Abstract
Background
Molecular classification of tumour clonality is currently not evaluated in multiple invasive breast carcinomas, despite evidence suggesting common clonal origins. There is no consensus about which type of data (e.g. copy number, mutation, histology) and especially which statistical method is most suitable to distinguish clonal recurrences from independent primary tumours.
Methods
Thirty-seven invasive breast tumour pairs were stratified according to laterality and time interval between the diagnoses of the two tumours. In a multi-omics approach, tumour clonality was analysed by integrating clinical characteristics (n = 37), DNA copy number (n = 37), DNA methylation (n = 8), gene expression microarray (n = 7), RNA sequencing (n = 3), and SNP genotyping data (n = 3). Different statistical methods, e.g. the diagnostic similarity index (SI), were used to classify the tumours as clonally related recurrences or independent primary tumours.
Results
The SI and hierarchical clustering showed similar tendencies and the highest concordance with the other methods. Concordant evidence for tumour clonality was found in 46% (17/37) of patients. Notably, no association was found between the current clinical guidelines and molecular tumour features.
Conclusions
A more accurate classification of clonal relatedness between multiple breast tumours may help to mitigate treatment failure and relapse by integrating tumour-associated molecular features, clinical parameters, and statistical methods. Guidelines need to be defined with exact thresholds to standardise clonality testing in a routine diagnostic setting.
Electronic supplementary material
The online version of this article (10.1186/s13058-018-1022-y) contains supplementary material, which is available to authorized users.
Keywords: Tumour clonality, Bilateral breast cancer, Ipsilateral breast cancer, Intertumour heterogeneity, Similarity index, Multiple breast cancer
Background
Approximately 2–15% of women previously diagnosed with breast cancer will develop a second primary carcinoma in the contralateral breast during their lifetime [1, 2]. Interestingly, the risk of developing a breast tumour in the contralateral breast is 2–6-fold higher in breast cancer patients than the risk of developing a first primary breast cancer in the general population [2]. These findings indicate a clonal relationship between bilateral breast cancers as well as a consequence of genetic predisposition and treatment [2, 3]. However, discordance in histologic patterns between bilateral tumours suggests that the majority of bilateral breast cancers have independent tumour origins [4]. Clonality is defined as two tumours deriving from the same progenitor cell that previously underwent malignant changes and gave rise to both of the detected tumours [5]. Consequently, in the early development of the two clones the driver events of the progenitor cell (e.g. copy number alteration (CNA), DNA methylation, mutation, and gene expression profiles) need to have been identical. Due to heterogeneity in subclonal drifts, the variability between the two clones results from the accumulation of diverse molecular changes associated with tumour progression [6]. Nevertheless, similarities in certain tumour features might be due to genetic predisposition and shared environment instead of indicating metastatic spread.
Ipsilateral (unilateral) secondary tumours occur in 10–15% of patients undergoing breast-conserving surgery and radiation therapy [7]. At present, the concordance of hormone receptor status in tumour pairs is the main factor when evaluating potential clonal relatedness of two breast tumours. Clinical characteristics of breast tumours with independent origin are the presence of an in situ component in the second tumour, different degrees of differentiation, different histological subtypes (e.g. invasive carcinoma no special type (NST), invasive lobular carcinoma, tubular, medullary, etc.), absence of locoregional or distant metastases, long time interval between the two tumours, and differences in stage and anatomic location [8, 9]. Determining the concordance of histopathological characteristics between multiple breast carcinomas is insufficient for discerning whether multiple tumours are true recurrences of the primary tumour (clonal) or a new unrelated primary lesion (independent tumour) [10]. Bilateral tumours are currently clinically diagnosed as two different entities, while ipsilateral tumours are classified as local recurrences [1]. Clonal recurrences can represent treatment failure of the first tumour, warranting a change of therapy for the second tumour. Contrastingly, two independent tumours with the same clinical features can be treated similarly since the treatment was successful for the first tumour.
Different techniques in the field of molecular genetics have been used to elucidate tumour clonality, e.g. allelic imbalances [11, 12], CGH (comparative genomic hybridization) [13, 14], array comparative genomic hybridization (aCGH) [15, 16], as well as whole exome and whole genome sequencing [17–19]. In addition, several analytical tools have been proposed to justify the routine clinical use of determining tumour clonality [5, 13, 15, 20–22].
In the present study, 74 invasive breast tumours corresponding to 37 patients were stratified by laterality (bilateral vs. ipsilateral) and the time interval between the diagnosis of the first and second tumour (synchronous vs. metachronous). Both tumours from the same patient were analysed using several genome-wide screening methods and statistical approaches to assess tumour clonality. The level of concordance among the different statistical techniques and molecular data might help to define clonality in multiple tumours and guide treatment decisions for clinicians.
Methods
Patients and clinicopathological data
Fresh-frozen tumour specimens for 74 invasive breast carcinomas, corresponding to 37 patients diagnosed in Western Sweden between 1988 and 1998 with multiple breast cancers, were selected from the tumour bank at the Sahlgrenska University Hospital Oncology Lab (Gothenburg, Sweden). The patients were stratified into four groups based on the anatomic location of the multiple breast cancers (ipsilateral or bilateral) and time interval between the diagnoses (synchronous or metachronous). Ipsilateral was defined as tumours occurring in the same breast while bilateral was defined as the occurrence of tumours in both breasts. Metachronicity was defined as a time interval greater than 6 months between the diagnoses of the first and second tumours, while synchronicity specified that the two tumours occurred concurrently. Clinicopathological information was obtained from Regional Cancer Centre West (Gothenburg, Sweden) and the Sympathy and Melior databases (Sahlgrenska University Hospital). A part of the dataset was stratified into the molecular breast cancer subtypes (normal-like, basal-like, luminal subtype A, luminal subtype B/human epidermal growth factor receptor 2 (HER2)+, luminal subtype B/HER2-, and HER2/oestrogen receptor (ER)-) as described elsewhere [23, 24]. Luminal subtype B was further stratified according to HER2 status as determined by aCGH; HER2+ was set to log2 ratio ≥ + 0.5 and HER2- was set to log2 ratio < + 0.5 [25]. Routine haematoxylin and eosin-stained slides from formalin-fixed paraffin-embedded (FFPE) blocks were revised by a board-certified breast pathologist. Classification of the subtypes based on immunohistochemistry was not possible due to the lack of information on the Ki-67 status. The patients had an average follow-up time of 7.2 years. None of the patients were diagnosed with distant metastasis at the time of diagnosis of either the first or second tumours. The selection criteria were to use samples from opposite quadrants for ipsilateral cases and no nipple involvement. Representative imprints from each tumour specimen were stained with May-Grünwald Giemsa (Chemicon, Temecula CA, USA) and evaluated for neoplastic cells. Tumour specimens with at least 70% neoplastic cell content were included in downstream analyses.
Array comparative genomic hybridization (aCGH) analysis
aCGH and data pre-processing was performed as previously described [24] and summarised in the Additional file 1: Supplementary Methods. Segmented data for segment analysis were generated using the “GLAD” package [26] in R (v3.4.3) [27]. The “Clonality” package [28] was used to define the likelihood ratio with individual comparisons (LR2) and LR2 p value and required copy number data procession with the “DNAcopy” package [29].
DNA methylation analysis
Sixteen samples were randomly selected to represent each clinical group with four samples corresponding to two patients per group. Purified genomic DNA was processed at the SNP&SEQ technology platform, Uppsala, Sweden, using Illumina Infinium MethylationEPIC BeadChips (MethylationEPIC_v-1-0; mapped to UCSC Feb 2009 hg19: GRCh37). Raw data (IDAT files) were processed in R using the “RnBeads” package [30]. The probes were normalised with the BMIQ method (beta mixture quantile dilation) [31]. Beta values were obtained with “RnBeads”. The intensity values were extracted using the “ChAMP” package to generate segmented copy number data for the segment analysis [32, 33]. The “conumee” package was used to extract unsegmented information of CNAs on the probe level [34]. The unsegmented CNAs were used for the similarity index (SI), the distance measure and the clustering analysis.
