Abstract
As the target organ for numerous pathogens, the lung epithelium exerts critical functions in health and disease. However, research in this area has been hampered by the quiescence of the alveolar epithelium under standard culture conditions. Here, we used human distal airway epithelial cells (DAECs) to generate alveolar epithelial cells. Long-term, robust growth of human DAECs was achieved using co-culture with feeder cells and supplementation with epidermal growth factor (EGF), Rho-associated protein kinase inhibitor Y27632, and the Notch pathway inhibitor dibenzazepine (DBZ). Removal of feeders and priming with DBZ and a cocktail of lung maturation factors prevented the spontaneous differentiation into airway club cells and instead induced differentiation to alveolar epithelial cells. We successfully transferred this approach to chicken distal airway cells, thus generating a zoonotic infection model that enables studies on influenza A virus replication. These cells are also amenable for gene knockdown using RNAi technology, indicating the suitability of the model for mechanistic studies into lung function and disease.
Keywords: Distal airway epithelial cells, Influenza A virus, Alveolar epithelial cells, Lung disease
Highlights
-
•
Long-term growth of DAECs was achieved using co-culture with feeder cells and compound supplementation.
-
•
DAECs treated with Notch inhibitors and growth factors differentiated towards alveolar epithelial cells.
-
•
Human and chicken DAECs support replication of different IAV strains.
Imai-Matsushima et al. describe a method for stable culture of human and chicken distal airway epithelial cells (DAECs). Upon Notch inhibitor and growth factor supplementation these cells can generate alveolar epithelial cells. DAECs support IAV virus infection and are amenable to gene knockdown, enabling studies into lung function and respiratory infections.
1. Introduction
Lower respiratory infections, chronic obstructive pulmonary disease and lung cancer are amongst the top ten causes of death worldwide. However, research into these conditions has been hampered by the absence of stable primary cell models.
The human alveolar epithelium is lined with non-proliferative type I alveolar epithelial (AEI) cells specialized for gas exchange, and proliferative type II alveolar epithelial (AEII) cells, which produce surfactant proteins with essential roles during innate immune responses [1, 2]. AEII cells are targeted by various pathogens that cause life-threatening pneumonia, several of which have a zoonotic origin [3]. This includes influenza A virus (IAV), whose natural reservoir is birds but which has undergone human adaptation causing serious pandemics [[4], [5], [6], [7], [8], [9]]. Comparative studies using available cell types to study IAV zoonosis, such as human lung adenocarcinoma cell line A549 and the chicken embryonic fibroblast cell line DF-1, are likely to confound species-, tissue-, and cell line-specific effects. Therefore, establishing human and avian primary cell models would enable more physiologically relevant comparison.
Although alveolar epithelial cell culture models have been established previously, these have been hampered by spontaneous transdifferentiation of type II to type I-like cells in vitro [10, 11]. Therefore, most experiments with primary human alveolar epithelial cells require isolation from fresh tissue and are limited by the supply of lung specimens, as well as the time and cost associated.
Organoid models allow long-term culture of cells derived from rapidly regenerating organs using dedicated adult stem cells [[12], [13], [14]]. However, the regeneration of adult lung epithelium is only activated upon injury, impeding the identification of the cell types involved. Most of the evidence appoints differentiated alveolar epithelial cells as the progenitor cells of the alveolar epithelium, although other reports suggest that specialized stem cells are recruited upon severe alveolar damage [15]. The potential to differentiate alveolar linages from human distal airway stem cells (DASCs) was addressed previously [16]. Human DASCs were found to express P63 and cytokeratin 5 (CK5), which are markers for progenitor cells of the stratified epithelium, and were able give rise to podoplanin+ AEI cells and CC10+ airway club cells, but not surfactant protein C+ AEII cells [16]. Bove and colleagues grew human AEII cells in culture with feeder cells and the rho kinase inhibitor Y-27632 for >30 population doublings [17]. However, markers of AEII cells were downregulated after the first passage and the phenotype of the cells after feeder removal was not extensively characterized.
The growth-promoting effect of feeders has been linked to activation of apoptosis and secretion of growth factors [18, 19]. The same mechanism has been suggested to orchestrate regeneration after tissue damage and it is likely to underlie the robust growth of lung epithelial cells in feeder co-culture [18]. Additionally, the overlapping marker profile of human distal airway epithelial cells (DAECs) with that of regenerating murine epithelial cells challenged with influenza virus supports this hypothesis [17, 20, 21]. As epithelial cell proliferation is usually followed by coordinated differentiation, we hypothesized that it may be possible to induce differentiation towards alveolar epithelial cells by using factors that induce terminal differentiation of lung progenitors derived from pluripotent stem cells [22].
The result is a novel method that allows expansion of human DAECs using feeder cells. Feeder removal induced a strong inflammatory response and differentiation into an airway club cell phenotype. Addition of small molecules and growth factors at the end of the expansion phase induced differentiation into AEII cells, followed by trans-differentiation into type I cells. We successfully adapted this method to chicken DAECs and compared the growth kinetics of different IAV strains between the two species. Additionally, our model supports siRNA transfection, enabling the application of advanced molecular techniques on primary DAECs to allow physiologically relevant research on various human and zoonotic lung diseases.
2. Material and Methods
2.1. Isolation and Culture of Primary DAECs
2.1.1. Human
Non-malignant tissue samples were obtained from pneumectomy specimens from the Clinic for Infectious Diseases and Pulmonary Medicine, Charité University Hospital, Berlin under signed informed consent. Scientific usage for experimental purposes was approved by the ethics committee of the Charité University Medicine, Berlin (EA2/079/13). Tissue pieces were processed according to the method by Daum et al. [23] with modifications. Briefly, they were washed with balanced salt solution buffer (BSSB:137 mM NaCl/5.0 mM KCl/0.7 mM Na2HPO4/10 mM HEPES/5.5 mM glucose/1.2 mM MgSO4/1.8 mM CaCl2, pH 7.4), minced finely, digested with trypsin (Serva) and elastase (Merck Millipore), passed through a 70 μm filter, centrifuged at 300 g for 5 min, washed twice with BSSB, resuspended in culture medium and plated into flasks previously seeded with irradiated NIH/3 T3-GFP feeders. The cell yield was donor dependent and ranged from 1.0–5.0 × 106 cells/g of tissue.
Expansion medium was based on F-medium [24] with modifications: 3:1 mixture of Ham's F-12 nutrient mix (Life Technologies) and DMEM supplemented with 5% fetal calf serum/0.4 μg/ml hydrocortisone (Sigma-Aldrich)/5 μg/ml recombinant human insulin (Sigma-Aldrich)/8.4 ng/ml cholera toxin (Sigma-Aldrich)/24 μg/ml adenine (Sigma-Aldrich)/10 ng/ml recombinant human epidermal growth factor (Life Technologies)/9 μM Y27632 (Miltenyi Biotec), supplemented with 10 μg/ml ciprofloxacin (Bayer Vital) and 10 μg/ml vancomycin (Serva) for the first three days, and 2.5 μg/ml amphotericin B (Cayman Chemical, Ann Arbor, USA) for the first seven days. Flasks were maintained in a humidified incubator at 37 °C with 5% CO2, and medium changed and 1.5–2 × 104/cm2 freshly irradiated feeders supplemented every 2–3 days.
2.1.2. Chicken
Lungs of 1-year-old roosters were provided by Annett Kannegießer (Albrecht-Daniel-Thaer-Institute of Agricultural and Horticultural Sciences, Berlin, Germany) in accordance with German and European regulations and with approval of the Berlin state authorities. DAECs were isolated as above, and cultured at 39 °C.
2.2. Induction of Differentiation to Alveolar Epithelial Cells
Four days after passaging, DAECs in co-culture were treated with small molecules and growth factors to induce differentiation toward alveolar epithelial cells: 3 μM CHIR99021 (Merck Millipore)/10 ng/ml keratinocyte growth factor (Peprotech)/100 μM 3-isobutyl-1-methylxanthine (Sigma-Aldrich)/100 μM 8-bromoadenosine 3′,5′-cyclic monophosphate (Sigma-Aldrich)/25 nM dexamethasone (Sigma-Aldrich)/10 ng/ml fibroblast growth factor-10 (Peprotech) [22] and 20 μM DBZ. Three days later cells were separated from feeders and subjected to downstream experiments. For further details see Supplementary Materials and Methods.
