Abstract
Backgrounds
Genome-wide association studies (GWASs) have identified several gastric cancer (GC) susceptibility loci in Asians, but their effects on disease outcome are still unknown. This study aimed to investigate whether these GWAS-identified genetic variants could serve as robust prognostic biomarkers for GC.
Methods
A multistage clinical cohort, including a total of 2432 GC patients in the Chinese population, was used to identify the association between GWAS-identified risk variants and overall survival of GC. Hazard ratios (HRs) and 95% confidence intervals (CIs) were computed by Cox regression analysis, and the log-rank P was calculated by the log-rank test with the Kaplan-Meier method.
Results
We found that rs2274223 A>G in PLCE1 was associated with increased GC survival in both training set (P = .011), which was independently replicated in validation set 1 (P = .045), but not in validation set 2. The area under the curve (AUC) from receiver-operator characteristic (ROC) curve showed this clinical relevance with onset age-dependence, especially in the subgroup of early-onset cases. Moreover, a significant improvement in overall survival prediction was identified when the rs2274223 genetic effect was included in the estimation; this result was also supported by the prognostic nomogram. In addition, patients with lower expression of PLCE1 showed benefits via longer survival, potentially due to the functional effect of rs2274223.
Interpretation
This preliminary study suggests that a GWAS-identified genetic variant in PLCE1 may serve as a potential biomarker for GC survival. Additional replication with larger samples size is warranted to further investigation.
Keywords: GWAS, Genetic variants, Gastric cancer, Survival
Research in context.
Considering the difference between the tumorigenesis and progression of gastric cancer, we investigated whether GWAS-identified susceptibility SNPs could affect gastric cancer survival. The rs2274223 A>G SNP, an independent protective factor, was significantly associated with increased gastric cancer survival, especially in the early-onset subgroup. Additionally, the genetic effect of rs2274223 improved overall survival prediction within a prognostic nomogram model, and patients with lower PLCE1 expression showed longer survival. These findings suggest that rs2274223 could act as a potential biomarker for gastric cancer prognosis.
1. Introduction
Gastric cancer is one of the most common malignancies dangerous to human health, being fifth in morbidity and third in mortality among cancers worldwide [1]. Notably, the Asian region has the highest rate of gastric cancer incidence, which is partly attributed to its differences in diverse hereditary backgrounds, behavioral factors and Helicobacter pylori infection [2, 3]. Although the diagnosis and therapy of gastric cancer have been greatly improved in recent years, the 5-year survival rate remains poor at approximately 30% [4]. To date, clinical staging has been widely applied to determine tumor aggression and prognosis, but a wide heterogeneity of prognosis still exists, mainly due to deficiencies in the staging system. Therefore, a number of studies have been devoted to discovering new biomarkers to combine with traditional tumor diagnosis, staging and prognosis and thus to improve early diagnosis and prognostic prediction [5].
In recent years, emerging evidence has demonstrated the significant genetic effects of single nucleotide polymorphisms (SNPs) on gastric cancer development and progression [6, 7]. Genome-wide association studies (GWASs) are now well known as a powerful approach to explore complex disease-risk-related variants. Recently, five significant gastric-cancer-related GWASs from Asian populations have identified a moderate number of independent loci and SNPs with genome-wide statistical significance, including rs2294008 in PSCA at 8q24 [8], rs2274223 in PLCE1 at 10q23 [9], rs4072037 in MUC1 at 1q21 [9], rs98401504 in ZBTB20 at 3q13 [10] and rs13361707 in PTGER4 at 5q13 [10]. These genetic variants have been further studied in diverse ethnic backgrounds, and some have been identified as high-quality biomarkers for screening gastric cancer susceptibility [11].
However, few studies have focused on the effects of genetic factors on gastric cancer clinical outcomes. Considering the difference between gastric cancer etiology and its developmental mechanism, we hypothesized that these GWAS-identified susceptibility SNPs were associated with survival time in gastric cancer patients. In this study, we evaluated the association between the risk variants for gastric cancer found in previous GWASs and patients' survival based on large, multistage clinical cohorts in Chinese populations, and we assessed the potential of these variants as prognostic biomarkers for gastric cancer.