Whole transcriptome RNA sequencing (RNA-seq)
Total RNA samples were processed at the Science for Life Laboratory (National Genomics Infrastructure, Stockholm, Sweden). Illumina TruSeq strand-specific RNA libraries (Ribosomal depletion using RiboZero human) containing 125 bp paired-end reads were obtained for each sample on a HiSeq2000 sequencer (Illumina, San Diego, CA, USA). The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) [35], as described in the Additional file 1: Supplementary Methods.
Genome-wide single nucleotide polymorphism (SNP) genotyping analysis
Genome-wide SNP genotyping analysis was processed with Illumina Infinium HumanOmni2.5–8 v1.3 Beadchips at the SCIBLU Genomics DNA Microarray Resource Center (SCIBLU), Sweden, as described in the Additional file 1: Supplementary Methods.
Statistical analyses
A p value cut-off of 0.05 was applied in all statistical tests.
Definition of tumour clonality
Tumours derived from a common precursor tumour cell should share certain features, i.e. similar CNAs, genetic variants, shared segments, DNA methylation and gene expression patterns, in addition to non-matching features that were acquired over time. We applied different statistical methods on different types of molecular data to identify similarities between the tumours that classify a tumour pair as clonal and reject the null hypothesis (different features due to independent development of primary tumours).
Similarity index (SI)
The SI assesses whether two tumours identified in the same patient are clonally related or two independent entities by identifying genetic aberrations that are patient-specific and non-recurrent aberrations frequently identified in cancer [21]. In brief, DNA copy number data were normalised and discretized (heterozygous loss (<− 0.3); normal; low-level gain (> 0.3)) and unique (NU), shared (NS), and opposite (NO) changes were calculated for each tumour pair to obtain the SI:
The SI ranges between 0 (completely different) and 1 (identical genomic profiles). The permutation-based PSI gives the percentage of similarities between two tumours that are not due to recurrent chromosomal aberrations or randomness.
The SI remained unchanged for the gene expression microarray data. The normalised log2 ratios were discretized using a 1.5 fold change cut-off (underexpressed (log2 ratio < − 0.58); neutral; overexpressed (log2 ratio > 0.58)).
Calculation of the SI was modified for the methylation data (SImet) because the SI for copy number data is based on measuring the amount of alterations from the biologically neutral state (two copies per allele). In DNA methylation, neither methylated nor unmethylated can be defined as the neutral state of a cytosine due to the dynamic of methylation. The SImet uses beta values discretized according to thresholds defined by Du et al. [36], where beta values > 0.8 are defined as methylated, and beta values < 0.2 as unmethylated, while the range from 0.2 to 0.8 is hemi-methylated. The SImet counts the number of all common states between the first and the second tumour per probe and divides it by the total number of probes, giving the percentage of shared methylation states. The main difference is that the SImet uses all probe states while the SI is based on the changes from the neutral state and therefore does not count two tumours that are normal as a shared state.
Hierarchical clustering
Unsupervised hierarchical clustering was applied using single linkage with Euclidean distance [37]. Clustering was performed using the basic “stats” package [27] for the aCGH-derived copy number data (imputed log2 ratios), the DNA methylation data (beta values and intensity values), the microarray-derived gene expression data (normalised log2 ratios), and the SNP array data (B allele frequency (BAF) and log R ratio (LRR) values). Two tumours of the same patient were defined as similar (clonal) if they clustered together in the terminal branch of the dendrogram.
Distance measure
The distance measure was used to compute the distance matrix of the Euclidean distances between different tumour samples to evaluate the similarity between two samples. The Euclidean distance was computed using the basic “stats” package [27] for the aCGH-derived copy number data (imputed log2 ratios), the DNA methylation data (beta values and intensity values), the microarray-derived gene expression data (normalised log2 ratios), and the SNP array data (LRR values). The distance measure was calculated for true tumour pairs which derive from the same patient and for all artificial combinations of tumour pairs from different patients (permutation). Tumour pairs that are more similar on the probe level will show a shorter distance from each other. Statistical significance for clonality was defined as the distance of a tumour pair of the same patient that is in the lower fifth percentile of the distribution of distances.
Shared segment analysis
In segmented copy number data, the breakpoints and the copy number of each segment was compared between the tumours. A shared segment was defined as an overlap of the exact loci in both ends of the segment where the change in status (loss or gain) occurred with the same direction (increase or decrease in copy numbers). The segment analysis was performed on segmented copy number data derived from aCGH (imputed log2 ratios), DNA methylation array (intensity values), and SNP array (LRR values). Shared segments were counted for true tumour pairs and all artificial pairs of the respective cohort. Clonality was defined as the number of shared segments above the 95th percentile.
Mutational changes (genetic variants) and fusion transcript analysis
Mutational changes that were identical in both tumours were counted for true tumour pairs and all artificial pairs of the cohort. Clonality was defined as the number of shared mutations above the 95th percentile of the permutation distribution. Shared mutations were counted for genomic and exonic RNA-seq data. In addition, a panel of 254 breast cancer and DNA repair-specific mutation spots proposed by Begg et al. was analysed [38]. The overlap of RNA-seq counts of the genomic and exonic data with the 254-gene panel was used to count the shared mutations of the true and artificial pairs of the cohort. Clonality was defined as the number of shared mutations above the 95th percentile. To test for clonality using profiles of somatic mutations in the “Clonality” package [28], loci-specific probabilities of observing a mutation were obtained from the TCGA breast cancer dataset [39]. Furthermore, fusion transcripts of all tumours were compared and transcripts with identical 5′ and 3′ fusion partner breakpoints were counted.
Cohen’s kappa
Cohen’s kappa measures the chance-corrected agreement for two observations [40]. Cohen’s kappa indices of agreement between different methods applied to estimate clonality were calculated using the R-package “rel” [41].
Results
Tumour synchronicity strongly associated with metastatic spread to the axillary lymph nodes
The 37 breast cancer patients were stratified into four clinical groups based on tumour laterality and the time interval between the diagnoses of the first and second tumours (BM: bilateral-metachronous; BS: bilateral-synchronous; IM: ipsilateral-metachronous; IS: ipsilateral-synchronous). The clinicopathological characteristics are shown in Additional file 2: Table S1. Metastatic spread to the axillary lymph nodes was more prevalent in the synchronous groups (BS: 100%; IS: 85.7%) as compared to the metachronous groups (BM: 61.5%; IM: 14.3%; P = 0.001).
Discordances in histopathological characteristics in 32% of the tumour pairs
For the clinical classification of clonality, several histopathological and molecular features were taken into consideration, including histological subtype, the status of ER and HER2, and the molecular subtype (Table 1). While the receptor status was available for most samples, the molecular subtype was only defined for about 40% of the tumours. Thirty-two percent of the patients (12/37) showed discordances between the first and the second tumour, with one-fourth of the 12 patients showing two discordant changes. Most changes were found in the histological subtypes (35%; 6/17 patients), while the molecular subtype differed in 25% (2/8 patients), ER status in 11% (4/35 patients), and HER2 status in 8% (3/37 patients). In patients with metachronous cancer, changes in receptor status from positive to negative were observed for patients BM6 and BM7. The discordant changes were equally distributed between the different clinical groups and showed no significance when stratified by group.
Table 1.