2.3. IAV Replication Assay
IAV strains A/England/195/2009 (H1N1pdm), A/WSN/1933 (H1N1), A/Mallard/Germany/439/2004 (H3N2) and A/Panama/2007/1999 (H3N2) were used to determine replication kinetics in human and chicken DAECs. Cells were infected at multiplicity of infection (MOI) 0.01. After 55 min cells were washed and culture medium containing 10 μg/ml TPCK-treated trypsin added. Cells were then incubated at 37 °C and supernatant samples collected at indicated times. Viral titer in supernatants was determined by plaque assay using Madin-Darby canine kidney (MDCK) cells.
A/Vietnam/1203/2004 (H5N1) was tested in a BSL3 laboratory. For titration of viral progeny 50 μl supernatant was transferred to MDCK cells seeded in 96-well plates. After 6 h cells were fixed with 3.7% formaldehyde, immunolabeled with anti-influenza A nucleoprotein (NP) antibody (BIO-RAD) followed by anti-mouse IgG secondary antibody (Dianova). Images were acquired using automated microscopy and analyzed by ScanR Analysis Software (Olympus).
2.4. siRNA Transfection
Purified human DAECs or A549 cells were subjected to reverse transfection using siRNAs targeting Lamin A/C, NP or the scrambled sequence Allstars (Qiagen) and HiperFect (Qiagen). After 20 min incubation, cells containing 9 μM Y27632 were mixed with the transfection complexes and incubated for 48 h at 37 °C.
2.5. Statistics
Statistical significance was determined by two-tailed paired t-test; experiments represent three biological replicates, unless otherwise indicated. Samples were not randomized and no sample size estimation was carried out.
3. Results
3.1. DBZ Improves In Vitro Proliferation of Primary Human DAECs
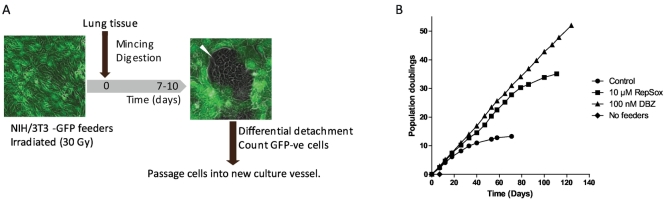
We first established co-culture with feeder cells to allow long-term propagation of human DAECs. NIH/3 T3 feeders were transduced with GFP to allow for removal by flow cytometry. The workflow and a colony of human DAECs in co-culture with GFP-expressing feeders are shown in Fig. 1A. Although cells successfully proliferated for 13 population doublings (PDs), growth was not as long-lasting as previously reported [17]. We thus tested a list of small molecules (Suppl. Table 1) and growth factors with effects on signaling pathways involved in lung development and/or maintenance of adult epithelial cells. We selected the TGF-β type I receptor inhibitor RepSox (10 μM) and the Notch pathway inhibitor DBZ (100 nM) (Suppl. Table 1), but only DBZ was able to maintain exponential growth beyond 130 PD (Fig. 1B) and was used for all subsequent experiments.
Fig. 1.
Successful culture of human distal airway epithelial cells (DAECs) with feeders.
(A) Initial workflow of co-culturing human DAECs with irradiated NIH3T3/GFP (feeders). Feeders were plated one day prior to epithelial cell isolation from tissues. A lung tissue sample was finely minced and digested with trypsin and elastase. Liberated cells were plated on feeders in expansion medium. The GFP-negative epithelial cells form discrete colonies (white arrowhead) in 7 to 10 days. Epithelial cells were harvested by differential trypsinization upon passaging. After counting under a fluorescent microscope 2 × 10 [5] GFP-negative cells were seeded in the next culture plate.
(B) The growth curves of human DAECs cultured in different conditions. Cells did not reach enough confluence (80%) after 8 days from initial seeding when they were cultured only with expansion medium (No feeders, ♦). Expansion medium and irradiated feeders (Control, ●) allowed the cells to expand up to approximately 75 population doublings (PDs). Additionally, supplementation with RepSox (10 μM RepSox, ■) and dibenzazepine (DBZ) (100 nM DBZ, ▲) contributed to the longevity of the DAECs compared with feeders alone. Results are representative of three independent experiments using cells from three different donors.
3.2. Proliferating Human DAECs Express Regenerating Cell-Like Molecular Markers
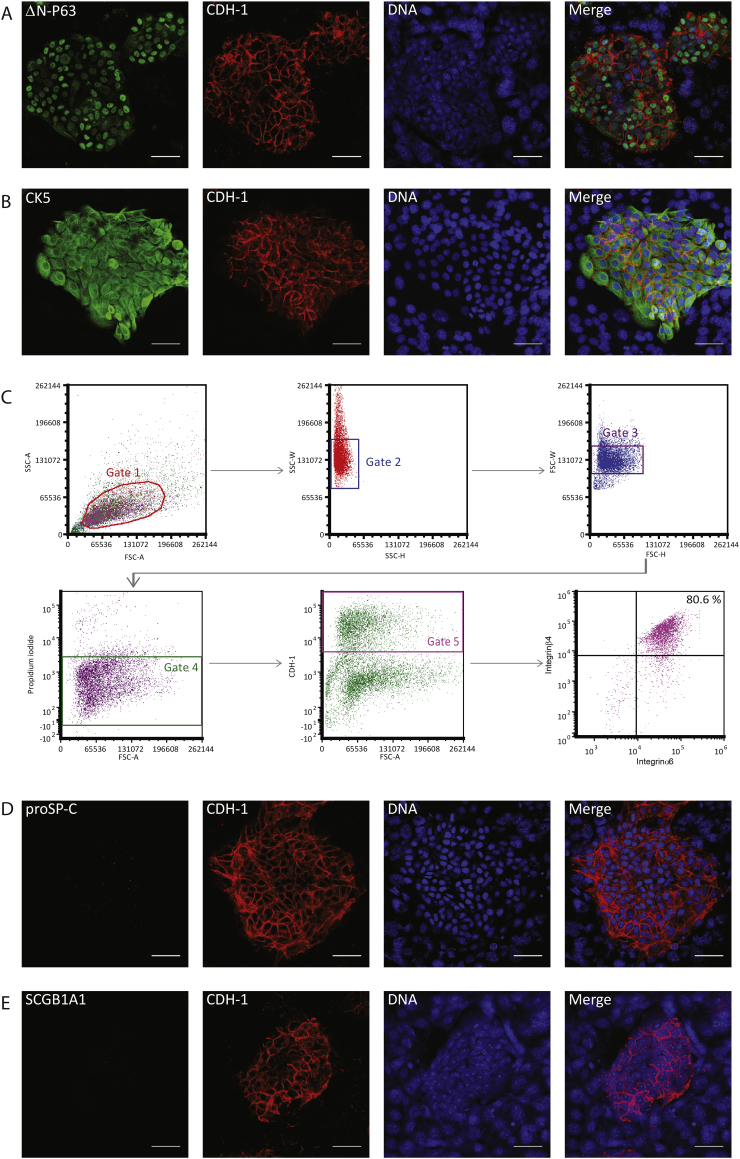
Immunofluorescence labeling showed that the majority of human DAECs expressed both cytokeratin 5 (CK5) and the N-terminal truncated form of P63 (ΔN-P63), considered the main markers of regenerating cells in the mouse lung after severe injury (Fig. 2A, B) [16]. Flow cytometry analyses further revealed that 80.6% of E-cadherin+ DAECs were double positive for α6 and β4 integrins (Fig. 2C), markers of regenerating cells in mouse lung [25]. Regenerating cells are negative for markers of AEII cells and airway club cells - the main mature epithelial cells with proliferation potential in the human distal lung. As expected, DAECs were negative for the AEII cell marker pro-surfactant protein C (proSP-C) at passage 4 (Fig. 2D), although some were positive at earlier passages (Suppl. Fig. 1). DAECs were also negative for the airway club cell marker secretoglobin1A1 (SCGB1A1) (Fig. 2E).
Fig. 2.
Human DAECs show regenerating cell-like phenotype in co-culture.
Immunofluorescent micrograph of human DAECs in co-culture with feeders. (A) Most of the cells in the CDH-1 (red) -positive epithelial colony show positive nuclear signal for the N-terminal-truncated form of P63 (ΔN-P63, green). (B) Cytokeratin 5 (CK5, green) is expressed in all epithelial cells. Red: CDH-1. Note that feeder cells are negative for CDH-1 and have large speckled nuclei. Scale bars: 50 μm.
(C) Flow cytometry analysis of integrin α6β4 expression on human DAECs in co-culture with feeders. After sequential gating to single cells (Gate 1-3), live (Gate 4) and CDH-1+ (Gate 5) epithelial cells were subjected to analysis. The integrin α6 + β4+ population (Gate 6) consisted of 80.6% of the epithelial cells.
(D–E) Immunofluorescence labelling of molecular markers of differentiation. CDH-1 (red)-positive epithelial cells are negative for both proSP-C (D) or SCGB1A1 (E). Scale bar: 50 μm. Results are representative of three independent experiments using cells from one donor.