2. Methods
2.1. Study Population
A two-stage follow-up study was designed to investigate the effect of gastric cancer risk SNPs on patients' survival. In the first stage, we enrolled patients from Yixing People's Hospital, Yixing city, as a training set, for which detailed population information has been described in our previous publication [7]. In the second stage, patients from Nantong city and Nanjing city were considered as validation sets 1 and 2, respectively. For validation set 1, a total of 480 patients were recruited from Nantong Tumor Hospital from December 2000 to July 2006, and 471 of them were successfully followed up, with 113.0 months for the maximum follow-up time and 41.1 for the median. For validation set 2, a total of 1021 patients with adequate follow-up information were enrolled from January 2005 to December 2009, with 87.8 months maximum follow-up time and 34.0 months median. In each cohort, the clinical pathological variables, including tumor size, tumor site (cardia or noncardia), histological type, invasion, lymph node, distant metastasis, and TNM stage (American Joint Commission for Cancer Staging, 6th ed., 2002), were collected from the medical records of the patients. All subjects signed an informed consent, and our study was approved by the Institutional Review Board of Nanjing Medical University, Nanjing, China.
2.2. SNP Genotyping
Genomic DNA was extracted from paraffin sections of tumor tissues according to the detailed method reported previously [7]. A TaqMan PCR Genotyping Assay using the ABI 7900HT Real Time PCR System (Applied Biosystems, Foster City, CA) was utilized to perform candidate SNP genotyping. For quality control, all genotype analyses were performed by two blinded individuals who did not know the subjects' status. Approximately 10% of all samples were selected randomly for genotype confirmation, and the results were 100% concordant.
2.3. Statistical Analysis
The overall survival time was the primary outcome in this study, and it was calculated from the day of gastric cancer diagnosis until death or the last follow-up. Median survival time (MST) was used to compare the life span associated with each variable; if the median was not available, the mean survival time was used as an alternative. A multivariate Cox regression analysis was utilized to evaluate the adjusted hazard ratios (HRs) and corresponding 95% confidence intervals (CIs) with adjustments for age, sex, tumor size, histological type and TNM stage. A Cox stepwise regression analysis was performed to determine what factors could be used as an independent factor for gastric cancer prognosis, with P < .05 for entering and P > .10 for removing the model. The genetic effects of each SNP were estimated using additive, dominant, recessive and codominant models. The association between survival time and each included variable was measured using the Kaplan-Meier method and the log-rank test. Subsequently, a survival model including genetic effect was built to assess the prognostic efficacy by using a time-dependent receiver-operator characteristic (ROC) curve analysis and calculating the area under the curve (AUC) of the ROC curve. In addition, a nomogram was formulated based on the results of the Cox stepwise regression analysis, and its performance was evaluated by the concordance index (C-index) and assessed by comparing nomogram predictions to Kaplan-Meier estimates of survival probability; bootstrap analyses with 1000 resamples were applied to these activities. All tests within two-sided were performed using the SAS software (version 9.2, SAS Institute, Cary, NC) and R version 3.1.3.
3. Results
3.1. Patient Characteristics
The demographic and clinical pathological characteristics of each cohort are shown in Supplementary Table 1. Briefly, a total of 2432 gastric cancer patients from three independent cohorts were enrolled for survival analysis, and in each group, patients with larger tumor size, diffuse type and late TNM stages (including invasion depth, lymph node and distant metastasis) had shorter survival times than other patients did (all log-rank P < .001).
3.2. Effects of GWAS-Identified SNPs on Gastric Cancer Survival
The flow chart of this study is shown in Fig. 1. Five candidate risk SNPs were evaluated for their associations with gastric cancer survival in the training set, and the analysis identified that rs2274223 in PLCE1 exhibited a significant association with gastric cancer survival (OR = 0.84, 95% CI = 0.72–0.98, P = .026 in additive model; OR = 0.78, 95% CI = 0.65–0.95, P = .011 in dominant model, Table 1 and Supplementary Table 2). Subsequently, we performed survival analysis on rs2274223 in two validation cohorts and found a similar protective effect of rs2274223 on gastric cancer survival. In line with the training set, rs2274223 in PLCE1 was significantly associated with increased gastric cancer survival in validation set 1 (OR = 0.80, 95% CI = 0.65–0.98, P = .028 in additive model; OR = 0.78, 95% CI = 0.62–0.99, P = .045 in dominant model, Table 1). Although rs2274223 was not associated with gastric cancer survival in validation set 2, the same direction of rs2274223 protective effect on gastric cancer survival was found (Table 1). Furthermore, the combined analysis of all enrolled patients identified an obvious elevation of gastric cancer survival associated with the genetic effect of rs2274223 (OR = 0.86, 95% CI = 0.78–0.95, P = .002 in additive model; OR = 0.82, 95% CI = 0.73–0.93, P = .001 in dominant model, Table 1).
