Abstract
Background
Activation of intrinsic p53 tumor-suppressor (TS) pathways is an important principle underlying cancer chemotherapy. It is necessary to elucidate the precise regulatory mechanisms of these networks to create new treatment strategies.
Methods
Comprehensive analyses were carried out by microarray. Expression of miR-101 was analyzed by clinical samples of lung adenocarcinomas.
Findings
We discovered a functional link between p53 and miR-101, which form a molecular circuit in response to nucleolar stress. Inhibition of RNA polymerase I (Pol I) transcription resulted in the post-transcriptional activation of miR-101 in a p53-dependent manner. miR-101 induced G2 phase–specific feedback regulation of p53 through direct repression of its target, EG5, resulting in elevated phosphorylation of ATM. In lung cancer patients, low expression of miR-101 was associated with significantly poorer prognosis exclusively in p53 WT cases. miR-101 sensitized cancer cells to Pol I transcription inhibitors and strongly repressed xenograft growth in mice. Interestingly, the most downstream targets of this circuit included the inhibitor of apoptosis proteins (IAPs). Repression of cIAP1 by a selective inhibitor, birinapant, promoted activation of the apoptosis induced by Pol I transcription inhibitor in p53 WT cancer cells.
Interpretation
Our findings indicate that the p53–miR-101 circuit is a component of an intrinsic TS network formed by nucleolar stress, and that mimicking activation of this circuit represents a promising strategy for cancer therapy.
Fund
National Institute of Biomedical Innovation, Ministry of Education, Culture, Sports & Technology of Japan, Japan Agency for Medical Research and Development.
Keywords: p53, Nucleolar stress, miR-101, Tumor-suppressor network
Research in context.
Activation of p53 tumor-suppressor (TS) pathways is an important principle underlying chemotherapeutic strategies for treating p53–wild-type (WT) cancers. Here, we describe the p53–miR-101 circuit as a component of a TS network. miR-101 is upregulated in a p53-dependent manner after exposure of cells to Pol I inhibitors, and is involved in positive-feedback regulation of p53 via repression of EG5, resulting in induction of apoptosis. Moreover, reduced expression of miR-101 is associated with poor prognosis in p53 WT lung adenocarcinoma (LADC) patients. The most downstream targets of this circuit included the inhibitor of apoptosis proteins (IAPs). Combination treatment with inhibitors of IAP and Pol I represents a promising strategy for efficient elimination of p53 WT cancer cells.
1. Introduction
The p53 tumor-suppressor (TS) protein, encoded by the TP53 gene, has been termed the “guardian of the genome” in recognition of its role in maintaining genome integrity in response to various oncogenic insults [1, 2]. TP53 is mutated and/or inactivated in half of human cancers, and dysfunction of p53 makes a critical contribution to the onset of carcinogenesis [3, 4]. On the other hand, nearly half of all tumors retain wild-type (WT) p53 function, but the effector networks downstream of p53 are disrupted in many tumors due to mutations in regulatory genes. In the context of therapeutics, inactivation or reduced activation of the downstream networks of p53 is a more difficult to address than mutation in p53 itself. Many chemotherapeutic agents activate p53 through various mechanisms, resulting in induction of the appropriate downstream networks by selective activation of p53 target genes. Consequently, even after activation of p53, incomplete activation of downstream pathways can dramatically decrease the efficacy of chemotherapy.
MicroRNAs (miRNAs), a class of small non-coding RNAs, act as intrinsic mediators in intracellular networks by regulating gene expression at the post-transcriptional level [5]. miRNA expression is altered in almost all human cancers, strongly suggesting that miRNA dysfunction is associated with cancer pathogenesis [[6], [7], [8]]. In addition, miRNAs are globally downregulated in many types of human cancers, suggesting that they function as intrinsic TSs [9, 10]. Consistent with this idea, multiple miRNAs are involved in the regulation of p53 TS pathways [11]. Moreover, p53 itself regulates multiple miRNAs, many of which have tumor-suppressive functions, at the transcriptional and post-transcriptional levels. p53 selectively transactivates tumor-suppressive miRNAs according to the type of stress experienced by the cell [12, 13]. Thus, it is clear that precise activation of intrinsic p53 networks, as well as control of the degree and duration of pathway activation, is fine-tuned by multiple miRNAs. Comprehensively understanding the molecular connections between p53 downstream networks and miRNAs is key to elucidating TS networks, and detailed analyses of these networks are expected to reveal crucial molecules and facilitate the formulation of novel strategies for effective therapy.
In this study, we discovered that a p53-dependent TS network triggered by nucleolar stress is tuned by miR-101. Activation of this network, the p53–miR-101 circuit, enables induction of apoptosis in p53 WT cancer cells by G2 phase–specific positive-feedback regulation of p53 mediated by direct repression of EG5. The importance of this circuit is highlighted by the observation that, in lung adenocarcinoma (LADC) patients, reduced expression of miR-101 is associated with significantly worse prognosis exclusively in p53 WT cases. We identified the inhibitor of apoptosis proteins (IAPs) as the most downstream target of this circuit. Repression of cellular inhibitor of apoptosis protein 1 (cIAP1; also known as BIRC2) by the molecularly targeted drug birinapant, in combination with the polymerase I (Pol I) transcription inhibitor CX-5461, promoted induction of apoptosis in p53 WT cancer cells, implying that this combination therapy mimics activation of the p53–miR-101 circuit. Our data provide molecular insights that could facilitate development of strategies for treating p53 WT cancer.
2. Materials and Methods
2.1. Cell Culture and Transfection
The colon cancer cell lines HCT116 and RKO and the lung cancer cell lines A549 and A427 were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS). All cell lines were grown at 37 °C in humidified air in 5% CO2. HCT116 p53−/− cells were kindly provided by Dr. Bert Vogelstein (The Johns Hopkins University, Baltimore, MD, USA). miRNAs and siRNAs used in this study are listed in Supplementary Table S1. miRNAs and siRNAs were introduced into cells at the indicated concentrations, individually or in combination, using the Lipofectamine RNAiMAX reagent (Life Technologies/Thermo Fisher Scientific, Waltham, MA, USA; Cat# 13778150).
2.2. Antibodies and Reagents
Anti-PARP (Cat# 9542; RRID: AB_2160739), anti-phospho-p53 (Ser15) (Cat# 9284; RRID: AB_331464), anti-phospho-Histone H3 (Ser10) (Cat# 3377; RRID: AB_1549592), anti-Histone H3 (Cat# 9715; RRID: AB_331563), anti-phospho-cdc2/CDK1 (Tyr15) (Cat# 9111S; RRID: AB_331460), anti-cdc2/CDK1 (Cat# 9112S; RRID: AB_10693432), anti-cyclin A2 (Cat# 4656S; RRID: AB_10691320), anti-p27 Kip1 (Cat# 3686S; RRID: AB_2077850), and anti-p21 (Cat# 2947; RRID: AB_823586) were purchased from Cell Signaling Technologies (Danvers, MA, USA). Anti-ATM (Cat# 1549-1; RRID: AB_725574), anti-phospho-ATM (Ser1981) (Cat# 2152-1; RRID: AB_991678), anti-DNA-PKcs (Cat# 3922-1), anti-stathmin1 (Cat# 1972-1; RRID: AB_991829), and anti-EG5 (Cat# S1765; RRID: AB_10640358) were purchased from Epitomics (Burlingame, CA, USA). Other antibodies included anti-ATR (Bethyl Laboratories, Montgomery, TX, USA; Cat# A300 – 138A; RRID: AB_2063318) and anti-NEK4 (Bethyl Laboratories, Cat# A302-673A; RRID: AB_10568794), anti-p53 (DO-1, Santa Cruz Biotechnology, Dallas, TX, USA; Cat# sc-126; RRID: AB_628082), anti-GAPDH (Chemicon/EMD Millipore, Burlington, MA, USA; Cat# MAB374; RRID: AB_2107445), anti-α-tubulin (Sigma-Aldrich, St. Louis, MO, USA; Cat# T6074; RRID: AB_477582), anti-EZH2 (BD Biosciences, San Jose, CA, USA; Cat# 612666; RRID: AB_2102429), anti-cyclin B (BD Biosciences, Cat# 610219; RRID: AB_397616), and anti-human Ciap-1/hiap-2 (R&D Systems, Minneapolis, MN, USA; Cat# AF8181, RRID: AB_2259001).
Actinomycin D (ActD; Cat# A9415; CAS: 50-76-0), 5-Fluorouracil (5-FU; Cat# F6627; CAS: 51-21-8), K858 (EG5 inhibitor) (Cat# K3644; CAS: 72926-24-0), and paclitaxel (PTX; Cat# T7402; CAS: 33069-62-4) were purchased from Sigma-Aldrich. CX-5461 (Cat# A11065; CAS: 1138549-36-6) was purchased from AdooQ BioScience (Irvine, CA, USA). ML-60218 (Cat# 557403; CAS: 577784-91-9) and RO-3306 (CDK1 inhibitor) (Cat# 217699; CAS: 872573-93-8) were from Calbiochem (San Diego, CA, USA). These reagents were dissolved in DMSO. Adriamycin (ADR; Sigma-Aldrich, Cat# D1515; CAS: 25316-40-9) and doxycycline (Dox; Sigma, Cat# D9891; CAS: 24390-14-5) and α-Amanitin (Wako Pure Chemical, Osaka, Japan; Cat# 010-229611; CAS: 23109-05-9) were dissolved in water. All reagents were used at the indicated concentrations.
2.3. Cell Proliferation Assay
Cell proliferation was measured by quantification of formazan product using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA; Cat# G3580). Cells transfected with either miRNA mimic or negative control were lysed in MTS tetrazolium solution and incubated for 1–4 h at 37 °C in a humidified 5% CO2 atmosphere. Absorbance at 490 nm was measured on an ARVO plate reader (PerkinElmer, Waltham, MA, USA).
2.4. Clinical Samples
Surgical specimens of human LADC were obtained from patients treated at the National Cancer Center Hospital (NCC cohort) or recruited through the NCI-MD case–control study OH98CN027 (NCI cohort). In the NCC cohort, a total of 200 LADC cases were selected from 608 consecutive cases, and samples were subjected to multi-omics analysis [14]. Data from 76 of 200 LADC cases for which clinical data and genomic, transcriptomic, and miRNA profiles were available were used. None of the patients had received prior treatment with chemotherapy before surgery. Patients in the NCI cohort (n = 82) were recruited from seven hospitals. All were histologically diagnosed as having non-small cell lung cancer (NSCLC), and were recruited within 24 months after diagnosis. Only patients with LADC were included in this study. At the time of surgery, a portion of the tumor specimen and noninvolved adjacent lung tissue were flash-frozen and stored at −80 °C until use. Documented informed consent was obtained in each case. Gene expression and miRNA expression analyses of NCC cohort samples were carried out on the Agilent platform [15]. miRNA expression profiling of NCI cohort samples was performed using NanoString nCounter [16]. TP53 mutation was confirmed by exome sequencing (NCC cohort) or targeted sequencing of TP53 (NCI cohort), targeting exons and proximal splice sites, on the Illumina platform. Institutional review board approval for the use of clinical samples was obtained from the ethics committees at each institution. Clinico-pathological features of patients in both cohorts are summarized in Table 1.
Table 1.
