Abstract
Background:
MicroRNAs (miRNAs) have been extensively studied over the decades and have been identified as potential molecular targets for cancer therapy. To date, many miRNAs have been found participating in the tumorigenesis of non-small cell lung cancer (NSCLC). The present study was designed to evaluate the functions of miR-125b-1-3p in NSCLC cells.
Methods:
MiR-125b-1-3p expression was detected in tissue samples from 21 NSCLC patients and in NSCLC cell lines using the real-time polymerase chain reaction. A549 cell lines were transfected with a miR-125b-1-3p mimic or miR-125b-1-3p antisense. Cell counting kit-8, wound healing, Matrigel invasion assays, and flow cytometry were used to assess the effects of these transfections on cell growth, migration, invasion, and apoptosis, respectively. Western blotting was used to detect apoptosis-related proteins, expression of S1PR1, and the phosphorylation status of STAT3. Significant differences between groups were estimated using Student's t-test or a one-way analysis of variance.
Results:
MiR-125b-1-3p was downregulated in NSCLC samples and cell lines. Overexpression of miR-125b-1-3p inhibited NSCLC cell proliferation (37.8 ± 9.1%, t = 3.191, P = 0.013), migration (42.3 ± 6.7%, t = 6.321, P = 0.003), and invasion (57.6 ± 11.3%, t = 4.112, P = 0.001) and simultaneously induced more NSCLC cell apoptosis (2.76 ± 0.78 folds, t = 3.772, P = 0.001). MiR-125b-1-3p antisense resulted in completely opposite results. S1PR1 was found as the target gene of miR-125b-1-3p. Overexpression of miR-125b-1-3p inhibited S1PR1 protein expression (27.4 ± 6.1% of control, t = 4.083, P = 0.007). In addition, S1PR1 siRNA decreased STAT3 phosphorylation (16.4 ± 0.14% of control, t = 3.023, P = 0.015), as in cells overexpressing miR-125b-1-3p (16.7 ± 0.17% of control, t = 4.162, P = 0.026).
Conclusion:
Our results suggest that miR-125b-1-3p exerts antitumor functions in NSCLC cells by targeting S1PR1.
Keywords: Apoptosis, miR-125b-1-3p, Non-Small Cell Lung Cancer, S1PR1, STAT3
摘要
背景:
微小RNA作为潜在的肿瘤治疗靶点目标,近年来引起了诸多研究者的关注。迄今为止,有诸多miRNA被发现在非小细胞肺癌(NSCLC)中起到重要作用。本次研究目的探索miR-125-1-3p在NSCLC中的作用及功能。
方法:
MiR-125b-1-3p在21个NSCLC患者组织以及NSCLC细胞系中的表达水平采用RT-PCR进行检测。利用miR-125b-1-3p mimic 或者 miR-125b-1-3p antisense转染NSCLC A549细胞系用来过表达或沉默miR-125b-1-3p。CCK-8, 划痕实验,侵袭实验以及流 式细胞分别用来检测miR-125b-1-3p对细胞增殖,迁移,侵入以及凋亡的影响。Western blotting用来检测凋亡相关蛋白,S1PR1 蛋白以及STAT3蛋白的磷酸化状态。
结果:
MiR-125b-1-3p在NSCLC组织以及细胞系中表达下调。过表达miR-125b-1-3p后可以抑制NSCLC细胞的增殖 (37.8 ± 9.1%, t = 3.191, P = 0.013),迁移 (42.3 ± 6.7%, t = 6.321, P = 0.003) 以及侵袭 (57.6 ± 11.3%, t = 4.112, P = 0.001)。与此同时,过 表达miR-125b-1-3p可以同时诱导NSCLC细胞凋亡 (2.76 ± 0.78 fold, t = 3.772, P = 0.001)。抑制miR-125b-1-3p则取得相反的表 型结果。S1PR1被发现是miR-125b-1-3p的调控基因。过表达miR-125b-1-3p能够抑制S1PR1蛋白的表达 (27.4 ± 6.1% of control, t = 4.083, P = 0.007)。此外过表达miR-125b-1-3p (16.7 ± 0.17% of control, t = 4.162, P = 0.026) 以及沉默S1PR1蛋白 (16.4 ± 0.14% of control, t = 3.023, P = 0.015) 后能够抑制STAT3蛋白的磷酸化。
结论:
我们的发现揭示了miR-125b-1-3能够通过抑制S1PR1,从而在NSCLC中起到抑癌基因的作用。
INTRODUCTION
Lung cancer is the leading cause of cancer-related mortality worldwide and represents a severe public health issue.[1] Non-small cell lung cancer (NSCLC) is the major form and accounts for over 80% of all cases of lung cancers.[2] Despite great treatment advances have been made over the past decades, the 5-year overall survival rate is only 16% for all stages of NSCLC.[3] The initiation and progression of NSCLC are due to dysregulation of oncogenes and tumor-repressor genes. However, the molecular mechanisms underlying NSCLC have not been fully elucidated yet. Hence, it is important to identify new molecular mechanisms underlying the development of NSCLC and uncover novel therapeutic targets to improve the prognosis of patients with this disease.
