Abstract
Cell migration is a highly conserved process involving cytoskeletal reorganization and restructuring of the surrounding extracellular matrix. Although there are many studies describing mechanisms underlying cell motility, little has been reported about the contribution of alternative isoform use toward cell migration. Here, we investigated whether alternative isoform use can affect cell migration focusing on reversion-inducing-cysteine-rich protein with Kazal motifs (RECK), an established inhibitor of cell migration. We found that a shorter isoform of RECK is more highly expressed in proliferating fibroblasts, in TGF-β–treated fibroblasts, and in tumors compared with differentiated tissue. Knockdown of this short RECK isoform reduces fibroblast migration through Matrigel. Thus, this short isoform of RECK generated by a combination of alternative splicing and alternative polyadenylation plays an opposing role to the canonical RECK isoform, as knockdown of canonical RECK results in faster cell migration through Matrigel. We show that the short RECK protein competes with matrix metalloprotease 9 (MMP9) for binding to the Kazal motifs of canonical RECK, thus liberating MMP9 from an inactivating interaction with canonical RECK. Our studies provide a new paradigm and a detailed mechanism for how alternative isoform use can regulate cell migration by producing two proteins with opposing effects from the same genetic locus.
INTRODUCTION
Alternative splicing is the incorporation of different exons from the same gene into the final transcript in different contexts (Kornblihtt et al., 2013). Alternative polyadenylation (APA) occurs when the same genetic locus produces multiple transcripts ending at different polyadenylation sites (Tian and Manley, 2013). Combinations of alternative splicing and alternative polyadenylation in the coding sequence of genes can result in the expression of distinct proteins from the same genetic locus (Li et al., 2017). Alternative terminal exon use can convert IgM heavy chain from its membrane-bound to its secreted form (Alt et al., 1980; Takagaki et al., 1996; Peterson, 2011), and receptor tyrosine kinases (RTKs) can be converted to soluble decoy RTKs (Vorlova et al., 2011). Proliferating cells, cancer cells, stem cells in the early stages of mouse development, and tumors tend to use proximal polyadenylation sites (Mayr and Bartel, 2009; Elkon et al., 2012; Morris et al., 2012; Masamha et al., 2014; Rehfeld et al., 2014), possibly due to higher levels of cleavage and polyadenylation factors (Morris et al., 2012).
Matrix metalloproteinases (MMPs) are important contributors to cell migration (Nabeshima et al., 2002). MMPs, structurally related zinc-dependent endopeptidases, degrade extracellular material. Some MMPs, such as MMP2 and MMP9, can be secreted into the extracellular environment, while others like MT1-MMP are membrane-anchored. The role of MMPs in promoting migration and invasion includes clearing away extracellular matrix that may occlude a cell’s advancement (Wolf et al., 2013). In addition, MMPs can generate epitopes that actively promote migration (Zuo et al., 1998). Further, MMPs can also digest proteins involved in adhesion and thereby promote cancer cell invasion (Belkin et al., 2001; Kajita et al., 2001).
Reversion-inducing-cysteine-rich protein with Kazal motifs (RECK), a glycosylphosphatidylinositol (GPI)-anchored MMP inhibitor, has been shown to inhibit cell migration (Oh et al., 2001; Morioka et al., 2009). RECK was originally isolated as a gene that reverts the morphology of transformed cells (Takahashi et al., 1998). RECK was later discovered to inhibit members of the MMP family (MMP-2, MMP-9, and MT1-MMP) and the A Disintegrin And Metalloproteinase (ADAM) family (ADAM10) (Takahashi et al., 1998; Oh et al., 2001, 2004; Miki et al., 2007; Chang et al., 2008; Hong et al., 2014). RECK’s ability to inhibit matrix metalloproteinases is important for its role in suppressing cell migration (Takahashi et al., 1998). RECK−/− mouse embryonic fibroblasts (MEFs) exhibit a lack of anterior-posterior polarity, faster migration, more rapid changes in direction as they migrate, and fewer focal adhesions and actin stress fibers compared with controls (Morioka et al., 2009). RECK−/− MEFs also contained increased levels of GTP-bound Rac1 and Cdc42 (Morioka et al., 2009).
We hypothesized that changes in the expression of different gene isoforms might be related to the migratory capacity of cells. We report here on our studies investigating the role of RECK isoforms in fibroblast migration. We propose a novel model in which changes in the capacity for migration are mediated through the expression of different isoforms of RECK.
RESULTS
The ratio of a short RECK isoform to canonical, long RECK is higher in proliferative and TGF-β–treated cells
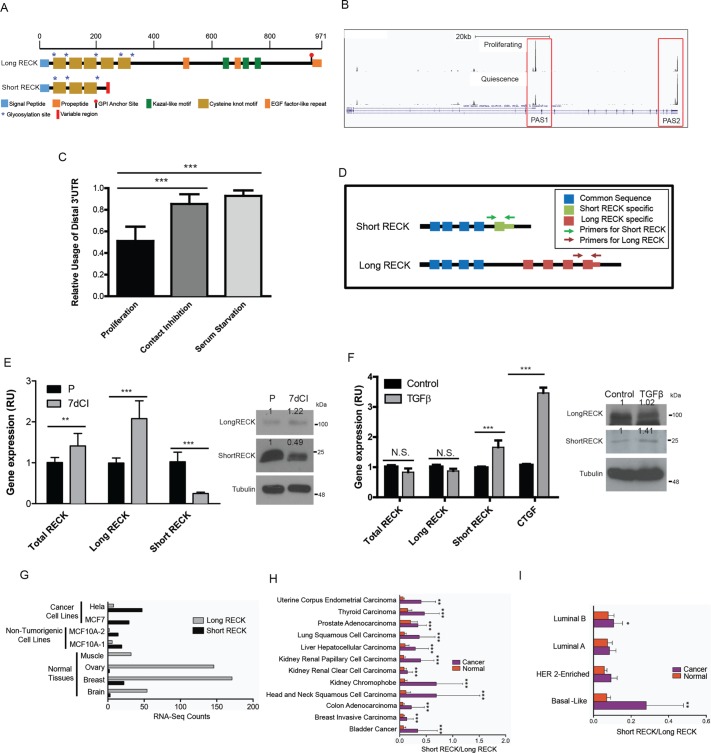
Our laboratory applied a targeted methodology, polyadenylation site–enriched RNA-Seq, to monitor the extent to which transcripts terminate at different possible polyadenylation sites in order to identify genes that undergo APA when primary human dermal fibroblasts exit the proliferative cell cycle and enter quiescence (unpublished data). The Gnomegen RNA-Seq Library Preparation Kit was used to create cDNA libraries containing fragments enriched for 3′ UTR ends. To generate these libraries, mRNA was fragmented; transcripts were incubated with poly(T) sequences attached to biotin; polyadenylated transcripts were captured with streptavidin beads; and polyadenylation sites were sequenced on an Illumina sequencer. Our data show that RECK can be expressed in dermal fibroblasts as either a long, canonical isoform (long RECK) or shorter isoforms generated by a combination of alternative splicing and APA (short RECK; Figure 1A). The transcript encoding the longer RECK isoform was more highly expressed in fibroblasts induced into quiescence by contact inhibition of proliferation or serum starvation, while the transcript encoding the shorter RECK isoform was expressed more highly in proliferating fibroblasts (Figure 1B). In three independent poly(A)-enriched RNA-Seq experiments, the full-length RECK isoform (long RECK) exhibited a statistically significant shift toward increased use of the distal polyadenylation site in fibroblasts induced into quiescence via contact inhibition of proliferation or serum starvation, compared with proliferating fibroblasts (p < 0.001, Student’s t test; Figure 1C). Because the short RECK transcript includes a 3′ UTR that is eliminated via splicing from the long RECK transcript, we could design real-time reverse transcriptase-PCR (RT-PCR) primers specific for the long or short RECK isoforms, in addition to primers that recognize both isoforms (Figure 1D). Real-time RT-PCR with isoform-specific primers confirmed increased expression of the long RECK isoform and decreased expression of the short RECK isoform in fibroblasts induced into quiescence by 7 d of contact inhibition (7dCI) compared with proliferating fibroblasts (P; Figure 1E). The short RECK isoform encodes a shorter protein (NM_001316348.1, 25 kDa), distinct from the protein encoded by the longer, canonical RECK (NM_021111.2, 110 kDa; Figure 1A). The final, 13-amino-acid exon of short RECK and the 3′ UTR of short RECK are not present in the mRNA encoding long RECK. These distinctions between the amino acid sequences of the proteins encoded by the short and long RECK isoforms allowed us to design short RECK-specific antibodies that recognize short RECK’s unique final exon (Supplemental Figure S1). Immunoblotting with this short RECK-specific polyclonal antibody confirmed that short RECK protein levels are lower in fibroblasts induced into quiescence by 7 d of contact inhibition than proliferating fibroblasts (Figure 1E).