Demographic and clinical characteristics of study cohorts.
| NCC (n = 76) | NCI (n = 82) | |
|---|---|---|
| Age, years | ||
| Mean (SD) | 58.2 (10.6) | 62.6 (10.4) |
| Range | 30–75 | 32–84 |
| Sex (%) | ||
| Male | 46 (60.5) | 48 (58.5) |
| Female | 30 (39.5) | 34 (41.5) |
| Race | ||
| Asian | 76 (100.0) | 0 (0.0) |
| Caucasian | 0 (0.0) | 49 (59.8) |
| African American | 0 (0.0) | 33 (40.2) |
| Smoking history (%) | ||
| Never smoker | 28 (36.8) | 3 (3.7) |
| <20 pack-years | 8 (10.5) | 13 (15.9) |
| ≥20 pack-years | 40 (52.6) | 59 (72.0) |
| Unknown pack-years | 0 (0.0) | 4 (4.9) |
| Unknown smoking history | 0 (0.0) | 3 (3.7) |
| Smoking pack-years | ||
| Mean (SD) | 28.1 (30.7) | 52.7 (33.9) |
| AJCC TNM 7th Stage (%) | ||
| I | 17 (22.4) | 43 (52.4) |
| II | 27 (35.5) | 24 (29.3) |
| III | 27 (35.5) | 13 (15.9) |
| IV | 5 (6.6) | 0 (0.0) |
| Unknown | 0 (0.0) | 2 (2.4) |
| Therapy after surgery (%) | ||
| None | 25 (32.9) | 36 (43.9) |
| Chemotherapy | 33 (43.4) | 9 (11.0) |
| Radiotherapy | 12 (15.8) | 6 (7.3) |
| Chemotherapy plus radiotherapy | 6 (7.9) | 22 (26.8) |
| Unknown | 0 (0.0) | 9 (11.0) |
2.5. Expression Microarray
HCT116 and RKO cells were transfected with either miR-101 or negative control miRNA (Ambion/Life Technologies) at a final concentration of 5 nM for 3 days. Total RNA was extracted using the miRNeasy Mini kit (Qiagen, Valencia, CA, USA; Cat# 217004). For each sample, 175 ng of total RNA was labeled with Cy3 using the Low Input Quick Amp Labeling Kit, one color (Agilent Technologies, Santa Clara, CA, USA; Cat# 5190-2305), and hybridized to Whole Human Genome Microarrays, 4x44K (Agilent Technologies, Cat# G4112F). Samples were analyzed in triplicate. After hybridization at 65 °C for 17 h, arrays were washed with washing buffer and scanned on an Agilent microarray scanner. The data were numerically converted with Feature Extraction Software (Agilent Technologies) and analyzed using the GeneSpring GX software (Agilent Technologies). Gene Ontology (GO) analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (http://david.abcc.ncifcrf.gov/home.jsp) with default settings for terms related to GO biological processes (BP_FAT). The data were deposited in the Gene Expression Omnibus (GEO) database with accession number GSE85696.
2.6. Cell-Cycle Analysis
Cells were washed twice in cold phosphate-buffered saline (PBS), fixed in cold 100% methanol, and stored at −20 °C until use. Prior to analysis, cells were incubated at 37 °C for 30 min in PBS containing 25 μg/ml RNase A (Nippon Gene, Cat# 312-01931), propidium iodide (PI) (Sigma, Cat# P4170) was added at a final concentration of 50 μg/ml, and the sample was incubated at 4 °C for 1 h in the dark. After incubation, cells were passed through a cell strainer and subjected to flow cytometry on a FACSCalibur instrument (BD Biosciences). Data were processed using the FlowJo software.
2.7. Apoptosis Assay
Apoptotic cells were detected using the Annexin V–FITC Apoptosis Detection kit I (BD Biosciences, Cat# 556547). Cells were harvested and washed with cold PBS and re-suspended with cold 1× binding buffer, followed by incubation with Annexin V–FITC at room temperature for 15 min in the dark. Immediately before analysis by flow cytometry, either 7-AAD (BD Biosciences, Cat# 559925) or PI was added to each sample, and apoptotic cells were detected on a SONY EC800.
2.8. Mitotic Index
Cells transfected with either miR-101 or negative control at a final concentration of 5 nM for 3 days were detached and fixed in cold 100% methanol, and stored at −20 °C. Cells were washed in PBS with 0·5% BSA, re-suspended in PBS with 0·25% Triton X-100, incubated on ice for 15 min, and then incubated at room temperature for 1 h with either anti-phospho-Histone H3 (Ser10) Rabbit mAb Alexa Fluor 488 conjugate (Cell Signaling Technologies, Cat# 3465S; RRID: AB_10695860) or Rabbit mAb IgG XP Isotype control Alexa Fluor 488 conjugate (Cell Signaling Technology, Cat# 2975S; RRID: AB_10699151). After washing with PBS/0·5% BSA, cells were incubated for 30 min at room temperature with PBS containing 50 μg/ml RNase A and 2·5 μg/ml 7-AAD. The proportion of mitotic cells was determined on a FACSCalibur instrument, and data were processed using the FlowJo software.
2.9. Separation by Cell-Cycle Phase
Cells were harvested with Accutase (Innovative Cell Technologies, Cat# AT-104) and passed through a cell strainer. After centrifugation, cells were washed once with cold staining buffer consisting of DMEM with 0·1 mM EDTA, 2% FBS, and 10 mM HEPES; re-suspended in staining buffer at a concentration of 5 × 106 cells/ml; and then incubated with Hoechst 33342 (5 μg/ml) (Invitrogen, Cat# H3570) and SYTOX Red (5 nM) (Life Technologies, Cat# S34859) for 1 h at 37 °C. Cells were separated according to cell-cycle phase on a FACSAria II (BD Biosciences) equipped with laser lines with wavelengths of 375, 488, and 633 nm. The sorted cells were collected in culture medium, washed twice with CELLOTION (TaKaRa, Shiga, Japan; Cat# CB051), and then processed for immunoblot (IB) analysis.
2.10. IB Analysis
Cells were lysed in ice-cold lysis buffer consisting of 25 mM Tris-HCl (pH 7·4), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0·1% SDS, 1× protease inhibitor cocktail, and 1× phosphatase inhibitor cocktail (Roche Diagnostics). Protein concentrations were determined using the Protein Assay Reagent (Bio-Rad Laboratories, Hercules, CA, USA; Cat# 500-0006). Equal concentrations of protein samples were loaded onto a 10–20% gradient polyacrylamide gel (ATTO, Tokyo, Japan), and separated proteins were transferred onto PVDF membrane (EMD Millipore). Membranes were incubated for 1 h in blocking buffer [5% non-fat dry milk in TBS-T buffer (0·05% Tween-20)], probed with diluted primary antibodies at 4 °C overnight, and then incubated with horseradish peroxidase–conjugated sheep anti–mouse IgG or donkey anti–rabbit IgG (GE Healthcare, Little Chalfont, UK). Signals were detected using SuperSignal WestDura Extended Duration Substrate (Pierce/Thermo Fisher Scientific, Cat# 34075).
2.11. Quantitative Real-Time PCR
Total RNA, including miRNA, was isolated using the miRNeasy Mini kit and treated with DNase I (Qiagen, Cat# 79254). For quantitative analysis of miRNA expression, 80 ng of total RNA was reverse-transcribed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA; Cat# 4366597) and miRNA-specific primers (Applied Biosystems), and PCR was performed using TaqMan Fast Advanced Master Mix and TaqMan microRNA assays (Applied Biosystems, Cat# 4444558). To measure primary miRNA levels, total RNA was reverse-transcribed with SuperScript VILO MasterMix (Invitrogen, Cat# 11755-250), and synthesized cDNA was quantified using TaqMan Fast Advanced Master Mix and TaqMan Pri-miRNA Assays (Applied Biosystems). RNU48 small nuclear RNA, TBP mRNA (TATA box–binding protein), or PPIA mRNA (peptidylprolyl isomerase A) was used as an internal control. Relative expression levels were calculated by the comparative cycle threshold (Ct) method. All PCRs were performed in triplicate on an Applied Biosystems ViiA7 Real-Time PCR System.
2.12. Reporter Plasmid Construction and Luciferase Assay
PCR amplification of the 3′-untranslated regions (3’-UTRs) of STMN1, NEK4, and EG5 cDNAs from HCT116 cells was performed using the primer sets shown in Table S1. Amplified products were sub-cloned into pCR2.1-TOPO TA vector (Invitrogen, Cat# 45-0641) and sequenced, and then cloned into pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Cat# E1330). Site-directed mutagenesis of the miR-101 target sites in the STMN1, NEK4, and EG5 mRNAs was performed using the PrimeSTAR Mutagenesis Basal Kit (TaKaRa, Cat# R046A) and the mutagenesis primer sets listed in Table S1. The nucleotide sequences of the 5′-seed of the miR-101 target site in the STMN1, NEK4, and EG5 mRNAs were replaced with the recognition sequences for EcoRV or KpnI (lowercase, underlined in Table S1). For the luciferase assay, HCT116 cells were seeded at a density of 5 × 104 cells per well in 24-well plates and co-transfected with 200 ng of reporter plasmid and either Pre-miR miRNA Precursor Molecules (Ambion) at a final concentration of 10 nM or miRCURY LNA microRNA Inhibitors (Exiqon) at a final concentration of 50 nM. Transfections were performed using Lipofectamine 2000 (Life Technologies, Cat# 11668019). Lysates of transfected cells were subjected to firefly and Renilla luciferase assays using the Dual-Luciferase Reporter assay system (Promega, Cat# E1910).
2.13. Generation of Lentiviruses for Inducible miRNA Expression and Virus-Infected Cells
The genomic region of the hsa-miR-101-1 gene was amplified from human female genomic DNA (Promega, Cat# G152A) using PrimeSTAR GXL DNA Polymerase (TaKaRa, Cat# R050A) and the primer sets listed in Table S1. Amplified products were cloned into pInducer10-mir-RUP-PheS (Addgene Plasmid #44011) and then sequenced. To generate the negative control vector, a non-targeting sequence was introduced into pInducer10-mir-RUP-PheS.
To produce lentiviral particles, 293FT cells (Invitrogen, Cat# R70007) were transiently transfected with viral vector using the Trans-Lentiviral Packaging System (Open Biosystems, Cat# TLP4614) and Lipofectamine 3000 (Life Technologies, Cat# L3000015), and the medium was replaced with fresh growth medium to produce virus particles. HCT116 or A549 cells were transduced with lentivirus by the standard method, incubated for 7–10 days in growth medium containing 1 μg/ml puromycin, and then cloned by limiting dilution. To induce expression of miR-101, cells were treated with 1 μg/ml Dox for the indicated times, and miR-101 expression was confirmed by qRT-PCR. Proliferation of inducible miR-101–expressing cells was assessed using a Countess Automated Cell Counter (Life Technologies). Cell numbers were averaged from three independent experiments.
2.14. Xenograft Model
Genetically engineered HCT116 cells (3 × 106 cells/mouse) or A549 cells (3 × 106 cells/mouse) were detached from the bottom of the culture dish using TrypLE Express (Life Technologies, Cat# 12604-013), passed through a cell strainer, and then re-suspended in 100 μl of PBS. The cells were then subcutaneously implanted into the backs of 6-week-old female nude mice (14 mice in HCT116 or 18 mice in A549) (Charles River Laboratories Japan Inc., Yokohama Japan).
