Abstract
Cytokines are critical mediators of inflammation and host immune defense. Cytokine production is regulated at both transcriptional and post-transcriptional levels. Post-transcriptional damping of inflammatory mRNAs is mediated by a set of RNA binding proteins (RBPs) interacting with cis-elements, such as AU-rich elements (ARE) and stem-loop structures. Whereas ARE-binding proteins such as tristetraprolin and a stem-loop recognizing protein, Roquin, downregulate cytokine mRNA abundance by recruiting a CCR4-NOT deadenylase complex, another stem-loop RBP, Regnase-1, acts as an endoribonuclease, directly degrading target cytokine mRNAs. These RBPs control translation-active or -inactive mRNAs in distinct intracellular locations. The presence of various RBPs regulating mRNAs in distinct locations enables elaborate control of cytokines under inflammatory conditions. Dysregulation of cytokine mRNA decay leads to pathologies such as the development of autoimmune diseases or impaired activation of immune responses. Here we review current knowledge about the post-transcriptional regulation of immune responses by RBPs and the importance of their alteration during inflammatory pathology and autoimmunity.
Keywords: innate immunity, cytokines, adaptive immunity, inflammation, mRNA decay, translation
Introduction
Inflammation is an essential component of immune responses that allows multicellular organisms to inhibit detrimental stimuli. Inflammation is mediated by pro-inflammatory cytokines such as tumor necrosis factor (TNF), interleukin-6 (IL-6) and IL-1β. Cytokine expression is tightly regulated in innate immune cells such as macrophages and dendritic cells (DCs), controlling their activation and maturation.1–3) Cytokine mRNA expression is controlled at both transcriptional and post-transcriptional levels. Cytokine mRNA is rapidly induced in response to a set of pattern-recognition receptors (PRRs) such as Toll-like receptors (TLRs), RIG-I-like receptors (RLRs) and Nod-like receptors (NLRs).3–5) PRRs recognize molecular structures that are broadly shared by pathogens, known as pathogen-associated molecular patterns (PAMPs).6,7) Upon PAMP recognition, PRRs initiate a series of signaling pathways that lead to the activation of transcription factors, including NF-κB, AP-1, interferon (IFN)-regulatory factors (IRFs) and CCAAT/enhancer-binding protein β (C/EBPβ). In addition to eliciting inflammation, PRR signaling simultaneously induces maturation of DCs, which are responsible for stimulating the second line of host defense, adaptive immunity.
Activated transcription factors transcribe a family of mRNAs encoding pro-inflammatory cytokines that are translated to proteins, resulting in the secretion of the cytokines. When stimulation ceases, the cytokine-mediated inflammatory response is rapidly terminated, and damaged tissues are repaired. Because overproduction of cytokines by immune cells leads to autoimmune pathogenesis,3,8) post-transcriptional damping of cytokine expression is a critical step for the resolution of inflammation and prevention of unintended tissue damage.9–14) Post-transcriptional regulation determines the fate of mRNA in association with RNA-binding proteins (RBPs), including tristetraprolin (TTP), ARE/poly-(U) binding degradation factor 1 (AUF1), Regnase-1 (also known as Zc3h12a and Mcpip1), Roquin and AT-rich interactive domain-containing 5a (Arid5a).10,15) In addition to direct RBP-mediated regulation, microRNAs, through cooperation with RBPs, are important regulators of gene expression in the immune system.16,17) Most of the regulatory RBPs are known to interact with cis-elements present in the 3′ untranslated regions (3′ UTRs) of mRNAs, and stabilize or destabilize the target transcript.18–20) AU-rich elements (AREs) and stem-loop structures in 3′ UTRs are well-studied examples of cis-elements recognized by a set of RBPs for controlling the stability of target mRNAs (Fig. 1). Recognition of AREs by ARE-binding proteins (ARE-BPs), including TTP, AUF1 and HuR, regulates the decay of mRNAs via the recruitment of the CCR4-NOT deadenylase complex. In contrast hand, stem-loop structures are recognized by distinct sets of RBPs, including Regnase-1, Roquin and Arid5a.9,21–26) Whereas Regnase-1 and Roquin are critical for the maintenance of immune homeostasis and prevention of autoimmunity in mice, Arid5a plays a role in sustaining immune responses. In this review, we will discuss the post-transcriptional mechanisms of immune responses by RBPs such as TTP, Roquin and Regnase-1, and also focus on how Regnase-1 regulates inflammatory mRNA stability to control immune responses.
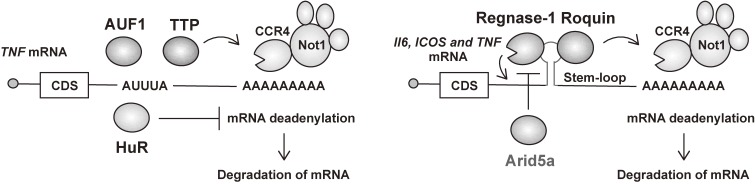
Figure 1.
Post-transcriptional regulation of inflammation-related mRNAs by RBPs. Regulatory RBPs target the 3′ UTRs of mRNAs to repress expression of the target transcript. AREs and stem-loop structures in 3′ UTRs are well-studied cis-elements recognized by a set of RBPs for controlling the stability of target mRNAs. Whereas TTP, AUF1 and HuR recognize AREs, Regnase-1, Roquin and Arid5a bind to stem-loop structures. CDS, coding sequence.
Cis-elements recognized by RBPs for regulation of inflammatory mRNAs
The regulation of mRNA stability is important for the control of gene expression, and is determined by cis-acting sequences in the 3′ UTRs that promote mRNA degradation. Two well-characterized motifs present within the 3′ UTR of mRNAs are the ARE and the stem-loop structure. Clusters of AREs in mRNAs encoding cytokines were first identified over 25 years ago,27) and subsequent studies showed that inflammatory mRNAs are subject to ARE-mediated decay.28,29) AREs provide binding sites for trans-acting RBPs such as TTP, AUF1 and HuR, which subsequently regulate the stability and translation of mRNA. ARE basic motifs include pentamers of AUUUA, nonamers of UUAUUUAUU, and AU-rich clusters composed of linked pentamers and/or nonamers.30) AREs are located at the 3′ UTR of mRNA transcripts for cytokines such as granulocyte-macrophage colony-stimulating factor (GM-CSF), TNF, IL-2, IL-3, IL-6 and IL-8, as well as pro-inflammatory factors like cyclooxygenase 2 (COX2). In addition to AREs, stem-loop structures with a pyrimidine-purine-pyrimidine (Py-Pu-Py) tri-loop sequence in 3′ UTR of inflammatory mRNAs are also recognized by a set of RBPs, including Regnase-1, Roquin and Arid5a, for controlling degradation of mRNA.23,24) Structural studies show that the N-terminal ROQ domain in Roquin tightly interacts with the RNA stem and its tri-loop in the TNF 3′ UTR.25,26) Stem-loop structures are located at the 3′ UTRs of immune-related mRNAs, including TNF, IL6, COX2 (also known as PTGS2), NFKBIZ and NFKBID.23,24) mRNAs harboring AREs and stem-loop structures are prone to quick degradation in immune cells. Thus, AREs and stem-loop structures are critical elements in controlling gene expression at the post-transcriptional level.
