Abstract
Modifying RNA through either splicing or editing is a fundamental biological process for creating protein diversity from the same genetic code. Developing novel chemical biology tools for RNA editing has potential to transiently edit genes and to provide a better understanding of RNA biochemistry. Current techniques used to modify RNA include the use of ribozymes, adenosine deaminase and tRNA endonucleases. Herein, we report a nanozyme that is capable of splicing of virtually any RNA stem-loop. This nanozyme is comprised of a gold nanoparticle functionalized with three enzymes: two catalytic DNA strands with ribonuclease function and an RNA ligase. The nanozyme cleaves and then ligates RNA targets, performing a splicing reaction that is akin to the function of the spliceosome. Our results show that the three-enzyme reaction can remove a 19 nt segment from a 67 nt RNA loop with up to 66% efficiency. The complete nanozyme can perform the same splice reaction at 10% efficiency. These splicing nanozymes represent a new promising approach for gene manipulation that has potential for applications in living cells.
Keywords: RtcB, deoxyribozyme, DNAzyme, nanozyme, gold nanoparticle, synthetic biology
Tools that can manipulate nucleic acids are very powerful, capable of controlling almost all cellular outcomes. RNA in particular is a desirable and accessible target, as it is in the cytoplasm, not bound by histones and chromatin, and is thus more accessible than DNA.1 Modulating RNA can have tremendous potential for elucidating RNA biology, gene knockdown and regulating splicing. Two major methods have been developed to manipulate RNA. The first operates by modulating the activity of the spliceosome,2–3 while the second approach employs RNA modifying enzymes1, 4 and ribozymes.6 Key examples of the latter approach include adenosine deaminase1, 7 and the tRNA endonuclease from Methanococcus jannaschii (MJ-EndA).4, 8 Adenosine deaminases that act on RNA (ADAR) have been shown to create A to G point mutations by converting adenosine to inosine,1 which can be used to correct RNA errors. For example, by coupling to an antisense RNA strand and a λ-phage RNA binding protein, it can target and correct nonsense mutations in the cystic fibrosis transmembrane conductance regulator (CFTR), restoring translation at 100% efficiency.1 Alternatively, MJ-EndA functions by cleaving bulge-helix-bulge (BHB) regions in RNA. Artificial BHBs can be created in trans by introducing a guide RNA strand that recruits MJ-EndA to these RNA sequences. The cleavage product is then repaired by cellular ligases. MJ-EndA has demonstrated activity for splicing in vitro.4 This approach requires delivery of a plasmid encoding the endonuclease, along with the guide RNA strand, and has shown an efficiency as high as 30% splicing.8
In addition to protein enzymes, ribozymes6, 9–10 or catalytic RNAs, are also actively used to control RNA splicing. Originally discovered as self-splicing group I introns,11 ribozymes have been modified and used for RNA knockdown,10 intron removal,6 as well as trans-splicing of 3′12 and 5′ segments.13 Thus far, ribozyme-based editing has shown 10–50% efficiency in mammalian cells under ideal conditions.14 Given the importance of manipulating RNA in cell and molecular biology and biochemistry, the development of new approaches to modify RNA is highly desirable.
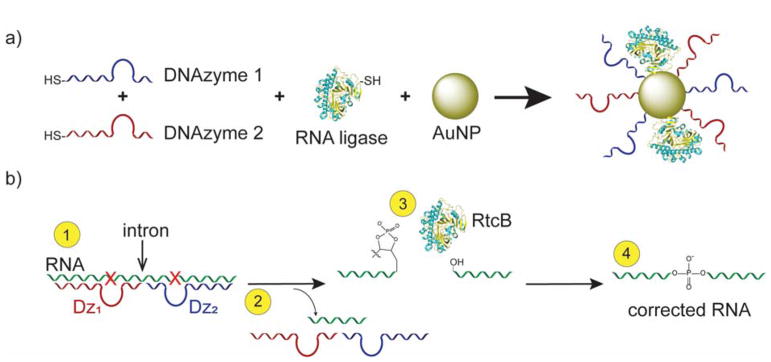
Herein, we developed a new method for RNA splicing by generating a nanozyme, (see Scheme 1) which generally refers to a particle that mimics enzymatic activity either through the property of the particle or its attached ligands.15–16 In our case, we built a splicing nanozyme by attaching RNA cleaving and ligating enzymes onto a gold nanoparticle (AuNP) scaffold. To the best of our knowledge, this technique is the first RNA splicing nanozyme.
Scheme 1.
DNAzymes and RtcB can work together in an RNA splicing reaction. (a) Nanozymes are constructed of two DNAzymes and an RNA ligase (RtcB) attached to a gold nanoparticle (AuNP) scaffold. (b) DNAzymes cleave target RNA at purine / pyrimidine junctions (red “X”), removing an intron and leaving 2′-3′-cyclic phosphates that RtcB can ligate to produce corrected RNA. Note: Crystal structure is from the P. horikoshii RtcB species [PDB=4ISZ].5
We chose the 10–23 DNAzyme17 as the site-specific RNA-cleaving component of this nanozyme (Figure S1), since mammalian cells also readily internalize DNAzyme-AuNP conjugates, allowing efficient gene knockdown in vitro and in vivo.18–19 DNAzymes are not found in nature, and are synthetic constructs generated through rounds of selection (SELEX) for a specific enzymatic activity – in this case, for RNA cleavage.17 The 10–23 DNAzyme is composed of a Mg/Mn2+ dependent20 15-nucleotide (nt) catalytic core flanked by two 6–10 nt binding arms. The binding arms can be tuned to bind any RNA target with high specificity. DNAzymes cleave at purine / pyrimidine junctions – most often AU or GU residues. After cleavage, the binding arms dissociate as they are no longer thermally stable, allowing for a new round of binding and cleavage to take place.17
RtcB was selected as the RNA ligating enzyme of the nanozyme because it is the only known ligase that can directly ligate the termini produced by DNAzymes: 2′3′-cyclic phosphate and 5′-OH.21 RtcB is well-conserved throughout bacteria, archaea and metazoa,22 having functions in bacteria for RNA repair,23 and in metazoa, for splicing of tRNAs24 and upregulating the unfolded protein response.25 RtcB’s natural substrate for ligation is hydrolyzed stem-loop RNA. Since it is expressed in most mammalian cells for RNA ligation and ligates the products of DNAzyme cleavage, it is well-suited for generating our nanozyme.
In principle, this nanozyme obviates the need for genetic engineering, providing a potential delivery vehicle into cells. It also sequesters its enzyme cargo onto the AuNP surface, away from degrading proteases and nucleases, increasing stability over current strategies.18, 26 The spliced product is not a substrate for the DNAzymes, helping to move the reaction toward completion. This technique opens the door to a new method of cellular splicing heretofore unexplored in the literature: that of coupling a natural and synthetic enzyme to splice RNA targets. Herein, we investigate and characterize the splicing of these combined enzymes, both separately and integrated onto a gold nanoparticle scaffold.
RESULTS AND DISCUSSION
RtcB is more active on stem-loops than linear RNA
To test the activity of RtcB, the ligase was isolated using a lacI inducible plasmid expressing E.coli N-terminal hexahistidine-tagged RtcB (Figure S2). Throughout this work, RtcB activity was assayed using fluorescein-labeled target RNAs and the products were quantified using 15% polyacrylamide gel electrophoresis (PAGE).
Inspired by Desai and Raines’s experiments, we first tested the action of RtcB using a 7 mer stem-loop tRNAglu mimic and found that RtcB ligated this substrate with 100% efficiency, while it ligated two 10 mer linear RNA strands with an efficiency of up to 46% (Figure S3–S4). Additionally, we found that the stem-loop target ligation was rapid, reaching completion within 2 min (Figure S5). Our results agree with Desai and Raines, who demonstrated that RtcB is more active on stem-loops than on linear RNA substrates27 and postulated that this selectivity is due to the proximity of the stem-loop termini. In subsequent studies on RtcB by Tanaka and Chakravarty, the majority of RtcB substrates tested were stem-loops or linear strands that were allowed to cyclize.23, 28 Our results further confirm that the stem loop is the preferred substrate for RtcB mediated ligation, suggesting that it will also be the preferred substrate for splicing reactions.
