Abstract
Despite reports implicating disrupted purine metabolism in causing a wide spectrum of neurological defects, the mechanistic details of purine biosynthesis in neurons are largely unknown. As an initial step in filling that gap, we examined the expression and subcellular distribution of three purine biosynthesis enzymes (PFAS, PAICS and ATIC) in rat hippocampal neurons. Using immunoblotting and high-resolution light and electron microscopic analysis, we find that all three enzymes are broadly distributed in hippocampal neurons with pools of these enzymes associated with mitochondria. These findings suggest a potential link between purine metabolism and mitochondrial function in neurons and provide an impetus for further studies.
Keywords: Purine biosynthesis, Hippocampus, Neuron, Mitochondria
Introduction
Purines are a component of many essential biomolecules in cells ranging from DNA and RNA to ATP and GTP. The cellular level of purines is maintained by coordinated actions of two pathways: the salvage pathway and the de novo biosynthesis pathway (for review, see Pedley and Benkovic 2017). The salvage pathway produces purines by recycling degraded bases in a single enzyme-catalyzed reaction (Murray 1971). The de novo biosynthesis pathway produces purines through a 10-step reaction catalyzed by six enzymes (Greenberg and Jaenicke 1957; Buchanan and Hartman 1959). Under physiological conditions, the salvage pathway is responsible for controlling the basal cellular purine level. Under conditions with higher purine demands such as active cell growth, the de novo biosynthesis pathway plays a dominant role in increasing purine synthesis (Pedley and Benkovic 2017). It has been reported that the regulatory capacity of the de novo biosynthesis is greater than that of the salvage pathway (Yamaoka et al. 1997).
Much of our understanding of purine synthesis comes from in vitro biochemical experiments (Hartman and Buchanan 1959) and studies of tumor cell lines such as HeLa ovarian carcinoma cells (for example, see An et al. 2008). These studies have provided molecular details of purine biosynthesis as well as a link between altered purine synthesis and cancer (Natsumeda et al. 1984; Pedley and Benkovic 2017). However, the basic biology of purine synthesis in neurons has not been explicitly investigated. In humans, mutations in the enzymes of the purine biosynthesis pathway result in a wide range of profound neurological abnormalities. For example, mutation in adenylosuccinate lyase (ASL), the enzyme that catalyzes the eighth step in the purine biosynthetic pathway, causes autism and psychomotor delay (Jaeken and Van den Berghe 1984). Mutation in ATIC, the enzyme that catalyzes the ninth and tenth steps of purine biosynthesis, causes severe mental retardation, epilepsy and blindness (Marie et al. 2004). These human cases highlight the importance of the purine biosynthesis pathway and its individual enzymes in the normal functioning of the brain and neurons. An important step toward understanding the purine biosynthesis pathway in neurons is to learn the subcellular locations of the enzymes for the pathway and their spatial relationship to cellular organelles. In this study, we used high-resolution light and electron microscopic immunocytochemistry to reveal the expression, distribution and spatial organization of purine biosynthesis enzymes in hippocampal neurons.
Results and Discussion
PFAS, PAICS and ATIC are Expressed in the Brain and Cultured Hippocampal Neurons
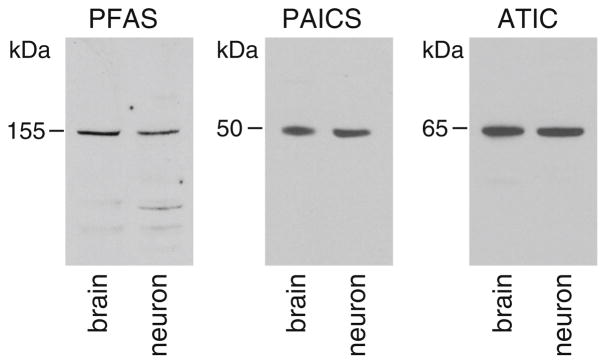
The purine biosynthesis pathway is composed of 10 chemical steps catalyzed by six enzymes. To begin understanding purine biosynthesis in the brain and in neurons, we examined the expression of the enzymes for the pathway in rat brains and cultured hippocampal neurons. We initially attempted to examine all six enzymes using commercially available antibodies, which have been characterized in HeLa cells (An et al. 2008; French et al. 2016; and personal communication with S.J. Benkovic et al.). Among the six antibodies, we found that the antibodies to PFAS (phosphoribosylformylglycinamidine synthase), PAICS (phosphoribosyl aminoimidazole succinocarboxamide synthetase) and ATIC (5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase) each reproducibly detected a major protein band at the expected molecular mass for the respective enzyme, in both extracts of brain tissues and lysates of cultured hippocampal neurons (Fig. 1). We therefore focused on examining the subcellular localization of PFAS, PAICS and ATIC in hippocampal neurons in vitro and in vivo.