Regulation of cytokine production by TTP
TTP is a well-characterized zinc-finger protein that regulates immune functions.31–34) TTP transcription is induced by inflammatory modulators such as TNF, LPS and IFN-γ. TTP-deficient mice develop arthritis, dermatitis and cachexia, caused by the spontaneous overexpression of pro-inflammatory cytokines, particularly TNF, suggesting that TTP is involved in the damping of inflammation. Treatment of these mice with TNF-specific antibodies reverses almost all the pathology, implicating dysregulated cytokine expression as a principal cause of this phenotype. Subsequent biochemical studies showed that TTP decreases TNF mRNA stability by binding to its ARE,32,35) which is further confirmed by structural analysis.36) TTP harbors two CCCH-type zinc-finger domains and associates with mRNAs via AREs present in the 3′ UTRs, leading to removal of the poly(A) tail by recruitment of a CCR4-NOT deadenylase complex.
Deadenylation promotes rapid mRNA degradation that is thought to occur in processing bodies (PBs), small cytoplasmic foci that contain many enzymes required for mRNA decay. Cells exposed to various stresses such as heat shock, oxidative stress or glucose deprivation promote the assembly of stress granules (SGs), which are small cytoplasmic foci that harbor translationally-arrested mRNAs, stalled translation initiation factors and ARE-binding proteins such as T-cell-restricted intracellular antigen-1 (TIA-1) and TIA-1-related protein (TIAR) that suppress translation.37) Under conditions of stress, TTP-bound mRNAs are recruited to SGs and the translational repressor TIA-1 stalls translation,38–40) and TTP facilitates delivery of selected mRNAs from SGs to PBs for degradation, suggesting a TTP-dependent dynamic relationship between SGs and PBs. Although the exact mechanism of cytoplasmic TTP-dependent mRNA turnover remains unclear, TTP is a key player in post-transcriptional gene regulation, particularly with regard to the regulation of ARE-mediated decay of cytokine mRNAs.
Regulation of T cell activation by Roquin
The Roquin family proteins, Roquin-1 and Roquin-2, bind to mRNAs involved in the activation of both innate and acquired immune systems, and induce their degradation. Roquin-1 was identified through an N-ethyl-N-nitrosourea (ENU)-induced mutagenesis screen in mice, in which a single point mutation changed Methionine 199 to Arginine (Arg).41) A loss of function mutation in the Roquin-1 ROQ domain (M199R) by the ENU mutagenesis in mice (the so-called sanroque strain, Roquin-1San/San mice) leads to the development of autoimmune disease characterized by an increase in follicular helper T (Tfh) cells.41) Roquin-1 harbors a ROQ domain and a CCCH-type zinc finger domain. Roquin-1 recognizes stem-loop motifs in the 3′ UTRs of its target mRNAs via its ROQ domain and adjacent CCCH-type zinc finger domain, which is further confirmed by structural analysis.25,26,42) Roquin-1 promotes mRNA decay by recruiting the CCR4-NOT deadenylase complex or recruiting a EDC4 decapping protein.23,43)
The second family member, Roquin-2, shares a similar structure and has some overlapping functions with Roquin-1. Unlike Roquin-1San/San mice, the ablation of Roquin-1 alone did not cause autoimmunity, although induced expansion of macrophages and T cells were observed.44) T cell-specific deletion of Roquin-2 does not affect immune cell homeostasis.45,46) However, T cell-specific Roquin-1/2-double-deficient mice develop an autoimmune phenotype with increased Tfh cell numbers, which phenotypically resembles Roquin-1San/San mice.45,46) Roquin proteins interact with and destabilize multiple mRNAs including inducible T-cell co-stimulator (Icos), Ox40 and A20.47–49) Among them, the lack of Icos in Roquin-1San/San mice rescued the autoimmunity, indicating that Roquin-1 and Roquin-2 regulate Icos expression in T cells to prevent an aberrant increase of Tfh cells.47) Taken together, these studies reveal the importance of Roquin-1- and Roquin-2-mediated post-transcriptional regulation in Tfh cell differentiation and the maintenance of immune homeostasis.
Post-transcriptional regulation through AREs is often influenced by non-ARE sequences in the same mRNA. Indeed, Roquin-1 was shown to have a role in the regulation of TNF mRNA decay via binding to a constitutive decay element (CDE), which forms a stem-loop structure.23) After Roquin-1 binds to the CDE through its unique ROQ domain, Roquin recruits the CCR4-NOT deadenylase complex for RNA degradation. The coexistence of multiple regulatory RNA elements in a single mRNA ensures that several RBPs can work together to regulate the mRNA expression levels. This is further supported by in vivo evidence that Roquin-1San/San mice develop TNF-driven inflammation and arthritis comparable to the disease that develops in TTP-deficient mice.46) These findings reveal a complex interplay of two regulatory RBPs in the regulation of TNF production.
Regnase-1 is an essential RNase for maintenance of immune homeostasis
Regnase-1 was identified as a protein containing a CCCH zinc finger domain and a PIN (PilT N terminus)-like RNase domain (Fig. 2), and is a lipopolysaccharide (LPS, TLR4 ligand)-inducible gene.50,51) Structural studies showed that the PIN domain harbors the RNase catalytic center52,53) and in vitro cleavage assays revealed that Regnase-1 possesses endonuclease activity as shown by the digestion of circular RNAs.24,51) Mice lacking Regnase-1 (Regnase-1−/−) spontaneously develop severe autoimmune inflammatory disease, suffer from severe anemia, and mostly die within 12 weeks. Regnase-1−/− mice also show augmented serum immunoglobulin levels and autoantibody production, together with a greatly increased number of plasma cells, as well as infiltration of plasma cells to the lung. The production of IL-6 and IL-12p40 in response to TLR ligands is greater in Regnase-1−/− macrophages compared with wild-type cells.51) Although the activation of TLR signaling pathways is normal, the decay of Il-6 mRNA is impaired in Regnase-1−/− macrophages. Biochemical studies showed that Regnase-1 destabilizes mRNA via a conserved non-not ARE element present in the 3′ UTR of Il-6 mRNA. Thus, Regnase-1-mediated control of mRNA expression has an essential role in maintaining homeostasis and Regnase-1 is an essential RNase that prevents immune disorders by directly controlling the stability of a set of inflammatory mRNAs (Fig. 2).