Overview of the clinical and histological characteristics of the primary and secondary tumours
| Primary tumour | Secondary tumour | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Laterality | Synchronicity | Group | Time interval (days) | Histology | ER | HER2 | Molecular subtype | Histology | ER | HER2 | Molecular subtype | Discordance | Clinical classification |
| BM1 | bilateral | metachronous | BM | 346 | Invasive carcinoma NST | pos | neg | ND | ND | pos | neg | ND | concordant | |
| BM2 | bilateral | metachronous | BM | 1694 | Invasive carcinoma NST | pos | neg | Luminal B | Invasive lobular carcinoma | pos | neg | ND | Histology | discordant |
| BM3 | bilateral | metachronous | BM | 1652 | ND | pos | neg | ND | Invasive carcinoma NST | pos | neg | ND | concordant | |
| BM4 | bilateral | metachronous | BM | 581 | Invasive carcinoma NST | neg | neg | Basal-like | Invasive carcinoma NST | neg | neg | ND | concordant | |
| BM5 | bilateral | metachronous | BM | 1954 | Invasive carcinoma NST | pos | neg | ND | Invasive lobular carcinoma | pos | neg | ND | Histology | discordant |
| BM6 | bilateral | metachronous | BM | 1417 | Invasive lobular carcinoma | pos | neg | ND | Invasive carcinoma NST | neg | neg | HER2/ER- | Histology; ER | discordant |
| BM7 | bilateral | metachronous | BM | 456 | Invasive carcinoma NST | pos | pos | ND | Invasive carcinoma NST | neg | pos | ND | ER | discordant |
| BM8 | bilateral | metachronous | BM | 1152 | Invasive lobular carcinoma | pos | neg | ND | Invasive lobular carcinoma | pos | neg | ND | concordant | |
| BM9 | bilateral | metachronous | BM | 972 | Invasive carcinoma NST | neg | neg | ND | Invasive carcinoma NST | neg | neg | ND | concordant | |
| BS1 | bilateral | synchronous | BS | 0 | Invasive carcinoma NST | pos | neg | ND | Invasive carcinoma NST | pos | neg | Luminal B | concordant | |
| BS2 | bilateral | synchronous | BS | 0 | Invasive lobular carcinoma | pos | neg | ND | Invasive carcinoma NST | pos | neg | Luminal B | Histology | discordant |
| BS3 | bilateral | synchronous | BS | 14 | Invasive carcinoma NST | pos | neg | ND | Invasive carcinoma NST | pos | neg | ND | concordant | |
| BS4 | bilateral | synchronous | BS | 0 | ND | pos | neg | ND | ND | pos | neg | ND | concordant | |
| BS5 | bilateral | synchronous | BS | 6 | Invasive carcinoma NST | pos | neg | ND | Invasive carcinoma NST | ND | neg | ND | concordant | |
| BS6 | bilateral | synchronous | BS | 0 | ND | neg | neg | ND | ND | neg | neg | ND | concordant | |
| BS7 | bilateral | synchronous | BS | 0 | Invasive carcinoma NOS | pos | neg | Luminal B | Invasive lobular carcinoma | neg | neg | ND | Histology; ER | discordant |
| BS8 | bilateral | synchronous | BS | 0 | Invasive carcinoma NST | pos | neg | ND | Invasive carcinoma NST | pos | neg | ND | concordant | |
| IM1 | ipsilateral | metachronous | IM | 1855 | ND | pos | neg | ND | ND | pos | neg | Luminal B | concordant | |
| IM2 | ipsilateral | metachronous | IM | 448 | ND | neg | neg | ND | ND | neg | neg | ND | concordant | |
| IM3 | ipsilateral | metachronous | IM | 1944 | Invasive carcinoma NST | pos | pos | Luminal B | ND | pos | pos | Luminal B | concordant | |
| IM4 | ipsilateral | metachronous | IM | 567 | Invasive carcinoma NST | pos | neg | Luminal B | ND | pos | neg | HER2/ER- | Subtype | discordant |
| IM5 | ipsilateral | metachronous | IM | 712 | Invasive carcinoma NST | neg | neg | Basal-like | ND | neg | neg | ND | concordant | |
| IM6 | ipsilateral | metachronous | IM | 664 | ND | pos | neg | ND | ND | pos | neg | ND | concordant | |
| IM7 | ipsilateral | metachronous | IM | 2454 | ND | pos | neg | ND | ND | pos | neg | ND | concordant | |
| IM8 | ipsilateral | metachronous | IM | 563 | Invasive carcinoma NST | pos | neg | ND | Invasive lobular carcinoma | pos | neg | Luminal B | Histology | discordant |
| IM9 | ipsilateral | metachronous | IM | 2142 | Invasive carcinoma NOS | ND | neg | Luminal B | Invasive carcinoma NOS | pos | neg | Luminal B | concordant | |
| IS1 | ipsilateral | synchronous | IS | 0 | Invasive carcinoma NST | neg | neg | Basal-like | Invasive carcinoma NST | neg | pos | Luminal B | HER2; subtype | discordant |
| IS2 | ipsilateral | synchronous | IS | 0 | Invasive carcinoma NST | pos | neg | ND | ND | pos | neg | Luminal B | concordant | |
| IS3 | ipsilateral | synchronous | IS | 0 | ND | neg | neg | Basal-like | ND | neg | pos | Basal-like | HER2 | discordant |
| IS4 | ipsilateral | synchronous | IS | 50 | ND | neg | neg | ND | ND | pos | neg | Luminal B | ER | discordant |
| IS5 | ipsilateral | synchronous | IS | 0 | Invasive carcinoma NST | pos | neg | Luminal B | Invasive carcinoma NST | pos | neg | Luminal B | concordant | |
| IS6 | ipsilateral | synchronous | IS | 0 | Invasive carcinoma NST | pos | neg | ND | ND | pos | neg | ND | concordant | |
| IS7 | ipsilateral | synchronous | IS | 0 | Invasive carcinoma NOS | pos | neg | ND | ND | pos | neg | ND | concordant | |
| IS8 | ipsilateral | synchronous | IS | 0 | Invasive carcinoma NST | pos | neg | Luminal B | ND | pos | neg | ND | concordant | |
| IS9 | ipsilateral | synchronous | IS | 0 | ND | neg | pos | Basal-like | ND | neg | neg | ND | HER2 | discordant |
| IS10 | ipsilateral | synchronous | IS | 0 | Invasive carcinoma NST | pos | neg | Luminal B | ND | pos | neg | Luminal B | concordant | |
| IS11 | ipsilateral | synchronous | IS | 0 | Invasive carcinoma NOS | neg | pos | HER2/ER- | ND | neg | pos | HER2/ER- | concordant | |
ER oestrogen receptor status, HER2 human epidermal growth factor receptor 2 status, ND not determined, NOS not otherwise specified, NST no special type
Stratification by laterality revealed differential copy number imbalances
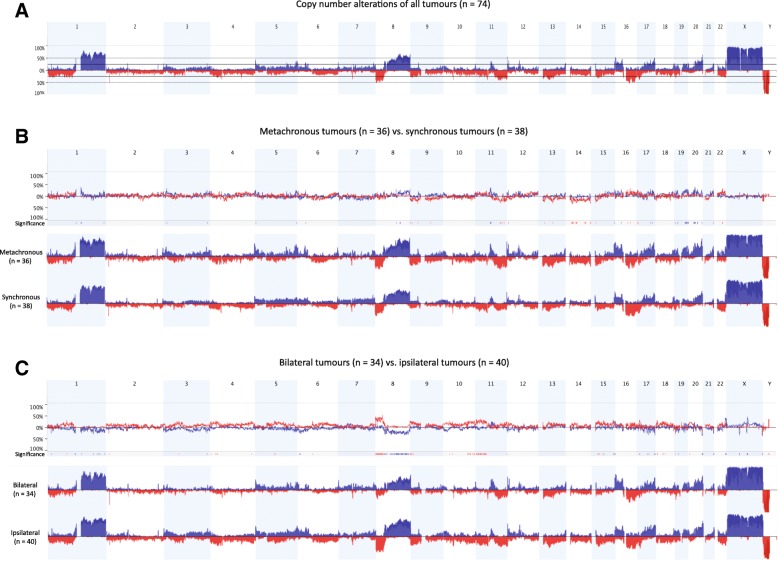
DNA copy number analysis using aCGH was performed to identify recurrent regions of DNA copy number gain (blue) and loss (red) in at least 25% of the tumours in the patient cohort. Recurrent DNA gains were identified on chromosomes 1q, 8q, 16p, 17q, and 20q, while DNA loss was detected on 1p, 8p, 11q, 13q, and 16q (Fig. 1a). These results were in line with DNA gains and losses frequently identified in breast cancer [42–44]. There was very little difference in the DNA copy number profiles when stratified by synchronicity (excluding copy number variations (CNVs) and probes from sex chromosomes) with 59 significantly different genomic regions displaying DNA copy number imbalances (Fig. 1b). Most noticeable were losses of the entire chromosome 14 and the long arm of chromosome 11 in the metachronous subgroup. In contrast, stratification by laterality yielded 134 statistically significant minimal common regions of copy number imbalances, including more fractions of genome altered in the ipsilateral subgroup with prominent losses on 8p and 11p (Fig. 1c).