MicroRNAs (miRNAs) are a class of short and noncoding RNA molecules that play critical roles in several biological processes such as proliferation, angiogenesis, apoptosis, cell differentiation, DNA damage repair, and stress responses.[4] Previous studies suggested that aberrant expression of miRNAs, such as miR-128-3p, miR-608, miR-15a, and miR-16, contributes to the pathogenesis of NSCLC.[5,6,7] However, articles about the role of iR-125b-1-3p in NSCLC are limited.
In this study, we evaluated the expression of miR-125b-1-3p in NSCLC tumor tissues and cell lines. Then, we forced expression of miR-125b-1-3p in NSCLC cells to investigate its functions. Bioinformatic analysis was applied to identify the potential targets of miR-125b-1-3p. Moreover, the effects of miR-125b-1-3p on STAT3 signaling pathway were evaluated as well.
METHODS
Ethical approval
This study was approved by the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University. Informed consents were obtained from all participants.
Patient samples and cell culture
From January 2015 to October 2017, 21 pairs of NSCLC biopsies and matched adjacent nontumor tissues were collected from patients at the First Affiliated Hospital of Wenzhou Medical University. All of the NSCLC patients were pathologically confirmed and tumor node metastasis (TNM) staging classifications were determined by the Union for International Cancer Control/American Joint Committee on Cancer 7th edition for the lung. None of the patients received antitumor treatment before. Clinical characteristics of the patients are summarized in Table 1. All the samples were divided into a miRNA high expression group (n = 10) and low expression group (n = 11) using the median miR-125b-1-3p as the cutoff point. The NSCLC cell lines such as A549, H450, H1299, and 16-HBE normal lung bronchus epithelial cells were purchased from the American Type Culture Collection. Cells were cultured in RPMI-1640 medium (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Rockford, IL, USA), 100 U/L penicillin, and 100 μg/ml streptomycin (Sigma-Aldrich, St. Louis, MO, USA). Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Table 1.
Correlation between miR-125-1-3p expression level and clinicopathological characteristics in NSCLC patients, n
| Characteristics | Low level (n = 11) | High level (n = 10) | t | P |
|---|---|---|---|---|
| Gender | ||||
| Male | 4 | 6 | 0.582 | 0.279 |
| Female | 7 | 4 | ||
| Age | ||||
| <60 years | 5 | 5 | 0.074 | 0.835 |
| ≥60 years | 6 | 5 | ||
| Tumor size | ||||
| <3 cm | 4 | 4 | 0.387 | 0.864 |
| ≥3 cm | 7 | 6 | ||
| TNM stage | ||||
| I, II | 4 | 5 | 0.489 | 0.528 |
| III, IV | 7 | 5 | ||
| Histological grade | ||||
| Well/moderate | 6 | 7 | 0.593 | 0.466 |
| Poor | 5 | 3 | ||
| T stage | ||||
| T0–T2 | 2 | 7 | 4.324 | 0.017 |
| T3–T4 | 9 | 3 | ||
| Lymph node metastasis | ||||
| Yes | 8 | 2 | 7.332 | 0.016 |
| No | 3 | 8 |
TNM: Tumor node metastasis; NSCLC: Non-small cell lung cancer.
RNA extraction, complementary DNA synthesis, and the quantitative real-time polymerase chain reaction
Total RNA was extracted using TRIZOL reagent (Invitrogen, Waltham, MA, USA) followed by reverse transcription. Complementary DNAs were then used as templates for quantitative real-time polymerase chain reaction with iQ™ SYBR® Green Supermix (Bio-Rad, Hercules, CA, USA). U6 was used as the control for normalization. Relative mRNA expression levels were calculated using the 2−ΔΔCT method. All experiments were repeated at least three times.
Oligonucleotides
MiR-125b-1-3p mimic, a mimic control, miR-125b-1-3p antisense, and an antisense control were synthesized by the GenePharma Company (Suzhou, Jiangsu, China). A549cells were plated in six-well plates (4 × 105 cells/well) for 24 h before transfection with miRNA or a plasmid using RNAiMAX (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions.
Cell Counting Kit-8 assay
The Cell Counting Kit-8 (CCK-8) assay was used to determine the viability of cells transfected with miR-125b-1-3p mimic, mimic control, miR-125b-1-3p antisense, and antisense control for 24, 48, or 72 h. All experiments were repeated at least three times.
Clonogenic assay
For the colony formation assay, 500 cells were plated in six-well plates and cultured in medium containing 10% fetal bovine serum. The medium was refreshed every 3 days. After incubation for 14 days, cells were fixed with methanol and stained with 0.1% crystal violet (Sigma-Aldrich, St. Louis, MO, USA). Visible colonies were counted manually. Triplicate wells were measured for each group.