FIGURE 1:
Short RECK isoform levels are elevated in proliferating and TGF-β–treated fibroblasts and cancer cells. (A) Schematic showing the short RECK isoform (NM_001316348.1) and the long RECK isoform (NM_021111.2). The short RECK isoform (molecular weight = 25 kDa) shares 212 amino acids with the long RECK isoform (molecular weight = 110 kDa) and contains a 13 amino acid–long sequence specific for the short RECK isoform at its C-terminus. (B) Poly(A) site–enriched RNA-Seq data from proliferating and serum-starved fibroblasts for RECK. PAS1 indicates the proximal polyadenylation site that produces the short RECK isoform, and PAS2 indicates the distal polyadenylation site that produces the long RECK isoform. (C) Average relative usage of the distal isoform (RUD) plotted for RECK in proliferating, 7-d contact inhibition of proliferation, and 7-d serum starvation fibroblasts with poly(A) site–enriched RNA-Seq. Data were generated in three independent biological replicates and error bars reflect SD. RECK RUD values for contact inhibition (p < 0.001, unpaired two-sided t test) and serum starvation (p < 0.001, unpaired two-sided t test) conditions are significantly higher than RUD values for proliferating cells. Averages and SD are shown. (D) Diagram illustrating primers targeting specific RECK isoforms. (E) RECK isoform expression in proliferating and contact-inhibited fibroblasts. Real-time RT-PCR analysis of total RECK, long RECK, and short RECK mRNA expression under proliferating (P) or 7-d contact inhibition (7dCI) conditions was performed. Data are shown as relative units (RUs) compared with the baseline state, total RECK in proliferating conditions, which is represented as 1 and indicates the target transcript divided by the internal control. Total RECK mRNA increases with quiescence induced by contact inhibition of proliferation (p < 0.01, unpaired two-sided Student’s t test). Long RECK mRNA expression increases in 7-d contact inhibition conditions (p < 0.001, unpaired two-sided Student’s t test), whereas short RECK mRNA expression decreases in the same condition (p < 0.001, unpaired two-sided Student’s t test). RECK was amplified with real-time primers and normalized to UBC control primers (Materials and Methods). Averages and SD are shown. On the right, immunoblotting of proliferating and 7-d contact-inhibited fibroblast protein lysates with an antibody against the long RECK isoform and a polyclonal antibody against the short RECK isoform is shown. Tubulin served as a loading control. (F) RECK isoform expression with TGF-β treatment. Real-time RT-PCR data for RECK isoform expression in fibroblasts treated with TGF-β. CTGF served as a positive control for TGF-β response. mRNA levels of total RECK or long RECK do not change with TGF-β treatment. mRNA expression of the short RECK isoform increases with TGF-β treatment (p < 0.001, unpaired two-sided Student’s t test). Data are shown as relative units compared with the control condition. There was no significant difference in long RECK mRNA expression levels when fibroblasts were treated with TGF-β (p = 0.18, Student’s t test). Averages and SD are shown. Immunoblotting for the long RECK isoform, the short RECK isoform, and tubulin are shown on the right for control fibroblasts and fibroblasts treated with TGF-β. Short RECK protein levels increase in response to TGF-β treatment. (G) Cancer cell lines have higher short-to-long RECK isoform ratios compared with differentiated tissues such as brain, breast, ovary, and muscle. Published data on the frequency of genome-wide polyadenylation site usage in multiple tissues and cancer cell lines (Lianoglou et al., 2013) was analyzed for the number of reads mapping to the last exon of the long or short RECK isoform. (H) A higher ratio of short to long RECK isoforms is consistently observed in tumors. Per-exon RNA-Seq read counts for tumor tissues and paired normal tissues along with any available clinical data were obtained from the TCGA public data portal. For comparison of RECK expression between tumor and paired normal tissue, read counts were normalized to the total number of reads per sample. RNA-Seq read counts assigned to the unique short RECK isoform exon and the final long RECK exon were used to infer expression of short and long RECK isoforms, respectively. Short-to-long RECK isoform ratios, as well as normalized short and long RECK read counts, were determined for tumor and paired normal samples. Two-tailed paired t tests were performed separately for each tumor type using GraphPad Prism 6.04. (I) To determine RECK isoform expression in breast cancers with different clinical characteristics, the relative use of the distal isoform (RUD) was calculated and the log2-fold change between tumor and paired normal was determined and plotted based on breast cancer subtype. Breast cancer subtypes (basal-like, HER2-enriched, Luminal A, or Luminal B) were determined based on the PAM50 subtype, a 50-gene breast cancer subtype predictor (Parker et al., 2009). The basal-like gene expression pattern overlaps most significantly with triple-negative breast cancers. Statistical significance was determined by one-way ANOVA followed by Tukey post-hoc test for two-way comparisons to control for multiple hypothesis testing. For all figures, *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Treating fibroblasts with TGF-β, a strong inducer of fibroblast migration (Cordeiro et al., 2000; Acharya et al., 2008), resulted in increased expression of short RECK isoform transcripts (Figure 1F) and short RECK protein (Figure 1F). We analyzed a published data set on the use of polyadenylation sites in tissues and cancer cell lines (Lianoglou et al., 2013). We counted the number of polyadenylation site–enriched RNA-Seq reads for each sample that mapped either to the final exon of the short RECK isoform or the final exon of the long RECK isoform. We found that differentiated tissues such as ovary, muscle, breast, and brain contained more RNA-Seq reads mapping to the final exon of the long RECK isoform and fewer reads mapping to the last exon of the short RECK isoform (Figure 1G). In contrast, the tumorigenic cancer cell lines (HeLa and MCF7) and the nontumorigenic MCF10a cell line contained more reads mapping to the last exon of the short RECK isoform and fewer reads mapping to the final exon of the long RECK isoform (Figure 1G). We also investigated RNA-Seq data from The Cancer Genome Atlas (TCGA). RNA-Seq read counts for each exon were obtained from tumor tissues and paired normal tissues through the public data portal. RNA-Seq read counts assigned to the unique short RECK isoform final exon and the long RECK final exon were used to infer expression of short and long RECK isoforms, respectively. This analysis revealed that the ratio of short RECK to long RECK increased in all 13 tumors analyzed compared with normal tissue from the same site (Figure 1H). Levels of long RECK decreased in eight types of cancer compared with normal tissue, while levels of short RECK increased in seven types of cancer compared with normal tissue (Supplemental Figure S2). When we investigated breast cancer in more depth, we discovered that the ratio of short to long RECK was particularly high in the basal-like breast cancer subtype, which largely overlaps with the highly metastatic triple negative breast cancer subtype (Figure 1I; Lin et al., 2008; Tseng et al., 2013). Altogether, the short isoform is induced in proliferating versus quiescent fibroblasts, TGF-β–treated versus control fibroblasts, and cancer cells versus differentiated cells.
Knockdown of short RECK reduces fibroblast migration
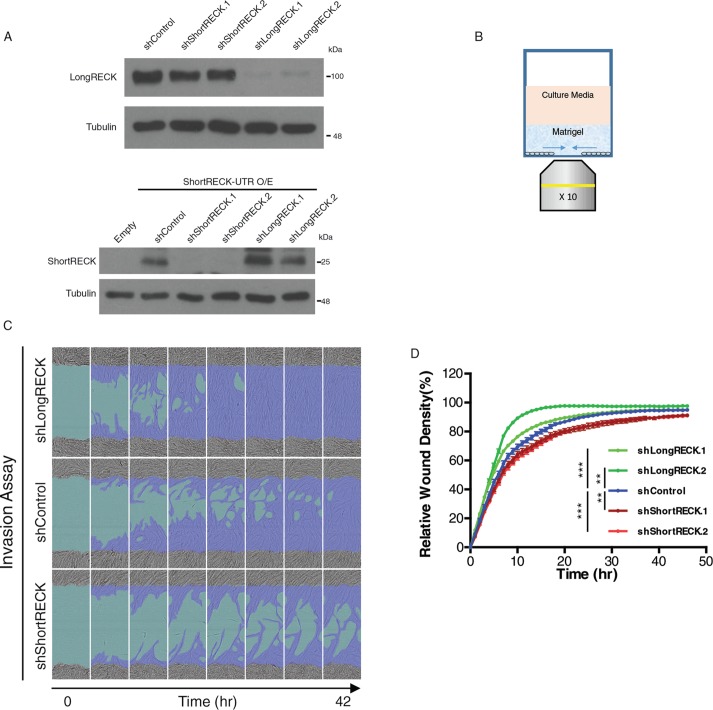
To understand the role of RECK isoforms in cell migration, we generated lentiviral vectors expressing short hairpin RNAs (shRNAs) that specifically target either short or long RECK and introduced them into HEK293T cells (Figure 2A). These isoform-specific shRNAs did not affect protein levels of the other isoform (Figure 2A). We used a real-time imaging instrument to monitor cell migration in fibroblasts expressing shRNAs that target RECK isoforms or fibroblasts expressing control shRNAs (Figure 2B). To specifically test the effects of RECK on the ability of cells to migrate through the extracellular matrix, we denuded an area on a tissue culture well, and coated the well with Matrigel (Figure 2B). By monitoring cell migration into Matrigel, we observed that knockdown of long RECK resulted in increased fibroblast migration (Figure 2, C and D, and Supplemental Movie S1), consistent with previous reports (Mahl et al., 2016). Strikingly, knockdown of short RECK resulted in the opposite result, decreased migration (Figure 2, C and D, and Supplemental Movie S1). To clarify whether these differences in cell migration could reflect effects of RECK isoforms on cell proliferation rather than a difference in the cells’ migration, we monitored cell proliferation rates using a WST-1 assay. No significant differences in proliferation rates were discovered over a 48 h time course after either short RECK or long RECK knockdown (Supplemental Figure S3A). Overexpression of the short RECK isoform, the long RECK isoform, or both together did not affect fibroblast migration rates through Matrigel (Supplemental Figure S3B). Taken together, our findings show that cell migration can be regulated through changes in RECK isoform expression. Our results further demonstrate that reducing the levels of the long RECK isoform promotes cell migration while reducing the levels of the short RECK isoform results in slower cell migration.
