05 | 0.000189 | 2.6276939 | 1.235565 | 1.2355646 | 1 |
| CDK14_7_90855907_90868783_90891759_90898655_FF | CDK14 | 0.3654475 | 0.3654475 | 7.6163383 | 3.65E-07 | 9.09E-06 | 6.6905099 | 1.288281 | 1.2882812 | 1 |
| CELSR1_22_46424151_46427901_46457142_46458718_FF | CELSR1 | 0.1775237 | 0.1775237 | 7.5038883 | 4.52E-07 | 1.07E-05 | 6.4755148 | 1.130941 | 1.130941 | 1 |
| CHAMP1_13_114265206_114269925_114317775_114320752_RR | CHAMP1 | −0.230721 | −0.230721 | −5.754561 | 1.57E-05 | 0.000152 | 2.9095095 | 0.852209 | −1.173421 | −1 |
| CHSY3_5_129929941_129937973_130119673_130124677_RF | CHSY3 | 0.2485594 | 0.2485594 | 8.4314793 | 8.14E-08 | 3.02E-06 | 8.1965796 | 1.18802 | 1.1880203 | 1 |
| CHSY3_5_130104704_130115924_130186143_130190278_RR | CHSY3 | 0.4537319 | 0.4537319 | 14.076616 | 1.91E-11 | 1.93E-08 | 16.432119 | 1.369578 | 1.3695785 | 1 |
| CNTN4_3_2273300_2281776_2314685_2325027_FR | CNTN4 | 0.3355172 | 0.3355172 | 15.964666 | 2.12E-12 | 1.04E-08 | 18.519694 | 1.26183 | 1.2618297 | 1 |
| CNTNAP2_7_146728706_146734820_146785878_146792823_RF | CNTNAP2 | −0.440614 | −0.440614 | −7.429603 | 5.21E-07 | 1.18E-05 | 6.332521 | 0.736821 | −1.357182 | −1 |
| CTNNA3_10_66299269_66302507_66496211_66513003_FF | CTNNA3 | −0.436585 | −0.436585 | −14.22858 | 1.58E-11 | 1.70E-08 | 16.610284 | 0.738882 | −1.353396 | −1 |
| CTNNA3_10_66496211_66513003_66783614_66787250_RR | CTNNA3 | −0.309997 | −0.309997 | −8.536173 | 6.76E-08 | 2.64E-06 | 8.3834114 | 0.806643 | −1.239705 | −1 |
| CTNNA3_10_66282876_66290806_66496211_66513003_RR | CTNNA3 | −0.309827 | −0.309827 | −5.790351 | 1.45E-05 | 0.000143 | 2.986383 | 0.806739 | −1.239559 | −1 |
| CTNND2_5_11851889_11854697_11917286_11928978_FR | CTNND2 | 0.2692826 | 0.2692826 | 13.156667 | 6.09E-11 | 3.43E-08 | 15.312047 | 1.205208 | 1.2052083 | 1 |
| DAO_12_108832598_108835352_108845485_108846981_FF | DAO | 0.2606268 | 0.2606268 | 7.576059 | 3.94E-07 | 9.68E-06 | 6.6137018 | 1.197999 | 1.197999 | 1 |
| DBF4B_17_44683672_44686134_44709459_44712660_RR | DBF4B | −0.269502 | −0.269502 | −8.674587 | 5.29E-08 | 2.23E-06 | 8.6281464 | 0.829606 | −1.205391 | −1 |
| DGKB_7_14197847_14209024_14254130_14267710_FR | DGKB | −0.43638 | −0.43638 | −9.791092 | 8.03E-09 | 5.98E-07 | 10.51043 | 0.738986 | −1.353205 | −1 |
| DGKB_7_14197847_14209024_14322087_14328928_FF | DGKB | −0.306194 | −0.306194 | −7.866591 | 2.28E-07 | 6.51E-06 | 7.162654 | 0.808772 | −1.236442 | −1 |
| DIO2_14_80195869_80197807_80255209_80263592_RR | DIO2 | −0.25104 | −0.25104 | −8.332113 | 9.73E-08 | 3.40E-06 | 8.0178794 | 0.84029 | −1.190065 | −1 |
| DIO2_14_80255209_80263592_80371554_80373780_RR | DIO2 | −0.307653 | −0.307653 | −13.98334 | 2.14E-11 | 1.99E-08 | 16.321825 | 0.807955 | −1.237693 | −1 |
| DPP10_2_115459376_115465174_115685306_115694818_FR | DPP10 | −0.327317 | −0.327317 | −8.420239 | 8.31E-08 | 3.06E-06 | 8.1764331 | 0.797017 | −1.254678 | −1 |
| DPP10_2_114901417_114910472_115087678_115094206_FF | DPP10 | 0.3377111 | 0.3377111 | 14.406874 | 1.28E-11 | 1.51E-08 | 16.816949 | 1.26375 | 1.26375 | 1 |
| DSCR4_21_37943561_37949090_38013989_38021392_FF | DSCR4 | −0.429276 | −0.429276 | −12.81368 | 9.54E-11 | 4.43E-08 | 14.875361 | 0.742634 | −1.346558 | −1 |
| ERBB4_2_212097934_212104780_212317329_212325591_RF | ERBB4 | −0.315689 | −0.315689 | −4.50592 | 0.000247 | 0.001323 | 0.1537846 | 0.803467 | −1.244606 | −1 |
| ERC1_12_1043264_1050801_1096484_1101318_FR | ERC1 | 0.233095 | 0.233095 | 13.553981 | 3.66E-11 | 2.79E-08 | 15.804721 | 1.175354 | 1.1753537 | 1 |
| FAM126A_7_22873341_22878517_22945935_22949410_RF | FAM126A | −0.177613 | −0.177613 | −4.427145 | 0.000295 | 0.001522 | −0.023251 | 0.884165 | −1.131011 | −1 |
| FARP1_13_98271575_98282700_98346930_98348486_FR | FARP1 | 0.2548222 | 0.2548222 | 14.739287 | 8.58E-12 | 1.48E-08 | 17.195539 | 1.193189 | 1.1931887 | 1 |
| FBXO8_4_174254227_174258882_174284851_174288977_RR | FBXO8 | −0.339902 | −0.339902 | −6.993433 | 1.22E-06 | 2.24E-05 | 5.4774396 | 0.790095 | −1.265671 | −1 |
| FER1L6_8_123963222_123969450_124085753_124093275_FR | FER1L6 | 0.350543 | 0.350543 | 5.6694435 | 1.88E-05 | 0.000175 | 2.726108 | 1.27504 | 1.2750404 | 1 |
| FHIT_3_61064178_61073078_61136784_61147623_RR | FHIT | 0.3193953 | 0.3193953 | 4.8524668 | 0.000113 | 0.000713 | 0.9299331 | 1.247807 | 1.2478074 | 1 |
| FRMD3_9_83388882_83396653_83414350_83418756_FR | FRMD3 | −0.383857 | −0.383857 | −11.50548 | 5.83E-10 | 1.14E-07 | 13.106219 | 0.766386 | −1.304826 | −1 |
| GALNTL6_4_172641518_172647332_172893415_172910203_RF | GALNTL6 | −0.310946 | −0.310946 | −14.25721 | 1.53E-11 | 1.70E-08 | 16.643649 | 0.806113 | −1.240521 | −1 |
| GFPT1_2_69307499_69311057_69383954_69393165_FR | GFPT1 | −0.317965 | −0.317965 | −6.644846 | 2.45E-06 | 3.70E-05 | 4.7752111 | 0.802201 | −1.246571 | −1 |
| GFRA1_10_116092148_116100672_116113263_116118675_FR | GFRA1 | −0.319379 | −0.319379 | −7.079082 | 1.03E-06 | 1.99E-05 | 5.6474303 | 0.801415 | −1.247793 | −1 |
| GLIS3_9_3998831_4010284_4144132_4146272_FF | GLIS3 | −0.309431 | −0.309431 | −6.997876 | 1.21E-06 | 2.23E-05 | 5.4862817 | 0.80696 | −1.239219 | −1 |
| GMDS_6_2030214_2038438_2217079_2225905_RF | GMDS | −0.412681 | −0.412681 | −9.027894 | 2.87E-08 | 1.47E-06 | 9.2412584 | 0.751226 | −1.331157 | −1 |
| GPC6_13_93573654_93584189_93748106_93754722_RF | GPC6 | 0.316254 | 0.316254 | 11.287633 | 8.00E-10 | 1.32E-07 | 12.794673 | 1.245093 | 1.2450934 | 1 |
| GRIK2_6_101912977_101914543_101980061_101996881_FF | GRIK2 | −0.412403 | −0.412403 | −10.64034 | 2.11E-09 | 2.34E-07 | 11.838361 | 0.751371 | −1.330901 | −1 |
| GRIP1_12_66761739_66764959_66791577_66801892_RR | GRIP1 | −0.333991 | −0.333991 | −10.33187 | 3.39E-09 | 3.23E-07 | 11.365922 | 0.793339 | −1.260496 | −1 |
| GRM3_7_86653882_86655924_86695387_86706974_RF | GRM3 | −0.311021 | −0.311021 | −9.568226 | 1.16E-08 | 7.66E-07 | 10.147448 | 0.806071 | −1.240585 | −1 |
| GRM7_3_6827121_6836161_7047731_7057814_RR | GRM7 | −0.359722 | −0.359722 | −7.091136 | 1.01E-06 | 1.96E-05 | 5.6712713 | 0.779315 | −1.283179 | −1 |
| HCFC2P1_13_108440737_108444674_108461634_108471282_RR | HCFC2P1 | 0.3137758 | 0.3137758 | 11.555559 | 5.42E-10 | 1.09E-07 | 13.177127 | 1.242957 | 1.2429565 | 1 |
| HCFC2P1_13_108440737_108444674_108461634_108471282_FR | HCFC2P1 | 0.329425 | 0.329425 | 11.346554 | 7.34E-10 | 1.27E-07 | 12.879437 | 1.256513 | 1.2565125 | 1 |
