Nonalcoholic steatohepatitis (NASH) is characterized by adipose tissue dysfunction with insulin resistance and the dysregulation of adipokines.1 Recent data indicate repartitioning of iron from the liver to adipocytes in obesity and a role for iron in the development of adipose tissue dysfunction.2, 3 However, the molecular mechanisms have not been established. To test the hypothesis that iron modulates adipokine release, we performed a quantitative proteomics analysis of the human Simpson-Golabi-Behmel Syndrome (SGBS) adipocyte secretome after 48 hours of treatment with ferric ammonium citrate (FAC). We used stable isotope-labeled amino acids in cell culture (SILAC) to characterize changes in the adipocyte secretome in response to iron. This technique has enabled direct comparison of quantities of individual proteins in the adipocyte secretome in response to iron using a proteomics approach as a tool for the identification of novel treatment targets in NASH. Detailed methodology is described in Supplementary Methods.
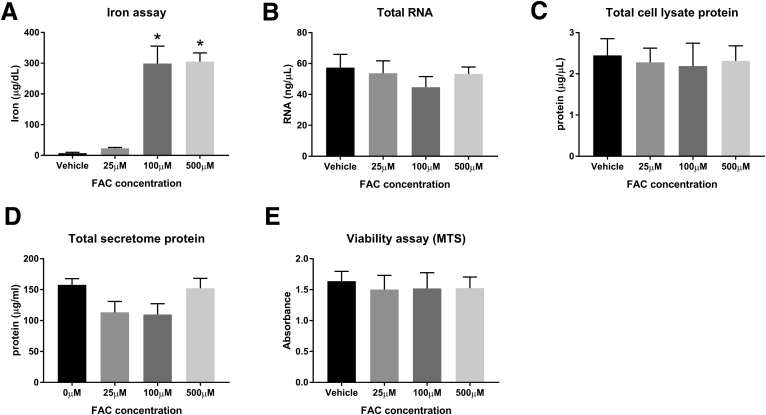
We first showed that 100 μmol/L FAC causes significant adipocyte iron loading without compromising cell viability. We found that compared with vehicle, both 100 μmol/L and 500 μmol/L FAC caused significant increases in cellular iron concentration (P = .007 and P = .006, respectively) (Supplementary Figure 1A). There was no effect of iron loading on cellular viability (MTS) assay, total messenger RNA (mRNA), total whole-cell lysate protein, or total secretome protein (Supplementary Figure 1B–E).
Supplementary Figure 1.
Optimization of iron loading in SGBS cells. (A) Iron assay, P = .004 (1-way analysis of variance [ANOVA]), *P < .01 (Dunnett multiple comparisons test compared with 0 μmol/L FAC, n = 2 per group). (B) Total RNA (P = NS by 1-way ANOVA, N = 3 per group). (C) Total lysate protein (P = NS by 1-way ANOVA, n = 3 per group). (D) Total secretome protein (P = NS by 1-way ANOVA, N = 3 per group). (E) Viability assay (MTS) (P = NS by 1-way ANOVA, N = 3 per group). Data are presented as means and SEM. MTS, [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium].
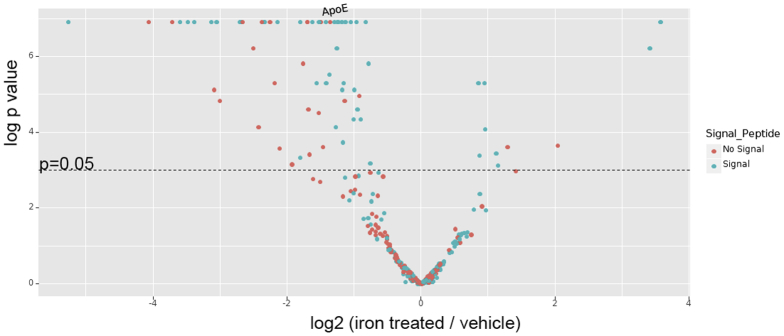
Given these findings, we selected 100 μmol/L FAC as the concentration to compare with vehicle in the secretome SILAC proteomic analysis. A total of 338 proteins were quantified in the adipocyte secretome by SILAC proteomics. These are represented by the volcano plot in Supplementary Figure 2 and the proteomics data have been deposited into the ProteomeXchange Consortium via the Proteomics Identifications (PRIDE) partner repository (www.proteomexchange.org) with the data set identifier PXD006341. Iron treatment led to significant differential secretion of 60 of these proteins (>2-fold change; P < .05). We then manually reviewed UniProt database descriptions of these 60 proteins.4 This generated a list of 20 proteins of interest (highlighted in bold in Supplementary Table 1). These proteins of interest and their synonyms then were entered into a PubMed title/abstract search in association with NASH and its synonyms. This identified 3 proteins as candidate intermediates for iron-induced adipose tissue dysfunction in NASH. These proteins were adiponectin, annexin A1, and apolipoprotein E (ApoE).
