Abstract
The budding yeast centrosome, often called the spindle pole body (SPB), nucleates microtubules for chromosome segregation during cell division. An appendage, called the half bridge, attaches to one side of the SPB and regulates SPB duplication and separation. Like DNA, the SPB is duplicated only once per cell cycle. During meiosis, however, after one round of DNA replication, two rounds of SPB duplication and separation are coupled with homologue segregation in meiosis I and sister-chromatid segregation in meiosis II. How SPB duplication and separation are regulated during meiosis remains to be elucidated, and whether regulation in meiosis differs from that in mitosis is unclear. Here we show that overproduction of the half-bridge component Kar1 leads to premature SPB separation during meiosis. Furthermore, excessive Kar1 induces SPB overduplication to form supernumerary SPBs, leading to chromosome missegregation and erroneous ascospore formation. Kar1-mediated SPB duplication bypasses the requirement of dephosphorylation of Sfi1, another half-bridge component previously identified as a licensing factor. Our results therefore reveal an unexpected role of Kar1 in licensing meiotic SPB duplication and suggest a unique mechanism of SPB regulation during budding yeast meiosis.
INTRODUCTION
The spindle pole body (SPB) of budding yeast is a microtubule-organizing center, equivalent to the centrosome in animal cells (Adams and Kilmartin, 2000; Jaspersen and Winey, 2004; Cavanaugh and Jaspersen, 2017). Embedded in the nuclear envelope, the SPB is formed by a three-layered structure, with the central plaque directly interacting with the nuclear membranes, whereas the outer and inner plaques nucleate cytoplasmic and nuclear microtubules, respectively (Moens and Rapport, 1971; Byers and Goetsch, 1974; Winey and Byers, 1993). During meiosis, the outer plaque is modified to nucleate prospore membranes instead of microtubules for ascospore development (Moens and Rapport, 1971; Neiman, 2011). In addition to the three plaques, the SPB has a membrane-associated appendage, called the half bridge, which is asymmetrically attached to one side of the SPB (Byers and Goetsch, 1975). The main function of the half bridge is to provide a platform for SPB duplication and to keep duplicated SPBs engaged before their separation (Jaspersen and Winey, 2004; Seybold et al., 2015; Cavanaugh and Jaspersen, 2017). How exactly SPB duplication and separation are regulated at the half bridge remains to be further elucidated.
The half bridge is made up of four proteins, Cdc31, Kar1, Sfi1, and Mps3 (Spang et al., 1993, 1995; Jaspersen et al., 2002; Kilmartin, 2003). The first three localize to the cytoplasmic side of the nuclear envelope, whereas Mps3 localizes to the nuclear side. Acting as the major structural component of the half bridge, Sfi1 is a filamentous protein that possesses ∼20 Cdc31-binding sites at its central alpha-helical domain (Kilmartin, 2003; Li et al., 2006), whereas Kar1 and Mps3 are integral membrane proteins (Vallen et al., 1992; Jaspersen et al., 2002) and thought to anchor the half bridge to the nuclear envelope (Seybold et al., 2015). Structural analysis has shown that the N-terminus of Sfi1 is attached to the central plaque of the mother SPB (Li et al., 2006; Burns et al., 2015). At the time of SPB duplication, the half bridge is elongated to become a full bridge, owing to the dimerization of Sfi1 molecules at their C-termini (Li et al., 2006; Seybold et al., 2015). Protein–protein interaction assays have shown that Kar1 binds to the C-terminus of Sfi1 and thus tethers Sfi1 and Cdc31 to the outer nuclear membrane (Seybold et al., 2015). These findings suggest that Kar1 plays an important role in maintaining the integrity of the half bridge.
Genetically identified as a key factor that regulates karyogamy during cell mating in budding yeast, Kar1 localizes to the half bridge and is also required for SPB duplication (Rose and Fink, 1987). Protein domain analysis has revealed that the Kar1 region important for karyogamy is separable from that required for SPB duplication (Vallen et al., 1992). To initiate SPB duplication, a precursor of the SPB, the satellite, made up of SPB components primarily of the central plaque, is deposited at the distal end of the bridge (Adams and Kilmartin, 1999; Jaspersen and Winey, 2004). Duplication of the SPB is controlled by the cyclin-dependent kinase Cdk1 and to a lesser extent by the pololike kinase, Cdc5 (Haase et al., 2001; Jaspersen et al., 2004; Crasta et al., 2008; Avena et al., 2014; Elserafy et al., 2014). Like the nuclear genome, the SPB is duplicated only once per cell cycle (Elserafy et al., 2014; Ruthnick and Schiebel, 2016). Recent studies have shown that licensing of SPB duplication is modulated by the state of Sfi1 phosphorylation at its C-terminus (Avena et al., 2014; Elserafy et al., 2014), to which Kar1 binds. Phosphorylation of Sfi1 at six key residues by Cdk1 promotes SPB separation, whereas dephosphorylation of Sfi1 at anaphase by the phosphatase Cdc14 is critical for the next round of SPB duplication (Avena et al., 2014; Elserafy et al., 2014; Fox et al., 2017). Surprisingly, removal of the C-terminus of Sfi1, which eliminates its dimerization domain as well as the six key phosphorylation sites, still maintains yeast cell growth at room temperature (Seybold et al., 2015). Therefore, other components of the half bridge, including Kar1, may play an alternative role for licensing SPB duplication.
We show here that overproduction of Kar1 in meiosis leads to supernumerary SPB formation and chromosome missegregation. During meiosis, after only one round of DNA replication, two continuous cell divisions occur to reduce the genome size by half in gametes. In budding yeast, the SPB is first duplicated in meiosis I to form a bipolar spindle for homologue segregation, and then SPBs reduplicate at interphase II to form two spindles for sister-chromatid segregation in meiosis II (Moens and Rapport, 1971). Therefore, the second round of SPB duplication is decoupled from DNA replication, but before the end of meiosis I, SPBs must be licensed for reduplication (Fox et al., 2017). Furthermore, during the early phase of meiosis I when homologous chromosomes pair and recombine, duplicated SPBs are engaged without separation, forming a side-by-side configuration for an extended period (Li et al., 2015). We have found that ectopically overproduced Kar1 promotes SPB separation at prophase I, but SPB reduplication occurs only after the onset of metaphase I. Our findings thereby are in contrast to those in mitosis, where high dosage of Kar1 arrests yeast cells with a single unduplicated SPB (Rose and Fink, 1987; Seybold et al., 2015). We also show that Kar1-mediated SPB duplication is independent of the phosphorylation status of Sfi1, indicating the existence of an alternative mechanism to license SPB duplication during yeast meiosis.
RESULTS
Kar1 is expressed in yeast meiosis and localizes to the SPB
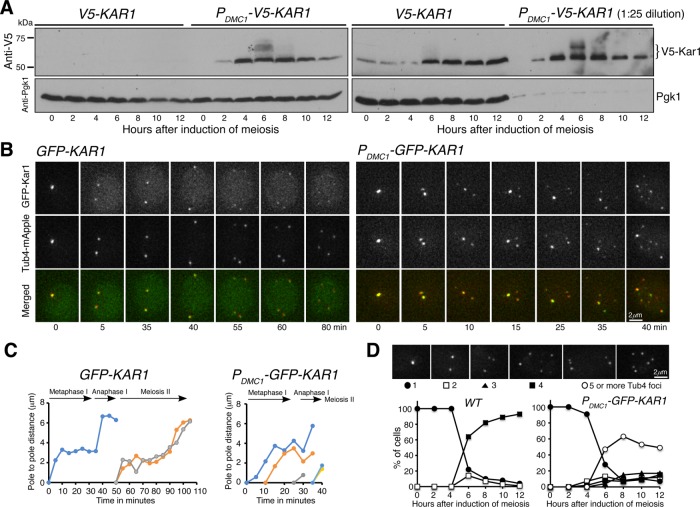
We seek to understand the function of Kar1 in yeast meiosis. Previously, we have shown that Kar1 is copurified with the SPB component Spc97 (Li et al., 2015), revealing that Kar1 is present at the SPB during budding yeast meiosis. To determine the protein level of Kar1, we generated an N-terminal V5-tagged KAR1 allele, which is under the control of its endogenous promoter (Figure 1A). By Western blotting, we found that Kar1 was present throughout meiosis, but its protein level increased approximately fourfold 6 h after the induction of meiosis (Figure 1A and Supplemental Figure 1), which approximately corresponded to meiosis I. The level of Kar1 remained high for the rest of meiosis (Figure 1A), supporting the finding of an upregulation of KAR1 gene expression during mid and late meiosis (Chu et al., 1998). This elevated level of Kar1 during meiosis is in sharp contrast to that of another half-bridge component of the SPB, Mps3, whose protein level decreases dramatically after meiosis I (Li et al., 2017). Of note, Kar1 appeared to be modified posttranslationally, forming multiple upper shifting bands (Figure 1A, t = 6 h, and Supplemental Figure 1) that are indicative of protein hyperphosphorylation. Taking these observations together, we conclude that KAR1 is upregulated and its gene product is present throughout yeast meiosis.