FIGURE 2:
The short RECK isoform is essential for cell migration. (A) Western blot results showing the effects of shRNAs that target RECK isoforms on RECK expression levels. On top, shRNAs that target the long RECK isoform expressed in HEK293T cells reduce levels of the long RECK isoform, while shRNAs that target the short RECK isoform have little effect on levels of the long RECK isoform. On the bottom, HEK293T cells overexpressing short RECK with its 3′ UTR were used to monitor the effects of knockdown of short and long RECK on the short RECK isoform. shRNAs that target the short RECK isoform resulted in down-regulation of the short RECK isoform, whereas shRNAs that target the long RECK isoform had little effect on levels of the short RECK isoform. (B) Schematic depicting the methodology for testing cell invasion. (C, D) Isoform-specific RECK knockdown affects fibroblast migration. Fibroblasts expressing shRNAs that target the long RECK isoform migrate more rapidly than fibroblasts expressing a control shRNA. (C) Still images taken from movies of migrating cells. (D) Quantifications of relative wound density (100 * density in wound/density outside wound) over time. shControl vs. shLongRECK.1 (p < 0.001, repeated measures two-way ANOVA with Dunnett’s multiple comparison test [simplified as ANOVA]), shControl vs. shLongRECK.2 (p < 0.01, ANOVA). Fibroblasts expressing shRNAs that target the short RECK isoform migrate more slowly than fibroblasts expressing a control shRNA. shControl vs. shShortRECK.1 (p < 0.01, ANOVA), shControl vs. shShortRECK.2 (p < 0.001, ANOVA). Averages and SEM are shown.
Movie S1.
Supplementary Movie 1
RECK isoforms do not change the activity of the AKT, ERK, or JNK signaling pathways
We then tested whether knockdown of a RECK isoform would affect the activity of signaling pathways established as regulators of cell migration (Klemke et al., 1997; Hunger-Glaser et al., 2003; Montero et al., 2003; Huang et al., 2004; Xue and Hemmings, 2013). We tested the activity of the AKT, ERK, and JNK signaling pathways using total and phosphospecific antibodies. We did not detect significant changes in the levels of total or phosphorylated AKT, ERK, or JNK in short or long RECK knockdown fibroblasts (Supplemental Figure S4). Any minor changes observed in the levels of the phosphorylated forms of these proteins did not support a role for any of these pathways in the changes in cell migration induced by RECK isoforms. These observations suggest that fibroblast cell migration can be regulated independently of these established cell migration signaling pathways.
Short RECK and long RECK interact with each other
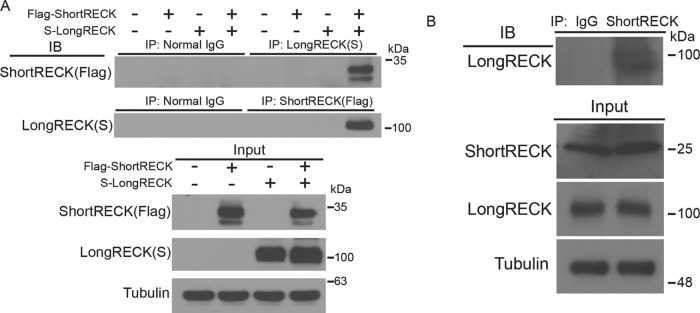
Alternative terminal exon use can result in the production of shorter, truncated protein isoforms that interfere with and inhibit the full-length protein’s activity (Yao et al., 2012). Previous studies have shown that long RECK proteins dimerize (Omura et al., 2009). With this in mind, we hypothesized that short RECK may inhibit the anti-migratory function of long RECK through protein–protein interactions. We introduced Flag-tagged-short RECK and S-tagged-long RECK into 293T cells and observed an interaction when we immunoprecipitated for either short RECK or long RECK (Figure 3A). Using coimmunoprecipitation, we also confirmed previous studies that the long RECK isoform can interact with itself (Omura et al., 2009) (Supplemental Figure S5A). In addition, we observed that the short RECK isoform can interact with itself as well (Supplemental Figure S5B).
FIGURE 3:
The short RECK isoform interacts with the long RECK isoform. (A) Coimmunoprecipitation revealed an interaction between the short and long RECK isoforms. Both Flag-tagged short RECK and S-tagged long RECK isoforms were overexpressed in 293T cells. Protein lysates were analyzed with coimmunoprecipitation, and an association was observed between short and long RECK isoforms. The association was observed when either the short RECK or long RECK isoform was used for immunoprecipitation. Expression levels of input proteins are shown at the bottom. (B) Coimmunoprecipitation with endogenous short RECK and long RECK isoforms. Cell lysates were collected from primary cultured human fibroblasts, proteins were pulled down using a short RECK-specific antibody, and immunoblotting was performed with a long RECK-specific antibody.
Coimmunoprecipitation of endogenous short and long RECK with a commercially available long RECK-specific antibody and the antibody we generated against short RECK revealed short RECK and long RECK interact in human fibroblasts (Figure 3B). In sum, our data show that short RECK generated via a combination of alternative splicing and alternative polyadenylation can bind to canonical long RECK in human fibroblasts.
Short and long RECK associate with each other in the secretory pathway and on the cell surface
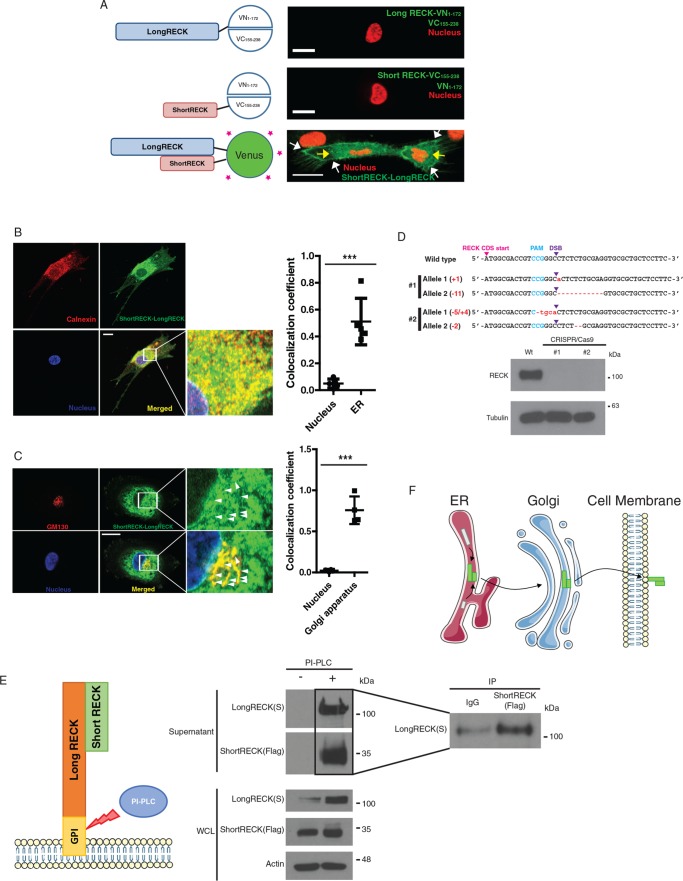
To understand how and where short RECK might affect the role of the long, canonical RECK protein as a binding partner, we visualized the interaction between short and long RECK using a bimolecular fluorescence complementation assay (BiFC; Figure 4A). Introduction of the N-terminus of Venus and the C-terminus of Venus with neither conjugated to RECK resulted in diffuse green fluorescence throughout the cell (Supplemental Figure 6A). Fibroblasts that were transduced with a lentiviral vector expressing the N-terminus of Venus (Supplemental Figure 6B), the C-terminus of Venus (Supplemental Figure 6B), long RECK fused to the N-terminus of Venus 1-172 (Supplemental Figure 6B), or short RECK fused to the C-terminus of Venus 155-238 (Supplemental Figure 6B) exhibited no fluorescent signal. Fibroblasts expressing the N-terminus of Venus and the C-terminus of Venus exhibited fluorescence throughout the cell (Supplemental Figure 6B). Fibroblasts expressing the long isoform of RECK fused to the N-terminus of Venus and the unlabeled C-terminus of Venus, or the short isoform of RECK fused to the C-terminus of Venus and the unlabeled N-terminus of Venus exhibited no fluorescence (Figure 4A, top and middle). Fibroblasts expressing both short RECK fused to the C-terminus of Venus and long RECK fused to the N-terminus of Venus (short RECK–Long RECK BiFC fibroblasts) exhibited fluorescent signals in the perinuclear region of the cell and on the cell surface (Figure 4A, bottom). To further investigate the subcellular location of the interaction between short and long RECK, we stained short RECK–Long RECK BiFC fibroblasts with GM130, a Golgi marker, or calnexin, an ER marker. We observed that ∼50% of the short RECK–long RECK interaction overlaps with calnexin (Figure 4B), and ∼70% of the signal from the Golgi marker overlaps with the short RECK–long RECK interaction signal (Figure 4C). The overlap between the localization of short RECK and long RECK was also observed on the cell surface using immunofluorescence staining without permeabilization (Supplemental Figure S6C).
