Abstract
To function in diverse cellular processes, the dynamic properties of microtubules must be tightly regulated. Cellular microtubules are influenced by a multitude of regulatory proteins, but how their activities are spatiotemporally coordinated within the cell, or on specific microtubules, remains mostly obscure. The conserved kinesin-8 motor proteins are important microtubule regulators, and family members from diverse species combine directed motility with the ability to modify microtubule dynamics. Yet how kinesin-8 activities are appropriately deployed in the cellular context is largely unknown. Here we reveal the importance of the nonmotor tail in differentially controlling the physiological functions of the budding yeast kinesin-8, Kip3. We demonstrate that the tailless Kip3 motor domain adequately governs microtubule dynamics at the bud tip to allow spindle positioning in early mitosis. Notably, discrete regions of the tail mediate specific functions of Kip3 on astral and spindle microtubules. The region proximal to the motor domain operates to spatially regulate astral microtubule stability, while the distal tail serves a previously unrecognized role to control the timing of mitotic spindle disassembly. These findings provide insights into how nonmotor tail domains differentially control kinesin functions in cells and the mechanisms that spatiotemporally control the stability of cellular microtubules.
INTRODUCTION
Microtubules (MTs) are essential cytoskeletal filaments, composed of polymerized tubulin, that play organizational and dynamic roles in eukaryotic cells (Nogales, 2000). MTs are intrinsically dynamic, and stochastically transition between extended periods of polymerization and depolymerization. When a MT switches into the depolymerizing state, the transition is termed “catastrophe,” and the transition out of depolymerization is called a “rescue” (Mitchison and Kirschner, 1984). They are polar filaments with the “minus end” typically associated with the MT organizing center, or centrosome, and the more dynamic “plus end” extending outward toward the cell periphery. MT-based structures can be complex and long lived yet also highly dynamic. Thus, cells must control the behavior of MTs to build networks that are mechanically robust while maintaining sufficient dynamicity and flexibility. For instance, the mitotic spindle persists throughout mitosis and undergoes dramatic morphological transitions that are essential for cell viability (Goshima and Scholey, 2010). In early mitosis, anti-parallel MTs emanating from two centrosomes are cross-linked by proteins of the Ase1/PRC1/MAP65 family to form a bipolar structure (Schuyler et al., 2003; Janson et al., 2007). Additional “kinetochore MTs” from opposite poles establish connections to sister chromatids. On anaphase, the cross-linked MTs must undergo net polymerization to maintain overlap in the midzone region as the spindle elongates. At the same time, kinetochore MTs depolymerize to pull the chromatids to opposite poles. Alongside these processes, dynamic astral MTs interact with regions of the cortex to position the spindle within the cell (Pearson and Bloom, 2004). It has been shown in the budding yeast Saccharomyces cerevisiae that this behavior of astral MTs is under tight spatial regulation (Fukuda et al., 2014). Defining how MTs are spatiotemporally controlled is central to understanding dynamic MT-dependent processes.
Kinesins are motor proteins that play critical roles in intracellular transport as well as the organization and dynamics of the MT network. Most kinesins utilize ATP-dependent motor activity to translocate along MTs or to cross-link and slide MTs relative to one another (Hirokawa, 2011). They can also transport MT regulatory proteins (Carvalho et al., 2004; Gandhi et al., 2011) and, in an expanding number of cases, can directly modify the dynamic behavior of the MT (Endow et al., 1994; Desai et al., 1999; Bringmann et al., 2004; Sproul et al., 2005; Gupta et al., 2006; Mayr et al., 2007; Du et al., 2010; Chen and Hancock, 2015; Hibbel et al., 2015; van Riel et al., 2017). Kinesins are typically composed of three components: a catalytic motor domain, a coiled-coil that mediates dimerization, and a “tail” region. Although some tails are known to bind MTs and form cross-links via the motor domain (Chandra et al., 1993; Mountain et al., 1999; Fink et al., 2009; Sturgill et al., 2014), they canonically bind cargos for molecular transport. In the absence of cargo, the tail can autoinhibit the motor domain to prevent unproductive transport activity (Coy et al., 1999; Friedman and Vale, 1999; Stock et al., 1999). Relative to the biochemical and mechanistic details of the motor domains, kinesin tails are less well studied, and comparatively little is known about how they contribute to kinesin functions when the motors operate in multiple cellular contexts.
Kinesin-8 are a specialized class of kinesins that are critical regulators of MT dynamics. They have N-terminal motor domains with plus end–directed motility (Pereira et al., 1997; Gupta et al., 2006; Varga et al., 2006). Family members from diverse species regulate the length of cellular MTs and structures including the spindle and cilia (Cottingham and Hoyt, 1997; Straight et al., 1998; West et al., 2001; Garcia et al., 2002; Gandhi et al., 2004; Goshima and Vale, 2005; Goshima et al., 2005; Wang et al., 2010; Weaver et al., 2011; Niwa et al., 2012), and several have been shown to directly control MT dynamics (Gupta et al., 2006; Varga et al., 2006; Mayr et al., 2007; Du et al., 2010; Stumpff et al., 2011; Wang et al., 2016; Locke et al., 2017). The budding yeast kinesin-8, Kip3, is a multifunctional regulator that contains four distinct activities: plus end–directed motility, depolymerase activity, anti-parallel MT cross-linking, and a poorly understood MT stabilizing property (Gupta et al., 2006; Varga et al., 2006; Su et al., 2011, 2013; Arellano-Santoyo et al., 2017). Kip3 also functions in discrete cellular contexts. During the process of anaphase spindle positioning, it destabilizes a subset of cellular MTs spatially at the bud tip and then stabilizes those same MTs near the bud neck (Fukuda et al., 2014). Kip3 also controls the length of the anaphase spindle by destabilizing MTs when they overgrow the spindle midzone region (Rizk et al., 2014). Moreover, although Kip3 does not destabilize MTs within the midzone during anaphase, it readily disassembles the midzone on mitotic exit (Woodruff et al., 2010). Currently, the molecular basis for the spatial and temporal regulation of Kip3 activities, or those of related kinesin-8s, is essentially unknown.
Here we investigate the role of the nonmotor tail in mediating the spatiotemporal functions of Kip3. We find that in the absence of the tail, the motor domain alone is sufficient to control astral microtubule dynamics at the bud tip to position the spindle in early mitosis. On the basis of structural features, we identify two distinct regions of the tail that confer opposite sensitivities to MT destabilizing compounds and show how they differentially mediate Kip3 function in specific cellular contexts. While one region operates to drive Kip3-dependent rescue of astral MTs spatially within the cytoplasm, analysis of the other reveals a novel role for the tail in temporally controlling spindle disassembly. These findings demonstrate the importance of the nonmotor regions in directing kinesin function in discrete physiological contexts and provide insight into the mechanisms that spatiotemporally control cellular microtubules.
RESULTS
Distinct regions of the Kip3 tail confer opposite sensitivities to MT destabilizing compounds
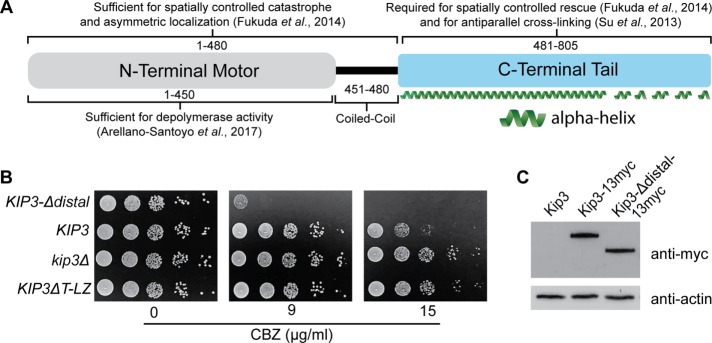
The C-terminal 481–805 residues, or “tail,” of Kip3 influences activities, including antiparallel MT cross-linking and MT stabilization (Figure 1A) (Su et al., 2013; Fukuda et al., 2014). The mechanisms underlying these activities, and whether specific activities are mediated by distinct regions of the tail, remain unknown. Analysis of the predicted structure of the Kip3 tail reveals regions with distinct structural features (Supplemental Figure S1) (Kelley et al., 2015). The region most proximal to the N-terminal motor and coiled coil, residues ∼481–690, is strongly predicted to be continuous α-helix. Following this, the distal ∼690–805 region is composed of several shorter α-helices interspersed among less ordered sections.
