Abstract
The functional role and specificity of tumor infiltrating lymphocytes (TIL) is generally not well characterized. Prominent lymphocyte infiltration is the hallmark of the most common form of hereditary colon cancer, hereditary nonpolyposis colon cancer (HNPCC) and the corresponding spontaneous colon cancers with the microsatellite instability (MSI) phenotype. These cancers are caused by inherited or acquired defects in the DNA mismatch–repair machinery. The molecular mechanism behind the MSI phenotype provides a clue to understanding the lymphocyte reaction by allowing reliable prediction of potential T cell epitopes created by frameshift mutations in candidate genes carrying nucleotide repeat sequences, such as TGFβRII and BAX. These tumors therefore represent an interesting human system for studying TIL and characterizing tumor-specific T cells. We here describe T cell reactivity against several T helper cell epitopes, representing a common frameshift mutation in TGFβRII, in TIL and peripheral blood lymphocytes from patients with MSI+ tumors. The peptide SLVRLSSCVPVALMSAMTTSSSQ was recognized by T cells from two of three patients with spontaneous MSI+ colon cancers and from all three patients with HNPCC. Because such mutations are present in 90% of cancers within this patient group, these newly characterized epitopes provide attractive targets for cancer vaccines, including a prophylactic vaccine for individuals carrying a genetic disposition for developing HNPCC.
Carcinoma of the colorectum is the second most common malignancy in both genders in the developed countries (1). Approximately 15% of colorectal carcinomas show microsatellite instability (MSI), which reflects an increased rate of mutations, primarily in short DNA repeats (2, 3). These cancers are mainly localized in the proximal part of the colon. MSI is caused by defects in the DNA mismatch–repair (MMR) machinery. This machinery corrects mismatched nucleotides and repairs incision/deletion mutations in DNA sequences. When such mutations occur in coding regions of proteins, the resulting shift in reading frame of the gene (frameshift) gives rise to the synthesis of truncated proteins that have lost their function, accelerating tumor progression (4). Germline mutations in the MMR genes have been identified in Lynch syndrome, also called hereditary nonpolyposis colon cancer (HNPCC), and patients with this syndrome develop tumors with MSI at an early age.
In cancers with the MSI phenotype, the important targets for mutation are microsatellite-like sequences in coding regions of various cancer-related genes including BAX and transforming growth factor β receptor type II (TGFβRII). TGFβRII is involved in growth inhibition of many cells, including epithelial cells. The gene encoding TGFβRII is mutated in 90% of colorectal cancers with MSI. Mutations occur as 1- or 2-base deletions or insertions in a poly(A)10 tract of the gene (5, 6). BAX encodes a protein that has a central role in the induction of apoptosis and harbors a poly(G)8 tract that is altered in more than 50% of MSI+ tumors (7). Such frameshift mutations result in previously undescribed amino acid sequences in the C-terminal part of the proteins, and early termination where a stop codon appears, as illustrated in TGFβRII in Fig. 1. These frameshift sequences only occur in tumor cells and their premalignant progenitors, and may accordingly constitute a previously unknown group of potential tumor specific antigens. Recently, several frameshift-derived HLA-A2-restricted CTL epitopes have been described (8), including a peptide derived from mutant TGFβRII also identified by us (24).
Figure 1.
Frameshift mutations in TGFβRII. The human gene coding for TGFβRII contains a poly(A) sequence (A10) from base no. 709 to base no. 718. Frameshift mutations in this A10 repeat have been observed, both as A9 (−1A) and A11 (+1A). These mutations result in mutant peptide sequences with the first amino acid of the altered sequence in either position 128 or 129 as compared with the normal TGFβRII protein. We have synthesized several of these mutant peptides, among them peptide 538. From the SYFPEITHI–MHC Database (http.//www.medizin.uni-tuebingen.de/sfb510/index.html) we have predicted possible peptide motifs from peptide 538 for three HLA class II molecules.
MSI+ colorectal cancers are characterized by a strong “Crohn type” lymphocyte infiltration. The degree of lymphocyte infiltration in the tumor has been found to correlate with a better prognosis in colorectal cancers (9), but so far no information is available on the function and specificity of these cells.
The purpose of the present study was to identify T cell epitopes encoded by common frameshift mutations and to test the hypothesis that one component of the TILs would be directed against frameshift peptides. To this end, we identified frameshift peptides derived from TGFβRII and BAX that carry several HLA class-II-binding motifs, and used these to stimulate T cells from patients and normal donors.
Materials and Methods
Patients.
The female patient, IMT, had an adenocarcinoma localized to the proximal colon. The HLA type of IMT was HLA-A1, 2; B7, 8; DR3, 4 (DRB1*0301, 1401); DQ1, 2. Donor 6947 was a normal healthy donor with the HLA type HLA-A2, 3; B7, 60; DR1, 2; DQ1. The HLA-DP molecule was genomically typed to be DPB1*0301. Six patients, EA, GAA, TT, LRT, EMH, and SJ, had adenocarcinomas localized to the proximal colon and were selected based on an abundant infiltrate of T cells in their tumors. The HNPCC patients PEW, RM, and NH, had previously undergone operation for colon cancer. All samples were obtained after informed consent by the patients.