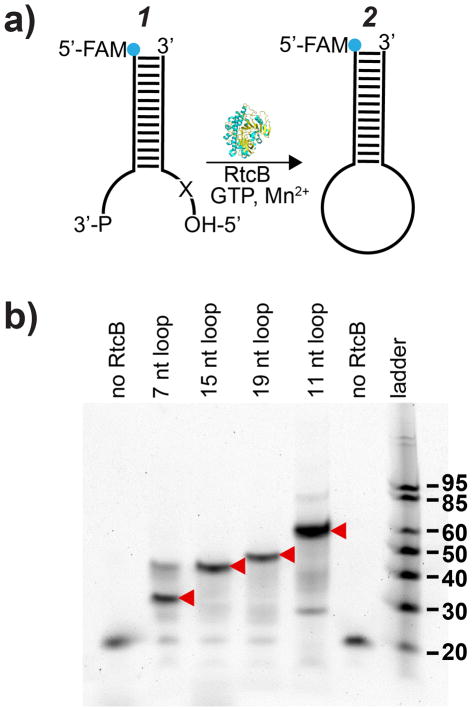
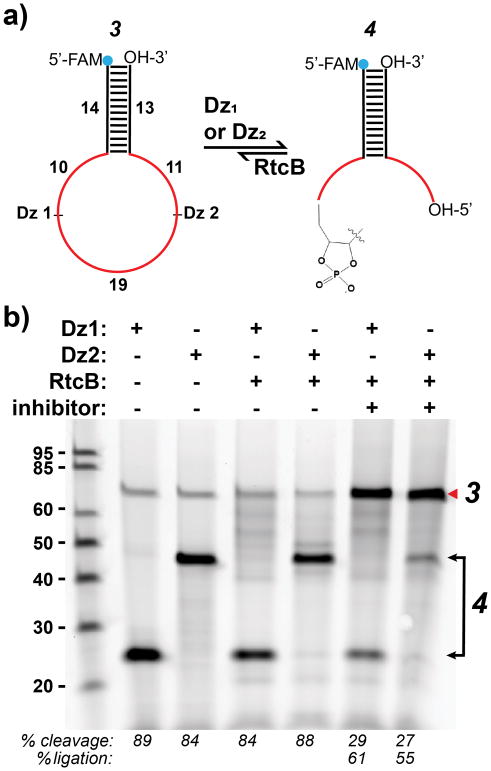
Given the strong dependence on substrate geometry, we next examined the efficiency of RtcB ligation as a function of stem-loop size (7, 11, 15, 19 nts), to determine if RtcB could ligate stem-loops larger than tRNA anticodon loops. We modified the tRNAglu stem-loop with increasing numbers of unpaired base pairs and introduced additional unpaired nucleotides on the 5′-end (Figure 1a, 1), increasing loop size. We found that all the stem-loops tested were ligated to near 100% efficiency (Figure 1b). To interrogate the ligation of loops larger than 19 nts, we used DNAzymes to cleave a DNA / RNA hybrid stem-loop target (Figure 2a, 3), producing single-stranded overhangs of 10 and 30 nts or 11 and 29 nts, with a total loop size of 40 nts. As proof-of-concept, we chose model DNAzymes known to have a relatively high kcat (Dz1)29 or operate at low Mg2+ concentration (Dz2)30 for use throughout this work (Figure S1). We also confirmed the specificity of Dz1 and Dz2 by introducing single and double nucleotide mutations in the binding arms and measured the nuclease activity (Figure S6). After DNAzyme cleavage of the RNA / DNA hybrid substrate for 2 hrs, an aliquot of the reactions was taken (Figure 2b, lanes 2–3) and an inhibitor strand complementary to the DNAzymes was introduced along with RtcB. The reaction was then allowed to proceed for another 1 hr (Figure 2b, lanes 6–7). Using PAGE, we assayed the efficiency of RtcB ligation of these cleavage products with and without inhibitor strand. Analysis of the resulting gel showed that when an inhibitor strand was present, these stem-loops were ligated back to substrate 3 with 55–60% efficiency (Figure 2b, lanes 6–7). The inhibitor strands to inactivate the DNAzymes were necessary to block DNAzyme action and allow for RtcB ligation. The reduction in efficiency is likely due to the enlarged loop, as well as the limited cyclic phosphodiesterase activity of RtcB (vide infra).31 Importantly, this data indicates that RtcB is amenable for ligation of stem-loops as large as 40 nts and shows that it can process the products of DNAzyme cleavage.
Figure 1.
Testing the role of loop size in ligation efficiency. (a) Schematic showing model RNA substrate for testing RtcB activity as a function of loop size. “X” indicates added nucleotides (see Table S2). (b) Reaction was conducted at 37 °C for 1 hr, 200 nM FAM-labeled strand, 250 nM unlabeled strand, 150 mM NaCl, 1 mM Mn2+, 0.1 mM GTP and 1 μM RtcB. Lane 1: 19 nt FAM-labeled strand alone; lane 6, stem-loop of 7 nt without RtcB to ligate. Red arrows indicate ligation products. Note that RtcB can ligate either 2′3′-cyclic phosphates or 3′ phosphates.
Figure 2.
(a) Schematic showing DNAzyme cleavage and ligation back to substrate 3. RNA is shown in red and DNA in black. (b) Gel showing single cleavage reactions and ligation back to substrate. Lanes 2–3: single Dz digests; lanes 4–5: single Dz digests after RtcB addition; lanes 6–7: same reaction shown in lanes 4–5 after addition of inhibitor strands complementary to the Dz(s). Ligation reactions contain 150 mM NaCl, 2 mM Mn2+, 0.4 μM substrate 3, 0.4 μM Dz1 or Dz2, 0.42 μM Dz inhibitor (lane 6–7), 0.4 mM GTP and 2.2 μM RtcB. Red arrow indicates ligation product. Note that lanes were loaded evenly.
DNAzymes and RtcB splice an RNA stem-loop
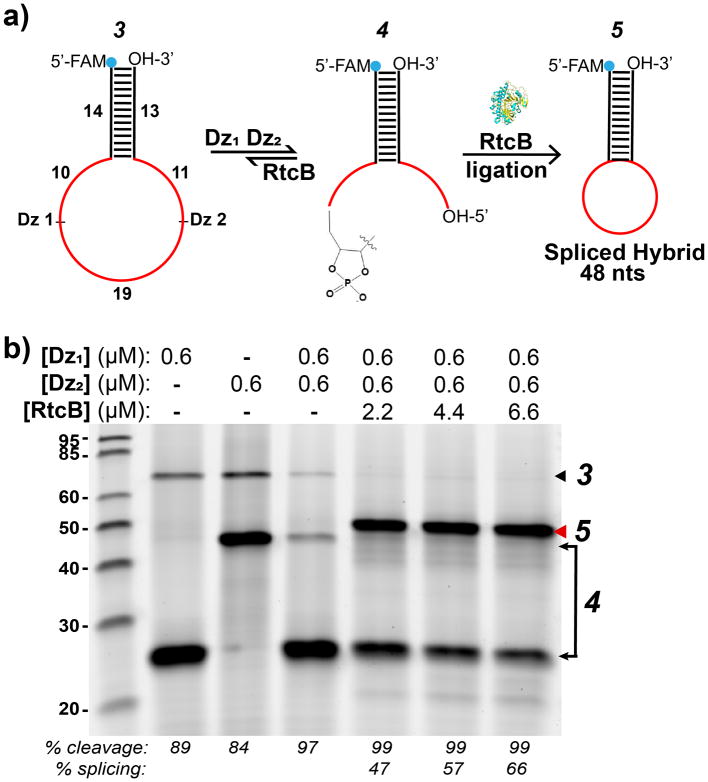
Next, we tested whether it was possible for DNAzymes and RtcB to splice in a one-pot reaction. We first incubated substrate 3 with an equimolar concentration (0.6 μM) of both Dz1 and Dz2 for 2 hrs in the presence of 2 mM Mn2+. The DNAzymes bound adjacent sites in the loop region of the stem-loop substrate 3 and cleaved, removing a 19 nt intron (Figure 3a, 4). After cleavage, RtcB was added to the reaction at different concentrations (2.2, 4.4 and 6.6 μM). Upon addition of RtcB, a smaller spliced stem-loop product 5 was produced (48 nts) at 47–66% yield, depending on the concentration of the enzyme (Figure 3b). In general, splicing yield varied between 45–68% with 2.2 μM of RtcB (data not shown). Yield was calculated by integrating the splice product band intensity and dividing by the integrated intensity of all the bands per lane. All values were background subtracted using the integrated intensity above and below each band (Figure S7). Note that in this case a hybrid RNA / DNA substrate was used for splicing; however, all RNA substrates can also be used (Figure 4). Interestingly, RtcB and DNAzymes spliced RNA targets both in sequential reactions (with DNAzyme addition followed by RtcB addition) and in one-pot reactions, with similar yields (Figure S8). As with the ligation of RNA stem-loops, the splice reaction is also relatively fast, and splice product was observed 5 min after adding RtcB enzyme (Figure S9).
Figure 3.