FIGURE 1:
The proximal and distal regions of the Kip3 tail confer opposite sensitivities to microtubule destabilizing compounds. (A) Molecular organization and associated activities of the budding yeast S. cerevisiae kinesin-8, Kip3. Numbers represent amino acid residues. (B) Relative carbendazim (CBZ) sensitivity of cells lacking the entire tail (KIP3ΔT-LZ) (Su et al., 2011) or only the distal tail region (KIP3-Δdistal) relative to kip3Δ and KIP3 control cells. Serial dilutions of each strain were plated onto rich media containing increasing concentrations of CBZ and grown at 24°C for 3 d. Relative to tailless Kip3 (KIP3ΔT-LZ), inclusion of the proximal tail results in high CBZ sensitivity (KIP3-Δdistal), while further addition of the distal tail increases resistance (KIP3). (C) Full-length Kip3 and Kip3-Δdistal are expressed at comparable steady-state levels. Western blot of strains expressing the indicated version of Kip3 from the endogenous KIP3 promotor. Kip3 and Kip3-Δdistal were detected by fusion to the myc epitope tag. Actin was blotted as a loading control. Kip3ΔT-LZ and Kip3 were previously shown to be expressed at similar levels (Su et al., 2011). In A, secondary structure was predicted as in Supplemental Figure S1.
We hypothesized that these structurally distinct “proximal” and “distal” regions of the Kip3 tail mediate specific cellular activities. To test this idea, we modified endogenous KIP3 to remove the region encoding residues 691–805 and express the truncated “Kip3-Δdistal” protein. Removal of the entire tail (481–805) in the “Kip3ΔT-LZ” protein produces resistance to the MT destabilizing drugs benomyl and carbendazim, indicating that the tail is needed for efficient MT destabilization in vivo (Figure 1B) (Su et al., 2011; Fukuda et al., 2014). Conversely, removal of only the distal region results in hypersensitivity, suggesting MTs are excessively destabilized in KIP3-Δdistal cells (Figure 1B). Expression levels of Kip3 and Kip3-Δdistal are indistinguishable (Figure 1C). Thus, the increased carbendazim sensitivity does not result from elevated Kip3-Δdistal levels but rather altered activity between Kip3 and Kip3-Δdistal. Notably, the proximal and distal regions of the tail confer opposite phenotypes. Relative to the tailless Kip3ΔT-LZ, inclusion of the proximal 481–690 region generates carbendazim hypersensitivity with Kip3-Δdistal (Figure 1B). Further addition of the distal region increases resistance with full-length Kip3 (Figure 1B). Together the results suggest that the proximal and distal tail regions mediate distinct cellular functions.
Kip3-Δdistal localizes to MT plus ends and regulates overall MT dynamics similarly to full-length Kip3
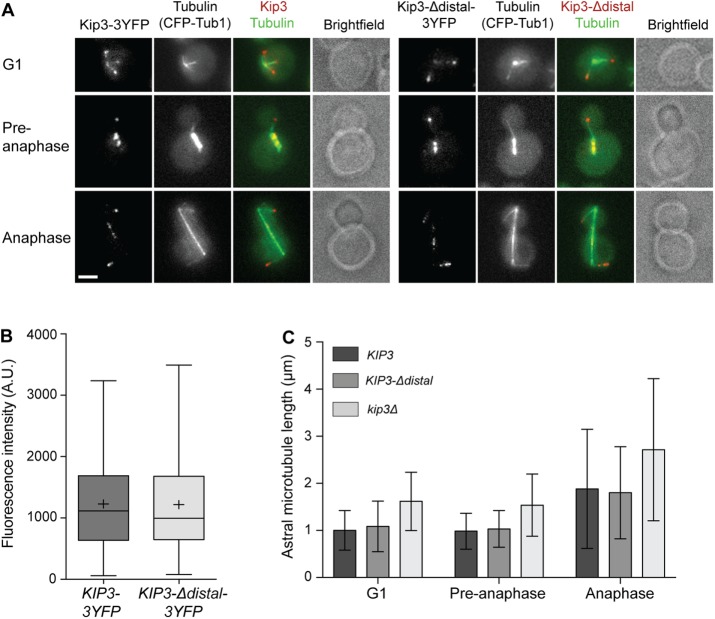
In G1, preanaphase, and anaphase cells with properly positioned spindles, Kip3-3YFP is observed as discontinuous speckles along the length, and prominent foci at the plus ends of polymerizing but not depolymerizing astral MTs (Gupta et al., 2006). Kip3 also localizes to the mitotic spindle during preanaphase and anaphase (Gupta et al., 2006). Throughout the cell cycle, the localization of Kip3-Δdistal closely resembles that of full-length Kip3 (Figure 2A). Additionally, quantitative analysis of astral MTs in G1 cells shows similar amounts of Kip3-3YFP and Kip3-Δdistal-3YFP localize to MT plus ends (Figure 2B).
FIGURE 2:
Effect of the Kip3 tail on localization and regulation of astral MT length. (A) Kip3 (left) and Kip3-Δdistal (right) localize similarly as prominent foci at the plus ends of astral MTs and to preanaphase and anaphase spindles. Representative images of cells expressing CFP-tubulin and YFP-labeled Kip3 or Kip3-Δdistal are shown. (B) Amount of Kip3-3YFP and Kip3-Δdistal-3YFP on MT plus ends in G1 cells. Center line is median, boxes represent the 25th to 75th percentiles, and whiskers encompass from 2.5 to 97.5 percentiles; (+) represents mean values. (C) Astral MT length in G1, preanaphase, and anaphase cells of the indicated genotype. For kip3Δ, p < 0.001 vs. KIP3 and KIP3-Δdistal at all stages. KIP3 vs. KIP3-Δdistal are not statistically significant. Mean ± SD. (A) Bar, 2 μm; (B) n = 118 for Kip3-3YFP and 111 for Kip3-Δdistal-3YFP; (C) n > 150 MTs for each cell type in each category.
We next examined how Kip3-Δdistal regulates astral MT dynamics. In both G1 and anaphase cells, MT polymerization and depolymerization rates are similar in cells harboring Kip3 or Kip3-Δdistal (Table 1). In contrast, MTs in kip3∆ cells depolymerize significantly faster than those in KIP3 or KIP3-Δdistal cells (Table 1). Kip3 regulates the frequency of MT catastrophe and rescue events. Although these transitions are regulated spatially in vivo (Gupta et al., 2006; Fukuda et al., 2014), the effect is reflected in the overall length of astral MTs (Cottingham and Hoyt, 1997). While MTs in kip3∆ cells are clearly longer than those in control cells harboring full-length Kip3, MT length in KIP3-Δdistal cells matches that in control cells throughout the cell cycle (Figure 2C). Overall, these data demonstrate that the distal tail region is required neither for the general localization to astral MTs nor for the overall regulation of their dynamics by Kip3.
TABLE 1:
In vivo parameters of microtubule dynamics for astral microtubules in KIP3, kip3Δ, and KIP3-Δdistal cells.
| KIP3 | kip3Δ | KIP3-Δdistal | |
|---|---|---|---|
| G1 | |||
| Growth rate, μm/min | 1.74 ± 0.77 (16) | 1.33 ± 0.49 (13) | 1.46 ± 0.34 (12) |
| Shortening rate, μm/min | –2.60 ± 0.86 (21) | –3.28 ± 0.94 (17)* | –2.53 ± 0.74 (19) |
| Anaphase | |||
| Growth rate, μm/min | 1.32 ± 0.34 (19) | 1.03 ± 0.14 (11)* | 1.47 ± 0.40 (22) |
| Shortening rate, μm/min | –2.21 ± 0.77 (18) | –2.88 ± 0.78 (10)* | –2.10 ± 0.35 (23) |
For KIP3, kip3Δ, and KIP3-Δdistal cells, a total of 1664 and 2058, 1266 and 2402, and 1578 and 1812 s of microtubule lifetime were analyzed for G1 and anaphase cells, respectively. Rates are reported as mean ± SD; number of events is in parentheses; *p < 0.05 by unpaired two-tailed Student’s t test.
Kip3-Δdistal regulates astral MT dynamics at the bud tip during Kar9-dependent early spindle positioning
The budding yeast spindle is positioned by two major, yet distinct, mechanisms (Pearson and Bloom, 2004). In early mitosis, the Kar9-dependent mechanism positions the spindle adjacent to the bud neck. During anaphase, the dynein-dependent mechanism pulls one spindle pole body (SPB; yeast centrosome) through the bud neck. Strains lacking a single mechanism remain viable. However, the loss of components from both pathways prevents spindle positioning and thus results in synthetic lethality.