When tumors reached 50–100 mm3 in size, mice were divided randomly into two groups (5–8 animals each) for subsequent analysis of tumor development with or without Dox treatment. Dox was delivered through the diet (625 mg/kg, Harlan Laboratories, Cat# TD.01306), and tumor volumes were measured with calipers. Administration of Dox was monitored by fluorescence imaging of turboRFP using an IVIS Lumina Living Image System (Xenogen). Xenograft tumors were dissected and stored in liquid nitrogen for RNA and protein extraction, or fixed in 4% paraformaldehyde for immunohistochemistry.
2.15. Histological Analyses
Dissected tissues were fixed in formalin, embedded in paraffin, and sectioned into 3 μm slices by standard procedures. Sections were deparaffinized/rehydrated and stained with hematoxylin–eosin. For antigen retrieval, sections were autoclaved for 15 min in 10 mM sodium citrate buffer (pH 6·0). Tissue sections were incubated with rabbit monoclonal anti-p53 antibody (clone 7F5, 1/160 dilution; Cell Signaling Technology, Cat# 2527; RRID: AB_331211) at 4 °C overnight, followed by incubation with SignalStain Boost IHC Detection Reagent (Cell Signaling Technology, Cat# 8114) for 30 min at room temperature. Immunoreactive signals were visualized with 3,3′-diaminobenzidine tetrahydrochloride solution (Vector, Cat# SK-4105), and nuclei were counterstained with hematoxylin.
2.16. Immunofluorescence
Cells transfected with either miR-101 or negative control (5 nM final concentration) for 2 days were grown on μ-dishes (Ibidi, Munich, Germany), and then fixed with 100% methanol for 30 min at −20 °C. After blocking with PBS–1% BSA (PBS-BSA), cells were incubated at room temperature for 1 h with a rabbit monoclonal anti-p53 antibody (clone E26) (Epitomics #1026-1, 1/100 dilution) and mouse monoclonal anti-γH2A.X (Ser139) antibody (Millipore #05-636, RRID: AB_309864, 1/500 dilution) in PBS-BSA, and then incubated for 1 h with secondary anti-rabbit IgG antibody coupled to Alexa Fluor 488 (Molecular Probes, Cat# A11008, RRID: AB_143165, 1/250 dilution) and anti-mouse IgG antibody coupled to Alexa Fluor 594 (Molecular Probe Cat# A21201; RRID: AB_2535787, 1/500 dilution). Nuclei were stained with Hoechst 33342 (1 μg/ml). Stained cells were subjected to fluorescence microscopy.
3. Statistical Analysis
Statistical analyses were performed using Prism (GraphPad Software, La Jolla, CA, USA). Differences in expression were evaluated by Student's t-test, and differences in relapse-free survival (RFS) were evaluated by log-rank test. p < 0·05 was considered statistically significant.
4. Results
4.1. miR-101 is an Activator of the p53 Pathway
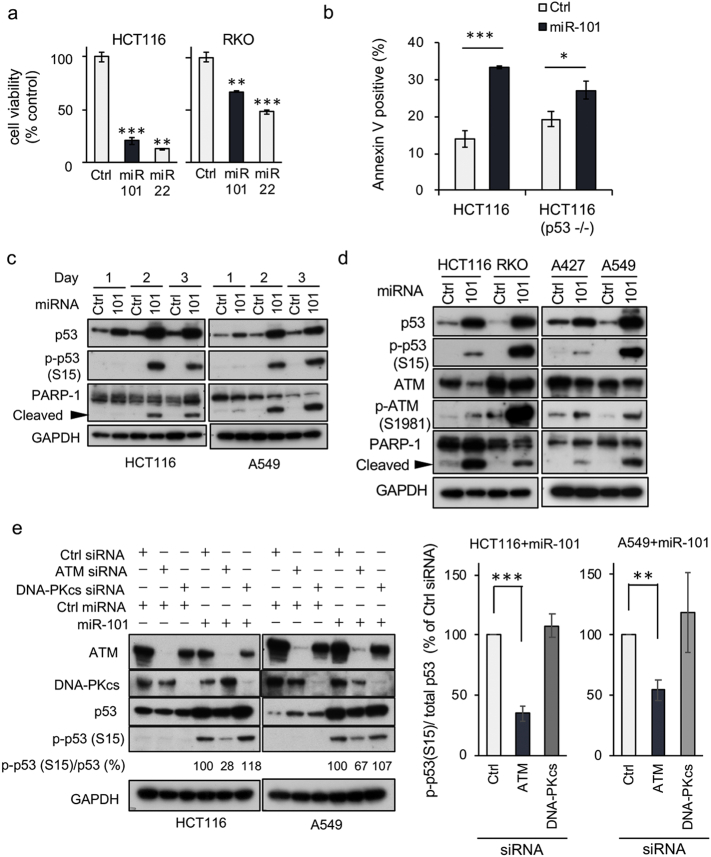
Using a previously established functional dropout screen [17], we identified miR-101 as a tumor-suppressive miRNA. In p53 WT (HCT116 and RKO) cell lines, miR-101 decreased cell proliferation dramatically, to a similar extent as miR-22, which induced apoptosis in p53 WT cancer cells [13] (Fig. 1a). Annexin V–positive cells were abundant after introduction of miR-101 into p53 WT cells, but the effect was weak in p53−/−, suggesting that miR-101 induces apoptosis dominantly in a p53 WT context (Fig. 1b). Microarray analysis of HCT116 and RKO in the presence of miR-101 (GEO accession number, GSE85696) confirmed that the miRNA induced an ATM-dependent DNA damage response pathway and affected expression levels of p53 downstream targets (Supplementary Fig. 1a and b). In both p53 WT cell lines, HCT116 and A549, elevated phosphorylation of p53 on Ser15 was observed 2 days after introduction of miR-101, concomitant with cleavage of PARP-1 (Fig. 1c), suggesting that miR-101 induces apoptosis through upregulation of p53 target genes. After introduction of miR-101, we observed phosphorylation of ATM, which phosphorylates p53 at Ser15 [18, 19], although phosphorylation levels varied among cell lines (Fig. 1d); in particular, the p53 phosphorylation level was lower in A427 cells than in the cell lines examined. Ser 15 phosphorylation of p53 was decreased by depletion of ATM with short interfering RNA (siRNA), but not by depletion of DNA-PKcs, another p53 kinase [20] (Fig. 1e). Furthermore, knockdown (KD) of ATR, an ATM-related kinase [21], decreased Ser15 phosphorylation in HCT116 cells, but not in A549 cells (Supplementary Fig. 1c). Therefore, we concluded that miR-101 induces activation of the ATM–p53 axis. We then stained cells for γH2AX, the primary target of ATM after DNA damage [22]. As expected, miR-101 induced nuclear accumulation of p53. However, in miR-101–expressing cells γH2AX did not exhibit the typical nuclear staining pattern induced by the DNA-damaging agent ADR, suggesting that miR-101 activates the ATM–p53 axis with an unknown mechanism (Supplementary Fig. 1d).
Fig. 1.
miR-101 is an activator of p53. (a) Cells were transfected with either miR-101 or negative control miRNA (Ctrl), and cell proliferation was monitored by MTS assay. miR-22 was used as a positive control for cell proliferation. Each experiment was performed in triplicate and statistical analysis was applied. Data are shown as means ± SD. Two independent experiments gave a similar result. A representative result is shown. ** and *** indicate p < 0·01 and p < 0·001, respectively. (b) Either 5 nM of miR-101 or Ctrl was transfected into the indicated cell lines for 3 days. Apoptotic cells were detected by staining with Annexin V and 7-amino actinomycin D (7-AAD), and then analyzed by flow cytometry. Percentages of Annexin V-positive cells, including 7-AAD positive and negative cells, were graphed. Statistical analysis was performed by Student's t-test. Data are expressed as means ± SD of three independent experiments. *** and * indicate p < 0·001 and p = 0·027, respectively. (c) Cells were transfected with either 5 nM miR-101 or Ctrl; protein samples were collected at the time points indicated at the top, and then subjected to immunoblot analysis. Data are representative of two independent experiments. (d) ATM phosphorylation after introduction of miR-101, analyzed by IB. Data are representative of two independent experiments. (e) Cells were co-transfected with the indicated combination of siRNA and miRNA (5 nM each) for 3 days, and then subjected to IB analysis. Data are representative of three independent experiments. Quantitative assessment of pS-p53 was performed using the Image Lab software (ver. 6, Bio-Rad). After normalization against total p53, the level of pS-p53 was expressed as a percentage relative to the level in Ctrl siRNA+ miR-101, which was defined as 100%. In the right graphs, data are shown as means ± SEM of three independent experiments. ** and *** indicate p < 0·01 and p < 0·001, respectively.
miR-101 was previously identified as a repressor of EZH2, a histone lysine methyltransferase that is a component of Polycomb repressive complex 2 [23]. Hence, we sought to determine whether repression of EZH2 was associated with miR-101–induced activation of the p53 network. When EZH2 was depleted by siRNAs, p53 accumulated in the presence, but not the absence, of miR-101 (Supplementary Fig. 1e), indicating that miR-101 activates the intracellular p53 network by repressing unknown targets.
4.2. miR-101 Blocks Entry into Mitosis and Induces p53-Dependent Apoptosis
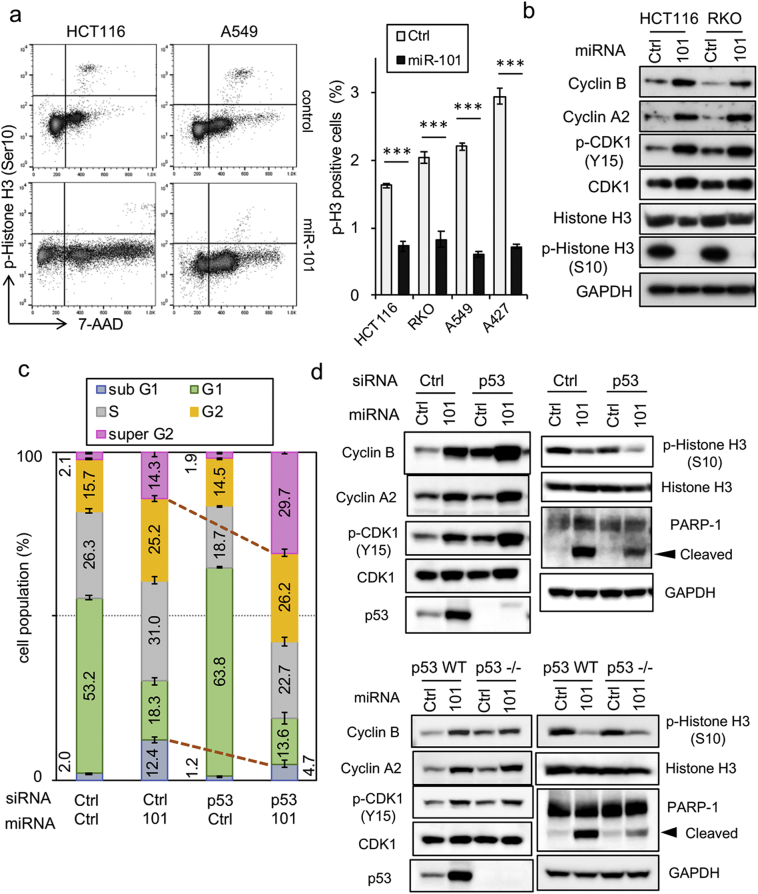
Flow cytometric analysis indicated that miR-101 caused an increase in the sub-G1 population (Supplementary Fig. 2a), as also shown in Fig. 1b. G2/M-phase cells also accumulated (Supplementary Fig. 2a), consistent with the function of the ATM–p53 axis as a G2/M checkpoint [24]. In this experiment, A427 exhibited a different pattern than other cell lines, which might be attributed to the inactivation of the p53 pathway despite the presence of a WT p53 gene [25]. The level of phosphorylated histone H3, a marker of M phase [26], was significantly reduced in miR-101–expressing cells (Fig. 2a and Supplementary Fig. 2b), indicating that miR-101 inhibits entry into M phase by triggering cell-cycle arrest in G2 phase. IB analysis revealed enrichment of the G2 population, reflected by accumulation of cyclin B and phosphorylated CDK1 [27] (Fig. 2c). To address the p53 dependence of these phenomena, we performed siRNA-mediated KD of endogenous p53. Intriguingly, in the absence of p53, the populations of sub-G1 and super-G2 cells were decreased and increased by miR-101, respectively, but the G2/M population was unchanged (Fig. 2c, columns 2 and 4). Furthermore, IB analysis revealed that accumulation of cyclin B, cyclin A2, and phospho-CDK1 was still detectable after p53 KD (Fig. 2d, upper left image), and also in p53−/− cells (Fig. 2d, bottom left image), indicating that p53 was not involved in the observed G2 arrest. As expected, the level of cleaved PARP-1 was reduced in both p53 KD and p53−/− cells (Fig. 2d, right images). Collectively, these results imply that p53 regulates a critical branchpoint in the decision of whether to undergo apoptosis following the G2-phase arrest induced by miR-101.