RtcB ligation of DNAzyme cleavage products and splicing. (a) Schematic showing reactions used to test splicing. 5′-FAM-labeled in vitro splice substrate 3 is cleaved, removing 19 nt intron to produce 4. RtcB addition produces spliced stem-loop 5. RNA is shown in red. (b) Cleavage and splice reaction with soluble Dz(s) and RtcB. Dz1 (lane 2) and Dz2 (lane 3) single cleavage produces bands at 24 and 43 nts. Addition of RtcB to a Dz1Dz2 double digest produces splice product (red arrow). Reaction conditions: 150 mM NaCl, 0.6 μM each Dz, 0.6 μM substrate 3, 2 mM Mn2+, 0.4 mM GTP, 2.2, 4.4 or 6.6 μM RtcB. DNAzyme cleavage (2 hrs) and RtcB ligation (1 hr) proceeded at 37°C.
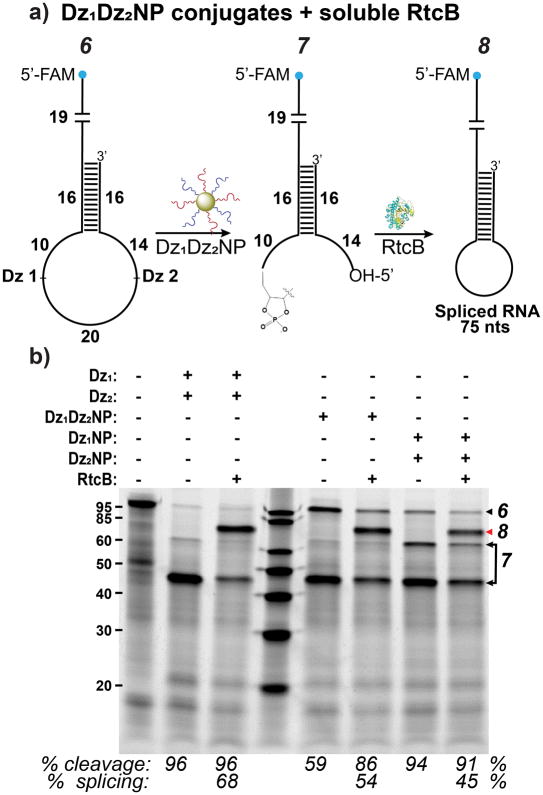
Figure 4.
DNAzyme conjugates (Dz1Dz2NPs) can splice with excess soluble RtcB. (a) Scheme showing splicing by Dz1Dz2NPs and RtcB on RNA substrate. (b) Splicing by soluble DNAzymes is compared to NPs with either both or single Dz’s attached. Red arrow indicates splice product. Reaction conditions: 150 mM NaCl, 1 mM Mn2+, 0.4 μM substrate 6, 0.4 mM GTP, 2.2 μM RtcB, lanes 2–3, 0.4 μM Dz(s), lanes 5–6, 9 nM Dz1Dz2NPs, lanes 7–8, 9 nM Dz1NP and Dz2NP. Cleavage was conducted at 37°C for 2 hrs and splicing at 37°C for 1 hr. Note that the cleavage yield increased in lane 6 compared to lane 5 likely due to the additional 1 hr incubation time following treatment with RtcB.
There are two possible reasons that likely limit splice yield. The first is that the DNAzymes could be binding to the target and not dissociating adequately, thus inhibiting RtcB binding and ligation. Additionally, stalled or inactive RtcB enzyme may sequester RNA ends, binding them but not effectively performing the ligation reaction. To test whether DNAzymes could be inhibiting the splice reaction, we bound the DNAzymes with a complementary strand after cleavage and then added RtcB. The yield of this reaction showed a 5% increase (Figure S10) over reactions with free DNAzymes. We thus shortened the DNAzyme arms from 9 nts to 8 and 7 nts, to decrease the Tm, reducing DNAzyme-target stability and enhancing product dissociation. We observed a 12% increase in splicing (from 48 to 60%) (Figure S11) upon using 8 nt arms over 9 nt arms. However, no additional improvement in splicing was observed for the 7 nt arm DNAzyme. Only Dz2 was tested with 7 nt arms, since Dz1’s Tm was already significantly reduced with 8 nt arms, and further shortening the arms would likely limit binding to the target (Table S1). Therefore, these experiments appear to justify our hypothesis that the DNAzyme arms inhibit splicing, but that factor alone cannot account for the observed lower yield.
Interestingly, ligation efficiency was nearly quantitative when 3′-P termini were used for the stem-loop ligation (Figure 1b), suggesting that the 2′3′-cyclic phosphates formed as the DNAzyme cleavage products reduce the yield of splicing. As RtcB first converts the 2′3′-cyclic phosphate to a 3′-P before ligation,31 it is possible the cyclic phosphatase reaction stalls splicing yield. Nevertheless, these splice reactions show that DNAzymes and RtcB are indeed able to function under identical conditions to splice RNA stem-loop targets, removing a 19 nt intron. These results confirm the potential for a splicing nanozyme system employing both classes of enzymes.
Dz1Dz2NP splicing using excess RtcB
As a first step toward splicing with a nanozyme composed of both DNAzymes and RtcB conjugated to a gold nanoparticle scaffold, we first tested whether soluble RtcB could splice in the presence of DNAzyme-functionalized gold nanoparticle conjugates (Dz1Dz2NPs). To produce Dz1Dz2NPs, we adopted previously reported protocols for single component DzNPs.18 Briefly, thiolated DNA was incubated with citrate stabilized AuNP and progressively salted.32–33 After washing to remove excess salt, mature Dz1Dz2NPs were then incubated with an RNA substrate (Figure 4, 6), allowing cleavage to proceed. We found that Dz1Dz2NPs cleaved target RNA producing 7 with an efficiency between 86–94%, in the presence of 1 mM Mn2+. When RtcB was added to these digests, a 54% splicing yield was observed (Figure 4, 8), similar to what was obtained for splicing with soluble DNAzymes and RtcB (Figure 3b; Figure 4b, lane 3). Additionally, mixing Dz1NPs and Dz2NPs also resulted in efficient cleavage. These products were spliced at a yield of 45% (Figure 4b, lanes 7–8). The yield was lower in this case possibly due to the added steric bulk of two AuNPs being involved in the cleavage reaction. For single component DzNPs, the number of Dz(s) per NP was quantified using a fluorescence assay, which showed that there were 102±9 strands of Dz1 and 54±6 strands of Dz2 on the AuNPs. Since for Dz1Dz2NPs each Dz was added in equimolar amounts, the number of strands of each on the NP may be estimated at half their number on single component DzNPs. Overall, these experiments show that splicing is effectively achieved with DNAzyme-nanoparticle conjugates and excess soluble RtcB.
Nanozyme synthesis
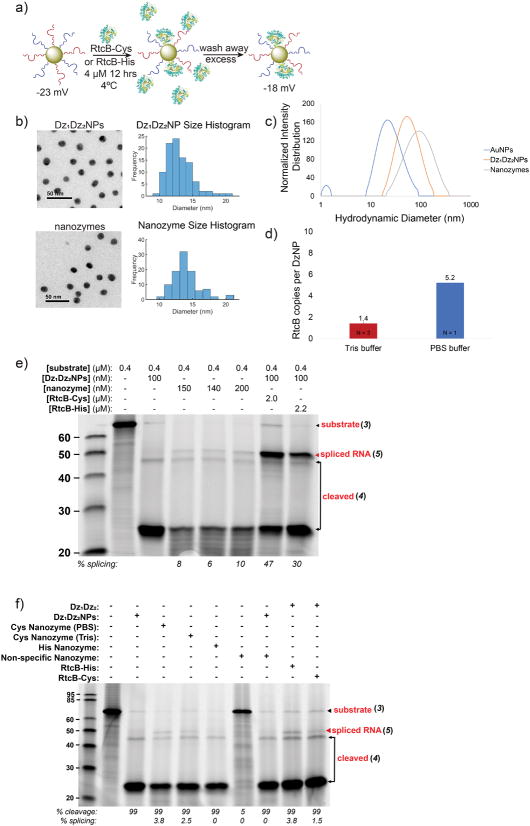
To produce complete nanozymes we needed to attach both DNAzymes and RtcB to a single particle. The RtcB enzyme was engineered with two cysteine residues at the N-terminus to enhance AuNP binding through thiol-Au chemistry. Dz1Dz2NPs were first synthesized as described (see Methods). Afterward, RtcB-Cys (4.7 μM) was attached to these Dz1Dz2NPs in an overnight incubation at 4°C in 100 mM Tris-HCl (Figure 5a), thus allowing RtcB binding by thiol exchange and production of nanozymes. The zeta potential of the Dz1Dz2NPs and nanozymes was also measured (Figure 5a). These measurements suggest that RtcB alters the zeta potential of the Dz1Dz2NP particles. A fluorescence assay indicated that an average of 1.4 RtcB molecules were bound to each Dz1Dz2NP. However, when the thiol exchange was performed in the presence of 1x PBS, 5.2 RtcB were bound per Dz1Dz2NP (Figure S12). This greater degree of binding is likely due to charge screening, as 1x PBS has a greater ionic strength than the Tris buffer used for binding studies. As expected, the cysteine residues were critical in the binding of the RtcB to the AuNPs to produce complete nanozymes, since His-tagged RtcB failed to bind to AuNPs, as shown by failure to generate splice product after washing the particles (Figure S13). Fluorescence lifetime spectroscopy was also used to confirm the association between the RtcB and the gold nanoparticle surface. Gold nanoparticles are known to reduce the fluorescence lifetime of organic dyes due to the process of nanometal surface energy transfer (NSET). To perform this experiment, we labeled RtcB-Cys and RtcB-His ligases with NHS-Alexa488. The fluorescence lifetime of Alexa488 was then measured when the enzyme was soluble and compared to that of enzyme incubated with the DNAzyme-functionalized gold nanoparticles. The measurements show a significant reduction in the fluorescence lifetime of RtcB-Cys upon incubation with gold nanoparticles, which is in contrast to RtcB-His enzymes that were indistinguishable regardless of nanoparticle interaction (Figure S14). This result confirms that the engineered Cys residues are important in mediated binding to the gold nanoparticle. Finally, the AuNPs, Dz1Dz2NPs and nanozymes were characterized by transmission electron microscopy (TEM) and dynamic light scattering (DLS) (Figure 5b, c). TEM shows that the Dz1Dz2NPs and nanozymes were not aggregated by addition of DNA and protein to the surface of the gold. Additionally, DLS provided another confirmation that these species were attached to the particle surface, in addition to the fluorescence and spectroscopy assays that we performed (Figure S12, S14).