3.3. Human DAECs Do Not Spontaneously Differentiate Into Alveolar Epithelial Cells
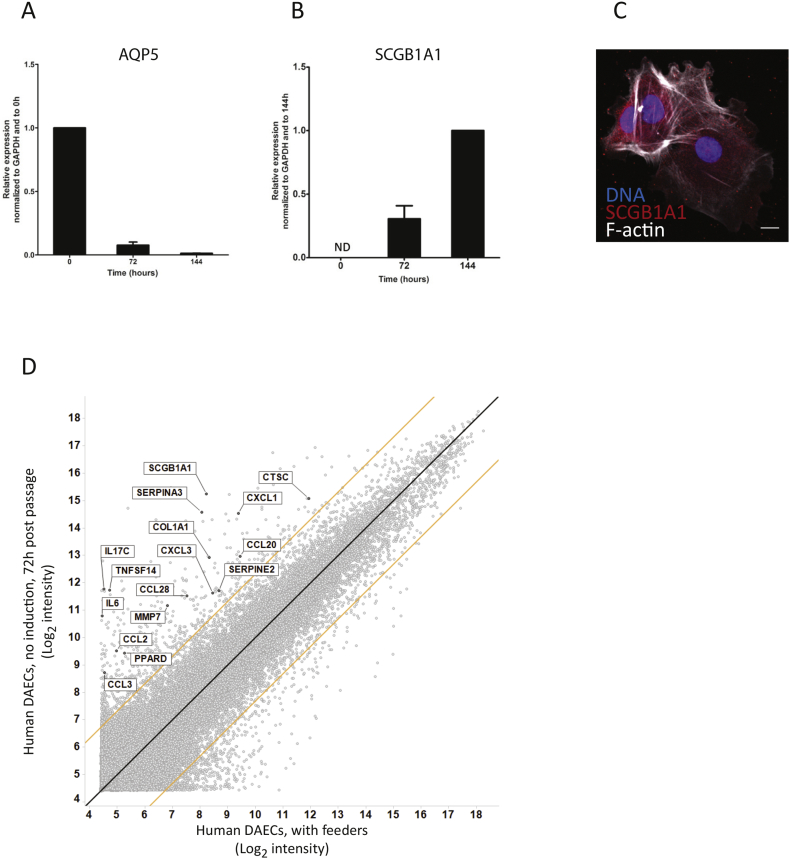
Removal of feeders and transfer to a conventional culture environment did not induce differentiation into mature alveolar epithelial cells, as qRT-PCR analyses revealed that the AEI cell marker AQP5 was downregulated 6 days later (Fig. 3A). Instead, SCGB1A1 was upregulated, suggesting differentiation toward an airway club cell phenotype (Fig. 3B). Immunofluorescence analysis at 72 h confirmed the presence of SCGB1A1 (Fig. 3C). Microarray analyses showed strong up-regulation of inflammatory cytokines, tissue remodeling-associated proteases, protease inhibitors, collagen 1A1 as well as SCGB1A1 (Fig. 3D). In contrast, RASSF2, a component of the NF-κB pathway, was downregulated.
Fig. 3.
SCGB1A1 and inflammation-related genes are up-regulated after human DAEC separation from feeders.
Comparison of gene expression by qRT-PCR at 0, 72 and 144 h after passaging to conventional cell culture environment. Aquaporin5 (haqp5) is strongly downregulated (A) and secretoglobin1a1 (scgb1a1) is upregulated after separation from feeders. Expression level was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (gapdh) at 0 h (haqp5) or 144 h (scgb1a1). Data are shown as mean ± SD of one experiment representative of three independent biological experiments using cells from one donor. ND: not detectable.
(C) Immunofluorescent micrograph of human DAECs 144 h after passage. Positive staining of SCGB1A1 (red) is seen in the cytoplasm of the cells. Scale bar: 50 μm. Images are representative of three independent experiments using cells from one donor.
(D) Microarray analysis of gene expression profile comparing human DAECs during co-culture and at 72 h post separation. Black line: y = x, orange lines: 5-fold change. Inflammatory cytokines (e.g. IL-6, CCL2, CCL3, CXC-motif chemokine ligands), tissue remodeling-associated proteases (e.g. MMP7, cathepsin C), protease inhibitors (e.g. SERPINs), collagen 1A1 and SCGB1A1 were strongly upregulated, while RASSF2 was downregulated 2.3-fold. Microarray was performed in cells derived from two independent donors.
3.4. Induction Treatment Directs Differentiation Into Alveolar Epithelial Cells
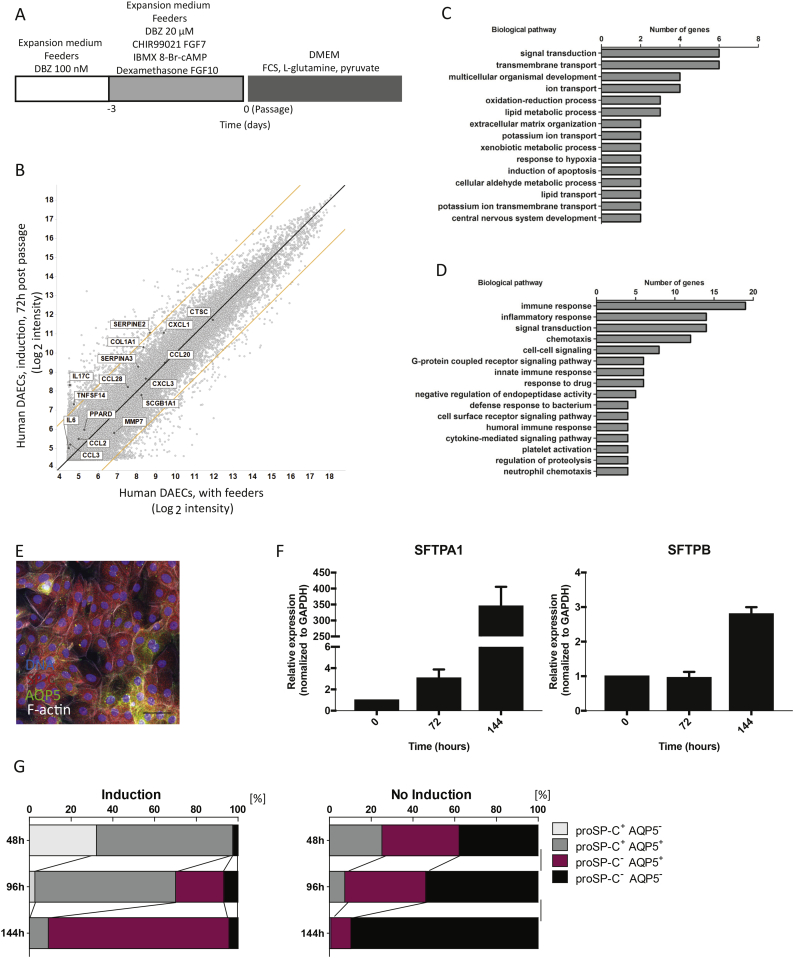
Failure of feeder removal to induce a phenotype towards alveolar epithelial cells implied that additional signals are required. We thus adopted an induction step published by Huang et al. [22]. Here, they used CHIR99021, keratinocyte growth factor, 3-isobutyl-1-methylxanthine (IBMX), 8-bromoadenosine 3′,5′-cyclic monophosphate (8-Br-cAMP), dexamethasone and FGF-10 to induce differentiation of human pluripotent stem cells into airway epithelial cells that express p63 but lack markers of more differentiated cells [16]. We also increased the concentration of DBZ during induction to suppress persistent Notch signaling, which causes differentiation toward airway club cells in mice. After three days of induction, cells were passaged and feeders removed to allow differentiation (Fig. 4A). This resulted in reduction of SCGB1A1 (Suppl. Fig. 2A), the goblet cell marker MUC5B, and the ciliated cell marker FOXJ1 (Suppl. Fig. 2B), and upregulation of AQP5 after 72 h (Suppl. Fig. 2C). To exclude epithelial to mesenchymal transition (EMT), we measured the expression of the epithelial marker CDH-1, the mesenchymal marker α-SMA, and the EMT regulatory factor Slug, after feeder removal with and without induction medium (Suppl. Fig. 2D). No substantial reduction of CDH-1 or increase of α-SMA or Slug were recorded, suggesting the epithelial phenotype is maintained.
Fig. 4.
A short term induction treatment directs human DAECs to an alveolar epithelial phenotype.