Cells and Media.
Peripheral blood mononuclear cells (PBMCs) were prepared by centrifugation over Lymphoprep (Nycomed, Oslo). B-lymphoblastoid cell lines (B-LCLs) used as antigen-presenting cells were from the 10th and 11th International Histocompatibility Workshop cell panels. Autologous B-LCLs were generated by Epstein–Barr virus transformation of B cells from the donors. Where nothing else is indicated, all cells were cultured in RPMI medium1640 (BioWhittaker) supplemented with gentamicin, 15% heat-inactivated human serum (T cells), or 10% heat-inactivated FCS (Life Technologies) (cell lines). Conditioned medium (CM) was produced as described by Romani et al. (10).
Peptides.
Peptides were purchased from Neosystem S.A (Strasbourg, France) or provided by Norsk Hydro Research Center. The peptides corresponded to amino acid sequences resulting from frameshifts in TGFβRII and BAX and were synthesized and purified as described (11). The amino acid sequences were as follows: Mutated TGFβRII peptides: p523, SLVRLSSCV; p573, RLSSCVPVA; p577, SSCVPVALM; p578, LSSCVPVAL; p579, VPVALMSAM; p537, AMTTSSSQKNITPAILTCC; p538, SLVRLSSCVPVALMSAMTTSSSQ; p539, ALMSAMTTSSSQKNITPAILTCC; p540, SPKCIMKEKKSLRLSSCVPVA; p541, PKCIMKEKKKSLVRLSSCV; p542, SPKCIMKEKKAW; p543, PKCIMKEKKKAW; p621, KSLVRLSSCVPVALMSAMT. Mutated Bax peptides: p517, RHPSWPWTRCLRMRPPRS; p518, IQDRAGRMGGRHPSWPWTRCLR; p519, GGTRAGPGPGASGCVHQEAERV; p520, ASGCVHQEAERVSQAHRGRTGQ; p521, IQDRAGRMGGGGTRAGPGPGAS.
Antibodies.
The following antibodies were used in blocking experiments: B7/21, anti-HLA-DP (a gift of F. Bach, University of Minneapolis, Minneapolis); SPV-L3, anti-HLA-DQ (a gift of H. Spits, The Netherlands Cancer Institute, Amsterdam) B8/11, anti-HLA-DR (a gift of B. Malissen, Centre d'Immunologie INSERM-Centre National de la Recherche Scientifique de Marseille-Luminy, Marseille, France).
Peptide-Specific T Cell Stimulation.
PBMCs from the normal donor 6947 were stimulated with frameshift peptides by using a protocol involving dendritic cells (DCs): DCs were generated essentially as described (12, 13). After 2 weeks of culture, DCs were loaded with combinations of two peptides at 25 μM of each peptide (p537/539, p538 + p540, p541 + p542, and p543) together with IL-4 and granulocyte–macrophage colony-stimulating factor (GM-CSF) and incubated overnight. The DC were matured with a combination of 10 ng/ml tumor necrosis factor (TNF)-α (BioSource International, Camarillo, CA) and 25% CM for 24 h. Responder T cells were prepared by using thawed, nonadherent autologous cells. Peptide-loaded DCs and 3 × 106 T cells were cocultured in a 24-well culture plate in Iscove's modified Dulbecco's medium (IMDM) containing 15% human serum, 25% CM, and 10 ng/ml TNF-α. After 10 days the bulk cultures were restimulated by using irradiated (30 Gy), autologous peptide pulsed PBMCs (25 μM of each peptide for 2 h at 37°C). TNF-α (10 ng/ml) was added together with the responding cells. IL-2 (10 units/ml; Amersham Pharmacia) was added after 2 days and the bulk cultures were tested for peptide specificity at day 10. Activated T cells from one responding bulk culture stimulated with p538 + p540 were isolated with anti-CD25 coated Dynabeads (Dynal, Oslo) and cloned by limiting dilution (5/1/0.3 cells per well) as described (14). Clones were obtained from 24 of 600 wells seeded. Twenty-two clones were expanded and tested for peptide specificity.
PBMCs from patients EA, GAA, TT, LRT, EMH, and SJ and from HNPCC patients PEW, RM, and NH were plated as 2–4 × 106 cells per well in 24-well plates and stimulated with 10–25 μM of each of the mutated TGFβRII peptides p537–p543, p621 (HNPCC patients) and the mutated Bax peptides p517-p521. IL-2 (10 units/ml) was added the next day. After 12–13 days, cultures were restimulated by using irradiated (30 Gy) peptide-pulsed (10–15 μM of each peptide) autologous PBMCs as feeder cells. IL-2 (10 units/ml) was added at day 1. The bulk cultures were tested for peptide specificity after 10 days.
Expansion and Testing of Tumor-Infiltrating Lymphocytes.