Supplementary Figure 2.
Volcano plot of relative signal intensity of proteins identified in the adipocyte secretome. The x-axis denotes log2 of the ratio of iron/vehicle-treated cells, with proteins to the left of zero representing those down-regulated by iron and those to the right representing up-regulation by iron. The y-axis denotes statistical significance with a line representing a P value of .05. Proteins above this line have a P value <.05. SILAC-labeled adipocytes generated 338 proteins that were identified in the secretome by mass spectrometry. Of these, 213 had reduced signal intensity in response to iron, whereas 125 had increased signal intensity. Of the 213 proteins with reduced signal intensity, 61 had a statistically significant (P < .05) down-regulation in response to iron. Of these, 53 had a greater than 2-fold decrease in response to iron. Of the 125 proteins with increased signal intensity, 11 had a statistically significant (P < .05) up-regulation in response to iron. Of these, 7 proteins had a greater than 2-fold increase response to iron. Those proteins containing signal peptide (as determined by signal peptide annotations on the UniProt database) are shown in red. Those without signal peptide are shown in blue.
Our SILAC analysis showed that iron treatment resulted in an 81% reduction in annexin A1 secretome signal intensity (P = .001). This may be important because annexinA1 knockout (KO) mice show greater degrees of hepatic lobular inflammation and fibrosis than controls when fed a methionine-choline–deficient diet.5 Adipocyte iron also previously has been shown to transcriptionally down-regulate serum adiponectin in mouse-derived adipocytes, 3T3-L1 cells.6 Our findings now support this in a human adipocyte cell line with a 55% reduction in adiponectin signal intensity in iron-treated SGBS cells (P = .005).
We next focused on the iron regulation of ApoE secretion. ApoE appears to protect against steatohepatitis in mice. In an ApoE KO model, unlike wild-type controls, ApoE KO mice fed 7 weeks of a Western diet developed impaired glucose tolerance, steatohepatitis, and hepatic fibrosis.7 ApoE is a component of lipoproteins, and promotes very low density lipoprotein–induced adipogenesis.8 ApoE knockout mice also readily develop atherosclerosis on an atherogenic diet.8
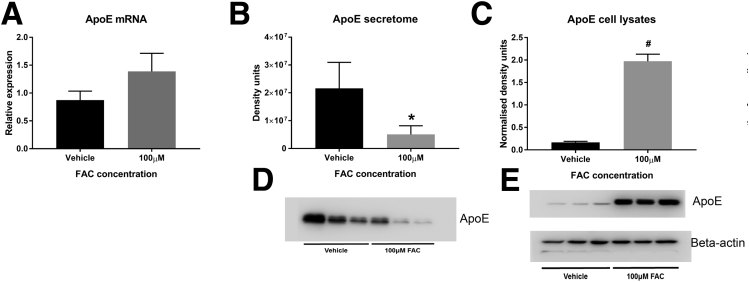
Iron reduced secreted ApoE by 58% (P = .001) and 76% (P = .007), as measured by SILAC and Western blot, respectively. Conversely, iron treatment increased intracellular ApoE levels by more than 11-fold (P = .0005), without causing a significant change in mRNA levels (Figure 1). It therefore appears that iron inhibits the secretion of ApoE from adipocytes, causing ApoE to become sequestered intracellularly.
Figure 1.
SGBS ApoE expression after FAC treatment. (A) ApoE mRNA (P = NS, ratio paired t test), (B) secretome ApoE densitometry (*P = .001, ratio paired t test), (C) lysate ApoE densitometry normalized to β-actin (#P = .0005, ratio paired t test), (D) secretome ApoE immunoblot, and (E) whole-cell lysate ApoE and β-actin immunoblots (N = 3 per group). Data are presented as means and SEM.