FIGURE 4:
A short RECK–long RECK complex is localized in the perinuclear region and on the cell surface. (A) The N-terminal half of Venus (1–172) was conjugated to the long RECK isoform, and the C-terminal half of Venus (155–238) was conjugated to the short RECK isoform for bimolecular fluorescence complementary (BiFC) assays. When either long RECK-VN and free VC or short RECK-VC and free VN were overexpressed, no green signal was observed (top and middle). When both long RECK-VN and short RECK-VC were overexpressed in the same cell, a green signal was observed on the cell surface (white arrows) and in the perinuclear region (yellow arrows; bottom). The scale bar indicates 20 μm. (B) Immunofluorescence staining of short RECK-long RECK BiFC cells (green) with calnexin, an ER marker (red). About 50% of the short RECK–long RECK interaction signal overlaps with calnexin. The scale bar indicates 20 μm. Averages and SD are shown. p < 0.001, unpaired two-sided Student’s t test. (C) Immunofluorescence staining of short RECK–long RECK BiFC cells with GM130, a Golgi apparatus marker. The green signal indicating tubule structures around the nucleus (white arrowheads) overlapped with the red GM130 signal to produce a merged yellow signal. About 70% of GM130 overlaps with the short RECK–long RECK interaction signal. The scale bar indicates 20 μm. Averages and SD are shown. p < 0.001, unpaired two-sided Student’s t test. (D) Characterization of a RECK knockout HEK293T cell line established by CRISPR/Cas9. Top panel, sequencing results from two different RECK knockout cell lines. Frameshift mutations were introduced into the RECK sequence by introducing Cas9 endonuclease and a guide RNA targeting RECK (PAM, protospacer adjacent motif; DSB, double-strand break). Bottom panel, Western blot results showing knockout of RECK in CRISPR/Cas9-engineered 293Ts. (E) Left panel, strategy to detect short RECK–long RECK complex on the cell surface. PI-PLC cleaves the GPI anchor attaching long RECK to the cell surface, releasing the complex into the supernatant. Middle panel, Western blot results showing the release of short RECK on the cell surface by PI-PLC treatment. The short RECK isoform, which does not have a GPI anchor, is liberated from the cell surface when long RECK is cleaved from the cell surface by PI-PLC. Right panel, liberated short RECK and long RECK isoforms in the supernatant interact based on coimmunoprecipitation. WCL, whole cell lysate. (F) Hypothesized model for the trafficking of a short RECK–long RECK complex. According to the model, short RECK–long RECK complexes are formed in the ER and then transported to the cell surface via the Golgi apparatus.
To further explore how long and short RECK interact with each other, we used CRISPR/Cas9 technology to specifically mutate and inactivate the gene encoding RECK in 293T cells. Sequencing revealed inactivating frameshifts in both RECK alleles (Figure 4D, top) and immunoblotting revealed a lack of expression of RECK (Figure 4D, bottom). Using RECK−/− 293T cells, we tested more specifically for short RECK–long RECK interaction on the cell surface. We overexpressed S-tagged long RECK and Flag-tagged short RECK and treated with phosphatidylinositol-specific phospholipase C (PI-PLC) to cleave the long RECK GPI anchor (Figure 4E, left). Immunoblotting of conditioned medium revealed increased levels of both short and long RECK upon PI-PLC treatment (Figure 4E, middle). Coimmunoprecipitation of conditioned media from PI-PLC–treated cells revealed the interaction between short and long RECK, further supporting our finding that short RECK can bind to long RECK on the cell surface (Figure 4E, right). Taken together, our data support a model in which there is an interaction between short and long RECK, that this interaction is initiated within the ER, and that a complex containing both protein isoforms follows the protein secretion pathway via the Golgi apparatus to finally localize on the cell surface (Figure 4F).
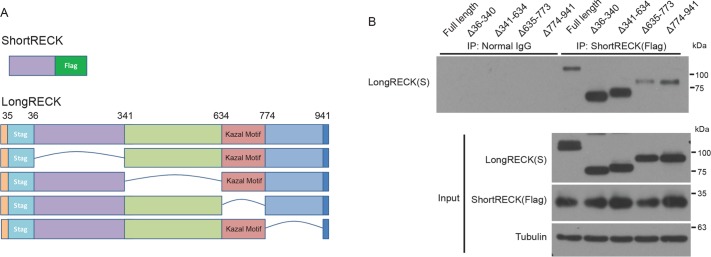
Short RECK interacts with long RECK at the Kazal motif region
To dissect the region within long RECK that mediates interaction with short RECK, we engineered a series of deletion mutants of long RECK (Figure 5A). All of the engineered RECK proteins were expressed (Figure 5B, input). As the deleted regions shifted closer to the GPI anchor, the efficiency with which they localized on the cell surface and were released by PI-PLCase treatment decreased (Supplemental Figure 7A). We introduced Flag-tagged short RECK and one of the deletion mutants of S-tagged long RECK into RECK-/- 293T cells and tested their interaction with coimmunoprecipitation. Immunoprecipitation of Flag-tagged–short RECK showed dramatically reduced binding for the long RECK ∆635–773 mutant, indicating that the region between amino acids 635 and 773 of long RECK is important for short RECK binding (Figure 5B). GST–pull down assays also showed similar results (Supplemental Figure S7B). Amino acids 635–773 of RECK include the Kazal motif region shown to be important for suppressing MMP activity through protein–protein interactions (Miki et al., 2007; Chang et al., 2008). We performed a similar experiment to compare the binding sites for short RECK–long RECK interaction with the region(s) important for long RECK–long RECK interaction. We cointroduced long RECK with a Myc tag and one of a series of deletion mutants of long RECK with an S-tag. Coimmunoprecipitation revealed that the C-terminal region of the long RECK isoform, amino acids 774–941, were important for long-RECK–long RECK interaction (Supplemental Figure S7C). This mutant was missing multiple C-terminal amino acids but retained the GPI anchor. The localization of long RECK–long RECK interaction to the C-terminus is consistent with an earlier report that came to a similar conclusion based on single-particle reconstruction (Omura et al., 2009). Taken together, our findings about the interaction among RECK isoforms support the importance of the Kazal motif region for long RECK–short RECK interaction.
FIGURE 5:
The Kazal-like motif domain is important for short RECK binding to long RECK. (A) Schematic depiction of the structure of the short RECK isoform conjugated to Flag and versions of the long RECK isoform with domain-specific deletions constructed for coimmunoprecipitation. (B) Results from a coimmunoprecipitation experiment with the constructs shown in A. Input samples are shown at the bottom. Short RECK–long RECK interaction was reduced when the domain containing the Kazal-like motif was deleted (∆635-773).
Short RECK competes with MMP9 for binding to long RECK
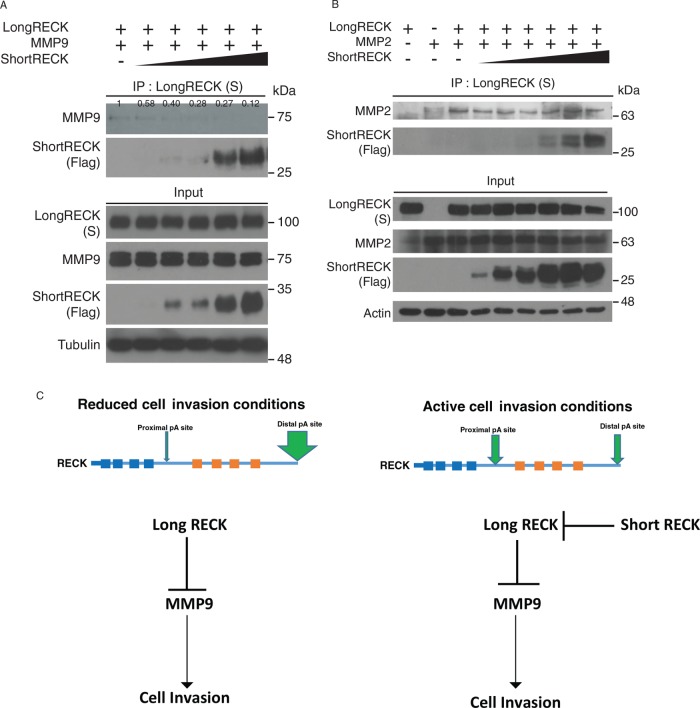
Because short RECK interacts with long RECK in a region that includes the Kazal motifs, we hypothesized that short RECK competes with MMPs for binding to long RECK. To test this model, we introduced long RECK, MMP2, or MMP9, and increasing concentrations of short RECK into RECK−/− 293T cells for competitive coimmunoprecipitation. Increasing short RECK expression reduced coimmunoprecipitation between MMP9 and long RECK by as much as 88% of the binding level observed without short RECK (Figure 6A). Binding to MMP2 was unaffected (Figure 6B). We also tested whether short RECK can disrupt dimerization of long RECK (Chang et al., 2008; Omura et al., 2009). Using coimmunoprecipitation, we found that introducing increasing amounts of short RECK did not affect the ability of long RECK to dimerize (Supplemental Figure S8). These results show that short RECK can compete with MMP9 for binding at the Kazal motif region of long RECK, and does not interfere with long RECK dimerization.
FIGURE 6: .
The short RECK isoform competes with MMP9 for binding to the long RECK isoform. (A) We generated RECK-null 293T cells that overexpress the long RECK isoform and MMP9. Increasing amounts of the short RECK isoform was transfected into cells as shown on the top of each lane. Western blots of input for immunoprecipitation are shown at the bottom. Samples were immunoprecipitated for the long RECK isoform with an antibody against S-tag and immunoblotted for the short RECK isoform with an anti-Flag antibody or an antibody against MMP9. The interaction between MMP9 and the long RECK isoform decreased with increasing levels of the short RECK isoform. The number above each band indicates relative expression level quantified with ImageJ. (B) RECK-null 293T cells overexpressing the long RECK isoform, MMP2, and increasing levels of the short RECK isoform were analyzed by coimmunoprecipitation. Western blots of input are shown at the bottom. Samples were immunoprecipitated with an antibody to S protein and analyzed with immunoblotting for the short RECK isoform and MMP2. The interaction between MMP2 and the long RECK isoform was not affected by increasing levels of the short RECK isoform. (C) Working model. In reduced cell migration conditions, the long RECK isoform tends to be expressed, resulting in a low short RECK:long RECK ratio. Under these conditions, the long RECK isoform suppresses MMP9 activity to inhibit cell migration. In contrast, under conditions of active cell migration, polyadenylation factors target proximal polyadenylation sites, resulting in a high short RECK:long RECK ratio. Short RECK inhibits the functions of long RECK through protein–protein interaction, leading to increased free MMP9, which can contribute to increased cell invasion.