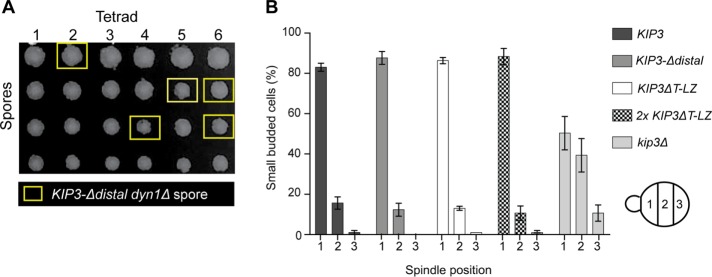
Kip3 functions in Kar9-dependent spindle positioning (Cottingham and Hoyt, 1997; DeZwaan et al., 1997; Miller et al., 1998), which involves myosin-dependent transport of MT plus ends along polarized actin cables to the bud tip (Pearson and Bloom, 2004). Once there, Kip3 halts MT polymerization and induces catastrophe (Gupta et al., 2006). In kip3∆ cells, prolonged MT interaction and excessive polymerization at the bud tip repeatedly pushes the spindle away from the bud neck and disrupts Kar9-mediated positioning (Gupta et al., 2006). As a result, kip3∆ is synthetic lethal with dyn1∆ (DeZwaan et al., 1997; Miller et al., 1998). When diploid cells heterozygous for both KIP3-Δdistal and dyn1∆ are sporulated, KIP3-Δdistal dyn1∆ double mutants are recovered at the expected frequency (27% double mutants from 26 tetrads), and display growth rates similarly to wild type (Figure 3A). In contrast to kip3∆, the truncated KIP3-Δdistal is not synthetic lethal with dyn1∆, suggesting that the distal tail is not required for Kar9-dependent spindle positioning.
FIGURE 3:
The Kip3 tail is not required for Kar9-dependent spindle positioning. (A) KIP3-Δdistal is not synthetic lethal with dyn1Δ. Dissection of tetrads resulting from a cross between KIP3-Δdistal and dyn1Δ haploid cells. Twenty-eight of 104 spores from 26 tetrads (∼1:4) possessed both KIP3-Δdistal and dyn1Δ mutations (yellow boxes). (B) Early mitotic spindles are properly positioned in cells harboring tailless Kip3ΔT-LZ or Kip3-Δdistal. Graph depicts position of the early mitotic spindle relative to the bud neck in small budded cells of the indicated genotype. For each category, kip3Δ is statistically different vs. each cell type. Graph shows three experiments where n = 239, 124, 127 for KIP3; 221, 111, 49 for KIP3-Δdistal; 182, 129, 150 for KIP3ΔT-LZ; 241, 140, 189 2x KIP3ΔT-LZ; and 180, 106, 117 for kip3Δ.
We next looked at spindle positioning in cells expressing green fluorescent protein (GFP)-tagged tubulin (GFP-Tub1). In control KIP3 cells, the early spindle in small budded cells is efficiently positioned in the mother cell adjacent to the bud neck (Figure 3B). In kip3∆ cells, they are positioned throughout the mother cell (Figure 3B). Similarly to control cells, the spindle in KIP3-Δdistal cells is also positioned near the neck (Figure 3B). This result prompted us to examine spindle positioning in the absence of the entire Kip3 tail. It was previously shown that, due to reduced processivity, cells require two copies of KIP3ΔT-LZ-YFP for complete localization to astral MT plus ends (Su et al., 2011). We found that early spindles are properly positioned in 2x KIP3ΔT-LZ cells (Figure 3B). Moreover, as in control KIP3 and KIP3-Δdistal cells, spindles are also positioned properly in cells harboring a single copy of KIP3ΔT-LZ (Figure 3B). Together these data show that the entire tail of Kip3 is not required and that the motor and coiled-coil alone control astral MT dynamics sufficiently to facilitate the Kar9-dependent spindle positioning mechanism.
The proximal region of the Kip3 tail spatially mediates astral MT rescue
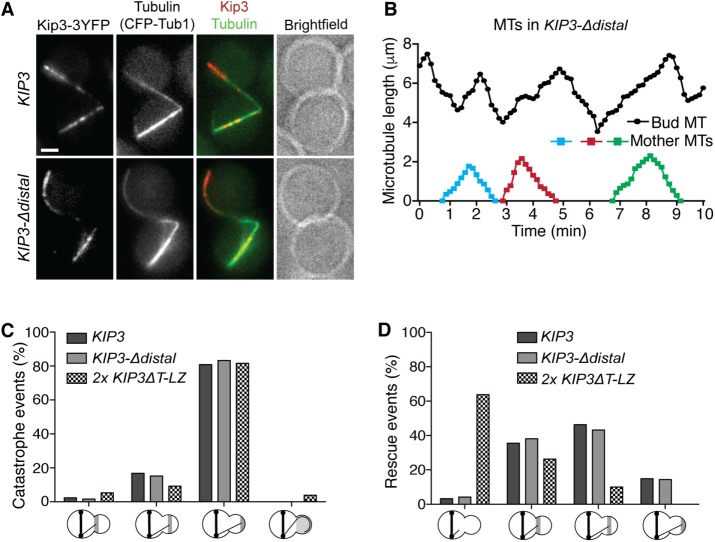
When anaphase spindles are mispositioned, dynein motors bound to astral MTs anchor on the bud cortex and pull the astral MT, with its associated SPB, through the bud neck to properly realign the spindle (Lee et al., 2003; Sheeman et al., 2003). In these cells, Kip3 localizes asymmetrically to specific subsets of astral MTs; although it continues to localize as foci at the plus ends of astral MTs in the mother, Kip3 localizes prominently along the length of those in the bud compartment (Fukuda et al., 2014). Additionally, Kip3 differentially regulates MT lifetime in the mother and bud and spatially controls MT stability within the bud (Fukuda et al., 2014). To reproducibly generate cells with mispositioned anaphase spindles, we inhibited dynein function (dyn1Δ). Similarly to full length, Kip3-Δdistal localizes to the plus ends of MTs in the mother and along the length of astral MTs specifically inside the bud (Figure 4A).
FIGURE 4:
The proximal region of the Kip3 tail spatially mediates astral MT rescue. (A) In anaphase cells with mispositioned spindles, Kip3 and Kip3-Δdistal localize asymmetrically to astral MTs in the bud compartment. Representative images of cells expressing CFP-tubulin and YFP-labeled Kip3 or Kip3-Δdistal. (B) Astral MT lifetime history plots from a representative KIP3-Δdistal cell with mispositioned anaphase spindle. Relative to those in the mother compartment, MTs in the bud (top trace) undergo extended cycles of polymerization and depolymerization. (C, D) Location of (C) catastrophe and (D) rescue events in the bud of anaphase cells with mispositioned spindles. In contrast to tailless Kip3ΔT-LZ, but similarly to full-length Kip3, Kip3-Δdistal promotes rescue of depolymerizing MTs near the bud neck. The x-axis represents events within each third of the bud. (C) The rightmost group represents MTs that overgrow the bud and curl around the bud tip prior to catastrophe. (D) The leftmost category depicts MTs that depolymerize out of the bud without undergoing rescue. Mispositioned anaphase spindles were generated by dyn1Δ. (A) Bar, 2 μm; (C, D) catastrophe and rescue events for KIP3 = 125 and 121, for 2x KIP3ΔT-LZ = 76 and 80, and for KIP3-Δdistal = 125 and 118, respectively.
The asymmetric localization of Kip3 is correlated with the spatial regulation of MT dynamics. Astral MTs in the mother display dynamic parameters similarly to typical yeast MTs and persist for an average lifetime of ∼2 min before depolymerizing back to the SPB. In the bud, Kip3 induces catastrophe specifically when MTs reach the bud tip and subsequently induces rescue of depolymerizing MTs near the bud neck, which dramatically extends MT lifetime within the bud (Fukuda et al., 2014). In KIP3-Δdistal cells, astral MTs in the mother display typical lifetimes of ∼2 min. However, those in the bud undergo repeated cycles of polymerization and depolymerization with dramatically extended lifetimes (Figure 4B). We previously showed that without the entire tail, the Kip3 motor and coiled coil are sufficient to induce catastrophe at the bud tip, similarly to full-length Kip3 (Fukuda et al., 2014). Consistent with this, Kip3-Δdistal also induces catastrophe almost exclusively when MTs reach the bud tip (Figure 4C). After undergoing catastrophe, nearly all depolymerizing MTs in KIP3 cells undergo rescue before they exit the bud compartment (Figure 4D). In contrast, the majority of MTs in the tailless 2x KIP3ΔT-LZ cells, which display KIP3ΔT-LZ localization similar to full-length Kip3 (Fukuda et al., 2014), fail to rescue and subsequently exit the bud, demonstrating that the tail region 481–805 is required for this function (Figure 4D). As in control KIP3 cells, depolymerizing MTs undergo rescue before exiting the bud in cells harboring Kip3-Δdistal (Figure 4D). Thus, the distal tail region 691–805 is not required for spatially induced rescue in the bud, and the proximal region from 481 to 690 is capable of carrying out this function of Kip3.