A tumor biopsy taken from patient IMT was finely cut with a scalpel and cultured in RPMI medium 1640 supplemented with gentamicin, 15% heat-inactivated human pool serum and 100 units/ml IL-2. The TILs that grew out of the biopsy were cultured with IL-2 for 17 days and then frozen. For generation of TIL clones, TILs were thawed, rested for 1 day in 15% human serum/RPMI medium 1640 and cloned by limiting dilution (3 cells/well) as described (14). Autologous PBMCs used as APC were pulsed for 3 h at 37°C with a mixture of 1 μM of each of the peptides p523, p573, p577, p578, p579, p538, p540, and p541. To enhance binding and induce DC maturation, 3 μg/ml of β2-microglobulin (β2m) (Sigma) and 10 ng/ml of TNF-α was also present. Clones were obtained from 14 of 1,200 wells seeded. Growing TLCs were expanded and tested for peptide specificity.
Proliferative Assays.
Irradiated (100 Gy) B-LCLs or irradiated (30 Gy) PBMCs were used as APC and were seeded (5 × 104 cells per well) in 96-well U-bottomed microtiter plates. Peptides were added at a final concentration of 10–25 μM before addition of T cells (2–5 × 104). In antibody blocking experiments, APCs were incubated with mAbs for 30 min at 37°C before addition of T cells. Final concentration of MAbs were 10 μg/ml. The concentration chosen had earlier been shown to inhibit HLA-DR and -DQ rectricted T cell responses (14) as well as HLA-DP restricted T cell responses (15). Proliferation was measured at day 3 after labeling with 3.7 × 104 Bq [3H]thymidine (NEN) overnight before harvesting. Values are given as mean cpm from triplicates or duplicates ± SD. SD was usually less than 10%.
Identification of TGFβRII Mutation.
Microdissection was performed on the HLA-DR+ and HLA-DR− areas of the tumor biopsy taken from donor IMT. DNA was extracted from the microdissected samples by adding 4 μg of proteinase K, 10 × PCR (Qiagen) buffer and distilled water to a final volume of 25 μl. The samples were heated at 65°C for 2 h, followed by inactivation of proteinase K at 96°C for 15 min. DNA from the patients EA, GAA, TT, LRT, EMH, and SJ, was extracted from paraffin embedded tissue by a standard phenol chloroform technique.
All PCR reactions were performed on a PTC-200 terminal cycler (MJ Research, Cambridge, MA). PCR was performed by adding 2.5 mM of each dNTP (Perkin–Elmer), 10× PCR buffer (Qiagen), 2 units of HotStarTaq Polymerase (Qiagen) and 10 pmol of each primer (MedProbe, Oslo) directly in the extraction tube or with 50 ng DNA. Final PCR volume was 50 μl. PCR primers were as follows: 5′-CTT TAT TCT GGA AGA TGC TGC T-3′ and 5′-fluorescein CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC GGA AGA AAG TCT CAC CAG GC-3′, and for sequencing 5′-TTC TCT CTC TCC CTC TCC CC-3′ and 5′-TGC ACT CAT CAG AGC TAC AGG-3′. Cycling parameters for the PCR reaction was 35 cycles with denaturation for 1 min at 94°C, annealing at 56°C for 1 min and elongation at 72°C for 1 min.
Automated constant denaturant capillary electrophoresis (ACDCE) analyses were performed to determine mutation status of the samples. An ABI Prism 310 Genetic analyzer (Applied Biosystems, Perkin–Elmer) with uncoated Capillaries (Applied Biosystems, part no. 402839) and denaturant gel POP4 (Applied Biosystems part no. 402838) was used for separation of fragments. Amplified PCR products were diluted 1:50 in distilled water before loading, which was achieved with 15.0 kV for 5 s. The fluorescein-labeled PCR products were run at 15.0 kV (319 V/cm) and the laser power was 9.9 mW. The optimal electrophoresis conditions for separation of mutant fragments were determined by running PCR products with known mutations at different temperatures with 1°C intervals and by calculating the theoretical melting profiles by using MACMELT computer program (MedProbe). During analysis of PCR fragments, the temperature of the denaturing zone was held constant to achieve optimal separation. Electropherograms were analyzed as described (16) with the GENE SCAN software supplied for the 310 ABI Genetic Analyzer. Electropherograms showing a tentative mutant sample with ACDCE was further investigated. By mixing the tentative mutant with a control sample with known mutation status followed by denaturation, reannealing and reanalyzing with ACDCE, the quality of the mutant was confirmed (17). Samples scored as mutants with ACDCE were also sequenced on the 310 Applied Biosystems genetic analyzer under conditions described by the manufacturer and with 5′-TGC ACT CAT CAG AGC TAC AGG-3′ as sequence primer.
Immunohistochemistry.
Sections cut from frozen material were used for immunohistochemistry. A standard panel containing antibodies against CD22, CD20, CD3, CD4, CD8, activated T-cells, IL-receptors, HLA-DQ, and HLA-DR antigens was applied. Positive and negative controls were included.
Results
Generation of T Cells Specific for TGFβRII Frameshift Peptides in Normal Donors.