Cell migration and invasion assays
For migration assay, cells were seeded in a serum-free medium in the upper Transwell chambers (Corning, New York, USA). Twenty-four hours later, cells were fixed with methanol and stained by 0.1% crystal violet. Migrated cells were counted under microscope and statistically analyzed. For invasion assay, Transwell chambers were coated with Matrigel (BD Bioscience, San Jose, CA, USA) to growth the cells for 24 h. Residual cells on the upper Transwell chamber were counted and statistically analyzed.
Cell apoptosis assays
An Annexin V-FITC/propidium iodide (PI) Double Staining Kit (BD Biosciences, San Jose, CA, USA) was used to detect cellular apoptosis according to the manufacturer's instructions. Cells were collected, washed with cold phosphate-buffered saline, and resuspended at 1 × 106 cells/ml in 1 ml binding buffer. Next, 5 μl Annexin V-FITC and 5 μl PI were added and incubated for 15 min at room temperature in the dark. Cells were detected by flow cytometry. All experiments were repeated three times.
Western blotting analyses
Cellular extracts were prepared by washing cells with phosphate-buffered saline and lysing in a lysis buffer containing a protease inhibitor. Protein concentrations were measured using Bradford protein assay kit (Beyotime, Haining, Jiangsu, China). Equal amounts of protein were loaded, separated by SDS-PAGE gel electrophoresis, and then transferred to PVDF membrane. Then, the membrane was blocked with 5% milk for 1 h at room temperature, they were incubated with following primary antibodies: caspase-3 (#14220), Bcl-2 (#2872), Mcl-1 (#94296), Bax (#2774), S1PR1 (#11424), p-STAT3 (#4113), STAT3 (#9196), and glyceraldehyde-3-Phosphate Dehydrogenase (#2118) (Cell Signaling Technology, Danvers, MA, USA), followed by incubation with horseradish peroxidase-conjugated secondary antibodies: antimouse (#7076) and antirabbit (#7074; Cell Signaling Technology, Danvers, MA, USA). Protein bands were visualized using an enhanced chemiluminescent substrate (Thermo Scientific, Rockford, IL, USA). Protein bands were quantified by densitometric analysis using Quantity One software (Bio-Rad Laboratories, San Diego, CA, USA).
Caspase-3 activity assay
Caspase-3 activity was determined using a Caspase-3 Colorimetric Activity Assay Kit (Beyotime, Haining, Jiangsu, China) according to the manufacturer's guidelines. Briefly, cells were collected, washed, lysed, and centrifuged. Sample lysates containing 50 μg of protein were assayed for caspase-3 activity. Absorbance was measured at 405 nm using a microplate reader (BioTek, Winooski, VT, USA).
Luciferase assays
The sequence in the 3’-untranslated region (UTR) of the human S1PR1 gene targeted by miR-125b-1-3p was predicted using microRNA.org (http://www.microrna.org/). The 3’-UTR of S1PR1 and a sequence with mutations of two nucleotides in the miR-125b-1-3p target site were cloned into a pGL3 promoter vector to generate the recombinant constructs: wild-type and mutant 3’-UTRs, respectively. For the luciferase assay, A549 cells were co-transfected with wild-type and mutant 3’-UTRs of S1PR1 and the miR-125b-1-3p mimic or scrambled controls (NC). Luciferase activity was analyzed using the Dual-Luciferase Reporter Assay System according to the manufacturer's instructions (Promega, Madison, WI, USA) at 48 h posttransfection.
Statistical analyses
All statistical analyses were performed using SPSS 18.0 software (IBM, Chicago, IL, USA). Data from at least three independent experiments, each performed in triplicate, were presented as means ± standard deviation (SD). Significant differences between groups were estimated using a one-way analysis of variance (ANOVA). A P < 0.05 was considered as statistically significant.
RESULTS
Expression levels of miR-125b-1-3p in non-small cell lung cancer samples and cell lines
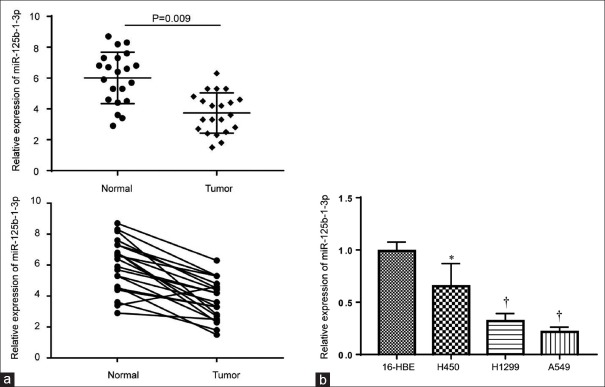
Twenty-one pairs of NSCLC biopsies and matched adjacent nontumor tissue were analyzed. In addition, we detected miR-125b-1-3p expression levels in NSCLC cell lines (A549, H450, and H1299) and in 16-HBE normal lung bronchus epithelial cells. The results showed that miR-125b-1-3p was downregulated significantly in the NSCLC samples (t = 5.112, P = 0.009; Figure 1a) and in cell lines compared to the control group (H450, t = 2.156, P = 0.036; H1299, t = 4.278, P = 0.007; and A549, t = 5.462, P = 0.006, respectively, Figure 1b). For further analysis, all the samples were divided into a miRNA high expression group (n = 10) and low expression group (n = 11) using the median miR-125b-1-3p as the cutoff point. The results showed that miR-125b-1-3p expression was not correlated with age (t = 0.074, P = 0.835), gender (t = 0.582, P = 0.279), tumor size (t = 0.387, P = 0.864), and TNM stages (t = 0.489, P = 0.528), but it was correlated with tumor stage (t = 4.324, P = 0.017) and lymph node metastasis (t = 7.332, P = 0.016; Table 1).