| HDAC4_2_239204078_239210374_239360246_239368872_FR | HDAC4 | 0.3383322 | 0.3383322 | 7.5918458 | 3.83E-07 | 9.44E-06 | 6.6438323 | 1.264294 | 1.2642941 | 1 |
| HPGD_4_174493630_174502964_174542132_174543999_RF | HPGD | 0.1857242 | 0.1857242 | 11.142003 | 9.91E-10 | 1.53E-07 | 12.583558 | 1.137388 | 1.1373878 | 1 |
| HUS1_7_47823192_47830325_47842954_47848362_RF | HUS1 | −0.174947 | −0.174947 | −4.911921 | 9.92E-05 | 0.000643 | 1.0624681 | 0.8858 | −1.128923 | −1 |
| IL1A_2_112795355_112798834_112816387_112823836_RR | IL1A | −0.103722 | −0.103722 | −3.037051 | 0.006829 | 0.019309 | −3.100991 | 0.930629 | −1.074542 | −1 |
| IL1A_2_112765786_112772711_112810765_112813086_RR | IL1A | 0.1305595 | 0.1305595 | 3.6789915 | 0.001613 | 0.006001 | −1.701299 | 1.094718 | 1.0947181 | 1 |
| INSR_19_7099584_7101451_7138185_7142897_RF | INSR | 0.1639687 | 0.1639687 | 10.303077 | 3.55E-09 | 3.32E-07 | 11.321255 | 1.120365 | 1.1203649 | 1 |
| IQGAP2_5_76698020_76702533_76717099_76725306_FF | IQGAP2 | −0.310676 | −0.310676 | −10.6174 | 2.18E-09 | 2.39E-07 | 11.803605 | 0.806264 | −1.240289 | −1 |
| IQGAP2_5_76475531_76481400_76717099_76725306_FR | IQGAP2 | 0.2854775 | 0.2854775 | 11.70784 | 4.36E-10 | 9.61E-08 | 13.391129 | 1.218814 | 1.2188136 | 1 |
| KCNMA1_10_77411418_77416164_77530837_77543678_RF | KCNMA1 | 0.3281284 | 0.3281284 | 8.0962784 | 1.49E-07 | 4.77E-06 | 7.5883505 | 1.255384 | 1.2553837 | 1 |
| KCNN2_5_114353357_114360896_114430044_114433040_RR | KCNN2 | −0.232264 | −0.232264 | −7.774872 | 2.71E-07 | 7.34E-06 | 6.9906211 | 0.851298 | −1.174677 | −1 |
| KCNS3_2_17835106_17846049_17968712_17974054_FR | KCNS3 | −0.288729 | −0.288729 | −11.37806 | 7.01E-10 | 1.24E-07 | 12.924607 | 0.818623 | −1.221564 | −1 |
| KIAA0513_16_85000072_85002703_85074194_85077477_RF | KIAA0513 | 0.2060683 | 0.2060683 | 5.4679401 | 2.91E-05 | 0.000245 | 2.2888169 | 1.15354 | 1.1535402 | 1 |
| KIFAP3_1_169911687_169919606_169986992_169993331_FR | KIFAP3 | 0.2934999 | 0.2934999 | 11.698018 | 4.42E-10 | 9.62E-08 | 13.377399 | 1.22561 | 1.22561 | 1 |
| LINGO2_9_28333777_28339631_28527863_28540481_FR | LINGO2 | −0.374728 | −0.374728 | −9.390644 | 1.55E-08 | 9.38E-07 | 9.8537519 | 0.771251 | −1.296595 | −1 |
| LINGO2_9_28314155_28333777_28522753_28525112_FR | LINGO2 | −0.463013 | −0.463013 | −7.06033 | 1.07E-06 | 2.04E-05 | 5.6102993 | 0.72547 | −1.378417 | −1 |
| LINGO2_9_28314155_28333777_28510932_28517950_FF | LINGO2 | −0.352913 | −0.352913 | −2.842326 | 0.010477 | 0.027372 | −3.509391 | 0.783001 | −1.277137 | −1 |
| LYPD3_19_43451257_43453307_43494229_43495321_FR | LYPD3 | −0.108357 | −0.108357 | −3.699682 | 0.001539 | 0.005775 | −1.655267 | 0.927644 | −1.078 | −1 |
| MACROD2_20_15750414_15763248_15953272_15965024_RF | MACROD2 | −0.439686 | −0.439686 | −10.55166 | 2.41E-09 | 2.56E-07 | 11.703674 | 0.737295 | −1.356309 | −1 |
| MAGI2_7_78371356_78378740_78502868_78511891_FR | MAGI2 | −0.309137 | −0.309137 | −7.781863 | 2.67E-07 | 7.28E-06 | 7.003776 | 0.807124 | −1.238966 | −1 |
| MAGI2_7_79009346_79018304_79275810_79284623_RF | MAGI2 | 0.349446 | 0.349446 | 10.577281 | 2.32E-09 | 2.48E-07 | 11.742684 | 1.274071 | 1.2740713 | 1 |
| MDGA2_14_47087312_47092151_47301555_47314511_FR | MDGA2 | −0.376564 | −0.376564 | −2.53523 | 0.020267 | 0.046875 | −4.13055 | 0.77027 | −1.298246 | −1 |
| MGLL_3_127731924_127736779_127863699_127870283_RF | MGLL | −0.174384 | −0.174384 | −4.247058 | 0.000443 | 0.002114 | −0.428303 | 0.886146 | −1.128482 | −1 |
| NAV2_11_19476194_19490077_19749138_19755031_FR | NAV2 | 0.3411694 | 0.3411694 | 9.3822413 | 1.57E-08 | 9.44E-07 | 9.839756 | 1.266783 | 1.266783 | 1 |
| NCKAP5_2_133209962_133217496_133394726_133400754_FF | NCKAP5 | −0.363475 | −0.363475 | −8.42102 | 8.30E-08 | 3.06E-06 | 8.1778322 | 0.77729 | −1.286521 | −1 |
| NCKAP5_2_133084614_133091683_133242680_133249277_RR | NCKAP5 | 0.3800747 | 0.3800747 | 11.195242 | 9.16E-10 | 1.46E-07 | 12.661003 | 1.301409 | 1.3014092 | 1 |
| NEFH_22_29442588_29445314_29482081_29484217_RR | NEFH | 0.1558858 | 0.1558858 | 12.070176 | 2.62E-10 | 7.50E-08 | 13.89074 | 1.114105 | 1.1141054 | 1 |
| NEFH_22_29467180_29469328_29482081_29484217_FR | NEFH | 0.3600069 | 0.3600069 | 5.6898359 | 1.80E-05 | 0.000169 | 2.7701205 | 1.283432 | 1.283432 | 1 |
| NEFH_22_29434926_29438399_29467180_29469328_RF | NEFH | 0.3191557 | 0.3191557 | 5.4943817 | 2.75E-05 | 0.000235 | 2.3464398 | 1.2476 | 1.2476002 | 1 |
| NEIL3_4_177308770_177311798_177363195_177365833_RR | NEIL3 | −0.440388 | −0.440388 | −4.173407 | 0.000524 | 0.00242 | −0.593968 | 0.736936 | −1.356969 | −1 |
| NELL1_11_21419712_21428422_21531472_21542215_RR | NELL1 | −0.312849 | −0.312849 | −9.035358 | 2.83E-08 | 1.46E-06 | 9.2540329 | 0.805051 | −1.242158 | −1 |
| NFIA_1_60869920_60875614_60989414_60996659_RR | NFIA | 0.3492172 | 0.3492172 | 14.019512 | 2.04E-11 | 1.98E-08 | 16.364679 | 1.273869 | 1.2738693 | 1 |
| NFIA_1_61228266_61235258_61246134_61250763_RF | NFIA | 0.3172619 | 0.3172619 | 12.061499 | 2.65E-10 | 7.53E-08 | 13.87893 | 1.245964 | 1.2459636 | 1 |
| NHSL1_6_138480977_138485998_138514855_138523394_RF | NHSL1 | 0.2467506 | 0.2467506 | 7.5585632 | 4.08E-07 | 9.94E-06 | 6.5802688 | 1.186532 | 1.1865317 | 1 |
| NOX4_11_89337353_89346448_89499668_89503122_RF | NOX4 | −0.321336 | −0.321336 | −12.36707 | 1.74E-10 | 6.04E-08 | 14.290321 | 0.800328 | −1.249487 | −1 |
| NXPH1_7_8466580_8469680_8501926_8510453_FR | NXPH1 | −0.343359 | −0.343359 | −9.44196 | 1.42E-08 | 8.81E-07 | 9.9390318 | 0.788204 | −1.268707 | −1 |
| OPCML_11_133291292_133302754_133378245_133382756_FR | OPCML | −0.380902 | −0.380902 | −12.74907 | 1.04E-10 | 4.70E-08 | 14.791885 | 0.767957 | −1.302156 | −1 |
| OSBP2_22_30729329_30730970_30763180_30773593_RR | OSBP2 | −0.432395 | −0.432395 | −14.48833 | 1.16E-11 | 1.51E-08 | 16.910523 | 0.741031 | −1.349472 | −1 |
| OSBP2_22_30729329_30730970_30817623_30822792_RR | OSBP2 | −0.366312 | −0.366312 | −11.45424 | 6.28E-10 | 1.19E-07 | 13.033392 | 0.775763 | −1.289053 | −1 |
| PA2G4P4_3_156818917_156822698_156846218_156854773_RR | PA2G4P4 | −0.174657 | −0.174657 | −5.663311 | 1.91E-05 | 0.000177 | 2.7128629 | 0.885978 | −1.128696 | −1 |
| PACRG_6_163022776_163030350_163324239_163328316_RR | PACRG | −0.352275 | −0.352275 | −9.207722 | 2.11E-08 | 1.20E-06 | 9.547019 | 0.783348 | −1.276572 | −1 |
| PARVB_22_43989557_43996453_44182513_44187012_FF | PARVB | −0.170178 | −0.170178 | −7.603815 | 3.74E-07 | 9.26E-06 | 6.6666544 | 0.888733 | −1.125198 | −1 |