Figure 5. Characterizing the splicing of nanozyme constructs.
(a) Scheme illustrating nanozyme synthesis. Zeta potential = −22.5±1.5 for Dz1Dz2NPs and −17.7±1.4 for nanozymes in 10 mM Tris-HCl, pH 7.4. (b) Transmission electron microscope (TEM) characterization of Dz1Dz2NPs and nanozymes. DNAzymes and RtcB are not visible in unstained TEM; however, no aggregation was observed. (c) DLS of unmodified AuNPs, Dz1Dz2NPs and nanozymes. (d) Plot showing the measured density of RtcB on nanozymes. Measurement was performed using fluorescence spectrometry. (e) Triplicate nanozyme splicing reactions. Lane 2: RNA / DNA hybrid substrate 3 only; lane 3: Negative control containing Dz1Dz2NPs and no RtcB; lane 4–6: nanozymes after washing 3 times yields splice product (red arrow); lane 7: positive control with Dz1Dz2NPs and 2 μM soluble RtcB-Cys; lane 8: positive control with Dz1Dz2NPs and 2.2 μM soluble RtcB-His. Reaction conditions: 0.4 μM substrate, 150 mM NaCl, 0.4 mM GTP, 2 mM Mn2+, 37°C for 2 hrs. (f) Gel showing the role of buffer and immobilization chemistry in tuning nanozyme efficiency. Lane 2: DNA / RNA hybrid substrate 3 alone; lane 3, negative control with Dz1Dz2NPs without RtcB; lane 4, nanozyme, produced by RtcB incubated in 1x PBS; lane 5, nanozyme produced by RtcB incubated in 100 mM Tris; lane 6, nanozymes produced by incubation with RtcB-His; lane 7, inactive nanozyme, produced with non-specific DzNPs incubated with RtcB-Cys in 100 mM Tris; lane 8, inactive nanozyme supplemented with active Dz1Dz2NP; lane 9, soluble 37.5 nM RtcB-His in presence of excess Dz1 / Dz2; lane 10, soluble 37.5 nM RtcB-Cys in presence of excess Dz1 / Dz2. Nanozyme concentration was 25 nM in all lanes. Red arrow indicates splice product.
Nanozyme conjugates splice RNA stem-loop targets
Despite the low copy number of RtcB molecules per nanozyme, we next tested the activity of our nanozyme in splicing an RNA substrate 3 (Table S2). Soluble DNAzymes with and without RtcB enzyme were used as positive and negative controls, respectively. After overnight RtcB-Cys incubation, the fresh nanozymes were washed three times with 100 mM Tris-HCl buffer, to remove excess soluble RtcB-Cys. After each wash, nanozyme aliquots were then mixed with substrate 3 and incubated at 37°C for 2 hrs (Figure S15). Nanozymes digested the RNA target and showed a splicing efficiency of 3% after the third wash (Figure S15, lane 6, 5) at 24 nM concentration of nanozyme. Another set of experiments was conducted, concentrating the nanozymes in each reaction up to ~200 nM. In this case, nanozyme splicing reactions were performed in triplicate and splice product was as high as 10% (Figure 5e), as determined by band intensities. We found that increasing the amount of nanozymes did increase the amount of splice product within the range of nanozyme concentrations tested, although the increase was not linear, due, in part, to decreasing activity of RtcB-Cys as it ages. Further experiments were then conducted with additional controls, such as creation of a nanozyme with RtcB-Cys and non-specific DNAzymes. This construct was unable to cleave or splice target RNA, as expected (Figure 5f, lane 7). Additionally, we found that RtcB-Cys must be located on the same particle as the target-specific DNAzymes to effect splicing. Non-specific nanozymes, that included RtcB-Cys, were unable to splice the target even after active Dz1Dz2NPs were introduced (Figure 5f, lane 8). To further explore the finding that the nanozyme splicing reaction is most optimal when the nuclease and ligase activity are localized to the same particle, we performed an additional set of experiments where we compared the activity of the complete nanozyme against that of a binary mixture of particles where the nuclease and ligase activities are isolated onto different particles (Figure S16). Taken together, the lower yield when using binary mixtures of particles is likely due to the substrate associating with the Dz1Dz2NPs and thus reducing the association with the RtcB. Finally, we tested splice reactions with excess DNAzymes and soluble RtcB equivalent to the amount of RtcB on nanozymes. Splicing was found to be equivalent to nanozyme splicing (Figure 5f, lane 9–10), indicating that RtcB activity is maintained on the AuNP surface. These experiments suggest that E.coli RtcB is likely a single-turnover enzyme34 and thus, the limiting reagent in the splice reaction.
Overall, this nanozyme shows utility in splicing RNA stem-loops. For cellular splicing, it is possible that RtcB does not need to be included on the nanozyme, since endogenous RtcB is already expressed and may be recruited to the particle for splicing. Future work will focus on increasing the efficiency of the nanozyme and transitioning toward in vivo splicing.
CONCLUSION
In summary, we demonstrate that RtcB and DNAzymes – natural and synthetic enzymes, respectively – can be coupled to a gold nanoparticle and work together to splice RNA targets with up to a 10% yield. This activity is observed despite direct coupling of the RtcB to the gold nanoparticle surface. This nanozyme is the first example of a splicing nanoparticle system and the only known combination of a natural and synthetic enzyme for splicing. We demonstrate that RtcB and DNAzymes must be on the same particle to achieve splicing and cannot complement each other on separate particles. We also demonstrate that the cysteine residues on RtcB are essential for RtcB binding to the Dz1Dz2NPs, without which, no nanozyme is formed; and after washing of Dz1Dz2RtcB-His-NPs, no detectable splicing is observed (Figure S12). Labeled His-nanozymes also do not show a change in their fluorescence lifetime (Figure S14c-d). Additionally, DNAzymes and RtcB also splice RNA targets up to 45–66% yield when including excess RtcB in solution. This system provides a completely new method of RNA splicing that adds to the toolkit for in vitro work. Since DNAzyme-NP conjugates and nanozymes readily enter cells in vitro18 (Figure S17) and DNAzyme-NP conjugates have shown uptake in vivo,19 it also provides a platform with which to conduct cellular splicing. In future work, we plan to increase splicing efficiency inside cells by arraying more RtcB around the particle using a linker strand, as well as recruiting endogenously expressed RtcB (HSPC117) to the particle surface for specific splicing.