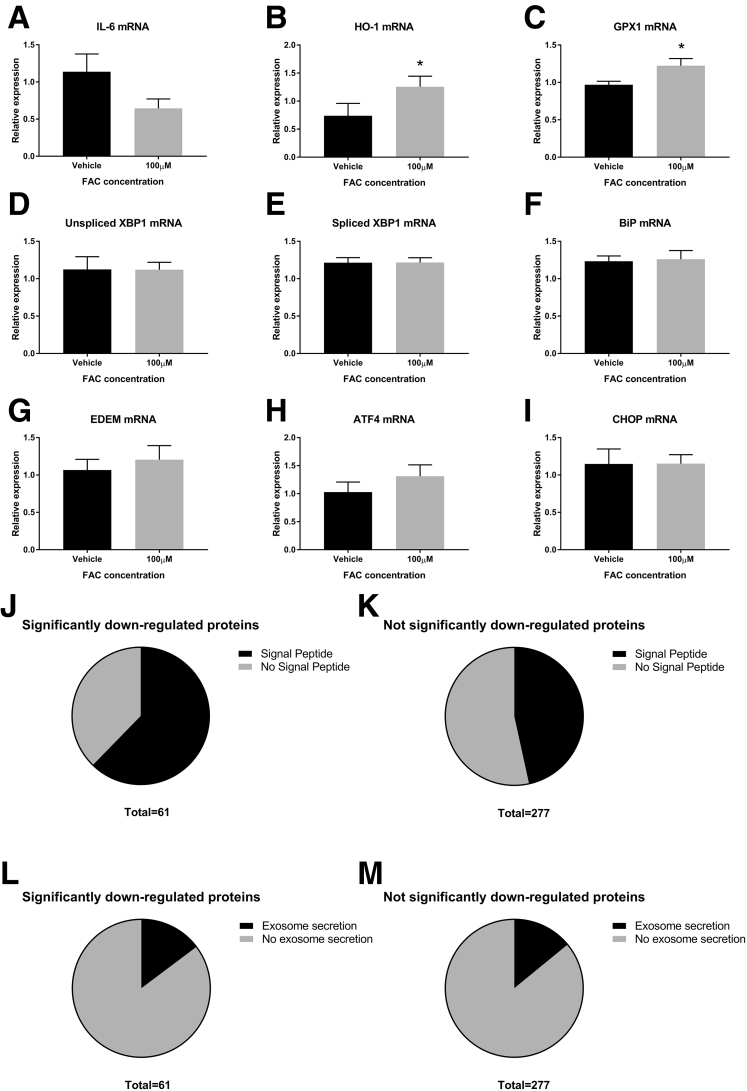
Similar effects on ApoE secretion have been shown with iron treatment in primary cultured astrocytes and cortical neurons.9 Taken together with our data, it seems possible that iron may have similar effects on a range of cell types and represents a clear target for further investigation. Treatment with iron in our study showed an up-regulation of anti-oxidant responses (heme-oxygenase-1 and glutathione peroxidase-1 mRNA), indicating the presence of oxidative stress. Interleukin 6 mRNA, however, was not increased with iron treatment, and there was no difference among multiple markers of endoplasmic reticulum stress (Supplementary Figure 3A–I).
Supplementary Figure 3.