FIGURE 1:
Protein level and localization of Kar1 in budding yeast meiosis. (A) Western blots showing the protein level of Kar1. Yeast cells were induced to undergo synchronous meiosis, and aliquots were withdrawn at indicated times and prepared for Western blotting. V5-Kar1 was probed by an anti-V5 antibody. The PDMC1-V5-KAR1 allele specifically overproduces Kar1 in meiosis. Note that in the gel shown to the right, samples from PDMC1-V5-KAR1 were diluted 25-fold. The level of Pgk1 serves as a loading control. (B) Time-lapse live-cell microscopy showing GFP-Kar1 localization in yeast meiosis. Tub4-mApple serves as a marker for the yeast SPB. Projected images from 12 optical sections are shown. Note that GFP-Kar1 colocalized with Tub4-mApple and that supernumerary Kar1 and Tub4 foci were formed in the PDMC1-GFP-KAR1 strain. Time zero refers to the point of SPB separation in meiosis I. (C) Pole-to-pole distance from the GFP-KAR1 and PDMC1-GFP-KAR1 strains shown in B. Note that ∼30 min after the formation of supernumerary Tub4 foci, it becomes difficult to track the corresponding spindle poles. (D) Quantification of supernumerary Tub4 foci formation. Yeast cells were induced to undergo synchronous meiosis as in A, and fluorescence microscopy was performed to determine the number of Tub4 foci at indicated time points. At least 100 cells were counted at each time point. Time-course experiments were repeated, and data from one representative experiment are shown.
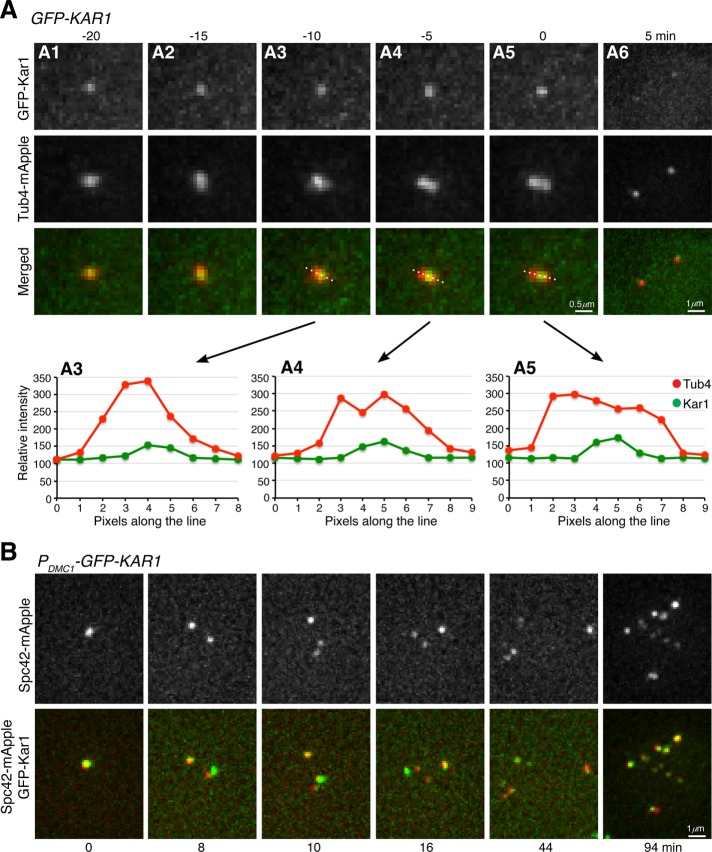
To localize Kar1 in meiotic cells, we generated a functional GFP-KAR1 allele, which served as the only copy of KAR1 in the yeast genome (Figure 1B). We used the SPB component, Tub4, which was fused to mApple, to serve as a marker for the SPB (Figure 1B and Supplemental Figure 1). By time-lapse live-cell fluorescence microscopy, we found that GFP-Kar1 was colocalized with Tub4-mApple throughout meiosis (Figure 1B and Figure 2A). Quantitative analysis of SPB separation parameters, including the duration of metaphase I (35 ± 13 min, n = 11) and the pole-to-pole distance at metaphase I and anaphase I, showed that cells with GFP-Kar1 appeared normal (Figure 1, B and C; Shirk et al., 2011 and see below). To pinpoint Kar1’s localization to the half-bridge area of the SPB, we focused on cell cycle transition from prophase I to metaphase I, when duplicated SPBs separated to form two distinctive Tub4 foci (Figure 2A). Line scan of fluorescence intensity revealed that Kar1 was concentrated at the interspace of the separating Tub4 foci (Figure 2D), indicating that Kar1 is localized to the half-bridge area, consistent with it being a half-bridge protein previously found in mitosis (Vallen et al., 1992; Burns et al., 2015; Seybold et al., 2015). Together, our data demonstrate that Kar1 is an SPB component and likely localizes to the region of the SPB half bridge during yeast meiosis.
FIGURE 2:
Localization of Kar1 to the half-bridge area during yeast meiosis. (A) Time-lapse live-cell microscopy was performed as in Figure 1B. Single optical sections are shown. Graphs of line-scan pixel intensity shown in A3 to A4 correspond to the top images. Time zero refers to the point of SPB separation in meiosis I. Note that GFP-Kar1 localizes to the central region of two separating SPBs, which are marked by Tub4-mApple. (B) Colocalization of Spc42 with Kar1 in yeast meiosis. Time-lapse live-cell microscopy was performed as in A. Time interval was set at 2 min; projected images from 12 optical sections are shown. Time zero refers to the point of the first round of Spc42 separation in meiosis I. Note that all Spc42-mApple foci colocalize with GFP-Kar1.
Increased level of Kar1 leads to supernumerary SPB formation in yeast meiosis
Because the expression of KAR1 is up-regulated (Chu et al., 1998) and Kar1 protein level increases during meiosis (Figure 1A), we hypothesized that Kar1 plays a regulatory role in meiotic SPB duplication and separation. To dramatically increase the protein level of Kar1 specifically during yeast meiosis, we used an enhanced DMC1 promoter that we developed (Fan et al., 2017) to generate the heterologous allele PDMC1-KAR1 (Figure 1A and Supplemental Figure 1). Compared to the endogenous level of Kar1 in wild-type cells, PDMC1-KAR1 cells produced ∼25-fold more Kar1 during yeast meiosis as determined by Western blotting and fluorescence-intensity-based assay (Figure 1A and Supplemental Figure 1). By Western blotting, we found that overproduced Kar1 also showed retarded migrating bands 6 h after the induction of meiosis, supporting the idea that Kar1 is modified posttranslationally during yeast meiosis.
To determine the consequence of Kar1 overproduction during meiosis, we performed time-lapse fluorescence microscopy to monitor SPB duplication and separation (Figure 1B). In wild-type cells, SPBs, marked by Tub4-mApple and GFP-Kar1, separated at the onset of metaphase I to form a bipolar spindle; this process had a duration of 35 min (Figures 1, B and C, and 3, A and B, and Supplemental Figure 2). At anaphase I, the pole-to-pole distance of the spindle increased to more than 6 μm (Figures 1, B and C, and 3A and Supplemental Figure 2). On entering into meiosis II, cells underwent a second round of SPB duplication and separation to generate four spores, each inheriting exactly one SPB (Figures 1D and 3, A–D). In contrast, in PDMC1-KAR1 cells, the third Tub4/Kar1 focus started to emerge ∼10 min after the first round of SPB separation in meiosis I (Figures 1, B and C, and 3, A–C), indicating an accelerated rate of SPB duplication and/or premature SPB separation when Kar1 is overproduced. Noticeably, 6 h after the induction of meiosis PDMC1-KAR1 cells formed 5 or more Tub4/Kar1 foci (Figures 1, B–D, and 3, A–C). The Tub4 foci formed in mutant cells always colocalized with Kar1, although their fluorescence intensity varied (Figure 1B). Twelve hours after induction of meiosis, more than half of the PDMC1-KAR1 cells formed five or more Tub4/Kar1 foci (Figures 1D and 3D). Because the wild-type copy of KAR1 was present in these cells, the phenotype caused by PDMC1-KAR1 therefore was hypermorphic. In addition, we found that the core component of the SPB central plaque, Spc42, also colocalized with the supernumerary Kar1 foci during meiosis (Figure 2B). Altogether, these results support the idea that, in the presence of excessive Kar1, meiotic cells overduplicate and form supernumerary SPBs.
FIGURE 3:
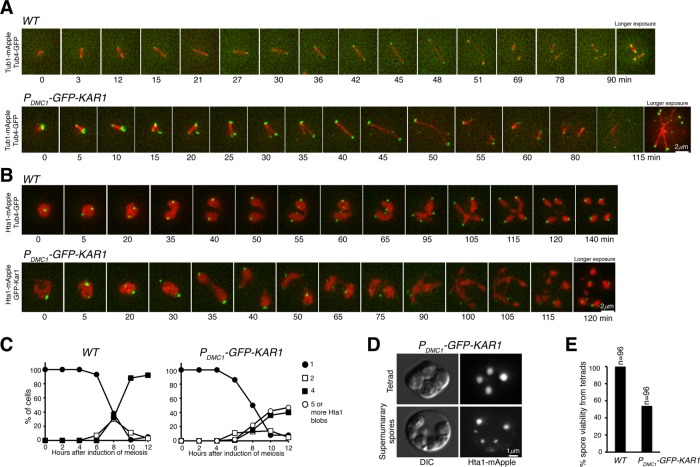
Supernumerary SPB and ascospore formation in Kar1-overproduced cells during yeast meiosis. (A) Spindle dynamics during yeast meiosis. Wild-type (WT, Tub1-mApple, Tub4-GFP) and PDMC1-GFP-KAR1 cells were induced to undergo synchronous meiosis, and live-cell fluorescence microscopy was performed to monitor microtubule and SPB movement in meiosis. Time interval was set at either 3 or 5 min; time zero refers to the point of SPB separation in meiosis I. Merged images from 12 optical sections were shown. Extended data of A are shown in Supplemental Figure 2. Note that numerous spindles were formed in the PDMC1-GFP-KAR1 cell. (B) Chromosome segregation in yeast meiosis. Wild-type (HTA1-mApple TUB4-GFP) and PDMC1-GFP-KAR1 cells were induced to undergo synchronous meiosis, and live-cell microscopy was performed as in A. (C) Quantification of chromosome segregation in cells shown in B. Note that five or more Hta1 blobs were formed in the PDMC1-GFP-KAR1 cell. (D) Ascospore formation during yeast meiosis. Selected images from PDMC1-GFP-KAR1 cells show abnormal ascospore formation (lower panel). (E) Quantification of spore viability. One-day-old tetrads were dissected and grown on YPD for 3 d to determine viability.