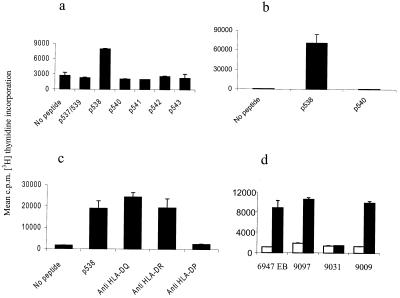
To generate frameshift peptide specific TLC in vitro, PBMCs from healthy donors were stimulated weekly with autologous peptide pulsed DCs or PBMCs in bulk cultures for up to 5 weeks. Of 16 donors tested, five or six showed some evidence of response after multiple stimulations. In donor 6947, T cell growth was observed after the third weekly stimulation in the bulk culture primed with the peptide combination p538 + p540. The responder cells were tested for peptide specific proliferation and were found to recognize the p538 peptide only (Fig. 2a). T cells from the bulk culture were cloned by limiting dilution. Only a single clone, TLC 6947–22, specific for the p538 peptide (Fig. 2b) and of the CD3+, CD4+, CD8− phenotype (not shown) could be generated. Antibody-blocking studies revealed that TLC 6947–22 was HLA class II restricted, and that the TGFβRII frameshift peptide was presented by HLA-DP antigens (Fig. 2c). Panel studies using homozygous B-LCLs as APC revealed that DPB1*0301 is the restriction molecule of TLC 6947–22 (Fig. 2d). Interestingly, the cell line 9009, which carries the HLA-DPB1*1401 molecule, only differing from HLA-DPB1*0301 by a Y→H substitution in position 9, is also capable of presenting p538 (Fig. 2d). This finding demonstrates that the T cell repertoire of normal donors contains T helper (Th) cells capable of recognizing a frameshift peptide from TGFβRII and further provides evidence for a role of HLA-DPB1*0301 molecules in presenting this tumor antigen.
Figure 2.
Specificity of T cells from donor 6947 for p538. (a) Peptide-specific proliferation of bulk culture 6947 against autologous PBMCs pulsed with TGFβRII frameshift peptides. (b) Peptide-specific proliferation of the TLC 6947-22 against the p538 peptide. Autologous B-LCLs (6947 EB) were used as APCs. (c) Blocking of the TGFβRII specific response by anti-HLA-DP mAb. (d) Panel studies using homozygous B-LCLs corresponding to DPB1 alleles of donor 6947. Solid bars, APC + p538; open bars, APC − p538. The HLA-DPB1 type of the different B-LCLs was 6947; EB: DPB1*0301, 9097: DPB1*0301/0401, 9031: DPB1*0401, 9009: DPB1*0401/1401.
T Cell Responses Against Frameshift Peptides in Patients with Proximal Colon Cancer.
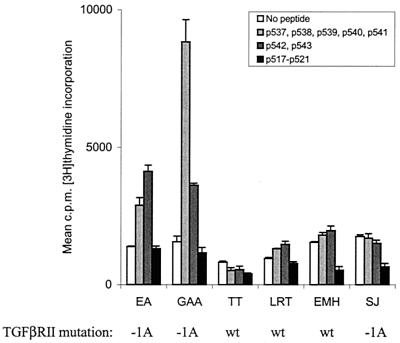
We investigated a consecutive series of patients with right-sided colon cancer operated during 1998 and selected a number of patients with strong tumor lymphocyte infiltration for further study. Bulk cultures from peripheral blood of six of these cancer patients were generated. After stimulation with peptide pulsed autologous PBMCs and one round of restimulation, T cell growth was observed and the responder cells were tested for peptide specific proliferation. Two of the six patients (EA and GAA) responded to mixtures of TGFβRII frameshift peptides (Fig. 3). Interestingly, a peptide mixture containing peptides other than p538 was also recognized by Th cells from the patients, indicating that other epitopes derived from TGFβRII frameshifts are also immunogenic. None of these patients responded against the mutated Bax peptides tested. PCR amplification and mutation analysis of the six patients showed that three samples had a (1A) deletion in the (A)10 polyadenine tract, the three other harbored a wild type sequence. The two patients who showed T cell responses were among the three with a (1A) deletion. These results indicate that T cells specific for TGFβRII frameshift peptides may indeed be a component of the immune response against the tumor seen in these patients. Because of limited access to blood from the patients, the peptides were tested in mixtures rather than as single peptides. Attempts to clone the responding cells were unsuccessful.
Figure 3.
Proliferative responses of bulk cultures generated from colorectal cancer patients. Bulk cultures generated from six patients with colorectal cancer localized to the proximal colon were tested for proliferation to TGFβRII and Bax frameshift peptides. Autologous B-LCLs or PBMCs were used as APCs. Patients' mutation status is indicated.
T Cell Responses Against Frameshift Peptides in HNPCC Patients.
