Abstract
DNA polymerase θ (Pol θ) is a multifunctional enzyme. It is nonessential in normal cells, but its upregulation in cancer cells correlates with cellular resistance to oxidative damage and poor prognosis. Pol θ possesses polymerase activity and poorly characterized lyase activity. We examined the Pol θ lyase activity on various abasic sites and determined that the enzyme is inactivated upon attempted removal of the oxidized abasic site commonly associated with C4′-oxidation (pC4-AP). Covalent modification of Pol θ by the DNA lesion enabled determination of the primary nucleophile (Lys2383) responsible for Schiff base formation in the lyase reaction. Unlike some other base excision repair polymerases, Pol θ uses a single active site for polymerase and lyase activity. Mutation of Lys2383 significantly reduces both enzyme activities but not DNA binding. Demonstration that Lys2383 is required for polymerase and lyase activities indicates that this residue is an Achilles heel for Pol θ and suggests a path forward for designing inhibitors of this attractive anticancer target.
Graphical Abstract
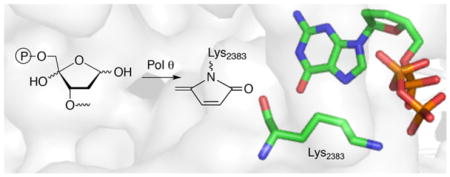
Of the 17 DNA polymerases so far identified in humans, 11 are involved in DNA repair and damage response.1 Five of these repair polymerases, as well as the mitochondrial DNA polymerase (Pol γ) possess lyase activity (Scheme 1A). Lyase activity is frequently associated with excising the remnant (dRP) resulting from abasic site (AP) incision by apurinic endonuclease 1 (Ape1) during base excision repair (BER).2–8 AP and oxidized variants, C4-AP and DOB are DNA lesions commonly produced as a result of oxidative damage by γ-radiolysis and chemotherapeutics, including bleomycin.9–13 pC4-AP produced from C4-AP by Ape1, and DOB, are possible lyase substrates. However, pC4-AP and DOB irreversibly inhibit DNA polymerase β (Pol β), the primary enzyme responsible for excising dRP (Scheme 1B, C).14,15 The oxidized abasic sites also inactivate DNA polymerase λ (Pol λ), a back-up of Pol β.16 DNA polymerase θ (Polθ) promotes resistance to bleomycin and ionizing radiation, suggesting that the interaction of Pol θ with these lesions may be clinically relevant.17,18 Pol θ is a nonessential enzyme in healthy cells, but homologous recombination-deficient cancers, including many ovarian cancers, are hyper-dependent upon Polθ expression.19,20 Interestingly, Pol θ expression is upregulated in breast, lung, and ovarian cancers, and this correlates with poor prognosis.21 Consequently, Pol θ is an attractive target for synthetic lethal therapy in BRCA-deficient cancers, along with other cancers containing DNA repair defects. Pol θ functions in translesion synthesis and double strand break repair in human cells, and has also been implicated in BER.17,22,23 Like Pol β, Pol θ possesses lyase activity, although little is known about this process.6 We wish to report details on Pol θ lyase activity that increases our understanding of this enzymatic process. The experiments also provide insight into whether Pol θ lyase activity is relevant to the enzyme’s ability to enhance cellular resistance to oxidative damage, its validation as an anticancer target, and direction for inhibitor design
Scheme 1.
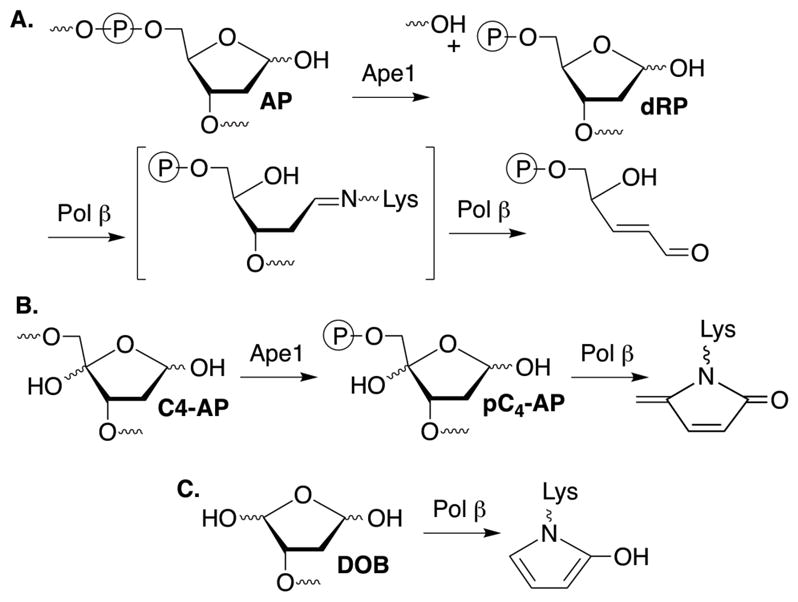
Base excision repair.