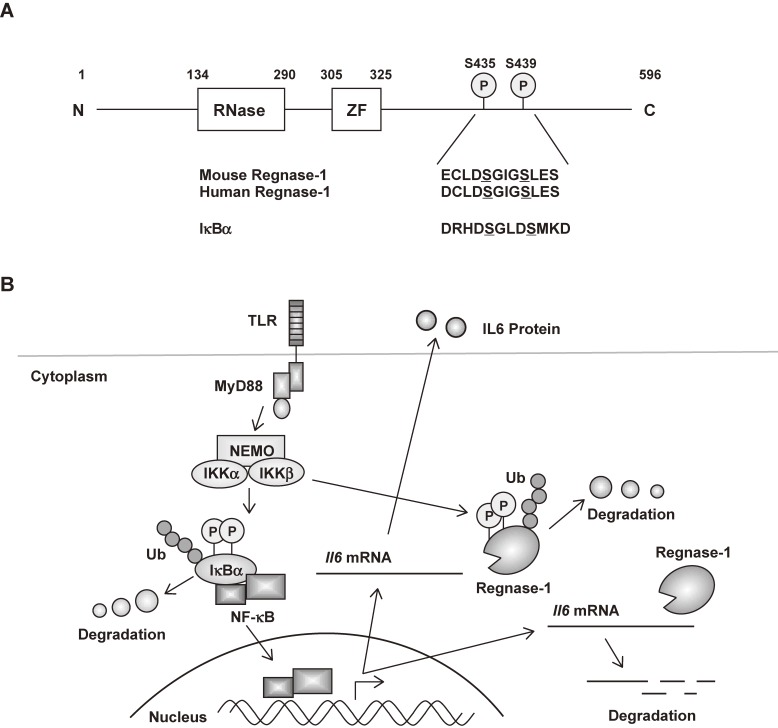
Figure 2.
Post-transcriptional regulation of Il-6 expression by Regnase-1 in innate immune cells. (A) Schematic representation of mouse Regnase-1. The sequence of the DSGXXS motif is also shown. ZF, CCCH type zinc finger. (B) In unstimulated cells, Regnase-1 suppresses Il-6 production by degrading Il-6 mRNA through its 3′ UTR. Upon TLR stimulation, IKK complex phosphorylates Regnase-1 at Ser435 and Ser439, which results in the rapid degradation of Regnase-1 protein similar to IκBα. The transcribed Il-6 mRNA is thereby stabilized, which facilitates robust Il-6 production. At a later stage, de novo synthesis of Regnase-1 leads to the degradation of Il-6 mRNA and its own mRNA, promoting the resolution of inflammation.
Phosphorylation of Regnase-1 by the IKK complex induces Regnase-1 degradation to control cytokine mRNAs
Although the expression of Regnase-1 mRNA is induced in response to stimulation with TLR ligands and IL-17A, Regnase-1 protein is expressed even in unstimulated macrophages, mouse embryonic fibroblasts (MEFs), thymus, spleen, lymph nodes and lungs.54,55) Regnase-1 protein is rapidly degraded in response to stimulation with IL-1β or TLR ligands but not TNF. Degradation of Regnase-1 protein is important for higher expression of Il-6 mRNA. Biological studies showed that inhibitors of transcription factor NF-κB (IκB) kinase (IKK) complex, which is composed of IKKα, IKKβ and IKKγ (NEMO) subunits, controls the stability of Il-6 mRNA by phosphorylating Regnase-1 in response to stimulation via IL-1 receptor (IL-1R) or TLR. Regnase-1 has the canonical DSGXXS motif known to be phosphorylated by the IKK complex, and Regnase-1 is phosphorylated by IKKβ at Ser435 and Ser439 in this motif. Phosphorylated Regnase-1 undergoes ubiquitin-proteasome-mediated degradation via the E3 ligase β-TrCP complex (also known as FBW1). Downstream of TLR/IL-1R, IRAK1 can interact with and phosphorylate Regnase-1, suggesting that modification of Regnase-1 via IRAK1 is required for inducing IKK-dependent degradation. Regnase-1 signaling is triggered by IL-17A as well as IL-36A, and both cytokines are reported to activate IKKs, inducing the degradation of Regnase-1 in keratinocytes.56) Whereas IL-36a signals through a receptor belonging to the IL-1R family, the IL-17 receptor triggers IKK activation independently of MyD88 and IRAKs. Nevertheless, it is not yet clear how IL-17 receptor signaling induces Regnase-1 degradation. Interestingly, Regnase-1 mRNA is targeted by Regnase-1 itself via a stem-loop region present in the Regnase-1 3′ UTR. These findings show that whereas the IKK complex phosphorylates IκBα to activate transcription, it also phosphorylates Regnase-1 to release a ‘brake’ on Il-6 mRNA expression (Fig. 2).