MATERIALS AND METHODS
Expression and Purification of RtcB-His
To express, isolate and purify hexahistidine-tagged RtcB enzyme, we adapted protocols from both the Raines and Shuman labs.23, 27 RtcB with an N-terminal hexahistidine tag under a lac-I inducible promoter (pQE-70) was obtained from the Raines lab27 and transformed into Keio JW5688-1 (ΔrtcA::kan) from the E. coli Genetic Stock Center (Yale). Briefly, 5 ml of LB (100 μg/ml Amp, 50 μg/ml Km) was inoculated with a single colony of the above transformant and grown at 37°C overnight. This culture was used to inoculate 1L of LB (100 μg/ml AMP, 50 μg/ml Km) which grew at 37°C shaking until OD600 = 0.6 – 0.8. Expression was then induced by addition of 0.5 mM IPTG and grown for 2.5 hours at 32°C. The culture was spun down at 3,100 g at 6°C for 30 min. Pellet was transferred to a 50 ml Falcon tube and resuspended in 10 ml lysis buffer. Lysis buffer was prepared by mixing 100 μl of protease inhibitor cocktail (PIC, Sigma Aldrich, P8849) in 50 ml buffer A [50 mM Tris-HCl, pH 7.4, 250 mM NaCl, 10% sucrose] on ice. Culture was sonicated on a probe sonicator, 10 sec pulse, 20 sec rest (11–14 mM, lvl 4) for 4 min. The lysate was cleared by centrifuging on an Optima XE-90 ultracentrifuge for 30 min at 24,100 rpm (67,000 g). The lysate was then added to a Falcon tube with 15 ml buffer A and 3 ml Ni:NTA beads (QIAGEN, cat. no. 30210) equilibrated in 15 ml buffer A. Lysate was then allowed to rock on a nutating mixer at 4°C for 1 hr. After incubation, Ni:NTA beads were centrifuged at 4,000 g for 15 min and resuspended in buffer B1 [50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10% glycerol, 25 mM imidazole]. This wash was repeated two more times. The beads were then added to a column (Bio-rad, #737-4156) and washed with ~20 ml wash buffer [50 mM Tris-HCl, pH 7.4, 2 M KCl], 10 ml buffer B1, 10 ml buffer B2 [buffer B1, 40 mM imidazole], 10 ml buffer B1 and eluted with 7 ml elution buffer [buffer B1, 300 mM imidazole]. Protein elution was estimated by the Bradford Assay and dialyzed against 500 ml of buffer C [10 mM Tris-HCl, pH 8.0, 350 mM NaCl, 1 mM DTT, 10% glycerol] for two hours. Buffer was changed four times at two hour intervals and finally dialyzed overnight at 4°C. After dialysis, aliquots of RtcB enzyme were frozen in liquid nitrogen and stored at −80°C.
Expression and Purification of Cysteine-modified RtcB
RtcB that had been modified with two cysteine residues before the N-terminal His-tag was expressed under a lac-I inducible promoter (pQE-70) and transformed into E.coli BL21. Briefly, 5 ml of LB (100 μg/ml Amp) was inoculated with a single colony of the above transformant and grown at 37°C overnight. This culture was then used to inoculate 250 mL of LB (100 μg/ml Amp) which grew at 37°C shaking until OD600 = 0.6. Expression was then induced by addition of 0.1 mM IPTG and grown for 4 hrs at 37°C. Culture was spun down at 4,000 rpm 6°C for 20 min and the pellet kept on ice. Cell pellets were resuspended in a total of 10 ml lysis buffer [50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole], supplemented with 100 μl Protease Inhibitor Cocktail (PIC), 60 μl lysozvme [20 mg/mL stock], 5–10 μl benzonase. The resuspended cells were incubated on ice for 20 min, then sonicated for 3.5 minutes: 15 sec pulse, 15 sec rest. The lysate was then centrifuged at 4°C, 4,000 rpm for 30 min. Ni-NTA beads (QIAGEN, cat. no. 30210) were prepared by adding 1.2 mL of the resin slurry into a 15 mL Falcon tube. Slurry was centrifuged at 4,000 rpm for 2 min, and the ethanol was decanted and the beads resuspended in 4 mL of lysis buffer. The tube was shaken for 2 min, centrifuged for 2 min, and decanted and rinsed with lysis buffer two more times. The cell lysate was added to the Ni-NTA beads and the beads were resuspended and rotated on a nutating mixer at 4°C for 0.5–1.5 hrs. The beads were then transferred to a 30 mL propylene column and the initial flow through was collected. The column was washed, with the elutions collected, as follows: 20 mL wash 1: 50 mM potassium phosphate (pH 7.8), 150 mM NaCl, 10 mM imidazole; 15 mL wash 2: 50 mM potassium phosphate (pH 7.8), 150 mM NaCl, 50 rnM imidazole; 1 ml elution 1: 50 mM potassium phosphate (pH 7.8), 150 mM NaCl, 250 mM imidazole; 1 mL elution 2: 50 mM potassium phosphate (pH 7.8), 150 mM NaCl, 500 mM imidazole; 1 mL elution 3: 50 mM potassium phosphate (pH 7.8), 150 mM NaCl, 1,000 mM imidazole. A 12% PAGE gel was run for each fraction to determine which fraction contained the majority of the protein. 10 μl of each fraction was mixed with 10 μl of SDS loading dye and boiled in a thermocycler on the boil cycle for 10 min at 95°C. 15 μl were loaded onto a 12% SDS-PAGE gel and run for 40 min at 220 V, 60 mA. The resulting gel was stained with Coomassie blue 30 min, then destained [40% methanol, 10% glacial acetic acid]. Fractions with protein were dialyzed against 1 L of cold storage buffer (50 mM HEPES / 10 mM MgCl2). The concentration of the protein was verified by a NanoDrop 2000c spectrophotometer. 50 μl aliquots of RtcB protein was flash frozen in liquid nitrogen and stored in a −80°C freezer.
DNAzyme Design
Two DNAzymes were adopted from the literature29–30 for our study. These were selected because of either relatively rapid catalysis or good activity at low Mg2+ concentration. The first DNAzyme (Dz1), DT-99, has 9 nt binding arms and is active against HPV, with a kobs of 0.21 min−1 at 10 mM Mg2+.29 The sequence is 5′-GTTTCTCTAGGCTAGCTACAACGAGTGTTCTTG-3′, with the catalytic core underlined. The second DNAzyme also has 9 nt binding arms and is active against the VEGF receptor, at 0.01 mM Mg2+.30 The sequence is 5′-TGCTCTCCA GGCTAGCTACAACGACCTGCACCT-3′.
Designing Construct 3
In order to confirm splicing on another platform, a 5′-FAM-labeled 67 nt synthetic DNA / RNA hybrid was ordered from IDT as shown (“r” indicates ribonucleotides): 5′-AGACGAGTCTCACGrCrArArGrArArCrArCrGrUrArGrArGrArArArCrArGrGrUrGrCrArGrGrGrUrGrGrArGrArGrCrAGTCGTGAGACTCGTC-3′. This sequence contained a DNA stem-loop and the recognition sites for both the DT-99 (italicized) and VEGFR DNAzymes (underlined) described previously, allowing the removal of a 19-bp intron. Cleavage sites for each DNAzyme are in bold (see Table S2, 3).
Synthesis of Gold Nanoparticles
Citrate-stabilized gold nanoparticles of 13.1 ± 1.9 nm were made using the Turkevich method as described by the Mirkin lab.32 Briefly, all glassware to be used was washed with aqua regia and rinsed with Nanopure water. First, 300 ml of 1 mM hydrogen tetrochloroaurate (III) trihydrate was prepared with Nanopure water in a three-prong 500-ml round-bottom flask. The flask was boiled with condenser attached until reflux was achieved at 1 drip / sec, stirring continuously. In a 50 mL Falcon tube, 30 ml of 38.8 mM sodium citrate tribasic dehydrate was prepared with Nanopure water. The sodium citrate was quickly dumped into the refluxing solution. The flask was re-sealed and refluxed for 15 min. The solution rapidly changed color: yellow to black, to purple to deep red. The reaction was taken off the heat and cooled to room temperature (~2–4 hrs). The particles were filtered through a 0.45-μm acetate filter, producing monodisperse AuNPs, and transferred into a clean flask. The absorbance of the particles was verified by a Nanodrop 2000c spectrophotometer. Correctly synthesized 13 nm particles should have a λmax of ~519 nm and a peak width of ~50 nm.