KIP3-Δdistal is a gain-of-function allele controlling anaphase spindle length
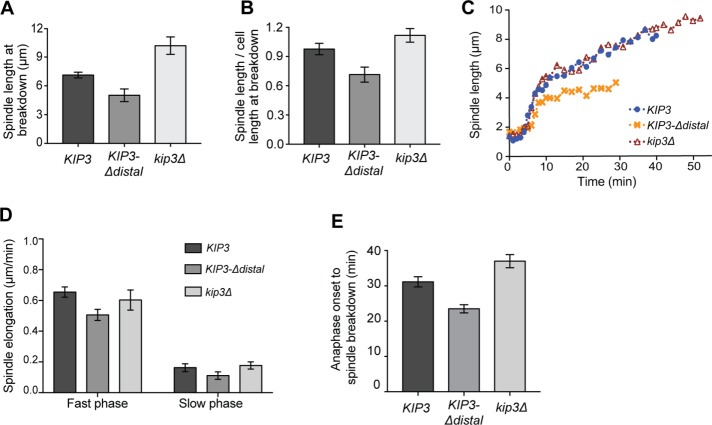
Since the distal tail is not required for the known functions of Kip3 on astral MTs, examined above, we investigated its role in the mitotic spindle. Kip3 regulates spindle length during anaphase by depolymerizing interpolar MTs that grow beyond the midzone, which governs the size of the midzone and limits the spindle elongation forces generated within (Rizk et al., 2014). As a result, spindles stop elongating once they have attained a length equal to that of the cell. On mitotic exit Kip3 also disassembles the spindle (Woodruff et al., 2010).
To test the idea that the distal tail mediates Kip3 activity on the spindle, we monitored anaphase spindle elongation in cells expressing GFP-Tub1 (α-tubulin). Spindle elongation in kip3Δ cells is not properly terminated and, as a result, spindles are significantly longer than in control cells and often exceed cell length at the time of disassembly (Figure 5, A and B). By contrast, spindles in KIP3-Δdistal cells do not attain the length observed in control cells (Figure 5A) and are disassembled when significantly shorter than the length of the cell (Figure 5B). The rate of spindle elongation during the initial fast period of anaphase is slower in KIP3-Δdistal cells relative to both KIP3 and kip3Δ cells (Figure 5, C and D). Although not statistically significant, elongation during the subsequent slow phase is also somewhat reduced in the KIP3-Δdistal cells (Figure 5, C and D). If the reduced elongation rates simply account for the shorter terminal spindle length in KIP3-Δdistal cells, then the timing from anaphase onset to spindle breakdown should remain similar in KIP3-Δdistal and KIP3 cells. This duration is significantly reduced in KIP3-Δdistal cells, however, indicating that spindles in these cells break down sooner than those in control cells (Figure 5E). In contrast, the duration from anaphase onset to spindle breakdown is substantially increased in kip3Δ cells (Figure 5E).
FIGURE 5:
KIP3-Δdistal is a gain of function allele controlling spindle length. (A) Anaphase spindle length and (B) length relative to cell length at breakdown in cells of the indicated genotype. Compared to spindles in control cells, those in kip3Δ and KIP3-Δdistal cells are significantly longer and shorter, respectively. Mean ± SD; p < 0.0001 for both kip3Δ or KIP3-Δdistal vs. KIP3. (C) Representative examples of spindle elongation in cells of the indicated genotype. (D) Anaphase spindle elongation rates. Mean ± SEM; p = 0.005 for KIP3 vs. KIP3-Δdistal fast phase; other comparisons are not statically significant. (E) Anaphase duration from onset of elongation to spindle breakdown. Mean ± SEM; p < 0.002 for KIP3 vs. KIP3-Δdistal and kip3Δ; p < 0.0001 for KIP3-Δdistal vs. kip3Δ. (A, B) n = 35, 44, and 30 for KIP3, KIP3-Δdistal, and kip3Δ, respectively; (D) n = 20 for each cell type from two independent days (n = 10/day); (E) total of n = 25, where n = 10 and 15 from two independent days for each cell type.
The distal tail prevents spindle midzone destabilization
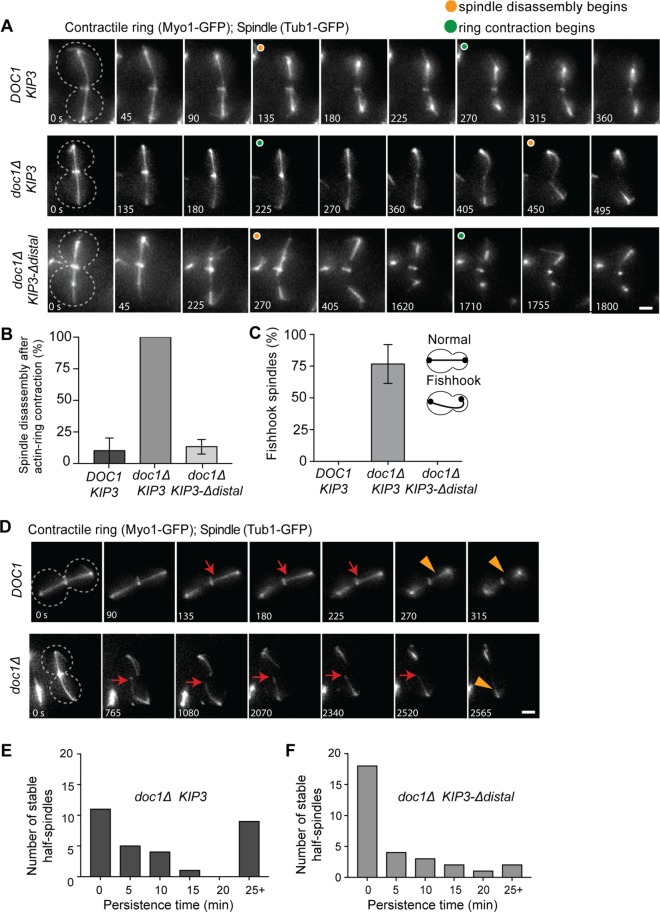
Our results indicate that spindles may be prematurely disassembled in KIP3-Δdistal cells. Thus, we examined whether the distal tail influences the timing of spindle disassembly. When cells exit mitosis, at least three mechanisms function to disassemble the spindle (Woodruff et al., 2010). In one mechanism, the kinase Aurora B (Ipl1) phosphorylates MT-associated proteins to destabilize spindle MTs. In a second, ubiquitin-mediated degradation of midzone proteins separate the spindle halves. And in the third mechanism, Kip3 functions independently of the other two mechanisms to drive depolymerization of the spindle MTs. We reasoned that if the distal tail functions to prevent spindle midzone destabilization, then the requirement for Kip3-independent mechanisms of spindle disassembly may be diminished in KIP3-Δdistal cells.
When cells exit mitosis, both the spindle is disassembled and the actinomyosin ring contracts at the bud neck to facilitate cytokinesis. To test the idea that spindle midzones are prematurely disassembled in KIP3-Δdistal cells, we monitored the timing of spindle disassembly relative to actinomyosin ring contraction in cells expressing both GFP-Tub1 and Myo1-GFP (actinomyosin ring marker). In the vast majority of control cells, the anaphase spindle is efficiently disassembled prior to actinomyosin ring contraction (Figure 6, A and B). Consistent with previous results (Woodruff et al., 2010), spindle disassembly is impaired in doc1∆ cells, which have diminished ubiquitin-mediated degradation of midzone proteins (Figure 6A). In 100% of these cells, the spindle is not disassembled until after actinomyosin ring contraction (Figure 6B). Strikingly, replacing Kip3 with Kip3-Δdistal effectively rescues the delayed spindle disassembly observed in doc1∆ cells (Figure 6, A and B). Thus, removal of the distal tail accelerates the timing of spindle disassembly.
FIGURE 6:
The distal tail region of Kip3 prevents spindle midzone destabilization. (A) Time-lapse images of the spindle (GFP-Tubulin) and actinomyosin ring (Myo1-GFP) during late anaphase/mitotic exit in control (DOC1 KIP3), doc1Δ KIP3, and doc1Δ KIP3-Δdistal cells. Cell outline indicated by dotted line, and orange and green circles denote when spindle breakdown and actinomyosin ring contraction begins, respectively. In doc1Δ cells spindle breakdown is delayed and the actinomyosin ring closes on the intact spindle. (B) Percentage of spindles that break down after actinomyosin ring contraction in cells of the indicated genotype. (C) Fishhook spindles in each genotype. For B and C p < 0.0001 for doc1Δ KIP3 vs. either cell type; mean ± SD. (D) Time-lapse images of DOC1 and doc1Δ cells expressing GFP-tubulin and Myo1-GFP during anaphase spindle breakdown. Cell outline is indicated by a dotted line; red arrows denote stable half-spindles, and orange arrowheads denote depolymerizing half-spindle. Note that the half-spindle in the doc1Δ cell is hyperstable. (E, F) Histograms showing the persistence time of hyperstable half-spindles in E doc1Δ KIP3 and (F) doc1Δ KIP3-Δdistal cells during spindle breakdown. p < 0.05 for doc1Δ KIP3 vs. doc1Δ KIP3-Δdistal. (B, C, E, F) n = 30 for each genotype, with 10 spindles observed in each of three experiments; (E, F) the half-spindle with the longest persistence time for each spindle is reported.