Figure 1.
Expression levels of miR-125b-1-3p in 21 pairs of NSCLC tissues and three different NSCLC cell lines. (a) Expression level of miR-125b-1-3p in 21 pairs of NSCLC biopsies and matched adjacent nontumor tissue. (b) Expression levels of miR-125b-1-3p in 16-HBE normal lung bronchus epithelial cells and NSCLC cell lines. *P < 0.05, †P < 0.01, compared to the 16-HBE normal lung bronchus epithelial cells. NSCLC: Non-small cell lung cancer.
Effect of miR-125b-1-3p on cell growth, migration, invasion, and apoptosis in vitro
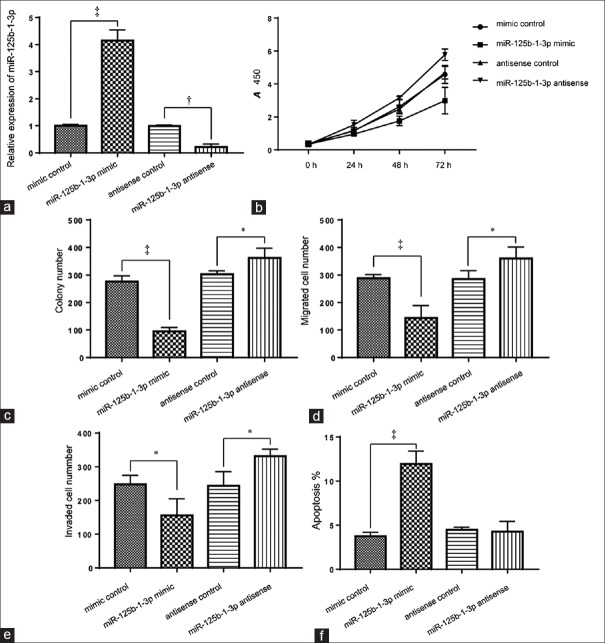
A549 cells were transfected successfully with miR-125b-1-3p mimic, mimic control, miR-125b-1-3p antisense, and antisense control. The expression levels of miR-125b-1-3p were upregulated (4.1 ± 0.23 folds vs. control, t = 5.125, P = 0.001) and decreased (15.20 ± 0.07% of control, t = 2.123, P = 0.021) after transfection with miR-125b-1-3p and miR-125b-1-3p antisense, respectively [Figure 2a]. The CCK-8 and colony formation assays revealed that overexpression of miR-125b-1-3p significantly inhibited the proliferation of A549 cells compared to the controls (37.8 ± 9.1%, t = 3.191, P = 0.013; Figure 2b and 2c). The wound healing and Transwell invasion assays showed the migration and invasion cell numbers of the miR-125b-1-3p mimic group were significantly decreased to 42.3 ± 6.7% (t = 6.321, P = 0.003) and 57.6 ± 11.3% (t = 4.112, P = 0.001) of control, respectively [Figure 2d and 2e]. In addition, overexpression of miR-125b-1-3p significantly induced more apoptosis compared with the control group (2.76 ± 0.78 folds, t = 3.772, P = 0.001; Figure 2f).
Figure 2.
Effect of miR-125b-1-3p on cell growth, migration, invasion, and apoptosis in vitro. A549 cells were transfected with miR-125b-1-3p mimic or miR-125b-1-3p antisense. (a) The relative expression of miR-125b-1-3p was detected by the quantitative real-time polymerase chain reaction. Cell viability (b), colony numbers (c), migration (d), invasion (e), and apoptosis (f) were assessed using the CCK-8, clonogenic, cell migration, invasion assays, and flow cytometry, respectively. *P < 0.05; †P < 0.01; ‡P < 0.001. CCK-8: Cell Counting Kit-8.
MiR-125b-1-3p regulates apoptosis-related proteins
Apoptosis-related proteins were detected using Western blotting to further reveal the mechanism by which miR-125b-1-3p-induced apoptosis. Overexpression of miR-125b-1-3p caused cleavage of caspase-3 and significantly increased its activity (1.81 ± 0.42 folds vs. control, t = 2.323, P = 0.012; Figure 3a and 3b). At the same time, miR-125b-1-3p overexpression downregulated the antiapoptotic proteins, Bcl-2 (47.1 ± 4.2% of control; t = 3.182, P = 0.007) and Mcl-1 (45.5 ± 2.8% of control, t = 4.112, P = 0.008) but upregulated the pro-apoptotic protein, Bax (2.72 ± 0.14 folds vs. control, t = 4.791, P = 0.001; Figure 3c).