Fig. 1.
Genome-wide frequency plots of DNA copy number gains (blue) and losses (red) for the entire cohort (a), as well as cohorts stratified by the time interval between the tumours (b; metachronous (n = 36) vs. synchronous (n = 38)) and the laterality (c; bilateral (n = 34) vs. ipsilateral (n = 40))
DNA methylation showed higher variability in synchronous tumours
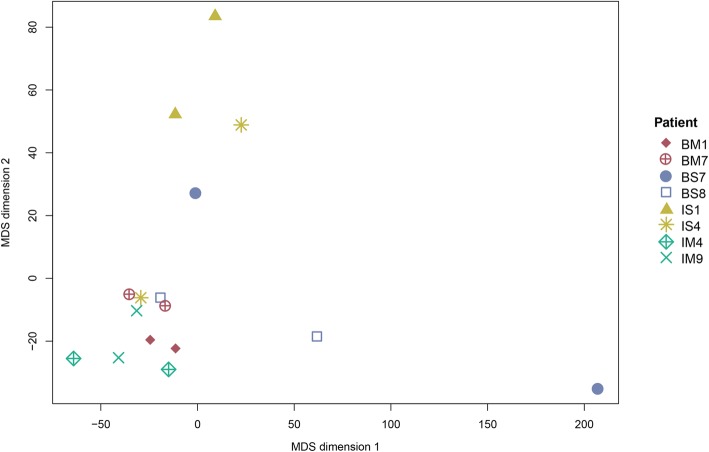
The variability of the beta values was the highest in the bilateral and synchronous groups and consequently in the BS group, which was in line with patients BS7 and BS8 having the highest variability in methylation patterns between the two respective tumour pairs (Additional file 3: Table S2). Principal component analysis of the methylation data showed a statistically significant association with synchronicity (P = 0.007), while no further associations to other variables were found. Kruskal’s non-metric multidimensional scaling (MDS) demonstrated that most of the synchronous samples were further away from each other, while the metachronous samples formed a distinct cluster, suggesting a higher variability of beta values in synchronous samples (Fig. 2).
Fig. 2.
Kruskal’s non-metric multidimensional scaling (MDS) plot of beta values from the DNA methylation cohort (n = 16). The MDS plot visualised similarities between the individual samples based on information from the distance matrix
Strong consensus in clonality could be found for the tumours of patients BM7, BS8, and IS1, while the tumour pairs for patients BS7 and IS4 were determined to be independent primary tumours (Table 2). In general, DNA methylation intensity values were a more liberal method for clonality classification, in particular the clustering analysis, and frequently classified tumour pairs as similar in comparison with other types of molecular data.
Table 2.
Summary of clonality tests for the methylation cohort (n = 8)
| Patients | BM1 | BM7 | BS7 | BS8 | IM4 | IM9 | IS1 | IS4 | |
|---|---|---|---|---|---|---|---|---|---|
| Laterality | Bilateral | Bilateral | Bilateral | Bilateral | Ipsilateral | Ipsilateral | Ipsilateral | Ipsilateral | |
| Synchronicity | Metachronous | Metachronous | Synchronous | Synchronous | Metachronous | Metachronous | Synchronous | Synchronous | |
| Group | BM | BM | BS | BS | IM | IM | IS | IS | |
| aCGH data | Similarity Index | ||||||||
| SI | 0.261 | 0.580 | 0.323 | 0.571 | 0.402 | 0.237 | 0.217 | 0.200 | |
| PSI | 25.600 | 66.450 | 39.810 | 65.930 | 51.650 | 17.890 | 10.510 | 2.710 | |
| P | 0.183 | 0.005 | 0.087 | 0.005 | 0.032 | 0.269 | 0.359 | 0.434 | |
| Clustering (Euclidean distance, single linkage) | |||||||||
| Clustering | different | similar | different | similar | similar | different | different | different | |
| Distance measure | |||||||||
| Euclidean | 46.801 | 19.823 | 59.036 | 16.362 | 43.821 | 60.220 | 63.932 | 66.099 | |
| 5th percentile | not sign. | significant | not sign. | significant | not sign. | not sign. | not sign. | not sign. | |
| Shared segments | |||||||||
| Segments | 34 | 37 | 10 | 32 | 24 | 3 | 53 | 38 | |
| 95th percentile | significant | significant | not sign. | significant | significant | not sign. | significant | significant | |
| Clonality package | |||||||||
| LR2 | 0.044 | 135.409 | 0.011 | 34,945,440 | 0.008 | 0.006 | 0.000 | 0.001 | |
| P | 0.262 | 0.009 | 0.455 | 0.000 | 0.519 | 0.552 | 0.912 | 0.879 | |
| Methylation data Beta values |
Similarity Index for methylation | ||||||||
| SImet | 0.879 | 0.880 | 0.685 | 0.815 | 0.868 | 0.871 | 0.911 | 0.833 | |
| PSI | 8.510 | 8.610 | 0.000 | 1.240 | 7.350 | 7.660 | 11.710 | 3.410 | |
| P | 0.018 | 0.018 | 0.947 | 0.526 | 0.018 | 0.018 | 0.018 | 0.333 | |
| Clustering (Euclidean distance, single linkage) | |||||||||
| Clustering | similar | different | different | different | similar | different | similar | different | |
| Distance measure | |||||||||
| Euclidean | 87.969 | 86.972 | 228.809 | 125.860 | 92.508 | 90.574 | 71.300 | 134.247 | |
| 5th percentile | significant | significant | not sign. | not sign. | not sign. | not sign. | significant | not sign. | |
| Methylation data Intensity values |
Similarity Index | ||||||||
| SI | 0.578 | 0.573 | 0.439 | 0.594 | 0.517 | 0.565 | 0.648 | 0.467 | |
| PSI | 20.360 | 19.770 | 0.000 | 22.520 | 11.010 | 18.650 | 29.070 | 1.410 | |
| P | 0.018 | 0.018 | 0.737 | 0.018 | 0.088 | 0.018 | 0.018 | 0.456 | |
| Clustering (Euclidean distance, single linkage) | |||||||||
| Clustering | similar | similar | different | similar | similar | similar | similar | similar | |
| Distance measure | |||||||||
| Euclidean | 148.829 | 130.060 | 194.202 | 158.604 | 160.988 | 147.229 | 124.268 | 171.320 | |
| 5th percentile | significant | significant | not sign. | significant | significant | significant | significant | not sign. | |
| Shared segments | |||||||||
| Segments | 64 | 14 | 9 | 18 | 6 | 11 | 47 | 7 | |
| 95th percentile | significant | significant | not sign. | significant | not sign. | not sign. | significant | not sign. | |
| Clonality package | |||||||||
| LR2 | 4.53 | 28.46 | 0.00 | 241,461,300 | 300,802.20 | 1.63 | 34,907,040,000,000 | 0.15 | |
| P | 0.080 | 0.036 | 0.795 | 0.000 | 0.009 | 0.107 | 0.000 | 0.241 | |
| Consensus in clonality: | 7/13 | 12/13 | 0/13 | 10/13 | 8/13 | 4/13 | 9/13 | 2/13 | |
PSI percentage of similarities between two tumours that are not due to recurrent chromosomal aberrations or randomness, LR2 final likelihood ratio with individual comparisons. Statistically significant variables (P < 0.05) are displayed in bold text
Ipsilateral synchronous tumours showed similar gene expression by microarray
The gene expression cohort consisted of seven patients with ipsilateral tumours (three metachronous and four synchronous). The clonality analyses based on gene expression microarray data showed strong concordance to the clinical groups with all four synchronous cases being similar for all analyses while 2/3 metachronous cases were classified as different entities (Additional file 4: Table S3). All analyses of the gene expression cohort were in line with the aCGH results except for patient IM4, whom was classified as independent in the gene expression analysis and equivocal in the aCGH data set. MDS demonstrated similar gene expression patterns between the tumour pairs of the patients IM3, IS3, and IS10 (Additional file 5: Figure S1A).