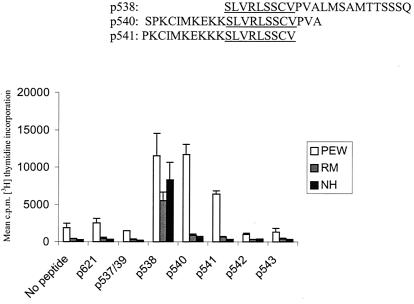
To search for memory T cell responses to the frameshift peptides in this patient group, bulk cultures from peripheral blood from three HNPCC patients were generated. Three normal donors were used as controls. After the first restimulation with peptide pulsed autologous PBMCs, T cell growth was observed and the responder cells were tested for peptide-specific proliferation. Although none of the controls showed peptide specific responses (data not shown), all three HNPCC patients responded to peptide p538 and donor PEW also responded well to p540 and p541 (Fig. 4), indicating that he had been primed in vivo against multiple epitopes derived from the common (1A) frameshift mutation. The amino acid sequences of peptides p538, p540, and p541 show a common motif probably involved in the specific response, and studies with a large number of Th cell clones derived from this patient are currently being performed to characterize his response in detail. Preliminary data from donor PEW indicate that his T cell clones recognize p538 presented by both HLA-DQ*0602 and -DQ*0603.
Figure 4.
Proliferative responses of bulk cultures generated from three HNPCC patients. PBMCs from HNPCC patients were stimulated with frameshift TGFβRII peptides and tested for peptide-specific response in a proliferation assay. Autologous PBMCs were used as APCs. The amino acid sequences of peptides p538, p540, and p541 are given, and the common/overlapping motif is underlined.
TIL Responses Against Frameshift Peptides.
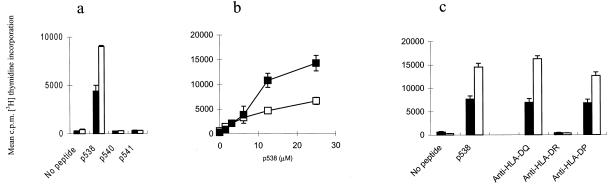
In a different approach, we studied T cell responses against the selected frameshift peptides in TILs, isolated from tumor biopsies. Seven right-sided colon cancers were tested. The MSI status of the tumor was not known before operation. Not all samples gave rise to growing T cells, and T cell clones were obtained from only one patient. From patient IMT, tumor infiltrating lymphocytes grew out of the biopsy when given IL-2 as a growth factor and were expanded in vitro on IL-2 and cloned by limiting dilution. Of the 14 clones that could be expanded, all were CD4+. Preliminary screening showed that 2 of the clones were reactive with the eight-peptide mixture used for cloning (data not shown). After expansion, the clones were tested for peptide-specific proliferation. TLC IMT8 and IMT9 both reacted specifically with the TGFβRII frameshift peptide p538, no reactivity was observed with the two other frameshift peptides tested (Fig. 5a). Both clones demonstrated a dose-dependent increase in the proliferative response to the peptide (Fig. 5b). Antibody-blocking studies revealed that IMT8 and IMT9 were HLA class II restricted, and that the transforming growth factor β peptide was presented by HLA-DR (Fig. 5c). Patient IMT carried DRB1*0301 and -*1401 as determined by genomic typing. By employing a panel of homozygous B-LCLs as APC, DRB1*1401 was identified as the restriction element of TLC IMT8 and IMT9 (Table 1). This adds an HLA-DRB1*1401 motif to the list of binding motifs already identified by using the SYFPEITI database (Fig. 1), and together with the HLA-DP DPB1*0301 restriction shown above indicates that p538 is a promiscuous HLA class II binding peptide. When tested with a panel of mAb specific for TCRVβ regions, both IMT8 and IMT9 were found to express TCR Vβ17 (not shown). Together these results indicate that frameshift peptides derived from mutant TGFβRII are tumor-specific antigens and that an HLA-DR-restricted epitope from mutant TGFβRII is generated during processing in vivo.
Figure 5.
Specificity of TLC IMT8 and IMT9 for the TGFβRII frameshift peptide p538. (a) Peptide-specific proliferation against autologous B-LCLs loaded with mutated TGFβRII peptides. (b) Dose–response curves of TLC IMT8 and IMT9 obtained by pulsing the cell line 9054 (HLA-DRB1*1401) with increasing amounts of the p538 peptide. (c) Blocking of the peptide-specific response by anti-HLA class II mAbs. The cell line 9054 was used as APC. Filled squares and bars, TLC IMT8; open squares and bars, TLC IMT 9.
Table 1.