During a typical anaphase, spindles obtain full length and are then disassembled on mitotic exit. Spindles with a “fishhook” morphology, in which their length is significantly longer than that of the cell and the spindle is forced to bend, are rare in control cells (Figure 6C). Fishhook spindles are common in cells lacking Kip3, which normally functions to terminate spindle elongation (Rizk et al., 2014). The majority of doc1∆ cells also display fishhook spindles during anaphase (Figure 6C). Notably, Kip3-Δdistal completely abolishes the fishhook spindles observed in doc1∆ cells (Figure 6C). When spindle halves separate during disassembly in control cells, the interpolar MTs are rapidly depolymerized, at least in part by Kip3 (Woodruff et al., 2010). Indeed, we found that all spindle halves were efficiently depolymerized within 5 min of midzone separation (Figure 6D, n = 30 spindles). By contrast, in doc1∆ cells the depolymerization of half spindles is significantly inhibited, with interpolar MTs often remaining stable for more than 25 min after separation of the spindle halves (Figure 6, D and E). As with cdh1∆, which also impairs ubiquitin-mediated degradation, this half spindle persistence phenotype likely results from the failure to remove and degrade stabilizing midzone proteins from the broken half spindles (Woodruff et al., 2010). In doc1∆ KIP3-Δdistal cells, the persistence time of half spindles is markedly reduced compared with doc1∆ cells. Most notably, Kip3-Δdistal reduces the number of half spindles that persist for over 25 min by 78% and increases those depolymerized within 5 min by 64% (Figure 6, E and F). These data reveal that in the presence of stabilizing midzone proteins, spindle MTs are disassembled more efficiently in cells harboring Kip3-Δdistal rather than full-length Kip3. Taken together, our results demonstrate that the distal tail mediates the timing of spindle disassembly, and that, in the absence of this region, the spindle midzone is prematurely disassembled during anaphase and/or more efficiently disassembled on mitotic exit.
The distal tail of Kip3 temporally regulates spindle disassembly during anaphase
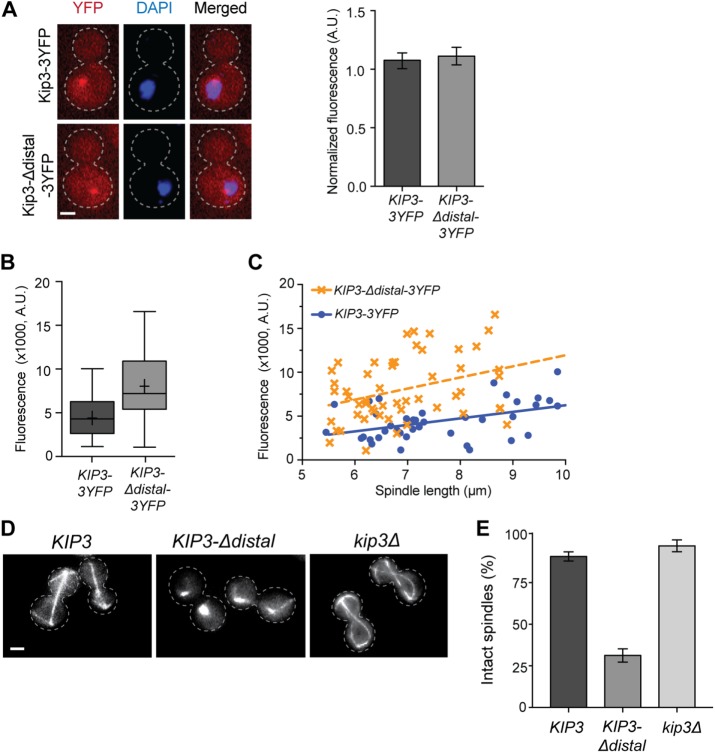
Yeast undergoes closed mitosis in which the nuclear envelope does not completely break down. As a result, altered concentrations of Kip3 within the nucleus could potentially affect spindle stability. Although PSORT analysis (Nakai and Horton, 1999) predicts the presence of nuclear localization signals in both the motor and distal tail regions of Kip3, we determined whether removal of the distal tail alters the balance between cytoplasmic and nuclear localized pools. We first removed the MT-bound pool by depolymerizing MTs with nocodazole and then quantified the relative amount of 3YFP-tagged Kip3 or Kip3-Δdistal in the cytoplasm and nucleus of mitotic cells (Figure 7A). In addition to having similar steady-state levels overall (Figure 1C), the accumulation in the nucleus relative to cytoplasm is similar for both Kip3 and Kip3-Δdistal, indicating that spindle stability is not perturbed due to altered nuclear concentration of Kip3-Δdistal.
FIGURE 7:
The distal tail of Kip3 temporally regulates spindle disassembly during anaphase. (A) Removal of the distal tail does not alter Kip3 distribution between cytoplasm and nucleus. Left, Single focal plane fluorescence images of cells expressing 3YFP-tagged Kip3 or Kip3-Δdistal (red) following microtubule depolymerization by 15-min nocodazole treatment. Cell outline is indicated by a dotted line; DNA is stained with DAPI (blue); the bright YFP foci is Kip3/Kip3-Δdistal associated with the spindle pole body and/or kinetochores. Right, graph depicting the nuclear-to-cytoplasmic ratio of fluorescence signal for Kip3-3YFP and Kip3-Δdistal-3YFP. Mean ± SD. (B) Quantification of Kip3-3YFP or Kip3-Δdistal-3YFP localized to the spindle midzone region. Center line is median, boxes represent the 25th to 75th percentiles, and whiskers encompass from minimum to maximum values; (+) represents mean values; p = 0.002. (C) Kip3-3YFP or Kip3-Δdistal-3YFP localization to the spindle midzone region relative to spindle length. Both Kip3 and Kip3-Δdistal are positively correlated with spindle length. Dashed lines represent best fit linear regression, and each symbol represents a separate spindle. R2 = 0.20 and 0.12 for KIP3 and KIP3-Δdistal; p < 0.0001 for the difference in elevation (fluorescence intensity) between the lines. The difference between slopes is not statistically significant. (D) Images of microtubules (GFP-Tub1) in KIP3, KIP3-Δdistal, and kip3Δ cells during anaphase arrest (cdc15-2, 2 h at 37°C). Note that spindles are disassembled in KIP3-Δdistal cells. Cell outline indicated by dotted line. (E) Quantification of spindle stability during anaphase arrest in cells of the indicated genotype. Mean ± SD; p < 0.0001 for KIP3-Δdistal vs. either cell type. (A, D) Bars, 2 μm; (A) n = 62 for both; (B, C) n = 39 for Kip3-3YFP and 53 for Kip3-Δdistal-3YFP; (E) graph shows three experiments where n = 313, 281, 175 for KIP3; 259, 266, 275 for KIP3-Δdistal; and 196, 95, 148 for kip3Δ.
We next examined whether the distal tail regulates Kip3 localization to the midzone. Quantification of Kip3 within the central 3 μm of anaphase spindles greater than 5.5 μm in length revealed that Kip3-Δdistal localization to the central spindle region is significantly increased relative to full-length Kip3 (Figure 7B). Kip3 is a highly processive motor, which allows it to localize in a length-dependent manner along MTs in vitro (Varga et al., 2006) and in the bud compartment of cells with mispositioned spindles (Fukuda et al., 2014). Whether midzone localization of Kip3 is dependent on spindle length, however, has not been determined. Although levels can vary between cells, full-length Kip3 accumulation in the central spindle region is positively correlated with spindle length (Figure 7C). As observed on astral MT plus ends (Gupta et al., 2006), this variation may result from increased localization to the ends of polymerizing versus depolymerizing spindle MTs. While Kip3-Δdistal localization is similarly length dependent, the amount on the central spindle is consistently increased relative to full-length Kip3 over the range of spindle lengths (Figure 7C). Thus, at comparable concentrations in the nucleoplasm, the distal tail limits Kip3 accumulation in the midzone region of anaphase spindles.