Preparation of DNAzyme-Functionalized Gold Nanoparticles (Dz1Dz2NPs)
To prepare maximally packed Dz1Dz2NPs, we followed previous protocols.18, 32–33 Briefly, the 3′-thiolated T10-linker DNAzymes were first ordered from Integrated DNA Technologies (IDT). Their sequences are as follows: for the DT-99 DNAzyme: GTT TCT CTA GGC TAG CTA CAA CGAGTG TTC TTGTTTTTTTTTT/3ThioMC3-D/, and for the VEGFR DNAzyme: TGC TCT CCA GGC TAG CTA CAA CGA CCT GCA CCT TTTTTTTTTT/3ThioMC3-D/. Next, 60 nmol of each DNAzyme were reduced in 1 ml of 0.1 M DTT in disulfide cleavage buffer [170 mM phosphate buffer (pH = 8.0)] and allowed to incubate at room temperature for 2 hrs with occasional vortexing. A Nap-25 column (GE Healthcare) was flushed with four column volumes of Nanopure water. 1 ml of reduced sample was applied to the column and allowed to flow through completely. Then, 1.5 ml of Nanopure water was allowed to enter the column completely. Samples were eluted with 2.5 ml Nanopure water, collecting 4 drops at a time in microcentrifuge tubes. Tubes were Nanodropped and fractions with DNAzymes were combined. The volume of the sample was recorded and DNAzymes concentrations were determined from UV absorbance. To each 1 ml of AuNPs, 2 nmol of each of the reduced DNAzymes were added in a cleaned EPA vial (4 nmol total DNAzymes). The vial was wrapped in foil and allowed to equilibrate on an orbital shaker overnight at room temperature. The following day, phosphate adjustment buffer [100 mM phosphate buffer (pH 7.0)] was added to the Dz1Dz2NPs to 9 mM final phosphate concentration. SDS was added to ~0.1% (wt/vol). The tubes were wrapped in foil and incubated on an orbital shaker for 30 min at room temperature. Afterward, NaCl was added to the Dz1Dz2NPs with salting buffer [10 mM phosphate buffer (pH 7.0), 2M NaCl] in eight increments of final concentration as follows: 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6 and 0.7 M. After each addition, the Dz1Dz2NPs were sonicated in a bath sonicator (VWR 97043-968) 20–30 sec, wrapped in foil and incubated on an orbital shaker 20 min. Salt additions were continued until 0.7 M NaCl was reached. Dz1Dz2NPs were stored in 4°C cold room until use.
Dz1Dz2NP Washing Procedure
In an Eppendorf tube, 1 mL of Dz1Dz2NPs were spun down in a table top centrifuge at 13,000 rpm 4°C. Supernatant was removed by pipetting and the pellet was resuspended in 500 μl of Nanopure water, with ≤ 1 min sonication. The wash was repeated two more times and the pellet resuspended in the desired volume and buffer. Sonication is only necessary for the first resuspension.
DNAzyme-Functionalized Gold Nanoparticle Splicing Assay
To test whether or not Dz1Dz2NPs plus soluble RtcB were active for splicing (Figure 3), 20 μl reactions were setup with either gold conjugates containing both DNAzymes, or single DNAzyme gold particles mixed as follows: 50 mM Tris-HCl (pH 7.4 at 37°C), 1.5 mM MnCl2,150 mM NaCl, 9.09 nM DNAzyme-conjugates (each conjugate) 0.4 μM 5′-FAM labeled stem-loop RNA (see SI). This reaction was incubated in a water bath at 37°C for 2 hours, after which 2.2 μM RtcB and 0.4 mM GTP were added, and the reaction continued for another 1 hr. After incubation, 10 μl samples were quenched in stop solution (5 μl 95% formamide, 10 mM EDTA and 5 μl Ultrapure water (Invitrogen)). Samples were run on a 15% polyacrylamide gel electrophoresis (PAGE) in 1x Tris-borate EDTA (TBE) buffer pre-heated to 70°C for 30 min. PAGE gel was imaged on a Typhoon TRIO Variable Mode Imager (Amersham Biosciences) at 600 PMT.
Denaturing RNA Polyacrylamide Gel Electrophoresis
RNA reactions were imaged on 8%, 10% or 15% polyacrylamide electrophoresis gels (PAGE) using a Mini-PROTEAN Tetra Cell (Bio-rad). To prepare a 15% gel, the following recipe was used (10 ml): 4.2 g urea, 1 ml 1x TBE, 5 ml 37.5:1 acrylamide/bis solution (#1610158, Bio-rad), 3.5 μl TEMED, 70 μl 10% APS. The ingredients were mixed except for TEMED and APS and allowed to dissolve, with stirring. The gel solution was filtered through a 0.20 μm filter (if the gel was being stained with SYBR gold or Diamond stain). Upon addition of TEMED and APS, the solution was mixed by stirring for 10 s and poured, setting for 45 min or until use. Gel plates were prepared by washing with antibacterial soap, rinsing in Nanopure water, and RNase zapped (Ambion) 5 min. They were then rinsed with regular water, Nanopure water and 100% ethanol and sonicated in isopropanol for 7 min. After sonication, the gel plates were rinsed in 100% ethanol and allowed to dry in a dust free area. To avoid leaking, plates were prepared with a thin layer of halocarbon grease on the sides and the bottom of the gel was parafilmed before pouring. Combs were RNase zapped for 5 min and rinsed in regular, then Nanopure water.
TEM Imaging of DzNPs and nanozymes
Samples were prepared by depositing 5 μl of 11 nM AuNPs, washed Dz1Dz2NPs or washed nanozymes, onto a carbon film 200 mesh copper grid (CF200-Cu, lot #150318, Electron Microscopy Sciences). Samples were then allowed to sit 5 min and excess liquid was wicked away with a tip of filter paper. Samples were imaged on a JEOL JEM-1210 transmission electron microscope. The extinction coefficient was estimated from the particle diameter and used to determine particle concentrations by a NanoDrop 2000C spectrophotometer.
Measurement of Dz1Dz2NP Activity
Activity of Dz1Dz2NPs was determined through digestion of a fluorescein labeled RNA / DNA hybrid substrate (construct 3), as follows (“r” indicates ribonucleotides): 5′-FAM-AGACGAGTCTCACGrCrArArGrArArCrArCrGrUrArG rArGrArArArCrArGrGrUrGrCrArGrGrGrUrGrGrArGrArGrCrAGTCGTGAGACTCGTC-3′. DNAzyme cleavage sites are in bold. Dz1 recognition site is italicized. Dz2 recognition site is underlined. Each digest was run on a 15% RNA polyacrylamide gel as described and imaged on a Typhoon TRIO Variable Mode Imager. Cleavage products were estimated in size compared to a ladder. Band intensity was determined using ImageJ, and percent cleavage products determined as indicated (Figure S6).
Calculation of DNAzymes per AuNP
The number of DNAzymes per AuNP was approximated using the Quant-iT™ OliGreen® ssDNA Reagent and Kit (ThermoFisher, Grand Island, NY), after releasing the DNAzymes from the gold core. Briefly, 100 μl of DzNPs were aliquoted in 0.2, 0.4, 0.6 and 0.8 nM amounts in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5), and the gold core dissolved with 1 μl of 5 M potassium cyanide (KCN), that was added to each well. Note that KCN is hazardous and must be kept away from acid to avoid producing noxious cyanide fumes. It should be used in the hood and disposed of separately. A well with TE buffer but no DzNPs served as the control. The AuNP core was allowed to dissolve for 30 min, releasing the DNAzymes. An equal volume (100 μl) of 1x OliGreen reagent made up in TE buffer was added to each well and pipetted up and down to mix. The resulting wells were imaged immediately on a Bio-Tek Synergy HT plate reader with an approximately 2 min lag time. The fluorescence intensity at 485/528 nm excitation/emission were compared to a standard curve of soluble DNAzymes. This standard curve was produced by diluting a stock of each DNAzyme (4 μg/ml) to known concentrations (0.1, 0.2, 0.5, 0.75, 1, and 2 μg/mL) in 100 μl of TE buffer. After adding 1x OliGreen reagent, fluorescence intensities at each concentration were measured and plotted. Using this plot, the fluorescence intensity corresponding to the number of DNAzymes per well could be determined and this number was divided by the AuNP concentration to approximate the number of DNAzymes per NP.
Nanozyme Synthesis
Salted Dz1Dz2NPs (1 mL) were washed as follows: The NPs were spun down at 13,000 rpm (15,871 g) for 20 min and resuspended in 500 μl Nanopure water. Afterward, they were sonicated in a bath sonicator 1 min, vortexed and spun down as above. This wash was repeated three more times. After the second wash, Dz1Dz2NPs were resuspended in 500 μl of 100 mM Tris-HCl pH 7.43 at 37°C rather than Nanopure water. Sonication is not necessary after the first wash. After the final wash, the Dz1Dz2NPs were resuspended in 100 mM Tris-HCl pH 7.43 at 37°C up to 100 μl total volume. They were then incubated with 20 μl of 28.3 μM cysteine-modified RtcB to a final concentration of 4.72 μM, and allowed to incubate at 4°C overnight. The following day, the nanozymes were spun down 13,000 rpm (15,871 g) in a microcentrifuge 20 min at 4°C and the supernatant was pipetted off. The particles were resuspended in 500 μl 100 mM Tris-HCl buffer, pH 7.43 at 37°C. This wash was repeated two more times. Note: Never sonicate the nanozymes. Pipetting up and down should be sufficient to resuspend. If the pellet does not resuspend, attempt to break it up with a pipette tip, wait 20 sec, then gently pipette up and down again.