Malt1-induced cleavage of Regnase-1 in CD4+ T cells regulates immune activation
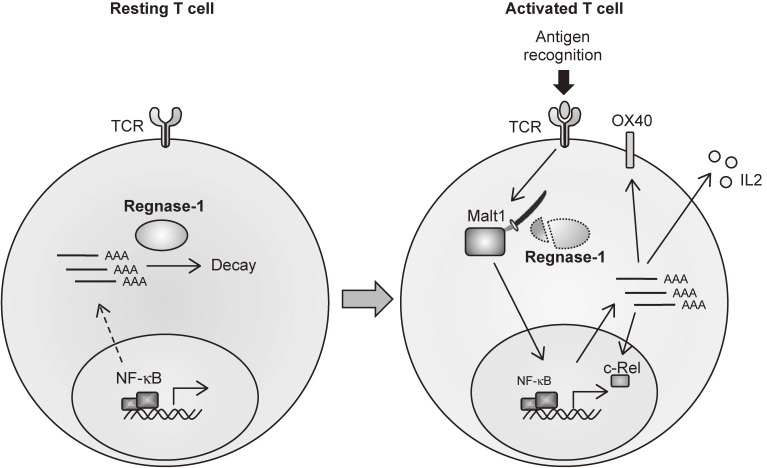
In addition to innate immune cells, Regnase-1 is also critical for the regulation of T cell activation by targeting genes such as ICOS and Ox40 for degradation.57) Most Regnase-1−/− splenic T cells show effector/memory characteristics and produce IFN-γ in response to T-cell receptor (TCR) stimulation. Lack of Regnase-1 in T cells alone leads to the development of autoimmune diseases. Aberrant activation of CD4+ T cells in peripheral tissues is responsible for the pathology of CD4-Cre+ Regnase-1fl/fl mice. In T cells, Regnase-1 directly regulates the degradation of immunoregulatory mRNAs, such as Il-2, Ox40 and c-Rel, which are important for the maintenance of normal effector functions. Lack of Regnase-1 in T cells results in the increase of Th subsets such as Th17, Th1 and Th2. Furthermore, the IL-17-mediated signaling is suppressed by Regnase-1, and heterozygous deletion of Regnase-1 in mice resulted in the exacerbation of experimental autoimmune encephalitis as well as psoriatic skin inflammation.58,59) Interestingly, Regnase-1 is cleaved at Arg111 by Malt1/paracaspase in response to TCR stimulation, facilitating T cell activation.57) These findings demonstrate that Regnase-1 is essential for suppressing unwanted T-cell-mediated immune reaction by targeting multiple mRNAs encoding transcription factors, surface molecules and cytokines (Fig. 3). Dynamic control of Regnase-1 expression in T cells is critical for controlling T cell activation. Manipulation of Malt1-mediated Regnase-1 expression appears to be an important control point for T-cell-mediated immune responses in vivo.
Figure 3.
Malt1-induced cleavage of Regnase-1 in T cells regulates immune activation. Regnase-1 regulates the mRNAs of a set of genes, including c-Rel, Ox40 and Il-2, through cleavage of their 3′ UTRs. TCR stimulation leads to cleavage of Regnase-1 at Arg111 by Malt1, resulting in a ‘brake’ of Regnase-1 function and leading to activation of T cells.
Regnase-1 and Roquin recognize mRNAs with common stem-loop structures in inflammatory mRNAs
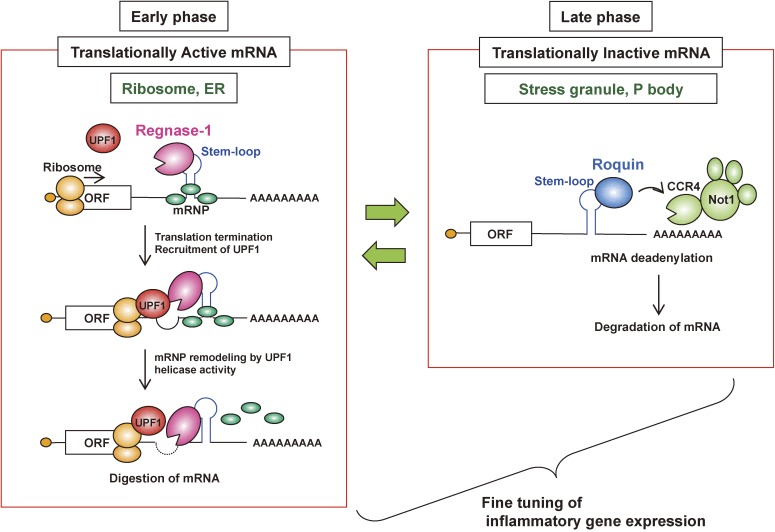
Transcriptome-wide analysis of Regnase-1-binding motifs by high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) showed that Regnase-1 recognizes stem-loop structures with varying stem lengths (3–7 nucleotides) with the Py-Pu-Py loop sequence located in their 3′ UTRs.24) Interestingly, this rule is consistent with that in the reported Roquin target mRNAs.23) Indeed, Regnase-1 and Roquin destabilize the same sets of mRNAs with the common target stem-loop sequences. Despite the presence of overlapping target mRNAs, Regnase-1 and Roquin are found to degrade inflammatory mRNAs via spatiotemporally distinct mechanisms.24) Whereas Roquin localizes to SGs and PBs, Regnase-1 localizes to the cytoplasm—especially the endoplasmic reticulum (ER)—but not to PBs and SGs. Regnase-1 destabilizes translationally active mRNAs in the polysome fraction, and translation termination is required for Regnase-1-mediated mRNA decay. Furthermore, Regnase-1 associates with UPF1, an RNA helicase essential for the mRNA quality control system called nonsense-mediated RNA decay (NMD).60) Furthermore, UPF1 helicase activity is critical for Regnase-1-mediated mRNA decay, similar to the NMD.
Regnase-1 and Roquin function non-redundantly in regulating innate and acquired immune cells. In LPS-stimulated MEFs, Regnase-1 and Roquin tend to control target inflammatory mRNAs in the early and late phase, respectively.61) Furthermore, lack of Regnase-1 and Roquin in T cells leads to the development of much severer inflammatory disease compared with mice lacking Regnase-1 or Roquin in T cells, indicating that Regnase-1 and Roquin function non-redundantly in T cells.62) In addition to Regnase-1, Roquin also undergoes Malt1-mediated degradation upon TCR stimulation, suggesting that both Regnase-1 and Roquin function as the molecular switch in controlling TCR-mediated T cell activation.25) These findings reveal that Regnase-1 and Roquin suppress aberrant immune cell activation in a non-redundant manner depending on their translation status and enable elaborate control of inflammation (Fig. 4). However, how Regnase-1 regulates translationally active mRNAs in UPF1-dependent manner requires further investigation.
Figure 4.
Regnase-1 and Roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Although Regnase-1 and Roquin regulate an overlapping set of mRNAs via a common stem-loop structure located in their 3′ UTRs, they function by spatiotemporally distinct mechanisms. Whereas Roquin destabilizes translationally inactive mRNAs by recruiting the CCR4-NOT deadenylase complex in processing-bodies (PBs) and stress granules (SGs), Regnase-1 localizes to the endoplasmic reticulum (ER) and ribosomes, destabilizes translationally active mRNAs, and requires the RNA helicase activity of UPF1, similar to NMD. Regnase-1 and Roquin tend to control mRNAs at the early and late phases of inflammation, respectively. mRNP complex, mRNA-protein complex.