Mechanistic aspects of iron-related dysregulation of protein secretion. (A) Interleukin 6 (IL6) mRNA (P = NS, paired t test, N = 3 per group). (B and C) Oxidative stress (*P < .05, both N = 3 per group). (B) Heme-oxygenase (HO-1) mRNA (P = .01, paired t test). (C) Glutathione peroxidase 1 (GPX1) mRNA (P = .049, paired t test). (D–I) Endoplasmic reticulum stress (all N = 3 per group). (D) Unspliced X-box binding protein (XBP1) mRNA (P = NS, paired t test). (E) Spliced XBP1 mRNA (P = NS, paired t test). (F) Immunoglobulin binding protein (BiP) mRNA (P = NS, paired t test). (G) Endoplasmic reticulum degradation-enhancing α-mannidose-like protein (EDEM) mRNA (P = NS, paired t test). (H) Activating transcription factor 4 (ATF4) mRNA (P = NS, paired t test). (I) CCAAT/enhancer-binding protein homologous protein (CHOP) mRNA (P = NS, paired t test). (J–M) Enrichment with signal peptide and exosome proteins. (J) Proportion of proteins down-regulated significantly by iron with signal peptide vs no signal peptide. (K) Proportion of proteins not down-regulated significantly by iron with signal peptide vs no signal peptide. (L) Proportion of proteins down-regulated significantly by iron with exosome secretion vs no exosome secretion. (M) Proportion of proteins not down-regulated significantly by iron with exosome secretion vs no exosome secretion. Of the 61 proteins down-regulated significantly, 62% (38 of 61) had signal peptide, whereas of the remaining proteins only 47% (129 of 277) had signal peptide. The 1-tailed Fisher exact test showed significant enrichment with signal peptide (P = .02) among the group down-regulated significantly. In contrast, there was no significant enrichment of the exosomal pathway (P = .51, 1-tailed Fisher exact test), because 15% (9 of 61) of the proteins down-regulated significantly and 14% (39 of 277) of the remaining secretome proteins had been reported previously in the high-confidence proteins from the EVpedia database.
We considered whether iron may have a generalized effect on pathways of protein secretion, used by a variety of proteins. We evaluated the role of iron in the secretion of proteins by the classic and exosomal pathways using the UniProt and EVpedia databases, respectively.4, 10 We found enrichment of signal peptide-containing (P = .02), but not exosome-secreted, proteins (P = .51) among the iron-dysregulated proteins, suggesting that iron may have a specific effect on proteins secreted via the classic pathway (Supplementary Figure 3J–M).
This research has characterized the effect of iron on the adipocyte secretome. These data provide a platform for multiple avenues for future research. In addition, we have been able to show that increased iron results in sequestration of ApoE within adipocytes, which may be of key importance in the regulation of insulin resistance and liver injury in NASH. Identifying the molecular mechanisms of iron-induced inhibition of ApoE secretion from adipocytes, particularly relating to the role of oxidative stress, may show novel therapeutic strategies for improving adipocyte function in NASH.
Acknowledgment
ProteomeXchange Consortium via the Proteomics Identifications (PRIDE) partner repository: http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=pxd006341 data set identifier PXD006341.
Footnotes
Author contributions L. J. Britton was responsible for the study concept and design, acquisition of data, analysis and interpretation of data, drafting of the manuscript, and statistical analysis; K. Bridle was responsible for the study concept and design, analysis and interpretation of data, critical revision of the manuscript for important intellectual content, administrative, technical, or material support, and study supervision; L. A. Jaskowski was responsible for the acquisition of data, analysis and interpretation of data, critical revision of the manuscript for important intellectual content, and administrative, technical, or material support; J. He was responsible for the acquisition of data, critical revision of the manuscript for important intellectual content, and administrative, technical, or material support; C. Ng was responsible for the acquisition of data, critical revision of the manuscript for important intellectual content, and administrative, technical, or material support; J. E. Ruelcke was responsible for the acquisition of data, analysis and interpretation of data, critical revision of the manuscript for important intellectual content, statistical analysis, and administrative, technical, or material support; A. Mohamed was responsible for the analysis and interpretation of data, critical revision of the manuscript for important intellectual content, and statistical analysis; J. Reiling was responsible for the study concept and design, analysis and interpretation of data, and critical revision of the manuscript for important intellectual content; N. Santrampurwala was responsible for the study concept and design, analysis and interpretation of data, and critical revision of the manuscript for important intellectual content; M. M. Hill was responsible for the study concept and design, analysis and interpretation of data, critical revision of the manuscript for important intellectual content, statistical analysis, administrative, technical, or material support, and study supervision; J. P. Whitehead was responsible for the study concept and design, analysis and interpretation of data, critical revision of the manuscript for important intellectual content, administrative, technical, or material support, and study supervision; V. N. Subramaniam was responsible for the study concept and design and critical revision of the manuscript for important intellectual content; and D. H. G. Crawford was responsible for the study concept and design, analysis and interpretation of data, critical revision of the manuscript for important intellectual content, obtained funding, administrative, technical, or material support, and study supervision.