DISCUSSION
Our studies support a model in which alternative isoform use can regulate cell migration through its effects on the protein RECK. Several previous studies have provided evidence for a correlation between isoform length and cell migration. For example, during early stages of mouse development, transcripts tend to undergo polyadenylation at proximal polyadenylation sites, resulting in shorter 3′ UTRs (Ji et al., 2009). Similarly, widespread shortening of the 3′ UTR region of oncogenic mRNA transcripts is also observed in cancer cells (Mayr and Bartel, 2009). Considering that enhanced cell migration is a characteristic of cells during early developmental stages and proliferative cancer cell conditions (Friedl and Gilmour, 2009; Aman and Piotrowski, 2010), these findings would support a correlation between isoform length and cell migration.
We discovered that the short isoform of RECK is expressed more highly during proliferation and after treatment with TGF-β. Moreover, when we analyzed tissue samples from cancer patients, we observed more short RECK expression in cancer tissue than normal tissue biopsies from the same site. The most aggressive breast cancer type, the basal type, had the highest ratio of short RECK to long RECK (Figure 1). We then demonstrated based on shRNA-mediated knockdown that short RECK reduces cell migration (Figure 2). This was especially surprising given that previous studies have reported that canonical RECK is a migration inhibitor. We considered two models for the effects of short RECK on cell migration: 1) short RECK could activate cell migration–related signaling pathways, or 2) short RECK could suppress the role of long RECK in cell migration. We examined three representative signaling pathways (AKT, ERK, and JNK) that have established roles in cell migration (Klemke et al., 1997; Hunger-Glaser et al., 2003; Montero et al., 2003; Huang et al., 2004; Xue and Hemmings, 2013), but did not observe any significant changes following short RECK knockdown (Supplemental Figure S4). Although it is possible that short RECK has the potential to activate other signaling pathways that we have not studied, we turned our attention toward the effects of short RECK on long RECK. We discovered that short and long RECK can interact with each other via protein–protein interactions (Figures 3 and 4). Using bimolecular fluorescence, we visualized this interaction as a complex that appeared in the ER, the Golgi apparatus, and the cell surface (Figure 4). We also demonstrated that short RECK binds to long RECK at the Kazal motif region using domain-specific deletion mutants of long RECK. Deletion of the Kazal motif region resulted in a reduced ability for the two isoforms to interact with each other (Figure 5). Finally, we demonstrate that short RECK competes with MMP9 for binding to the Kazal motif of long RECK (Figure 6). Through the generation of short versus long RECK in proliferative and migratory cells, alternative isoform use can affect MMP activity, the quality of the extracellular matrix, and the ability of cells to migrate.
A previous study has shown that receptor tyrosine kinase 1 (RTK1) can undergo APA and the short isoform of RTK1 can act as a dominant negative and as a decoy for active RTK1 (Vorlova et al., 2011). This situation is similar to the model we propose for RECK in that a shorter isoform can inhibit the activity of the full-length isoform, but in the case of RECK, we do not have evidence that short RECK is displacing one of the versions of long RECK in a dimer. In fact, our competition assays showed no effects on canonical RECK dimerization as short RECK was introduced (Supplemental Figure S8).
Previous studies have shown that the activity of MMP2 or MMP9 is reduced by interaction with long RECK (Oh et al., 2001; Chang et al., 2008). In our study, interactions among RECK, MMP9, and MMP2 were confirmed. One interesting finding in our study was that, while the short RECK isoform outcompetes MMP9 in binding to long RECK, short RECK cannot effectively compete with MMP2 for binding to the long RECK isoform. This might be attributed to the fact that among all secreted MMPs, only MMP9 forms a homodimer (Dufour et al., 2010), which could affect its interaction with other binding partners. Further studies on the structure of these complexes will be required to assess this possibility.
The short RECK isoform that we have been studying is not the only shorter isoform of the RECK gene. In a recent publication, the authors reported on two short isoforms of RECK, RECK-B, and RECK-I (Trombetta-Lima et al., 2015). Neither of these is the same as the isoform we have been studying. The study did not observe any significant effects of overexpressing either of these two short RECK isoforms in cultured glioblastoma cell lines for their role in cell migration or invasion, but did discover that overexpressing RECK-B promoted anchorage-independent growth. In our studies, our polyadenylation site–enriched RNA-Seq methodology would detect all three short isoforms. The real-time PCR we performed was specific for the short RECK isoform that we have been investigating. The polyclonal antibodies we developed are also specific for the isoform we are studying. One of the shRNAs directed against the short RECK isoform we have been investigating would target the isoform we are studying and RECK-B. The other shRNA would target the isoform we have been studying, RECK-B and RECK-I.
Our findings raise the possibility that short RECK is an important and previously unrecognized contributor to cancer cell invasion. Although long RECK is considered a tumor suppressor (Takemoto et al., 2007; Chang et al., 2008; Kim et al., 2008), long RECK is infrequently mutated in tumors (cBioportal; Cerami et al., 2012), raising the possibility that other isoforms generated from the same locus promote cancer. The increased levels of the short RECK to long RECK ratio observed in multiple tumors are consistent with a possible role for RECK alternative isoform expression in tumor progression. Our studies also raise the possibility that when fibroblasts are activated to proliferate, for instance, in a wound, that the short RECK isoform is induced and contributes to fibroblast migration.
MATERIALS AND METHODS
Cell culture and treatments
Human foreskin samples for establishing primary fibroblast cell lines were obtained by the National Disease Research Interchange. All protocols were approved by the Princeton University IRB (protocol number 3134) and the University of California, Los Angeles, IRB (protocol number 14-000145). BJ5ta (immortalized fibroblasts), 293T cells, and HT1080 were purchased from the America Type Culture Collection (ATCC). These cells were maintained in 10% fetal bovine serum (FBS) in DMEM in a 37°C humid incubator under 5% CO2. 7dCI fibroblasts were collected 7 d after plating, or at an equivalent density, while 7dSS fibroblasts were seeded in full serum medium (10% FBS in DMEM), changed to reduced serum medium (0.1% FBS in DMEM), and collected 7 d after adding the reduced serum medium.
Polyadenylation site–enriched RNA-Seq
cDNA libraries containing fragments enriched for 3′ UTR ends were created with the Gnomegen RNA-Seq Library Preparation Kit for RNA profiling (Gnomegen, San Diego, CA). In short, mRNA was fragmented to a size range of 100–2000 base pairs using metal cations. Transcripts were incubated with a poly(T) sequence attached to biotin and polyadenylated transcripts were captured with streptavidin beads. RNA adapters were ligated to the 5′ end of the fragmented RNA and incubated at 37°C for 2 h. Agencourt Ampure XP beads (Beckman Coulter, Indianapolis, IN) were used to remove ligation reaction products. First-strand cDNA was synthesized by incubation with Superscript III reverse transcriptase (Life Technologies, Grand Island, NY) at 42°C for 40 min. Amgen Ampure XP beads were used to remove reverse transcription products from first-strand cDNA. Second-strand cDNA synthesis and Illumina sequencing adapter ligations were achieved through PCR amplification of first-strand cDNA product. Common forward primers were used for all samples; reverse primers with a unique barcode sequence were specific for each sample. The size distribution of the cDNA library was confirmed using a High Sensitivity DNA chip (Agilent Technologies) on a Bioanalyzer 2100 Instrument (Agilent Technologies). Libraries with a uniform size distribution between 200 and 1000 base pairs were subjected to gel size selection to enrich for 200–300 base pair–sized fragments. Single-end 140 base pair reads were generated on an Illumina HiSeq 2000 Instrument. The sequencing reaction was run for 147 cycles. Reads from polyadenylation site–enriched cDNA libraries were aligned to the human genome (hg19) using the STAR algorithm, which is designed to align untemplated adenosines (Dobin et al., 2013).
TCGA analysis
Data were obtained from The Cancer Genome Atlas (Cancer Genome Atlas, 2012a,b; Cancer Genome Atlas Research, 2012, 2013, 2014a,b; Cancer Genome Atlas Research et al., 2013). Per-exon RNA-Seq read counts for tumor tissues and paired normal tissues along with any available clinical data were obtained from the TCGA public data portal. For comparison of RECK expression between tumor and paired normal tissue, read counts were normalized to the total number of reads per sample. RNA-Seq read counts assigned to the unique short RECK exon and the final long RECK exon were used to infer expression of short and long RECK isoforms, respectively. Short-to-long RECK isoform ratios, as well as normalized short and long RECK read counts, were determined for tumor and paired normal samples. Two-tailed paired t tests were performed separately for each tumor type using GraphPad Prism 6.04. To determine RECK isoform expression in breast cancers with different clinical characteristics, the relative use of the distal isoform (RUD) was calculated and the log2-fold change between tumor and paired normal was determined and plotted based on PAM50 subtype. Statistical significance was determined by one-way analysis of variance (ANOVA) followed by Tukey post-hoc test for two-way comparisons to control for multiple hypothesis testing.