Figure 3.
miR-125b-1-3p regulates apoptosis-related proteins. Caspase-3 (a) activity, cleaved caspase-3 levels (b), and protein levels of Bcl-2, Mcl-1, and Bax (c) were detected in A549 cells transfected with miR-125b-1-3p mimic or miR-125b-1-3p antisense. *P < 0.01. †P < 0.001. compared to the control group.
MiR-125b-1-3p directly targets the S1PR1 gene
Potential targets of miR-125b-1-3p were predicted by TargetScan. Figure 4a shows the potential miR-125b-1-3p binding sites in the S1PR1 mRNA 3’-UTR. We constructed mutations of the miR-125b-1-3p binding site in the S1PR1 3’-UTR of mRNA luciferase reporter. The miR-125b-1-3p mimic significantly reduced the relative luciferase activity of the wild-type 3’-UTR of S1PR1 compared with the scrambled control (34.3 ± 2.4% of control, t = 5.143, P = 0.000) but did not affect the mutated luciferase reporter [Figure 4b]. In addition, protein levels of S1PR1 were reduced significantly in the miR-125b-1-3p mimic group compared to the mimic control (27.4 ± 6.1% of control, t = 4.081, P = 0.007), while the miR-125b-1-3p antisense group showed the opposite effect (1.22 ± 0.63 folds vs. control, t = 2.181, P = 0.032). This demonstrated that miR-125b-1-3p directly targeted the S1PR1 gene [Figure 4c].
Figure 4.
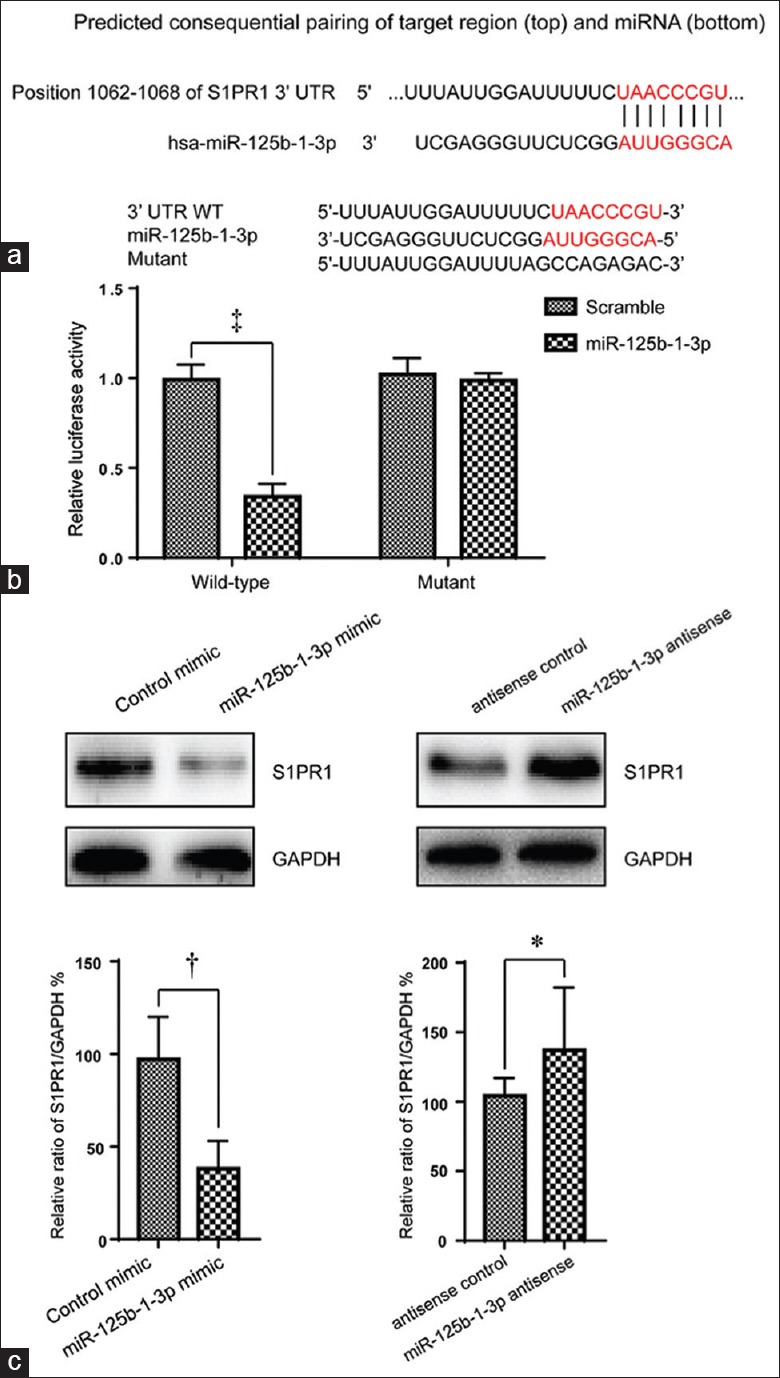
miR-125b-1-3p directly targets the S1PR1 gene. (a) Wild-type and mutations of miR-125b-1-3p binding sites in the S1PR1 mRNA 3′-UTR (red mark: seed region). (b) The luciferase assay was performed in A549 cells transfected with wild-type and mutant 3′-UTRs in the S1PR1 gene together with miR-125b-1-3p mimic or scrambled controls. (c) S1PR1 protein levels were detected by Western blotting in A549 cells transfected with miR-125b-1-3p mimic or miR-125b-1-3p antisense. *P < 0.05, †P < 0.01, ‡P < 0.001. UTR: Untranslated region.