Kar1-mediated supernumerary SPBs are functional in nucleating microtubules
To determine whether the supernumerary SPBs formed in PDMC1-KAR1 cells are functional in nucleating microtubules and thus regulate meiotic cell division, we monitored spindle formation and chromosome segregation by time-lapse fluorescence microscopy (Figure 3). To visualize microtubules, one copy of Tub1, the alpha-tubulin in budding yeast, was tagged by mApple (Figure 3A). In wild-type cells, a bipolar spindle formed at the onset of metaphase I, elongated at anaphase I, and then disassembled before the second round of SPB duplication and separation in meiosis II (Figure 3A). After SPB separation at the onset of metaphase I, spindles formed in PDMC1-KAR1 cells and elongated at anaphase I to a length that was comparable to those of the wild-type cells (Figures 1 and 3). Noticeably, the third and sometimes the fourth Kar1 focus emerged well before spindle elongation (Figure 3A, t = 10, and Supplemental Figure 2B). Essentially all of the supernumerary Kar1 foci in PDMC1-KAR1 cells were associated with microtubules, leading to multipolar spindle formation (Figure 3A). These findings demonstrate that these Kar1 foci represent functional microtubule-organizing centers in mutant cells.
To visualize chromosome segregation, we observed the dynamics of histone H2A with an HTA1-mApple allele (Figure 3B). Compared to those in wild-type cells, meiotic chromosome segregation appeared on time in PDMC1-KAR1 cells, of which two distinctive Hta1 masses formed around 35 min after SPB separation in meiosis I (Figure 3B). Wild-type cells formed four separate Hta1 masses at the end of meiosis, whereas ∼40% of mutant cells formed five or more Hta1 blobs, each associated with one or more Kar1 foci (Figure 3C). As a consequence, more than four ascospores formed in PDMC1-KAR1 asci (Figure 3D), and even in the asci with four spores, less than 60% of the spores were viable (Figure 3E), indicating massive chromosome missegregation in mutant cells. The above observations further support the idea that in the presence of excessive Kar1, yeast cells overduplicate the SPB in meiosis.
We used serial-section electron microscopy (EM) to determine the ultrastructure of Kar1-mediated SPBs (Figure 4). Morphologically, SPBs in PDMC1-KAR1 cells appeared normal, because they were imbedded into the nuclear membranes, formed layered structures, and nucleated microtubules, as those in wild-type cells (Figure 4). At prophase I, duplicated SPBs engaged and formed a side-by-side SPB configuration, but the bridge that linked the SPBs appeared smaller (p < 0.05, n = 15) with less electron density in PDMC1-KAR1 cells (Figure 4A). Importantly, at metaphase I before spindle elongation, wild-type cells always formed bipolar spindles (Figures 3A and 4B), but two of the 6 PDMC1-KAR1 metaphase I cells that we examined by serial-section EM possessed three functional SPBs and appeared to form multipolar spindles (Figure 4C shows one of them). This finding supports our light microscopy observation of premature SPB duplication and separation in cells with excessive Kar1 (Figure 3A and Supplemental Figure 2). Of note, in 14 wild-type and 10 mutant meiosis II cells that were examined by serial-section EM, only three or four SPBs in each cell could be determined with certainty, on the basis of their association with microtubules. Nevertheless, our light and electron microscopy data together show that the supernumerary SPBs formed in the presence of excessive Kar1 are capable of nucleating microtubules and forming spindles and therefore are functional in mediating chromosome segregation. Because excessive and often multipolar spindles form in these cells, chromosome segregation becomes erroneous, leading to inviable spores (Figure 3E).
FIGURE 4:
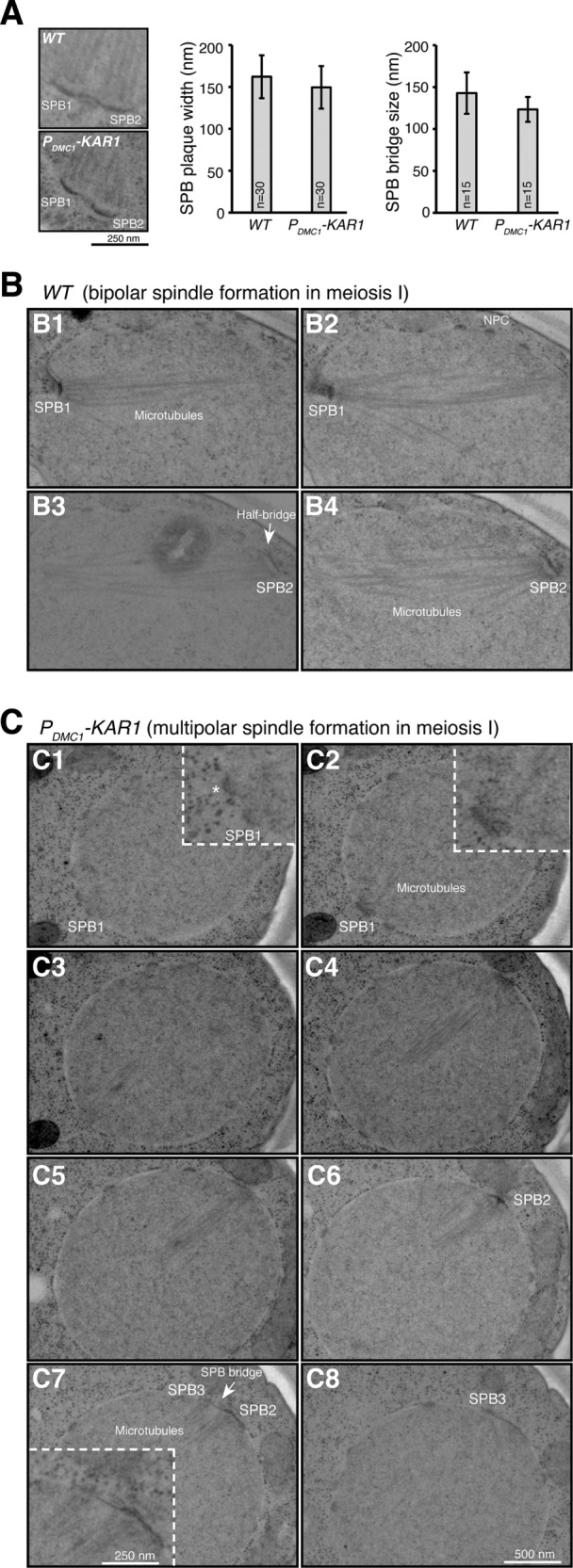
SPB ultrastructure visualized by electron microscopy (EM). (A) Side-by-side SPB configuration in meiosis I. Yeast cells were induced to undergo synchronous meiosis, and serial sectioning was performed to observe SPB morphology by EM. Representative images from the wild type (WT) and mutant are shown. Quantifications of SPB plaque width and bridge size are shown in the graphs to the right. For the t test for SPB plaque width, p = 0.035; bridge size p = 0.011. (B, C) Spindle formation in meiosis I. (B) A bipolar spindle in a WT cell captured in four serial sections. NPC, nuclear pore complex. (C) A tripolar spindle in a PDMC1-KAR1 cell captured in eight serial sections. Insets show 2× enlarged images. Note that in the mutant cell, the number 2 SPB has already reduplicated before spindle elongation at anaphase I. The asterisk indicates that this nuclear structure may represent another SPB in the mutant cell.
SPBs separate prematurely at prophase I in the excess of Kar1
To address the execution point of excessive Kar1-mediated supernumerary SPB formation, we arrested yeast cells at prophase I by deleting NDT80, which encodes a transcription factor that regulates the expression of mid- and late meiotic genes (Xu et al., 1995), and observed SPB duplication and separation when Kar1 was overproduced (Figure 5). In ndt80Δ cells, SPBs were duplicated but remained cohesive, most of the time forming only one Tub4 focus visible by fluorescence microscopy (Figure 5 and Shirk et al., 2011). In contrast, more than 25% of PDMC1-KAR1 ndt80Δ cells formed two Tub4 foci 8 h after the induction of meiosis (Figure 5, A and B). However, the double mutant rarely yielded more than two Tub4 foci (Figure 5B). Our findings demonstrate that SPBs separate prematurely at prophase I in the presence of an increased level of Kar1.