Fig. 2.
Induction of G2 arrest and p53-dependent apoptosis by miR-101. (a) HCT116 and A549 cells were transfected with either 5 nM miR-101 or Ctrl for 3 days, and the level of phosphorylated histone H3 was determined by flow cytometry. Positivity for phosphorylated histone H3 is indicated in the graph at the right. Data are expressed as means ± SD of three independent experiments. *** indicates p < 0·001. (b) HCT116 and RKO cells, transfected with miR-101, were incubated for 3 days, and cell extracts were subjected to IB analysis. Data are representative of two independent experiments. (c) miR-101 and p53 siRNA (5 nM each) were introduced into HCT116 cells in the indicated combinations, and cell-cycle analysis was carried out on a flow cytometer. Data are shown as means ± SD of three replicates and are representative of two independent experiments. (d) G2 arrest and apoptosis-related proteins were monitored by immunoblot analysis of p53-KD HCT116 cells or isogenic p53 KO cells. The data are representative of two independent experiments.
4.3. Identification of the miR-101 Targets Responsible for G2 Arrest and Apoptosis
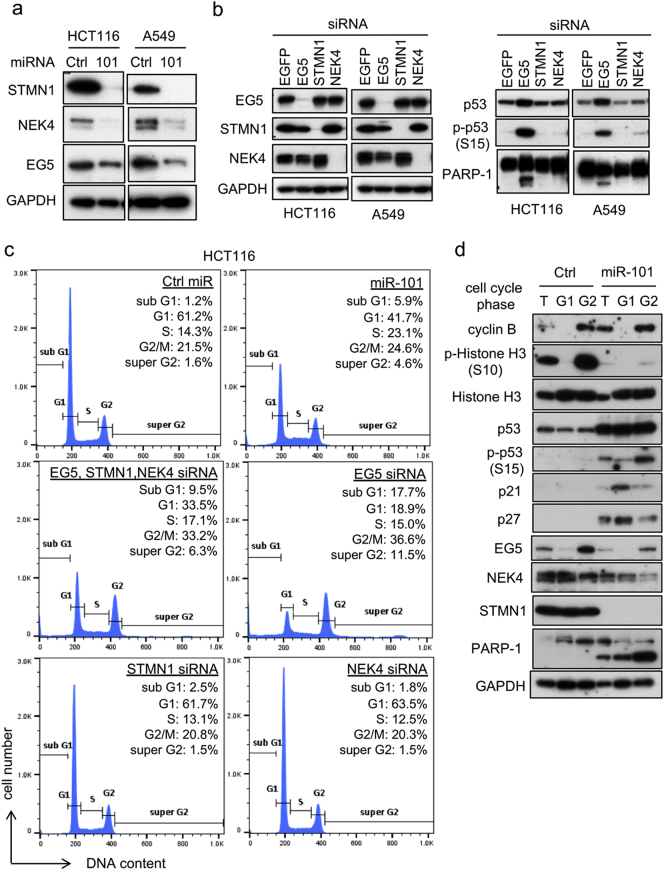
To identify the target genes responsible for miR-101–induced phenotypes, we analyzed microarray data. First, we applied the GO database to select G2/M regulators that were downregulated (i.e., fold change <1·0) by miR-101. Potential miR-101–binding sites in the full mRNA sequences of candidate genes were predicted in silico using miRWalk [28]. The list of candidate genes was sorted by the probability of being a target (p value), and the three top-ranked genes, KIF11 (EG5), STMN1 (stathmin), and NEK4, were chosen for further analyses (Supplementary Fig. 3a). NEK4 and EG5 regulate centrosome positioning at G2 phase during the onset of mitosis [29, 30], whereas stathmin is a microtubule regulator [31]. All three proteins were downregulated following introduction of miR-101 (Fig. 3a), and reporter gene assays confirmed that all were directly regulated by miR-101 (Supplementary Fig. 3b).
Fig. 3.
Identification of miR-101 targets. (a) HCT116 or A549 cells, transfected with miR-101 for 3 days, were subjected to immunoblot analysis. Data are representative of two independent experiments. (b) Accumulation of p53 was analyzed by siRNA knockdown of miR-101 targets. EGFP siRNA was used as a negative control. Cells transfected with siRNA (5 nM each) for 2 days were subjected to IB analysis. Data are representative of three independent experiments. (c) HCT116 cells were transfected with the indicated combination of siRNAs (total 5 nM) against target genes or miRNA (5 nM) for 2 days, and cell-cycle distribution was determined by flow cytometry. Proportions of cells in each cell-cycle phase, expressed as percentages, are shown as the means of three replicates. Histograms are representative of two independent experiments. (d) HCT116 cells, transfected with miR-101 or Ctrl (5 nM) for 3 days, were sorted by FACS. G1 and G2 cells were collected and subjected to immunoblot analysis.
We then investigated the association of these targets with the ATM–p53 axis by performing siRNA-mediated depletion of each gene. Interestingly, although single KD of NEK4 or STMN1 did not affect either total p53 or phospho-Ser15 p53 levels, EG5 depletion clearly induced accumulation of phospho-Ser15 p53 and increased PARP-1 cleavage (Fig. 3b, right images). In addition, EG5 KD induced phosphorylation of ATM (Supplementary Fig. 3c), suggesting that downregulation of EG5 is one of the causative events underlying activation of the ATM–p53 axis by miR-101. Single KD of these targets had different effects on the cell-cycle profile that were different from those of miR-101 expression. On the other hand, KD of all three genes induced a cell-cycle profile similar to that of miR-101 expression in HCT116 or A549 cells (Fig. 3c and Supplementary Fig. 3d). Although we did not obtain the optimum degree of downregulation of the target genes responsible for the miR-101–induced phenotypes, these results suggest that repression of multiple targets, including EG5, STMN1 and NEK4, is required for these phenotypes.
Next, we sorted cells by cell-cycle phase in the presence or absence of miR-101, and then subjected the collected cells to IB analysis. Cells were successfully separated by this method, as reflected by accumulation of phosphorylated histone H3 and cyclin B in the G2/M-phase sample of control cells (Fig. 3d, Ctrl, G2). When cells were transfected with miR-101, the level of phosphorylated histone H3 in G2 cells was clearly reduced (Fig. 3d, miR-101). Surprisingly, Ser15 phosphorylation of p53 was observed predominantly in G2 phase, whereas accumulation of total p53 was observed in G1 phase (Fig. 3d, miR-101, G2). Furthermore, EG5, whose expression is maximal from S to G2 phase [32], was also repressed in G2 cells, suggesting that inhibition of mitotic onset could be ascribed to repression of EG5 at G2 phase. Notably, strong cleavage of PARP-1 was observed in G2-phase cells (Fig. 3d). Together, these results indicate that miR-101 induces apoptosis via activation of the ATM–p53 axis, which can be attributed to G2-specific repression of EG5.
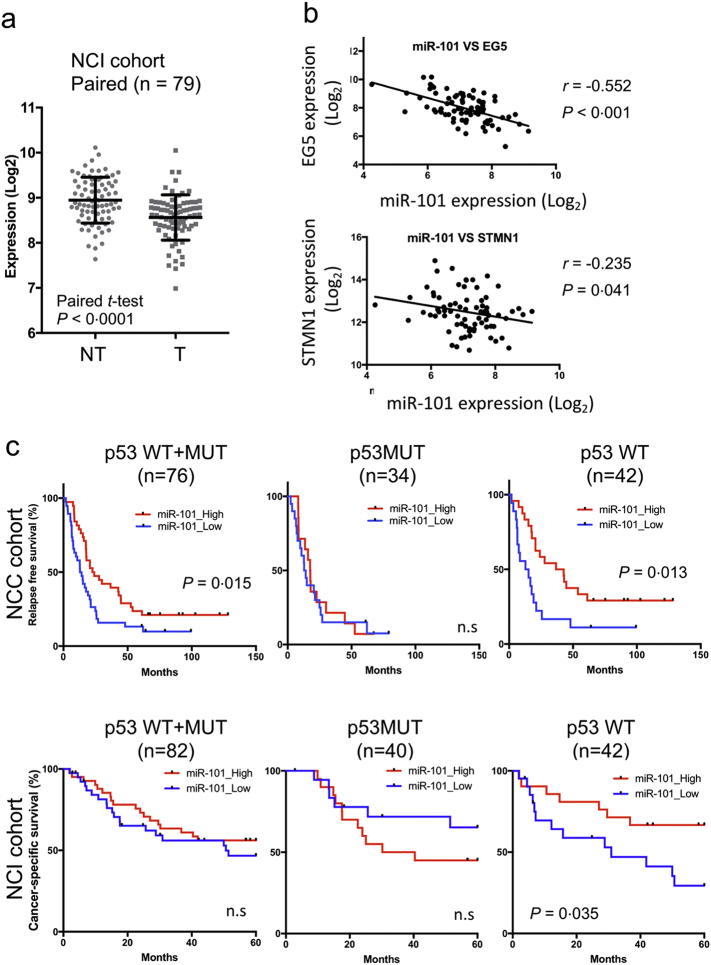
4.4. miR-101 Downregulation in Lung Cancer
We compared expression of miR-101 in clinical samples using two different LADC cohorts from the NCI (US) and NCC (Japan). Clinico-pathological characteristics of patients enrolled in these cohorts are provided in Table 1. As shown in Fig. 4a, miR-101 levels were significantly lower in tumor tissue (T) than in non-cancerous counterpart (NT), and about 70% of cases in the NCI cohort exhibited reduced expression of miR-101 (Fig. 4a and Supplementary Fig. 4a). Similarly, reduced expression of miR-101 was also observed in the NCC cohort (Supplementary Fig. 4b). Expression of EG5 (KIF11) was significantly higher in tumor tissues than in normal lung tissues, and STMN1 expression was slightly, but not significantly, elevated (Supplementary Fig. 4b). By contrast, NEK4 was significantly downregulated in tumor tissues (Supplementary Fig. 4b), indicating that NEK4 is not an important target in the context of lung cancer development; consequently, this factor was omitted from further analyses. Importantly, EG5 expression was significantly negatively correlated with expression of miR-101 (r = −0·552, p < 0·001), and STMN1 was weakly negatively correlated (r = −0·235, p = 0·041) (Fig. 4b), suggesting that both genes are physiological targets of miR-101 during the development of LADC.