Conflicts of interest The authors disclose no conflicts.
Funding This research was supported by Australian National Health and Medical Research Council grant APP1029574.
Supplementary Methods
Simpson-Golabi-Behmel Syndrome Pre-Adipocyte Differentiation and Iron Treatment
SGBS pre-adipocytes were a gift from Martin Wabitsch (University of Ulm, Ulm, Germany).1, 2 SGBS cells were passaged, proliferated, and differentiated at less than 50 generations in 12-well plates and 100-mm dishes as previously described.3 Cells were treated with 90 μg/mL heparin and 1 ng/mL fibroblast growth factor-1 (both Sigma-Aldrich, St. Louis, MO) throughout proliferation and differentiation. After 14 days of differentiation, cells were incubated with 0, 25, 100, or 500 μmol/L FAC (Sigma-Aldrich) for 24 hours. Media then was replaced with the same for a further 24 hours until the end of the experiment.
RNA Extraction and Real-Time Quantitative Polymerase Chain Reaction
RNA was extracted from treated SGBS adipocytes using a PureLink RNA mini kit (Invitrogen, Carlsbad, CA). Complementary DNA was synthesized from 1 μg RNA using a Sensifast complementary DNA synthesis kit (Bioline, London, UK) after treatment with DNase 1 (Invitrogen). Samples underwent thermal cycling using a ViiA7 real-time polymerase chain reaction machine (Invitrogen) with a Sensifast SYBR Lo-ROX Kit (Bioline). The following protocol was used: 2 minutes at 95°C, then 40 cycles of 5 seconds at 95°C, alternating with 20 seconds at 63°C, followed by a melt curve analysis. Relative mRNA quantities were determined by calibration of cycle threshold values to the standard curve of pooled complementary DNA samples. Results were normalized to cycle threshold values of cyclophilin.
Iron, Cellular Viability (MTS), and Protein Assays
Iron levels were quantified using a chromagen reagent method.4 Cellular viability was assessed using a CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI) according to the manufacturer’s instructions. Whole-cell lysate and secretome samples underwent protein estimation using a Pierce BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA).
Stable Isotope-Labeled Amino Acids in Cell Culture Proteomics
SILAC incorporates stable amino acid isotopes, without altering cellular biology, allowing direct comparison of the secretome by mass spectrometry between treatment groups.5 SGBS pre-adipocytes were grown in SILAC Dulbecco's modified Eagle medium:F12 media (Thermo Fisher Scientific) supplemented with dialyzed fetal bovine serum (Thermo Fisher Scientific) and 22.81 mg/L 2H4-lysine and 36.88 mg/L 13C6-arginine (K4R6) or 22.81 mg/L 13C615N2-lysine and 36.88 mg/L 13C615N4-arginine (K8R10). Incorporation of labeled amino acids was confirmed by liquid chromatography tandem mass spectrometry on tryptic peptides prepared from whole-cell lysates. Cell pellets were lysed in 8 mol/L urea in 100 mmol/L triethylamonium bicarbonate, and protein concentration was estimated using the Bradford assay (BioRad, Hercules, CA). Thirty micrograms of cell lysate was reduced and alkylated by incubating samples for 30 minutes at 37°C with 2.5 mmol/L tris(2-carboxyethyl)phosphine and then 5 mmol/L 2-chloroacetamide. The urea concentration was diluted to 1 mol/L with 100 mmol/L triethylamonium bicarbonate before adding 0.6 μg of trypsin. Samples were incubated overnight then acidified to 1% trifluoroacetic acid and cleaned with OMIX C18 tips according to the manufacturers’ protocol (Agilent, Santa Clara, CA). Liquid chromatography tandem mass spectrometry was performed as described later.