Fig. 1.
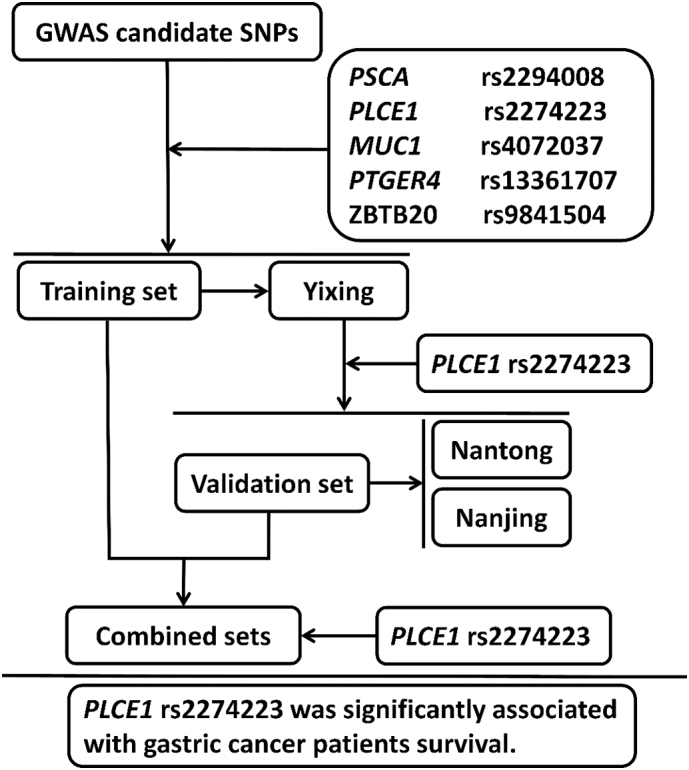
The flow chart for association analysis of the GWAS-identified SNPs and gastric cancer survival.
Table 1.
The association between rs2274223 in PLCE1 and gastric cancer patients' survival from three independent cohorts.
| Variation | Stages | Cohorts | AA/AG/GG |
Genetic models | log-rank P | HR (95% CI)b | Pb | ||
|---|---|---|---|---|---|---|---|---|---|
| Patients | Deaths | MST (months) | |||||||
| PLCE1 rs2274223 | Training set | Yixing | 509/361/68 | 256/150/31 | 52.9/96.4/68.5 | Additive model | 0.045 | 0.84 (0.72–0.98) | 0.026 |
| Dominant model | 0.014 | 0.78 (0.65–0.95) | 0.011 | ||||||
| Ptrend | 0.036 | ||||||||
| Validation set1 | Nantong | 262/178/29 | 179/111/17 | 39.6/44.5/42.3 | Additive model | 0.359 | 0.80 (0.65–0.98) | 0.028 | |
| Dominant model | 0.166 | 0.78 (0.62–0.99) | 0.045 | ||||||
| Ptrend | 0.156 | ||||||||
| Validation set2 | Nanjing | 578/363/80 | 234/140/29 | 50.2a/55.7a/36.5a | Additive model | 0.493 | 0.91 (0.77–1.06) | 0.224 | |
| Dominant model | 0.263 | 0.86 (0.70–1.05) | 0.140 | ||||||
| Ptrend | 0.236 | ||||||||
| Combined sets | 1349/902/177 | 669/401/77 | 53.5/85.2/79.1 | Additive model | 0.014 | 0.86 (0.78–0.95) | 0.002 | ||
| Dominant model | 0.004 | 0.82 (0.73–0.93) | 0.001 | ||||||
| Ptrend | 0.005 | ||||||||
Mean survival time was calculated when Median survival time (MST) could not be performed.
Adjusted for age, sex, tumor size, histological type and TNM stage.
3.3. Determination of the Independent Survival Effect of rs2274223 for Gastric Cancer
To avoid the impact of confounding factors acting on the hereditary effects, a stepwise Cox regression analysis was performed to evaluate whether rs2274223 had an independent effect on gastric cancer survival. We enrolled demographic variables (age and sex), pathological features (tumor size, histological type, tumor site, and TNM stage) and an rs2274223 dominant genetic effect into the regression model, and we observed that rs2274223 could act as an independent protective factor for gastric cancer survival with −0.196 of β (HR = 0.82, 95% CI = 0.73–0.93, P = .001, Table 2).