Fig. 4.
Expression of miR-101 and its target genes in LADC. (a) Expression levels of miR-101 were determined in LADC and matched non-cancerous regions in the NCI cohort (n = 79). Expression of miR-101 was analyzed on a NanoString. Significance was evaluated by paired t-test. (b) Correlation of miR-101 and target gene expression in NCC samples. (c) Kaplan–Meier survival curves representing relapse-free survival (RFS) and cancer-specific survival of LADC in the NCC (n = 76) and NCI (n = 82) cohorts, respectively. LADC cases, in which miR-101 levels were higher or lower than the median of all cases, were defined as miR-101–high (red) or –low (blue), respectively. Horizontal axis indicates duration (months). Statistical analysis was carried out by log-rank test. n.s., not significant.
Next, we investigated the dependence of miR-101 expression on TP53 status in LADCs. As indicated in Supplementary Fig. 4c, we observed no significant differences in miR-101 expression in either cohort. When we divided LADC cases according to miR-101 level (high: > median of total cases; low: < median) and compared their prognoses, we found that RFS was shorter in miR-101–low cases in the NCC cohort, indicating that a lower level of miR-101 is associated with poorer prognosis in LADC (Fig. 4c, left upper panel). Importantly, low expression of miR-101 was associated with poor prognosis only in p53 WT cases, but not in p53 MUT LADCs (Fig. 4c, upper middle and right panels). Moreover, miR-101–low cases in the NCI cohort exhibited shorter cancer-specific survival exclusively in p53 WT LADCs (Fig. 4c, lower right panel). These results strongly suggest that miR-101 contributes to intrinsic TS networks governed by p53, and that disruption of this axis promotes malignant growth of p53 WT tumors.
4.5. Activation of miR-101 by Pol I Transcription Inhibitors in a p53-Dependent Manner
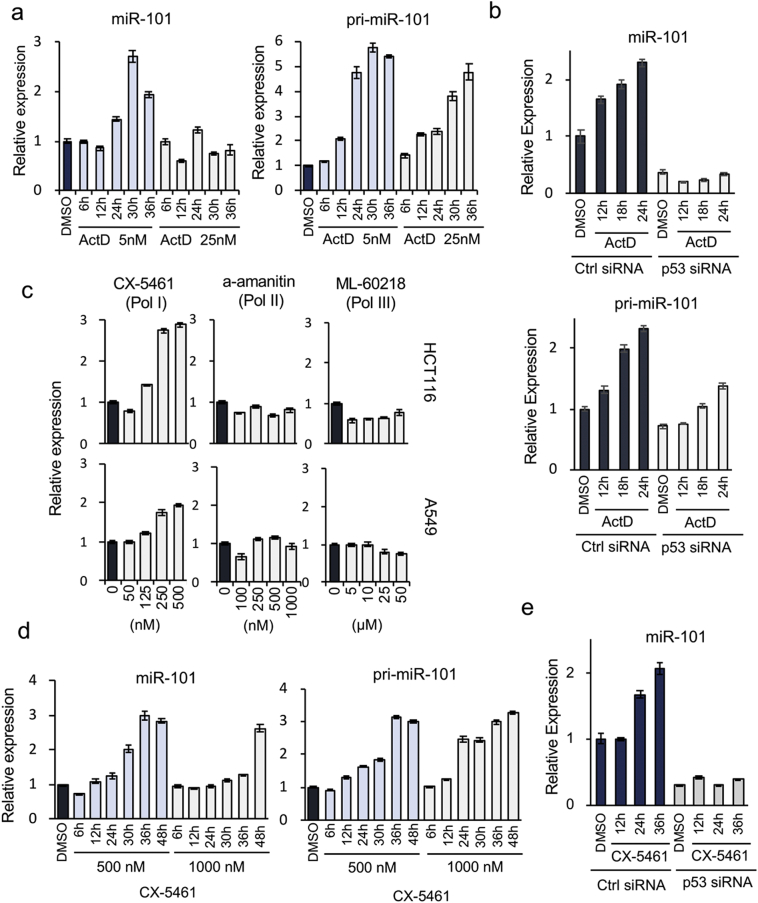
To further analyze the molecular link between miR-101 and the p53-dependent TS network, we searched for extracellular stimuli that induced the activation of both p53 and miR-101 in p53 WT HCT116 cancer cells. We used the following agents as p53-activating stresses: ActD, an inhibitor of RNA polymerases [33]; ADR, a topoisomerase inhibitor [34]; 5-FU, an inhibitor of DNA synthesis [35]; RO-3306 and PTX, inhibitors of mitotic function [36, 37]; and K858, an EG5 inhibitor [38]. In addition, we employed UV and γ-ray irradiation as DNA-damaging stresses. Interestingly, only 24 h of treatment with ActD caused reproducible induction of expression of miR-101 in p53 WT cancer cells (Fig. 5a and Supplementary Fig. 5a and b). The primary transcript level of miR-101 (pri-miR-101) increased prior to upregulation of its mature form (Fig. 5a, right graph). Importantly, induction of miR-101 was evident at a lower concentration of ActD, implicating inhibition of Pol I in the activation [39]. Although the mature form of miR-101 was clearly upregulated only after cells were exposed to the lower concentration of ActD, the level of pri-miR-101 was elevated by treatment at both concentrations (Fig. 5a, right graph), suggesting that activation of miR-101 is controlled at the post-transcriptional level. Moreover, production of mature miR-101 was markedly decreased by p53 KD in both HCT116 and A549 cells, whereas pri-miR-101 levels were elevated after exposure to ActD even in the KD cells, providing further evidence that activation of miR-101 in response to ActD treatment is mainly regulated by p53 at the post-transcriptional level (Fig. 5b and Supplementary Fig. 5c). Together, these results confirm that p53-dependent elevation of mature miR-101 is regulated post-transcriptionally.
Fig. 5.
Activation of miR-101 expression by inhibition of RNA polymerase I. (a) Left and right graphs show time courses of expression of miR-101 and pri-miR-101, respectively, in HCT116 cells following treatment with the indicated concentrations of ActD. Data are expressed as means ± SD of triplicate reactions after normalization against the corresponding level of TBP mRNA, and are representative of at least two independent experiments. Incubation times are provided below the graphs. (b) miR-101 and pri-miR-101 expression, as determined by qRT-PCR, after treatment with 5 nM ActD in the presence or absence of p53. Data are expressed as means ± SD of triplicate reactions after normalization against the corresponding level of PPIA mRNA, and are representative of two independent experiments. (c) miR-101 activation by the indicated concentrations of specific RNA polymerase inhibitors. Upper and lower graphs depict results for HCT116 and A549, respectively. CX-5461, alpha-amanitin, and ML-60218 are inhibitors of Pol I, Pol II, and Pol III, respectively. Concentrations of inhibitors used for the assay are listed below the lower graphs. Data are expressed as means ± SD of triplicate reactions after normalization against the corresponding level of TBP mRNA, and are representative of at least two independent experiments. (d) miR-101 and pri-miR-101 expression in HCT116 cells after treatment with two concentrations of CX-5461 were measured at the time points indicated below the graph. Data are expressed as means ± SD of triplicate reactions after normalization against the corresponding level of TBP mRNA, and are representative of at least two independent experiments. (e) Expression of miR-101 after exposure of HCT116 cells to CX-5461 in the p53 KD cells. Data are expressed as means ± SD of triplicate reactions after normalization against the corresponding level of PPIA mRNA, and are representative of two independent experiments.
Next, we treated cells with specific inhibitors of each class of RNA polymerase: CX-5461 for Pol I, α-amanitin for Pol II, and ML-60218 for Pol III [[40], [41], [42]]. Only Pol I transcription inhibition by CX-5461 activated miR-101 expression in both HCT116 and A549 cells (Fig. 5c). Treatment with CX-5461 also induced upregulation of pri-miR-101 (Fig. 5d). Upregulation of the mature form of the miR-101 gene was clearly abolished by p53 KD (Fig. 5e). Collectively, our data demonstrate that WT p53 is required for activation of miR-101 in response to inhibition of Pol I transcription, which causes nucleolar stress, and that a positive-feedback circuit between p53 and miR-101 functions as an intrinsic network that drives this response.
4.6. miR-101 is Involved in Regulation of the Late Stage of Nucleolar Stress
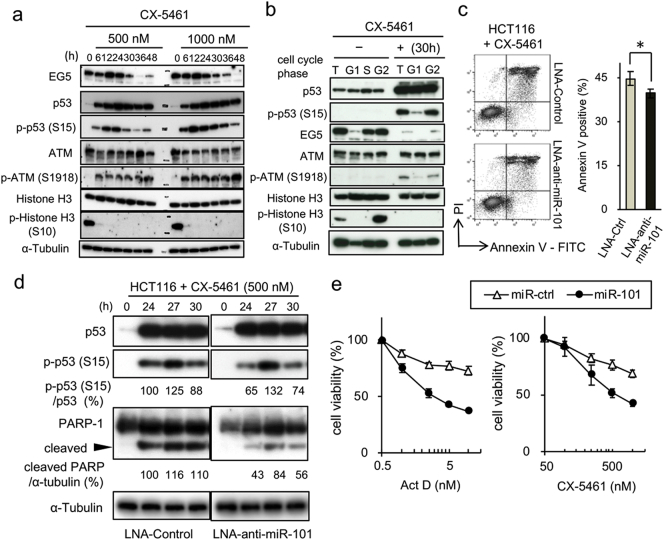
To further investigate the roles of miR-101 in the p53-dependent nucleolar stress response, we analyzed protein levels of EG5 and activation of the ATM–p53 axis. When cells were treated with CX-5461 for 24 h, the cell cycle was predominantly arrested in G2 phase (Supplementary Fig. 6a). EG5 was downregulated 36 h after treatment with CX-5461 (Fig. 6a), when miR-101 expression reached its peak (Fig. 5d). Although the p53 phosphorylation level was reduced, it persisted from 30 to 36 h after exposure to CX-5461. Phosphorylation of ATM occurred immediately and continued throughout the incubation period.