Real-time RT-PCR
Cells were harvested in TRIzol reagent (Invitrogen). Chloroform was added and the lysate was centrifuged. The aqueous supernatant containing RNA was collected. Isopropanol was added to precipitate the RNA. The precipitated RNA pellet was washed with 70% ethanol, then dissolved in water and quantified by NanoDrop 2000C (Thermo Scientific). RNA (50 ng) was used for one reaction of reverse transcription and amplified using a SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (Invitrogen). A reaction of 25 µl was amplified in 96-well plates (USA Scientific) in triplicate for each gene. Real-time RT-PCR was performed in a CFX96 real-time system (Bio-Rad) and all results were normalized to the expression level of UBC in the same sample. Gene expression levels were calculated using the 2−∆∆Ct method. We removed outliers, which were defined as values that were ±100% of the average of the other values. Primer sequences are as follows: short RECK isoform, 5′-TTGCGCCTCTATTAGTCCACAA-3′ and 5′-AGTAATAATTGCTCATTGGCAAAA-3′; long RECK isoform 5′-CAGAAATTGTGATCCTGATCATTCC-3′ and 5′-AAAGTCGGGCTGTCAGAGTTGATA-3′; total RECK 5′-AAATGTGCCGTGATGTATGTGAAC-3′ and 5′-TTAAACACACCTGGCAAAGATGAA-3′; UBC 5′-TCTTGTTTGTGGATCGCTGTGA-3′ and 5′-CAGGAGGGATGCCTTCCTTATC-3′; CTGF 5′-CTGCAGGCTAGAGAAGCAGAGC-3′ and 5′-GCTCAAACTTGATAGGCTTGGAGA-3′.
Antibodies
A monoclonal antibody against RECK (long isoform) and an antibody against tubulin were obtained from Thermo Fisher Scientific. For detection of FLAG, S-tag, GST, and HA, antibodies were purchased from Sigma-Aldrich, Cell Signaling Technology, Santa Cruz Biotechnology, and One World Lab, respectively. Antibodies against phospho-AKT, total AKT, phospho-ERK, total ERK, phospho-JNK, and total JNK were obtained from Cell Signaling Technology. To detect MMP2 and MMP9, antibodies were obtained from Abcam and Thermo Fisher Scientific. Antibodies against tubulin and actin used for loading controls were purchased from Sigma-Aldrich and Cell Signal Technology, respectively. Custom rabbit anti-human short RECK polyclonal antibody was generated twice. The first polyclonal was generated by GenScript (Piscataway, NJ) with experiments approved by the GenScript IACUC. The peptide CTDMFEFFANEQL was conjugated to KLH at the N-terminus. This peptide sequence was chosen to maximize antigenicity while maintaining predicted specificity to the short isoform of RECK. The peptide-KLH conjugate was used as an immunogen for New Zealand rabbits. Lyophilized antigen, preimmune serum, and affinity-purified antibody dissolved in phosphate-buffered saline (PBS; pH 7.4) with 0.02% sodium azide were provided by GenScript. Indirect enzyme-linked immunosorbent assays (ELISAs) were performed to validate the antibody. Free peptide (4 µg/ml) was plated at 100 µl/well in PBS, pH 7.4. A goat-raised anti-rabbit immunoglobulin G (H&L) secondary antibody conjugated to peroxidase was used as a secondary antibody. When the purified antibody was used as the primary antibody in these ELISAs, a clear signal above background, defined as either blank or preimmune serum, was observed at a 1:512,000 dilution. The titer (defined as the highest dilution with signal/blank ≥ 2.1) was determined as 1:512,000. For purification, Flag-tagged short RECK protein from 293T lysate was covalently immobilized onto beads conjugated with an anti-Flag antibody (Sigma-Aldrich). After mixing polyclonal short RECK antibody with the beads, the bound antibody was eluted using glycine (pH 2.5).
The second polyclonal antibody was generated by New England Peptide. The procedures were approved by the IACUC for SDIX LLC. The immunizing antigen was Ac-KKKC(dPEG4)DMFEFFANEQLLLL-OH. The peptide was conjugated to KLH and used to immunize rabbits multiple times. Serum from the rabbits was confirmed to react against the immunizing peptide conjugated to bovine serum albumin (BSA) by ELISA and to react more strongly to the immunizing peptide than to an irrelevant peptide conjugated to BSA.
Immunoblotting
Harvested cells were lysed in RIPA buffer (10 mM Tris-Cl, pH 8.0, 0.1% sodium deoxycholate, 0.5 mM EGTA, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 140 mM NaCl) containing protease inhibitor (Roche Applied Science) for 15 min on ice. After centrifugation, the supernatant was collected and the concentration was measured using a Thermo Scientific Pierce BCA Protein Assay kit (Thermo Fisher Scientific). Samples of 20 µg per well were loaded on SDS–PAGE gels and electrophoresed for 1 h and 25 min. The electrophoresed samples were transferred to 0.45 µm polyvinylidene difluoride membrane (Millipore) and blocked in 5% skim milk in 0.1% PBS-Tween (PBST) for 30 min. The primary and secondary antibody incubation steps were performed under conditions recommended by the antibody provider. Immunoblot data were quantified using ImageJ.
Coimmunoprecipitation and GST–pull down assay
Plasmids engineered to express the short RECK isoform or the long RECK isoform (4 µg) were introduced into 293T cells using Lipofectamine 2000 according to the manufacturer’s instructions. After transfection (48 h), cells were harvested and lysed in NETN buffer (100 mM NaCl, 0.5 mM EDTA, 20 mM Tris-Cl, pH 8.0, and 0.5% NP-40) containing protease inhibitor (Roche). Protein concentrations were measured by a BCA protein assay kit. Cell lysates (1 mg) were incubated overnight at 4°C with 2 µg of antibody. The next day, washed Sepharose A and G mixture (Sigma-Aldrich) was added and incubated for 1 h at 4°C. The protein mixture precipitated with the antibody and the precipitate was analyzed by Western blot using SDS–PAGE gels. For GST–pull down assays, short RECK conjugated to a GST tag and long RECK isoforms with domain-specific deletions were introduced into cells. Protein lysates were collected and GST-containing proteins were precipitated with GST•Bind Resin (EMD Millipore).
Plasmids and vectors
For short and long RECK isoforms, the short RECK isoform was synthesized by Genewiz (South Plainfield, NJ), and a cDNA encoding the long RECK isoform was purchased from Thermo Scientific. 3X Flag-tagged short RECK and long RECK isoforms were subcloned into pRRL-CMV-MCS-IRES-Hygro (UCLA Virus Core Laboratory) for mammalian cell overexpression. A vector expressing the long RECK isoform was obtained from Dharmacon. Plasmids containing MMP2 or MMP9 were purchased from DNASU, Arizona State University, and subcloned into a pRRL-CMV-MCS-IRES-Hygro vector for overexpression in mammalian cells. pBiFC-VC155 and pBiFC-VN173 were obtained from Addgene and subcloned into pRRL-CMV-MCS-IRES-Hygro for overexpression or lentivirus production.
RECK isoform-specific shRNAs were designed using the Broad Institute TRC design pipeline (https://www.broadinstitute.org/rnai/public/seq/search) and BLOCK-iT RNAi Designer (https://rnaidesigner.thermofisher.com/rnaiexpress). Knockdown shRNAs were cloned into the BamHI and EcoRI sites of the lentiviral plasmid pGreenPuro (Systems Biosciences) and expressed under control of the H1 promoter. Sequences are provided: shControl.1, 5′- GATCCCCTAAGGTTAAGTCGCCCTCGTTCAAGAGACGAGGGCGACTTAACCTTAGGTTTTTG-3′ and 5′-AATTCAAAAACCTAAGGTTAAGTCGCCCTCGTCTCTTGAACGAGGGCGACTTAACCTTAGGG-3′; shControl.2 5′-GATCCCAACAAGATGAAGAGCACCAACTTCCTGTCAGATTGGTGCTCTTCATCTTGTTGTTTTTG-3′ and 5′-AATTCAAAAACAACAAGATGAAGAGCACCAATCTGACAGGAAGTTGGTGCTCTTCATCTTGTTGG-3′; shShortRECK.1 5′-GATCCGCTTTGTACATCCTGGAATCCCTTCCTGTCAGAGGATTCCAGGATGTACAAAGCTTTTTG-3′ and 5′-AATTCAAAAAGCTTTGTACATCCTGGAATCCTCTGACAGGAAGGGATTCCAGGATGTACAAAGCG-3′; shShortRECK.2 5′-GATCCGATCTGTGCTCTCTGACATTTCTTCCTGTCAGAAAATGTCAGAGAGCACAGATCTTTTTG-3′ and 5′-AATTCAAAAAGATCTGTGCTCTCTGACATTTTCTGACAGGAAGAAATGTCAGAGAGCACAGATCG-3′; shLongRECK.1 5′-GATCCGCGTGGCAGTCGATTACTATGTTCAAGAGACATAGTAATCGACTGCCACGCTTTTTG
-3′ and 5′-AATTCAAAAAGCGTGGCAGTCGATTACTATGTCTCTTGAACATAGTAATCGACTGCCACGCG-3′; shLongRECK.2 5′-GATCCGCATCAAAGCATCTTGCATTACTTCCTGTCAGATAATGCAAGATGCTTTGATGCTTTTTG-3′ and 5′-AATTCAAAAAGCATCAAAGCATCTTGCATTATCTGACAGGAAGTAATGCAAGATGCTTTGATGCG-3′; lentiviral plasmids were cotransfected with pLP1, pLP2, and pLP/VSV.G packaging plasmids (Systems Biosciences) into 293T cells using Lipofectamine2000 (Life Technologies). Supernatants containing lentivirus were collected 48 and 72 h posttransfection and used to infect human fibroblasts. Stable lines were generated by adding selectable markers puromycin (for 5 d at 500 μg/ml) or hygromycin (for 3 d at 2 μg/ml).