FIGURE 5:
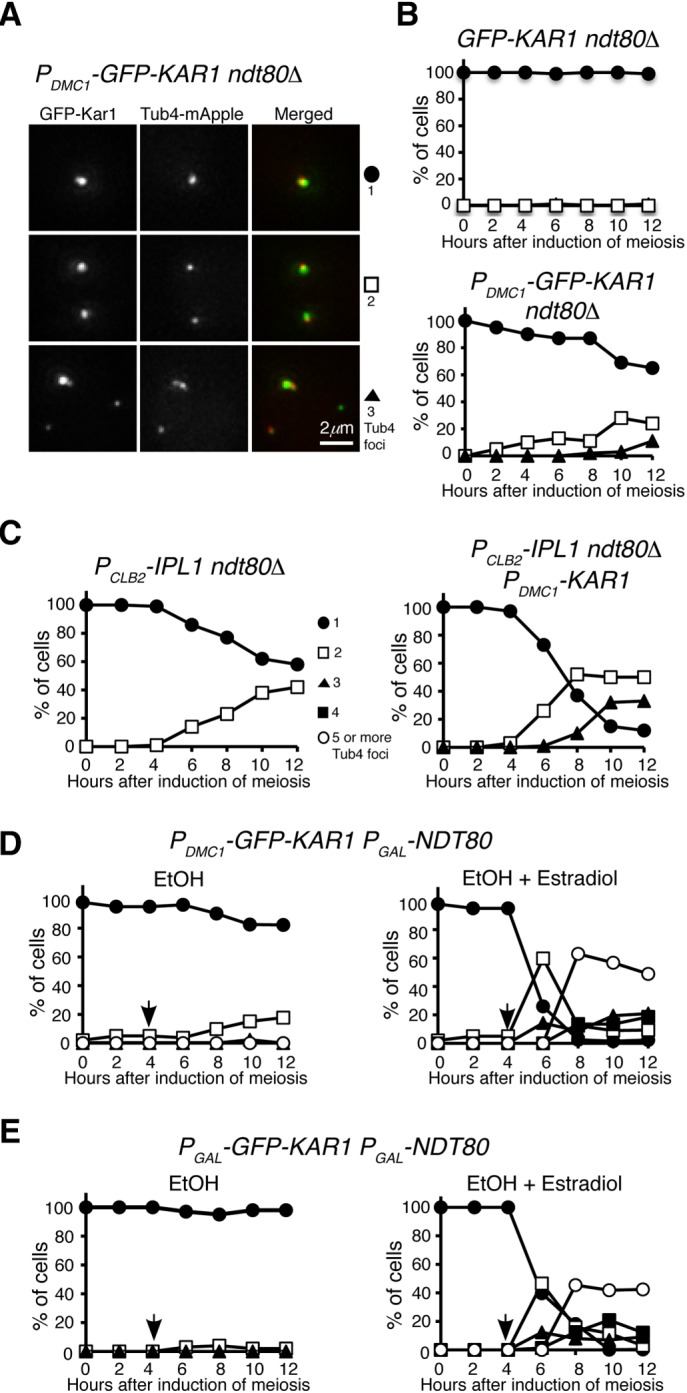
Kar1 promotes premature SPB separation at prophase I. (A) Representative images showing GFP-Kar1 localization at prophase I. Projected images are shown. (B) Quantification of SPB separation in GFP-KAR1 ndt80Δ and PDMC1-GFP-KAR1 ndt80Δ cells. (C) Quantification of SPB separation in PCLB2-IPL1 ndt80Δ and PCLB2-IPL1 ndt80Δ PDMC1-KAR1 cells. (D) Quantification of SPB separation in PDMC1-GFP-KAR1 PGAL1-NDT80 cells. Arrows indicate the time of addition of EtOH and estradiol, which was dissolved in EtOH. (E) Quantification of SPB separation in PGAL1-GFP-KAR1 PGAL1-NDT80 cells. As in D, arrows indicate the time of addition of EtOH and estradiol. The time-course experiments were repeated, and data from one representative experiment are shown.
We and others have shown previously that in the absence of Ipl1, the Aurora kinase in budding yeast, SPBs separate prematurely at prophase I (Shirk et al., 2011; Kim et al., 2013). To determine whether Kar1 and Ipl1 regulate SPB separation in a similar manner, we generated a double mutant PDMC1-KAR1 PCLB2-IPL1, which overproduced Kar1 but depleted Ipl1 in meiotic cells (Figure 5C). As we found previously, ∼40% of PCLB2-IPL1 ndt80Δ cells prematurely separated their SPBs at prophase I 12 h after the induction of meiosis, whereas more than 50% of PDMC1-KAR1 PCLB2-IPL1 ndt80Δ cells separated prematurely (Figure 5C). More importantly, around 30% of PDMC1-KAR1 PCLB2-IPL1 ndt80Δ cells formed three Tub4 foci (Figure 5C). This result is in contrast to that of PDMC1-KAR1 ndt80Δ, for which cells with three Tub4 foci were rarely observed (Figure 5B). Together, our findings support the idea that Kar1 promotes SPB separation, whereas Ipl1 inhibits SPB separation and to a lesser extent prevents SPB duplication.
Kar1 protein domains required for SPB duplication and separation in meiosis
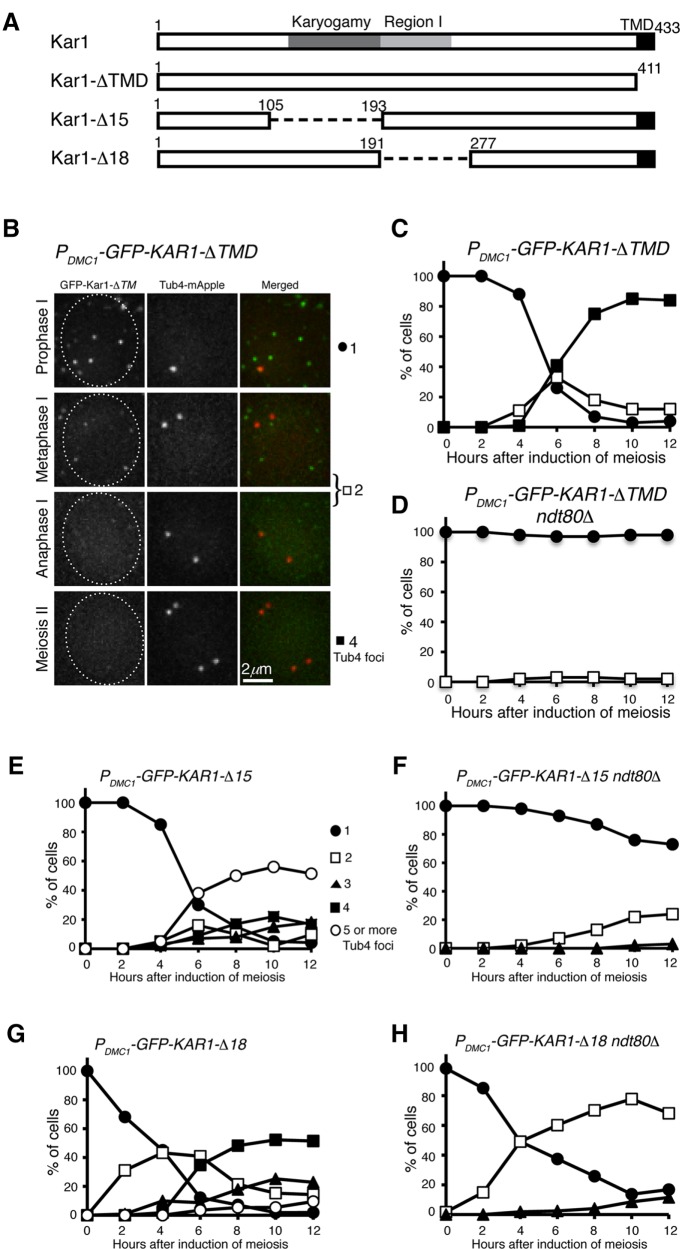
Because Kar1 is an integral membrane protein of the outer nuclear envelope, we asked whether the transmembrane domain of Kar1, which is located at its very C-terminus, is required for mediating supernumerary SPB formation. We generated a PDMC1-KAR1-ΔTMD allele, of which the C-terminal 22 amino acids were removed (Figure 6A). As shown in Figure 6B, GFP-Kar1-ΔTMD formed fluorescent foci inside the cell, but they failed to colocalize with Tub4, supporting the idea that the transmembrane domain is required for its localization to the SPB (Vallen et al., 1992). Importantly, SPB duplication and separation, as visualized by Tub4-mApple, was on schedule in PDMC1-KAR1-ΔTMD cells (Figure 6B). Of note, the endogenous wild-type copy of KAR1 was present in cells with PDMC1-driven deletion alleles (Figure 6, B–H). Therefore, overproduction of Kar1-ΔTMD has no discernable negative effect on meiotic SPB duplication and cell progression, and we conclude that the transmembrane domain of Kar1 is critical for its function in regulating SPB duplication and separation in meiosis as shown previously in mitosis (Vallen et al., 1992).
FIGURE 6:
Protein domains of Kar1 required for yeast meiosis. (A) Diagram showing the Kar1 protein structure. Deletions of kar1-Δ15 and -Δ18 have been described previously (Vallen et al., 1992). (B) Representative images showing the localization of GFP-Kar1-ΔTMD in yeast meiosis. Note that without its transmembrane domain (TMD), Kar1 fails to localize to the SPB, which is marked by Tub4-mApple. (C, D) Quantification of SPB separation in PDMC1-GFP-KAR1-ΔTMD cells. The ndt80Δ allele was used to arrest yeast cells at prophase I. (E, F) Quantification of SPB separation in PDMC1-GFP-KAR1-Δ15 cells. (G, H) Quantification of SPB separation in PDMC1-GFP-KAR1-Δ18 cells. At least 100 cells were counted at each time point. The time-course experiments were repeated, and data from one representative experiment are shown.