Varying tendencies for clonality within RNA-seq and SNP data
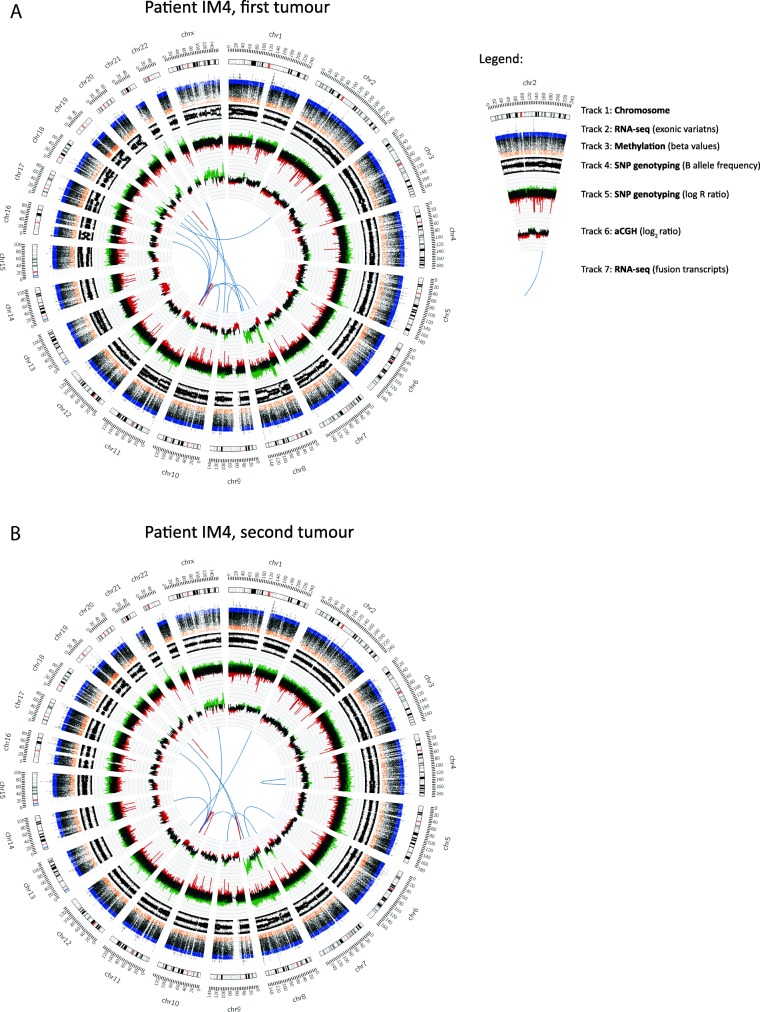
RNA-seq and SNP genotyping were performed for both tumours of patients IM4, IS10, and IS11. A total of 64 fusion transcripts were detected in the two tumours of patient IM4, with five fusion transcripts (7.8%) containing the same fusion breakpoints in the 5′- and 3′-gene partners in both tumours (Additional file 6: Table S4). For patients IS10 and IS11, 1/836 (0.1%) and 5/153 (3.3%) fusion transcripts were identical between the two tumours, respectively. No other shared fusion transcripts were found between different tumours. The RNA-seq data was then evaluated to identify shared genetic variants in genomic and exonic (coding) regions. Shared genomic variants (genome-wide and the 254-gene panel) showed similar tendencies that were in line with the aCGH distance measure and SNP shared segment (LRR) data (Additional file 7: Table S5). The shared exonic variants in the 254-gene panel only found two shared mutations in patient IM4, which contradicted most other RNA-seq results. The shared segment and clustering analyses of the SNP array data classified patient IS10 as clonal, which was in line with the aCGH results but contradicted the distance measure and MDS, which classified the LRRs of the tumour pair IM4 as most similar (Additional file 5: Figure S1B). The “Clonality” package applied on the exonic variants classified all tumour pairs as clonal. A circos plot summarising the results of patient IM4 visualised the similarities in copy number profiles of both aCGH-derived log2 ratio and SNP array-derived LRR and fusion transcripts (Fig. 3).
Fig. 3.
Circos plots depicting aCGH-derived DNA copy number profiles, genome-wide SNP genotyping, DNA methylation beta values, and RNA-seq data in the first (a) and second (b) tumour of breast carcinoma patient IM4. Circos plot Track 1: Chromosome cytobands from pter to qter. The centromere is shown as a red bar. Track 2: Mutations in exonic regions (exonic variants) identified with RNA-seq data are shown as dark grey bars. Track 3: Beta values of DNA methylation data. Track 4: B allele frequency of SNP genotyping data. Track 5: Log R ratio of SNP genotyping data, where copy number gains and losses are depicted in green and red, respectively. Track 6: Log2 ratio of aCGH data, where copy number gains and losses are depicted in green and red, respectively. Track 7: Gene fusions identified with RNA-seq data. Intrachromosomal and interchromosomal gene fusions are shown in red and blue lines, respectively
Tumour clonality defined in 46% of the patients
Calculation of Cohen’s kappa indices was applied to detect the highest agreement between the different statistical methods used to estimate clonality. For the aCGH data, hierarchical clustering and the similarity index (SI) were identified as the most appropriate (0.659 and 0.630, respectively). Since the SI is easier to interpret as a measure and independent of the cohort, it presented the most reasonable definition of clonality and determined 46% (17/37) of the tumour pairs as clonal (Fig. 4). No statistical significance was found to associate the tumour clonality defined by the SI with the clinical classification (Wilcoxon rank sum test: PLaterality = 0.247; PSynchronicity = 0.095; Analysis of variance (ANOVA): PClinical groups = 0.229), highlighting the alarming reality that there is very little connection between current clinical guidelines and the biology underlying tumour clonality.
Fig. 4.
Overview of the different statistical methods applied sorted by the type of data. Red boxes indicate that the analysis defined the tumour pair as clonal and blue boxes indicate independence of the tumours. BAF B allele frequency, BM bilateral-metachronous, BS bilateral-synchronous, IM ipsilateral-metachronous, IS ipsilateral-synchronous, LRR log R ratio, SI similarity index, SImet modified SI for methylation data
The majority of the analyses conducted were in agreement with the SI except for patients BM1, IM4, IM7, IS1, and IS7 (Fig. 4). Interestingly, the histopathological concordances often showed opposite tendencies compared to the aCGH analysis. The different methods applied to the DNA methylation, gene expression and SNP array data sets displayed strong homogeneity within their type of data regardless of the method applied. The results for the SI and hierarchical clustering were consistent in most data sets. The distance measure also overlapped with these results but seemed to be a more conservative measure since fewer tumour pairs were classified as clonal. The shared segment analysis with the aCGH data clearly favoured the clonality hypothesis with defining 21/37 tumour pairs as clonal along with 4/8 cases in the methylation intensity data and 1/3 in the LRR data. The shared segment analysis was in most cases consistent between the different types of data.
Discussion
Here, we show that molecular and statistical analyses are powerful tools for classifying clonal recurrences and independent primary tumours. This study provides valuable insight into which molecular technologies were most informative for investigating clonal relatedness in tumour pairs. Although tumour clonality should govern the choice of treatment, bilateral breast tumours are generally treated as different primary tumours and not as potential failure of the previous treatment. Tumour characteristics such as histological subtype, molecular subtype, presence of ductal carcinoma in situ (DCIS), and receptor status are currently used to choose treatment strategies for patients with multiple breast tumours. However, to fully comprehend the association between multiple tumours, routine clinical and diagnostic testing needs to be conducted in conjunction with molecular and bioinformatics methods.
In the majority of the analyses, the type of molecular data analysed had a stronger impact on clonality determination than the analytical method used. This raises the question of which biological phenomenon provides the most stable evidence for clonality. DNA methylation and gene expression are more dynamic than DNA mutations and CNAs, and might therefore be more similar due to environmental factors. CNAs are acquired at early stages of tumourigenesis [45, 46] making them the most stable type of biological data in this study. An overlap of tendencies in clonality between the aCGH and DNA methylation data was seen for only 50% of the cohort (BM7, BS7, BS8, and IS4), giving a less optimistic view on using DNA methylation as a clonality tool compared to results from other reported studies [47, 48]. In the DNA methylation data, synchronicity accounted for more variation than metachronicity, providing further evidence that synchronous tumours are more different from each other with regard to DNA methylation patterns. However, the small cohort size limited the conclusions that can be drawn. The overlap of results between gene expression and copy number data was surprisingly high since gene expression is more unstable than DNA alterations. Gene expression-based analyses defined all IS cases as clonal indicating that gene expression patterns are very similar for tumour cells arising in the same breast at the same time, possibly due to their adjacent microenvironment.