Cell migration assays
Cells were plated in IncuCyte ImageLock 96-well microplates (Essen Bioscience) at a concentration of 25,000 cells/well the day before the experiment. Plates were coated with Matrigel (Corning) at 100 µg/ml before cell seeding. The next day, a scratch wound of 700–800 µm width was generated in the center of the well using the IncuCyte WoundMaker (Essen Bioscience). After the wounds were generated, they were filled with 50 μl of 0.5 mg/ml Matrigel on ice. Matrigel was allowed to solidify at 37°C for 30 min. Complete medium (100 ml) was poured on top of the Matrigel, and wound recovery was monitored with live imaging for up to 96 h. The results were analyzed with Incucyte Zoom software.
WST-1 assay
Cells (20,000) were plated in 24-well plates. After cell plating (48 h), cell proliferation was compared using the Premixed WST-1 cell proliferation assay system (Clonetech) as described in the manufacturer’s protocol. The WST-1 solution was added to complete media and incubated at 37°C for 2 h. The plate was analyzed for absorbance at 440 nm.
Immunofluorescence staining
Cells grown on coverslips were rinsed with PBS and then fixed with 4% paraformaldehyde in PBS, pH 7.4 for 15 min. For permeabilization, cells were incubated for 10 min in 0.25% Triton X-100 in PBS. After blocking for 30 min using 1% BSA in PBST, cells were incubated with primary antibody (diluted 1:100) in 1% BSA in PBST overnight at 4°C. Cells were stained with secondary antibody (diluted 1:200) for 1 h at room temperature and counterstained with 4′,6-diamidino-2-phenylindole to visualize nuclei. When observing the cell surface, the permeabilization step was omitted.
RECK knockout by CRISPR/Cas9
By participating in Horizon’s free CRISPR Guide RNA program, we were able to test five different guide RNAs for their efficiency in knocking out RECK. The double-strand-break efficiency was tested using the GeneArt Genomic Cleavage Detection Kit (Life Technologies). A plasmid containing Cas9 and 5′-GCGCACCTCGCAGAGAGGCC-3′ showed the highest efficiency and was introduced into 293T cells to introduce DNA breaks via nonhomologous end joining. Individual cells were plated at limiting dilution and clones were grown. Cell lines derived from a single cell were monitored with immunoblotting for RECK levels. Individual clones with reduced levels of the long RECK isoform were selected for further analysis. The DNA sequence near the region targeted by the RECK guide RNA was amplified by PCR (5′-AACAAAGAGCCCTGGTACGG-3′ and 5′-CATCTAAACCTCGCGACGAT-3′) and the PCR product was cloned into pCR4Blunt-TOPO plasmid using Zero Blunt TOPO PCR Cloning Kits (Invitrogen). Sequencing was performed by Genewiz using primers targeting the T3 priming site 5′-ATTAACCCTCACTAAAGGGA-3′.
PI-PLC treatment to cleave the GPI anchor
On the day before the transfection, RECK knockout 293T cells were seeded with 4,400,000 cells per 10 cm dish. For transfection, 4 µg of short RECK and long RECK overexpression vectors were introduced into the RECK knockout 293T cell line using Lipofectamine 2000 (Invitrogen). After transfection (24 h), cells were rinsed twice with PBS and then incubated in 5 ml of PI-PLC (Sigma-Aldrich) in DMEM at 100 mU/ml or DMEM without PI-PLC for the control. After 24 h of PI-PLC treatment, the supernatant and cells were separately harvested for analysis. The supernatant was filtered through a 0.45 μm syringe filter and used for Western blotting.
Cytokines
Recombinant human TGF-β (BioLegend) was prepared in PBS at 5 μg/ml for a final concentration of 5 ng/ml.
Data availability
The full alternative polyadenylation data set described in this study is available through the Gene Express Omnibus database as accession number GSE117337 deposited on July 18, 2018.
Supplementary Material
Acknowledgments
We thank the Eli and Edythe Broad Center for Regenerative Medicine and Stem Cell Research for Core Facilities that house a confocal microscope and IncuCyte instrument. We acknowledge Jeff Long (University of California, Los Angeles [UCLA]) for sharing equipment for cloning and a confocal microscope. H.A.C. was the Milton E. Cassel scholar of the Rita Allen Foundation ( http://www.ritaallenfoundation.org). H.N.L. acknowledges a Whitcome Fellowship from the UCLA Molecular Biology Institute. E.L.J. was supported in part by National Science Foundation Graduate Research Fellowship DGE-0646086. This work was funded by grants to H.A.C. from National Institute of General Medical Sciences Center of Excellence Grant no. P50 GM071508, PhRMA Foundation Grant no. 2007RSGl9572, National Institute of General Medical Sciences R01 GM081686, National Institute of General Medical Sciences R01 GM0866465, the Eli and Edythe Broad Center for Regenerative Medicine and Stem Cell Research, the Iris Cantor Women’s Health Center/UCLA, National Cancer Institute P50 CA092131, the UCLA Life Science Innovation Fund, UCLA Clinical and Translational Science Institute Grant no. UL1TR000124, the Leukemia Lymphoma Society, and funds from Rockefeller University. H.A.C. is a member of the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research, the Jonsson Comprehensive Cancer Center, the UCLA Molecular Biology Institute, and the UCLA Bioinformatics Interdepartmental Program. We thank Luisa Iruela-Arispe, Tracy Johnson, William Lowry, Jorge Torres, and Jose Rodriguez for helpful input, Barry Coller for suggestions on the choice of peptides for immunization to prepare the antibody to the short form of RECK, and Hina Zafar for assistance with RECK ELISA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Some images in this paper were modified from Servier Medical Art ( http://smart.servier.com/).
Abbreviations used:
- 7dCI
7 d contact inhibited
- 7dSS
7 d serum starved
- ADAM
a disintegrin and metalloproteinase
- ADAM10
A Disintegrin and MetalloProteinase family
- ANOVA
analysis of variance
- APA
alternative polyadenylation
- ATCC
American Type Culture Collection
- BCA
bicinchoninic acid
- BiFC
bimolecular fluorescence complementation
- BSA
bovine serum albumin
- CRISPR
clustered regularly interspaced short palindromic repeats
- DSB
double-strand break
- ELISA
enzyme-linked immunosorbent assay
- ER
endoplasmic reticulum
- ERK
extracellular signal-regulated kinase
- FBS
fetal bovine serum
- GPI
glycosylphosphatidylinositol
- GST
glutathione- S-transferase
- GTP
guanosine triphosphate
- HA
hemagglutinin
- HEK293
human embryonic kidney
- HER2
human epidermal growth factor receptor 2
- IACUC
Institutional Animal Care and Use Committee
- IRB
institutional review board
- JNK
c-Jun N-terminal kinase
- KLH
keyhole limpet hemocyanin
- long RECK
long RECK isoform
- MCF
Michigan Cancer Foundation
- MEF
mouse embryonic fibroblast
- MMP
matrix metalloproteinase
- MMP2
matrix metalloproteinase 2
- MMP9
matrix metalloproteinase 9
- MT1-MMP
membrane type I matrix metalloproteinase
- P
proliferating
- PAM
protospacer adjacent motif
- PAS
polyadenylation site
- PBS
phosphate-buffered saline
- PBS-T
phosphate-buffered saline-Tween
- PI-PLC
phosphatidylinositol-specific phospholipase C
- RECK
reversion-inducing-cysteine-rich protein with Kazal motifs
- RIPA
radioimmunoprecipitation assay
- RNA-Seq
RNA sequencing
- RTK
receptor tyrosine kinase
- RTK1
receptor tyrosine kinase 1
- RT-PCR
reverse transcriptase-PCR
- RU
relative units
- RUD
relative use of the distal polyadenylation site
- shRNA
short hairpin RNA
- short RECK
short RECK isoform
- TCGA
The Cancer Genome Atlas
- TGF-β
transforming growth factor β
- UBC
ubiquitin c
- WCL
whole cell lysate
- WST-1
water soluble tetrazolium salt-1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-12-0708) on June 6, 2018.