| PASD1_X_151600201_151608969_151676020_151678489_RF | PASD1 | 0.3220619 | 0.3220619 | 5.8035886 | 1.41E-05 | 0.00014 | 3.0147787 | 1.250116 | 1.2501159 | 1 |
| PASD1_X_151608969_151613880_151648877_151652329_RR | PASD1 | 0.3167815 | 0.3167815 | 2.5872149 | 0.018153 | 0.042868 | −4.027661 | 1.245549 | 1.2455488 | 1 |
| PLCB1_20_8599928_8617739_8698163_8700449_FR | PLCB1 | −0.364492 | −0.364492 | −11.16982 | 9.51E-10 | 1.49E-07 | 12.624056 | 0.776742 | −1.287428 | −1 |
| PLCB1_20_8599928_8617739_8856413_8858679_FF | PLCB1 | −0.316603 | −0.316603 | −8.251608 | 1.13E-07 | 3.85E-06 | 7.8721116 | 0.802958 | −1.245395 | −1 |
| PLEKHM3_2_207850576_207856308_208003089_208006543_FF | PLEKHM3 | −0.385734 | −0.385734 | −8.300103 | 1.03E-07 | 3.58E-06 | 7.9600253 | 0.76539 | −1.306524 | −1 |
| PON2_7_95405100_95420940_95465337_95474032_FR | PON2 | −0.178367 | −0.178367 | −5.268811 | 4.50E-05 | 0.000344 | 1.8526765 | 0.883703 | −1.131602 | −1 |
| PPP2R5E_14_63423316_63431065_63511598_63515339_FF | PPP2R5E | 0.3121838 | 0.3121838 | 7.9196338 | 2.07E-07 | 6.02E-06 | 7.2616101 | 1.241586 | 1.2415857 | 1 |
| PRKCA_17_66441276_66447067_66475597_66481312_RF | PRKCA | 0.2425179 | 0.2425179 | 7.4515733 | 5.00E-07 | 1.15E-05 | 6.3748921 | 1.183056 | 1.1830556 | 1 |
| PTPRD_9_9798181_9808250_9882262_9891784_RR | PTPRD | −0.453093 | −0.453093 | −4.232884 | 0.000458 | 0.002168 | −0.460189 | 0.730475 | −1.368972 | −1 |
| PTPRD_9_9551379_9564487_9756141_9761726_RF | PTPRD | 0.3282945 | 0.3282945 | 10.781263 | 1.70E-09 | 2.09E-07 | 12.050556 | 1.255528 | 1.2555283 | 1 |
| RAP1GAP2_17_2837827_2840795_2974137_2976800_RF | RAP1GAP2 | −0.145079 | −0.145079 | −4.529412 | 0.000234 | 0.001268 | 0.2065487 | 0.90433 | −1.105791 | −1 |
| RERGL_12_18254365_18255916_18350532_18364514_RF | RERGL | 0.5204704 | 0.5204704 | 17.245127 | 5.42E-13 | 8.31E-09 | 19.793409 | 1.434423 | 1.4344229 | 1 |
| RNF6_13_26220822_26221931_26255215_26259295_RR | RNF6 | 0.3776285 | 0.3776285 | 8.7845474 | 4.37E-08 | 1.95E-06 | 8.8207418 | 1.299204 | 1.2992044 | 1 |
| RNU6-1264P_17_6162286_6163870_6195952_6199184_FR | RNU6-1264P | −0.152642 | −0.152642 | −9.038985 | 2.81E-08 | 1.46E-06 | 9.2602376 | 0.899602 | −1.111603 | −1 |
| RORA_15_60977475_60986445_61019417_61030714_RR | RORA | 0.3713685 | 0.3713685 | 14.275336 | 1.50E-11 | 1.70E-08 | 16.664729 | 1.293579 | 1.2935793 | 1 |
| RPL9P15_3_154664590_154668268_154687949_154695597_RF | RPL9P15 | 0.2007438 | 0.2007438 | 5.5395118 | 2.49E-05 | 0.000218 | 2.4446246 | 1.149291 | 1.1492908 | 1 |
| SCNN1B_16_23299279_23301635_23325818_23330880_FF | SCNN1B | −0.143058 | −0.143058 | −6.057416 | 8.25E-06 | 9.28E-05 | 3.5552838 | 0.905598 | −1.104243 | −1 |
| SETBP1_18_44770146_44772357_44885635_44895879_RF | SETBP1 | 0.355569 | 0.355569 | 11.916137 | 3.25E-10 | 8.20E-08 | 13.679971 | 1.27949 | 1.2794901 | 1 |
| SETBP1_18_44885635_44895879_45067644_45082246_FR | SETBP1 | 0.3703696 | 0.3703696 | 11.300548 | 7.85E-10 | 1.31E-07 | 12.813285 | 1.292684 | 1.2926839 | 1 |
| SLC9A8_20_49784052_49786161_49876581_49883310_RR | SLC9A8 | 0.1512545 | 0.1512545 | 7.2660355 | 7.15E-07 | 1.50E-05 | 6.0149546 | 1.110535 | 1.1105347 | 1 |
| SOD1_21_31645974_31648479_31675373_31681286_RF | SOD1 | −0.11317 | −0.11317 | −2.698249 | 0.014316 | 0.03529 | −3.804687 | 0.924554 | −1.081602 | −1 |
| SORCS2_4_7241158_7248673_7449931_7456800_FR | SORCS2 | 0.3595133 | 0.3595133 | 10.619505 | 2.18E-09 | 2.39E-07 | 11.806804 | 1.282993 | 1.282993 | 1 |
| SPAG16_2_213413840_213427575_213513566_213522278_FF | SPAG16 | −0.390916 | −0.390916 | −12.83763 | 9.25E-11 | 4.35E-08 | 14.906206 | 0.762645 | −1.311225 | −1 |
| SPTLC3_20_13133531_13136394_13174631_13182869_FF | SPTLC3 | 0.2023548 | 0.2023548 | 12.546312 | 1.36E-10 | 5.33E-08 | 14.527397 | 1.150575 | 1.1505748 | 1 |
| STX7_6_132435332_132445259_132542354_132547678_FR | STX7 | 0.3463849 | 0.3463849 | 12.466212 | 1.52E-10 | 5.81E-08 | 14.421833 | 1.271371 | 1.2713708 | 1 |
| STX7_6_132435332_132445259_132511475_132513451_FR | STX7 | 0.3187939 | 0.3187939 | 10.825435 | 1.59E-09 | 2.01E-07 | 12.116602 | 1.247287 | 1.2472874 | 1 |
| SYN3_22_32593146_32596862_32844795_32854272_RF | SYN3 | 0.3341746 | 0.3341746 | 17.115411 | 6.20E-13 | 8.31E-09 | 19.669129 | 1.260656 | 1.2606559 | 1 |
| SYN3_22_32723620_32730327_32844795_32854272_RF | SYN3 | 0.3345379 | 0.3345379 | 11.01189 | 1.20E-09 | 1.70E-07 | 12.392979 | 1.260973 | 1.2609735 | 1 |
| TANGO6_16_68932310_68938132_69047292_69051982_FF | TANGO6 | 0.1825419 | 0.1825419 | 6.0822581 | 7.83E-06 | 8.95E-05 | 3.6077649 | 1.134882 | 1.1348817 | 1 |
| TANGO6_16_68852020_68856905_68932310_68938132_RF | TANGO6 | 0.3129309 | 0.3129309 | 9.4084105 | 1.51E-08 | 9.16E-07 | 9.8833156 | 1.242229 | 1.2422288 | 1 |
| TARDBP_1_10989562_10991726_11014552_11017016_FF | TARDBP | −0.096493 | −0.096493 | −5.268967 | 4.50E-05 | 0.000344 | 1.8530201 | 0.935304 | −1.069171 | −1 |
| THSD7A_7_11420798_11426394_11712489_11724603_RF | THSD7A | 0.3135925 | 0.3135925 | 7.2665721 | 7.14E-07 | 1.50E-05 | 6.0160025 | 1.242799 | 1.2427986 | 1 |
| TMTC1_12_29532107_29533497_29647358_29657646_RF | TMTC1 | 0.3355121 | 0.3355121 | 11.641723 | 4.79E-10 | 1.00E-07 | 13.298511 | 1.261825 | 1.2618252 | 1 |
| TMTC1_12_29647358_29657646_29765279_29767497_FF | TMTC1 | 0.3639387 | 0.3639387 | 3.4118158 | 0.002954 | 0.009869 | −2.291571 | 1.286935 | 1.2869345 | 1 |
| TP63_3_189677768_189688253_189719534_189721726_FR | TP63 | −0.179873 | −0.179873 | −6.360572 | 4.39E-06 | 5.73E-05 | 4.1904365 | 0.88278 | −1.132784 | −1 |
| UBQLN2_X_56536168_56538402_56553450_56557221_FR | UBQLN2 | 0.2784451 | 0.2784451 | 3.1075781 | 0.00584 | 0.016979 | −2.950784 | 1.212887 | 1.212887 | 1 |
| UBQLN2_X_56536168_56538402_56570114_56575112_RR | UBQLN2 | 0.3839068 | 0.3839068 | 3.3139007 | 0.003682 | 0.011761 | −2.505503 | 1.304871 | 1.3048707 | 1 |
| UBQLN2_X_56536168_56538402_56570114_56575112_FF | UBQLN2 | 0.3937521 | 0.3937521 | 3.2929041 | 0.00386 | 0.012196 | −2.551177 | 1.313806 | 1.3138058 | 1 |
| UBQLN2_X_56536168_56538402_56570114_56575112_FR | UBQLN2 | 0.3538926 | 0.3538926 | 2.693743 | 0.014455 | 0.035577 | −3.813818 | 1.278004 | 1.2780042 | 1 |
| VSNL1_2_17493823_17506407_17651961_17655968_FR | VSNL1 | 0.3086278 | 0.3086278 | 8.9568305 | 3.24E-08 | 1.60E-06 | 9.1192635 | 1.238529 | 1.2385291 | 1 |