Hierarchical clustering has been used, among other methods, in several studies to define clonality [15, 47, 49]. Clustering is designed as an unsupervised classification tool to discover underlying structures of a data set under the assumption that the number of clusters and their members are unknown. The disadvantage of clustering is that clonality depends on the relationship between individual tumours and the linkage between tumour clusters. Using Euclidean distance with single linkage is the only way to circumvent these disadvantages [37]. The results from the SI and hierarchical clustering analyses exhibited a strong overlap in their classification. Calculation of Cohen’s kappa showed the highest agreement of the different analyses for the SI and the clustering. Thus, the SI represented the most suitable approach in defining clonality since it is a specialised technique specifically developed for this purpose and provides easy interpretation.
In the DNA methylation cohort, clustering of the intensity values classified 7/8 tumour pairs as clonal and therefore did not provide a precise segregation between clonally related tumours and independent tumours. The aCGH, DNA methylation intensity and LRR data should biologically refer to the same phenomenon (CNAs) and consequently show the same tendencies for different genomic loci. Therefore, it was unexpected that the results of the clustering and shared segment analysis for those data sets did not show stronger concordance. Furthermore, it was anticipated that the results from the clustering and the distance measure were more in agreement since the first step of clustering is the Euclidean distance. In most cases, the distance measure seemed to be a stricter method than the SI and clustering.
In comparison with genomic variants, mutation analyses based on exonic variants or gene panels represent a subset of the full picture. The different tendencies between the methods represent a drawback for potential applications of sequencing panels in the clinic. The fusion transcript analysis was the only method that did not show any overlaps between patients. Moreover, unspliced fusion transcripts provide the transcribed level of CNAs, which highlights the functional consequences of CNAs and makes them an important tool to assess tumour clonality. Our RNA-seq-based mutation approach had several limitations starting with the lack of matched normal samples to exclude germline mutations and normal DNA nucleotide variations. However, common genetic variants found in the human population were removed. Furthermore, our approach did not account for the frequency of mutations in breast cancer since rare mutations give much stronger evidence for clonality than common mutations [22]. In the frequency-based approach of the “Clonality” package, a further limitation was that RNA-seq data was compared with whole exome sequencing data from TCGA. In addition, the RNA-seq cohort was too small to perform meaningful statistics regarding the 95th percentile, which is a general limitation of using permutation-based approaches. Therefore, the results from this cohort have to be viewed with caution and in context to the other results. Tumours from patient IS10, for example were clonal regarding all other analyses except the RNA-seq and SNP genotyping array.
Whole genome sequencing (WGS) is the more appropriate method to evaluate mutations in comparison with RNA-seq, which does not give information on untranscribed DNA sequences. Hence, the lack of common mutations cannot be considered as a guarantee that tumour pairs are independent. However, intratumour heterogeneity complicates clonality analyses due to biological differences in different parts of a tumour and subclone evolution. In aCGH, contamination with normal cells could diminish the intensity of detected CNAs and small cell populations might not be detected. However, by using only samples that showed a tumour cell content of at least 70%, we ruled out that a lack of clonal relatedness could be due to a lack of tumour cells.
Few studies based on molecular approaches have been conducted to define clonality in multiple breast tumours and there is no consensus on which type of data and analysis method provides the most stable definition of clonality. A direct comparison of these studies to the findings presented here might, however, not be justified due to differences in the study set-up, methods and statistics. In a study on a contralateral cohort using low-coverage WGS, Alkner et al. demonstrated clonal relatedness in 10% (1/10) of the patients [19], which was lower than the clonal relatedness of bilateral tumours in our study (29%, 5/17 patients). Klevebring et al. found 12% (3/25) of their BM cohort to be clonally related using whole exome sequencing (WES) [18], which was also lower than the clonal relatedness of BM tumours in our study (22%, 2/9 patients). Desmedt et al. studied IS tumours and defined 67% (24/36) of the patients as clonal using a targeted mutation screening and 100% (8/8) of the patients as clonal using low-coverage WGS [50]. Our IS cohort showed clonality in 64% (7/11) of the patients, which is surprisingly closer to the mutational approach than the copy number-based approach. Our report is the first, to our knowledge, to compare different approaches (type of molecular data and statistical method) and clinical groups (BM, BS, IM, and IS) between each other.
Conclusions
There are many studies published on tumour clonality using different types of data and statistical methods. Most studies defined their own methods and cohort-specific cut-offs. Currently, there is no consensus about which type of data and especially which statistical analysis is the most suitable and there are surprisingly few studies that compare and evaluate the feasibility of these different approaches. Nonetheless, extremely similar or different tumour pairs (BM7, BS7, IM3, IS4, and IS5) showed consistent results regardless of the statistical analysis or biological data used, but clinic guidelines need to be defined with exact thresholds in order to standardise clonality testing in a routine diagnostic setting. In metachronous cancer, clonality between the first and second tumour may indicate an insufficient effect of the treatment for the first tumour and the patient could benefit from a change in treatment. An independent new primary tumour would indicate a more favourable prognosis than a recurrence. Hence, the discrimination between a clonal and independent origin of the second tumour is of high importance for the patient. In our study, the distance measure proved to be the most conservative method for defining clonality and the shared segment analysis the most liberal. Gene expression data classified all ipsilateral-synchronous cases as clonal, demonstrating that gene expression strongly depends on the nearby tumour microenvironment. The SI using aCGH data was found to be the most suitable method to classify tumour clonality, as it had the highest concordance with all results and can be easily integrated into clinic routine using FFPE samples to obtain copy number data. But most importantly, the definition of tumour clonality based on the current clinicopathological markers needs to be revised due to the limited intersects between current clinical guidelines and the underlying biology of tumour clonality.
Additional files
Supplementary Methods. Description of nucleic acid isolation and purification, aCGH gene expression microarray, RNA-seq and SNP array analysis. (DOCX 37 kb)
Table S1. Overview of clinical characteristics of the patient and tumour information stratified by the clinical groups (BM, BS, IM, and IS). (XLSX 16 kb)
Table S2. Variabilities of the studied sample groups with the variability spanning between 5th and 95th percentile of the beta values. (XLSX 10 kb)
Table S3. Summary of clonality tests for the gene expression microarray cohort (n = 7). (XLSX 14 kb)
Figure S1. Non-metric multidimensional scaling (MDS) plot of (A) normalised log2 ratios from gene expression data, and (B) LRR values from SNP array data. The MDS plot visualised similarities between the individual samples based on information from the distance matrix. (TIF 1784 kb)
Table S4. Overview of the shared fusion transcripts in patient IM4, IS10, and IS11. (XLSX 13 kb)
Table S5.Summary of clonality tests for the RNA-seq and SNP genotyping cohort (n = 6). (XLSX 14 kb)
Acknowledgements
We are grateful to BILS (Bioinformatics Infrastructure for Life Sciences) and NBIS (National Bioinformatics Infrastructure Sweden) for their bioinformatics support.
Funding
This work was supported by grants from the Stiftelsen Assar Gabrielssons Fond (FB 17–09; JB), the Swedish Cancer Society (CAN 2012/406; CAN 2015/311; K H), the King Gustav V Jubilee Clinic Cancer Research Foundation (2016:65; KH), and the LUA/ALF-agreement in West of Sweden healthcare region (PK).
Availability of data and materials
The aCGH and methylation data sets supporting the conclusions of this article are available in the NCBI Gene Expression Omnibus repository, accessible through GEO Series accession number GSE108985 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108985). The RNA-seq, SNP array and gene expression microarray data are accessible through GSE97293 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97293) and GSE97177 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97177).