Ability of different HLA homozygous cell lines to present the p538 peptide to TLC IMT 8 and IMT 9
| Cell lines | APC | DRB1 | IMT 8
|
IMT 9
|
||
|---|---|---|---|---|---|---|
| cpm* | SI† | cpm* | SI† | |||
| L0081785 | 9018 | 0301 | 833 | 1 | 905 | 1 |
| AMALA | 9064 | 1402 | 1,010 | 1 | 839 | 1 |
| OMW | 9058 | 1301 | 927 | 2 | 838 | 1 |
| 31227AB0 | 9061 | 1401 | 8,918 | 20 | 20,793 | 63 |
| LZL | 9099 | 1402 | 1,197 | 1 | 986 | 1 |
| H0301 | 9055 | 1302 | 589 | 2 | 462 | 1 |
| HHKB | 9065 | 1301 | 999 | 1 | 897 | 1 |
| BM92 | 9092 | 0404 | 1,262 | 1 | 1,218 | 1 |
| EK | 9054 | 1401 | 9,643 | 19 | 15,508 | 44 |
| WDV | 9062 | 1301 | 551 | 1 | 431 | 1 |
| HOM2 | 9005 | 0101 | 508 | 1 | 865 | 1 |
| D0208915 | 9008 | 1501 | 755 | 1 | 546 | 1 |
| SCHU | 9013 | 1501 | 960 | 1 | 1,051 | 1 |
| GB | – | 1404 | 857 | 1 | 841 | 1 |
| IMT EB‡ | – | 0301/1401 | 4,406 | 15 | 9,006 | 22 |
Figures in bold type show HLA types shared with the patient.
cpm, mean values of duplicates.
Stimulatory index; defined as mean cpm in duplicates with antigen divided by mean cpm in cultures without antigen.
Autologous B-LCL.
Because we used fresh biopsies in this series of experiments, no information on lymphocyte infiltration or MSI of the tumors was available at the onset of in vitro culture. The finding of a, HLA-DR-restricted CD4+ T cell response against a TGFβRII frameshift peptide in patient IMT suggested that the patient would have a tumor with MSI and a TGFβRII frameshift mutation, and that CD4 lymphocytes would be present in the lymphocyte infiltrate.
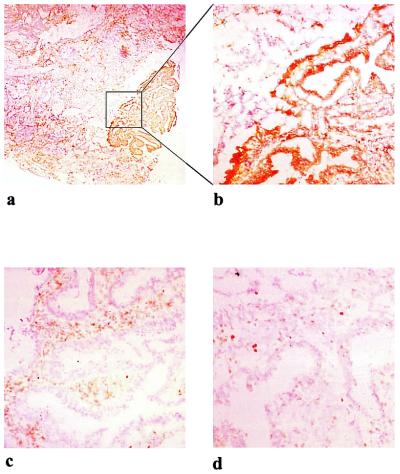
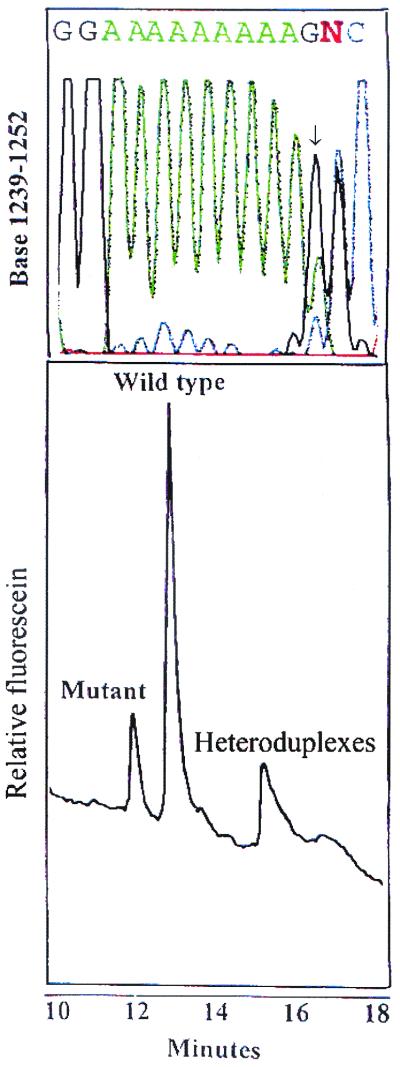
To verify this, we performed immunohistochemistry with several different antibodies. As is evident from Fig. 6, the tumor showed patchy expression of HLA-DR (Fig. 6 a and b), whereas HLA-DQ and -DP was not expressed (not shown). Prominent CD4+ T cell infiltration was observed in the same areas (Fig. 6c), but relatively few CD8+ T cells were present (Fig. 6d). Many of the T cells displayed an activated phenotype and the tumor also contained dendritic cells scattered in the same area as the T cells (not shown). These findings fit very well with the isolation of HLA-DR restricted T cells from the tumor, and indicate that the cells cloned are representative of the in situ activity. To determine the mutation status in the polyadenine tract of TGFβRII, automated CDCE was adapted from Bjorheim et al. (16). ACDCE has been reported to detect 0.6% of mutated alleles in a background of wild-type alleles. To further increase the tentative mutant allele fraction in the tumor tissue, laser capture microdissection of the HLA-DR positive and negative areas was performed before PCR. We found evidence for the presence of a mutation in both the DR− and the DR+ areas. In Fig. 7, a sample with a heterozygous (1A) deletion is illustrated with electropherogram from the ACDCE analysis. The sequence confirming the deletion is shown in Fig. 7 Upper.
Figure 6.
Photomicrographs of stained sections of the tumor biopsy obtained from donor IMT. Immunostaining with mAbs against HLA-DR (a) and same area magnified in b, CD4 (c) and CD8 (d).
Figure 7.