Table 2.
The evaluation of PLCE1 rs2274223 SNP as an independent factor for gastric cancer survival by stepwise Cox regression analysis.
| Variables | β | SE | HR (95% CI) | P |
|---|---|---|---|---|
| Age | ||||
| >60 vs. ≤60 | 0.172 | 0.061 | 1.18 (1.05–1.34) | 0.005 |
| Tumor size | ||||
| >5 vs. ≤5 | 0.331 | 0.064 | 1.39 (1.23–1.58) | <0.001 |
| Histological type | ||||
| Intestinal vs. Diffuse | −0.222 | 0.069 | 0.80 (0.70–0.92) | 0.001 |
| TNM stage | ||||
| I-II vs. III-IV | 0.947 | 0.074 | 2.58 (2.23–2.98) | <0.001 |
| PLCE1 rs2274223 | ||||
| AG/GG vs. AA | −0.196 | 0.061 | 0.82 (0.73–0.93) | 0.001 |
3.4. Stratified Analysis of the Effect of rs2274223 on Gastric Cancer Survival
We further evaluated the effect of rs2274223 on gastric cancer survival by stratifying demographic features (age and sex) and clinical phenotypes (tumor size, location, histological type and TNM stage). As shown in Supplementary Table 3, the protective effect of the rs2274223 G allele on patients' survival was statistically significant in younger and female patients and in patients with larger tumor size, noncardia type, diffuse type or later TNM stage (all P < .05).
3.5. Effect of rs2274223 on Early-Onset patients' Survival
Considering that young gastric cancer patients have a different molecular genetic profile from those with tumors occurring at greater ages [12], we hypothesized that rs2274223 could exert different genetic effects on early-onset patients and older patients. Fig. 2a shows the age distribution of gastric cancer onset, revealing that approximately 8.5% of gastric cancer patients fell into the early-onset gastric cancer category (onset age ≤ 45), which was in accordance with the results of previous population studies [13]. Interestingly, a distinct dose-response effect of rs2274223 A>G on gastric cancer survival was identified when patients were divided by diagnostic age; the G allele of rs2274223 played a significant protective role in younger patients, especially early-onset cases (HR = 0.53, 95% CI = 0.34–0.83, P = .001, Fig. 2b).
Fig. 2.
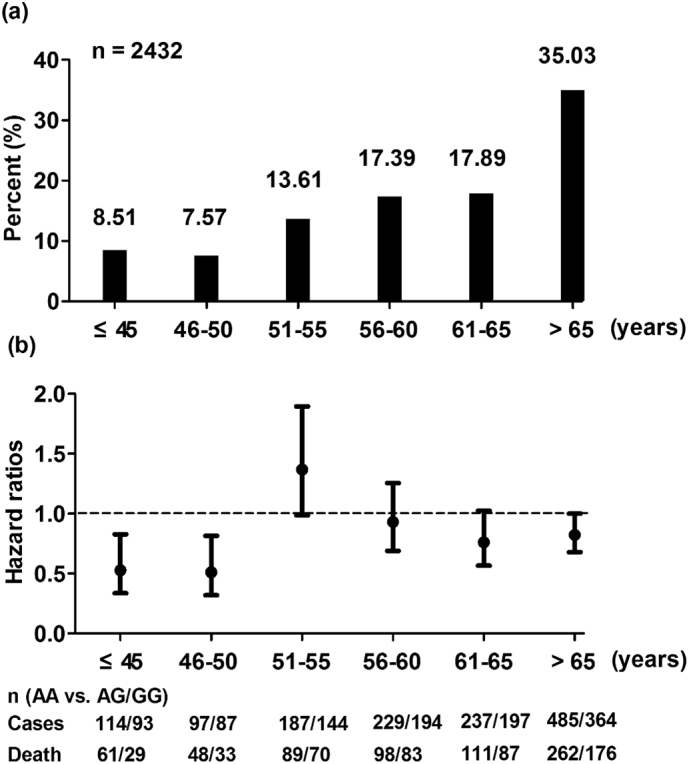
Distribution of onset age and the stratified effect of rs2274223 on gastric cancer survival. (a) shows the distribution of gastric cancer onset age in the combined cohorts; the early-onset age was defined as an age <45 years; (b) represents the association between rs2274223 and gastric cancer survival stratified by onset age; the hazard ratios were calculated by the Cox regression analysis with an adjustment for sex, tumor size, histological type and TNM stage in the dominant model.