Abbreviations
- aCGH
Array comparative genomic hybridization
- ANOVA
Analysis of variance
- BAF
B allele frequency
- BM
bilateral-metachronous
- BS
bilateral-synchronous
- CGH
Comparative genomic hybridization
- CNA
Copy number alteration
- CNV
Copy number variation
- DCIS
Ductal carcinoma in situ
- ER
Oestrogen receptor status
- FFPE
Formalin-fixed paraffin-embedded
- HER2
Human epidermal growth factor receptor 2 status
- IM
Ipsilateral-metachronous
- IS
Ipsilateral-synchronous
- LR2
Likelihood ratio with individual comparisons
- LRR
Log R ratio
- MDS
Kruskal’s non-metric multidimensional scaling
- NOS
Not otherwise specified
- NST
No special type
- RNA-seq
RNA sequencing
- SI
Similarity index
- SImet
Modified SI for methylation data
- SNP
Single nucleotide polymorphism
- WES
Whole exome sequencing
- WGS
Whole genome sequencing
Authors’ contributions
KH and PK were responsible for overall study concept. JB, TZP, and AD were responsible for the design of experiments. PK, AD, JB, AK, and EWR collected the clinical data. TZP, and HE contributed to the bioinformatics analyses. SN contributed to the statistical analyses. EWR and EFA provided technical and material support. AD, TZP and JB performed the experiments. JB analysed the data, performed the statistical analyses, and wrote the manuscript. All authors reviewed, edited, and approved the final manuscript.
KH and PK share last author status.
The study was ethically approved by the Sahlgrenska Academy Medical Faculty Research Committee, Gothenburg, Sweden (S164–02).
Not applicable.
The authors declare that they have no competing interests.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jana Biermann, Phone: (+46)-31-7866751, Email: jana.biermann@gu.se.
Toshima Z. Parris, Email: toshima.parris@oncology.gu.se
Szilárd Nemes, Email: Szilard.Nemes@registercentrum.se.
Anna Danielsson, Email: anna.danielsson@oncology.gu.se.
Hanna Engqvist, Email: hanna.engqvist@gu.se.
Elisabeth Werner Rönnerman, Email: elisabeth.ronnerman@vgregion.se.
Eva Forssell-Aronsson, Email: eva.forssell_aronsson@radfys.gu.se.
Anikó Kovács, Email: aniko.kovacs@vgregion.se.
Per Karlsson, Email: per.karlsson@oncology.gu.se.
Khalil Helou, Email: khalil.helou@oncology.gu.se.
References
- 1.Raymond JS, Hogue CJ. Multiple primary tumours in women following breast cancer, 1973-2000. Br J Cancer. 2006;94(11):1745–1750. doi: 10.1038/sj.bjc.6603172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y, Thompson W, Semenciw R, Mao Y. Epidemiology of contralateral breast cancer. Cancer Epidemiol Biomark Prev. 1999;8(10):855–861. [PubMed] [Google Scholar]
- 3.Vaittinen P, Hemminki K. Risk factors and age-incidence relationships for contralateral breast cancer. Int J Cancer. 2000;88(6):998–1002. doi: 10.1002/1097-0215(20001215)88:6<998::AID-IJC25>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 4.Dawson PJ, Maloney T, Gimotty P, Juneau P, Ownby H, Wolman SR. Bilateral breast cancer: one disease or two? Breast Cancer Res Treat. 1991;19(3):233–244. doi: 10.1007/BF01961160. [DOI] [PubMed] [Google Scholar]
- 5.Begg CB, Eng KH, Hummer AJ. Statistical tests for clonality. Biometrics. 2007;63(2):522–530. doi: 10.1111/j.1541-0420.2006.00681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 7.Lannin DR, Haffty BG. End results of salvage therapy after failure of breast-conservation surgery. Oncology (Williston Park) 2004;18(3):272–279. [PubMed] [Google Scholar]
- 8.Chaudary MA, Millis RR, Hoskins EO, Halder M, Bulbrook RD, Cuzick J, Hayward JL. Bilateral primary breast cancer: a prospective study of disease incidence. Br J Surg. 1984;71(9):711–714. doi: 10.1002/bjs.1800710924. [DOI] [PubMed] [Google Scholar]
- 9.Noguchi S, Motomura K, Inaji H, Imaoka S, Koyama H. Differentiation of primary and secondary breast cancer with clonal analysis. Surgery. 1994;115(4):458–462. [PubMed] [Google Scholar]
- 10.Intra M, Rotmensz N, Viale G, Mariani L, Bonanni B, Mastropasqua MG, Galimberti V, Gennari R, Veronesi P, Colleoni M, et al. Clinicopathologic characteristics of 143 patients with synchronous bilateral invasive breast carcinomas treated in a single institution. Cancer. 2004;101(5):905–912. doi: 10.1002/cncr.20452. [DOI] [PubMed] [Google Scholar]
- 11.Imyanitov EN, Suspitsin EN, Grigoriev MY, Togo AV, Kuligina E, Belogubova EV, Pozharisski KM, Turkevich EA, Rodriquez C, Cornelisse CJ, et al. Concordance of allelic imbalance profiles in synchronous and metachronous bilateral breast carcinomas. Int J Cancer. 2002;100(5):557–564. doi: 10.1002/ijc.10530. [DOI] [PubMed] [Google Scholar]
- 12.Saad RS, Denning KL, Finkelstein SD, Liu Y, Pereira TC, Lin X, Silverman JF. Diagnostic and prognostic utility of molecular markers in synchronous bilateral breast carcinoma. Mod Pathol. 2008;21(10):1200–1207. doi: 10.1038/modpathol.2008.35. [DOI] [PubMed] [Google Scholar]
- 13.Waldman FM, DeVries S, Chew KL, Moore DH, 2nd, Kerlikowske K, Ljung BM. Chromosomal alterations in ductal carcinomas in situ and their in situ recurrences. J Natl Cancer Inst. 2000;92(4):313–320. doi: 10.1093/jnci/92.4.313. [DOI] [PubMed] [Google Scholar]
- 14.Park SC, Hwang UK, Ahn SH, Gong GY, Yoon HS. Genetic changes in bilateral breast cancer by comparative genomic hybridisation. Clin Exp Med. 2007;7(1):1–5. doi: 10.1007/s10238-007-0123-1. [DOI] [PubMed] [Google Scholar]
- 15.Bollet MA, Servant N, Neuvial P, Decraene C, Lebigot I, Meyniel JP, De Rycke Y, Savignoni A, Rigaill G, Hupe P, et al. High-resolution mapping of DNA breakpoints to define true recurrences among ipsilateral breast cancers. J Natl Cancer Inst. 2008;100(1):48–58. doi: 10.1093/jnci/djm266. [DOI] [PubMed] [Google Scholar]
- 16.Brommesson S, Jonsson G, Strand C, Grabau D, Malmstrom P, Ringner M, Ferno M, Hedenfalk I. Tiling array-CGH for the assessment of genomic similarities among synchronous unilateral and bilateral invasive breast cancer tumor pairs. BMC Clin Pathol. 2008;8:6. doi: 10.1186/1472-6890-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castellarin M, Milne K, Zeng T, Tse K, Mayo M, Zhao Y, Webb JR, Watson PH, Nelson BH, Holt RA. Clonal evolution of high-grade serous ovarian carcinoma from primary to recurrent disease. J Pathol. 2013;229(4):515–524. doi: 10.1002/path.4105. [DOI] [PubMed] [Google Scholar]
- 18.Klevebring D, Lindberg J, Rockberg J, Hilliges C, Hall P, Sandberg M, Czene K. Exome sequencing of contralateral breast cancer identifies metastatic disease. Breast Cancer Res Treat. 2015;151(2):319–324. doi: 10.1007/s10549-015-3403-6. [DOI] [PubMed] [Google Scholar]
- 19.Alkner S, Tang MH, Brueffer C, Dahlgren M, Chen Y, Olsson E, Winter C, Baker S, Ehinger A, Ryden L, et al. Contralateral breast cancer can represent a metastatic spread of the first primary tumor: determination of clonal relationship between contralateral breast cancers using next-generation whole genome sequencing. Breast Cancer Res. 2015;17:102. doi: 10.1186/s13058-015-0608-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostrovnaya I, Seshan VE, Begg CB. Comparison of properties of tests for assessing tumor clonality. Biometrics. 2008;64(4):1018–1022. doi: 10.1111/j.1541-0420.2008.00988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nemes S, Danielsson A, Parris TZ, Jonasson JM, Bulow E, Karlsson P, Steineck G, Helou K. A diagnostic algorithm to identify paired tumors with clonal origin. Genes Chromosomes Cancer. 2013;52(11):1007–1016. doi: 10.1002/gcc.22096. [DOI] [PubMed] [Google Scholar]
- 22.Ostrovnaya I, Seshan VE, Begg CB. Using somatic mutation data to test tumors for clonal relatedness. Ann Appl Stat. 2015;9(3):1533–48. [DOI] [PMC free article] [PubMed]
- 23.Hu H, Li J, Plank A, Wang H, Daggard G. Comparative Study of Classification Methods for Microarray Data Analysis. In: Proceedings of the Fifth Australasian Conference on Data Mining and Analystics: 2006; Sydney, Australia edn, vol. 2006: Inc: Australian Computer Society. 2006;61:33–37.