The ability of the 98 kDa Pol θ fragment to excise pC4-AP or DOB was compared to its reactivity with a comparable DNA substrate containing dRP. Under single turnover conditions pC4-AP (kobs = 0.93 ± 0.11 min−1) was excised the most rapidly of the 3 substrates, but DOB (kobs = 0.32 ± 0.01 min−1) was removed approximately twice as fast as dRP (kobs = 0.17 ± 0.02 min−1) (Figure S1). The dRP excision rate constant by Pol θ is comparable to a previous report and is also similar to those reported for two other polymerases (Rev1 and Pol ι) but ~1,000-fold slower than Pol β.7,15,24,25 More significantly, under multiple turnover conditions (Figure 1A), it appeared that pC4-AP excision ceased following 3–4 turnovers. Additional evidence that pC4-AP inactivates Pol θ was obtained by carrying the reaction out under multiple turnover conditions in which additional aliquots of enzyme were periodically added (Figure 1B). A burst of activity (3–4 turnovers) was observed after the addition of each aliquot, followed by cessation of conversion, consistent with inactivation of Pol θ.
Figure 1.
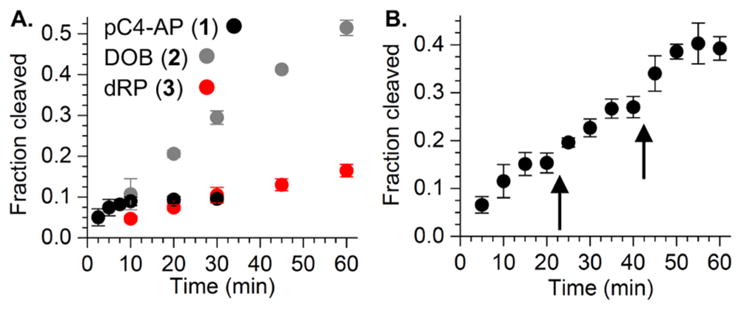
Pol θ inactivation by pC4-AP. (A.) dRP, DOB, and pC4-AP excision (100 nM) by Pol θ (2.5 nM). (B.) Repeated loss of Pol θ lyase activity following addition (5 nM) to pC4-AP (100 nM). Timing of additional Pol θ aliquots indicated by arrows.

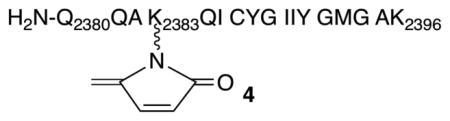
Of the DNA polymerases (Pol β, Pol λ) that are inactivated by oxidized abasic sites (DOB, pC4-AP), Pol θ is the first polymerase that is inactivated by just one. The effect of pC4-AP on the enzyme provided an opportunity to identify the source of its lyase activity. The location of Pol θ lyase activity was localized to a 24 kDa region of the polymerase domain, but the specific lysine responsible for Schiff base formation was unknown.6 Pol θ was subjected to trypsin digestion following incubation with excess 1. A single modified peptide (Figure 2A), whose mass (z = 3) corresponded to 4 (Figure 2B), was identified by LC-MS/MS. The peptide encompasses amino acids 2380–2396 in Pol θ (observed mass = 1948.9606 Da, calculated mass = 1948.9540 Da), and contains two tyrosine residues in addition to a more nucleophilic, internal lysine (Lys2383). Fragmentation of 4 (Figure S2) is consistent with Lys2383 modification.
Figure 2.