Moreover, we conducted a time-dependent ROC analysis to estimate the predictive ability of rs2274223 for gastric cancer survival. As shown in Supplementary Fig. 1, higher predictive accuracy was identified for the integration of rs2274223 genotype with clinical features (tumor size, histological type, depth of invasion, lymph node and distant metastasis) than for only the clinical features, especially in the subgroup of early-onset cases; in addition, the effect of the rs2274223 genotype took time to accrue.
3.6. Clinical Application of the rs2274223 Genetic Effect for Gastric Cancer Survival Prediction
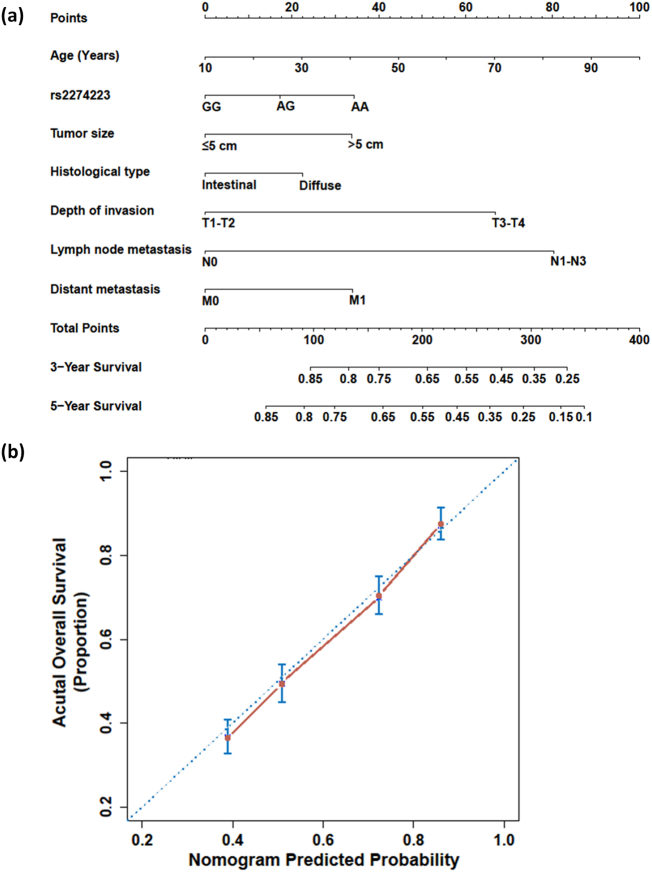
Currently, nomograms have been widely proposed as an alternative to the traditional staging systems, or even as a new standard, in many cancer types [14]. In this study, we established a prognostic nomogram for gastric cancer survival based on independent prognostic factors including rs2274223 genotype and the variables diagnostic age, tumor size, histological type, depth of invasion, lymph node metastasis and distant metastasis in a multivariate regression analysis (Fig. 3a). The bootstrap-corrected C-index for overall survival prediction was significantly increased from 0.513 to 0.612 in each independent factor to 0.662 in this nomogram model. The calibration plot for the probability of overall survival illustrated an optimal agreement between the prediction by nomogram and actual observation (Fig. 3b).
Fig. 3.
Gastric cancer survival nomogram and corresponding calibration curve. (a) The nomogram allows the user to obtain the probability of three- and five-year overall survival corresponding to a patient's combination of covariates: locate patient's features on each axis, and compare to the “Point” axis to determine how many points are attributed to each feature; and then, locate the sum of the points for all variables on the “Total Points” line to determine the individual probability of gastric cancer on the “3-Year Survival” or “5-Year Survival” line. (b) The calibration curve of the nomogram for predicting gastric cancer overall survival. Actual overall survival is plotted on the y-axis, and predicted is on the x-axis.
3.7. Functional Annotation of rs2274223 and PLCE1
In addition, by analyzing the Encyclopedia of DNA Elements (ENCODE) and the Roadmap Epigenomics Project database as implemented in RegulomeDB (http://regulome.stanford.edu/) and HaploReg v4 (http://www.broadinstitute.org/mammals/haploreg), we observed that SNP rs2274223 was located at some motifs regions referring to specific transcription factors, and thus involved in chromatin structure and histone modification (Supplementary Table 4). This finding indicates that the potential biological effect of rs2274223 on gastric cancer risk and prognosis.