Regnase-1 is critical for the maintenance of iron homeostasis
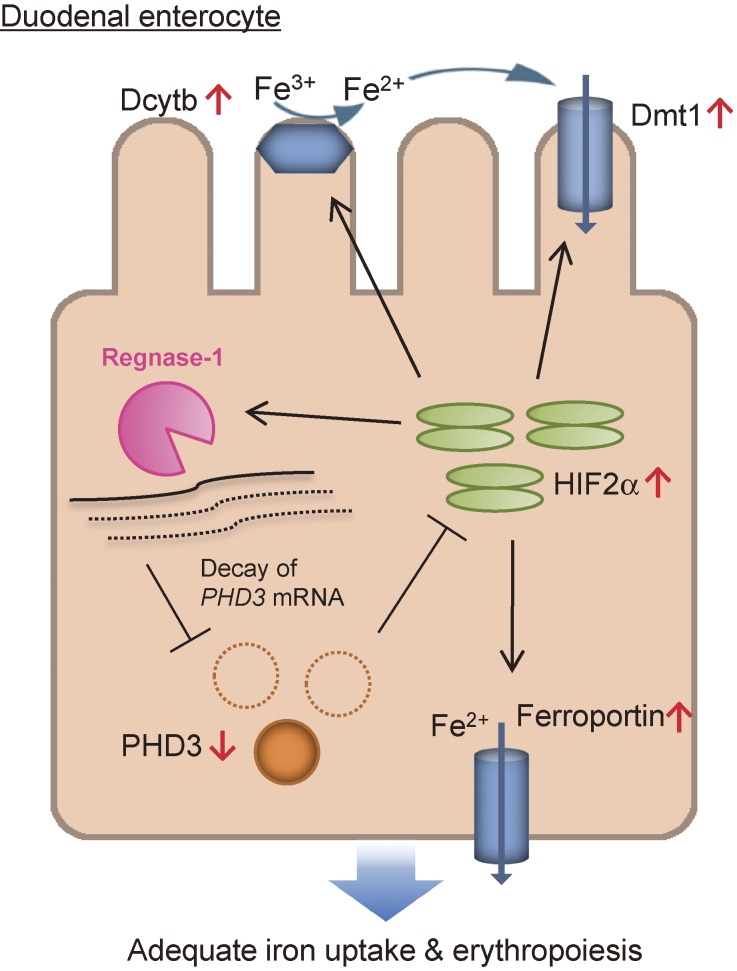
It is well known that iron metabolism is altered in the course of inflammation.63,64) We found that Regnase-1 is also critical for the degradation of mRNAs involved in iron metabolism in vivo.21) Regnase-1−/− mice suffer from severe anemia as well as autoimmune disease. Duodenal Regnase-1 regulates the expression of prolyl-hydroxylase-domain-containing protein 3 (PHD3) by targeting its 3′ UTRs. PHD3 impairs duodenal iron uptake via suppression of transcription factor hypoxia-inducible factor-2α (HIF2α), which promotes efficient duodenal iron uptake under iron-deficient conditions.65) Interestingly, HIF2α binds to the Regnase-1 promoter and induces Regnase-1 expression, providing a positive feedback loop for HIF2α activation via PHD3. In addition, Transferrin receptor (Tfrc) mRNA was found to be degraded by Regnase-1, and the competition between Regnase-1 and Iron-response proteins is critical for the control of Tfrc mRNA levels.21) These results demonstrate that Regnase-1-mediated regulation of iron-related transcripts is essential for the maintenance of iron homeostasis (Fig. 5).
Figure 5.
Model of Regnase-1-mediated regulation of iron uptake in the duodenum. Regnase-1 destabilizes PHD3 mRNA, thereby increasing the activity of HIF2α, a critical regulator of iron homeostasis in the duodenum. Accumulated HIF2α then induces the up-regulation of iron transporters and reductases (Dmt1, Ferroportin and Dcytb), leading to adequate iron uptake. Moreover, Regnase-1 itself is transcriptionally up-regulated via HIF2α under iron-deficient conditions. Therefore, the Regnase-1-PHD3-HIF2α axis forms a positive feedback loop for HIF2α amplification to regulate iron uptake in the duodenum.
Critical roles of Arid5a in stabilizing stem-loop mRNA for enhancing inflammation
Stem-loop structures with a Py-Pu-Py loop in 3′ UTR are also recognized by another RBP, Arid5a.66) In contrast to Regnase-1 and Roquin, Arid5a stabilizes target mRNAs by inhibiting the actions of Regnase-1 and Roquin. Arid5a stabilizes Il-6 mRNA in innate immune cells.66) Furthermore, Arid5a is critical for Th17 and Th1 cell polarization by stabilizing mRNAs encoding Stat3 and T-bet, respectively.67,68) Mice lacking Arid5a are resistant to LPS-induced shock due to impaired production of IFN-γ and Il-6.68) Whereas Arid5a mRNA is rapidly induced in response to LPS, p38 MAP kinase phosphorylates Arid5a resulting in its degradation during the late phases of inflammation.69) These results suggest that inflammatory mRNAs with stem-loop structures are recognized by both stabilizing and destabilizing RBPs, and their amounts are regulated by the balance of action of these RBPs.
Conclusion and perspectives
Regulation of RNA metabolism is a critical step for gene expression that facilitates the fine-tuning of mRNA expression levels during steady state as well as inflammatory conditions. Dysregulation of RNA metabolism can result in various diseases, including inflammatory and autoimmune diseases.70) RBPs have emerged as important regulators of multiple facets of RNA metabolism and immune responses. During the innate immune responses, several RBPs, such as TTP, Roquin and Regnase-1, promote the resolution of inflammation by eliminating the pro-inflammatory cytokine mRNAs. During the activation of T cells, these RBPs control the adaptive immune responses and maintain immune homeostasis. Failure of these regulatory controls leads to autoimmune syndromes.
The post-transcriptional regulation of inflammation is controlled by Regnase-1 and Roquin in a spatiotemporally distinct manner, although they recognize common stem-loop structures. It is interesting that the mechanism for the NMD quality control system is shared with that for the degradation of cytokine mRNAs. Although UPF1 is essential for Regnase-1-mediated mRNA decay, the molecular mechanisms of UPF1 involvement are still unclear. Because excess and prolonged production of cytokines leads to the onset of inflammatory diseases, prolonged stability of inflammatory cytokine mRNAs may be considered aberrant, and thus targeted by a similar mechanism of the NMD. Strict control over the life of mRNA by RBPs is a key strategic step by which immune cells determine their phenotypes and functions, and differential regulation of Regnase-1- and Roquin-mediated mRNA degradation are thus necessary for the precise control of inflammation.