ACDCE electropherogram and sequencing results of mutation analysis in the (10A) tract of TGFβRII in patient IMT. (Lower) The mutant and wild-type peaks of a heterozygous one base deletion in TGFβRII polyadenine tract. To verify the results the sample was sequenced and the result is illustrated Upper. Note that the sequence is out of frame after the polyadenine tract because of a heterozygous one base deletion in the sequence.
Discussion
In some cancers, notably malignant melanomas, there is good clinical and experimental evidence for some level of immunological control of tumor growth (reviewed in ref. 18). In colorectal tumors, mainly circumstantial evidence exists for tumor immunogenicity. MSI+ colon cancer represents a subtype of colorectal cancer, and has a reputation for better survival that is associated with a pronounced stromal inflammatory reaction including the presence of activated cytotoxic intraepithelial lymphocytes. Significantly, tumor cells in close proximity with these activated lymphocytes undergo apoptosis (19). The antigens presumably recognized by the tumor-infiltrating T cells in MSI+ tumors have so far not been characterized. We here identify a highly immunogenic peptide (p538) derived from a frameshift mutation in TGFβRII as a target for tumor infiltrating Th cells in MSI+ tumors. This peptide, which results from a common (1A) deletion in the poly(A)10 nucleotide tract in the TGFβRII gene, was selected based on the presence of multiple good HLA class II and class I binding motifs. The finding of Th cells with specificity for this mutant TGFβRII peptide in the TIL population and the ready generation of such Th cells from the blood of patients with MSI+ tumors, but not from MSI− tumor patients or healthy donors, strongly indicate that a corresponding peptide is generated by natural processing of aberrant TGFβRII protein in vivo. Our data also demonstrate that T cell responses against several other peptides that correspond to the same mutation are present in some of the patients. Together these data provide evidence for immune surveillance against TGFβRII frameshift mutations at the level of Th cells in cancer patients with MSI+ tumors. Expression of HLA class II molecules in colon cancer is a favorable prognostic marker probably indicating an ongoing immune reaction (20). The observation of dense infiltration of CD4+ lymphocytes in association with patchy areas of the tumor that had turned on HLA-DR expression (Fig. 6) indicates local cytokine production, and may be a direct manifestation of immune surveillance. These findings are potentially of great importance because they provides a direct link between the molecular mechanism giving rise to the mutator phenotype and the prominent lymphocyte infiltration characteristic for such tumors. Peptide p538 also contains the newly described HLA-A2-restricted CTL epitope RLSSCVPVA (8), and it is conceivable that in the HLA-A2-positive patients, the helper T cell response may also contribute to the generation of an efficient CTL response against the mutant TGFβRII protein. This remains to be investigated. The concerted action of a Th and a CTL response against the same mutant antigen could provide a particularly efficient form of immune surveillance. Based mainly on the observation that colon cancer cell lines derived from MSI+ tumors frequently have lost HLA class I expression because of mutations in β2m, it has been hypothesized that such tumors are under a particularly strong selective pressure (21). We are now in the position to test this hypothesis by correlating HLA class I and class II expression of tumor cells in situ with T cell responses against defined epitopes that are derived from some of the key molecular alterations that take place in MSI+ tumors.
In the experiments reported here, we did not find evidence for T cells specific for mutant Bax peptides in the patients. The absence of a memory response against the peptides tested may have several causes. BAX mutations are less frequent than TGFβRII mutations, and the mutations representing the Bax peptides with the best binding characteristics may be even less frequent; thus, the number of patients tested in this study may be too small. Alternatively, the candidate peptides selected for this study may not correspond to the peptides derived from processing of mutant BAX in tumor cells or antigen-presenting cells. Our findings do not exclude the possibility that T cells specific for frameshift mutations other than those found in TGFβRII may form part of the patients' T cell response in MSI+ tumors. In general, genetically altered proteins provide a source of tumor-specific peptides that can result in antitumor antigen T cell responses. In tumors with deficient DNA mismatch repair, the very high spontaneous mutation rate may give rise to numerous unique peptides that may be recognized as tumor antigens. An increasing number of genes mutated in MSI+ are being characterized (22). Many of these genes have presumably been selected during carcinogenesis, and therefore are expressed in the majority of tumors with this phenotype. We are presently screening patient lymphocyte responses against a large panel of potential epitopes from such genes. In addition, we also expect that a large number of individual, nonselected “bystander” genes will be mutated in MSI+ cancers. These mutations may be impossible to identify by the “reverse immunology” approach used here, but may be picked up occasionally by using the cDNA expression approach pioneered by Boon et al. (23).
The molecular mechanism behind the mutator phenotype has provided a clue to the understanding of the lymphocyte reaction in MSI+ tumors by allowing reliable prediction of potential T cell epitopes created by frameshift mutations in candidate genes carrying nucleotide repeat sequences. This finding offers an opportunity for new vaccination approaches in patients with MSI+ tumors or at risk of developing such tumors. The T cell epitopes described here have a number of properties that make them well suited for a prophylactic vaccine for healthy HNPCC carriers and for patients with sporadic MSI+ cancers. The corresponding frameshift mutations are predictable, and occur frequently (90%) in tumors resulting from acquired MMR defects. They frequently elicit T cell responses in patients and their HLA binding is promiscuous.