We then investigated the prognostic significance of PLCE1 expression on gastric cancer survival by Kaplan-Meier Plotter (http://kmplot.com/analysis). Patients were split into two groups (high- and low-expression groups) with the option auto select best cutoff. The log-rank result of the Kaplan-Meier Plotter analysis showed that patients with low expression of PLCE1 could benefit from longer survival times than those with high expression (HR = 1.30, 95% CI = 1.07–1.58, P = .007, Supplementary Fig. 2).
4. Discussion
An increasing number of gastric cancer risk SNPs have been widely reported as potential susceptibility biomarkers for gastric cancer [11]. However, whether these risk SNPs are also associated with gastric cancer outcomes has not been comprehensively elucidated. In this study, we utilized three independent clinical cohorts to evaluate the effects of these newly GWAS-identified SNPs on gastric cancer survival (i.e., rs2294008 at 8q24, rs2274223 at 10q23, rs4072037 at 1q21, rs98401504 at 3q13 and rs13361707 at 5q13). We found that rs2274223 A>G in PLCE1 was associated with increased gastric cancer survival, especially in early-onset gastric cancer patients, and that the integration of the rs2274223 genetic effect with clinical features could obviously increase the prediction efficacy of gastric cancer prognosis. Unlike what we observed in validation set 1, no significant association was observed between rs2274223 in PLCE1 and gastric cancer survival in validation set 2, which led us to comprehensively investigate the association in further validation with larger samples.
The SNP rs2274223 A>G, first reported by Chinese gastric cancer GWASs [9, 15], was found to have a risk effect for almost all digestive system malignancies, including esophageal squamous cell carcinoma [16], colorectal cancer [17], head and neck cancer [18] and gallbladder cancer [19]. Intriguingly, the G allele of rs2274223 tended to show a risk effect on the subgroup of gastric cardia cancer (GCC) rather than gastric noncardia cancer (GNCC) in both Asian and Caucasian populations [11]. This effect may be attributable to the diversity of molecular and genetic signatures in the development and progression of gastric cancer [20]. We thus performed a stratified analysis to detect whether this site-specific relationship existed for survival. When assessing clinical feature subtypes, we observed that the G allele of rs2274223 was remarkably associated with longer survival in GNCC patients, rather than in GCC patients. This interesting finding was completely different from the susceptibility studies, suggesting that there might be distinctly diverse molecular and hereditary mechanisms between the occurrence and development of gastric cancer within different types.
Moreover, when we stratified by onset age, we found that the younger patients (onset age < 60 years) with the G allele showed longer survival than those with the A allele, but no similar effect was found in the older group. Previous studies have indicated that approximately 10% of gastric cancer patients fall into the early-onset category (onset at the age of 45 or younger) [13], and these patients suffer from greater effects of inherited genetic factors and less exposure to environmental carcinogens [21]. Thus, by performing a more precise subgroup analysis with respect to onset age, we found an obvious onset-age-based dose-response effect of rs2274223 on gastric cancer survival, in which the protective effect of the G allele was significant in the subgroup of patients with onset age <50 years, especially in the early-onset group. Similarly, the time-dependent analysis demonstrated the higher prognostic accuracy in the integration of rs2274223 genetic effect and clinical evaluation index.
Considering clinical applications, we further fitted a prognostic nomogram for gastric cancer survival including rs2274223 genotype and other prognostic indicators. As expected, an obvious improvement of overall survival prediction was identified, and this nomogram might be used in management of mortality risk by means of therapy modification. All these findings indicated the possibility and feasibility of clinical application of inherited genetic factors to gastric cancer survival evaluation. Obviously, however, these findings require further investigation in large prospective studies.