Labeled (>97%) cells underwent differentiation to adipocytes as described earlier. At day 14 after differentiation, cells were treated with vehicle (media) (K4R6, medium-weight cells) or 100 μmol/L FAC (in media) (K8R10, heavy-weight cells) for a further 48 hours using exactly equal volumes of media, replacing the media after 24 hours. Media for secretome analysis was collected from K4R6 and K8R10 cells and mixed 1:1 (vol/vol) before centrifugation at 600 × g at 4°C for 10 minutes to remove cell debris. Supernatant then was concentrated using Amicon Ultra 15 mL 10-kilodalton centrifugal filter units (Merck Millipore, Burlington, MA) in a fixed-angle centrifuge at 5000 × g to provide approximately 1-mL samples of concentrated mixed secretome. Thirty micrograms of protein was separated on 10% sodium dodecyl sulfate–polyacrylamide electrophoresis gels to 10 mm. Protein visualization, excision of bands, and in-gel trypsin digestion were performed using a semiautomated method as described.6 A band corresponding to the same molecular weight as transferrin (media additive) was removed before digestion to provide a protein sample exclusively secreted from cultured adipocytes.
Mass Spectrometry
A Q Exactive Plus Orbitrap Mass Spectrometer (Thermo Fisher Scientific), coupled with Easy-nLC 1000 and EASY-spray ion source (both Thermo Fisher Scientific), was used to analyze the digested peptides. Samples were loaded onto an EASY-Spray PepMap RSLC C18 2-μm column (50 cm × 75 μm ID), with a Nanoviper Acclaim C18 guard (75 μm × 2 cm) (both Thermo Fisher Scientific). A 90-minute method was run using a combination of buffer A (0.1% formic acid) and buffer B (0.1% formic acid:acetonitrile). A 2-step gradient was run comprising a 60-minute gradient from 3% to 25% buffer B and a 12-minute gradient from 25% to 40% buffer B. The flow rate was 250 nL/min. The mass spectrometer was programmed to acquire a full mass spectrometry resolution of 70,000 with an ACG target of 3 × 106, with a maximum injection time of 100 ms. The mass spectrometry scan range was from 350 to 1400 m/z. Tandem mass spectrometry was set to acquire a resolution of 17,500 with an ACG target of 5 × 105 and a maximum injection time of 55 ms. The loop count was set to 20 with a dynamic exclusion after 30 seconds. Raw data were processed with Spectrum mill (Rev B.05.00.181 SP1; Agilent). Selected modifications included fixed carbamidomethylation of cysteine and SILAC labels (Arg 0-6-10 daltons Lys 0-4-8 daltons) and variable oxidized methionine. Results were searched against the Human UniProt database (downloaded June 1, 2015).7 Trypsin was selected as the digestion enzyme, with 2 maximum missed cleavages allowed. The precursor mass tolerance was set at ±20 ppm and product mass tolerance was ±20 ppm. Data were analyzed using the online Quantitative Proteomics P value Calculator using no normalization and nonadjusted P values.8
Immunoblotting
Western blot using whole-cell lysate samples (10 μg) and 5-μL concentrated secretome samples was performed as previously described.9 A 1:500 dilution of primary antibody against ApoE (sc-53570; Santa Cruz, Dallas, TX) was applied to the membranes. A 1:100,000 dilution of goat anti-mouse horseradish peroxidase antibody (Invitrogen) was applied as secondary antibody. ApoE whole-cell lysate densitometry was normalized against densitometry using β-actin as a reference protein (1:2000 primary antibody) (cat no. 4967, Cell Signaling, Danvers, MA) and 1:20,000 goat anti-rabbit horseradish-peroxidase antibody (Invitrogen).
Supplementary Table 1.