MiR-125b-1-3p affecting the STAT3 phosphorylation status
The effects of miR-125b-1-3p on the phosphorylation level of STAT3 were determined by Western blotting. We first transfected A549 cells with S1PR1 siRNA to knock down S1PR1 expression. Figure 5a shows that S1PR1 was successfully downregulated in the S1PR1 siRNA group (12.33 ± 0.14% of control, t = 3.621, P = 0.004) and the miR-125b-1-3p mimic group (48.38 ± 0.12% of control, t = 2.163, P = 0.022). At the same time, the phosphorylation level of STAT3 was obviously decreased in the S1PR1 siRNA (16.42 ± 0.14% of control, t = 3.021, P = 0.015) and miR-125b-1-3p mimic groups (16.71 ± 0.17% of control, t = 4.162, P = 0.026), respectively [Figure 5a and 5b]. Conversely, the level of phosphorylated STAT3 obviously increased in the anti-miR-125b-1-3p group compared with the control groups (2.64 ± 0.84 folds, t = 5.332, P = 0.004). However, the total levels of STAT3 were not affected. Thus, miR-125b-1-3p affects the phosphorylation status of STAT3.
Figure 5.
miR-125b-1-3p affects the phosphorylation status of STAT3. (a) Protein levels of S1PR1, and phosphorylated and total levels of STAT3, were detected by Western blotting in A549 cells transfected with S1PR1 siRNA, miR-125b-1-3p mimic, miR-125b-1-3p antisense, or their corresponding control vectors. (b) Relative ratios of S1PR1to GAPDH and phospho (p)-STAT3 to total STAT3, *P < 0.05, †P < 0.01. GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
DISCUSSION
The prevalence of NSCLC is increasing worldwide, and the prognosis of NSCLC patients remains poor. Therefore, improved therapeutic strategies for NSCLC patients are vital for the management of NSCLC. In recent years, miRNAs have emerged as potential antitumor agents due to its ability to affect the development of various types of cancers including NSCLC.[9] Indeed, a better clarification of the functions of miRNAs in the pathogenesis of NSCLC may help in the search for more effective NSCLC therapies.
MiR-125b-1-3p has been found to play an important role in cancer and immunosuppression.[10] MiR-125b-1-3p was reportedly downregulated significantly in Kaposi's sarcoma.[11] In contrast, miR-125b-1-3p was found elevated in DICER1-mutated pleuropulmonary blastoma.[12] This study found that miR-125b-1-3p was downregulated in NSCLC cells. Those discrepancies may be due to different cancer types, since mounting evidences indicate that miRNAs are highly tissue specific and can act as either oncogenes or tumor suppressors.[13,14] For instance, miR-205 can play a dual role, depending on the specific tumor type and target genes.[15] Therefore, further investigations of miR-125b-1-3p in more cancer types are necessary. Noteworthy, we also revealed that miR-125b-1-3p exerts antitumor effects at least partly through induction of apoptosis. Dysregulated apoptosis is a common cause of cancer, and the treatment of NSCLC is usually impeded by the intrinsic or acquired resistance to apoptosis induced by anticancer agents.[16] Many studies revealed that miRNAs could overcome the resistance to chemotherapy in NSCLC. For example, miR-101 could enhance paclitaxel (PTX)-induced apoptosis in NSCLC.[17] MiR-216b was able to sensitize NSCLC cells to cisplatin-induced apoptosis.[18] Moreover, miR-129-5p could also overcome resistance to cisplatin in NSCLC cells.[19] Hence, it would be interesting to investigate whether miR-125-1-3p also possesses the ability to overcome chemoresistance in NSCLC.
To understand the functional mechanism of miRNAs, it is critical to identify targets involved in their regulation. SIPR1 was further identified as a direct functional target of miR-125-1-3p in NSCLC cells. S1PR1 is a G-protein-coupled receptor of the bioactive lipid, sphingosine-1-phosphate (S1P).[20,21] Tumor cells secrete S1P, promoting growth, survival, motility, and metastasis through binding to S1PRs in a autocrine/paracrine manner.[22] A very recent study showed that upregulation of S1PR1 could lead to the proliferation and invasion of NSCLC cells.[23] Considering S1PR1 is the predominant one compared with other S1PRs in NSCLC cells, the ability of miR-125-1-3p to repress S1PR1 might be applied as a promising strategy against NSCLC.