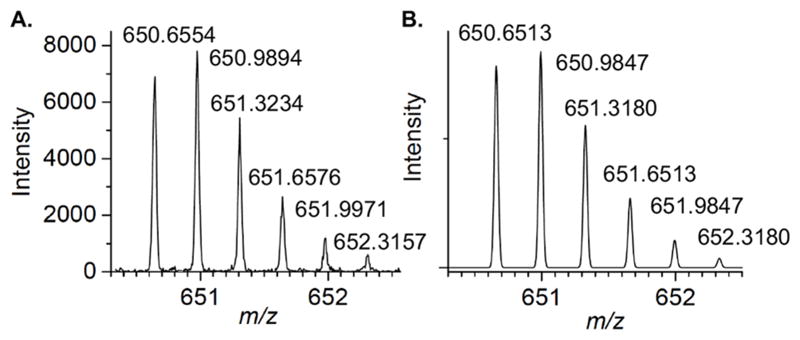
Modified peptide (4) detected in tryptic digest of Pol θ (1 μM) incubated with pC4-AP (1, 10 μM). (A.) Observed spectrum (z = 3) (B.) Calculated spectrum (z = 3)
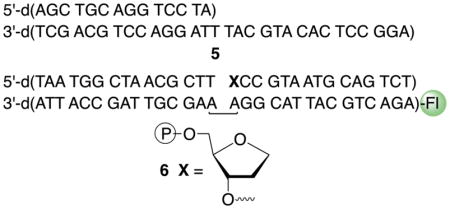
The X-ray co-crystal structure of Pol θ with DNA and ddGTP reveals that Lys2383 complexes the incoming nucleotide triphosphate within the polymerase active site (Figure 3).26 Formation of 4 suggests that unlike Pol β and Pol λ, at least one amino acid in Pol θ is involved in polymerase and lyase reactions.4,27 The function of Lys2383 was investigated further by mutating it to alanine (K2383A) and arginine (K2383R) (Table 1). The Km for dA incorporation in 5 by K2383A is more than 50-times higher than wild type Pol θ and kcat is reduced >100-fold. The polymerase activity (kcat/Km) of K2383R, which retains positive charge for possible dNTP binding, is ~10-fold greater than the alanine mutant but is still more than 3,000-times less active than wild type enzyme (Figure S3). However, DNA binding, as measured via fluorescence anisotropy on 6 (Figure 4, Figure S4), is unaffected by either mutation.
Figure 3.
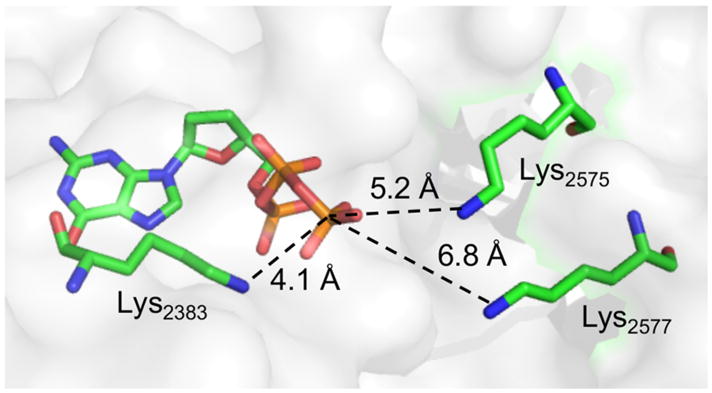
Pol θ structure showing Lys2383 and other potential nitrogen nucleophiles. (PDB: 4X0Q)
Table 1.
Polymerase activity of wild type Pol θ and mutants.a
| Pol θ variant | Km (μM)b | kcat (min−1)b | kcat/Km (μM•min) −1 |
|---|---|---|---|
| wt29 | 5 ± 1 | 65 ± 12 | 13.6 |
| K2383A | 1094 ± 37 | 0.5 ± 0.1 | 4.8 × 10−4 |
| K2383R | 311 ± 24 | 1.4 ± 0.2 | 4.3 × 10−3 |
Kinetics of dA incorporation in 5 were measured.
Data are the ave. ± std. dev. of 2 experiments each consisting of 3 replicates.
Figure 4.
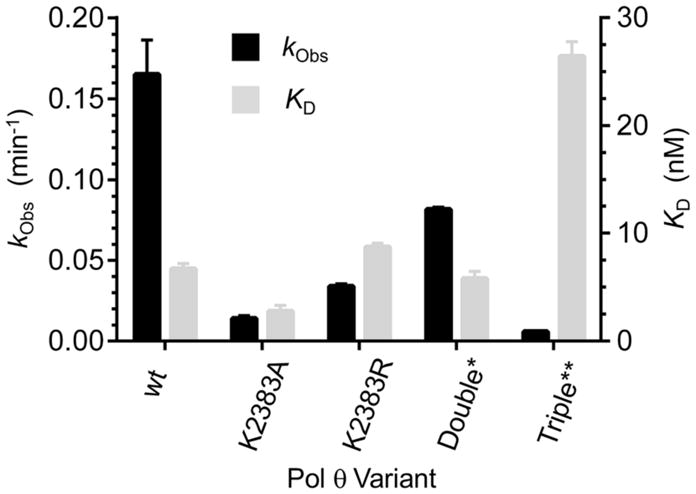
DNA binding (KD) and lyase activity (kObs) of wild type Pol θ and various Lys mutants. *Double = K2575A/K2577A, **Triple = K2383A/K2575A/K2577A. KD values are the ave. ± std. dev. of 3 experiments utilizing different samples, and were determined using 6 via fluorescence anisotropy. kObs are the ave. ± std. dev. of at least 2 experiments determined using 3, each consisting of 3 replicates.