In addition to its role in SPB duplication, Kar1 is required for nucleating microtubules at the half bridge during karyogamy (Rose and Fink, 1987; Vallen et al., 1992). To determine whether Kar1’s role in microtubule nucleation is also required for regulating SPB duplication and separation in meiosis, we first used a separation-of-function mutant, kar1-Δ15, which abolishes microtubule nucleation and thus is defective in karyogamy but is fully competent for SPB duplication and cell growth (Figure 6A and Vallen et al., 1992). When Kar1-Δ15 was overproduced in meiosis by way of PDMC1- kar1-Δ15, SPBs overduplicated to form more than four Tub4 foci just as observed in PDMC1-KAR1 cells (Figures 1B and 6E), indicating that microtubule-nucleating at the half bridge mediated by Kar1 is not necessary for Kar1-mediated SPB duplication in meiosis. Of note, the karyogamy domain of Kar1 appears to play a role in spore wall formation and maturation (Gordon et al., 2006). Next, we determined whether deletion of region 1 (Kar1-Δ18), which abolishes SPB duplication in mitosis (Vallen et al., 1992), also inhibits Kar1’s activity in promoting SPB overduplication (Figure 6, A and G). Indeed, ectopically expressing PDMC1- kar1-Δ18 had little effect on supernumerary SPB formation (Figure 6, A and G). Similarly, we found that PDMC1-KAR1-Δ15 ndt80Δ but not PDMC1-KAR1-ΔTMD ndt80Δ cells separated SPB prematurely at prophase I (Figure 6, F and H). Intriguingly, deletion of region 1 (PDMC1- kar1-Δ18) increased the rate of premature SPB separation approximately fourfold (Figure 6H), indicating that region 1 plays an inhibitory role in SPB separation. Together, our findings indicate that the transmembrane domain and region 1 but not the karyogamy domain of Kar1 are critical for generating the hypermorph of PDMC1-KAR1.
Supernumerary SPBs form after prophase I in the excess of Kar1
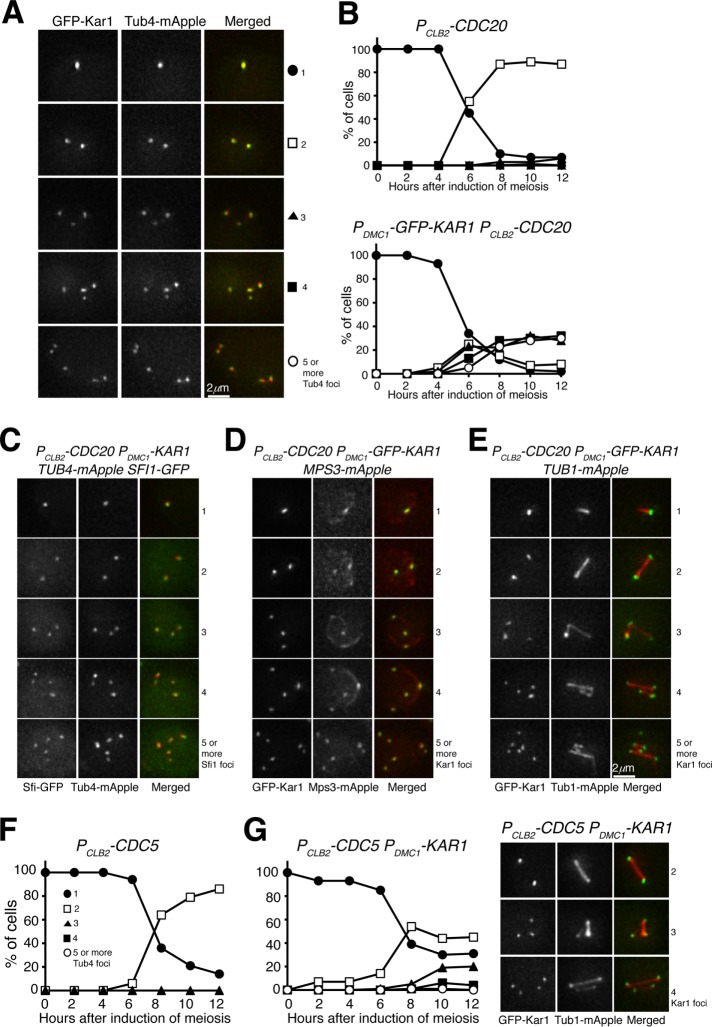
To test the hypothesis that supernumerary SPBs form only after prophase I, we used an inducible PGAL-NDT80 allele in meiosis, first to arrest cells at prophase I and then to release yeast cells from prophase I arrest by inducing the expression of PGAL-NDT80 with estradiol (Carlile and Amon, 2008). In PDMC1-KAR1 PGAL-NDT80 cells that were arrested at prophase I, we found that only two Tub4 foci were formed 12 h after induction of meiosis (Figure 5D). This observation is consistent with the idea that SPBs separate prematurely when Kar1 is overproduced. On the addition of estradiol, yeast cells were released from prophase I arrest and continued meiosis (Figure 5D). As expected, supernumerary Tub4 foci were formed in PDMC1-KAR1 PGAL-NDT80 cells (Figure 5D). In addition, we constructed a PGAL-KAR1 PGAL-NDT80 strain, in which both KAR1 and NDT80 were under the control of the GAL promoter (Figure 5E). On the addition of estradiol, both Kar1 and Ndt80 were induced, leading to the formation of supernumerary Tub4 foci in ∼40% of the cells (Figure 5E). The above findings demonstrate that in the presence of excessive Kar1, formation of supernumerary SPBs occurs only after prophase I.
To further determine the execution point of Kar1-mediated SPB duplication, we arrested yeast cells at metaphase I by way of PCLB2-CDC20 (Lee and Amon, 2003), overproduced Kar1 by PDMC1-KAR1, and determined the formation of supernumerary SPBs by observing Tub4 and/or Kar1 foci formation (Figure 7A). After the induction of meiosis, PCLB2-CDC20 cells separated their SPBs and formed two Tub4 foci, but these cells never entered into anaphase I (Figure 7B). In contrast, in PDMC1-KAR1 PCLB2-CDC20 cells, supernumerary Tub4 and Kar1 foci formed in ∼60% of the cells, most of them with colocalizing Tub4 and Kar1 signals (Figure 7A). Additionally, in PDMC1-KAR1 PCLB2-CDC20 cells, the supernumerary Tub4 foci were also associated with Sfi1, another half-bridge component, and the Kar1 foci were colocalized with Mps3 as well (Figures 6C and 7D). More importantly, the supernumerary Kar1 foci in PDMC1-KAR1 PCLB2-CDC20 cells formed numerous spindles (Figure 7E), demonstrating their ability in microtubule nucleation. From these observations, we conclude that in the presence of an elevated level of Kar1, supernumerary SPBs can form only after prophase I but before the onset of anaphase I.
FIGURE 7:
The execution point of Kar1-mediated SPB duplication in yeast meiosis. (A) Representative images showing GFP-Kar1 and Tub4 localization in PDMC1-GFP-KAR1 PCLB2-CDC20 cells. Depletion of Cdc20 arrests yeast cells at metaphase I. (B) Quantification of SPB separation in PCLB2-CDC20 and PDMC1-GFP-KAR1 PCLB2-CDC20 cells. Note the formation of supernumerary SPBs in PDMC1-GFP-KAR1 PCLB2-CDC20 cells. (C) Representative images showing Sfi1 localization in PDMC1-GFP-KAR1 cells at metaphase I. Note that Sfi1-GFP colocalizes with Tub4-mApple. (D) Representative images showing Mps3 localization in PDMC1-GFP-KAR1 cells at metaphase I. (E) Representative images showing Tub1 localization in PDMC1-GFP-KAR1 cells at metaphase I. Projected images are shown (C–E). (F) Quantification of SPB separation in Cdc5-depleted (PCLB2-CDC5) cells in meiosis. (G) Quantification of SPB separation in PDMC1-GFP-KAR1 PCLB2-CDC5 cells. Representative images of GFP-Kar1 and Tub1-mApple are shown to the right. The time-course experiments were repeated, and data from one representative experiment are shown.
Cdc5 regulates Kar1-mediated SPB duplication
We have shown previously that depletion of the pololike kinase, Cdc5, in budding yeast meiosis inhibits SPB duplication during meiosis (Shirk et al., 2011). To determine whether Kar1-mediated SPB overduplication requires Cdc5 activity, we used a meiosis-specific Cdc5 depletion mutant, PCLB2-CDC5 (Lee and Amon, 2003). After induction to undergo meiosis, PCLB2-CDC5 cells were arrested at a metaphase-I-like stage with two separated SPBs (Figure 7F and Shirk et al., 2011). When Kar1 was overproduced in Cdc5-depleted (PDMC1-KAR1 PCLB2-CDC5) cells, SPBs separated, but their duplication was largely inhibited (Figure 7G), supporting the idea that Cdc5 is required for SPB duplication in meiosis (Shirk et al., 2011).
Kar1 plays a role in licensing SPB duplication during yeast meiosis
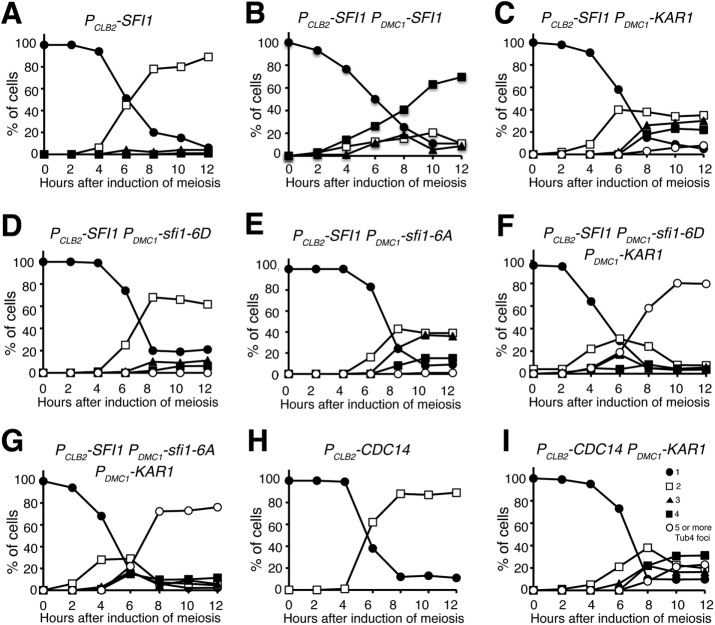
Previous work with vegetative yeast cells has shown that phosphorylation at the C-terminus of the half-bridge component Sfi1, mediated by both Cdk1 and Cdc5, regulates SPB separation (Avena et al., 2014; Elserafy et al., 2014), whereas dephosphorylation of Sfi1, presumably by the phosphatase Cdc14 (Fox et al., 2017), licenses SPB duplication. To determine the requirement of Sfi1 in meiotic cell division, we generated a meiosis-specific allele, PCLB2-SFI1, to deplete Sfi1 in meiosis (Figure 8A). By fluorescence microscopy, we found that in PCLB2-SFI1 cells, SPBs, marked by Tub4-mApple, separated and formed an anaphase-I-like elongated spindle (Figure 8A), but mutant cells never underwent the second round of SPB duplication and separation (Figure 8A). Of note, 12 h after the induction of meiosis, more than 60% PCLB2-SFI1 cells formed dyads, therefore exiting meiosis prematurely (Figure 8A). We constructed PDMC1-SFI1 to express SFI1 and its mutant forms specifically in meiosis (Figure 8B and see below). Importantly, we found that meiotic Sfi1 produced by PDMC1-SFI1 suppressed the phenotype of PCLB2-SFI1, because cells of PCLB2-SFI1 PDMC1-SFI1 completed meiosis with two rounds of SPB duplication and separation and formed tetrads at the end of meiosis (Figure 8B). We therefore conclude that Sfi1 is an essential SPB component required for SPB duplication in meiosis, as it is in mitosis (Kilmartin, 2003).