Kip3 disassembles spindles on mitotic exit, but not during prolonged delays in anaphase, and our data indicate that the distal tail may be critical for this temporal regulation of anaphase spindle stability. To test this, we held cells in late anaphase with a cdc15-2 mediated arrest and monitored midzone stability (Figure 7D). In most control cells, the midzone remained intact and spindles maintained a length approximately equal to that of the cell (Figure 7E). Similarly, anaphase spindles remained intact in kip3∆ cells. In sharp contrast, spindles were prematurely disassembled during anaphase in cells harboring Kip3-Δdistal (Figure 7, D and E). All together these results demonstrate that the distal region of the Kip3 tail temporally regulates the stability of the spindle midzone during anaphase.
DISCUSSION
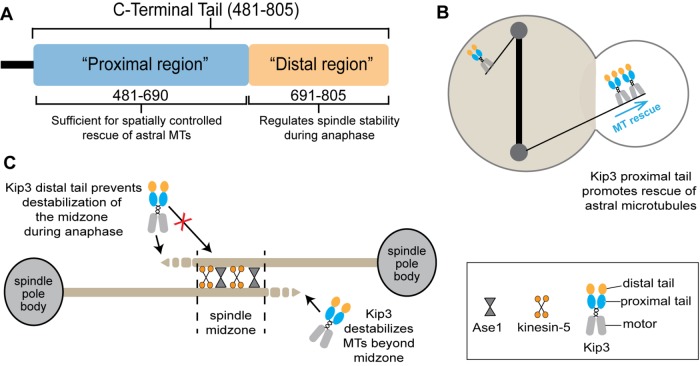
To accomplish diverse cellular functions, the dynamic behavior of MTs must be regulated in both space and time. Kip3 combines several activities into a single motor protein (Gupta et al., 2006; Varga et al., 2006; Su et al., 2011, 2013) and regulates multiple aspects of MT behavior in cells. Our data reveal how the proximal and distal regions of the nonmotor tail section are functionally distinct and play important roles in the spatiotemporal control of specific cellular activities (Figure 8). Although the entire tail is dispensable for the regulation of MT growth at the bud tip during Kar9-dependent spindle positioning, the proximal tail functions to spatially stabilize astral MTs (Figure 8B), and the distal region is critical to control the temporal stability of the anaphase spindle (Figure 8C). Overall, our findings provide fundamental insights into how nonmotor regions contribute to context specific kinesin function and control the spatial and temporal stability of cellular MTs.
FIGURE 8:
Distinct cellular roles of Kip3 are mediated by specific regions of the kinesin tail. (A) Molecular organization of the Kip3 tail. (B) The proximal tail region promotes astral MT rescue spatially near the bud neck in cells with mispositioned anaphase spindles. (C) The distal tail region functions to temporally regulate anaphase spindle stability.
During anaphase, midzone MTs must remain dynamic to allow spindle elongation, yet be sufficiently stable to maintain a robust bipolar structure. Kip3 controls anaphase spindle length by depolymerizing MTs that grow beyond the midzone (Rizk et al., 2014). When spindles are mispositioned, or cells with properly positioned spindles are arrested in late anaphase, Kip3 continues to regulate spindle length. Notably, during these delays the midzone integrity is robust and spindles remain intact. How is it that over these extended periods Kip3 readily destabilizes MTs when they overgrow the midzone yet does not destabilize those within the midzone itself? Moreover, when anaphase is complete, Kip3 functions to efficiently disassemble the spindle (Woodruff et al., 2010). Our data now show that Kip3 activity within the midzone is temporally regulated to control anaphase spindle stability. One model is that the distal tail prevents Kip3 from destabilizing MTs within the midzone during anaphase. Alternatively, the distal tail may promote stabilization of midzone MTs, perhaps via antiparallel MT cross-linking (Su et al., 2013). Removal of only the distal tail (Kip3-Δdistal) induces premature midzone destabilization. If this were simply due to the loss of a stabilizing activity conferred by the distal tail, then removal of Kip3 altogether (kip3Δ) should have a similar effect. Yet removal of Kip3 results in the opposite outcome, a hyperstable midzone and delayed spindle disassembly. Thus, removal of the distal tail results in increased midzone destabilization that is absent when Kip3 is fully removed. This destabilizing activity is therefore likely mediated by Kip3-Δdistal. It remains possible that Kip3 anti-parallel MT cross-linking contributes to midzone stabilization, but in this case it would also serve to prevent destabilization by Kip3. Our results reveal the role of the distal tail region in regulating the timing of spindle disassembly and highlight the need for spatial and temporal mechanisms to ensure the stability of critical MT structures, such as the spindle midzone.
The distal tail of Kip3 could regulate spindle midzone stability through several potential mechanisms. Notably, we show that Kip3 accumulates in the midzone region in a length-dependent manner. A length-dependent increase in midzone MT depolymerization by Kip3 has been proposed as a mechanism to balance spindle elongation with disassembly (Su et al., 2013). A length-dependent increase in Kip3 and its associated depolymerase activity alone, however, does not control spindle disassembly because full-length spindles remain stable during prolonged anaphase arrest. Additional distal tail-dependent mechanism(s) must protect the midzone from Kip3-mediated destabilization during anaphase. Kip3-Δdistal localization to spindles is increased relative to Kip3, and yet it is unlikely the distal tail inhibits MT binding in general because the level on astral MTs remains unchanged. Two potential mechanisms through which the distal tail could regulate Kip3 localization to spindles specifically would be if Kip3 interacts differently with astral and spindle MTs or if midzone-associated factor(s) inhibit Kip3 from accumulating on and/or depolymerizing MTs in that region. It is also possible that the distal tail directly inhibits the activity of the motor domain within the nucleus or spindle midzone.
How Kip3 stabilizes MTs is poorly understood. Tailless Kip3 does not support astral MT rescue, but we find rescue is efficiently promoted by Kip3-Δdistal. Thus, the proximal tail section is sufficient to carry out this MT stabilizing function. This activity may include interactions with the motor domain. Alternatively, the proximal tail could work independently of the motor by interacting directly with the MT (Su et al., 2011) or with additional regulatory factors. Such a mechanism would likely be transferable and may operate in MT regulators other than Kip3/kinesin-8. Similarly to full-length Kip3, Kip3-Δdistal slows MT depolymerization, suggesting that this activity may be coupled to the rescue mechanism. Further defining the region that mediates rescue will allow tests of potential models to elucidate how Kip3 selectively stabilizes a subset of cellular MTs (Fukuda et al., 2014).
Kinesin-8 proteins from diverse species combine motility with various effects on MT dynamics, either in vivo or in vitro (Garcia et al., 2002; West et al., 2002; Gatt et al., 2005; Gupta et al., 2006; Mayr et al., 2007; Du et al., 2010; Kim et al., 2014; Gergely et al., 2016; Walczak et al., 2016; Wang et al., 2016; Möckel et al., 2017). Yet the extent to which MT destabilizing and stabilizing activities are shared among family members has not been firmly established. For example, human cells encode three kinesin-8 proteins, with differing activities, that mediate processes including astral MT dynamics (Stout et al., 2011; Tanenbaum et al., 2011; Walczak et al., 2016), chromosome segregation (Mayr et al., 2007; Stumpff et al., 2008, 2012; Häfner et al., 2014; Kim et al., 2014), and ciliary length (Niwa et al., 2012; Wang et al., 2016). One possibility is that, as the sole kinein-8 in budding yeast, Kip3 combines activities that can be dispersed among multiple family members in higher eukaryotes. However, differences in activity may also reflect the context or regulatory state of specific motors. This study highlights the need to both define the biochemical activities of MT regulators, as well as determine how they may be differentially controlled and integrated into specific cellular contexts.
MATERIALS AND METHODS
Yeast strains and basic assays
Yeast strains are derivatives of the S288C background. Yeast strains and plasmids used and generated in this study are detailed in Supplemental Table S1. Specific mutations and gene deletions were introduced by genetic crossing or fragment mediated homologous recombination. Tailless KIP3ΔT-LZ contains Kip3 residues 1–480 followed by a 31-residue leucine zipper (LZ) motif to reinforce dimerization and is identical to the construct previously reported by Su and colleagues (2011). Strains harboring two copies of KIP3ΔT-LZ (2x KIP3ΔT-LZ) were created by integrating a second copy of KIP3ΔT-LZ and associated regulatory elements downstream of the endogenous locus as previously reported (Fukuda et al., 2014). Kip3 truncations and modifications were verified by DNA sequencing. For tetrad analysis, complete tetrads were verified by proper segregation of the markers in each strain and by mating type testing. Carbendazim sensitivity was assayed by spotting serial dilutions of log phase cultures onto carbendazim-containing plates as described previously (Luchniak et al., 2013).