Measurement of RtcB on Nanozyme Conjugates
To 100 μl of 50.5 μM RtcB-Cys in 1x PBS, 0.1 mg of dried Alexa488 was added and allowed to react 2.5 hrs on ice. The resulting mixture was run through a P4 gel in 1x PBS. The degree of labeling (DOL) was calculated with the equation , where MW = the molecular weight of the protein, εdye = the extinction coefficient of the dye at its absorbance maximum 488 nm, and the protein concentration is in mg/mL. Dz1Dz2NPs (1 mL) were washed as previously described, and resuspended in 100 μl of PBS (30 μl Nanopure, 70 μl 1x PBS). To this sample, 13.6 μl of 41.8 μM (DOL=1.4) Alexa488-RtcB-Cys was added and allowed to incubate overnight at 4°C. The Dz1Dz2NP-Alexa488-RtcB-Cys in 100 mM Tris-HCl (pH 7.43 at 37°C) was washed three times by first voluming the sample to 500 μl, spinning down at 13,000 rpm in a table top centrifuge, removing the supernatant and repeating twice more. After the last removal of the supernatant, 40 μl 100 mM Tris-HCl were added and the sample absorbance at 520 nm was measured with a NanoDrop 2000c spectrophotometer. Next, 5 μl of 5 M potassium cyanide (KCN) was added to a final volume of 52 μl and incubated on ice 45 min. The sample was volumed to 100 μl in 100 mM Tris-HCl. Additionally, a standard curve was then aliquoted of Alexa488-RtcB-Cys in 0, 10, 50, 100, 200 and 400 nM concentrations. The fluorescence emission of Alexa488-RtcB-Cys samples was measured in a Horiba Scientific Dual-FL fluorometer with 10 accumulations. The data was plotted in Excel and the emission at 488 nm for each sample was recorded. Using the standard curve, the average number of RtcB on each Dz1Dz2NP was calculated.
Dynamic Light Scattering and Zeta Potential of Dz1Dz2NPs and nanozymes
Dynamic light scattering and zeta potential experiments were performed on a NanoPlus zeta/nano particle analyzer. For DLS, Dz1Dz2NPs and nanozymes were synthesized as described, washed 3 times in 100 mM Tris-HCl buffer, pH 7.43 at 37°C, and 60 μl samples were run at room temperature using 150 accumulations. Samples were kept on ice until collecting the DLS spectrum. The same procedure was used to collect zeta potential measurements, except nanozymes were created in 1x PBS buffer and washed in 10 mM Tris-HCl, pH 7.43 at 37°C as described, and 150 μl sample was measured.
Supplementary Material
Table S1. DNAzymes used in this study.
Table S2. Sequences used in this study.
Figure S1. DNAzymes used in this study (TIF)
Figure S2. SDS-PAGE of RtcB (TIF)
Figure S3. Activity of RtcB on linear and stem-loop strands (TIF)
Figure S4. Quantifying RtcB ligation yield on linear strands (TIF)
Figure S5. Quantifying RtcB kinetics on stem-loop substrate (TIF)
Figure S6. Dz1NPs and Dz2NPs with select mutations show lowered activity (TIF)
Figure S7. Image analysis for calculating splicing yield (TIF)
Figure S8. Sequential and one-pot splice reactions show similar yield (TIF)
Figure S9. RtcB splicing kinetics on substrate 3 (TIF)
Figure S10. RtcB splicing as a function of ligase concentration and DNAzyme inhibition (TIF)
Figure S11. RtcB splicing upon digestion with DNAzymes of decreasing arm length (TIF)
Figure S12. Calibration curve used to determine RtcB bound nanozyme (TIF)
Figure S13. His-tagged RtcB cannot form a splicing nanozyme (TIF)
Figure S14. RtcB is specifically bound to Dz1-2NPs (TIF)
Figure S15. The effect of multiple washes on nanozyme yield (TIF)
Figure S16. RtcB on non-specific NPs is still active on pre-cleaved substrate (TIF)
Figure S17. Flow cytometry of Dz1Dz2NPs and nanozymes entering MDA-MB-231 cells (TIF)
Acknowledgments
Funding Sources
NSF CAREER Award and NIH
K.S. would like to thank the NIH (R01-GM097399) and the NSF CAREER Award (1350829) for financial support. J.P. would like to thank the ARCS Foundation and S. and F. Burke for their generous support. R.G. would like to thank NIH (NIGMS GM124472) and the NSF Fellowship for financial support. J.P. and K.S. would like to thank K. Desai and R. Raines for the gift of the RtcB-lacI plasmid. We thank O. Laur for technical expertise in construction and synthesis of plasmid constructs. We thank K. Yehl, E. Weinert and G. Conn for helpful discussions about the data. We thank V. Ma for production of the cy3b-labeled construct for nanozyme cell uptake experiments. We thank H. Su, B. Deal and N. Baker for assistance with nanozyme uptake experiment and flow cytometry. We thank G. Raghunath for assistance with DLS and zeta potential measurements. We thank J. Brockman for production of particle construct size histograms using MatLab. We thank S. Druzak for technical expertise during the early stages of this project. We acknowledge the work of I. Bolin, G. Thomsen, B. Bohannon and Z. Mousavi – students trained during this work. We thank K. Clarke, V. Ma and B. Fontaine for critical reading of the manuscript.
ABBREVIATIONS
- AuNP
gold nanoparticle
- DNAzyme
deoxyribozyme
- DzNP
Deoxyribozyme nanoparticle
- Dz
DNAzyme
Footnotes
Author Contributions
J.P. and K.S. were responsible for writing this manuscript. J.P., K.S. and K.Y. discussed experiments and controls. J.P. performed experiments. K.G. expressed RtcB-Cys shown in Figure 5f. Brendan performed flow cytometry on Dz1Dz2NPs and nanozymes shown in Figure S17. Roxanne performed FLIM on nanozymes shown in Figure S14. All authors have given approval to the final version of the manuscript.
Notes
The authors declare no competing financial interest
ASSOCIATED CONTENT
Supporting Information. The following files are available free of charge via the Internet at http://pubs.acs.org.
SI Methods: Design and construction of substrate 6. Uptake of nanozymes into MDA-MB-231.