The K2383A and K2383R mutants also exhibit significantly decreased lyase activity on dRP (Figure 4, Figure S5). dRP excision is reduced by 90% when Lys2383 is replaced by alanine, whereas K2383R retains ~20% of the wild type enzyme’s lyase activity. A previous study showed that the polymerase and lyase activities of Pol θ reside in a common domain.6 Mutation of Asp2540 and Glu2541 eliminated polymerase activity but not lyase activity, suggesting that the two are independent of one another.6 However, mutation of Lys2383 to alanine (K2383A) or arginine (K2383R) reveals that the two activities share a common residue.
Based on the observation of residual lyase activity in the K2383A and K2383R mutants, we considered whether other lysine residues were involved in the lyase reaction. DNA-protein cross-links (DPCs) were detected when mixtures of K2383A or K2383R and 3 were incubated in the presence of NaBH4 (Figure S6). The observation of DPCs in experiments with mutant proteins could indicate that an additional nucleophile(s) was present in the enzyme, or that Lys2383 may not even be the primary nucleophile responsible for Schiff base formation. For instance, Pol β utilizes a backup residue, proposed to be Lys84, for this function when the primary nucleophile, Lys72 is removed.27,30 Alternatively, there is also precedent for the NaBH4 experiments providing misleading information regarding nucleophilic lysine residues in repair enzymes.31
To further investigate the proposed function of Lys2383 as the major nucleophile, additional Pol θ mutants were prepared. A crystal structure for Pol θ bound to a BER substrate is unavailable, so the crystal structure of Pol θ in a ternary complex with a primer-template and incoming ddGTP (PDB: 4X0Q) was utilized to identify potential alternative nucleophiles.26 Using the phosphate coordinated to Lys2383 as a point of reference, we determined that Lys2575 and Lys2577 were the only nucleophilic residues within the active site within 10 Å. Mutation of these two residues in addition to Lys2383 (K2383A/K2575A/K2577A) reduced lyase activity to 3% of the wild type and reduced Schiff base trapping by 80% (Figure 4, Figure S3). This suggests that Lys2575and Lys2577 may compensate for lyase activity in the absence of Lys2383; however, binding of 6 by the triple mutant was ~4-fold weaker than wild type Pol θ, indicating that other factors may contribute to this observation. Despite the possible role for Lys2575 or Lys2577 in lyase activity, neither residue is the primary nucleophile as the double mutant (K2575A/K2577A) showed only 50% reduction in lyase activity while binding 6 as strongly as wild type Polθ (Figure 4). Interestingly, Schiff base trapping was increased more than two-fold for the K2575A/K2577A mutant (Figure S6), suggesting a possible role for one or both of these residues in a step following Schiff base formation, perhaps in deprotonation of the C2′-position.
Several polymerases also possess the ability to excise dRP via Schiff base formation and may participate in BER. The X-family polymerases Pol β and Pol λ contain an 8-kDa lyase domain, separate from the polymerase domain, where dRP excision is conducted.4,27,32 Four additional polymerases, Pol θ, Pol ι, Rev1, and the mitochondrial polymerase Pol γ, possess lyase activity, yet they lack a similar 8-kDa lyase domain.6–8,24 Unlike Pol β and Pol λ, the major catalytic nucleophile is unknown for each of these polymerases. Inactivation of Pol θ by pC4-AP served two purposes. Firstly, it suggests that Pol θ induced cellular resistance to bleomycin is not due to the enzyme assisting in excision of C4-AP in addition to its established role in double-strand break repair.17 Moreover, inactivation of the enzyme enabled us to determine that Lys2383 is the major nucleophile for Pol θ lyase activity. Importantly, this residue is also essential for efficient polymerase activity, consistent with the crystal structure of Pol θ, where Lys2383 coordinates the γ-phosphate of the incoming dNTP.26 This is the first demonstration that a single residue functions in both lyase and polymerase activities for any BER polymerase. A previous study on Pol ι showed that mutation of a single residue, Lys207, reduced both polymerase and lyase activity.28 However, this residue was proposed to be important for DNA binding28 as opposed to a direct role in lyase and polymerase activities, as demonstrated for Lys2383 of Pol θ. Oligonucleotide substrates containing lesions (e.g. DOB, pC4-AP) capable of inactivating the lyases may be generally useful tools for identifying the key nucleophilic residues in such enzymes.