REFERENCES
- Acharya PS, Majumdar S, Jacob M, Hayden J, Mrass P, Weninger W, Assoian RK, Pure E. (2008). Fibroblast migration is mediated by CD44-dependent TGFβ activation. J Cell Sci , 1393–1402. [DOI] [PubMed] [Google Scholar]
- Alt FW, Bothwell AL, Knapp M, Siden E, Mather E, Koshland M, Baltimore D. (1980). Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3′ ends. Cell , 293–301. [DOI] [PubMed] [Google Scholar]
- Aman A, Piotrowski T. (2010). Cell migration during morphogenesis. Dev Biol , 20–33. [DOI] [PubMed] [Google Scholar]
- Belkin AM, Akimov SS, Zaritskaya LS, Ratnikov BI, Deryugina EI, Strongin AY. (2001). Matrix-dependent proteolysis of surface transglutaminase by membrane-type metalloproteinase regulates cancer cell adhesion and locomotion. J Biol Chem , 18415–18422. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N (2012a). Comprehensive molecular characterization of human colon and rectal cancer. Nature , 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N (2012b). Comprehensive molecular portraits of human breast tumours. Nature , 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N (2012). Comprehensive genomic characterization of squamous cell lung cancers. Nature , 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N (2013). Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature , 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. (2013). The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet , 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N (2014a). Comprehensive molecular characterization of urothelial bladder carcinoma. Nature , 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N (2014b). Comprehensive molecular profiling of lung adenocarcinoma. Nature , 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov , 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CK, Hung WC, Chang HC. (2008). The Kazal motifs of RECK protein inhibit MMP-9 secretion and activity and reduce metastasis of lung cancer cells in vitro and in vivo. J Cell Mol Med , 2781–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro MF, Bhattacharya SS, Schultz GS, Khaw PT. (2000). TGF-β1, -β2, and -β3 in vitro: biphasic effects on Tenon’s fibroblast contraction, proliferation, and migration. Invest Ophthalmol Vis Sci , 756–763. [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics , 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour A, Zucker S, Sampson NS, Kuscu C, Cao J. (2010). Role of matrix metalloproteinase-9 dimers in cell migration: design of inhibitory peptides. J Biol Chem , 35944–35956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Drost J, van Haaften G, Jenal M, Schrier M, Vrielink JA, Agami R. (2012). E2F mediates enhanced alternative polyadenylation in proliferation. Genome Biol , R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Gilmour D. (2009). Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol , 445–457. [DOI] [PubMed] [Google Scholar]
- Hong KJ, Wu DC, Cheng KH, Chen LT, Hung WC. (2014). RECK inhibits stemness gene expression and tumorigenicity of gastric cancer cells by suppressing ADAM-mediated Notch1 activation. J Cell Physiol , 191–201. [DOI] [PubMed] [Google Scholar]
- Huang C, Jacobson K, Schaller MD. (2004). MAP kinases and cell migration. J Cell Sci , 4619–4628. [DOI] [PubMed] [Google Scholar]
- Hunger-Glaser I, Salazar EP, Sinnett-Smith J, Rozengurt E. (2003). Bombesin, lysophosphatidic acid, and epidermal growth factor rapidly stimulate focal adhesion kinase phosphorylation at Ser-910: requirement for ERK activation. J Biol Chem , 22631–22643. [DOI] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. (2009). Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA , 7028–7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita M, Itoh Y, Chiba T, Mori H, Okada A, Kinoh H, Seiki M. (2001). Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J Cell Biol , 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NY, Lee JE, Chang HJ, Lim CS, Nam DH, Min BH, Park GH, Oh JS. (2008). Gamma-irradiation enhances RECK protein levels in Panc-1 pancreatic cancer cells. Mol Cells , 105–111. [PubMed] [Google Scholar]
- Klemke RL, Cai S, Giannini AL, Gallagher PJ, de Lanerolle P, Cheresh DA. (1997). Regulation of cell motility by mitogen-activated protein kinase. J Cell Biol , 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, Allo M, Dujardin G, Petrillo E, Munoz MJ. (2013). Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol , 153–165. [DOI] [PubMed] [Google Scholar]
- Li QQ, Liu Z, Lu W, Liu M. (2017). Interplay between alternative splicing and alternative polyadenylation defines the expression outcome of the plant unique OXIDATIVE TOLERANT-6 gene. Sci Rep , 2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. (2013). Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev , 2380–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin NU, Claus E, Sohl J, Razzak AR, Arnaout A, Winer EP. (2008). Sites of distant recurrence and clinical outcomes in patients with metastatic triple-negative breast cancer: high incidence of central nervous system metastases. Cancer , 2638–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahl C, Egea V, Megens RT, Pitsch T, Santovito D, Weber C, Ries C. (2016). RECK (reversion-inducing cysteine-rich protein with Kazal motifs) regulates migration, differentiation and Wnt/β-catenin signaling in human mesenchymal stem cells. Cell Mol Life Sci , 1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, Li W, Wagner EJ. (2014). CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature , 412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. (2009). Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell , 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Takegami Y, Okawa K, Muraguchi T, Noda M, Takahashi C. (2007). The reversion-inducing cysteine-rich protein with Kazal motifs (RECK) interacts with membrane type 1 matrix metalloproteinase and CD13/aminopeptidase N and modulates their endocytic pathways. J Biol Chem , 12341–12352. [DOI] [PubMed] [Google Scholar]
- Montero JA, Kilian B, Chan J, Bayliss PE, Heisenberg CP. (2003). Phosphoinositide 3-kinase is required for process outgrowth and cell polarization of gastrulating mesendodermal cells. Curr Biol , 1279–1289. [DOI] [PubMed] [Google Scholar]
- Morioka Y, Monypenny J, Matsuzaki T, Shi S, Alexander DB, Kitayama H, Noda M. (2009). The membrane-anchored metalloproteinase regulator RECK stabilizes focal adhesions and anterior-posterior polarity in fibroblasts. Oncogene , 1454–1464. [DOI] [PubMed] [Google Scholar]
- Morris AR, Bos A, Diosdado B, Rooijers K, Elkon R, Bolijn AS, Carvalho B, Meijer GA, Agami R. (2012). Alternative cleavage and polyadenylation during colorectal cancer development. Clin Cancer Res , 5256–5266. [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Inoue T, Shimao Y, Sameshima T. (2002). Matrix metalloproteinases in tumor invasion: role for cell migration. Pathol Int , 255–264. [DOI] [PubMed] [Google Scholar]
- Oh J, Seo DW, Diaz T, Wei B, Ward Y, Ray JM, Morioka Y, Shi S, Kitayama H, Takahashi C, et al. (2004). Tissue inhibitors of metalloproteinase 2 inhibits endothelial cell migration through increased expression of RECK. Cancer Res , 9062–9069. [DOI] [PubMed] [Google Scholar]
- Oh J, Takahashi R, Kondo S, Mizoguchi A, Adachi E, Sasahara RM, Nishimura S, Imamura Y, Kitayama H, Alexander DB, et al. (2001). The membrane-anchored MMP inhibitor RECK is a key regulator of extracellular matrix integrity and angiogenesis. Cell , 789–800. [DOI] [PubMed] [Google Scholar]
- Omura A, Matsuzaki T, Mio K, Ogura T, Yamamoto M, Fujita A, Okawa K, Kitayama H, Takahashi C, Sato C, Noda M. (2009). RECK forms cowbell-shaped dimers and inhibits matrix metalloproteinase-catalyzed cleavage of fibronectin. J Biol Chem , 3461–3469. [DOI] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, et al. (2009). Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol , 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson ML. (2011). Immunoglobulin heavy chain gene regulation through polyadenylation and splicing competition. Wiley Interdiscip Rev RNA , 92–105. [DOI] [PubMed] [Google Scholar]
- Rehfeld A, Plass M, Dossing K, Knigge U, Kjaer A, Krogh A, Friis-Hansen L. (2014). Alternative polyadenylation of tumor suppressor genes in small intestinal neuroendocrine tumors. Front Endocrinol (Lausanne) , 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL. (1996). The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell , 941–952. [DOI] [PubMed] [Google Scholar]
- Takahashi C, Sheng Z, Horan TP, Kitayama H, Maki M, Hitomi K, Kitaura Y, Takai S, Sasahara RM, Horimoto A, et al. (1998). Regulation of matrix metalloproteinase-9 and inhibition of tumor invasion by the membrane-anchored glycoprotein RECK. Proc Natl Acad Sci USA , 13221–13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto N, Tada M, Hida Y, Asano T, Cheng S, Kuramae T, Hamada J, Miyamoto M, Kondo S, Moriuchi T. (2007). Low expression of reversion-inducing cysteine-rich protein with Kazal motifs (RECK) indicates a shorter survival after resection in patients with adenocarcinoma of the lung. Lung Cancer , 376–383. [DOI] [PubMed] [Google Scholar]
- Tian B, Manley JL. (2013). Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci , 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta-Lima M, Winnischofer SM, Demasi MA, Astorino Filho R, Carreira AC, Wei B, de Assis-Ribas T, Konig MS, Bowman-Colin C, Oba-Shinjo SM, et al. (2015). Isolation and characterization of novel RECK tumor suppressor gene splice variants. Oncotarget , 33120–33133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng LM, Hsu NC, Chen SC, Lu YS, Lin CH, Chang DY, Li H, Lin YC, Chang HK, Chao TC, et al. (2013). Distant metastasis in triple-negative breast cancer. Neoplasma , 290–294. [DOI] [PubMed] [Google Scholar]
- Vorlova S, Rocco G, Lefave CV, Jodelka FM, Hess K, Hastings ML, Henke E, Cartegni L. (2011). Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol Cell , 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. (2013). Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol , 1069–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G, Hemmings BA. (2013). PKB/Akt-dependent regulation of cell motility. J Natl Cancer Inst , 393–404. [DOI] [PubMed] [Google Scholar]
- Yao P, Potdar AA, Arif A, Ray PS, Mukhopadhyay R, Willard B, Xu Y, Yan J, Saidel GM, Fox PL. (2012). Coding region polyadenylation generates a truncated tRNA synthetase that counters translation repression. Cell , 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Ferguson TA, Hernandez YJ, Stetler-Stevenson WG, Muir D. (1998). Neuronal matrix metalloproteinase-2 degrades and inactivates a neurite-inhibiting chondroitin sulfate proteoglycan. J Neurosci , 5203–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full alternative polyadenylation data set described in this study is available through the Gene Express Omnibus database as accession number GSE117337 deposited on July 18, 2018.