Fig. 6.
miR-101 is involved in regulation of the late stage of nucleolar stress. (a) Expression of miR-101 target molecules in HCT116 cells after exposure to CX-5461, analyzed by immunoblot analysis at the indicated time points. Concentrations of CX-5461 and incubation times are shown at the top of the image. Data are representative of two independent experiments. (b) G1- and G2-phase cells from CX-5461–treated and –untreated populations of HCT116 cells were separated by FACS, and the levels of target proteins were determined by immunoblot analysis. (c) Apoptotic cells induced by CX-5461 (500 nM) after treatment with miR-101 inhibitor (LNA-anti-miR-101) were analyzed by flow cytometry. Percentages of Annexin V–positive cells are indicated in the right graph. Each experiment was performed in triplicate and statistical analysis was applied. Data are shown as means ± SD. Two independent experiments gave a similar result. A representative result is shown. One-side t-test, *p < 0·05. (d) Cells, transfected with either LNA-anti-miR-101 or LNA-control, were incubated with 500 nM CX-5461 at the indicated time points. Total cell lysates were subjected to IB analysis. pS-p53 and cleaved PARP bands were quantified relative to total p53 and α-tubulin, respectively, using the ImageLab software (Bio-Rad); relative quantifications are shown. Data are representative of two independent experiments. (e) HCT116 cells transfected with 0·5 nM of either miR-101 or Ctrl were treated with the specified concentration of chemicals. After 24 h, cell viability was analyzed by MTS assay. Error bars indicate means ± SEM of three independent experiments.
We then performed cell-sorting analysis in the presence of CX-5461. As expected, EG5 was downregulated in G2 cells 30 h after exposure to CX-5461 (Fig. 6b). Importantly, phosphorylated p53 and ATM were predominantly detectable in the G2 population (Fig. 6b), consistent with our observation that introduction of miR-101 induces the p53 phosphorylation specifically in G2 phase (see Fig. 3d). Furthermore, inhibition of miR-101 by a locked nucleic acid (LNA), whose activity was confirmed in a reporter gene assay (Supplementary Fig. 6b), significantly decreased the number of apoptotic cells induced by CX-5461 (Fig. 6c). IB analysis also revealed that the phosphorylation level of p53 was lower in cells transfected with LNA-anti-miR-101 than in control cells (Fig. 6d), and the level of cleaved PARP-1 was also reduced (Fig. 6d). Taken together, these results strongly suggest that miR-101 is a component of a p53 network involved in responding to nucleolar stress, and that the miRNA enables full activation of this pathway, which is required for determination of cellular fate.
To further confirm the involvement of miR-101 in the nucleolar stress pathway, we analyzed the effect of miR-101 on the sensitivity of cells to M-phase inhibitors. Pre-introduction of a low concentration of miR-101 (0·5 nM) did not increase sensitivity to PTX and RO-3306 (Supplementary Fig. 6c). Similarly, the cytotoxicity of the EG5 inhibitor K858 was not affected by the cellular miR-101 level. On the other hand, miR-101 increased the sensitivity of cells treated to CX-5461 and ActD (Fig. 6e). The cells were also sensitized to 5-FU, which induces ribosomal stress by blocking RNA synthesis (Supplementary Fig. 6c). These results suggest that miR-101 is a crucial component in an intrinsic surveillance system that monitors aberrations of ribosomal biogenesis triggered by Pol I inhibition.
4.7. Inhibition of Tumor Growth by miR-101 in vivo
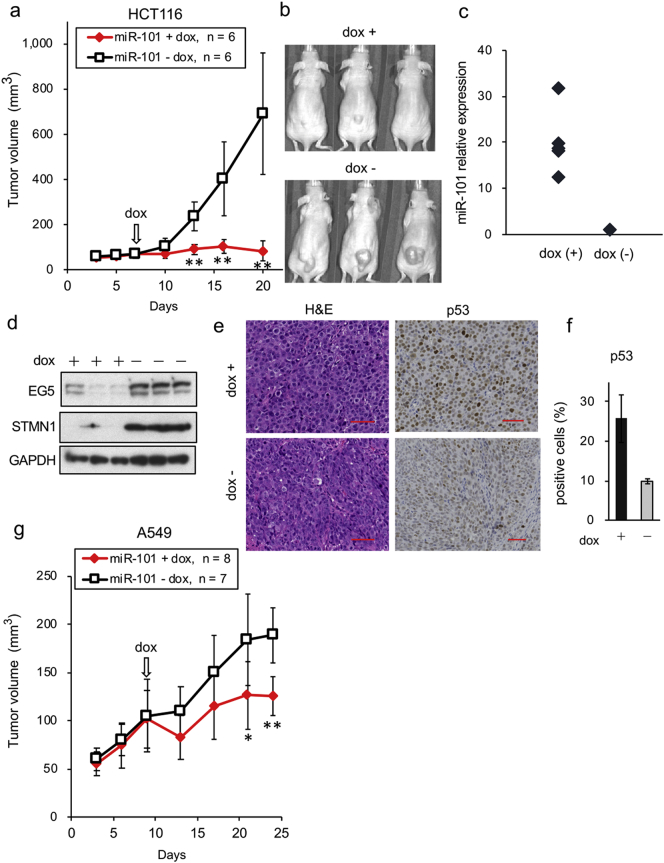
We next investigated the in vivo effect of miR-101 on growth and pathological changes of xenografts in the mice. For this purpose, we established HCT116 cell lines stably expressing miR-101 under the control of a Tet-inducible promoter. Expression of miR-101 could be monitored by the expression of RFP (Supplementary Fig. 7a and e). As indicated in Supplementary Fig. 7b, expression of miR-101 was markedly upregulated after addition of Dox, but very low in control cells. We then mixed equal numbers of cells from each clone (clones 4, 5, and 7) and monitored cell proliferation and target gene expression. After Dox treatment, cell proliferation was clearly inhibited, and expression of the important target EG5 was suppressed. In addition, accumulation of p53 and enhancement of PARP-1 cleavage were only detectable in miR-101–expressing cells (Supplementary Fig. 7c, d, and e).
These cells were subcutaneously injected into the back of BALB/c-nu mice, and tumors were allowed to form for 6 days in the absence of Dox. Delivery of Dox to xenografts was assessed by monitoring the RFP signal after the dietary change (Supplementary Fig. 7f). Xenografts of vector control cells did not change in size or growth rate after administration of Dox (Supplementary Fig. 7g and h). By contrast, induction of miR-101 by Dox strongly inhibited the growth of tumor xenografts (Fig. 7a and b). The level of miR-101 in the resultant tumors was clearly higher in Dox+ tumors than in controls (Fig. 7c), whereas EG5 and STMN1 were downregulated (Fig. 7d). Pathological analyses of the resultant xenografts revealed that cells in Dox− tumors had spindle shapes (Fig. 7e, bottom left), whereas those in Dox+ tumors had a polygonal structure reminiscent of epithelial growth (Fig. 7e, top left). In addition, p53-positive cells were abundant in the Dox+ tumors, as determined by immunohistochemistry and cell counting (Fig. 7e and f). Cells transduced with control virus did not exhibit changes in either miR-101 expression or pathological characteristics (Supplementary Fig. 7i and j).
Fig. 7.
Repression of xenograft growth by miR-101. (a) In vivo tumor growth of miR-101–expressing cells. HCT116-miR-101 cells (3 × 106 cells/mouse) were implanted into the backs of nude mice (n = 14). After tumor size reached to 50–100 mm3, mice were separated into two groups (5–8 mice per group). The number of mice in each group is indicated at the top of the graph. Dox administration was started at the time point indicated by the arrow. Tumor size was measured twice per week. Data are shown as means ± SD of six tumor xenografts. Representative data of two independent experiments, carried out on the same number of mice, are indicated. **p < 0·01. (b) Representative image of in vivo tumors obtained from three to six mice on day 20. (c) Expression levels of miR-101 in the resultant tumors. Tumor tissues were removed from mice at day 20, and miR-101 levels were quantified by qRT-PCR. Number of samples: Dox (+), n = 5; Dox (−), n = 6. Data are expressed as the mean of triplicate reactions after normalization against the corresponding level of the RNU48 snRNA. (d) Protein levels of miR-101 targets in xenograft tumors. Data are representative of two independent experiments. (e) Sections were obtained from the resultant tumors on day 20 and analyzed by H&E staining (left two images) and p53 immunostaining (right two images). Scale bars, 50 μm. (f) Strongly p53-positive cells were counted using the Hybrid Cell Count software. Data are expressed as the percentage of total cells (2 × 104 cells in each sample) and are shown as means ± SD. (g) In vivo tumor growth of miR-101–expressing A549 cells. A549-miR-101 cells (3 × 106 cells/mouse) were implanted into the backs of nude mice (n = 18). After the size of xenograft tumors reached 50–100 mm3, mice were divided into two groups (6–8 mice in each group). The number of mice for each group is indicated at the top of the graph. Dox administration was started at the time point indicated by the arrow. Data are representative of two independent experiments, carried out on the same number of mice. * and ** indicate p < 0·05 and p < 0·01, respectively.
We confirmed in vivo repression of xenograft tumors using lung cancer cell line A549. As expected, miR-101–expressing xenograft tumors were significantly smaller than controls (Fig. 7g). Collectively, these results indicate that miR-101 represses in vivo tumor growth by activating the p53 pathway.
4.8. Combined Treatment with CX-5461 and IAP Inhibitor Mimics Activation of the p53–miR-101 Circuit
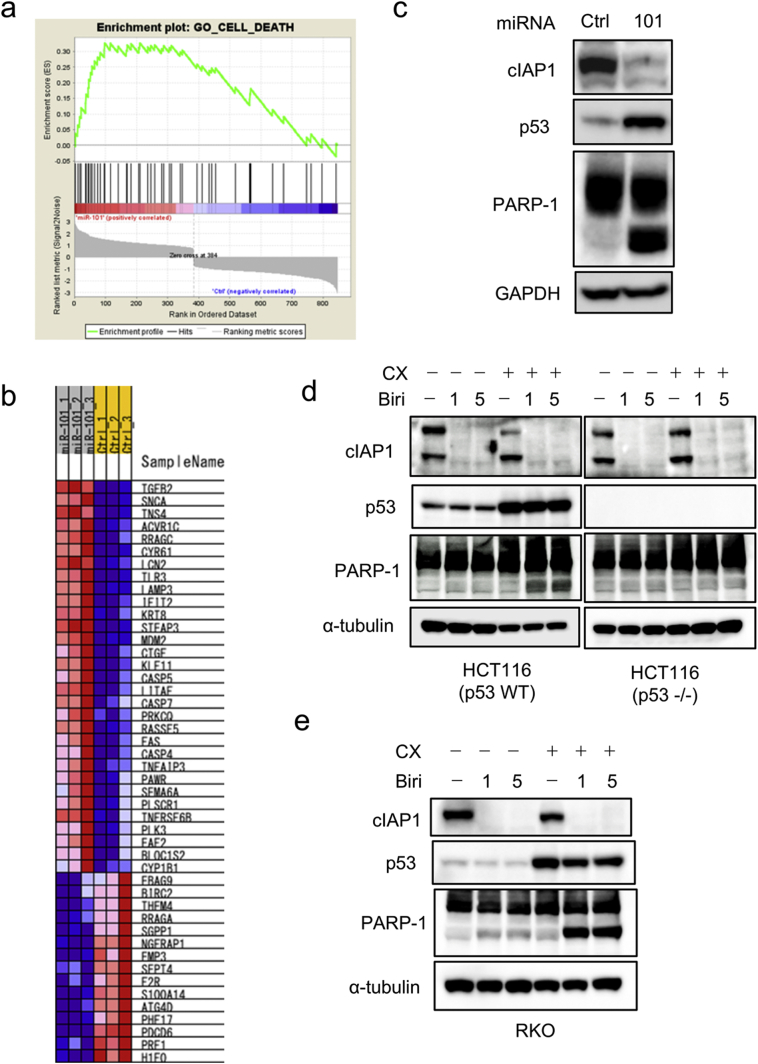
Our findings led us to hypothesize that a key molecule or molecules whose repression induces apoptosis in p53 WT cells function downstream of this circuit. If so, full activation of the p53–miR-101 circuit could be mimicked by combination treatment with Pol I transcription inhibitor and a molecularly targeted drug. To explore this possibility, we performed gene set enrichment analysis (GSEA) of the microarray data, and found that expression levels of many apoptosis regulators were altered by miR-101 (Fig. 8a and b). Due to the availability of a selective inhibitor [43], we focused on cIAP1 (also known as BIRC2), an endogenous inhibitor of caspase activation [44]. Microarray data revealed that cIAP1 was downregulated by miR-101 in HCT116 cells, whereas expression of other BIRC family proteins and their regulators was not significantly altered. On the other hand, miR-101 marginally suppressed cIAP1 expression and up-regulated of cIAP2 expression in RKO cells. (Supplementary Fig. 8a). Notably, miR-101-induced apoptosis in RKO cells was lower than that in HCT116 cells, although ATM and p53 were phosphorylated (Fig. 1d). Therefore, it is possible that downregulation of cIAPs (cIAP1 and cIAP2) is one of the crucial factors that determine apoptotic cell death after activation of the p53–miR-101 circuit. Indeed, IB analysis revealed that cIAP1 was repressed in HCT116 cells after introduction of miR-101, but only slightly repressed in RKO cells (Fig. 8c and Supplementary Fig. 8b).