Acknowledgments
We thank Dr. Jan Delabie for performing laser microdissection and Ms. Janne M. Røe for excellent technical assistance. This study was supported by the Norwegian Cancer Society.
Abbreviations
- MSI
microsatellite instability
- MMR
mismatch repair
- HNPCC
hereditary nonpolyposis colon cancer
- PBMC
peripheral blood mononuclear cells
- B-LCL
B-lymphoblastoid cell lines
- CM
conditioned medium
- DC
dendritic cells
- ACDCE
automated constant denaturant capillary electrophoresis
- Th
T helper
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Parkin D M, Pisani P, Ferlay J. Int J Cancer. 1999;80:827–841. doi: 10.1002/(sici)1097-0215(19990315)80:6<827::aid-ijc6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 2.Ionov Y, Peinado M A, Malkhosyan S, Shibata D, Perucho M. Nature (London) 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 3.Thibodeau S N, Bren G, Schaid D. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 4.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 5.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 6.Parsons R, Myeroff L L, Liu B, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- 7.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed J C, Perucho M. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 8.Linnebacher M, Gebert J, Rudy W, Woerner S, Yuan Y P, Bork P, von Knebel D M. Int J Cancer. 2001;93:6–11. doi: 10.1002/ijc.1298. [DOI] [PubMed] [Google Scholar]
- 9.Di Giorgio A, Botti C, Tocchi A, Mingazzini P, Flammia M. Int Surg. 1992;77:256–260. [PubMed] [Google Scholar]
- 10.Romani N, Reider D, Heuer M, Ebner S, Kampgen E, Eibl B, Niederwieser D, Schuler G. J Immunol Methods. 1996;196:137–151. doi: 10.1016/0022-1759(96)00078-6. [DOI] [PubMed] [Google Scholar]
- 11.Gedde-Dahl T, III, Eriksen J A, Thorsby E, Gaudernack G. Hum Immunol. 1992;33:266–274. doi: 10.1016/0198-8859(92)90334-j. [DOI] [PubMed] [Google Scholar]
- 12.Bakker A B, Marland G, de Boer A J, Huijbens R J, Danen E H, Adema G J, Figdor C G. Cancer Res. 1995;55:5330–5334. [PubMed] [Google Scholar]
- 13.Saeterdal I, thor Straten P, Myklebust J H, Kirkin A F, Gjertsen M K, Gaudernack G. Int J Cancer. 1998;75:794–803. doi: 10.1002/(sici)1097-0215(19980302)75:5<794::aid-ijc21>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 14.Gjertsen M K, Saeterdal I, Thorsby E, Gaudernack G. Br J Cancer. 1996;74:1828–1833. doi: 10.1038/bjc.1996.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fossum B, Gedde-Dahl T, III, Hansen T, Eriksen J A, Thorsby E, Gaudernack G. Eur J Immunol. 1993;23:2687–2691. doi: 10.1002/eji.1830231045. [DOI] [PubMed] [Google Scholar]
- 16.Bjorheim J, Ekstrom P O, Fossberg E, A L, Gaudernack G. BioTechniques. 2001;30:972–975. doi: 10.2144/01305st01. [DOI] [PubMed] [Google Scholar]
- 17.Guldberg P, Guttler F. Nucleic Acids Res. 1993;21:2261–2262. doi: 10.1093/nar/21.9.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawakami Y, Suzuki Y, Shofuda T, Kiniwa Y, Inozume T, Dan K, Sakurai T, Fujita T. Pigment Cell Res. 2000;13, Suppl. 8:163–169. doi: 10.1034/j.1600-0749.13.s8.29.x. [DOI] [PubMed] [Google Scholar]
- 19.Dolcetti R, Viel A, Doglioni C, Russo A, Guidoboni M, Capozzi E, Vecchiato N, Macri E, Fornasarig M, Boiocchi M. Am J Pathol. 1999;154:1805–1813. doi: 10.1016/S0002-9440(10)65436-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen S N, Rognum T O, Lund E, Meling G I, Hauge S. Br J Cancer. 1993;68:80–85. doi: 10.1038/bjc.1993.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Branch P, Bicknell D C, Rowan A, Bodmer W F, Karran P. Nat Genet. 1995;9:231–232. doi: 10.1038/ng0395-231. [DOI] [PubMed] [Google Scholar]
- 22.Woerner S M, Gebert J, Yuan Y P, Sutter C, Ridder R, Bork P, von Knebel D M. Int J Cancer. 2001;93:12–19. doi: 10.1002/ijc.1299. [DOI] [PubMed] [Google Scholar]
- 23.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den E B, Knuth A, Boon T. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 24.Sæterdal, I., Gjertsen, M. K., thor Straten, P., Eriksen, J. A. & Gaudernack, G. A. Cancer Immunol. Immunother., in press. [DOI] [PMC free article] [PubMed]