| WBSCR17_7_71656523_71666682_71682593_71686909_FR | WBSCR17 | 0.2893541 | 0.2893541 | 8.541732 | 6.69E-08 | 2.63E-06 | 8.3932893 | 1.222093 | 1.222093 | 1 |
| WWOX_16_78559412_78566930_78777764_78783921_FF | WWOX | −0.312268 | −0.312268 | −7.888672 | 2.19E-07 | 6.31E-06 | 7.203896 | 0.805375 | −1.241658 | −1 |
| XRCC1_19_43573185_43575618_43595713_43600345_RF | XRCC1 | −0.126515 | −0.126515 | −7.497073 | 4.58E-07 | 1.08E-05 | 6.4624281 | 0.916042 | −1.091653 | −1 |
| XRCC1_19_43573185_43575618_43595713_43600345_FR | XRCC1 | 0.1880671 | 0.1880671 | 8.9348232 | 3.36E-08 | 1.63E-06 | 9.0813491 | 1.139236 | 1.1392364 | 1 |
| ZBTB20_3_114403907_114406568_114458618_114478185_RR | ZBTB20 | −0.370005 | −0.370005 | −11.36464 | 7.15E-10 | 1.25E-07 | 12.90538 | 0.77378 | −1.292358 | −1 |
| ZFPM2_8_105632010_105638904_105814873_105824107_FR | ZFPM2 | −0.425339 | −0.425339 | −10.49363 | 2.64E-09 | 2.70E-07 | 11.615048 | 0.744664 | −1.342888 | −1 |
| ZFPM2_8_105525568_105531254_105736941_105746180_RR | ZFPM2 | −0.360993 | −0.360993 | −3.724595 | 0.001455 | 0.005518 | −1.599789 | 0.778628 | −1.28431 | −1 |
| ZFPM2_8_105572686_105580151_105814873_105824107_FR | ZFPM2 | −0.324519 | −0.324519 | −3.057007 | 0.006534 | 0.018627 | −3.058604 | 0.798565 | −1.252247 | −1 |
| ZNF804B_7_88937416_88946263_88973548_88984178_RR | ZNF804B | 0.3913267 | 0.3913267 | 16.279963 | 1.50E-12 | 1.04E-08 | 18.843266 | 1.311599 | 1.311599 | 1 |
| PDE4B_1_66194325_66201588_66342345_66350066_RF | PDE4B | −0.283024 | −0.283024 | −6.650398 | 2.43E-06 | 3.67E-05 | 4.7865237 | 0.821866 | −1.216743 | −1 |
Abbreviations. logFC: logarithm of the fold change; AveExpr: Average expression; adj.P·Val: Adjusted p-value; B: B-statistic (log-odds that that gene is differentially expressed); FC: Fold change; FC_1: Fold change centered around 1; Binary: Binary call for loop presence/absence.
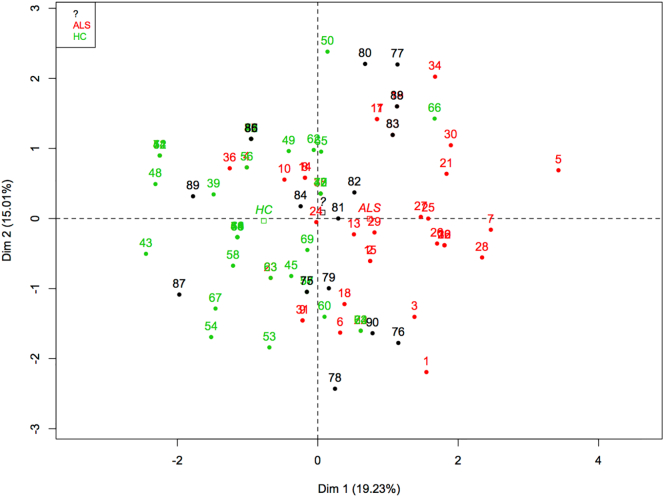
For the second step, the evaluation stage, the 153 biomarkers selected from the array analysis were translated into EpiSwitch™ PCR based-detection probes and used in multiple rounds of biomarker evaluation on an increasing number of patient samples. PCR primers were selected according to their ability to distinguish between ALS and healthy controls. Exact Fisher's P-value, GLMNET (alpha 0.5, penalized score) and standard logistic modeling scores including Coef, SE, Wald S and P-value were used to select the top eight biomarkers (Table 6). This selected chromosomal-conformation signature-biomarker set was then tested on a known (n = 74) and a blinded cohort (n = 16). Principal component analysis was also used to determine abundance levels and to identify potential outliers (Fig. 2). With EpiSwitch, initial screens identify significant markers using a small subset of patient samples, while the larger sample cohort sizes in later screens provide the statistical power to allow for the results to more closely approximate real-world populations.
Table 6.
The genomic loci contained in the chromosome conformation signature (CCS) used to inform an ALS diagnosis.
| Gene | Fisher's P value | Coefficient | SE |
|---|---|---|---|
| CD36 | 0.003 | −1.7788 | 0.8 |
| TAB2 | 0.098 | −0.8568 | 0.94 |
| GLYCAM1 | 0.213 | −0.9582 | 0.94 |
| GRB2 | 0.055 | −0.811 | 0.91 |
| FYN | 0.276 | −2.0869 | 0.93 |
| PTPRC | 0.027 | −1.7059 | 1.42 |
| DNM3 | 0.142 | −2.0227 | 0.92 |
| IKBKB | 0.117 | −1.395 | 1.32 |
Abbreviations. SE: Standard error.
Fig. 2.
Principal component analysis for the 8 markers applied to 74 known samples (ALS samples in red and healthy controls (HC) in green) and 16 unknown blinded samples (black). The blinded samples appear as a mixture of ALS and control samples.
The sample cohort sizes in this study were progressively increased to enable selection of the optimal markers for discriminating ALS samples from healthy controls. Cohort sizes were statistically powered to a level of sensitivity and specificity needed for clinical application. More specifically, in the first screening series, six ALS and six healthy control samples were used. In the subsequent screening step, 24 ALS and 24 controls samples were used. For the final screening, a panel consisting of the eight top biomarkers (Table 6) was applied to a separate cohort of 74 samples from the NEALS Biofluid Repository and Oxford University. Statistical analysis was carried out on the final screen of the binary data results. (See Results, (Table 7)).
Table 7.
Sensitivity and specificity of the ALS chromosome conformation signature (CCS) when used to classify a set of clinical samples (n = 74) taken from two ALS clinical trials.
| Statistic | Value | 95% Cl |
|---|---|---|
| Sensitivity | 0.8333 | 51.59% to 97.91% |
| Specificity | 0.7692 | 46.19% to 94.96% |
| Positive likelihood ratio | 3.61 | 1.30 to 10.06 |
| Negative likelihood ratio | 0.22 | 0.06 to 0.79 |
| Disease prevalence | 48% (*) | 27.80% to 68.69% |
| Positive predictive value | 76.92% (*) | 46.19% to 94.96% |
| Negative predictive value | 83.33% (*) | 51.59% to 97.91% |
Abbreviations. 95% CI: 95% confidence interval.
To further validate the ALS CCS, the panel was tested on a blinded, independent (n = 16) cohort of blood samples supplied by Oxford University. The results were analyzed using Bayesian Logistic modeling, p-value null hypothesis (Pr(>|z|) analysis, Fisher-Exact P test and Glmnet (Table 8).
Table 8.
Results from classification of blinded Oxford University samples (N = 16).
| Statistic | Value | 95% Cl |
|---|---|---|
| Sensitivity | 0.875 | 47.35% to 99.68% |
| Specificity | 0.75 | 34.91% to 96.81% |
| Positive likelihood ratio | 3.5 | 1.02 to 11.96 |
| Negative likelihood ratio | 0.17 | 0.03 to 1.09 |
| Disease prevalence | 50% (*) | 24.65% to 75.35% |
| Positive predictive value | 77.78% (*) | 39.99% to 97.19% |
| Negative predictive value | 85.71% (*) | 42.13% to 99.64% |
Abbreviations. 95% CI: 95% confidence interval.
Last, we explored the biological relevance of the biomarkers by using a second array with 171,408 potential chromosome conformations across 467 loci that were functionally related to ALS [15] (Table 9). The list of 150 top performing loci from this screen were uploaded to the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database containing over 9 million known and predicted protein-protein interactions (https://string-db.org) to create a network of ALS regulation.
Table 9.