(A) Time course of the treatment to induce alveolar epithelial phenotype for human DAECs. The treatment begins three days before separation from feeders. Concentration of dibenzazepine (DBZ) was increased to 20 μM in parallel with addition of different small molecule inhibitors and cytokines. Abbreviations: C; CHIR99021, F7; fibroblast growth factor 7, I; 3-Isobutyl-1-methylxanthine, A; 8-Br-cAMP, Dex; Dexamathasone, F10; fibroblast growth factor 10.
(B) Microarray analysis of gene expression profile during co-culture and after induction and separation from feeders. Black line: y = x, orange lines: 5-fold change. Microarray was performed in cells derived from two independent donors.
Gene ontology (GO) analysis of differentially expressed genes between induced- (C) and non-induced (D) hDAECs at 72 h after separation.
(E) Immunofluorescent staining of human DAECs 48 h after feeder removal. Green: AQP5, red: SP-C, gray: F-actin, blue: DNA. Scale bar: 50 μm. (F) Human DAECs were expanded in co-culture with feeder cells and treated with CHIR99021, keratinocyte growth factor, IBMX, 8-Br-cAMP, dexamethasone and FGF-10. At three days post-treatment feeders were removed and the expression of SFTPA1 and SFTPB was investigated using qRT-PCR over the course of 6 days (0, 72 and 144 h). (G) Quantification is shown as percentage of cells positive/negative for SP-C and AQP5 with (left) and without (right) induction treatment. Immunofluorescent labelling images at each time point were converted to binary by standardized procedure. Y axis indicates the time points after feeder removal. Images are representative of three independent experiments carried out using cells from one donor.
Microarray analysis comparing induced and non-induced human DAECs 72 h after feeder removal showed that SCGB1A1 and inflammation-related genes were up-regulated only in non-induced cells (Figs. 3D and 4B). Gene ontology (GO) analysis revealed that genes related to transmembrane transport and lipid metabolism were enriched in the induced cells (Fig. 4C), while immune and inflammatory response-related genes were upregulated in non-treated cells (Fig. 4D).
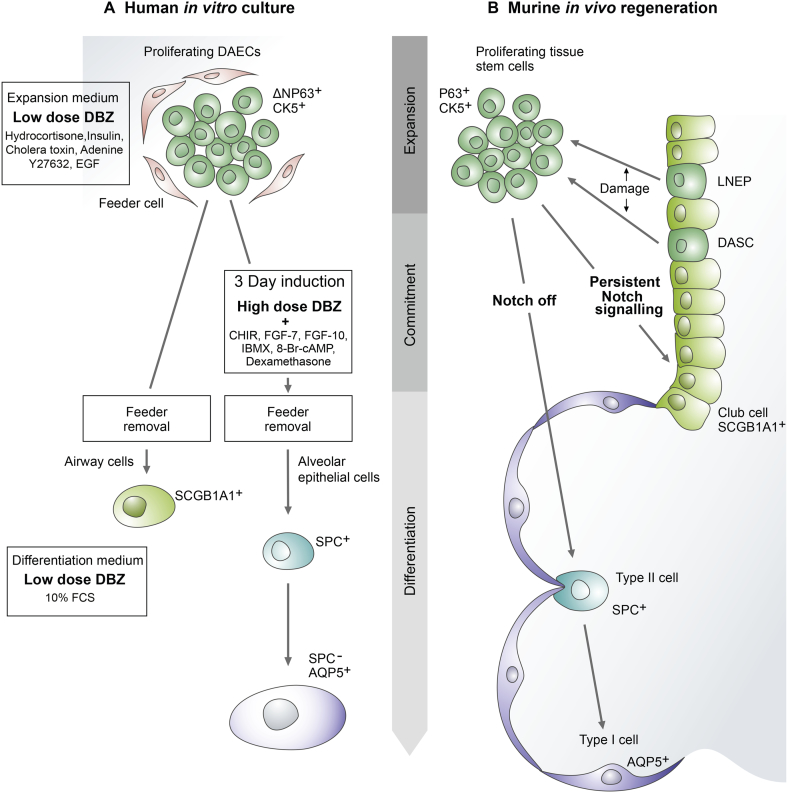
Next, we examined alveolar epithelial cell markers by immunofluorescence staining of AQP5 and SP-C (Fig. 4E). We also detected increased expression of surfactant proteins over time (Fig. 4F). The induced cells showed a spectrum of populations with different staining intensities of ProSP-C and AQP5 (Fig. 4G), shifting from mainly ProSP-C+cells at 48 h to mainly ProSP-C-/AQP5+ cells at 144 h of differentiation. These results are in congruence with the view that type II cells spontaneously differentiate into type I cells [26, 27]. Cellular morphology also changed from a type II-like to a large, flat type I-like phenotype (Suppl. Fig. 3A and B). By contrast, in non-induced cultures the proportion of ProSP-C−/AQP− cells increased to 89.7% by 144 h, and ProSP-C+/AQP5− cells were not detected without induction (Fig. 4G). A comprehensive schematic illustration of primary DAEC expansion and differentiation is included in Fig. 8.
Fig. 8.
Schematic illustration of primary DAEC expansion and differentiation.
(A) Proliferating human DAECs and their fate after induction and feeder removal in vitro. Treating the cells with high doses of the Notch pathway inhibitor dibenzazepine (DBZ) and other factors during the expansion phase induces commitment to an alveolar epithelial cell phenotype. (B) Lineage-negative epithelial progenitors (LNEPs) and distal airway stem cells (DASCs) are the specialized tissue stem cells that proliferate upon tissue damage in mice (e.g. influenza infection) [16, 20, 21]. Vaughan et al. proposed that murine LNEPs require Notch signalling to activate the ΔNp63 and CK5 program, and conversely Notch blockade promotes generation of alveolar epithelial cells [21].
Abbreviations: CHIR, CHIR99021; FGF, fibroblast growth factor; IBMX, 3-isobutyl-1-methylxanthine; 8-Br-cAMP, 8-bromoadenosine 3′,5′-cyclic monophosphate.
3.5. Expansion of Chicken DAECs in Co-Culture
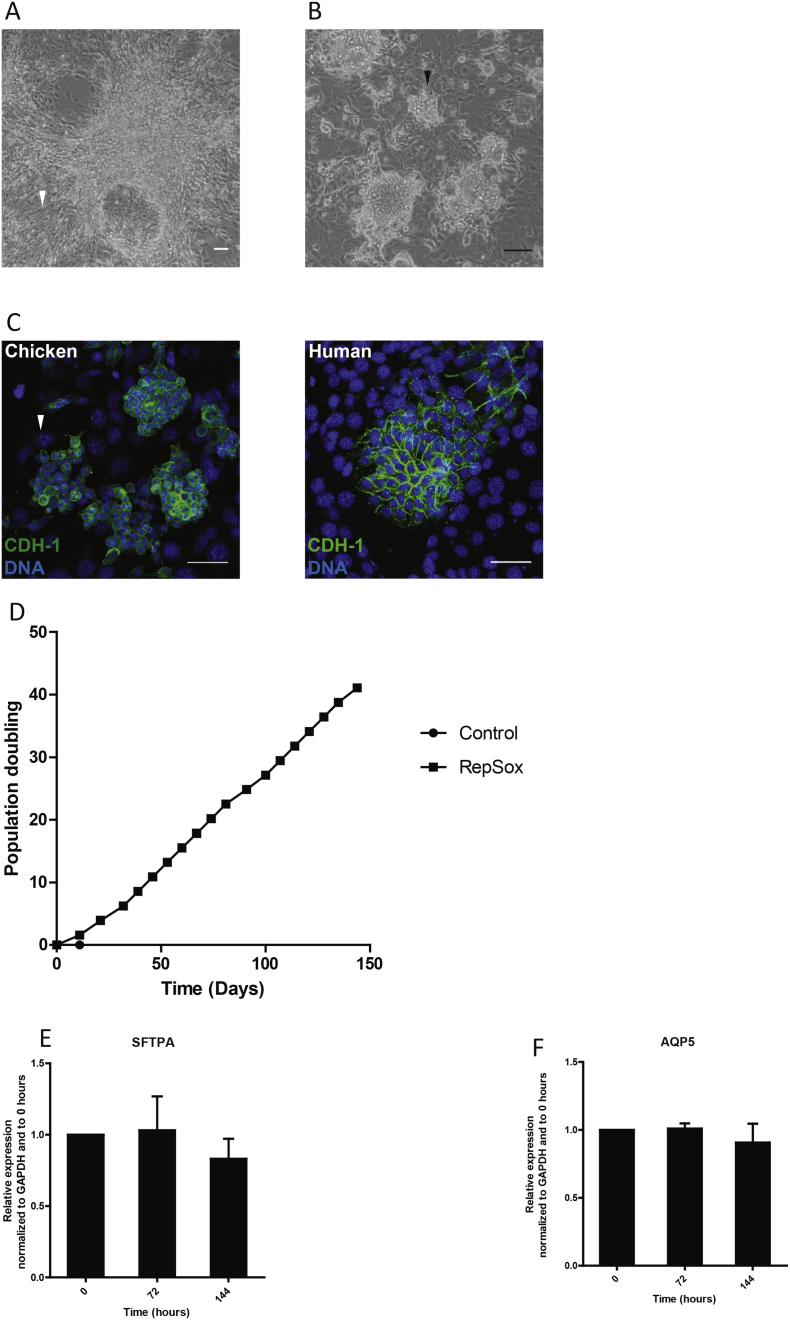
We next applied our method to chicken DAECs. Expansion medium alone did not support proliferation after passage 1. When co-cultured with feeders, numerous spindle-shaped cells appeared within 10-14 days, eventually converging on one another to leave behind bare areas (Fig. 5A). The addition of 10 μM RepSox prevented this, leading to the formation of DAEC colonies that were smaller and rounder than human DAECs (Fig. 5B and C). Cells grew for >150 days and reached over 40 population doublings (Fig. 5D). Other reagents, including DBZ, did not improve growth.