FIGURE 8:
Kar1 bypasses the requirement of Sfi1 dephosphorylation for SPB duplication in meiosis. Yeast cells were induced to undergo synchronous meiosis, and cell aliquots were withdrawn at indicated times. Live-cell fluorescence microscopy was performed to determine Tub4-marked SPB dynamics. (A, B) Quantification of SPB separation in Sfi1-depleted (PCLB2-SFI1) meiotic cells. Note that in the absence of Sfi1, SPBs were separated, but yeast cells were arrested in meiosis I. Ectopically produced Sfi1 permitted a second round of SPB duplication and cell progression in meiosis. (C) Quantification of SPB separation in PCLB2-SFI1 PDMC1-KAR1 cells. Note that more than 50% of cells underwent a second round of SPB duplication. (D, E) Quantification of SPB separation in PCLB2-SFI1 PDMC1-sfi1-6D and PCLB2-SFI1 PDMC1-sfi1-6D cells. Note that ectopically produced Sfi1-6A, but not Sfi1-6D, partially suppressed the phenotype of PCLB2-SFI1 in meiosis. (F, G) Quantification of SPB separation in PCLB2-SFI1 PDMC1-sfi1-6D PDMC1-KAR1 and PCLB2-SFI1 PDMC1-sfi1-6D PDMC1-KAR1 cells. Note that five or more SPBs formed in the above strains in meiosis. (H, I) Quantification of SPB separation in PCLB2-CDC14 and PCLB2-CDC14 PDMC1-KAR1 cells. Depletion of Cdc14 (PCLB2-CDC14) in meiosis prevents cell exit from meiosis I. At least 100 cells were counted at each time point. The time-course experiments were repeated, and data from one representative experiment are shown.
To determine the interplay between Kar1 and Sfi1 in meiotic SPB duplication, we overexpressed Kar1 in cells that were depleted of Sfi1 (PDMC1-KAR1 PCLB2-SFI1) and observed SPB dynamics using Tub4-mApple as a marker (Figure 8C). Surprisingly, more than 40% of PDMC1-KAR1 PCLB2-SFI1 cells formed three or four Tub4 foci 8 h after the induction of meiosis (Figure 8C), demonstrating that overproduced Kar1 promotes SPB duplication even when Sfi1 is limited in quantity. However, five or more Tub4 foci were rarely formed in PDMC1-KAR1 PCLB2-SFI1 cells, indicating that Sfi1, a structural component of the half bridge, is required for supernumerary SPB formation when Kar1 is overproduced.
To determine whether Kar1 can override the requirement of Sfi1 for licensing SPB duplication, we generated meiotic alleles sfi1-6A and sfi1-6D, in which the six phosphorylation sites located at the C-terminus of Sfi1 were mutated to alanine or aspartic acid to eliminate phosphorylation or mimic phosphorylation, respectively (Figure 8, D–G). Dephosphorylation of Sfi1 is required for licensing SPB duplication, because the phosphomimetic sfi1-6D mutant fails to duplicate SPB (Elserafy et al., 2014). We found that PCLB2-SFI1 PDMC1-sfi1-6D cells mostly retained two Tub4 foci during meiosis (Figure 8D), indicating that dephosphorylation of Sfi1 is critical for SPB duplication in meiosis. Supporting this idea, only 50% PCLB2-SFI1 PDMC1-sfi1-6A cells formed three or four Tub4 foci (Figure 8E). If phosphoregulation of Sfi1 is the rate-limiting factor for SPB duplication, then we would expect overproducing Kar1 to have no effect on SPB duplication in the sfi1-6D background. In contrast, when Kar1 was overproduced in both PDMC1-KAR1 PCLB2-SFI1 PDMC1-sfi1-6A and PDMC1-KAR1 PCLB2-SFI1 PDMC1-sfi1-6D cells, SPBs overduplicated, forming five or more Tub4-mApple foci (Figures 8, F and G). These findings demonstrate that Kar1 can override the requirement of Sfi1 dephosphorylation in licensing SPB duplication during meiosis.
To provide further evidence that Kar1 licenses SPB duplication in meiosis, we constructed the PCLB2-CDC14 mutant (Figure 8, H and I), which depleted Cdc14 in meiosis, thereby interfering with Sfi1 dephosphorylation (Fox et al., 2017). After being induced to undergo meiosis, PCLB2-CDC14 cells were arrested at an anaphase-I-like stage with separated SPBs and elongated spindles (Figure 8H and unpublished data). Overproducing Kar1 in PDMC1-KAR1 PCLB2-CDC14 cells led to the formation of five or more Tub4-mApple foci (Figure 8I). Therefore, we conclude that Kar1 plays an important role in licensing SPB duplication in yeast meiosis.
DISCUSSION
In this article, we have revealed an unexpected function of Kar1, which promotes SPB separation and licenses SPB duplication during budding yeast meiosis. We have shown that Kar1, when produced at an elevated level, induces SPB overduplication, resulting in supernumerary SPB formation during meiosis. In the absence of Kar1 or when Kar1 is overproduced, yeast cells fail to initiate SPB duplication during vegetative growth (Rose and Fink, 1987; Vallen et al., 1994). Similarly, we have found that depletion of Kar1 in meiosis leads to defective SPB duplication and spore formation (our published data). However, overproduced Kar1 leads to premature SPB separation at meiotic prophase I and supernumerary SPB formation on the onset of metaphase I. Kar1-induced supernumerary SPBs are functional because they can nucleate microtubules and form spindles that necessitate meiotic chromosome segregation. With excessive spindles formed in cells with overproduced Kar1, chromosome segregation becomes erroneous, forming inviable spores. During ascospore development, membranes are nucleated at the meiotic plaque, a modified outer plaque of the SPB, and then they extend to enclose the entire spore on the completion of meiosis (Neiman, 2011). Our observation that five or more ascospores frequently form in Kar1-overproduced cells further supports the idea that the supernumerary SPBs formed in these cells are functional in nucleating both microtubules for spindle formation and membranes for ascospore development. Finally, cells with excessive Kar1 overduplicate the SPB regardless of the phosphorylation status of Sfi1, demonstrating that in addition to Sfi1, Kar1 plays a role in licensing SPB duplication during yeast meiosis.
How does Kar1 promote SPB duplication?
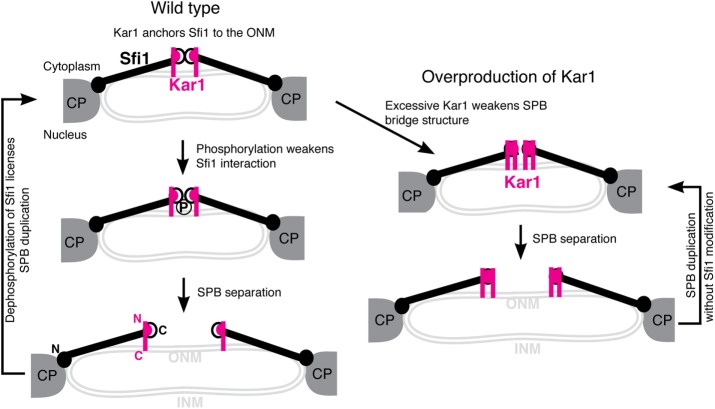
We have shown that excessive Kar1 leads to SPB premature separation at prophase I, indicating that Kar1 destabilizes the SPB bridge, which links duplicated SPBs. Kar1 binds to the C-terminus of Sfi1, where dimerization of Sfi1 is achieved (Li et al., 2006). Dimerized Sfi1 molecules provide the structural support of the SPB bridge (Li et al., 2006; Anderson et al., 2007; Seybold et al., 2015). We favor a model in which excessive Kar1 interferes with the Sfi1 dimerization domain and therefore weakens the bridge structure and relieves the inhibitory signal for SPB duplication (Figure 9). Meiotic Kar1 appears to act differently from its mitotic version, because in vegetative yeast cells, overexpression of Kar1 forms a highly elongated SPB bridge (Seybold et al., 2015). Alternatively, excessive Kar1 in meiosis could activate motor proteins, such as Cin8 (Hoyt et al., 1992), to promote SPB separation. We note that the above two scenarios are not mutually exclusive. Finally, a possibility currently has not been ruled out that excessive Kar1 may cause SPB fragmentation and thereby leads to the formation of multinucleated sporelike structures. Nevertheless, removal of the karyogamy domain, Kar1-Δ15, retains Kar1’s activity in promoting SPB separation, indicating that Kar1’s role in promoting SPB disjunction is separable from its role in nucleating microtubules. On the other hand, deletion of the region 1 domain, which binds to Sfi1 (Seybold et al., 2015), abolishes Kar1’s function in SPB duplication, consistent with the previous finding that region 1 is critical for SPB duplication (Vallen et al., 1992; Seybold et al., 2015).