Fig. 8.
Combined treatment with CX-5461 and birinapant mimics activation of the p53–miR-101 circuit. (a) GSEA analysis. (b) Heat map of expression levels of apoptosis regulators in the presence of miR-101. (c) Immunoblot analysis of cIAP1 after introduction of 5 nM miR-101 for 3 days. Data are representative of two independent experiments. (d and e) Cells, seeded at 3·6 × 105 cells/60 mm plate, were treated with or without 250 nM CX-5461 and the indicated concentrations of birinapant (μM) for 24 h. After harvesting, cell extracts were subjected to IB analysis. Data are representative of two independent experiments.
Birinapant, a small mimetic peptide of second mitochondria-derived activator of caspases (SMAC), is a selective inhibitor of IAP proteins [43] Treatment with birinapant is expected to inhibit cIAP1 and cIAP2. We analyzed the effect of combination treatment with CX-5461 and birinapant on induction of apoptosis in p53 WT cancer cells. As indicated in Fig. 8d, single treatment with birinapant did not activate p53 or affect cleavage of PARP-1. By contrast, combination treatment with CX-5461 and birinapant induced PARP-1 cleavage in p53 WT cells, but not in p53 KO cells (Fig. 8d). Importantly, strong cleavage of PARP-1 was also induced in RKO cells by the combination treatment (Fig. 8e). As expected, we observed no additional effect of this combination treatment in A549 cells, which is resistant to birinapant [25]. These results revealed that the cIAP family is one of the most downstream molecules in the p53–miR-101 circuit, and that its inhibition by a molecularly targeted drug can mimic activation of the circuit.
5. Discussion
miR-101 was initially shown to regulate epigenetic status by targeting EZH2 [23]. Later work revealed that miR-101 also controls autophagy through repression of STMN1 [45]. The results of this study indicate that activation of an intrinsic p53 TS pathway by miR-101 is not dependent on downregulation of EZH2 (Supplementary Fig. 1e). Furthermore, in our system, we detected no change in autophagy (data not shown). Therefore, although numerous miRNAs are involved in the regulation of p53 networks [46], miR-101 is unique in that it specifically activates the p53-dependent nucleolar stress response pathway.
From the standpoint of p53 activation, miR-101 inhibits proteasome formation by targeting the proteasome maturation factor POMP, leading to stabilization of p53 and activation of its downstream target genes, including p21 [47]. Our data revealed that miR-101–induced phosphorylation of p53 occurred only in G2 phase, even though p53 accumulated during all phases of the cell cycle (Fig. 3d). Accumulation of p21 and p27, which were induced by miR-101–mediated repression of POMP, was predominantly observed in G1 phase. On the other hand, G2 phase–specific phosphorylation of p53 induced by miR-101 was triggered by activation of ATM via repression of EG5 (Fig. 1e and Supplementary Fig. 3c). These findings indicate that miR-101 is linked to alternative pathways that promote efficient and/or appropriate activation of different p53 networks throughout the cell cycle, and that cell-cycle phase is the most important determinant of the specific TS network induced by the p53–miR-101 circuitry.
We sought to identify the cellular events that explain the intrinsic tumor-suppressive roles of the p53–miR-101 circuitry. These efforts revealed that inhibition of RNA Pol I, but not other RNA polymerases, is associated with activation of miR-101, implying that nucleolar dysfunction activates miR-101 expression in a physiological setting. Furthermore, we found that induction of miR-101 biogenesis by nucleolar stress is regulated at the post-transcriptional level. Moreover, chromatin immunoprecipitation (ChIP) assay using anti-p53 antibody revealed no binding of p53 to the 5 kb region around the miR-101 gene (data not shown). A previous study reported that miRNA biogenesis is modulated by p53 in response to DNA damage [10], but did not identify miR-101 among the class of miRNAs regulated by DNA damage– and p53-dependent processing, consistent with our finding that p53-dependent regulation of miR-101 biogenesis occurs only in the context of nucleolar stress. These results suggest that stress- and p53-dependent regulation of miRNA expression at the post-transcriptional level is crucial for the proper and appropriate connection of intrinsic TS networks.
Recently, the nucleolus, a key site of ribosome biogenesis, has attracted a great deal of interest as a target for cancer therapeutics [48]. Ribosome biogenesis, which is intimately linked to cell growth and proliferation [49], is tightly regulated by key signaling networks consisting of oncogenes and TS genes [50]. Therefore, nucleolar dysfunction may trigger activation of p53 and miR-101, with the goal of preventing malignant transformation of damaged cells. CX-5461, a small molecule that inhibits binding of SL1 to the promoter region of ribosomal DNA, activates the p53 network, resulting in induction of apoptosis [51]. A clinical trial of this compound is anticipated to demonstrate a potent anti-tumor effect [52]. Interestingly, CX-5461 activates the non-canonical ATM pathway in the absence of DNA damage [53], and induces G2 arrest and apoptosis in lymphoblastic leukemia cells [54]. These phenotypes are very similar to those induced by miR-101. In addition, miR-101 expression selectively increased cellular sensitivity to ActD and CX-5461 (Fig. 6e and Supplementary Fig. 6c). These findings strongly suggest that the molecular circuitry formed by miR-101 and p53 is a crucial component of the nucleolar stress response pathway.
The kinetics of activation and/or upregulation of mature miR-101 after treatment with CX-5461 were slow, and EG5 repression was required for 30 h. It is possible that miR-101 controls the late phase in the nucleolar stress pathway induced by CX-5461. Notably in this regard, recent works showed that CX-5461 binds to G4 DNA structures in the genome and induces DNA damage at these sites [55]. By contrast, we observed no activation of miR-101 in HCT116 cells after exposure to DNA-damaging stresses, including ADR, UV, and γ-irradiation. Therefore, it is reasonable to speculate that CX-5461 first induces a p53-dependent DNA damage response pathway, and subsequently activates the nucleolar stress pathway by inhibiting Pol I transcription. Meanwhile, miR-101 might contribute to continuous activation of the ATM–p53 axis in the absence of DNA damage.
The observed sensitization to Pol I transcription inhibitors and the phenotypes induced by miR-101 suggest that full activation of the p53–miR-101 circuit is necessary for effective therapy using Pol I transcription inhibitors. Our data demonstrate that mimicry of activation of this circuit using an IAP inhibitor represents a promising strategy for improving the efficacy of p53 activation therapy by Pol I inhibition. Expression of IAP family members is frequently altered in many types of tumors, reflecting the important roles of these proteins in preventing apoptosis of cancer cells [44]. Birinapant, a peptide mimetic of SMAC, promotes proteasomal degradation of IAP proteins; SMAC is an endogenous antagonist of IAP proteins that activates caspases. Therefore, birinapant is being investigated as a molecularly targeted drug capable of reactivating caspase. Although the safety of birinapant has been confirmed by a phase I clinical study [56], its efficacy as a standalone drug appears to be limited. Therefore, combination treatment with other drugs, including platinum-based chemotherapy and anti-PD1 drugs, will be investigated [57, 58]. We found that apoptosis induction by miR-101 was lower in RKO cells than in HCT116 cells (Fig. 1d). This is likely due to the weak repression of cIAP1 or the upregulation of cIAP2 (Supplementary Fig. 8a and b), suggesting that IAP proteins are among the targets of apoptosis induction by the p53–miR-101 circuit. Indeed, mimicry of the activation of this circuit by combination treatment with CX-5461 and birinapant drastically increased caspase activity in RKO cells (Fig. 8e). On the other hand, miR-101 also repressed cIAP1 in A549 cells (Supplementary Fig. 8b), although apoptosis induction was lower than in HCT116 cells. Combination treatment did not activate the apoptosis pathway (data not shown), consistent with a previous report that A549 cells are resistant to birinapant [59]. This finding suggests that alternative pathways act downstream of this circuit. Although our findings suggest that the regulation of apoptosis by the p53–miR-101 circuit is complex, the success of combination treatment suggests a strategy for the elimination of p53 WT cancers. Notably in this regard, analyses of clinical samples revealed that low expression of miR-101 was significantly associated with reduced RFS and cancer-specific survival exclusively in p53 WT LADC patients. Therefore, a certain subset of patients are the optimal targets for this combination treatment.
The molecular network and possible functions of the p53–miR-101 circuit are depicted in Supplementary Fig. 8c. When cells sense nucleolar stress triggered by Pol I inhibition, p53 is stabilized, leading to post-transcriptional activation of miR-101. Via downregulation of EG5 in G2 phase of the cell cycle, miR-101 contributes to continuous activation of positive-feedback regulation of the ATM–p53 axis. This circuitry is disrupted in LADC patients, resulting in a poor prognosis. This circuit suppresses several anti-apoptosis proteins, including members of the cIAP family. Mimicry of activation of the p53–miR-101 circuit by combination treatment with CX-5461 and a cIAP inhibitor represents a promising strategy for efficient elimination of p53 WT cancer cells.
Funding Sources
This work was partly supported by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO) (12-01); a Grant-in-Aid from Japan Society for the Promotion of Science (15H04305); a grant from the New Energy and Industrial Technology Development Organization of Japan Agency for Medical Research and Development (AMED) (JP17ae0101011h0004); Grants from AMED for the practical Research for Innovative Cancer Control (JP17ck0106168h0003 and JP18ck0106405h0001).
Declaration of Interests
The authors have no financial conflicts to declare.
Author Contributions
NT designed the research and wrote the paper. YF performed all experiments and analyzed data. MN and MW assisted with microarray analysis. MS, AIR, JY, TK, and CCH analyzed clinical samples. FT and TO supported the animal experiments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.06.031.