Loci that are functionally related to ALS used for the second array.
| Gene name | |||||
|---|---|---|---|---|---|
| DPP6 | AGBL1 | DNAH2 | KCNN3 | RAMP3 | ZMYND8 |
| C9orf72 | AGPAT5 | DNAH9 | KCNQ1 | RAP1GAP2 | ZNF407 |
| ITPR2 | AK8 | DOCK8 | KDM4B | RAPGEF4 | ZNF423 |
| UNC13A | AKAP6 | DOCK9 | KDR | RAPGEF5 | ZNF485 |
| MOB3B | AKAP7 | DPF3 | KIAA0040 | RBFOX3 | ZNF519 |
| ALS2 | AMD1 | DPP10 | KIAA0513 | RDH14 | ZNF710 |
| CDH13 | ANG | DSC3 | KIAA1644 | RERGL | ZNF804B |
| SOD1 | ANKDD1A | DSCR4 | KLHL29 | RETSAT | |
| ALK | ANKRD29 | DSTNP5 | KLHL38 | RGS17 | |
| BARD1 | ANKS1B | E2F7 | KRT18P3 | RNA5SP142 | |
| BTBD11 | ANO1 | ELK3 | KRT18P64 | RNA5SP158 | |
| FAM19A5 | ANXA5 | ELMSAN1 | KSR2 | RNF14 | |
| GRIP1 | AP2A2 | ELP3 | LAMA3 | RNF17 | |
| MACROD2 | APBB2 | EN1 | LHFP | RNF6 | |
| RBFOX1 | APP | EPB41L3 | LINGO2 | RNGTT | |
| SHROOM3 | ARHGEF3 | ERG | LIPC | RNU6-1264P | |
| TARDBP | ARMS2 | ERGIC1 | LMO2 | RNU6-242P | |
| AGAP1 | ARNT2 | ERMARD | LOX | RNU6ATAC32P | |
| AGBL4 | ARSG | ESRRG | LRIG1 | RORA | |
| ALDH1A2 | ASIC2 | EVC2 | LRP1B | RPF2 | |
| ATP2C2 | ATP2A3 | EXO1 | LSAMP | RPL9P15 | |
| ATXN2 | ATXN1 | EXOC2 | LYPD3 | RSPO4 | |
| BANK1 | ATXN7L1 | EYA1 | MAPK1 | SAMD5 | |
| BTBD16 | AVEN | FAM126A | MAST4 | SARM1 | |
| CACNA2D3 | B4GALT6 | FAM13B | MDGA2 | SCN8A | |
| CALN1 | BCL6 | FAM149A | MED13L | SCNN1B | |
| CDC42BPA | BEND7 | FAM155A | MGLL | SETBP1 | |
| CHSY3 | BMPR2 | FAM169B | MICAL3 | SEZ6L | |
| CNTN6 | BTLA | FAM189A1 | MICB | SGMS1 | |
| CNTNAP2 | C15orf32 | FAM194B | MKL2 | SGSM1 | |
| CSMD1 | C2CD2 | FBXO32 | MKLN1 | SHISA3 | |
| DAB1 | C4orf19 | FER | MORN5 | SIGLEC12 | |
| DIO2 | C6orf132 | FER1L6 | MTMR7 | SIRPG | |
| DNM1L | C6orf58 | FGF1 | MTUS2 | SLC24A2 | |
| DOCK1 | C7orf57 | FGF12 | MUC6 | SLC35A3 | |
| DSCAM | C8orf47 | FGGY | MYH11 | SLC35F3 | |
| ERBB4 | C9orf170 | FHDC1 | MYO10 | SLC41A1 | |
| ERC1 | CABIN1 | FHIT | MYO18B | SLC9A8 | |
| FARP1 | CADM2 | FHOD3 | MYO1D | SLIT3 | |
| FBXO8 | CAMK1D | FIG4 | NAALADL2 | SMIM13 | |
| FMN2 | CAPN9 | FMN1 | NAV2 | SNORD113–2 | |
| FTO | CAPZA1 | FNDC3B | NAV3 | SNRPD3 | |
| FUS | CASC10 | FRMD3 | NCKAP5 | SNTG2 | |
| GLI2 | CAST | GABBR2 | NEIL3 | SORBS2 | |
| HIATL1 | CCDC3 | GALNT2 | NELL1 | SORCS2 | |
| IL18RAP | CCDC81 | GALNTL6 | NFIA | SOX5 | |
| IQGAP2 | CD1A | GAS6 | NLRP7 | SOX7 | |
| KALRN | CD1B | GCH1 | NOTCH4 | SPAG16 | |
| KCNS3 | CD1E | GFPT1 | NOX4 | SPATA13 | |
| KIFAP3 | CDH12 | GFRA1 | NPR3 | SPG11 | |
| KRT18P55 | CDH23 | GLIS3 | NPTXR | SPTLC3 | |
| LARGE | CDH4 | GLT8D2 | NRXN1 | SRGAP3 | |
| LOXHD1 | CDH8 | GMDS | NRXN3 | SRRM4 | |
| LRPPRC | CDK14 | GNG7 | NUDT12 | STAC | |
| MAGI2 | CDRT4 | GNPDA1 | NXPH1 | STK36 | |
| NEFH | CELF2 | GPC6 | OPTN | STX7 | |
| NHSL1 | CELF4 | GPR176 | ORC5 | SUN3 | |
| NKAIN2 | CELSR1 | GRB14 | OSBP2 | SYN3 | |
| NSMAF | CEP44 | GRIK2 | OTP | SYNJ2 | |
| OPCML | CGNL1 | GRIK4 | PA2G4P4 | SYT9 | |
| PCDH15 | CHAF1A | GRIN2B | PACRG | SYTL3 | |
| PCSK6 | CHAMP1 | GRM3 | PALLD | TACR2 | |
| PDE4B | CHGB | GRM7 | PARK2 | TANGO6 | |
| PLXDC2 | CHRM5 | GRM8 | PARVB | TANK | |
| PON2 | CHST1 | GRN | PASD1 | TCEA1 | |
| PTPRN2 | CNTN3 | HABP2 | PAX7 | TCF7L1 | |
| PTPRT | CNTN4 | HCFC2P1 | PBX1 | TCL1B | |
| RGS6 | COL14A1 | HDAC4 | PCDH12 | TIAM2 | |
| ROBO2 | COL1A1 | HFE | PDCL3P1 | TMEM132C | |
| SLC25A26 | COL27A1 | HHAT | PDE7B | TMEM135 | |
| SNX29 | COL28A1 | HLA-DOA | PDZD2 | TMEM91 | |
| SPATA22 | COLGALT2 | HLA-DPB2 | PEPD | TMPRSS13 | |
| SPTLC1P2 | CREB3L2 | HLA-DRB9 | PFKP | TMTC1 | |
| SUSD1 | CREB5 | HMCN2 | PHC1 | TNPO3 | |
| THSD7A | CRHBP | HNF1B | PIGL | TP63 | |
| TIAM1 | CRYGGP | HNRNPA1P32 | PLA2G12B | TRAK2 | |
| TLR1 | CSRP1 | HPGD | PLCB1 | TUFT1 | |
| TLR10 | CST5 | HS3ST4 | PLEKHM3 | TULP4 | |
| TMEM132D | CTNNA3 | HSCB | PLGRKT | TXNRD1 | |
| TMEM163 | CTNND2 | HUS1 | PON1 | UBQLN2 | |
| TMTC2 | CTSC | IFI44L | PPP1R14C | VAPB | |
| UNC13C | CX3CR1 | IFT74 | PPP2R5E | VRK2 | |
| UPF2 | CXCL12 | IL1A | PRDM16 | VSNL1 | |
| VEGFA | DAO | IL20RA | PRKAG2 | WBSCR17 | |
| ZFP64 | DBF4B | INSIG2 | PRKCA | WDFY3 | |
| ACOXL | DCC | INSR | PRKCQ | WNT9A | |
| ADAMTS20 | DCLK1 | IQCJ-SCHIP1 | PRPH | WWOX | |
| ADARB2 | DCTN1 | JARID2 | PRR9 | XRCC1 | |
| ADCY1 | DGKB | KCNIP1 | PSD3 | YPEL1 | |
| ADH7 | DIAPH3 | KCNMA1 | PTPRD | ZBTB20 | |
| ADRBK2 | DIRC3 | KCNMB3 | PXDNL | ZFP36L1 | |
| AFTPH | DISC1 | KCNN2 | RAB3C | ZFPM2 | |
3. Results
3.1. Patient Clinical Characteristics
In order to develop, test and validate the CCS biomarker panel, we used blood samples from two main cohorts. The first cohort (Discovery) consisted of 50 ALS samples and 42 healthy controls provided by NEALS and Oxford BioDynamics. The second cohort (Validation) consisted of 16 samples (8 ALS and 8 healthy familial controls) provided by the University of Oxford. For the ALS patients, samples in both cohorts were sex matched (approximately 2/3 male and 1/3 female), with the majority of cases being sporadic ALS (84% in the Discovery Cohort and 75% in the Validation cohort) (Table 3). Average ALSFRS-R scores (35.9 in Discovery and 36.4 in Validation), average age at diagnosis (53.4 years in Discovery and 54.9 years in Validation) and disease duration (53.4 in Discovery and 54.9 in Validation) were similar in both cohorts (Table 3). Although ethnicity and race were not available for the Validation cohort, the vast majority of patients in the Discovery (90%) were non-Hispanic or Latino Whites.