Aberrant STAT3 signaling is often observed in various cancers including the NSCLC. Inhibition of STAT3 signals would reduce cell proliferation and increase apoptosis of cancer cells. Many studies have been conducted to investigate the correlation between miRNAs and STAT3. For instance, miR-206 could inhibit STAT3 signaling and thereby attenuates the growth and angiogenesis of NSCLC cells.[24] MiR-124 inhibits the proliferation of NSCLC cells and functions as a tumor suppressor through targeting STAT3.[25] MiR-9600 regulates tumor progression and promotes PTX sensitivity in NSCLC through altering expression of STAT3.[26] These studies suggest that STAT3 is subjected to the regulation of various miRNAs. In this study, we found that miR-125-1-3p was also involved in the STAT3 signaling pathway. Although the mechanisms of how miR-125-1-3p affects the STAT3 signaling are not fully revealed in this study. Given that S1PR1 is crucial for the activation of STAT3 in cancer cell.[27] MiR-125-1-3p may affect the STAT3 signaling through the regulation of S1PR1, and further investigations are needed to test this hypothesis.
There are limitations of this study. First, the number of samples in this analysis is relatively small. Second, there is a lack of survival analysis due to unable to collect the survival data of patients. Third, the investigation was performed based on cell lines and future in vivo analysis is needed. In summary, we revealed that miR-125b-1-3p was downregulated and function as a tumor suppressor in NSCLC. S1PR1 was identified as a direct target of miR-125b-1-3p in NSCLC cells. Moreover, STAT3 signaling pathway was also affected by the miR-125b-1-3p. Our findings may help the development of novel therapeutic targets for the treatment of NSCLC patients.
Financial support and sponsorship
This work was supported by the grants from the Natural Science Foundation of Zhejiang Province (No. LY14H160042) and Wenzhou Municipal Foundation of Science and Technology (No. Y20160049).
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Ning-Ning Wang
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: New biological insights and recent therapeutic advances. CA Cancer J Clin. 2011;61:91–112. doi: 10.3322/caac.20102. doi: 10.3322/caac.20102. [DOI] [PubMed] [Google Scholar]
- 3.Laskin JJ, Sandler AB. State of the art in therapy for non-small cell lung cancer. Cancer Invest. 2005;23:427–42. doi: 10.1081/cnv-67172. doi: 10.1081/CNV-67172. [DOI] [PubMed] [Google Scholar]
- 4.Grimaldi A, Zarone MR, Irace C, Zappavigna S, Lombardi A, Kawasaki H, et al. Non-coding RNAs as a new dawn in tumor diagnosis. Semin Cell Dev Biol. 2017;78:37–50. doi: 10.1016/j.semcdb.2017.07.035. doi: 10.1016/j.semcdb.2017.07.035. [DOI] [PubMed] [Google Scholar]
- 5.Frixa T, Sacconi A, Cioce M, Roscilli G, Ferrara FF, Aurisicchio L, et al. MicroRNA-128-3p-mediated depletion of drosha promotes lung cancer cell migration. Carcinogenesis. 2018;39:293–304. doi: 10.1093/carcin/bgx134. doi: 10.1093/carcin/bgx134. [DOI] [PubMed] [Google Scholar]
- 6.Othman N, Nagoor NH. MiR-608 regulates apoptosis in human lung adenocarcinoma via regulation of AKT2. Int J Oncol. 2017;51:1757–64. doi: 10.3892/ijo.2017.4174. doi: 10.3892/ijo.2017.4174. [DOI] [PubMed] [Google Scholar]
- 7.Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, et al. MiR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553–9. doi: 10.1158/0008-5472.CAN-08-4277. doi: 10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- 8.Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1:49–52. doi: 10.1016/s1535-6108(02)00027-2. doi: 10.1016/S1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 9.Legras A, Pécuchet N, Imbeaud S, Pallier K, Didelot A, Roussel H, et al. Epithelial-to-mesenchymal transition and microRNAs in lung cancer. Cancers (Basel) 2017;9 doi: 10.3390/cancers9080101. pii:E101 doi: 10.3390/cancers9080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun YM, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 2013;6:6. doi: 10.1186/1756-8722-6-6. doi: 10.1186/1756-8722-6- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu XJ, Pu XM, Zhao ZF, Zhao YN, Kang XJ, Wu WD, et al. The expression profiles of microRNAs in kaposi's sarcoma. Tumour Biol. 2015;36:437–46. doi: 10.1007/s13277-014-2626-1. doi: 10.1007/s13277-014-2626-1. [DOI] [PubMed] [Google Scholar]