Microscopy
Imaging of yeast cells was performed on an AxioImager M2 microscope (Carl Zeiss) using a 63 × 1.4 NA Plan-APOCHROMAT oil immersion objective with a piezoelectric-driven Z-stage and a Coolsnap HQ2 charged-coupled device (CCD) camera (Photometrics). Specific fluorophores were imaged using Semrock filter sets (Semrock). Microscope automation, image capture, and analysis were done using SlideBook software (Intelligent Imaging Innovations). Unless otherwise noted, time-lapse movies were acquired using log-phase yeast cultures grown in synthetic complete (SC) media at 24°C. Cells were mounted on 1.2% agarose pads in the same media (Luchniak et al., 2013). For imaging temperature-sensitive mutants, cells were maintained at restrictive temperature using an ASI 400 Air Stream Stage Incubator (Nevtek).
Localization of Kip3 and Kip3-Δdistal
Kip3 was tagged with 3YFP as described previously (Gupta et al., 2006). Kip3-Δdistal was tagged using plasmid pMG162, which contains 500 base pairs of KIP3 coding sequence from amino acids 524–690 fused to a Gly-Ala-Gly-Ala-Gly-Asp-Pro-Pro-Val-Ala-Thr linker to three tandem copies of yellow fluorescent protein (YFP) followed by a stop codon. pMG162 was digested within the KIP3 coding region with SwaI. Transformation of cells with the linear plasmid truncates and tags endogenous KIP3 to create KIP3-Δdistal-3YFP.
To determine localization of Kip3-3YFP and Kip3-Δdistal-3YFP on cyan fluorescent protein–(CFP)MTs, images of YFP and CFP were captured with 10 z-planes spaced at 0.75 μm. Astral MT localization was measured with a 12-pixel spot placed at the MT plus end in the sum projection of Z-series images. Localization to the spindle midzone region was determined with a 4-pixel-wide line covering the center 3 μm of anaphase spindles. Total Kip3 signal in each region was determined by subtracting the neighboring cellular background fluorescence intensity from each pixel within the region being measured. To determine nuclear to cytoplasmic ratios, MTs were depolymerized by 15 min incubation in 35 μg/ml nocodazole followed by fixation in 4% formaldehyde. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI), and fluorescence intensity was measured using two 12-pixel spots inside and outside the nucleus in the single plane that contained the best-focused CFP-Tub1 SPB signal. Cells with a single nucleus and a medium-large sized bud, which represent predominantly anaphase cells were selected for analysis, but because spindles were depolymerized prior to scoring, this category may also include some preanaphase cells.
Microtubule dynamics analysis
Astral MT dynamics were calculated essentially as described previously (Entwistle et al., 2012). Images were captured at 6-s intervals for 6 min, and MT length was calculated as the average of two independent measurements of the three-dimensional length at each timepoint. Based on MT length over time, polymerization and depolymerization was identified as a line through at least four points (24 s) with a length change >0.4 μm and an R2 of at least 0.84. Attenuation was defined as similar durations with net length changes <±0.02 μm. Brief periods that did not fit these criteria were omitted. Rescue events were scored as transition out of depolymerization into polymerization or attenuation, and only time spent depolymerizing was used to calculate the frequency. Catastrophes were scored as transition from polymerization or attenuation into depolymerization, and total time spent polymerizing and attenuated was considered to determine frequency.
To analyze astral MT dynamics in the bud of cells with mispositioned spindles, images were captured at 10-s intervals for 10 min. The location of rescue and catastrophe events in the bud were calculated by dividing the bud into three equal regions as described previously (Fukuda et al., 2014). Only cells with normal morphologies that contained a mispositioned anaphase spindle that was straight and spanned the diameter of the mother cell, with neither pole in direct proximity to the bud neck, were analyzed.
Spindle stability analysis
To score spindle stability during anaphase, cells were grown to log phase and shifted to cdc15-2 restrictive temperature (37°C) for 2 h and imaged on a microscope stage maintained at 37°C using 12 z-planes at 0.75-μm intervals. Spindles were scored as no longer intact when the spindle midzone was visibly disconnected. For time-lapse imaging of spindles, cell images were collected every 45 s for 90 min. Spindle length was measured in three dimensions across z-planes. Spindle length at the time of breakdown was measured using the timepoint 1.5 min prior to breakdown. The rate of spindle elongation was determined by linear regression of the fast and slow phases. Fishhook spindles were defined as those that exceeded cell length, necessitating a bent morphology that wrapped along the cortex ≥25% circumference of the mother or bud compartment (Straight et al., 1998). To score spindle breakdown relative to actinomyosin ring contraction, cells expressing GFP-Tub1 and Myo1-GFP were imaged every 45 s for 60 min using 12 z-planes at 0.75-μm intervals. The stability of each half-spindle was defined as the timepoint at which the half-spindle MTs initiated sustained depolymerization.
Spindle position assay
To score spindle position, cells expressing GFP-Tub1 were imaged using 12 z-planes at 0.75-μm intervals. Only small budded cells with unseparated SPBs were counted. The mother compartment was partitioned into three equal areas with respect to bud position, and spindles were scored according to which area the center of the SPB was located.
Western blotting
Protein was extracted from midlog phase cultures grown at 30°C (OD600 ≈ 0.4). Cells were pelleted (2 min at 4000 × g), resuspended in ice-cold NaOH (0.1 M, 500 μl), incubated for 10 min on ice, and then pelleted again. Cells were resupended in Laemmli buffer and heated at 95°C for 10 min. Samples were incubated 10 min on ice and pelleted again, and supernatant was taken for SDS–PAGE (8% Tris–gly gel, semidry transfer). Antibodies: anti-myc clone 9e10 from mouse (EMD Millipore Corp, Billenca, MA); anti-beta actin from mouse (Abcam ab8224, Cambridge, MA); goat anti-mouse immunoglobulin G horseradish peroxidase conjugate (Pierce Biotechnology, Rockford, IL).
Statistical analyses
Unless otherwise noted, the unpaired, two-tailed student’s t test was used to determine p values. In Figure 5B, R2 values were determined by Pearson correlation coefficient; one-way analysis of covariance was utilized to analyze the difference between the slope and elevation of the two lines representing fluorescence intensity over spindle length.
Supplementary Material
Acknowledgments
We thank G. Barnes and D. Pellman for helpful reagents. This work was supported by a National Institutes of Health grant (R01GM094313) to M.L.G.
Abbreviations used:
- CBZ
carbendazim
- CFP
cyan fluorescent protein
- DAPI
4′,6-diamidino-2-phenylindole
- GFP
green fluorescent protein
- MT
microtubule
- SPB
spindle pole body
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-03-0199) on June 6, 2018.