3.2. Identity of the Markers in the Signature
The EpiSwitch three-step biomarker selection process yielded a distinct chromosome conformational disease classification signature for ALS comprised of chromosomal interactions in eight genomic loci. The loci contained in the signature are CD36, TAB2, GLYCAM1, GRB2, FYN, PTPRC, DNM3 and IKBKB (Table 6). The final ALS CCS was derived from high-throughput analysis using the EpiSwitch discovery platform initially identifying 153 potential biomarker interactions. Statistical analysis results from the binary data analysis are shown in Table 5.
3.3. Sensitivity and Specificity Analysis
The discriminating power of the ALS CCS are shown in Table 6, Table 7. Sensitivity and specificity for ALS detection in the 74 unblinded-tissue samples using the ALS CCS was 83∙33% (CI 51∙59 to 97∙91%) and 76∙92 (46∙19 to 94∙96%), respectively. In an independent, blinded cohort, sensitivity of the ALS CCS reached 87∙50% (CI 47∙35 to 99∙68%) and specificity was 75∙0% (34∙91 to 96∙81%).
3.4. Biological Relevance in ALS
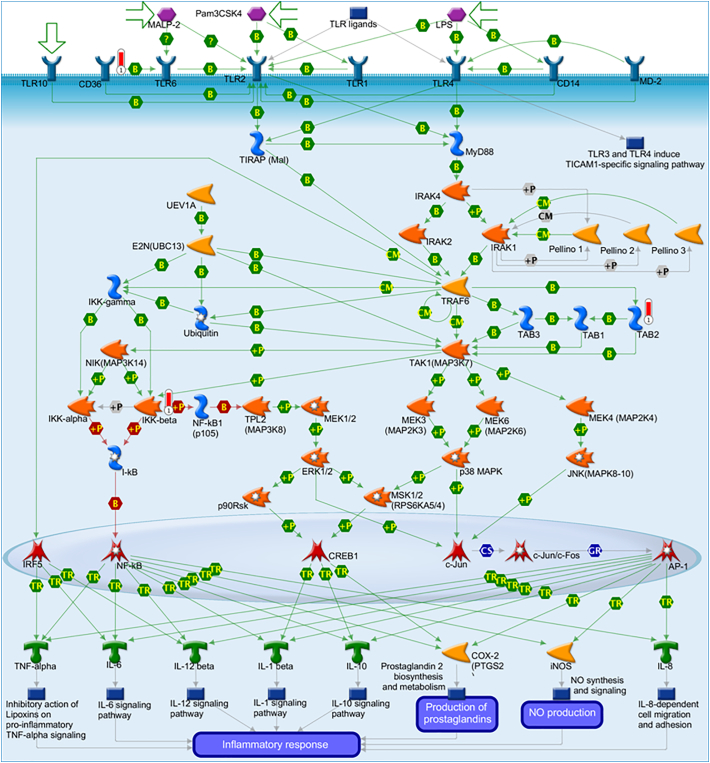
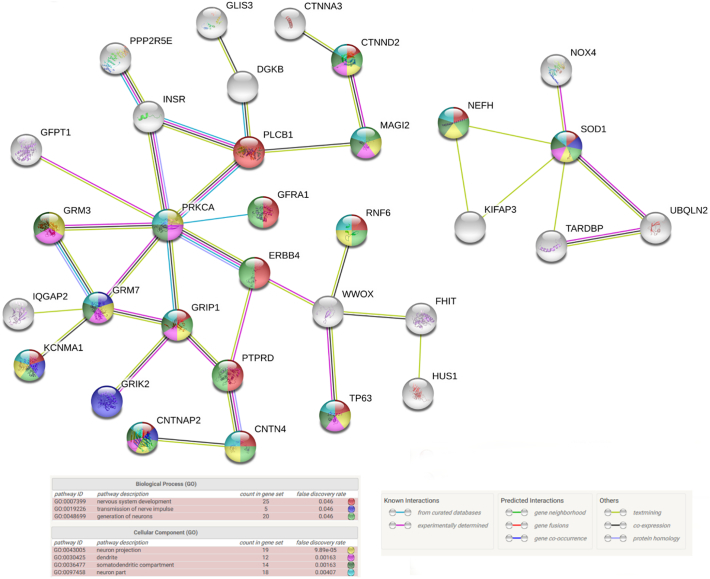
When we mapped the markers in the ALS CCS to the Metacore™ signaling pathway database, the TLR2 and 4 signaling pathways showed significant enrichment with three genomic loci (CD36, TAB2 and IKBKB) mapping to this signaling cascade (Fig. 3). To acquire additional insights into how the loci identified in this study contribute to the pathophysiology of ALS, we expanded our analysis to include an array-based comparison of ALS patients versus healthy controls using a set of loci that have been previously associated with ALS as an initial screen. Based on comparison of 16 ALS patients and 16 controls, 150 statistically disseminating markers were identified (Table 9). Genetic loci enriched with significant epigenetic deregulation were used to build a protein regulatory network using the STRING database (Fig. 4). When analyzing the resulting network (Additional File 1, Additional File 2), key hubs included proteins with known links to the pathophysiology of ALS including SOD1, TARDBP (TDP-43), NEFH, and UBQLN2 [[16], [17], [18]] (Fig. 4). In addition to the well-studied loci with known links to ALS, the network analysis confirmed the involvement of emerging and lesser-studied genomic loci in the development of ALS including KIFAP3 which has been recently identified as a potential risk locus for ALS and GRIP1 which was shown to be altered in ALS2 deficient spinal motor neurons leading to neuronal degeneration [[19], [20], [21]]. This indicates that consistent epigenetic deregulation is observed in key genetic loci in the largely sporadic ALS patient population used in this study.
Fig. 3.
Toll-like receptor signaling cascade showing the biological involvement of three of the eight chromosome conformation signature loci; CD36, TAB2 and IKKB (red thermometers) in the regulation of the inflammatory response. Image generated using Metacore™.
Fig. 4.
Protein STRING network of ALS regulation. The gene loci for the top 150 chromosome conformations that could best discriminate between ALS samples and healthy controls in the second array screen were uploaded as proteins to the STRING database and a resulting interaction network was generated. The two main networks are shown. Network nodes represent proteins and edges represent protein-protein associations. All nodes shown are query proteins and the first shell of interactors. Nodes are colored according to their association with the top gene ontology (GO) terms for Biological Process and Cellular Component. Edges are colored according to their interactions, either known, predicted or other.
4. Discussion
For the majority of patients with ALS, the etiology of the disorder is unknown. Mutations in a number of genes such as: C9orf72, Cu, Zn superoxide dismutase 1 (SOD1) and TAR-DNA binding protein (TDP-43), found in familial ALS occur in only about 10% of the patient population [[22], [23], [24], [25]]. Other studies suggest common pathogenic mechanisms for both the non-genetic and the genetic forms of ALS, as well as, similar clinical courses and dysfunctional features [2, 25]. A hexanucleotide expansion in the C9orf72 gene is the most common genetic mutation [26, 27]. The discovery of the repeat expansion of the C9orf72 hexanucleotide provides bridge between familial ALS and sporadic ALS [[27], [28], [29]]. The mutation is detectable in about 40% of familial ALS patients and 8–10% of sporadic ALS. The rapidly progressive nature of ALS means any improvement in diagnosis would be of great value to patients, their families and the professionals who treat them.
Epigenetics is the conduit for interactions between the environment and the genome. Epigenetic regulation of the genome can be used to explain complex diseases especially in the absence of any genetic mutations or patterns to explain pathology [14]. The foundation for diagnostic biomarker discovery using epigenetic frameworks rests with the detection and validation of conditional chromosome conformational changes at genetic loci of interest. Discovery is feasible and possible because chromosomal conformation comprises the smallest unit of regulated genome-linked to phenotype and asserts a high-level of regulation [14]. The binary quality (either the change is there or not), high biochemical stability and other characteristics of conditional chromosomal conformations make these genomic interactions highly advantageous as a source for potential diagnostic biomarkers [14].
OBD's platform technology and methodology, EpiSwitch, employs chromosome conformation capture and algorithmic analysis to detect and define a panel of epigenetic differences capable of discerning between diseased tissue samples and healthy controls. In this study, we identified, defined and evaluated chromosome conformations as biologically-distinguishing markers that comprise the first example of a non-invasive blood-based epigenetic signature for ALS. The study also provides the first indication that chromosomal conformational biomarker discovery may also provide a way to explore pathogenic pathways and mechanisms. The top performing EpiSwitch chromosome conformation biomarker maps to CD36, which encodes a fatty acid transport protein and has been shown to be linked to mitochondrial function. CD36 is also involved in the Toll Like Receptor (TLR) 2 and TLR4 signaling pathways. These toll-like receptors are activated in microglia in response to damage-associated molecular patterns. Interestingly, microglia have been shown to have a protective effect in early stage motor neuron degeneration [30]. Four biomarkers (i.e., Fyn, GRB2, IKBKB and CD45) appear in the pathways that map to the Major Histocompatibility Complex (MHC). Fyn plays a key role in initiating myelination by myelin-forming glial cells [31]. Three biomarker proteins (CD36, TAB2, IKBKB) are involved in pathways associated with the innate immune system. The IKK kinase complex is found in the regulation of gene expression in response to neurotransmission [32]. The location of these biomarkers in key pathways regulating the immune response point to the involvement of neuroinflammatory mechanisms in the pathogenesis of the disease [33].