References
- 1.Montiel-Gonzalez MF, Vallecillo-Viejo I, Yudowski GA, Rosenthal JJ. Correction of Mutations within the Cystic Fibrosis Transmembrane Conductance Regulator by Site-Directed Rna Editing. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:18285–90. doi: 10.1073/pnas.1306243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evers MM, Toonen LJ, van Roon-Mom WM. Antisense Oligonucleotides in Therapy for Neurodegenerative Disorders. Advanced drug delivery reviews. 2015;87:90–103. doi: 10.1016/j.addr.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Havens MA, Duelli DM, Hastings ML. Targeting Rna Splicing for Disease Therapy. Wiley interdisciplinary reviews. RNA. 2013;4:247–66. doi: 10.1002/wrna.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deidda G, Rossi N, Tocchini-Valentini GP. An Archaeal Endoribonuclease Catalyzes Cis- and Trans- Nonspliceosomal Splicing in Mouse Cells. Nature biotechnology. 2003;21:1499–504. doi: 10.1038/nbt908. [DOI] [PubMed] [Google Scholar]
- 5.Desai KK, Bingman CA, Phillips GN, Jr, Raines RT. Structures of the Noncanonical Rna Ligase Rtcb Reveal the Mechanism of Histidine Guanylylation. Biochemistry. 2013;52:2518–25. doi: 10.1021/bi4002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amini ZN, Olson Karen E, Muller Ulrich F. Spliceozymes: Ribozymes That Remove Introns from Pre-Mrnas in Trans. PloS one. 2014;9:1–11. doi: 10.1371/journal.pone.0101932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reenan R. Correcting Mutations by Rna Repair. The New England Journal of Medicine. 2014;370:172–174. doi: 10.1056/NEJMcibr1313514. [DOI] [PubMed] [Google Scholar]
- 8.Putti S, Calandra P, Rossi N, Scarabino D, Deidda G, Tocchini-Valentini GP. Highly Efficient, in Vivo Optimized, Archaeal Endonuclease for Controlled Rna Splicing in Mammalian Cells. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:3466–77. doi: 10.1096/fj.13-231993. [DOI] [PubMed] [Google Scholar]
- 9.Lewin AS, Hauswirth WW. Ribozyme Gene Therapy: Applications for Molecular Medicine. Trends in molecular medicine. 2001;7:221–228. doi: 10.1016/s1471-4914(01)01965-7. [DOI] [PubMed] [Google Scholar]
- 10.Mitsuyasu RT, Merigan TC, Carr A, Zack JA, Winters MA, Workman C, Bloch M, Lalezari J, Becker S, Thornton L, Akil B, Khanlou H, Finlayson R, McFarlane R, Smith DE, Garsia R, Ma D, Law M, Murray JM, von Kalle C, Ely JA, Patino SM, Knop AE, Wong P, Todd AV, Haughton M, Fuery C, Macpherson JL, Symonds GP, Evans LA, Pond SM, Cooper DA. Phase 2 Gene Therapy Trial of an Anti-Hiv Ribozyme in Autologous Cd34+ Cells. Nature medicine. 2009;15:285–92. doi: 10.1038/nm.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-Splicing Rna: Autoexcision and Autocyclization of the Ribosomal Rna Intervening Sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 12.Fiskaa T, Birgisdottir AB. Rna Reprogramming and Repair Based on Trans-Splicing Group I Ribozymes. New biotechnology. 2010;27:194–203. doi: 10.1016/j.nbt.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Alexander RC, Baum DA, Testa SM. 5′ Transcript Replacement in Vitro Catalyzed by a Group I Intron-Derived Ribozyme. Biochemistry. 2005;44:7796–7804. doi: 10.1021/bi047284a. [DOI] [PubMed] [Google Scholar]
- 14.Olson KE, Muller UF. An in Vivo Selection Method to Optimize Trans-Splicing Ribozymes. Rna. 2012;18:581–9. doi: 10.1261/rna.028472.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y, Ren J, Qu X. Nano-Gold as Artificial Enzymes: Hidden Talents. Advanced materials. 2014;26:4200–17. doi: 10.1002/adma.201400238. [DOI] [PubMed] [Google Scholar]
- 16.Lin Y, Ren J, Qu X. Catalytically Active Nanomaterials: A Promising Candidate for Artificial Enzymes. Accounts of chemical research. 2014;47:1097–105. doi: 10.1021/ar400250z. [DOI] [PubMed] [Google Scholar]
- 17.Santoro SW, Joyce Gerald F. A General Purpose Rna-Cleaving Dnazyme. Proceedings of the National Academy of Sciences of the United States of America. 1999;49:1262–1266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yehl K, Joshi JP, Greene BL, Dyer B, Nahta R, Salaita K. Catalytic Deoxyribozyme-Modified Nanoparticles for Rnai-Independent Gene Regulation. ACS nano. 2012;6:9150–9157. doi: 10.1021/nn3034265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Somasuntharam I, Yehl K, Carroll SL, Maxwell JT, Martinez MD, Che PL, Brown ME, Salaita K, Davis ME. Knockdown of Tnf-Alpha by Dnazyme Gold Nanoparticles as an Anti-Inflammatory Therapy for Myocardial Infarction. Biomaterials. 2016;83:12–22. doi: 10.1016/j.biomaterials.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silverman SK, Baum DA. Use of Deoxyribozymes in Rna Research. Methods Enzymol. 2009;469:95–117. doi: 10.1016/S0076-6879(09)69005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakravarty AK, Subbotin R, Chait BT, Shuman S. Rna Ligase Rtcb Splices 3′-Phosphate and 5′-Oh Ends Via Covalent Rtcb-(Histidinyl)-Gmp and Polynucleotide-(3′)Pp(5′)G Intermediates. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6072–7. doi: 10.1073/pnas.1201207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka N, Meineke B, Shuman S. Rtcb, a Novel Rna Ligase, Can Catalyze Trna Splicing and Hac1 Mrna Splicing in Vivo. The Journal of biological chemistry. 2011;286:30253–7. doi: 10.1074/jbc.C111.274597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka N, Shuman S. Rtcb Is the Rna Ligase Component of an Escherichia Coli Rna Repair Operon. The Journal of biological chemistry. 2011;286:7727–31. doi: 10.1074/jbc.C111.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Luhrmann R, Soll D, Martinez J. Hspc117 Is the Essential Subunit of a Human Trna Splicing Ligase Complex. Science. 2011;331:760–4. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- 25.Kosmaczewski SG, Edwards TJ, Han SM, Eckwahl MJ, Meyer BI, Peach S, Hesselberth JR, Wolin SL, Hammarlund M. The Rtcb Rna Ligase Is an Essential Component of the Metazoan Unfolded Protein Response. EMBO reports. 2014;15:1278–85. doi: 10.15252/embr.201439531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wadhwani P, Heidenreich N, Podeyn B, Burck J, Ulrich AS. Antibiotic Gold: Tethering of Antimicrobial Peptides to Gold Nanoparticles Maintains Conformational Flexibility of Peptides and Improves Trypsin Susceptibility. Biomater Sci. 2017;5:817–827. doi: 10.1039/c7bm00069c. [DOI] [PubMed] [Google Scholar]
- 27.Desai KK, Raines RT. Trna Ligase Catalyzes the Gtp-Dependent Ligation of Rna with 3′-Phosphate and 5′-Hydroxyl Termini. Biochemistry. 2012;51:1333–5. doi: 10.1021/bi201921a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakravarty AK, Shuman S. The Sequential 2′,3′-Cyclic Phosphodiesterase and 3′-Phosphate/5′-Oh Ligation Steps of the Rtcb Rna Splicing Pathway Are Gtp-Dependent. Nucleic acids research. 2012;40:8558–67. doi: 10.1093/nar/gks558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cairns MJ, Hopkins Toni M, Witherington Craig, Li Wang, Lun-Quan Sun. Target Site Selection for an Rna-Cleaving Catalytic DNA. Nature biotechnology. 1999;17:480–486. doi: 10.1038/8658. [DOI] [PubMed] [Google Scholar]
- 30.Ruble BK, Richards JL, Cheung-Lau JC, Dmochowski IJ. Mismatch Discrimination and Efficient Photomodulation with Split 10–23 Dnazymes. Inorganica Chim Acta. 2012;380:386–391. doi: 10.1016/j.ica.2011.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka N, Chakravarty AK, Maughan B, Shuman S. Novel Mechanism of Rna Repair by Rtcb Via Sequential 2′,3′-Cyclic Phosphodiesterase and 3′-Phosphate/5′-Hydroxyl Ligation Reactions. The Journal of biological chemistry. 2011;286:43134–43. doi: 10.1074/jbc.M111.302133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill HD, Mirkin CA. The Bio-Barcode Assay for the Detection of Protein and Nucleic Acid Targets Using Dtt-Induced Ligand Exchange. Nature protocols. 2006;1:324–36. doi: 10.1038/nprot.2006.51. [DOI] [PubMed] [Google Scholar]
- 33.Hurst SJ, Lytton-Jean AK, Mirkin CA. Maximizing DNA Loading on a Range of Gold Nanoparticle Sizes. Anal Chem. 2006;78:8313–8318. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desai KK, Beltrame AL, Raines RT. Coevolution of Rtcb and Archease Created a Multiple-Turnover Rna Ligase. Rna. 2015;21:1866–72. doi: 10.1261/rna.052639.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. DNAzymes used in this study.
Table S2. Sequences used in this study.
Figure S1. DNAzymes used in this study (TIF)
Figure S2. SDS-PAGE of RtcB (TIF)
Figure S3. Activity of RtcB on linear and stem-loop strands (TIF)
Figure S4. Quantifying RtcB ligation yield on linear strands (TIF)
Figure S5. Quantifying RtcB kinetics on stem-loop substrate (TIF)
Figure S6. Dz1NPs and Dz2NPs with select mutations show lowered activity (TIF)
Figure S7. Image analysis for calculating splicing yield (TIF)
Figure S8. Sequential and one-pot splice reactions show similar yield (TIF)
Figure S9. RtcB splicing kinetics on substrate 3 (TIF)
Figure S10. RtcB splicing as a function of ligase concentration and DNAzyme inhibition (TIF)
Figure S11. RtcB splicing upon digestion with DNAzymes of decreasing arm length (TIF)
Figure S12. Calibration curve used to determine RtcB bound nanozyme (TIF)
Figure S13. His-tagged RtcB cannot form a splicing nanozyme (TIF)
Figure S14. RtcB is specifically bound to Dz1-2NPs (TIF)
Figure S15. The effect of multiple washes on nanozyme yield (TIF)
Figure S16. RtcB on non-specific NPs is still active on pre-cleaved substrate (TIF)
Figure S17. Flow cytometry of Dz1Dz2NPs and nanozymes entering MDA-MB-231 cells (TIF)