FIGURE 9:
Model for excessive Kar1-mediated SPB separation and duplication in yeast meiosis. For simplicity, Cdc31 and Mps3 are omitted from the SPB half bridge. CP, central plaque; INM, inner nuclear membrane; ONM, out nuclear membrane; N, the N-terminus; C, the C-terminus.
We have found that overproduced Kar1 promotes SPB duplication to form supernumerary SPBs in meiosis, but SPB overduplicates only after metaphase I. These findings indicate that the execution point of Kar1-mediated SPB duplication occurs after the onset of metaphase I. Our EM analysis of the meiotic SPB reveals that morphologically, SPBs appear normal in Kar1-overproduced cells. This observation is consistent with the idea that the supernumerary SPBs found in Kar1-overproduced cells are competent in nucleating nuclear microtubules to form spindles and mediate chromosome segregation. But the rate of SPB duplication is accelerated in Kar1-overproduced cells, leading to supernumerary SPB formation. In Kar1-overproduced cells, the third SPB emerges within ∼15 min after the first round of SPB separation (this article), which is about one-fourth of the duration of normal SPB duplication that is observed for meiosis II in wild-type cells (Shirk et al., 2011). Duplication of the SPB starts with the deposition of the satellite material at the distal end of the SPB bridge (Cavanaugh and Jaspersen, 2017); we speculate that increased protein level of Kar1 also promotes the nucleation of the satellite materials at the distal tip of the bridge.
We have shown that Sfi1 is essential for meiotic SPB duplication. However, in the presence of excessive Kar1, dephosphorylation of Sfi1 becomes less important to the regulation of SPB duplication. Our finding therefore is in contrast to that previously observed in mitotic cells where dephosphorylation of Sfi1 at anaphase licenses SPB duplication (Avena et al., 2014; Elserafy et al., 2014). Excessive Kar1 appears to override the requirement of changing the phosphorylation status of Sfi1 in meiosis. Supporting this idea, the SPB bridge appears intact in cells without the Sfi1 C-terminus, to which Kar1 binds and where the key Sfi1 phosphorylation sites are located (Avena et al., 2014; Elserafy et al., 2014; Seybold et al., 2015). Importantly, removal of the C-terminus of Sfi1 maintains yeast cell viability at room temperature (Seybold et al., 2015). We speculate that the C-terminus of Sfi1 becomes dispensable in the presence of excessive Kar1 in meiosis. Alternatively, independently of Sfi1, Kar1 may play a separate role in licensing SPB duplication in meiosis, which remains to be further determined.
Is meiosis unique for SPB duplication?
How, then, is SPB duplicated in meiosis? As in mitosis, SPB duplication requires Cdk1’s kinase activity in meiosis (Carlile and Amon, 2008; Miller et al., 2012). Activation of the B-type cyclin Cdk1 activity occurs early during the vegetative cell cycle; in contrast, the B-type cyclins, including Clb1, Clb3, Clb4, Clb5, and Clb6, are uniquely regulated in meiosis to ensure proper meiotic cell progression (Marston and Amon, 2004; Carlile and Amon, 2008; Miller et al., 2012). Clb5- and Clb6-mediated Cdk1 activity is required for DNA replication and meiotic recombination and acts early in meiosis, whereas Clb1, Clb3, and Clb4 activate Cdk1 after the onset of metaphase I (Marston and Amon, 2004). In particular, CLB3 is expressed in meiosis I, but its protein product only appears in meiosis II (Carlile and Amon, 2008). Our finding that supernumerary SPBs only form after the onset of metaphase I in Kar1-overproduced cells supports the idea that Clb1/3/4-Cdk1 activity is required for SPB duplication in meiosis (Miller et al., 2012). In Kar1-overproduced cells, SPBs separate prematurely, and Kar1 licenses SPB duplication, therefore supernumerary SPB formation ensues. Finally, the SPB outer plaque is dramatically modified with meiosis-specific factors early on during meiosis (Neiman, 2011), which could exert a unique impact on SPB duplication in yeast meiosis.
The Kar1 protein appears to be highly modified during meiosis on the basis of its protein migration pattern (this work). Our preliminary study indicates that Kar1 is phosphorylated at 13 different sites in meiosis (unpublished data). Whether Kar1 phosphorylation plays a role in meiotic SPB duplication and separation will be determined in future studies.
Summary
This study reveals an unexpected role of Kar1 in licensing SPB duplication during yeast meiosis. We have shown that excessive Kar1 promotes SPB separation and induces supernumerary SPB formation, which causes chromosome missegregation. Kar1-mediated SPB duplication overrides the requirement of changing the phosphorylation status of Sfi1, demonstrating that Kar1 serves as an alternative licensing factor for SPB duplication during yeast meiosis.
MATERIALS AND METHODS
Yeast strains
All strains used in this study (Supplemental Table S1) are diploids isogenic to the SK1 genetic background. To generate meiosis-specific depletion alleles, including PCLB2-KAR1, PCLB2-SFI1, and PCLB2-CDC14, we used a PCR-based strategy to replace the endogenous promoters with the mitosis-specific promoter from CLB2 (Lee and Amon, 2003). A similar PCR-based strategy was used to tag Sfi1 with GFP to its C-terminus (Longtine et al., 1998). The following alleles have been reported previously: PCLB2-IPL1, PCLB2-CDC5, PCLB2-CDC20, GAL4.ER, PGAL-NDT80, TUB4-mApple, TUB4-GFP, MPS3-mApple, and TUB1-mApple (Lee and Amon, 2003; Carlile and Amon, 2008; Shirk et al., 2011; Li et al., 2015). These gene mutations and fluorescent-protein–tagged alleles were first generated in Mata and Matα haploid cells; homozygous diploids (zygotes) were obtained by mating the corresponding haploids.
Plasmids
Plasmids used in this study are included in Supplemental Table S2. To overproduce Kar1 in meiosis, we used the enhanced DMC1 promoter (Fan et al., 2017) to construct PDMC1-GFP-KAR1 (pHG433) and its variants (plasmids pHG465, pHG524, pHG549, and pHG614) by placing the KAR1 open reading frame under the control of the DMC1 promoter. The deletion alleles of kar1-∆15 and kar1-∆18 have been previously reported (Vallen et al., 1992). To delete Kar1’s transmembrane domain, we generated PDMC1-GFP-KAR1-ΔTMD (pHG549) by removing the coding sequence of the C-terminal 22 amino acids of Kar1. The PDMC1-GFP-KAR1 allele and its variants were linearized by AflII and incorporated at the LEU2 locus. Yeast strains with PDMC1-GFP-KAR1 and its variants retain the wild-type copy of the KAR1 gene. To construct PKAR1-GFP-KAR1 (pHG465), we cloned a 418–base pair fragment upstream of the KAR1 open reading frame to replace the DMC1 promoter in pHG433. The plasmid pHG465, which harbors the URA3 gene, was linearized by BglII and inserted at the KAR1 locus. URA3 positive colonies were counterselected in the 5′-FOA medium to remove the untagged copy of KAR1 (Guthrie and Fink, 1991). Therefore, GFP-KAR1, which is under the control of its endogenous promoter, serves as the only copy of KAR1 in the yeast genome (Figures 1B and 2 and Supplemental Figure S1B). Similarly, the PKAR1-V5-KAR1 allele (pHG559, Figure 1A and Supplemental Figure S1A) was under the control of its endogenous promoter and serves as the only copy of KAR1 in the yeast genome. The SFI1 open reading frame was cloned and placed under the control the DMC1 promoter to generate PDMC1-SFI1 (pHG254). The alleles of sfi1-6A and sfi1-6D have been previously reported (Elserafy et al., 2014), and they were under the control of PDMC1 for expression in meiosis (pHG434 and pHG435).
Yeast culture methods
A previously described method was used to induce yeast cells to undergo synchronous meiosis (Li et al., 2015). Briefly, yeast colonies were inoculated in the yeast extract, peptone, dextrose (YPD) medium overnight and then diluted in the yeast extract, peptone, potassium acetate (YPA) medium to reach an OD600 of 0.15–0.2. Yeast cells in YPA were vigorously shaken for ∼16 h to reach an OD600 of 1.4–1.6 and then transferred to the sporulation medium (2% potassium acetate); the time of transfer is referred to as t = 0 of meiosis in our time-course experiments. To induce the expression of the GAL promoter in meiosis, 2 µM β-estradiol (final concentration) was added to the culture medium 5 h after the transferring to the sporulation medium. An equal volume of ethanol was added to the control strains. All yeast cultures were grown at 30°C.
Live-cell fluorescence microscopy
A DeltaVision microscope system was used to acquire live-cell fluorescence images as before (Li et al., 2015). Briefly, an agarose pad with 2% potassium acetate was prepared on a concave slide, and a small aliquot of yeast cells was placed on the agarose, sealed with a coverslip, and scoped for desired time duration (Shirk et al., 2011). Images were acquired with a 63× (NA = 1.40) objective lens on an inverted microscope (IX-71, Olympus) equipped with a CoolSNAP HQ2 charge-coupled device camera (Photometrics). For time-lapse experiments, time interval was set at either 3 or 5 min, and 12 optical sections with 0.5-µm thickness were acquired at each time point. To reduce phototoxicity, we used a neutral density filter to reduce the intensity of the excitation light to ∼30% or less of the equipment output.