Fig. 1.
Expression of PFAS, PAICS and ATIC in rat brain (postnatal day 30) and hippocampal neurons (14–22 days in culture). PFAS, phosphoribosylformylglycinamidine synthase; PAICS, phosphoribosyl aminoimidazole succinocarboxamide synthetase; ATIC, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase. Total protein (20 μg) from brain tissue or neuron lysates was used in the blots. The antibody dilutions used were: PFAS 1:2500, PAICS 1:5000, and ATIC 1:5000. The blots were repeated three times
Distribution Pattern of PFAS, PAICS and ATIC in Hippocampal Neurons in Vitro
We examined the distribution of PFAS, PAICS and ATIC in cultured hippocampal neurons by immunofluorescence light microscopy. At relatively low magnification of microscopic view, the three enzymes similarly displayed a broad distribution pattern with small immunolabeled puncta dispersed throughout all parts of the neuron (Fig. 2A; supplemental figure 1). When the primary antibodies were omitted from the immunolabeling protocol, we observed no detectable fluorescence.
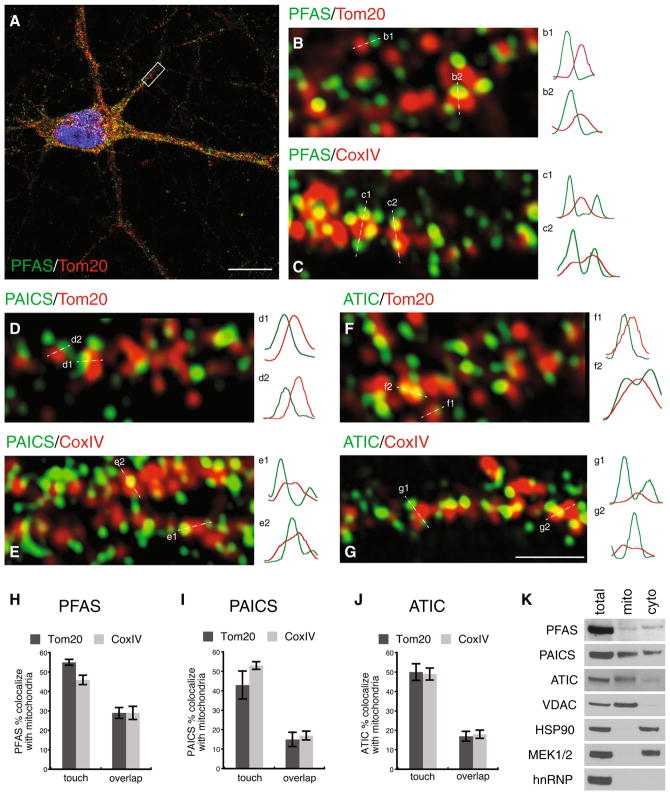
Fig. 2.