Regnase-1 is also proposed to harbor the potential to control virus infection by degrading viral RNAs.71,72) In addition, Regnase-1-like RNases are conserved among species, and the Caenorhabditis elegans homologue, REGE-1, is involved in the control of body fat by degrading an mRNA for transcription factor ETS-4.73) Although further in vivo studies in mammals are required, it is possible that Regnase-1-mediated degradation of RNAs controls broad biological phenomena beyond the control of immunity.
Recent studies on post-transcriptional regulation have uncovered novel players in the control of immune reactions. However, recent RBPome studies revealed that there are more than 1500 RBPs in humans, the functions of most of which are yet to be uncovered.74,75) For instance, the roles of Regnase-1 family proteins, Zc3h12b, c and d, in immune reactions are still poorly understood. Post-transcriptional regulation is also critical for the cell fate decision in immune cell development,15) indicating that the involvement of RBPs in the entire immune system is highly complex. Further studies may reveal totally novel regulatory mechanisms underlying the post-transcriptional regulation of immune reactions.
Given that Regnase-1 is critical for regulating inflammation and adaptive immunity, it is plausible that immune responses can be manipulated by interfering with or activating the functions of Regnase-1 and Roquin. Exploration of small molecules affecting Regnase-1 activity might be beneficial for the development of novel vaccine adjuvants or anti-tumor immunotherapy drugs.
Acknowledgments
This work was supported by AMED-CREST, AMED under Grant Number JP16gm0410017, and the JSPS through Core-to-Core Program. T.M. was funded by JSPS KAKENHI (16K08832), Grant-in-Aid for Scientific Research on Innovative Areas “Genome Science” (221S0002 and 16H06279), Takeda Science Foundation, the Uehara Memorial Foundation, Shimizu Foundation for Immunology and Neuroscience, Naito Foundation, Senri Life Science Foundation, Nakajima Foundation and Mochida Memorial Foundation for Medical and Pharmaceutical Research. O.T. was funded by Daiichi Sankyo Foundation of Life Science, Takeda Science Foundation and the Uehara Memorial Foundation.
Abbreviations
- Regnase-1
Regulatory RNase 1
- ARE
adenine- and uridine-rich element
- AUF1
ARE/poly-(U) binding degradation factor 1
- Dendritic cells
DCs
- TLRs
Toll-like receptors
- TNF
tumor necrosis factor
- interleukin-6
IL6
- TTP
tristetraprolin
- PAMPs
pathogen-associated molecular patterns
- PRRs
pattern-recognition receptors
- RBPs
RNA-binding proteins
- RLRs
Retinoic acid-Inducible Gene-I-like receptors
- IFN
Interferon
- IRFs
IFN-regulatory factors
- C/EBPβ
CCAAT/enhancer-binding protein β
- TIA-1
T-cell-restricted intracellular antigen-1
- TIAR
TIA-1-related protein
- Tfh cells
Follicular helper T cells
- PBs
Processing bodies
- SGs
Stress granules
- LPS
Lipopolysaccharide
- Py-Pu-Py
pyrimidine-purine-pyrimidine
- Arid5a
AT-rich interactive domain-containing 5a
Profile
Osamu Takeuchi was born in Fukui Prefecture in 1970 and graduated from Osaka University Medical School in 1995. After clinical training in the third department of internal medicine in Osaka University, he entered the graduate school of medicine, Osaka University, where he started working on innate immunity under the supervision of S. Akira. He discovered that different Toll-like receptors (TLRs) recognize different microbial components, and received Ph.D. degree in 2001. He subsequently completed postdoctoral training at Dana-Farber Cancer Institute under S. Korsmeyer. Then he became an assistant professor at Osaka University, and worked on the functional roles of TLR signaling molecules and RIG-I-like receptors in inflammation. In 2012, he moved to the Institute for Virus Research, Kyoto University, as a full professor. He is currently focusing on the posttranscriptional regulation of inflammation by a set of RNA binding proteins at the Institute for Frontier Life and Medical Sciences, Kyoto University. He received the JSPS Prize and Japan Academy Medal on 2016. He has been listed in Highly Cited Researchers from 2014 to 2017.
References
- 1).Chovatiya R., Medzhitov R. (2014) Stress, inflammation, and defense of homeostasis. Mol. Cell 54, 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Moresco E.M., LaVine D., Beutler B. (2011) Toll-like receptors. Curr. Biol. 21, R488–R493. [DOI] [PubMed] [Google Scholar]
- 3).Takeuchi O., Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820. [DOI] [PubMed] [Google Scholar]
- 4).Beutler B. (2009) Microbe sensing, positive feedback loops, and the pathogenesis of inflammatory diseases. Immunol. Rev. 227, 248–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Medzhitov R., Horng T. (2009) Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 9, 692–703. [DOI] [PubMed] [Google Scholar]
- 6).Janeway C.A., Jr. (1989) Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54 (Pt 1), 1–13. [DOI] [PubMed] [Google Scholar]
- 7).Akira S., Uematsu S., Takeuchi O. (2006) Pathogen recognition and innate immunity. Cell 124, 783–801. [DOI] [PubMed] [Google Scholar]
- 8).Marshak-Rothstein A. (2006) Toll-like receptors in systemic autoimmune disease. Nat. Rev. Immunol. 6, 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Fu M., Blackshear P.J. (2017) RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat. Rev. Immunol. 17, 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Anderson P. (2010) Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat. Rev. Immunol. 10, 24–35. [DOI] [PubMed] [Google Scholar]
- 11).Kafasla P., Skliris A., Kontoyiannis D.L. (2014) Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat. Immunol. 15, 492–502. [DOI] [PubMed] [Google Scholar]
- 12).Mino T., Takeuchi O. (2013) Post-transcriptional regulation of cytokine mRNA controls the initiation and resolution of inflammation. Biotechnol. Genet. Eng. Rev. 29, 49–60. [DOI] [PubMed] [Google Scholar]
- 13).Uehata T., Akira S. (2013) mRNA degradation by the endoribonuclease Regnase-1/ZC3H12a/MCPIP-1. Biochim. Biophys. Acta 1829, 708–713. [DOI] [PubMed] [Google Scholar]
- 14).Uehata T., Takeuchi O. (2017) Regnase-1 is an endoribonuclease essential for the maintenance of immune homeostasis. J. Interferon Cytokine Res. 37, 220–229. [DOI] [PubMed] [Google Scholar]