The findings reported here add an additional clinical tool to aid in ALS diagnosis and further increase understanding of the disease Support for those concepts can be found in the panel of biomarkers discovered during this investigation. The gene loci related to the biomarkers in the ALS CCS, and their protein products, feature in cellular pathways linked to the phenotypic manifestations of ALS including: fatty acid transport, mitochondrial dysfunction, and alterations in both immune function and glial activation. Future studies using this approach and assessing blood samples collected longitudinally can also be applied as a new approach to monitor disease progression. In fact, recent studies indicate that the prediction of progression in ALS is possible using a set of patient clinical data (e.g. ALSFRS-R, ALSFRS slope, Trunk sub-score, time since diagnosis, systolic blood pressure, predicted survival) [34, 35]. Although the clinical annotations available for many of the samples used in this study were limited, future analysis of samples from longitudinal studies using EpiSwitch in combination with clinical metadata analysis and predictive algorithms are warranted.
Additionally, the CCS identified during this investigation aligns with more recent evidence that points to the concept of non-cell-autonomous disease pathogenesis and the contribution of microglia in ALS [36]. A number of investigations have indicated that the cells lose their surveillance and neuro-protective capacity by switching from an activated neuroprotective to a neurodegenerative phenotype as the disease progresses. Under normal conditions microglia activation results in upregulation of MHC class 2 proteins involved in presentation of antigens to T lymphocytes. Microglia also express a diverse set of pattern recognition receptors, including TLRs, for sensing pathogen-associated molecular patterns and endogenous ligands derived from cellular injury.
In addition to the eight loci that make up the CCS, expanded array analysis and overlay of a STRING protein network with genetic loci enriched in epigenetic deregulation shows evidence for strong biological concordance with genetic cases based on familial SOD1, TDP-43 and UBQLN2 (ALS subtype 15) genetic variants [37]. Epigenetic deregulation of SOD1, TDP-43, ERBB4 (ALS19), UBQLN2, INSR– all present themselves as significant events related to ALS etiology in sporadic cases investigated in this study. In addition to the STRING network identifying interconnected nodes with known link to disease, the analysis is also useful in the identification of novel disease mechanisms for target discovery. For example, the most interconnected node in the network was the alpha subunit of Protein Kinase C (PRKCA) (Fig. 4). While a role for Protein Kinase C has been known for decades in ALS ([38, 39]), our results implicate a supportive role for the glutamate metabotropic receptors GRM3 and GRM7 as potential intermediaries that can serve as novel disease targets [40].
Some of the caveats associated with our findings are the relatively modest sample size and the clinical homogeneity of the samples. According to a recently published global epidemiology analysis of published studies, ALS affects approximately 100,000 people worldwide [41]. In this study, we used 50 ALS patient samples and 42 healthy controls to develop the CCS and validated the signature on an additional 16 samples (8 ALS and 8 healthy controls), which represents a very small fraction of global cases. It is important to note however, that historical studies seeking to identify fluid-based biomarkers of ALS diagnosis have been developed on the analysis of between 28 and 103 ALS patients and between 12 and 43 healthy controls [42], sample numbers that are within the range of this study. Perhaps more importantly is the relative homogeneity of the samples in this study. The patients in the discovery cohort were overwhelmingly (>90%) Non-Hispanic or Latino Whites. While in the United States it has been recognized that there is a higher rate of ALS among Whites, a recent analysis of worldwide ALS incidence confirms the observation that the disease can affect people from a wide range of racial and ethnic backgrounds [43, 44]. Future studies looking at larger patient sets and the inclusion of a greater diversity of ethnic representation are warranted. Last, it is important to acknowledge that the benefits and limitations of using any surrogate non-invasive readout in a liquid biopsy, including this study, remain an important point of inquiry. Interestingly, recent studies have demonstrated that while distinct differences in gene expression profiles are observed in different tissue and cellular types; epigenetic differences, from DNA methylation to histone modifications to high order chromatin structures, certain markers could show local synchronization across cellular types. This includes macrophages and dendritic cells involved in immune surveillance which show concordant epigenetic signals between the primary site of pathology and in surrogate blood-based readouts [8, 45]. The current understanding of this phenomenon involves the exosome-mediated resetting of selective targeted cellular populations, described as a horizontal transfer [46, 47]. This is particularly relevant to exosome-based transfer of non-coding RNA, such as miRNA, long implicated in epigenetic resetting of secondary cellular targets in peripheral blood and distant tissues and directly associated with resetting of specific chromosome conformations in individual cells. Further epigenetic-based biomarker studies and extended validation on independent cohorts will help to better understand the features of these observed systemic epigenetic sub-signatures.
Finally, we believe that epigenetic insights may help to provide biomarkers directly related to ALS. EMG and conductivity tests have increased the sensitivity of ALS diagnosis by providing the first biological information to inform ALS diagnosis. The sensitivity of our assay using patient samples approaches the sensitivity results reported using Awaji Criteria and those reported for CSF neurofilaments, another biomarker in development for diagnosing ALS [48, 49]. Ultimately, complex and heterogeneous diseases like ALS will require an integrative multi-omics approach to better characterize disease onset and progression and the results presented here offer an additional molecular approach to understand disease pathology.
One of the major challenges in the current clinical practice of diagnosing ALS is the time is takes to make a definitive diagnosis. Under the current “rule-out” paradigm, it can take months to confirm a diagnosis of ALS. In addition, many of the current tools for aiding a diagnosis are expensive and can themselves take days to weeks to interpret. The ALS CCS described here is based on simple, inexpensive and well-accepted molecular biology techniques and technical readouts are available within 24 h, offering a substantial time and cost savings to physicians and payors. While the application of the ALS CCS in the clinical setting will require further investigation, the potential use of a chromosome conformation signature to diagnose ALS from a simple to collect, non-invasive biofluid and simple, rapidly available clinical readouts available to physicians and caregivers promise to help fill the gap in the current methods for diagnosing ALS.
The following are the supplementary data related to this article.
ALS STRING network in .csv format.
ALS STRING network in XML format.
Clinical annotations for the blind independent sample cohort provided by Oxford University.
Acknowledgments
Acknowledgements
Oxford BioDynamics would like to thank Barbara Nasto for her assistance in the preparation of this manuscript, which was funded by Oxford BioDynamics.
Oxford BioDynamics holds ethical approval information for all patient samples. Oxford University holds written consents for individual patient samples. Consents for NEALS Biofluid Repository samples are held at NEALS Biofluid Repository.
Sources of Funding
This research was funded by Oxford BioDynamics as well as Innovate UK. Work in the Oxford Motor Neurone Disease Care and Research Centre is supported by grants from the Motor Neurone Disease Association and the Medical Research Council. MRT is funded by the Medical Research Council & Motor Neurone Disease Association Lady Edith Wolfson Senior Clinical Fellowship (MR/K01014X/1).
Author Contributions
The study was conceived and designed by M. Cudkowicz, J. Berry, E. Hunter and A. Akoulitchev. The manuscript was prepared by M. Salter (main author matthew.salter@oxfordbiodynamics.com), J. Green, J. Westra, A. Akoulitchev, M. Cudkowicz, L. Ossher and K. Talbot. Methodology development and data acquisition was done by M. Salter, J. Green, E. Corfield, L. Ossher and K. Talbot. Analysis and data interpretation were done by M. Salter, J. Green, J. Westra, E. Hunter, A. Ramadass, A. Akoulitchev and F. Grand.
Declaration of Interests
All authors (1) are employees of Oxford BioDynamics and are shareholders within the company. A. Akoulitchev and A. Ramadass are company directors. Oxford BioDynamics holds patents on the EpiSwitch™ technology. Authors (2) are employees of the University of Oxford, Nuffield Department of Clinical Sciences and Authors (3) is from Massachusetts General Hospital, Amyotrophic Lateral Sclerosis clinic and the Neurological Clinical Research Institute.