List of SGBS Secretome Proteins With Significantly Altered Signal Intensity in Response to Iron
| Accession number | Gene name | Protein name | Mean signal intensity ratio, iron/vehicle | SD | P value |
|---|---|---|---|---|---|
| Q8IX30 | SCUBE3 | Signal peptide, CUB and EGF-like domain-containing protein 3 | 0.026 | 0.016 | .001 |
| P61353 | RPL27 | 60S ribosomal protein L27 | 0.060 | 0.005 | .001 |
| Q9NQH7 | XPNPEP3 | Probable Xaa-Pro aminopeptidase 3 | 0.076 | 0.004 | .001 |
| P07996 | THBS1 | Thrombospondin-1 | 0.083 | 0.047 | .001 |
| Q76M96 | CCDC80 | Coiled-coil domain-containing protein 80 | 0.090 | 0.031 | .001 |
| P78539 | SRPX | Sushi repeat-containing protein SRPX | 0.096 | 0.620 | .001 |
| Q9UHI8 | ADAMTS1 | A disintegrin and metalloproteinase with thrombospondin motifs 1 | 0.114 | 0.090 | .002 |
| Q92538 | GBF1 | Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 | 0.118 | 0.024 | .004 |
| Q15063 | POSTN | Periostin | 0.121 | 0.149 | .001 |
| P08238 | HSP90AB1 | Heat shock protein (HSP) 90-β | 0.125 | 0.038 | .004 |
| P24593 | IGFBP5 | Insulin-like growth factor-binding protein 5 | 0.154 | 0.069 | .001 |
| Q15113 | PCOLCE | Procollagen C-endopeptidase enhancer 1 | 0.155 | 0.088 | .001 |
| Q9NTX5 | ECHDC1 | Ethylmalonyl-CoA decarboxylase | 0.158 | 0.068 | .001 |
| P25788 | PSMA3 | Proteasome subunit α type 3 | 0.176 | 0.442 | .001 |
| Q12931 | TRAP1 | Heat shock protein 75 kilodaltons, mitochondrial | 0.186 | 0.028 | .018 |
| P04083 | ANXA1 | Annexin A1 | 0.192 | 0.049 | .001 |
| P30101 | PDIA3 | Protein disulfide-isomerase A3 | 0.198 | 0.059 | .001 |
| Q99985 | SEMA3C | Semaphorin-3C | 0.199 | 0.225 | .001 |
| Q6NZI2 | PTRF | Polymerase I and transcript release factor | 0.210 | 0.134 | .001 |
| Q8TAV4 | STOML3 | Stomatin-like protein 3 | 0.220 | 0.084 | .006 |
| Q9UKZ9 | PCOLCE2 | Procollagen C-endopeptidase enhancer 2 | 0.226 | 0.030 | .001 |
| Q05469 | LIPE | Hormone-sensitive lipase | 0.231 | 0.196 | .023 |
| Q13642 | FHL1 | Four and a half LIM domains protein 1 | 0.263 | 0.006 | .029 |
| P02749 | APOH | β2-glycoprotein 1 | 0.286 | 0.310 | .048 |
| P02462 | COL4A1 | Collagen α-1(IV) chain | 0.286 | 0.351 | .001 |
| P15311 | EZR | Ezrin | 0.296 | 0.077 | .003 |
| P42765 | ACAA2 | 3-Ketoacyl-CoA thiolase, mitochondrial | 0.308 | 0.136 | .001 |
| P68104 | EEF1A1 | Elongation factor 1-α 1 | 0.309 | 0.215 | .001 |
| Q9NQC3 | RTN4 | Reticulon-4 | 0.312 | 0.119 | .007 |
| Q8IY17 | PNPLA6 | Neuropathy target esterase | 0.316 | 0.028 | .026 |
| Q92743 | HTRA1 | Serine protease HTRA1 | 0.324 | 0.075 | .001 |
| P08294 | SOD3 | Extracellular superoxide dismutase (Cu-Zn) | 0.339 | 0.027 | .005 |
| Q16836 | HADH | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | 0.348 | 0.207 | .011 |
| Q99715 | COL12A1 | Collagen α-1(XII) chain | 0.348 | 0.183 | .001 |
| P07355 | ANXA2 | Annexin A2 | 0.354 | 0.111 | .001 |
| P26038 | MSN | Moesin | 0.362 | 0.110 | .034 |
| P23284 | PPIB | Peptidyl-prolyl cis-trans isomerase B | 0.374 | 0.122 | .001 |
| O94769 | ECM2 | Extracellular matrix protein 2 | 0.375 | 0.060 | .005 |
| Q9NRN5 | OLFML3 | Olfactomedin-like protein 3 | 0.389 | 0.066 | .002 |
| P53396 | ACLY | Adenosine triphosphate–citrate synthase | 0.389 | 0.066 | .001 |
| Q16363 | LAMA4 | Laminin subunit α-4 | 0.407 | 0.091 | .001 |
| P14625 | HSP90B1 | Endoplasmin | 0.414 | 0.068 | .025 |
| Q9BU40 | CHRDL1 | Chordin-like protein 1 | 0.419 | 0.264 | .001 |
| P02649 | APOE | Apolipoprotein E | 0.421 | 0.053 | .001 |
| Q9NS98 | SEMA3G | Semaphorin-3G | 0.425 | 0.161 | .001 |