- 15).Turner M., Diaz-Munoz M.D. (2018) RNA-binding proteins control gene expression and cell fate in the immune system. Nat. Immunol. 19, 120–129. [DOI] [PubMed] [Google Scholar]
- 16).Alam M.M., O’Neill L.A. (2011) MicroRNAs and the resolution phase of inflammation in macrophages. Eur. J. Immunol. 41, 2482–2485. [DOI] [PubMed] [Google Scholar]
- 17).Turner M., Galloway A., Vigorito E. (2014) Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat. Immunol. 15, 484–491. [DOI] [PubMed] [Google Scholar]
- 18).Anderson P. (2008) Post-transcriptional control of cytokine production. Nat. Immunol. 9, 353–359. [DOI] [PubMed] [Google Scholar]
- 19).von Roretz C., Gallouzi I.E. (2008) Decoding ARE-mediated decay: is microRNA part of the equation? J. Cell Biol. 181, 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Shyu A.B., Wilkinson M.F., van Hoof A. (2008) Messenger RNA regulation: to translate or to degrade. EMBO J. 27, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Yoshinaga M., Nakatsuka Y., Vandenbon A., Ori D., Uehata T., Tsujimura T., et al. (2017) Regnase-1 maintains iron homeostasis via the degradation of transferrin receptor 1 and prolyl-hydroxylase-domain-containing protein 3 mRNAs. Cell Rep. 19, 1614–1630. [DOI] [PubMed] [Google Scholar]
- 22).Codutti L., Leppek K., Zalesak J., Windeisen V., Masiewicz P., Stoecklin G., et al. (2015) A distinct, sequence-induced conformation is required for recognition of the constitutive decay element RNA by Roquin. Structure 23, 1437–1447. [DOI] [PubMed] [Google Scholar]
- 23).Leppek K., Schott J., Reitter S., Poetz F., Hammond M.C., Stoecklin G. (2013) Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell 153, 869–881. [DOI] [PubMed] [Google Scholar]
- 24).Mino T., Murakawa Y., Fukao A., Vandenbon A., Wessels H.H., Ori D., et al. (2015) Regnase-1 and Roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell 161, 1058–1073. [DOI] [PubMed] [Google Scholar]
- 25).Schlundt A., Heinz G.A., Janowski R., Geerlof A., Stehle R., Heissmeyer V., et al. (2014) Structural basis for RNA recognition in roquin-mediated post-transcriptional gene regulation. Nat. Struct. Mol. Biol. 21, 671–678. [DOI] [PubMed] [Google Scholar]
- 26).Tan D., Zhou M., Kiledjian M., Tong L. (2014) The ROQ domain of Roquin recognizes mRNA constitutive-decay element and double-stranded RNA. Nat. Struct. Mol. Biol. 21, 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A. (1986) Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl. Acad. Sci. U.S.A. 83, 1670–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Shaw G., Kamen R. (1986) A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46, 659–667. [DOI] [PubMed] [Google Scholar]
- 29).Hamilton T.A., Novotny M., Datta S., Mandal P., Hartupee J., Tebo J., et al. (2007) Chemokine and chemoattractant receptor expression: post-transcriptional regulation. J. Leukoc. Biol. 82, 213–219. [DOI] [PubMed] [Google Scholar]
- 30).Wilusz C.J., Wormington M., Peltz S.W. (2001) The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2, 237–246. [DOI] [PubMed] [Google Scholar]
- 31).Sanduja S., Blanco F.F., Dixon D.A. (2011) The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip. Rev. RNA 2, 42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Carballo E., Lai W.S., Blackshear P.J. (1998) Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281, 1001–1005. [DOI] [PubMed] [Google Scholar]
- 33).Lai W.S., Carballo E., Strum J.R., Kennington E.A., Phillips R.S., Blackshear P.J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 19, 4311–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Taylor G.A., Carballo E., Lee D.M., Lai W.S., Thompson M.J., Patel D.D., et al. (1996) A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4, 445–454. [DOI] [PubMed] [Google Scholar]
- 35).Lai W.S., Carballo E., Thorn J.M., Kennington E.A., Blackshear P.J. (2000) Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J. Biol. Chem. 275, 17827–17837. [DOI] [PubMed] [Google Scholar]
- 36).Hudson B.P., Martinez-Yamout M.A., Dyson H.J., Wright P.E. (2004) Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 11, 257–264. [DOI] [PubMed] [Google Scholar]
- 37).Anderson P., Kedersha N. (2008) Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141–150. [DOI] [PubMed] [Google Scholar]
- 38).Phillips K., Kedersha N., Shen L., Blackshear P.J., Anderson P. (2004) Arthritis suppressor genes TIA-1 and TTP dampen the expression of tumor necrosis factor alpha, cyclooxygenase 2, and inflammatory arthritis. Proc. Natl. Acad. Sci. U.S.A. 101, 2011–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Murata T., Morita N., Hikita K., Kiuchi K., Kaneda N. (2005) Recruitment of mRNA-destabilizing protein TIS11 to stress granules is mediated by its zinc finger domain. Exp. Cell Res. 303, 287–299. [DOI] [PubMed] [Google Scholar]
- 40).Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., et al. (2005) Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Vinuesa C.G., Cook M.C., Angelucci C., Athanasopoulos V., Rui L., Hill K.M., et al. (2005) A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature 435, 452–458. [DOI] [PubMed] [Google Scholar]
- 42).Schuetz A., Murakawa Y., Rosenbaum E., Landthaler M., Heinemann U. (2014) Roquin binding to target mRNAs involves a winged helix-turn-helix motif. Nat. Commun. 5, 5701. [DOI] [PubMed] [Google Scholar]
- 43).Glasmacher E., Hoefig K.P., Vogel K.U., Rath N., Du L., Wolf C., et al. (2010) Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat. Immunol. 11, 725–733. [DOI] [PubMed] [Google Scholar]
- 44).Bertossi A., Aichinger M., Sansonetti P., Lech M., Neff F., Pal M., et al. (2011) Loss of Roquin induces early death and immune deregulation but not autoimmunity. J. Exp. Med. 208, 1749–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Vogel K.U., Edelmann S.L., Jeltsch K.M., Bertossi A., Heger K., Heinz G.A., et al. (2013) Roquin paralogs 1 and 2 redundantly repress the Icos and Ox40 costimulator mRNAs and control follicular helper T cell differentiation. Immunity 38, 655–668. [DOI] [PubMed] [Google Scholar]
- 46).Pratama A., Ramiscal R.R., Silva D.G., Das S.K., Athanasopoulos V., Fitch J., et al. (2013) Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity 38, 669–680. [DOI] [PubMed] [Google Scholar]