Appendix A. Supplementary data
Supplementary material
References
- 1.Levine A.J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88(3):323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Muller P.A., Vousden K.H. p53 mutations in cancer. Nat Cell Biol. 2013;15(1):2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 3.Gao J., Aksoy B.A., Dogrusoz U. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petitjean A., Mathe E., Kato S. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28(6):622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 5.Kloosterman W.P., Plasterk R.H. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Adams B.D., Kasinski A.L., Slack F.J. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24(16):R762–R776. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Leva G., Garofalo M., Croce C.M. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He L., He X., Lowe S.W., Hannon G.J. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7(11):819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang T.C., Yu D., Lee Y.S. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40(1):43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki H.I., Yamagata K., Sugimoto K., Iwamoto T., Kato S., Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460(7254):529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 11.Lujambio A., Lowe S.W. The microcosmos of cancer. Nature. 2012;482(7385):347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tazawa H., Tsuchiya N., Izumiya M., Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104(39):15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuchiya N., Izumiya M., Ogata-Kawata H. Tumor suppressor miR-22 determines p53-dependent cellular fate through post-transcriptional regulation of p21. Cancer Res. 2011;71(13):4628–4639. doi: 10.1158/0008-5472.CAN-10-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito M., Shimada Y., Shiraishi K. Development of lung adenocarcinomas with exclusive dependence on oncogene fusions. Cancer Res. 2015;75(11):2264–2271. doi: 10.1158/0008-5472.CAN-14-3282. [DOI] [PubMed] [Google Scholar]
- 15.Saito M., Shiraishi K., Matsumoto K. A three-microRNA signature predicts responses to platinum-based doublet chemotherapy in patients with lung adenocarcinoma. Clin Cancer Res. 2014;20(18):4784–4793. doi: 10.1158/1078-0432.CCR-14-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robles A.I., Arai E., Mathe E.A. An integrated prognostic classifier for stage I lung adenocarcinoma based on mRNA, microRNA, and DNA methylation biomarkers. J Thorac Oncol. 2015;10(7):1037–1048. doi: 10.1097/JTO.0000000000000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izumiya M., Okamoto K., Tsuchiya N., Nakagama H. Functional screening using a microRNA virus library and microarrays: a new high-throughput assay to identify tumor-suppressive microRNAs. Carcinogenesis. 2010;31(8):1354–1359. doi: 10.1093/carcin/bgq112. [DOI] [PubMed] [Google Scholar]
- 18.Banin S., Moyal L., Shieh S. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281(5383):1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 19.Canman C.E., Lim D.S., Cimprich K.A. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281(5383):1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 20.Lees-Miller S.P., Sakaguchi K., Ullrich S.J., Appella E., Anderson C.W. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol. 1992;12(11):5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tibbetts R.S., Brumbaugh K.M., Williams J.M. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13(2):152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burma S., Chen B.P., Murphy M., Kurimasa A., Chen D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276(45):42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 23.Varambally S., Cao Q., Mani R.S. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322(5908):1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor W.R., Stark G.R. Regulation of the G2/M transition by p53. Oncogene. 2001;20(15):1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 25.Strobeck M.W., Reisman D.N., Gunawardena R.W. Compensation of BRG-1 function by Brm insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J Biol Chem. 2002;277(7):4782–4789. doi: 10.1074/jbc.M109532200. [DOI] [PubMed] [Google Scholar]
- 26.Paulson J.R., Taylor S.S. Phosphorylation of histones 1 and 3 and nonhistone high mobility group 14 by an endogenous kinase in HeLa metaphase chromosomes. J Biol Chem. 1982;257(11):6064–6072. [PubMed] [Google Scholar]
- 27.Castedo M., Perfettini J.L., Roumier T., Kroemer G. Cyclin-dependent kinase-1: linking apoptosis to cell cycle and mitotic catastrophe. Cell Death Differ. 2002;9(12):1287–1293. doi: 10.1038/sj.cdd.4401130. [DOI] [PubMed] [Google Scholar]
- 28.Dweep H., Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 29.Blangy A., Lane H.A., D'Herin P., Harper M., Kress M., Nigg E.A. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83(7):1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 30.Doles J., Hemann M.T. Nek4 status differentially alters sensitivity to distinct microtubule poisons. Cancer Res. 2010;70(3):1033–1041. doi: 10.1158/0008-5472.CAN-09-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marklund U., Larsson N., Gradin H.M., Brattsand G., Gullberg M. Oncoprotein 18 is a phosphorylation-responsive regulator of microtubule dynamics. EMBO J. 1996;15(19):5290–5298. [PMC free article] [PubMed] [Google Scholar]
- 32.Uzbekov R., Prigent C., Arlot-Bonnemains Y. Cell cycle analysis and synchronization of the Xenopus laevis XL2 cell line: study of the kinesin related protein XlEg5. Microsc Res Tech. 1999;45(1):31–42. doi: 10.1002/(SICI)1097-0029(19990401)45:1<31::AID-JEMT3>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 33.Hurwitz J., Furth J.J., Malamy M., Alexander M. The role of deoxyribonucleic acid in ribonucleic acid synthesis. III. The inhibition of the enzymatic synthesis of ribonucleic acid and deoxyribonucleic acid by actinomycin D and proflavin. Proc Natl Acad Sci U S A. 1962;48:1222–1230. doi: 10.1073/pnas.48.7.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrence T.S. Reduction of doxorubicin cytotoxicity by ouabain: correlation with topoisomerase-induced DNA strand breakage in human and hamster cells. Cancer Res. 1988;48(3):725–730. [PubMed] [Google Scholar]
- 35.Heidelberger C., Ansfield F.J. Experimental and clinical use of fluorinated pyrimidines in cancer chemotherapy. Cancer Res. 1963;23:1226–1243. [PubMed] [Google Scholar]
- 36.Schiff P.B., Horwitz S.B. Taxol stabilizes microtubules in mouse fibroblast cells. Proc Natl Acad Sci U S A. 1980;77(3):1561–1565. doi: 10.1073/pnas.77.3.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vassilev L.T., Tovar C., Chen S. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci U S A. 2006;103(28):10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakai R., Iida S., Takahashi T. K858, a novel inhibitor of mitotic kinesin Eg5 and antitumor agent, induces cell death in cancer cells. Cancer Res. 2009;69(9):3901–3909. doi: 10.1158/0008-5472.CAN-08-4373. [DOI] [PubMed] [Google Scholar]
- 39.Choong M.L., Yang H., Lee M.A., Lane D.P. Specific activation of the p53 pathway by low dose actinomycin D: a new route to p53 based cyclotherapy. Cell Cycle. 2009;8(17):2810–2818. doi: 10.4161/cc.8.17.9503. [DOI] [PubMed] [Google Scholar]
- 40.Lindell T.J., Weinberg F., Morris P.W., Roeder R.G., Rutter W.J. Specific inhibition of nuclear RNA polymerase II by alpha-amanitin. Science. 1970;170(3956):447–449. doi: 10.1126/science.170.3956.447. [DOI] [PubMed] [Google Scholar]
- 41.Wu L., Pan J., Thoroddsen V. Novel small-molecule inhibitors of RNA polymerase III. Eukaryot Cell. 2003;2(2):256–264. doi: 10.1128/EC.2.2.256-264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drygin D., Lin A., Bliesath J. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011;71(4):1418–1430. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]
- 43.Allensworth J.L., Sauer S.J., Lyerly H.K., Morse M.A., Devi G.R. Smac mimetic Birinapant induces apoptosis and enhances TRAIL potency in inflammatory breast cancer cells in an IAP-dependent and TNF-alpha-independent mechanism. Breast Cancer Res Treat. 2013;137(2):359–371. doi: 10.1007/s10549-012-2352-6. [DOI] [PubMed] [Google Scholar]
- 44.Fulda S., Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat Rev Drug Discov. 2012;11(2):109–124. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- 45.Frankel L.B., Wen J., Lees M. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 2011;30(22):4628–4641. doi: 10.1038/emboj.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao J.M., Cao B., Zhou X., Lu H. New insights into p53 functions through its target microRNAs. J Mol Cell Biol. 2014;6(3):206–213. doi: 10.1093/jmcb/mju018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X., Schulz R., Edmunds S. MicroRNA-101 suppresses tumor cell proliferation by acting as an endogenous proteasome inhibitor via targeting the proteasome assembly factor POMP. Mol Cell. 2015;59(2):243–257. doi: 10.1016/j.molcel.2015.05.036. [DOI] [PubMed] [Google Scholar]
- 48.Burger K., Eick D. Functional ribosome biogenesis is a prerequisite for p53 destabilization: impact of chemotherapy on nucleolar functions and RNA metabolism. Biol Chem. 2013;394(9):1133–1143. doi: 10.1515/hsz-2013-0153. [DOI] [PubMed] [Google Scholar]
- 49.Dez C., Tollervey D. Ribosome synthesis meets the cell cycle. Curr Opin Microbiol. 2004;7(6):631–637. doi: 10.1016/j.mib.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 50.Golomb L., Volarevic S., Oren M. p53 and ribosome biogenesis stress: the essentials. FEBS Lett. 2014;588(16):2571–2579. doi: 10.1016/j.febslet.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 51.Bywater M.J., Poortinga G., Sanij E. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell. 2012;22(1):51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Devlin J.R., Hannan K.M., Hein N. Combination therapy targeting ribosome biogenesis and mRNA translation synergistically extends survival in MYC-driven lymphoma. Cancer Discov. 2016;6(1):59–70. doi: 10.1158/2159-8290.CD-14-0673. [DOI] [PubMed] [Google Scholar]
- 53.Quin J., Chan K.T., Devlin J.R. Inhibition of RNA polymerase I transcription initiation by CX-5461 activates non-canonical ATM/ATR signaling. Oncotarget. 2016;7(31):49800–49818. doi: 10.18632/oncotarget.10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Negi S.S., Brown P. rRNA synthesis inhibitor, CX-5461, activates ATM/ATR pathway in acute lymphoblastic leukemia, arrests cells in G2 phase and induces apoptosis. Oncotarget. 2015;6(20):18094–18104. doi: 10.18632/oncotarget.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu H., Di Antonio M., McKinney S. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat Commun. 2017;8 doi: 10.1038/ncomms14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amaravadi R.K., Schilder R.J., Martin L.P. A phase I study of the SMAC-mimetic Birinapant in adults with refractory solid tumors or lymphoma. Mol Cancer Ther. 2015;14(11):2569–2575. doi: 10.1158/1535-7163.MCT-15-0475. [DOI] [PubMed] [Google Scholar]
- 57.La V., Fujikawa R., Janzen D.M. Birinapant sensitizes platinum-resistant carcinomas with high levels of cIAP to carboplatin therapy. NPJ Precis Oncol. 2017;1 doi: 10.1038/s41698-017-0008-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Kearney C.J., Lalaoui N., Freeman A.J., Ramsbottom K.M., Silke J., Oliaro J. PD-L1 and IAPs co-operate to protect tumors from cytotoxic lymphocyte-derived TNF. Cell Death Differ. 2017;24(10):1705–1716. doi: 10.1038/cdd.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riley J.S., Hutchinson R., McArt D.G. Prognostic and therapeutic relevance of FLIP and procaspase-8 overexpression in non-small cell lung cancer. Cell Death Dis. 2013;4:e951. doi: 10.1038/cddis.2013.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material