Fig. 5.
Successful culture of chicken DAECs with feeders.
(A) Left: A phase contrast image of chicken DAECs cultured in expansion medium with feeders for 20 days after isolation from tissue. Note bundles of spindle-shaped cells and an adjacent bare area (arrowhead). (B) Clusters of chicken DAECs (arrowhead) grown in medium supplemented with 10 μM RepSox upon co-culture with feeders. Scale bar: 100 μm. Results are representative of three independent experiments using lungs from three different chickens.
(C) Immunofluorescent labeling of chicken and human DAECs in co-culture with feeders. Note that feeders are negative for CDH-1 (green) and have large speckled nuclei (white arrowhead).
(D) Growth curve showing that sustained growth was achieved with feeders and RepSox. Results are representative of three independent experiments using lungs from three different chickens.
qRT-PCR shows that expression of surfactant-associated protein A gene (sftpa) (E) and aquaporin 5 (aqp5) (F) at different time points is relatively stable. The expression level was normalized to GAPDH at 0 h. Data are shown as mean ± SD of three biological replicates derived from one chicken lung.
After removal of feeders, qRT-PCR analysis showed expression of surfactant-associated protein A (SFTPA) (Fig. 5E) and AQP5 (Fig. 5F), key markers of chicken pneumocytes. In contrast to human cells, chicken DAECs did not require additional factors to induce these genes, and their expression level remained relatively stable over time (Fig. 5E, F).
3.6. Replication Kinetics of Different Influenza Virus Strains in Human and Chicken DAECs
To assess the potential of this model for influenza A virus research, we infected human and chicken DAECs with different IAV strains that have either known or predicted receptor-binding specificities, based on published data. A/WSN/1933 (H1N1) was reported to have α2-6 sialic acid receptor binding preference, which is the linkage most abundantly expressed in the human upper respiratory tract [28], A/England/195/2009 (H1N1pdm) is predicted to have preference for α2-6 sialic acid receptors but might also bind avian-like α2-3 linkages based on the reported binding preferences of another representative pandemic H1N1 2009 virus [29], A/Panama/2007/1999 (H3N2) has been reported to have α2-6 sialic acid receptor binding [30], A/Vietnam/1203/2004 (H5N1) has been reported to have α2-3 sialic acid receptor binding preference [31], and A/Mallard/Germany/439/2004 (H3N2) is predicted to have α2-3 receptor binding [32].
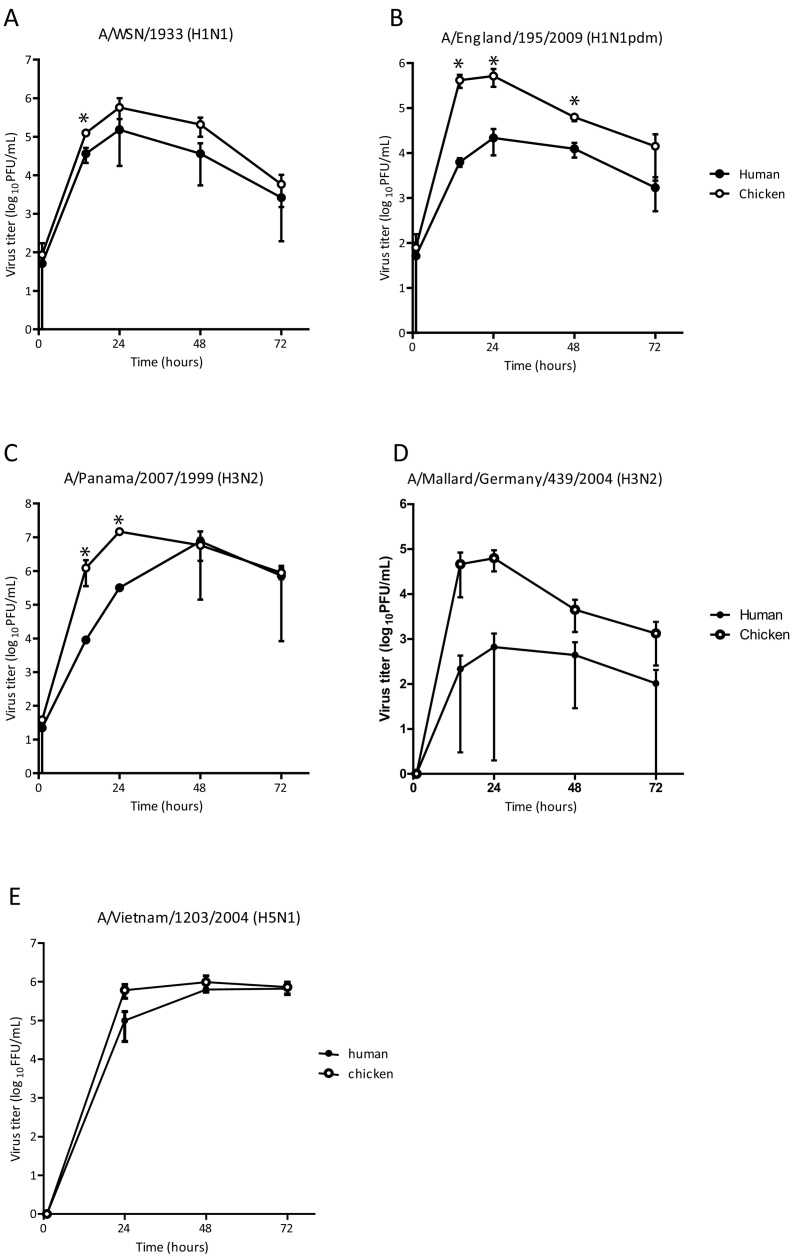
The lab-adapted A/WSN/1933(H1N1), the seasonal A/Panama/2007/1999(H3N2) and the pandemic A/England/195/2009(H1N1pdm), generated as a triple reassortant of human, porcine and avian viruses, replicated better in chicken DAECs (Fig. 6A–C). A/Mallard/Germany/439/2004(H3N2) exhibited low replication efficiency, especially in human DAECs (Fig. 6D), while the highly pathogenic A/Vietnam/1203/2004 (H5N1) replicated equally well in both species (Fig. 6E).
Fig. 6.
Replication kinetics of different IAV strains in human and chicken DAECs.
Human DAECs induced to differentiate to alveolar epithelial cells and chicken DAECs were separated from feeders, seeded in plastic culture vessels and infected with (A) A/WSN/1933 (H1N1), (B) A/England/195/2009 (H1N1pdm), (C) A/Panama/2007/1999 (H3N2), (D) A/Mallard/Germany/439/2004 (H3N2) at multiplicity of infection (MOI) of 0.01 plaque forming units (PFU)/cell. Supernatants were collected at 1, 14, 24, 48 and 72 h post-infection. The virus titres were determined by standard plaque assay in Madin-Darby canine kidney (MDCK) cells. Data are shown as mean ± SD from three independent experiments carried out in cells from one human donor or chicken lung. Statistical analysis was done by two-tailed paired t-test; *p < 0.1; **p < 0.05. (E) Human and chicken DAECs were infected with A/Vietnam/1203/2004 (H5N1) at MOI 0.05. The supernatants were harvested 24, 48 and 72 h post-infection. Viral progeny in the supernatant was titrated by infecting MDCK cells and subsequently detecting viral nucleoprotein by immunofluorescent staining. Data are shown as mean ± SD of three technical replicates representative of three independent experiments.