- 24.Parris TZ, Danielsson A, Nemes S, Kovacs A, Delle U, Fallenius G, Mollerstrom E, Karlsson P, Helou K. Clinical implications of gene dosage and gene expression patterns in diploid breast carcinoma. Clin Cancer Res. 2010;16(15):3860–3874. doi: 10.1158/1078-0432.CCR-10-0889. [DOI] [PubMed] [Google Scholar]
- 25.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen international expert consensus on the primary therapy of early breast Cancer 2011. Ann Oncol. 2011;22(8):1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hupe P, Stransky N, Thiery JP, Radvanyi F, Barillot E. Analysis of array CGH data: from signal ratio to gain and loss of DNA regions. Bioinformatics. 2004;20(18):3413–3422. doi: 10.1093/bioinformatics/bth418. [DOI] [PubMed] [Google Scholar]
- 27.R Core Team. R: A Language and Environment for Statistical Computing: R Foundation for Statistical Computing; 2018.
- 28.Ostrovnaya I, Seshan VE, Olshen A, Begg CB. Clonality: an R package for testing clonal relatedness of two tumors from the same patient based on their genomic profiles. Bioinformatics. 2011;27:1698–1699. doi: 10.1093/bioinformatics/btr267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seshan and Olshen . DNAcopy: A Package for Analyzing DNA Copy Data. 2010. [Google Scholar]
- 30.Assenov Y, Muller F, Lutsik P, Walter J, Lengauer T, Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat Meth. 2014;11(11):1138–1140. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feber A, Guilhamon P, Lechner M, Fenton T, Wilson GA, Thirlwell C, Morris TJ, Flanagan AM, Teschendorff AE, Kelly JD, et al. Using high-density DNA methylation arrays to profile copy number alterations. Genome Biol. 2014;15(2):R30. doi: 10.1186/gb-2014-15-2-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, Beck S. ChAMP: 450k Chip analysis methylation pipeline. Bioinformatics. 2014;30(3):428–430. doi: 10.1093/bioinformatics/btt684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hovestadt V, Zapatka M. R package version 1.9.0 edn. 2017. conumee: Enhanced copy-number variation analysis using Illumina DNA methylation arrays. [Google Scholar]
- 35.Lampa S, Dahlo M, Olason PI, Hagberg J, Spjuth O. Lessons learned from implementing a national infrastructure in Sweden for storage and analysis of next-generation sequencing data. Gigascience. 2013;2(1):9. doi: 10.1186/2047-217X-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du P, Zhang X, Huang C-C, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ostrovnaya I, Begg CB. Testing clonal relatedness of tumors using Array comparative genomic hybridization: a statistical challenge. Clin Cancer Res. 2010;16(5):1358. doi: 10.1158/1078-0432.CCR-09-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Begg CB, Ostrovnaya I, Geyer FC, Papanastasiou AD, Ng CKY, Sakr RA, Bernstein JL, Burke KA, King TA, Piscuoglio S, et al. Contralateral breast cancers: independent cancers or metastases? Int J Cancer. 2018;142(2):347–56. 10.1002/ijc.31051. [DOI] [PMC free article] [PubMed]
- 39.TCGA. The Cancer Genome Atlas (TCGA). https://cancergenome.nih.gov/.
- 40.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20(1):37–46. doi: 10.1177/001316446002000104. [DOI] [Google Scholar]
- 41.Lo Martire R. rel: Reliability Coefficients. version 1.3.1 2017. [Google Scholar]
- 42.Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, Esposito D, Alexander J, Troge J, Grubor V, et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006;16(12):1465–1479. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fridlyand J, Snijders AM, Ylstra B, Li H, Olshen A, Segraves R, Dairkee S, Tokuyasu T, Ljung BM, Jain AN, et al. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6(1):96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haverty PM, Fridlyand J, Li L, Getz G, Beroukhim R, Lohr S, Wu TD, Cavet G, Zhang Z, Chant J. High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer. 2008;47(6):530–542. doi: 10.1002/gcc.20558. [DOI] [PubMed] [Google Scholar]
- 45.Gao R, Davis A, McDonald TO. Sei E, Shi X, Wang Y, Tsai PC, Casasent A, Waters J, Zhang H, et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat Genet. 2016;48(10):1119–1130. doi: 10.1038/ng.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, Liang H, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512(7513):155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moarii M, Pinheiro A, Sigal-Zafrani B, Fourquet A, Caly M, Servant N, Stoven V, Vert JP, Reyal F. Epigenomic alterations in breast carcinoma from primary tumor to locoregional recurrences. PLoS One. 2014;9(8):e103986. doi: 10.1371/journal.pone.0103986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang KT, Mikeska T, Li J, Takano EA, Millar EK, Graham PH, Boyle SE, Campbell IG, Speed TP, Dobrovic A, et al. Assessment of DNA methylation profiling and copy number variation as indications of clonal relationship in ipsilateral and contralateral breast cancers to distinguish recurrent breast cancer from a second primary tumour. BMC Cancer. 2015;15:669. doi: 10.1186/s12885-015-1676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song F, Li X, Song F, Zhao Y, Li H, Zheng H, Gao Z, Wang J, Zhang W, Chen K. Comparative genomic analysis reveals bilateral breast cancers are genetically independent. Oncotarget. 2015;6(31):31820–31829. doi: 10.18632/oncotarget.5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desmedt C, Fumagalli D, Pietri E, Zoppoli G, Brown D, Nik-Zainal S, Gundem G, Rothe F, Majjaj S, Garuti A, et al. Uncovering the genomic heterogeneity of multifocal breast cancer. J Pathol. 2015;236(4):457–466. doi: 10.1002/path.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods. Description of nucleic acid isolation and purification, aCGH gene expression microarray, RNA-seq and SNP array analysis. (DOCX 37 kb)
Table S1. Overview of clinical characteristics of the patient and tumour information stratified by the clinical groups (BM, BS, IM, and IS). (XLSX 16 kb)
Table S2. Variabilities of the studied sample groups with the variability spanning between 5th and 95th percentile of the beta values. (XLSX 10 kb)
Table S3. Summary of clonality tests for the gene expression microarray cohort (n = 7). (XLSX 14 kb)
Figure S1. Non-metric multidimensional scaling (MDS) plot of (A) normalised log2 ratios from gene expression data, and (B) LRR values from SNP array data. The MDS plot visualised similarities between the individual samples based on information from the distance matrix. (TIF 1784 kb)
Table S4. Overview of the shared fusion transcripts in patient IM4, IS10, and IS11. (XLSX 13 kb)
Table S5.Summary of clonality tests for the RNA-seq and SNP genotyping cohort (n = 6). (XLSX 14 kb)
Data Availability Statement
The aCGH and methylation data sets supporting the conclusions of this article are available in the NCBI Gene Expression Omnibus repository, accessible through GEO Series accession number GSE108985 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108985). The RNA-seq, SNP array and gene expression microarray data are accessible through GSE97293 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97293) and GSE97177 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97177).