In addition, the molecular mechanism of the PLCE1 gene, harboring rs2274223, in gastric cancer etiology and progression has seldom been investigated. PLCE1, a member of the phospholipase C protein family, can be epigenetically regulated by microRNAs and thus involved in cancer cell behaviors, including cell growth, apoptosis and angiogenesis [[22], [23], [24]], which strongly supports our finding that lower expression of PLCE1 was associated with longer survival. However, the functional significance of SNP rs2274223 in gastric cancer susceptibility and development remains unclear. It is plausible that rs2274223 as a non-synonymous variant can result in the substitution of a histidine for an arginine, which might affect the conformation and the expression of PLCE1 protein, and even the molecular mechanism. In our study, the in-silico analysis showed that the SNP rs2274223 could alter the binding ability of some specific transcription factors to the motif region where it resides, and thus, it could be involved in chromatin structure and histone modification. Because of the emerging evidence that some specific SNPs can have long-range regulatory effects on nearby genes [25, 26], it is quite likely that rs2274223, as a regulatory quantitative trait locus (regQTL) SNP [27], could affect the function of nearby candidate genes and thus be involved in gastric cancer etiology and outcome. Notably, during our investigation, two novel loci at 5q14.3 and 1q22 were identified as significantly associated with risk of GNCC [28], but these SNPs were not enrolled in our study. These findings could be further evaluated for their association with gastric cancer survival.
In summary, this is the first study to investigate the association between gastric cancer risk SNPs and survival in multiple clinical cohorts. This preliminary study support rs2274223 as a potential prognostic biomarker for gastric cancer survival prediction, and this SNP should therefore be validated in diverse ethnic populations and prospective studies. Additionally, experimental studies are warranted to elucidate the mechanism of genetic variants in PLCE1 in gastric cancer development and progression.
Founding Sources
This study was supported by The National Key Research and Development Program of China (2017YFC1308900), National Natural Science Foundation of China (81773516), Distinguished Young Scholars of Nanjing (JQX13005) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine).
Conflicts of Interest
The authors disclose no potential conflicts of interest.
Authors Contributions
Jinfei Chen, Mulong Du and Meilin Wang conceived and designed the experiments. Dongying Gu, Rui Zheng and Junyi Xin wrote the paper. Shuwei Li, Haiyan Chu, Weida Gong, Fulin Qiang and Zhengdong Zhang contributed reagents/materials/analysis tools. Dongying Gu, Shuwei Li, Weida Gong and Fulin Qiang recruited samples. All authors reviewed the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.06.028.
Contributor Information
Meilin Wang, Email: mwang@njmu.edu.cn.
Mulong Du, Email: drdumulong@njmu.edu.cn.
Jinfei Chen, Email: jinfeichen@sohu.com.
Appendix A. Supplementary data
Supplementary Tables
Supplementary Figures
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Zheng R., Zeng H., Zhang S., Chen W. Estimates of cancer incidence and mortality in China, 2013. Chin J Cancer. 2017;36:66. doi: 10.1186/s40880-017-0234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desantis C.E., Lin C.C., Mariotto A.B., Siegel R.L., Stein K.D., Kramer J.L. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 5.Ludwig J.A., Weinstein J.N. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 6.Graziano F., Galluccio N., Lorenzini P., Ruzzo A., Canestrari E., D'Emidio S. Genetic activation of the MET pathway and prognosis of patients with high-risk, radically resected gastric cancer. J Clin Oncol. 2011;29:4789–4795. doi: 10.1200/JCO.2011.36.7706. [DOI] [PubMed] [Google Scholar]
- 7.Wang M., Bai J., Tan Y., Wang S., Tian Y., Gong W. Genetic variant in PSCA predicts survival of diffuse-type gastric cancer in a Chinese population. Int J Cancer. 2011;129:1207–1213. doi: 10.1002/ijc.25740. [DOI] [PubMed] [Google Scholar]
- 8.Sakamoto H., Yoshimura K., Saeki N., Katai H., Shimoda T., Matsuno Y. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet. 2008;40:730–740. doi: 10.1038/ng.152. [DOI] [PubMed] [Google Scholar]