3.7. Human DAECs Support Efficient Gene Knockdown Using RNAi Technology
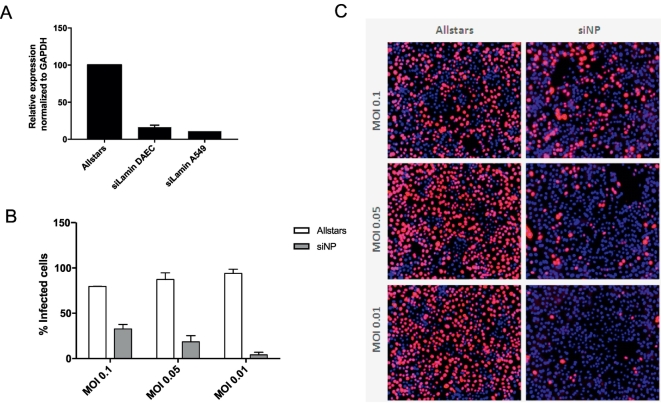
To facilitate downstream applications, we optimized delivery of siRNA to achieve comparable knockdown efficiencies for human DAECs and A549 cells (Fig. 7A) and assessed whether this technology could be used to determine the impact of a given knockdown on viral replication. Knockdown of the influenza virus factor nucleoprotein (NP) resulted in reduced replication of the A/WSN/1933 (H1N1) strain at various MOIs (Fig. 7B and C), demonstrating the potential of our model for infection biology and other downstream applications.
Fig. 7.
Human DAECs support siRNA transfection in vitro.
(A) Relative Lamin A/C mRNA levels after siRNA-mediated knockdown of Lamin A/C or the negative control siRNA Allstars in A549 or human DAECs as determined by qRT-PCR analyses. Expression levels were normalized to GAPDH. (B) Human DAECs transfected with siRNA against NP or Allstars were subjected to replication assay with A/WSN/1933 (H1N1) virus at indicated MOIs. Quantification reflects the percentage of cells infected after 72 h p.i. (C) Representative images. Data are shown as mean ± SD of three technical replicates representative of three independent experiments carried out in cells from one donor.
4. Discussion
We introduce here a feeder-based primary DAEC culture system and its application to lung infection research. Human DAECs maintained long-term growth and could be induced to generate alveolar epithelial cells. During expansion their marker profile was similar to that of regenerating cells in the mouse lung: ΔN-P63+/CK5+/integrin α6β4+/proSP-C−/SCGB1A1−. It has been suggested that mouse DAECs derive from specialized stem cells [20, 21] or include progenitors such as basal or mature alveolar epithelial cells [17]. Clarifying the origin of DAECs in the human lung has been impeded by the lack of molecular markers, which would not only allow assessment of clonal proliferation capacity, but also contribute to a better understanding of homeostasis and regeneration of the human lung.
The human distal airway epithelium includes airway epithelial cells at the proximal region and alveolar epithelial cells at the distal part. High Notch signalling activity has been linked to differentiation of progenitor cells towards airway club cells and inhibition of differentiation into alveolar cells [21, 33, 34]. While upon feeder removal, DAECs differentiated towards airway club cells, as shown by increased expression of SCGB1A1 and downregulation of AQP5 over time, no changes in genes regulated by canonical Notch signalling were recorded by our microarray experiment. Instead, we found rather robust inflammatory and proliferation pathway signatures confined to STAT3 and NF-κB signalling [35]. This might reflect involvement of non-canonical Notch activity, since non-canonical Notch signalling regulated by NF-κB or IL-6 expression has been reported previously for different tissues [36, 37].
In contrast to a previous report carried out in influenza-infected mice, in which differentiated cells expressed markers of airway club as well as ciliated and mucin-producing cells [16], our method induces efficient differentiation of human DAECs into proSP-C, SFTPA1, SFTPB and AQP5 positive alveolar epithelial cells. Although these factors have frequently been considered as main markers of mature alveolar cells [21, 38, 39], surfactant proteins are also expressed in immature cells, thus definitive confirmation that the cells are fully mature would require electron microscopy to confirm the presence of multilamellar bodies. It is interesting that our observations, including the molecular markers expressed in the proliferating DAECs and the induction method with Notch pathway inhibition, are consistent with the repair mechanism described for mouse lung epithelium (Fig. 8) [16, 21]. The morphological and phenotypical shift from type II toward type I-like cells after 48 h of feeder removal also closely resembles that of freshly isolated alveolar epithelial cells.
By contrast, we found many genes upregulated during default differentiation that have been associated with idiopathic pulmonary fibrosis, e.g. CCL2 [40], CCL3 [41], SERPINE2 [42], [43] and MMP7 [44, 45]. Current tools to support research in this field include animal models, cell lines such as the AEII cell line A549, non-epithelial models, as well as primary human bronchial epithelial, airway and alveolar epithelial cells [[46], [47], [48], [49], [50]]. However, the limited replicative lifespan of the latter cell types still remains a technical challenge to investigate these diseases. In this regard, our model might be of value to advance research in this field. However, future studies will be required to determine its applicability for research into lung diseases linked to aberrant regeneration, such as chronic obstructive pulmonary disease or idiophatic pulmonary fibrosis.
The fact that our model requires only three days of induction with growth factors and signalling molecules paves the way for downstream experimentation. The high cell density (>105/cm2) further reduces costs for platforms that require large numbers of cells. This enables the establishment of in vitro testing systems with cells from different genetic backgrounds or with genetic modifications introduced by lentivirus-mediated gene transduction or other genetic tools [51]. In addition, we can successfully cryopreserve both minced fresh tissue and DAECs, enabling transportation of these cells.
We also adapted the new model to avian DAECs – an important host species for zoonotic viruses affecting the lungs. Here, we achieved efficient long-term growth of chicken DAECs and differentiation towards cells positive for SFTPA and AQP5, molecular markers of chicken pneumocytes. The localization of these molecular markers in vivo strongly suggests that they are epithelial cells of the parabronchi, the site of gas exchange in birds [52, 53]. In contrast to human DAECs, chicken DAECs required the TGF-β receptor kinase inhibitor RepSox, not the Notch inhibitor DBZ to maintain proliferation, which closely resembles the signalling pathways activated in mammalian airway epithelial cells in vitro [[54], [55], [56]]. In addition, SFTPA and AQP5 were expressed regardless of induction treatment and remained relatively stable after feeder removal. While different cell types corresponding to the mammalian type I and II alveolar epithelial cells have been reported in chicken, the relationship between them remains unknown.
Chickens have been appointed as potential intermediate hosts for the transmission of avian influenza viruses to humans [57, 58]. Establishing models for avian and human primary DAECs enables direct comparison of influenza A virus replication in different relevant host cells, and ultimately allows deeper studies of the underlying biology. Various mutations that improve the replication efficiency of avian IAVs in mammalian hosts have been identified [59, 60]. Of these, the most extensively studied mutation is the single point mutation at influenza polymerase basic 2 (PB2) 627 residue from glutamic acid (E) to lysine (K). This has been linked to greater replication rates of avian viruses in human cells. Accordingly, the avian-like 627E-expressing virus A/Mallard/Germany/439/2004(H3N2) replicated better in chicken than in human DAECs. Although A/England/195/2009(H1N1pdm) also harbours 627E, compensatory mutations that allow for efficient replication in human cells have been identified [61]. Nonetheless, chicken DAECs still replicated A/England/195/2009(H1N1pdm) more productively than human DAECs. One possible explanation is that these viruses were generated in eggs and they might have acquired mutations that enable higher replication rates in chicken cells. For the other strains tested, these harbour the mammalian-like 627 K mutation in PB2 and still replicated equally well or better in chicken DAECs, suggesting that in addition to E627K PB2, other viral factors might affect host range [[62], [63], [64]]. Human-adapted viruses, such as H1N1pdm or H3N2, were found unable to infect poultry in previous studies. This discrepancy might be due to the absence of additional cell types that are required to mount an efficient immune response against some strains of influenza A virus in our ex vivo model. Along these lines, establishment of lung organoid cultures will benefit further investigations. Interestingly, the innate immune response of chickens against viruses is weak [65]. Chicken cells lack the retinoic acid-inducible gene (RIG-I) receptor, which can sense IAV, resulting in the induction of type-I interferon (IFN) responses. In contrast, strong RIG-I expression is observed in ducks, potentially explaining duck resistance to most influenza virus strains [66]. In vitro replication kinetics of IAVs in DAECs of aquatic birds, the natural reservoir of influenza virus, could help understand the tropism of influenza virus. However, the establishment of such models will depend on the availability of lungs from these species and the development of molecular tools to allow further characterization.