| P02751 | FN1 | Fibronectin | 0.441 | 0.146 | .001 |
| P14543 | NID1 | Nidogen-1 | 0.442 | 0.190 | .006 |
| Q08431 | MFGE8 | Lactadherin | 0.446 | 0.167 | .026 |
| Q15848 | ADIPOQ | Adiponectin | 0.449 | 0.652 | .005 |
| O75390 | CS | Citrate synthase, mitochondrial | 0.454 | 0.183 | .012 |
| Q92626 | PXDN | Peroxidasin homolog | 0.457 | 0.200 | .001 |
| P07942 | LAMB1 | Laminin subunit β1 | 0.484 | 0.110 | .001 |
| Q08629 | SPOCK1 | Testican-1 | 0.497 | 0.055 | .014 |
| O00462 | MANBA | β-mannosidase | 2.180 | 2.560 | .034 |
| Q13510 | ASAH1 | Acid ceramidase | 2.222 | 3.135 | .048 |
| P02794 | FTH1 | Ferritin heavy chain | 2.451 | 0.769 | .030 |
| Q02952 | AKAP12 | A-kinase anchor protein 12 | 2.665 | 0.572 | .047 |
| A6NCN2 | KRT87P | Putative keratin-87 protein | 4.113 | 0.011 | .022 |
| P23468 | PTPRD | Receptor-type tyrosine-protein phosphatase δ | 10.687 | 2.199 | .003 |
| P10586 | PTPRF | Receptor-type tyrosine-protein phosphatase F | 11.935 | 3.955 | .001 |
NOTE. Proteins shown had a greater than 2-fold change in signal intensity in response to iron, with P < .05. Proteins highlighted in bold represent the 20 proteins of interest after review of the UniProt protein descriptions. Data were analyzed using the online Quantitative Proteomics P value Calculator using no normalization and nonadjusted P values (N = 3 per group).
CUB, C1r/C1s, Uegf, bone morphogenetic protein-1; EGF, epidermal growth factor; HTRA, High-Temperature Requirement A.
References
- 1.Hebbard L. Nat Rev Gastroenterol Hepatol. 2011;8:35–44. doi: 10.1038/nrgastro.2010.191. [DOI] [PubMed] [Google Scholar]
- 2.Orr J.S. Diabetes. 2014;63:421–432. doi: 10.2337/db13-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simcox J.A. Cell Metab. 2013:329–341. doi: 10.1016/j.cmet.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The UniProt Consortium Nucleic Acids Res. 2017:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Locatelli I. Hepatology. 2014;60:531–544. doi: 10.1002/hep.27141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabrielsen J.S. J Clin Invest. 2012;122:3529–3540. doi: 10.1172/JCI44421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schierwagen R. Sci Rep. 2015;5:12931. doi: 10.1038/srep12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pendse A.A. J Lipid Res. 2009;50:S178–S182. doi: 10.1194/jlr.R800070-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H. J Alzheimers Dis. 2016;31:471–487. [Google Scholar]
- 10.Kim D.K. Bioinformatics. 2015;31:933–939. doi: 10.1093/bioinformatics/btu741. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Wabitsch M. Int J Obes Relat Metab Disord. 2001;25:8–15. doi: 10.1038/sj.ijo.0801520. [DOI] [PubMed] [Google Scholar]
- 2.Fischer-Posovszky P. Obes Facts. 2008:184–189. doi: 10.1159/000145784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo X. Diabetes. 2012;61:124–136. doi: 10.2337/db11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohyama M. Nature. 2009;457:318–321. doi: 10.1038/nature07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ong S.E. Nat Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 6.Ruelcke J.E. J Proteomics. 2016;149:3–6. doi: 10.1016/j.jprot.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 7.The UniProt C. Nucleic Acids Res. 2017:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen D. J Proteome Res. 2014;13:4184–4191. doi: 10.1021/pr500525e. [DOI] [PubMed] [Google Scholar]
- 9.Britton L. Physiol Rep. 2016:e12837. doi: 10.14814/phy2.12837. [DOI] [PMC free article] [PubMed] [Google Scholar]