Light microscopic data analysis
Microscopy images obtained from DeltaVision were deconvolved using SoftWorx. To determine the pole-to-pole distance in three dimensions (Figure 1C and Supplemental Figure S2), we used the point-to-point measurement tool provided by SoftWorx. To determine the fluorescence intensity of GFP-Kar1 and Tub4-mApple at the SPB (Supplemental Figure 1), we defined a 6 × 6 pixel area and obtained the total fluorescence intensity from single optical sections. The net intensity was determined by subtracting the mean background from the total. We also used SoftWorx to generate the line-scan-intensity graphs shown in Figure 2. Projected images from multiple optical sections were used for display. For time-course experiments, we repeated at least twice, and data from one representative experiment are used for display.
Electron microscopy
Yeast cells were induced to undergo synchronous meiosis as described above. Eight hours after induction, cells were harvested by centrifugation and frozen on the Leica EM-PACT high-pressure freezer (Wetzlar, Germany) at ∼2050 bar and then transferred to the Leica EM AFS for freeze substitution under liquid nitrogen into 2% osmium tetroxide/0.2% uranyl acetate/2% water/acetone. We used the following procedure for freeze substitution: –90°C to –80°C over 60 h, –80°C to –60°C over 5 h, −60°C for 4 h, –60°C to –20°C over 8 h, and –20°C to 0°C over 5 h, and, finally, samples were held at 0°C for 3 h. Samples were then removed from the AFS and brought to room temperature. Samples went through four changes of acetone over 1 h and were removed from the planchettes. They were embedded in acetone/Spurr’s mixtures to a final concentration of 100% Spurr’s over several days in a stepwise procedure as previously described (McDonald, 1999). Seventy-five-nanometer serial thin sections were cut on a Leica UC6 ultramicrotome, stained with uranyl acetate and Sato’s lead, and imaged on a Zeiss Merlin SEM (Oberkochen, Germany) equipped with a STEM detector and Atlas software. The pixel size of acquired images was set at 4 nm. SPB plaque width and bridge size were determined on the basis of their pixel numbers. Original EM data can be downloaded from the Stowers Original Data Repository at http://www.stowers.org/pubs/LIBPB-1305.
Western blotting
We used a similar Western blotting procedure as described previously (Li et al., 2015). Briefly, yeast cells were induced to undergo synchronous meiosis, cell aliquots were collected, and protein extracts were prepared for SDS–PAGE analysis. To determine the level of V5-Kar1, we used an anti-V5 antibody (1:10,000; Thermo Fisher Scientific, cat#CAB1001). The level of Pgk1 served as a loading control.
Supplementary Material
Acknowledgments
We thank Yanchang Wang and Robert Tomko for discussions. Mark Rose and Elmar Schiebel provided plasmids and yeast strains. Jen Kennedy assisted with text editing. Research in the Jaspersen lab is supported by the Stowers Institute for Medical Research and the National Institute of General Medicine of the National Institutes of Health (NIH) under award number R01 GM121443 (to S.L.J.). Research in the Yu lab is supported by NIH GM117102 and National Science Foundation MCB1121771 (to H.Y.).
Abbreviations used:
- EM
electron microscopy
- GFP
green fluorescent protein
- SPB
spindle pole body
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-03-0163) on May 30, 2018.
REFERENCES
- Adams IR, Kilmartin JV. (1999). Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J Cell Biol , 809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams IR, Kilmartin JV. (2000). Spindle pole body duplication: a model for centrosome duplication? Trends Cell Biol , 329–335. [DOI] [PubMed] [Google Scholar]
- Anderson VE, Prudden J, Prochnik S, Giddings TH, Jr, Hardwick KG. (2007). Novel sfi1 alleles uncover additional functions for Sfi1p in bipolar spindle assembly and function. Mol Biol Cell , 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avena JS, Burns S, Yu Z, Ebmeier CC, Old WM, Jaspersen SL, Winey M. (2014). Licensing of yeast centrosome duplication requires phosphoregulation of sfi1. PLoS Genet , e1004666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns S, Avena JS, Unruh JR, Yu Z, Smith SE, Slaughter BD, Winey M, Jaspersen SL. (2015). Structured illumination with particle averaging reveals novel roles for yeast centrosome components during duplication. Elife , e08586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. (1974). Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb Symp Quant Biol , 123–131. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L. (1975). Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol , 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Amon A. (2008). Meiosis I is established through division-specific translational control of a cyclin. Cell , 280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh AM, Jaspersen SL. (2017). Big lessons from little yeast: budding and fission yeast centrosome structure, duplication, and function. Annu Rev Genet , 361–383. [DOI] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. (1998). The transcriptional program of sporulation in budding yeast. Science , 699–705. [DOI] [PubMed] [Google Scholar]
- Crasta K, Lim HH, Giddings TH, Jr, Winey M, Surana U. (2008). Inactivation of Cdh1 by synergistic action of Cdk1 and polo kinase is necessary for proper assembly of the mitotic spindle. Nat Cell Biol , 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elserafy M, Saric M, Neuner A, Lin TC, Zhang W, Seybold C, Sivashanmugam L, Schiebel E. (2014). Molecular mechanisms that restrict yeast centrosome duplication to one event per cell cycle. Curr Biol , 1456–1466. [DOI] [PubMed] [Google Scholar]
- Fan J, Jin H, Yu HG. (2017). A dual-color reporter assay of cohesin-mediated gene regulation in budding yeast meiosis. Methods Mol Biol , 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox C, Zou J, Rappsilber J, Marston AL. (2017). Cdc14 phosphatase directs centrosome re-duplication at the meiosis I to meiosis II transition in budding yeast. Wellcome Open Res , 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon O, Taxis C, Keller PJ, Benjak A, Stelzer EH, Simchen G, Knop M. (2006). Nud1p, the yeast homolog of Centriolin, regulates spindle pole body inheritance in meiosis. EMBO J , 3856–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. (1991). Guide to yeast genetics and molecular biology. Methods Enzymol . [PubMed] [Google Scholar]
- Haase SB, Winey M, Reed SI. (2001). Multi-step control of spindle pole body duplication by cyclin-dependent kinase. Nat Cell Biol , 38–42. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. (1992). Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol , 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Giddings TH, Jr, Winey M. (2002). Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J Cell Biol , 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Huneycutt BJ, Giddings TH, Jr, Resing KA, Ahn NG, Winey M. (2004). Cdc28/Cdk1 regulates spindle pole body duplication through phosphorylation of Spc42 and Mps1. Dev Cell , 263–274. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. (2004). The budding yeast spindle pole body: structure, duplication, and function. Annu Rev Cell Dev Biol , 1–28. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV. (2003). Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J Cell Biol , 1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Meyer R, Chuong H, Dawson DS. (2013). Dual mechanisms prevent premature chromosome segregation during meiosis. Genes Dev , 2139–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Amon A. (2003). Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science , 482–486. [DOI] [PubMed] [Google Scholar]
- Li P, Jin H, Koch BA, Abblett RL, Han X, Yates JR, 3rd, Yu HG. (2017). Cleavage of the SUN-domain protein Mps3 at its N-terminus regulates centrosome disjunction in budding yeast meiosis. PLoS Genet , e1006830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Shao Y, Jin H, Yu HG. (2015). Ndj1, a telomere-associated protein, regulates centrosome separation in budding yeast meiosis. J Cell Biol , 247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Sandercock AM, Conduit P, Robinson CV, Williams RL, Kilmartin JV. (2006). Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J Cell Biol , 867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast , 953–961. [DOI] [PubMed] [Google Scholar]
- Marston AL, Amon A. (2004). Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol , 983–997. [DOI] [PubMed] [Google Scholar]
- McDonald K. (1999). High-pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabeling. Methods Mol Biol , 77–97. [DOI] [PubMed] [Google Scholar]
- Miller MP, Unal E, Brar GA, Amon A. (2012). Meiosis I chromosome segregation is established through regulation of microtubule-kinetochore interactions. Elife , e00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Rapport E. (1971). Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen). J Cell Biol , 344–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM. (2011). Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics , 737–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Fink GR. (1987). KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell , 1047–1060. [DOI] [PubMed] [Google Scholar]
- Ruthnick D, Schiebel E. (2016). Duplication of the yeast spindle pole body once per cell cycle. Mol Cell Biol , 1324–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybold C, Elserafy M, Ruthnick D, Ozboyaci M, Neuner A, Flottmann B, Heilemann M, Wade RC, Schiebel E. (2015). Kar1 binding to Sfi1 C-terminal regions anchors the SPB bridge to the nuclear envelope. J Cell Biol , 843–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirk K, Jin H, Giddings TH, Jr, Winey M, Yu HG. (2011). The Aurora kinase Ipl1 is necessary for spindle pole body cohesion during budding yeast meiosis. J Cell Sci , 2891–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Courtney I, Fackler U, Matzner M, Schiebel E. (1993). The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J Cell Biol , 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Courtney I, Grein K, Matzner M, Schiebel E. (1995). The Cdc31p-binding protein Kar1p is a component of the half bridge of the yeast spindle pole body. J Cell Biol , 863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen EA, Hiller MA, Scherson TY, Rose MD. (1992). Separate domains of KAR1 mediate distinct functions in mitosis and nuclear fusion. J Cell Biol , 1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen EA, Ho W, Winey M, Rose MD. (1994). Genetic interactions between CDC31 and KAR1, two genes required for duplication of the microtubule organizing center in Saccharomyces cerevisiae. Genetics , 407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Byers B. (1993). Assembly and functions of the spindle pole body in budding yeast. Trends Genet , 300–304. [DOI] [PubMed] [Google Scholar]
- Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. (1995). NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol , 6572–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.