Our method is likely adaptable to other species and our protocol for efficient siRNA transfection will further enable study of host factor dependencies through RNAi analyses. We thus expect that our model will enable investigation of the role of genetic determinants in lung health and disease, as well as enhance our understanding of zoonotic diseases.
Funding Sources
LMS was a fellow of the FP7 MSCA Project EIMID ITN (GA No. 264388) and FP7 Health Project ANTIFLU (GA No. 259842), both granted to TFM. ACH received funding through the German Research Foundation (DFG SFB-TR84 – B6). The funding sources had no role in writing, data collection, analysis, or interpretation, or any aspect pertinent to this study.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
AIM, LMS and TFM contributed to the study conception and design. AIM, LMS, SI and HJM performed the experiment and contributed to data acquisition. AH contributed clinical specimens. AIM, LMS, AK, TZ, AH, HB and TFM contributed to the analysis and interpretation of data. AIM, LMS and TFM wrote the manuscript.
Data Availability
Microarray data have been deposited in the Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) of the National Center for Biotechnology Information and can be assessed with the GEO accession number GSE86449.
Acknowledgements
The authors would like to thank Kathrin Lättig, Dagmar Frahm, Isabella Gravenstein, Anja Greiser, Marina Drabkina, Ina Wagner, Jenny Kirsch and Toralf Kaiser for technical support, and Rike Zietlow for editing the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.05.032.
Appendix A. Supplementary data
Supplementary material
References
- 1.Qian Z., Travanty E.A., Oko L., Edeen K., Berglund A., Wang J. Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am J Respir Cell Mol Biol. 2013;48:742–748. doi: 10.1165/rcmb.2012-0339OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crapo J.D., Young S.L., Fram E.K., Pinkerton K.E., Barry B.E., Crapo R.O. Morphometric characteristics of cells in the alveolar region of mammalian lungs. Am Rev Respir Dis. 1983;128:S42–S46. doi: 10.1164/arrd.1983.128.2P2.S42. [DOI] [PubMed] [Google Scholar]
- 3.Weinheimer V.K., Becher A., Tönnies M., Holland G., Knepper J., Bauer T.T. Influenza A viruses target type II pneumocytes in the human lung. J Infect Dis. 2012;206:1685–1694. doi: 10.1093/infdis/jis455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang R., Min Jou W., Huylebroeck D., Devos R., Fiers W. Complete structure of A/duck/Ukraine/63 influenza hemagglutinin gene: animal virus as progenitor of human H3 Hong Kong 1968 influenza hemagglutinin. Cell. 1981;25:315–323. doi: 10.1016/0092-8674(81)90049-0. [DOI] [PubMed] [Google Scholar]
- 5.Garten R.J., Davis C.T., Russell C.A., Shu B., Lindstrom S., Balish A. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawaoka Y., Krauss S., Webster R.G. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scholtissek C., Rohde W., Von Hoyningen V., Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 8.Worobey M., Han G.Z., Rambaut A. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc Natl Acad Sci U S A. 2014;111:8107–8112. doi: 10.1073/pnas.1324197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worobey M., Han G.Z., Rambaut A. A synchronized global sweep of the internal genes of modern avian influenza virus. Nature. 2014;508:254–257. doi: 10.1038/nature13016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster C.D., Varghese L.S., Skalina R.B., Gonzales L.W., Guttentag S.H. In vitro transdifferentiation of human fetal type II cells toward a type I-like cell. Pediatr Res. 2007;61:404–409. doi: 10.1203/pdr.0b013e3180332c6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs S., Hollins A.J., Laue M., Schaefer U.F., Roemer K., Gumbleton M. Differentiation of human alveolar epithelial cells in primary culture: morphological characterization and synthesis of caveolin-1 and surfactant protein-C. Cell Tissue Res. 2003;311:31–45. doi: 10.1007/s00441-002-0653-5. [DOI] [PubMed] [Google Scholar]
- 12.Kessler M., Hoffmann K., Brinkmann V., Thieck O., Jackisch S., Toelle B. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat Commun. 2015;6:8989. doi: 10.1038/ncomms9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 14.Schlaermann P., Toelle B., Berger H., Schmidt S.C., Glanemann M., Ordemann J. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut. 2016;65:202–213. doi: 10.1136/gutjnl-2014-307949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uhal B.D. Cell cycle kinetics in the alveolar epithelium. Am J Physiol. 1997;272:L1031–L1045. doi: 10.1152/ajplung.1997.272.6.L1031. [DOI] [PubMed] [Google Scholar]
- 16.Kumar P.A., Hu Y., Yamamoto Y., Hoe N.B., Wei T.S., Mu D. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell. 2011;147:525–538. doi: 10.1016/j.cell.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bove P.F., Dang H., Cheluvaraju C., Jones L.C., Liu X., O'Neal W.K. Breaking the in vitro alveolar type II cell proliferation barrier while retaining ion transport properties. Am J Respir Cell Mol Biol. 2014;50:767–776. doi: 10.1165/rcmb.2013-0071OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li F., Huang Q., Chen J., Peng Y., Roop D.R., Bedford J.S. Apoptotic cells activate the "phoenix rising" pathway to promote wound healing and tissue regeneration. Sci Signal. 2010;3 doi: 10.1126/scisignal.2000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palechor-Ceron N., Suprynowicz F.A., Upadhyay G., Dakic A., Minas T., Simic V. Radiation induces diffusible feeder cell factor(s) that cooperate with ROCK inhibitor to conditionally reprogram and immortalize epithelial cells. Am J Pathol. 2013;183:1862–1870. doi: 10.1016/j.ajpath.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuo W., Zhang T., Wu D.Z., Guan S.P., Liew A.A., Yamamoto Y. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature. 2015;517:616–620. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaughan A.E., Brumwell A.N., Xi Y., Gotts J.E., Brownfield D.G., Treutlein B. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517:621–625. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang S.X., Islam M.N., O'Neill J., Hu Z., Yang Y.G., Chen Y.W. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat Biotechnol. 2014;32:84–91. doi: 10.1038/nbt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daum N., Kuehn A., Hein S., Schaefer U.F., Huwer H., Lehr C.-M. Isolation, cultivation, and application of human alveolar epithelial cells. Methods Mol Biol. 2012;806:31–42. doi: 10.1007/978-1-61779-367-7_3. [DOI] [PubMed] [Google Scholar]
- 24.Liu X., Ory V., Chapman S., Yuan H., Albanese C., Kallakury B. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chapman H.A., Li X., Alexander J.P., Brumwell A., Lorizio W., Tan K. Integrin alpha6beta4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest. 2011;121:2855–2862. doi: 10.1172/JCI57673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotton D.N., Morrisey E.E. Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat Med. 2014;20:822–832. doi: 10.1038/nm.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason R., Williams M.C., Clements J.A. Isolation and identification of type 2 alveolar epithelial cells. Chest. 1975;67:36s–37s. doi: 10.1378/chest.67.2_supplement.36s. [DOI] [PubMed] [Google Scholar]
- 28.Leung H.S., Li O.T., Chan R.W., Chan M.C., Nicholls J.M., Poon L.L. Entry of influenza A Virus with a alpha2,6-linked sialic acid binding preference requires host fibronectin. J Virol. 2012;86:10704–10713. doi: 10.1128/JVI.01166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Childs R.A., Palma A.S., Wharton S., Matrosovich T., Liu Y., Chai W. Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nat Biotechnol. 2009;27:797–799. doi: 10.1038/nbt0909-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao Y., Koh X., Dong L., Du X., Wu A., Ding X. Rapid estimation of binding activity of influenza virus hemagglutinin to human and avian receptors. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stevens J., Blixt O., Tumpey T.M., Taubenberger J.K., Paulson J.C., Wilson I.A. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 32.Zhu N., Zhao J., Li Y., Ding C., Xia H., Tang S. Molecular characterization of H3N2 and H4N6 subtypes avian influenza viruses isolated from mallards in Poyang Lake, China in 2010. Chin Sci Bull. 2012;57:3586–3594. [Google Scholar]
- 33.Dang T.P., Eichenberger S., Gonzalez A., Olson S., Carbone D.P. Constitutive activation of Notch3 inhibits terminal epithelial differentiation in lungs of transgenic mice. Oncogene. 2003;22:1988–1997. doi: 10.1038/sj.onc.1206230. [DOI] [PubMed] [Google Scholar]