REFERENCES
- Arellano-Santoyo H, Geyer EA, Stokasimov E, Chen G-Y, Su X, Hancock W, Rice LM, Pellman D. (2017). A tubulin binding switch underlies Kip3/Kinesin-8 depolymerase activity. Dev Cell , 37–51. e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann H, Skiniotis G, Spilker A, Kandels-Lewis S, Vernos I, Surrey T. (2004). A kinesin-like motor inhibits microtubule dynamic instability. Science , 1519–1522. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Gupta ML, Jr, Hoyt MA, Pellman D. (2004). Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev Cell , 815–829. [DOI] [PubMed] [Google Scholar]
- Chandra R, Salmon ED, Erickson HP, Lockhart A, Endow SA. (1993). Structural and functional domains of the Drosophila ncd microtubule motor protein. J Biol Chem , 9005–9013. [PubMed] [Google Scholar]
- Chen Y, Hancock WO. (2015). Kinesin-5 is a microtubule polymerase. Nat Commun , 8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. (1997). Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J Cell Biol , 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy DL, Hancock WO, Wagenbach M, Howard J. (1999). Kinesin’s tail domain is an inhibitory regulator of the motor domain. Nat Cell Biol , 288–292. [DOI] [PubMed] [Google Scholar]
- Desai A, Verma S, Mitchison TJ, Walczak CE. (1999). Kin I kinesins are microtubule-destabilizing enzymes. Cell , 69–78. [DOI] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. (1997). Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J Cell Biol , 1023–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, English CA, Ohi R. (2010). The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr Biol , 374–380. [DOI] [PubMed] [Google Scholar]
- Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED. (1994). Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. Embo J , 2708–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entwistle RA, Rizk RS, Cheng DM, Lushington GH, Himes RH, Gupta ML., Jr (2012). Differentiating between models of epothilone binding to microtubules using tubulin mutagenesis, cytotoxicity, and molecular modeling. ChemMedChem , 1580–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G, Hajdo L, Skowronek KJ, Reuther C, Kasprzak AA, Diez S. (2009). The mitotic kinesin-14 Ncd drives directional microtubule-microtubule sliding. Nat Cell Biol , 717–723. [DOI] [PubMed] [Google Scholar]
- Friedman DS, Vale RD. (1999). Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat Cell Biol , 293–297. [DOI] [PubMed] [Google Scholar]
- Fukuda Y, Luchniak A, Murphy ER, Gupta ML., Jr (2014). Spatial control of microtubule length and lifetime by opposing stabilizing and destabilizing functions of Kinesin-8. Curr Biol , 1826–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Bonaccorsi S, Wentworth D, Doxsey S, Gatti M, Pereira A. (2004). The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol Biol Cell , 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi SR, Gierliński M, Mino A, Tanaka K, Kitamura E, Clayton L, Tanaka TU. (2011). Kinetochore-dependent microtubule rescue ensures their efficient and sustained interactions in early mitosis. Dev Cell , 920–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Koonrugsa N, Toda T. (2002). Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr Biol , 610–621. [DOI] [PubMed] [Google Scholar]
- Gatt MK, Savoian MS, Riparbelli MG, Massarelli C, Callaini G, Glover DM. (2005). Klp67A destabilises pre-anaphase microtubules but subsequently is required to stabilise the central spindle. J Cell Sci , 2671–2682. [DOI] [PubMed] [Google Scholar]
- Gergely ZR, Crapo A, Hough LE, McIntosh JR, Betterton MD. (2016). Kinesin-8 effects on mitotic microtubule dynamics contribute to spindle function in fission yeast. Mol Biol Cell , 3490–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Scholey JM. (2010). Control of mitotic spindle length. Annu Rev Cell Dev Biol , 21–57. [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD. (2005). Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol Biol Cell , 3896–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. (2005). Length control of the metaphase spindle. Curr Biol , 1979–1988. [DOI] [PubMed] [Google Scholar]
- Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. (2006). Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol , 913–923. [DOI] [PubMed] [Google Scholar]
- Häfner J, Mayr MI, Möckel MM, Mayer TU. (2014). Pre-anaphase chromosome oscillations are regulated by the antagonistic activities of Cdk1 and PP1 on Kif18A. Nat Commun , 4397. [DOI] [PubMed] [Google Scholar]
- Hibbel A, Bogdanova A, Mahamdeh M, Jannasch A, Storch M, Schäffer E, Liakopoulos D, Howard J. (2015). Kinesin Kip2 enhances microtubule growth in vitro through length-dependent feedback on polymerization and catastrophe. Elife , e10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. (2011). From electron microscopy to molecular cell biology, molecular genetics and structural biology: intracellular transport and kinesin superfamily proteins, KIFs: genes, structure, dynamics and functions. J Electron Microsc (Tokyo) (Suppl 1), S63–S92. [DOI] [PubMed] [Google Scholar]
- Janson ME, Loughlin R, Loïodice I, Fu C, Brunner D, Nédélec FJ, Tran PT. (2007). Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell , 357–368. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc , 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Fonseca C, Stumpff J. (2014). A unique kinesin-8 surface loop provides specificity for chromosome alignment. Mol Biol Cell , 3319–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W-L, Oberle JR, Cooper JA. (2003). The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol , 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J, Joseph AP, Peña A, Möckel MM, Mayer TU, Topf M, Moores CA. (2017). Structural basis of human kinesin-8 function and inhibition. Proc Natl Acad Sci USA , E9539–E9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchniak A, Fukuda Y, Gupta ML., Jr (2013). Structure-function analysis of yeast tubulin. Methods Cell Biol , 355–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MI, Hümmer S, Bormann J, Grüner T, Adio S, Woehlke G, Mayer TU. (2007). The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol , 488–498. [DOI] [PubMed] [Google Scholar]
- Miller RK, Heller KK, Frisèn L, Wallack DL, Loayza D, Gammie AE, Rose MD. (1998). The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol Biol Cell , 2051–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner M. (1984). Dynamic instability of microtubule growth. Nature , 237–242. [DOI] [PubMed] [Google Scholar]
- Möckel MM, Heim A, Tischer T, Mayer TU. (2017). Xenopus laevis Kif18A is a highly processive kinesin required for meiotic spindle integrity. Biol Open , 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton DA. (1999). The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J Cell Biol , 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. (1999). PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci , 34–36. [DOI] [PubMed] [Google Scholar]
- Niwa S, Nakajima K, Miki H, Minato Y, Wang D, Hirokawa N. (2012). KIF19A is a microtubule-depolymerizing kinesin for ciliary length control. Dev Cell , 1167–1175. [DOI] [PubMed] [Google Scholar]
- Nogales E. (2000). Structural insights into microtubule function. Annu Rev Biochem , 277–302. [DOI] [PubMed] [Google Scholar]
- Pearson CG, Bloom K. (2004). Dynamic microtubules lead the way for spindle positioning. Nat Rev Mol Cell Biol , 481–492. [DOI] [PubMed] [Google Scholar]
- Pereira AJ, Dalby B, Stewart RJ, Doxsey SJ, Goldstein LS. (1997). Mitochondrial association of a plus end-directed microtubule motor expressed during mitosis in Drosophila. J Cell Biol , 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk RS, Discipio KA, Proudfoot KG, Gupta ML., Jr (2014). The kinesin-8 Kip3 scales anaphase spindle length by suppression of midzone microtubule polymerization. J Cell Biol , 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler SC, Liu JY, Pellman D. (2003). The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J Cell Biol , 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D. (2003). Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol , 364–372. [DOI] [PubMed] [Google Scholar]
- Sproul LR, Anderson DJ, Mackey AT, Saunders WS, Gilbert SP. (2005). Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus ends. Curr Biol , 1420–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock MF, Guerrero J, Cobb B, Eggers CT, Huang TG, Li X, Hackney DD. (1999). Formation of the compact confomer of kinesin requires a COOH-terminal heavy chain domain and inhibits microtubule-stimulated ATPase activity. J Biol Chem , 14617–14623. [DOI] [PubMed] [Google Scholar]
- Stout JR, Yount AL, Powers JA, Leblanc C, Ems-McClung SC, Walczak CE. (2011). Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Mol Biol Cell , 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Sedat JW, Murray AW. (1998). Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol , 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Dassow von G, Wagenbach M, Asbury C, Wordeman L. (2008). The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell , 252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Du Y, English CA, Maliga Z, Wagenbach M, Asbury CL, Wordeman L, Ohi R. (2011). A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol Cell , 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Wagenbach M, Franck A, Asbury CL, Wordeman L. (2012). Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev Cell , 1017–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Das DK, Takizawa Y, Shin Y, Collier SE, Ohi MD, Hwang W, Lang MJ, Ohi R. (2014). Kinesin-12 Kif15 targets kinetochore fibers through an intrinsic two-step mechanism. Curr Biol , 2307–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Arellano-Santoyo H, Portran D, Gaillard J, Vantard M, Thery M, Pellman D. (2013). Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat Cell Biol , 948–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Qiu W, Gupta ML, Jr, Pereira-Leal JB, Reck-Peterson SL, Pellman D. (2011). Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/Kinesin-8. Mol Cell , 751–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, van der Vaart B, Galli M, Akhmanova A, Medema RH. (2011). A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr Biol , 1356–1365. [DOI] [PubMed] [Google Scholar]
- van Riel WE, Rai A, Bianchi S, Katrukha EA, Liu Q, Heck AJ, Hoogenraad CC, Steinmetz MO, Kapitein LC, Akhmanova A. (2017). Kinesin-4 KIF21B is a potent microtubule pausing factor. Elife , e24746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. (2006). Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol , 957–962. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Zong H, Jain S, Stout JR. (2016). Spatial regulation of astral microtubule dynamics by Kif18B in PtK cells. Mol Biol Cell , 3021–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Nitta R, Morikawa M, Yajima H, Inoue S, Shigematsu H, Kikkawa M, Hirokawa N. (2016). Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A. Elife , 2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Brust-Mascher I, Cheerambathur D, Scholey JM. (2010). Coupling between microtubule sliding, plus-end growth and spindle length revealed by kinesin-8 depletion. Cytoskeleton (Hoboken) , 715–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LN, Ems-McClung SC, Stout JR, Leblanc C, Shaw SL, Gardner MK, Walczak CE. (2011). Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr Biol , 1500–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RR, Malmstrom T, McIntosh JR. (2002). Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J Cell Sci , 931–940. [DOI] [PubMed] [Google Scholar]
- West RR, Malmstrom T, Troxell CL, McIntosh JR. (2001). Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol Biol Cell , 3919–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Drubin DG, Barnes G. (2010). Mitotic spindle disassembly occurs via distinct subprocesses driven by the anaphase-promoting complex, Aurora B kinase, and kinesin-8. J Cell Biol , 795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.