In addition, identification of Lys2383 as the residue responsible for Schiff base formation may have ramifications beyond its fundamental biochemical significance. DNA repair enzymes are active anticancer targets.33–38 Pol θ is especially interesting in this regard because it is nonessential for healthy cells but provides therapeutic resistance in cancer cells. The potential of Pol θ as an anticancer target was recently compared to poly(ADP-ribose) polymerase, a target that has attracted a great amount of attention, resulting in clinically successful inhibitors.19,39,40 Determination that modification of Lys2383 inactivates Pol θ suggests this residue represents an Achilles heel of the enzyme, the modification of which eviscerates its polymerase and lyase functions. The identification of a single nucleophilic residue within the active site of Pol θ suggests that a previously reported approach used for identifying Pol β inhibitors will be useful against this enzyme.36
Supplementary Material
Acknowledgments
We are grateful for support from the National Institute of General Medical Sciences (GM-063028). We thank Dr. Kun Yang for helpful discussions and assistance in preparing Figures 3 and 4. We thank the Center for Molecular Biophysics at Johns Hopkins University (Dr. Katherine Tripp) for the use of instrumentation for fluorescence anisotropy.
Footnotes
Representative kinetic plots, fluorescence anisotropy plots, expanded LC-MS/MS data analysis, MS characterization of oligonucleotides containing modified nucleotides. The Supporting Information is available free of charge on the ACS Publications website.
References
- 1.Wu WJ, Yang W, Tsai MD. Nature Rev Chem. 2017;1:0068. [Google Scholar]
- 2.Greenberg MM. Acc Chem Res. 2014;47:646–655. doi: 10.1021/ar400229d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsumoto Y, Kim K. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Diaz M, Bebenek K, Kunkel TA, Blanco L. J Biol Chem. 2001;276:34659–34663. doi: 10.1074/jbc.M106336200. [DOI] [PubMed] [Google Scholar]
- 5.Bebenek K, Tissier A, Frank EG, McDonald JP, Prasad R, Wilson SH, Woodgate R, Kunkel TA. Science. 2001;291:2156. doi: 10.1126/science.1058386. [DOI] [PubMed] [Google Scholar]
- 6.Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH. Nucleic Acids Res. 2009;37:1868–1877. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad R, Poltoratsky V, Hou EW, Wilson SH. Nucleic Acids Res. 2016;44:10824–10833. doi: 10.1093/nar/gkw869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Proc Nat Acad Sci USA. 1998;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dizdaroglu M. Mutat Res Rev Mutagenesis. 2015;763:212–245. doi: 10.1016/j.mrrev.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Dizdaroglu M, Jaruga P. Free Rad Res. 2012;46:382–419. doi: 10.3109/10715762.2011.653969. [DOI] [PubMed] [Google Scholar]
- 11.Regulus P, Duroux B, Bayle PA, Favier A, Cadet J, Ravanat JL. Proc Nat Acad Sci USA. 2007;104:14032–14037. doi: 10.1073/pnas.0706044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burger RM. Chem Rev. 1998;98:1153–1169. doi: 10.1021/cr960438a. [DOI] [PubMed] [Google Scholar]
- 13.Rabow LE, Stubbe J, Kozarich JW. J Am Chem Soc. 1990;112:3196–3203. [Google Scholar]
- 14.Guan L, Greenberg MM. J Am Chem Soc. 2010;132:5004–5005. doi: 10.1021/ja101372c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobs AC, Kreller CR, Greenberg MM. Biochemistry. 2011;50:136–143. doi: 10.1021/bi1017667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevens AJ, Guan L, Bebenek K, Kunkel TA, Greenberg MM. Biochemistry. 2013;52:975–983. doi: 10.1021/bi301592x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yousefzadeh MJ, Wyatt DW, Takata K-i, Mu Y, Hensley SC, Tomida J, Bylund GO, Doublié S, Johansson E, Ramsden DA, McBride KM, Wood RD. PLoS Genet. 2014;10:e1004654. doi: 10.1371/journal.pgen.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goff JP, Shields DS, Seki M, Choi S, Epperly MW, Dixon T, Wang H, Bakkenist CJ, Dertinger SD, Torous DK, Wittschieben J, Wood RD, Greenberger JS. Radiat Res. 2009;172:165–174. doi: 10.1667/RR1598.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higgins GS, Boulton SJ. Science. 2018;359:1217. doi: 10.1126/science.aar5149. [DOI] [PubMed] [Google Scholar]