Distribution of PFAS, PAICS and ATIC in cultured rat hippocampal neurons. A Representative example of a hippocampal neuron co-labeled with PFAS (green) and a mitochondrial marker Tom20 (red). Scale bar, 10 μm. B Enlarged view of the boxed region (dendrite) in A illustrating close proximity or co-localization between PFAS (green) and mitochondria (red). Image shown is a single middle section of z-scans of the dendrite. Sample fluorescence intensity profiles along dashed lines are shown on the right. C Example of a segment of a dendrite co-labeled with PFAS (green) and another mitochondrial marker, CoxIV (red). D, E Examples of dendrites from neurons co-labeled with PAICS (green) and either Tom20 (red) or CoxIV (red). F, G Examples of dendrites co-labeled with ATIC (green) and either Tom20 (red) or CoxIV (red). Fluorescence intensity profiles show various extents of overlaps between green PFAS, PAICS or ATIC and red mitochondria. Scale bar in G is 1 μm and applies to B–G See also Supplemental figure 1. H–J Quantification of PFAS, PAICS or ATIC puncta in close contact (pixel touching to<50 pixel overlapping) or co-localized (≥50 pixel overlapping) with Tom20- or CoxIV-labeled mitochondria. K Immunoblots showing the presence of PFAS, PAICS and ATIC in mitochondria isolated from cultured hippocampal neurons. Fifteen micrograms of proteins from neuronal lysates (total), isolated mitochondria (mito) and mitochondria-depleted cytosol (cyto) were analyzed by immunoblot with the indicated antibodies. Note that the purity of isolated mitochondria is illustrated by the enrichment of a mitochondrial marker VDAC, and the absence of cytosolic marker HSP90 and MEK1/2 as well as nuclear marker hnRNP. See also Supplemental figure 2 (Color figure online)
In HeLa cells, the expression pattern of the purine biosynthetic enzymes is influenced by the level of purine in culture medium: large clusters in purine-depleted medium and non-cluster diffused pattern in purine-rich medium (An et al. 2008). We cultured the hippocampal neurons in the medium designed and standardized for culturing neurons with normal physiology (Brewer et al. 1993; Kaech and Banker 2006). Under this culture condition, hippocampal neurons exhibited the small puncta pattern for PFAS, PAICS and ATIC, and this pattern was seen consistently in the neurons of different culture ages and different culture batches (Fig. 2A, supplemental figure 1 and data not shown).
Prompted by the observations that in HeLa cells the purine biosynthesis enzymes co-localize with mitochondria (An et al. 2008; French et al. 2016), we examined whether in neurons these enzymes are also associated with mitochondria. We performed double immunolabeling with PFAS, PAICS or ATIC antibody, in combination with an antibody against Tom20, a translocase which resides in the mitochondrial outer membrane (Endo and Kohda 2002; Hoogenraad et al. 2002). Using confocal microscopy with Airyscan (Zeiss 880, with ~140-nm xy and 200-nm z resolution), we found that large pools of the purine synthesis enzymes were closely apposed to Tom20-labeled mitochondria (Fig. 2B, D, F; supplemental figure 1). We quantified the immunolabeled puncta for PFAS, PAICS and ATIC that were associated with Tom20-immunolabeled mitochondria. We found that 55% ± 2 (n = 4000 puncta from 20 cells; all results are given as mean ± SEM) of PFAS puncta were in close contact with Tom20-labeled mitochondria (pixel touching to <50 pixel overlapping), and 29% ± 3 (n = 4000 puncta from 20 cells) of PFAS puncta co-localized (≥50 pixel overlapping) with Tom20-labeled mitochondria (Fig. 2H). Similar to PFAS, 40–50% of PAICS- and ATIC-immunolabeled puncta were scored as touching Tom20-labeled mitochondria (PAICS 43% ± 7, n = 2000 puncta from 10 cells; ATIC, 50% ± 4, n = 4000 puncta from 20 cells), while fewer PAICS- and ATIC-immunolabeled puncta were scored as overlapping/co-localizing with Tom20 puncta (Fig. 2I, J). We examined another mitochondrial marker, an inner mitochondrial membrane protein cytochrome C oxidase complex IV (CoxIV; Fig. 2C, E, G; supplemental figure 1). Quantitative image analysis of the three purine biosynthesis enzymes with respect to CoxIV is shown in Fig. 2H, I, J. Overall, the results of the two mitochondrial markers, Tom20 and CoxIV, were in agreement with each other. Therefore, the three enzymes for the purine biosynthesis are expressed in hippocampal neurons, with pools of these proteins associated with mitochondria.