References
- 1.Mitchell J.D., Callagher P., Gardham J., Mitchell C., Dixon M., Addison-Jones R. Timelines in the diagnostic evaluation of people with suspected amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND) – a 20-year review: can we do better? Amyotroph Lateral Scler. 2010;11:537–541. doi: 10.3109/17482968.2010.495158. [DOI] [PubMed] [Google Scholar]
- 2.Chen S., Sayana P., Zhang X., Le W. Genetics of amyotrophic lateral sclerosis: an update. Mol Neurodegener. 2013;8:28. doi: 10.1186/1750-1326-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson J.O., Mandrioli J., Benatar M., Abramzon Y., Van Deerlin V.M., Trojanowski J.Q. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heyn H., Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 5.Qureshi I.A., Mehler M.F. Understanding neurological disease mechanisms in the era of epigenetics. JAMA Neurol. 2013;70:703–710. doi: 10.1001/jamaneurol.2013.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter E., Ramadass A., Womersley H., Akoulitchev A. The Lancet Neurology Conference. 2016. Epigenetic footprints for neurodegenerative and autoimmune conditions: a comparative analysis. [Google Scholar]
- 7.Sherman A., Bowser R., Grasso D., Power B., Milligan C., Jaffa M. Proposed BioRepository platform solution for the ALS research community. Amyotroph Lateral Scler. 2011;12:11–16. doi: 10.3109/17482968.2010.539233. [DOI] [PubMed] [Google Scholar]
- 8.Bastonini E., Jeznach M., Field M., Juszczyk K., Corfield E., Dezfouli M. Chromatin barcodes as biomarkers for melanoma. Pigment Cell Melanoma Res. 2014;27:788–800. doi: 10.1111/pcmr.12258. [DOI] [PubMed] [Google Scholar]
- 9.Carini C., Hunter E., Ramadass A.S., Green J., Akoulitchev A., Mcinnes I.B. Chromosome conformation signatures define predictive markers of inadequate response to methotrexate in early rheumatoid arthritis. J Transl Med. 2018 doi: 10.1186/s12967-018-1387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakub J.W., Grotz T.E., Jordan P., Hunter E., Pittelkow M., Ramadass A. A pilot study of chromosomal aberrations and epigenetic changes in peripheral blood samples to identify patients with melanoma. Melanoma Res. 2015 doi: 10.1097/CMR.0000000000000182. [DOI] [PubMed] [Google Scholar]
- 11.McCord R., Sandmann T., Field M., Jordan P., Hunter E., Akoulitchev A. ACR Annual Meeting. 2014. Chromatin signatures of DLBCL subtypes. [Google Scholar]
- 12.Salter M., Elvidge W., Ramadass A., Womersley H., Grand F., Green J. The Lancet Neurology Conference. The Lancet Neurology Conference; 2016. Epigenetic signatures and early detection of neurodegenerative diseases. [Google Scholar]
- 13.Sangurdekar D., Plavina T., Singh C., Innis B., Enayetallah A., Subramanyam M. 32nd Congress of the European Committee for Treatment and Research in Multiple Sclerosis. 2016. Chromosome conformation capture to discover candidate epigenetic markers of multiple sclerosis disease severity. [Google Scholar]
- 14.Crutchley J.L., Wang X.Q.D., Ferraiuolo M. a, Dostie J. Chromatin conformation signatures: ideal human disease biomarkers? Biomark Med. 2010;4:611–629. doi: 10.2217/bmm.10.68. [DOI] [PubMed] [Google Scholar]
- 15.Chen X., Shang H.-F. New developments and future opportunities in biomarkers for amyotrophic lateral sclerosis. Transl Neurodegener. 2015;4:17. doi: 10.1186/s40035-015-0040-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen D.K.H., Thombre R., Wang J. Autophagy as a common pathway in amyotrophic lateral sclerosis. Neurosci Lett. 2018 doi: 10.1016/j.neulet.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su X.W., Broach J.R., Connor J.R., Gerhard G.S., Simmons Z. Genetic heterogeneity of amyotrophic lateral sclerosis: implications for clinical practice and research. Muscle Nerve. 2014;49:786–803. doi: 10.1002/mus.24198. [DOI] [PubMed] [Google Scholar]
- 18.Vajda A., McLaughlin R.L., Heverin M., Thorpe O., Abrahams S., Al-Chalabi A. Genetic testing in ALS: a survey of current practices. Neurology. 2017 doi: 10.1212/WNL.0000000000003686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai C., Xie C., McCormack S.G., Chiang H.-C., Michalak M.K., Lin X. Amyotrophic lateral sclerosis 2-deficiency leads to neuronal degeneration in amyotrophic lateral sclerosis through altered AMPA receptor trafficking. J Neurosci. 2006;26:11798–11806. doi: 10.1523/JNEUROSCI.2084-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landers J.E., Melki J., Meininger V., Glass J.D., van den Berg L.H., van Es M.A. Reduced expression of the kinesin-associated protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2009;106:9004–9009. doi: 10.1073/pnas.0812937106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Doormaal P.T.C., Ticozzi N., Gellera C., Ratti A., Taroni F., Chiò A. Analysis of the KIFAP3 gene in amyotrophic lateral sclerosis: a multicenter survival study. Neurobiol Aging. 2014;35 doi: 10.1016/j.neurobiolaging.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakkar N., Boehringer A., Bowser R. Use of biomarkers in ALS drug development and clinical trials. Brain Res. 2015 doi: 10.1016/j.brainres.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byrne S., Walsh C., Lynch C., Bede P., Elamin M., Kenna K. Rate of familial amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry. 2011;82:623–627. doi: 10.1136/jnnp.2010.224501. [DOI] [PubMed] [Google Scholar]
- 24.Simon N.G., Turner M.R., Vucic S., Al-Chalabi A., Shefner J., Lomen-Hoerth C. Quantifying disease progression in amyotrophic lateral sclerosis. Ann Neurol. 2014 doi: 10.1002/ana.24273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traynor B.J., Zhang H., Shefner J.M., Schoenfeld D., Cudkowicz M.E. Functional outcome measures as clinical trial endpoints in ALS. Neurology. 2004;63:1933–1935. doi: 10.1212/01.wnl.0000144345.49510.4e. [DOI] [PubMed] [Google Scholar]
- 26.Debray S., Race V., Crabbé V., Herdewyn S., Matthijs G., Goris A. Frequency of C9Orf72 repeat expansions in amyotrophic lateral sclerosis: a Belgian cohort study. Neurobiol Aging. 2013;34 doi: 10.1016/j.neurobiolaging.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 27.Renton A.E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J.R. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dejesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majounie E., Renton A.E., Mok K., Dopper E.G.P., Waite A., Rollinson S. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brites D., Vaz A.R. Microglia centered pathogenesis in ALS: insights in cell interconnectivity. Front Cell Neurosci. 2014;8 doi: 10.3389/fncel.2014.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyamoto Y., Tamano M., Torii T., Kawahara K., Nakamura K., Tanoue A. Data supporting the role of Fyn in initiating myelination in the peripheral nervous system. Data Br. 2016;7:1098–1105. doi: 10.1016/j.dib.2016.03.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mihalas A.B., Meffert M.K. IKK kinase assay for assessment of canonical NF-κB activation in neurons. Methods Mol Biol. 2015;1280:61–74. doi: 10.1007/978-1-4939-2422-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J., Hyeon S.J., Im H., Ryu H., Kim Y., Ryu H. Astrocytes and microglia as non-cell autonomous players in the pathogenesis of ALS. Exp Neurobiol. 2016;25:233–240. doi: 10.5607/en.2016.25.5.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berry J.D., Taylor A.A., Beaulieu D., Meng L., Bian A., Andrews J. Improved stratification of ALS clinical trials using predicted survival. Ann Clin Transl Neurol. 2018 doi: 10.1002/acn3.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor A.A., Fournier C., Polak M., Wang L., Zach N., Keymer M. Predicting disease progression in amyotrophic lateral sclerosis. Ann Clin Transl Neurol. 2016 doi: 10.1002/acn3.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Julien J.P. Amyotrophic lateral sclerosis: unfolding the toxicity of the misfolded. Cell. 2001 doi: 10.1016/s0092-8674(01)00244-6. [DOI] [PubMed] [Google Scholar]
- 37.Bowerman M., Vincent T., Scamps F., Perrin F.E., Camu W., Raoul C. Neuroimmunity dynamics and the development of therapeutic strategies for amyotrophic lateral sclerosis. Front Cell Neurosci. 2013;7:214. doi: 10.3389/fncel.2013.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krieger C., Lanius R.A., Pelech S.L., Shaw C.A. Amyotrophic lateral sclerosis: the involvement of intracellular Ca2+ and protein kinase C. Trends Pharmacol Sci. 1996;17:114–120. doi: 10.1016/0165-6147(96)10004-3. [DOI] [PubMed] [Google Scholar]
- 39.Lanius R.A., Paddon H.B., Mezei M., Wagey R., Krieger C., Pelech S.L. A role for amplified protein kinase C activity in the pathogenesis of amyotrophic lateral sclerosis. J Neurochem. 1995;65:927–930. doi: 10.1046/j.1471-4159.1995.65020927.x. [DOI] [PubMed] [Google Scholar]
- 40.Battaglia G., Bruno V. Metabotropic glutamate receptor involvement in the pathophysiology of amyotrophic lateral sclerosis: new potential drug targets for therapeutic applications. Curr Opin Pharmacol. 2018 doi: 10.1016/j.coph.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 41.Chiò A., Logroscino G., Traynor B., Collins J., Simeone J., White L.A. Global epidemiology of amyotrophic lateral sclerosis (ALS): a systematic review of the literature. Amyotroph Lateral Scler. 2012;13:126. [Google Scholar]
- 42.Vu L.T., Bowser R. Fluid-based biomarkers for amyotrophic lateral sclerosis. Neurotherapeutics. 2017 doi: 10.1007/s13311-016-0503-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marin B., Boumédiene F., Logroscino G., Couratier P., Babron M.-C., Leutenegger A.L. Variation in worldwide incidence of amyotrophic lateral sclerosis: a meta-analyss. Int J Epidemiol. 2016 doi: 10.1093/ije/dyw061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts A.L., Johnson N.J., Chen J.T., Cudkowicz M.E., Weisskopf M.G. Race/ethnicity, socioeconomic status, and ALS mortality in the United States. Neurology. 2016;87:2300–2308. doi: 10.1212/WNL.0000000000003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feinberg A.P. The key role of epigenetics in human disease prevention and mitigation. N Engl J Med. 2018;378:1323–1334. doi: 10.1056/NEJMra1402513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prattichizzo F., Micolucci L., Cricca M., De Carolis S., Mensà E., Ceriello A. Exosome-based immunomodulation during aging: a nano-perspective on inflamm-aging. Mech Ageing Dev. 2017 doi: 10.1016/j.mad.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 47.Ratajczak M.Z., Ratajczak J. Horizontal transfer of RNA and proteins between cells by extracellular microvesicles: 14 years later. Clin Transl Med. 2016 doi: 10.1186/s40169-016-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li D.W., Liu M., Cui B., Fang J., Guan Y.Z., Ding Q. The Awaji criteria increases the diagnostic sensitivity of the revised El Escorial criteria for amyotrophic lateral sclerosis diagnosis in a Chinese population. PLoS One. 2017:12. doi: 10.1371/journal.pone.0171522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oeckl P., Jardel C., Salachas F., Lamari F., Andersen P.M., Bowser R. Multicenter validation of CSF neurofilaments as diagnostic biomarkers for ALS. Amyotroph Lateral Scler Front Degener. 2016;17:404–413. doi: 10.3109/21678421.2016.1167913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ALS STRING network in .csv format.
ALS STRING network in XML format.
Clinical annotations for the blind independent sample cohort provided by Oxford University.