- 20.Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MIR, O’Connor KW, Konstantinopoulos PA, Elledge SJ, Boulton SJ, Yusufzai T, D’Andrea AD. Nature. 2015;518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood RD, Doublié S. DNA Repair. 2016;44:22–32. doi: 10.1016/j.dnarep.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshimura M, Kohzaki M, Nakamura J, Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A, Lan L, Seki M, Wood RD, Arakawa H, Buerstedde J-M, Hochegger H, Okada T, Hiraoka M, Takeda S. Mol Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon J-H, Roy Choudhury J, Park J, Prakash S, Prakash L. J Biol Chem. 2014;289:13177–13185. doi: 10.1074/jbc.M114.556977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasad R, Bebenek K, Hou E, Shock DD, Beard WA, Woodgate R, Kunkel TA, Wilson SH. J Biol Chem. 2003;278:29649–29654. doi: 10.1074/jbc.M305399200. [DOI] [PubMed] [Google Scholar]
- 25.Prasad R, Shock DD, Beard WA, Wilson SH. J Biol Chem. 2010;285:40479–40488. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zahn KE, Averill AM, Aller P, Wood RD, Doublié S. Nat Struct Mol Biol. 2015;22:304–311. doi: 10.1038/nsmb.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prasad R, Beard WA, Chyan JY, Maciejewski MW, Mullen GP, Wilson SH. J Biol Chem. 1998;273:11121–11126. doi: 10.1074/jbc.273.18.11121. [DOI] [PubMed] [Google Scholar]
- 28.Miropolskaya N, Petushkov I, Kulbachinskiy A, Makarova AV. Sci Rep. 2017;7:10194. doi: 10.1038/s41598-017-10668-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laverty DJ, Greenberg MM. Biochemistry. 2017;56:6726–6733. doi: 10.1021/acs.biochem.7b01093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prasad R, Batra VK, Yang XP, Krahn JM, Pedersen LC, Beard WA, Wilson SH. DNA Repair. 2005;4:1347–1357. doi: 10.1016/j.dnarep.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Williams SD, David SS. Biochemistry. 1999;38:15417–15424. doi: 10.1021/bi992013z. [DOI] [PubMed] [Google Scholar]
- 32.Belousova EA, Lavrik OI. DNA Repair. 2015;29:112–126. doi: 10.1016/j.dnarep.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Tahara Y-k, Auld D, Ji D, Beharry AA, Kietrys AM, Wilson DL, Jimenez M, King D, Nguyen Z, Kool ET. J Am Chem Soc. 2018;140:2105–2114. doi: 10.1021/jacs.7b09316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zafar MK, Eoff RL. Chem Res Toxicol. 2017;30:1942–1955. doi: 10.1021/acs.chemrestox.7b00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gowda ASP, Suo Z, Spratt TE. Chem Res Toxicol. 2017;30:715–725. doi: 10.1021/acs.chemrestox.6b00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul R, Banerjee S, Greenberg MM. ACS Chem Biol. 2017;12:1576–1583. doi: 10.1021/acschembio.7b00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donley N, Jaruga P, Coskun E, Dizdaroglu M, McCullough AK, Lloyd RS. ACS Chem Biol. 2015;10:2334–2343. doi: 10.1021/acschembio.5b00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zafar MK, Maddukuri L, Ketkar A, Penthala NR, Reed MR, Eddy S, Crooks PA, Eoff RL. Biochemistry. 2018;57:1262–1273. doi: 10.1021/acs.biochem.7b01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muvarak Nidal E, Chowdhury K, Xia L, Robert C, Choi Eun Y, Cai Y, Bellani M, Zou Y, Singh Zeba N, Duong Vu H, Rutherford T, Nagaria P, Bentzen Søren M, Seidman Michael M, Baer Maria R, Lapidus Rena G, Baylin Stephen B, Rassool Feyruz V. Cancer Cell. 2016;30:637–650. doi: 10.1016/j.ccell.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murai J, Huang S-yN, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y. Cancer Res. 2012;72:5588. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