To evaluate the mitochondrial association of PFAS, PAICS and ATIC biochemically, we measured the protein levels of these enzymes in mitochondria purified from neurons. PFAS, PAICS and ATIC were all detected in purified mitochondria in addition to the mitochondria-depleted cytosol (Fig. 2K; supplemental figure 2). The mitochondria-depleted cytosol was validated by the absence of the mitochondrial protein VDAC (Fig. 2K; supplemental figure 2). The purity of mitochondria was demonstrated by the absence of cytosolic protein HSP90 (heat shock protein) and MEK1/2 (MAP kinase or ERK kinase) (Fig. 2K). For ATIC, the protein band had a slightly higher molecular mass (Fig. 2K; supplemental figure 2). It is not clear why mitochondria-associated ATIC displayed a small shift in molecular mass, but it is possible that the shift is caused by a posttranslational modification; further studies are required to address this question. Together, our results of immunofluorescence light microscopy and immunoblotting are in line with previous findings of HeLa cells (An et al. 2008; French et al. 2016), showing that the purine biosynthesis enzymes can co-localize with mitochondria.
Subcellular Location of PFAS, ATIC and PAICS in Hippocampal Neurons in Vivo
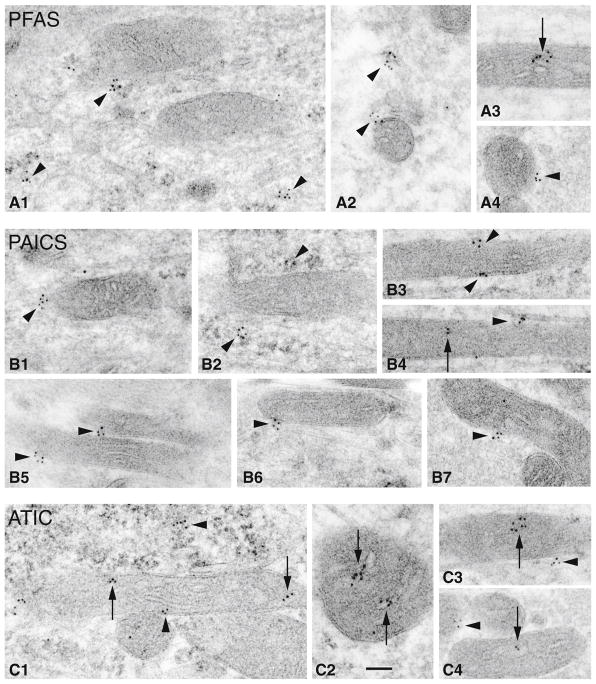
To reveal more precisely the physical relationship between the purine biosynthesis enzymes and mitochondria in neurons in the in vivo environment, we performed immunogold electron microscopy (immunoEM) analysis of the adult rat hippocampus. We focused on examining the neurons in the CA1 and CA3 areas of the hippocampus. We observed clusters of PFAS-associated immunogold particles positioned either in the immediate vicinity of mitochondria or directly on their surface (Fig. 3A1, A2 and A4). Fewer PFAS gold particles were found inside of mitochondria (Fig. 3A3). Similarly, we found more PAICS immunogold particles located near or on the surface of mitochondria (Fig. 3B1–B7), and some gold particles resided within mitochondria (Fig. 3B4; supplemental figure 3). With ATIC, the immunogold clusters were found more commonly inside the mitochondria (Fig. 3C1–C4) and less frequently near or on the surface of the mitochondria (Fig. 3C3, C4).
Fig. 3.