- 9.Abnet C.C., Freedman N.D., Hu N., Wang Z., Yu K., Shu X.O. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet. 2010;42:764–767. doi: 10.1038/ng.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y., Hu Z., Wu C., Dai J., Li H., Dong J. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet. 2011;43:1215–1218. doi: 10.1038/ng.978. [DOI] [PubMed] [Google Scholar]
- 11.Mocellin S., Verdi D., Pooley K.A., Nitti D. Genetic variation and gastric cancer risk: a field synopsis and meta-analysis. Gut. 2015;64:1209–1219. doi: 10.1136/gutjnl-2015-309168. [DOI] [PubMed] [Google Scholar]
- 12.Milne A.N., Sitarz R., Carvalho R., Carneiro F., Offerhaus G.J. Early onset gastric cancer: on the road to unraveling gastric carcinogenesis. Curr Mol Med. 2007;7:15–28. doi: 10.2174/156652407779940503. [DOI] [PubMed] [Google Scholar]
- 13.Kokkola A., Sipponen P. Gastric carcinoma in young adults. Hepatogastroenterology. 2001;48:1552–1555. [PubMed] [Google Scholar]
- 14.Jiang Y., Li T., Liang X., Hu Y., Huang L., Liao Z. Association of adjuvant chemotherapy with survival in patients with stage II or III gastric cancer. JAMA Surg. 2017;152 doi: 10.1001/jamasurg.2017.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L.D., Zhou F.Y., Li X.M., Sun L.D., Song X., Jin Y. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet. 2010;42:759–763. doi: 10.1038/ng.648. [DOI] [PubMed] [Google Scholar]
- 16.Jia X., Liu P., Zhang M., Feng T., Tang H., Tang Z. Genetic variants at 6p21, 10q23, 16q21 and 22q12 are associated with esophageal cancer risk in a Chinese Han population. Int J Clin Exp Med. 2015;8:19381–19387. [PMC free article] [PubMed] [Google Scholar]
- 17.Ezgi O., Merve A., Hakan Y.T., Gul O. Genetic variations in phospholipase C-epsilon 1 (PLCE1) and susceptibility to colorectal cancer risk. Biochem Genet. 2016;54:826–829. doi: 10.1007/s10528-016-9759-4. [DOI] [PubMed] [Google Scholar]
- 18.Yuan Z., Yuan H., Ma H., Chu M., Wang Y., Hu Z. Genetic variants at 10q23 are associated with risk of head and neck cancer in a Chinese population. Oral Oncol. 2013;49:332–335. doi: 10.1016/j.oraloncology.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Sharma K.L., Umar M., Pandey M., Misra S., Kumar A., Kumar V. Association of potentially functional genetic variants of PLCE1 with gallbladder cancer susceptibility in north Indian population. J Gastrointest Cancer. 2013;44:436–443. doi: 10.1007/s12029-013-9537-z. [DOI] [PubMed] [Google Scholar]
- 20.He Y., Wang C., Wang Z., Zhou Z. Genetic variant PLCE1 rs2274223 and gastric cancer: more to be explored? Gut. 2016;65:359–360. doi: 10.1136/gutjnl-2015-309968. [DOI] [PubMed] [Google Scholar]
- 21.Milne A.N., Offerhaus G.J. Early-onset gastric cancer: Learning lessons from the young. World J Gastrointest Oncol. 2010;2:59–64. doi: 10.4251/wjgo.v2.i2.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui X.B., Peng H., Li R.R., Mu J.Q., Yang L., Li N. MicroRNA-34a functions as a tumor suppressor by directly targeting oncogenic PLCE1 in Kazakh esophageal squamous cell carcinoma. Oncotarget. 2017;8:92454–92469. doi: 10.18632/oncotarget.21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han N., Zhao W., Zhang Z., Zheng P. MiR-328 suppresses the survival of esophageal cancer cells by targeting PLCE1. Biochem Biophys Res Commun. 2016;470:175–180. doi: 10.1016/j.bbrc.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 24.Zhai S., Liu C., Zhang L., Zhu J., Guo J., Zhang J. PLCE1 promotes esophageal Cancer cell progression by maintaining the transcriptional activity of snail. Neoplasia. 2017;19:154–164. doi: 10.1016/j.neo.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi J., Xu M., Makowski M.M., Zhang T., Law M.H., Kovacs M.A. A common intronic variant of PARP1 confers melanoma risk and mediates melanocyte growth via regulation of MITF. Nat Genet. 2017;49:1326–1335. doi: 10.1038/ng.3927. [DOI] [PubMed] [Google Scholar]
- 26.Luo Z., Rhie S.K., Lay F.D., Farnham P.J. A prostate cancer risk element functions as a repressive loop that regulates HOXA13. Cell Rep. 2017;21:1411–1417. doi: 10.1016/j.celrep.2017.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pai A.A., Pritchard J.K., Gilad Y. The genetic and mechanistic basis for variation in gene regulation. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1004857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z., Dai J., Hu N., Miao X., Abnet C.C., Yang M. Identification of new susceptibility loci for gastric non-cardia adenocarcinoma: pooled results from two Chinese genome-wide association studies. Gut. 2017;66:581–587. doi: 10.1136/gutjnl-2015-310612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables
Supplementary Figures