- 47).Yu D., Tan A.H., Hu X., Athanasopoulos V., Simpson N., Silva D.G., et al. (2007) Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature 450, 299–303. [DOI] [PubMed] [Google Scholar]
- 48).Janowski R., Heinz G.A., Schlundt A., Wommelsdorf N., Brenner S., Gruber A.R., et al. (2016) Roquin recognizes a non-canonical hexaloop structure in the 3′-UTR of Ox40. Nat. Commun. 7, 11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Murakawa Y., Hinz M., Mothes J., Schuetz A., Uhl M., Wyler E., et al. (2015) RC3H1 post-transcriptionally regulates A20 mRNA and modulates the activity of the IKK/NF-kappaB pathway. Nat. Commun. 6, 7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Takeuchi O. (2018) Endonuclease Regnase-1/Monocyte chemotactic protein-1-induced protein-1 (MCPIP1) in controlling immune responses and beyond. Wiley Interdiscip. Rev. RNA 9, doi:10.1002/wrna.1449. [DOI] [PubMed] [Google Scholar]
- 51).Matsushita K., Takeuchi O., Standley D.M., Kumagai Y., Kawagoe T., Miyake T., et al. (2009) Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458, 1185–1190. [DOI] [PubMed] [Google Scholar]
- 52).Xu J., Peng W., Sun Y., Wang X., Xu Y., Li X., et al. (2012) Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase. Nucleic Acids Res. 40, 6957–6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Yokogawa M., Tsushima T., Noda N.N., Kumeta H., Enokizono Y., Yamashita K., et al. (2016) Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions. Sci. Rep. 6, 22324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54).Iwasaki H., Takeuchi O., Teraguchi S., Matsushita K., Uehata T., Kuniyoshi K., et al. (2011) The IkappaB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat. Immunol. 12, 1167–1175. [DOI] [PubMed] [Google Scholar]
- 55).Ruiz-Romeu E., Ferran M., Gimenez-Arnau A., Bugara B., Lipert B., Jura J., et al. (2016) MCPIP1 RNase is aberrantly distributed in psoriatic epidermis and rapidly induced by IL-17A. J. Invest. Dermatol. 136, 1599–1607. [DOI] [PubMed] [Google Scholar]
- 56).Takaishi M., Satoh T., Akira S., Sano S. (2018) Regnase-1, an immuno-modulator, limits the IL-36/IL-36R auto-stimulatory loop in keratinocytes to suppress skin inflammation. J. Invest. Dermatol. doi:10.1002/wrna.1449. [DOI] [PubMed] [Google Scholar]
- 57).Uehata T., Iwasaki H., Vandenbon A., Matsushita K., Hernandez-Cuellar E., Kuniyoshi K., et al. (2013) Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell 153, 1036–1049. [DOI] [PubMed] [Google Scholar]
- 58).Garg A.V., Amatya N., Chen K., Cruz J.A., Grover P., Whibley N., et al. (2015) MCPIP1 endoribonuclease activity negatively regulates Interleukin-17-mediated signaling and inflammation. Immunity 43, 475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59).Monin L., Gudjonsson J.E., Childs E.E., Amatya N., Xing X.Y., Verma A.H., et al. (2017) MCPIP1/Regnase-1 restricts IL-17A- and IL-17C-dependent skin inflammation. J. Immunol. 198, 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60).Popp M.W., Maquat L.E. (2013) Organizing principles of mammalian nonsense-mediated mRNA decay. Annu. Rev. Genet. 47, 139–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61).Mino T., Takeuchi O. (2015) Regnase-1 and Roquin regulate inflammatory mRNAs. Oncotarget 6, 17869–17870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62).Cui X., Mino T., Yoshinaga M., Nakatsuka Y., Hia F., Yamasoba D., et al. (2017) Regnase-1 and Roquin Nonredundantly Regulate Th1 Differentiation Causing Cardiac Inflammation and Fibrosis. J. Immunol. 199, 4066–4077. [DOI] [PubMed] [Google Scholar]
- 63).Ganz T., Nemeth E. (2015) Iron homeostasis in host defence and inflammation. Nat. Rev. Immunol. 15, 500–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64).Muckenthaler M.U., Rivella S., Hentze M.W., Galy B. (2017) A red carpet for iron metabolism. Cell 168, 344–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65).Shah Y.M., Xie L. (2014) Hypoxia-inducible factors link iron homeostasis and erythropoiesis. Gastroenterology 146, 630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).Masuda K., Ripley B., Nishimura R., Mino T., Takeuchi O., Shioi G., et al. (2013) Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc. Natl. Acad. Sci. U.S.A. 110, 9409–9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67).Masuda K., Ripley B., Nyati K.K., Dubey P.K., Zaman M.M., Hanieh H., et al. (2016) Arid5a regulates naive CD4+ T cell fate through selective stabilization of Stat3 mRNA. J. Exp. Med. 213, 605–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68).Zaman M.M., Masuda K., Nyati K.K., Dubey P.K., Ripley B., Wang K., et al. (2016) Arid5a exacerbates IFN-gamma-mediated septic shock by stabilizing T-bet mRNA. Proc. Natl. Acad. Sci. U.S.A. 113, 11543–11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69).Nyati K.K., Masuda K., Zaman M.M., Dubey P.K., Millrine D., Chalise J.P., et al. (2017) TLR4-induced NF-kappaB and MAPK signaling regulate the IL-6 mRNA stabilizing protein Arid5a. Nucleic Acids Res. 45, 2687–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70).Rigby R.E., Rehwinkel J. (2015) RNA degradation in antiviral immunity and autoimmunity. Trends Immunol. 36, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71).Lin R.J., Chien H.L., Lin S.Y., Chang B.L., Yu H.P., Tang W.C., et al. (2013) MCPIP1 ribonuclease exhibits broad-spectrum antiviral effects through viral RNA binding and degradation. Nucleic Acids Res. 41, 3314–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72).Liu S., Qiu C., Miao R., Zhou J., Lee A., Liu B., et al. (2013) MCPIP1 restricts HIV infection and is rapidly degraded in activated CD4+ T cells. Proc. Natl. Acad. Sci. U.S.A. 110, 19083–19088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73).Habacher C., Guo Y., Venz R., Kumari P., Neagu A., Gaidatzis D., et al. (2016) Ribonuclease-mediated control of body fat. Dev. Cell 39, 359–369. [DOI] [PubMed] [Google Scholar]
- 74).Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., et al. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406. [DOI] [PubMed] [Google Scholar]
- 75).Gerstberger S., Hafner M., Tuschl T. (2014) A census of human RNA-binding proteins. Nat. Rev. Genet. 15, 829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]