- 34.Guseh J.S., Bores S.A., Stanger B.Z., Zhou Q., Anderson W.J., Melton D.A. Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development. 2009;136:1751–1759. doi: 10.1242/dev.029249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bollrath J., Greten F.R. IKK/NF-kappaB and STAT3 pathways: central signalling hubs in inflammation-mediated tumour promotion and metastasis. EMBO Rep. 2009;10:1314–1319. doi: 10.1038/embor.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dongre A., Surampudi L., Lawlor R.G., Fauq A.H., Miele L., Golde T.E. Non-canonical notch signaling drives activation and differentiation of peripheral CD4(+) T cells. Front Immunol. 2014;5:54. doi: 10.3389/fimmu.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin S., Mutvei A.P., Chivukula I.V., Andersson E.R., Ramskold D., Sandberg R. Non-canonical Notch signaling activates IL-6/JAK/STAT signaling in breast tumor cells and is controlled by p53 and IKKalpha/IKKbeta. Oncogene. 2013;32:4892–4902. doi: 10.1038/onc.2012.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Desai T.J., Brownfield D.G., Krasnow M.A. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507:190–194. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng D., Soh B.S., Yin L., Hu G., Chen Q., Choi H. Differentiation of club cells to alveolar epithelial cells in vitro. Sci Rep. 2017;7:41661. doi: 10.1038/srep41661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antoniades H.N., Neville-Golden J., Galanopoulos T., Kradin R.L., Valente A.J., Graves D.T. Expression of monocyte chemoattractant protein 1 mRNA in human idiopathic pulmonary fibrosis. Proc Natl Acad Sci U S A. 1992;89:5371–5375. doi: 10.1073/pnas.89.12.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Standiford T.J., Rolfe M.W., Kunkel S.L., Lynch J.P., 3rd, Burdick M.D., Gilbert A.R. Macrophage inflammatory protein-1 alpha expression in interstitial lung disease. J Immunol. 1993;151:2852–2863. [PubMed] [Google Scholar]
- 42.Steele M.P., Luna L.G., Coldren C.D., Murphy E., Hennessy C.E., Heinz D. Relationship between gene expression and lung function in Idiopathic Interstitial Pneumonias. BMC Genomics. 2015;16:869. doi: 10.1186/s12864-015-2102-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eitzman D.T., McCoy R.D., Zheng X., Fay W.P., Shen T., Ginsburg D. Bleomycin-induced pulmonary fibrosis in transgenic mice that either lack or overexpress the murine plasminogen activator inhibitor-1 gene. J Clin Invest. 1996;97:232–237. doi: 10.1172/JCI118396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richards T.J., Kaminski N., Baribaud F., Flavin S., Brodmerkel C., Horowitz D. Peripheral blood proteins predict mortality in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;185:67–76. doi: 10.1164/rccm.201101-0058OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song J.W., Do K.H., Jang S.J., Colby T.V., Han S., Kim D.S. Blood biomarkers MMP-7 and SP-A: predictors of outcome in idiopathic pulmonary fibrosis. Chest. 2013;143:1422–1429. doi: 10.1378/chest.11-2735. [DOI] [PubMed] [Google Scholar]
- 46.Izykowski N., Kuehnel M., Hussein K., Mitschke K., Gunn M., Janciauskiene S. Organizing pneumonia in mice and men. J Transl Med. 2016;14:169. doi: 10.1186/s12967-016-0933-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X., Fang S., Liu H., Wang X., Dai X., Yin Q. Role of human pulmonary fibroblast-derived MCP-1 in cell activation and migration in experimental silicosis. Toxicol Appl Pharmacol. 2015;288:152–160. doi: 10.1016/j.taap.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 48.Elbert K.J., Schafer U.F., Schafers H.J., Kim K.J., Lee V.H., Lehr C.M. Monolayers of human alveolar epithelial cells in primary culture for pulmonary absorption and transport studies. Pharm Res. 1999;16:601–608. doi: 10.1023/a:1018887501927. [DOI] [PubMed] [Google Scholar]
- 49.Forbes B., Ehrhardt C. Human respiratory epithelial cell culture for drug delivery applications. Eur J Pharm Biopharm. 2005;60:193–205. doi: 10.1016/j.ejpb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 50.Witherden I.R., Vanden Bon E.J., Goldstraw P., Ratcliffe C., Pastorino U., Tetley T.D. Primary human alveolar type II epithelial cell chemokine release: effects of cigarette smoke and neutrophil elastase. Am J Respir Cell Mol Biol. 2004;30:500–509. doi: 10.1165/rcmb.4890. [DOI] [PubMed] [Google Scholar]
- 51.van den Bogaard E.H., Rodijk-Olthuis D., Jansen P.A., van Vlijmen-Willems I.M., van Erp P.E., Joosten I. Rho kinase inhibitor Y-27632 prolongs the life span of adult human keratinocytes, enhances skin equivalent development, and facilitates lentiviral transduction. Tissue Eng Part A. 2012;18:1827–1836. doi: 10.1089/ten.tea.2011.0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bjornstad S., Paulsen R.E., Erichsen A., Glover J.C., Roald B. Type I and II pneumocyte differentiation in the developing fetal chicken lung: conservation of pivotal proteins from birds to human in the struggle for life at birth. Neonatology. 2014;105:112–120. doi: 10.1159/000355346. [DOI] [PubMed] [Google Scholar]
- 53.Zhang W., Cuperus T., van Dijk A., Skjodt K., Hansen S., Haagsman H.P. Developmental regulation of chicken surfactant protein A and its localization in lung. Dev Comp Immunol. 2016;61:80–87. doi: 10.1016/j.dci.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 54.Mou H., Vinarsky V., Tata P.R., Brazauskas K., Choi S.H., Crooke A.K. Dual SMAD signaling inhibition enables long-term expansion of diverse epithelial basal cells. Cell Stem Cell. 2016;19:217–231. doi: 10.1016/j.stem.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rock J.R., Gao X., Xue Y., Randell S.H., Kong Y.Y., Hogan B.L. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell. 2011;8:639–648. doi: 10.1016/j.stem.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rundhaug J.E., Nettesheim P. Growth regulation of primary rat tracheal epithelial cell cultures by endogenous transforming growth factor-beta s. J Cell Physiol. 1993;155:483–493. doi: 10.1002/jcp.1041550307. [DOI] [PubMed] [Google Scholar]
- 57.Gambaryan A., Webster R., Matrosovich M. Differences between influenza virus receptors on target cells of duck and chicken. Arch Virol. 2002;147:1197–1208. doi: 10.1007/s00705-002-0796-4. [DOI] [PubMed] [Google Scholar]
- 58.Kuchipudi S.V., Nelli R., White G.A., Bain M., Chang K.C., Dunham S. Differences in influenza virus receptors in chickens and ducks: Implications for interspecies transmission. J Mol Genet Med. 2009;3:143–151. doi: 10.4172/1747-0862.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manz B., Schwemmle M., Brunotte L. Adaptation of avian influenza A virus polymerase in mammals to overcome the host species barrier. J Virol. 2013;87:7200–7209. doi: 10.1128/JVI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Almond J.W. A single gene determines the host range of influenza virus. Nature. 1977;270:617–618. doi: 10.1038/270617a0. [DOI] [PubMed] [Google Scholar]
- 61.Otte A., Marriott A.C., Dreier C., Dove B., Mooren K., Klingen T.R. Evolution of 2009 H1N1 influenza viruses during the pandemic correlates with increased viral pathogenicity and transmissibility in the ferret model. Sci Rep. 2016;6:28583. doi: 10.1038/srep28583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Min J.Y., Santos C., Fitch A., Twaddle A., Toyoda Y., DePasse J.V. Mammalian adaptation in the PB2 gene of avian H5N1 influenza virus. J Virol. 2013;87:10884–10888. doi: 10.1128/JVI.01016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Makarova N.V., Ozaki H., Kida H., Webster R.G., Perez D.R. Replication and transmission of influenza viruses in Japanese quail. Virology. 2003;310:8–15. doi: 10.1016/s0042-6822(03)00094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Swayne D.E., Pantin-Jackwood M., Kapczynski D., Spackman E., Suarez D.L. Susceptibility of poultry to pandemic (H1N1) 2009 Virus. Emerg Infect Dis. 2009;15:2061–2063. doi: 10.3201/eid1512.091060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith J., Smith N., Yu L., Paton I.R., Gutowska M.W., Forrest H.L. A comparative analysis of host responses to avian influenza infection in ducks and chickens highlights a role for the interferon-induced transmembrane proteins in viral resistance. BMC Genomics. 2015;16:574. doi: 10.1186/s12864-015-1778-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barber M.R., Aldridge J.R., Jr., Webster R.G., Magor K.E. Association of RIG-I with innate immunity of ducks to influenza. Proc Natl Acad Sci U S A. 2010;107:5913–5918. doi: 10.1073/pnas.1001755107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Microarray data have been deposited in the Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo/) of the National Center for Biotechnology Information and can be assessed with the GEO accession number GSE86449.