- 12.Murray MJ, Bailey S, Raby KL, Saini HK, de Kock L, Burke GA, et al. Serum levels of mature microRNAs in DICER1-mutated pleuropulmonary blastoma. Oncogenesis. 2014;3:e87. doi: 10.1038/oncsis.2014.1. doi: 10.1038/oncsis.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vannini I, Fanini F, Fabbri M. Emerging roles of microRNAs in cancer. Curr Opin Genet Dev. 2018;48:128–33. doi: 10.1016/j.gde.2018.01.001. doi: 10.1016/j.gde.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Wang L, Chen C, Chu X. New insights into the regulatory role of microRNA in tumor angiogenesis and clinical implications. Mol Cancer. 2018;17:22. doi: 10.1186/s12943-018-0766-4. doi: 10.1186/s12943-018-0766-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin AY, Zhang XW, Liu L, Yu JP, Li H, Wang SZ, et al. MiR-205 in cancer: An angel or a devil? Eur J Cell Biol. 2013;92:54–60. doi: 10.1016/j.ejcb.2012.11.002. doi: 10.1016/j.ejcb.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Giménez-Bonafé P, Tortosa A, Pérez-Tomás R. Overcoming drug resistance by enhancing apoptosis of tumor cells. Curr Cancer Drug Targets. 2009;9:320–40. doi: 10.2174/156800909788166600. doi: 10.2174/156800909788166600. [DOI] [PubMed] [Google Scholar]
- 17.Zhang JG, Guo JF, Liu DL, Liu Q, Wang JJ. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J Thorac Oncol. 2011;6:671–8. doi: 10.1097/JTO.0b013e318208eb35. doi: 10.1097/JTO.0b013e318208eb35. [DOI] [PubMed] [Google Scholar]
- 18.Huang G, Pan J, Ye Z, Fang B, Cheng W, Cao Z, et al. Overexpression of miR-216b sensitizes NSCLC cells to cisplatin-induced apoptosis by targeting c-jun. Oncotarget. 2017;8:104206–15. doi: 10.18632/oncotarget.22171. doi: 10.18632/oncotarget.22171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Z, Cai H, Zhang Y, Chang L, Cui Y. MiR-129-5p inhibits non-small cell lung cancer cell stemness and chemoresistance through targeting DLK1. Biochem Biophys Res Commun. 2017;490:309–16. doi: 10.1016/j.bbrc.2017.06.041. doi: 10.1016/j.bbrc.2017.06.041. [DOI] [PubMed] [Google Scholar]
- 20.Meissner A. S1PR (Sphingosine-1-phosphate receptor) signaling in the regulation of vascular tone and blood pressure: Is S1PR1 doing the trick? Hypertension. 2017;70:232–4. doi: 10.1161/HYPERTENSIONAHA.117.09200. doi: 10.1161/HYPERTENSIONAHA.117.09200. [DOI] [PubMed] [Google Scholar]
- 21.Cantalupo A, Gargiulo A, Dautaj E, Liu C, Zhang Y, Hla T, et al. S1PR1 (Sphingosine-1-phosphate receptor 1) signaling regulates blood flow and pressure. Hypertension. 2017;70:426–34. doi: 10.1161/HYPERTENSIONAHA.117.09088. doi: 10.1161/HYPERTENSIONAHA.117.09088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anelli V, Gault CR, Snider AJ, Obeid LM. Role of sphingosine kinase-1 in paracrine/transcellular angiogenesis and lymphangiogenesis in vitro. FASEB J. 2010;24:2727–38. doi: 10.1096/fj.09-150540. doi: 10.1096/fj.09-150540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Y, Luo G, Jiang B, Yu M, Feng Y, Wang M, et al. Apolipoprotein M promotes proliferation and invasion in non-small cell lung cancers via upregulating S1PR1 and activating the ERK1/2 and PI3K/AKT signaling pathways. Biochem Biophys Res Commun. 2018;501:520–6. doi: 10.1016/j.bbrc.2018.05.029. doi: 10.1016/j.bbrc.2018.05.029. [DOI] [PubMed] [Google Scholar]
- 24.Xue D, Yang Y, Liu Y, Wang P, Dai Y, Liu Q, et al. MicroRNA-206 attenuates the growth and angiogenesis in non-small cell lung cancer cells by blocking the 14-3-3ς/STAT3/HIF-1α/VEGF signaling. Oncotarget. 2016;7:79805–13. doi: 10.18632/oncotarget.12972. doi: 10.18632/oncotarget.12972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Yu Z, Li Y, Liu S, Gao C, Hou X, et al. The tumor suppressor miR-124 inhibits cell proliferation by targeting STAT3 and functions as a prognostic marker for postoperative NSCLC patients. Int J Oncol. 2015;46:798–808. doi: 10.3892/ijo.2014.2786. doi: 10.3892/ijo.2014.2786. [DOI] [PubMed] [Google Scholar]
- 26.Sun CC, Li SJ, Zhang F, Zhang YD, Zuo ZY, Xi YY, et al. The novel miR-9600 suppresses tumor progression and promotes paclitaxel sensitivity in non-small-cell lung cancer through altering STAT3 expression. Mol Ther Nucleic Acids. 2016;5:e387. doi: 10.1038/mtna.2016.96. doi: 10.1038/mtna.2016.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Priceman SJ, Shen S, Wang L, Deng J, Yue C, Kujawski M, et al. S1PR1 is crucial for accumulation of regulatory T cells in tumors via STAT3. Cell Rep. 2014;6:992–9. doi: 10.1016/j.celrep.2014.02.016. doi: 10.1016/j.celrep.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]