Subcellular localization of PFAS, ATIC and PAICS in rat hippocampus. Immunogold localization (10 nm gold) of purine biosynthesis enzymes in the hippocampus of two rats (postnatal day 37). A PFAS; B PAICS; C ATIC. Note how small clusters of gold labeling (arrowheads) are formed near or directly on the sides or surfaces of mitochondria. Immunogold particles within a mitochondrion (arrows) are seen occasionally with PFAS and PAICS, but more commonly with ATIC. Most micrographs show mitochondria from dendrites and somas of neurons (A3, B3, 4, C2 from CA1; A1, 2, B1, 2, C1 from CA3); A4, B5–7 and C4 are mitochondria in mossy terminals of CA3. Scale bar in C2 is 100 nm and applies to all micrographs. See also Supplemental figure 3
We have shown that, under normal physiological conditions, the purine biosynthesis enzymes are present in all compartments of the hippocampal neuron and pools of these enzymes localize to the mitochondria. Among the three enzymes we have examined, the prevalence of ATIC within mitochondria is consistent with a previous report of mitochondrial proteomics that detected the presence of ATIC in mitochondria (Pagliarini et al. 2008). An association of PFAS with mitochondria has also been reported in HeLa cells (French et al. 2016), wherein PFAS was in the form of purinosomes. Purinosomes are dynamic macromolecular complexes of the purine biosynthetic enzymes (Pedley and Benkovic 2017). It is unknown whether PFAS or any of the purine biosynthesis enzymes organize into purinosomes in normal neurons. Nevertheless, the physical association of the individual enzymes with mitochondria observed in the present study suggests that a link between purine metabolism and mitochondrial functions likely exists in neurons. Our characterization of the subcellular location of the three purine biosynthesis enzymes serves as a lead for future in-depth research into further understanding purine biosynthesis pathways in neurons. We anticipate that knowledge of purine biosynthesis in the brain and in neurons should provide insight into the molecular underpinnings of severe neurological phenotypes (Jaeken and Van den Berghe 1984; Marie et al. 2004) and brain cancer (Wang et al. 2017), resulting from disrupted purine metabolism.
Materials and Methods
Animals
Timed pregnant rats were used as the source of embryonic brains to establish cultures of hippocampal neurons. Brain tissues from postnatal day 30 or 37 rats were used for immunoblotting or immunogold labeling, respectively. All animal procedures were approved by the NIA and the NIDCD Animal Care and Use Committees and complied with the NIH Guide for Care and Use of Laboratory Animals.
Antibodies
Commercial antibodies against the following proteins were used: PFAS rabbit polyclonal antibody (Cat# A304-218, RRID: AB_2620415; 1:2500 immunoblotting, 1:2000 immunofluorescence, 1:2000 immunogold), PAICS rabbit polyclonal antibody (Cat# A304-546, RRID: AB_2620741; 1:5000 immunoblotting, 1:5000 immunofluorescence, 1:2000 immunogold) and ATIC rabbit polyclonal antibody (Cat# A304-271, RRID: 2620467; 1:5000 immunoblotting, 1:5000 immunofluorescence, 1:2000 immunogold) were from Bethyl Laboratories. Tom20 mouse monoclonal antibody (Cat# WH0009804M1, RRID: 1843992; 1:1000 immunofluorescence) was from Sigma. CoxIV mouse monoclonal antibody (Cat# 4D11-B3-E8; 1:500 immunofluorescence), VDAC rabbit monoclonal antibody (Cat#4661, RRID: AB_10557420; 1:1000 immunoblotting), HSP90 rabbit monoclonal antibody (Cat#4877), MEK1/2 rabbit monoclonal antibody (Cat#8727) were from Cell Signaling. hnRNP rabbit polyclonal antibody was from Santa Cruz Biotechnology, Inc. (Cat# sc-15386).
Primary Culture of Rat Hippocampal Neurons
Cultures of hippocampal neurons were prepared from embryonic day 18 rat brains as described (Mattson et al. 1989; Kaech and Banker 2006; Yao et al. 2017). Dissociated neurons were plated at a density of ~100 cells/mm2. The neurons were grown in neurobasal medium supplemented with B27 (Invitrogen). For immunoblotting, the neurons were grown in polylysine (0.1 mg/ml)-coated plastic dishes. For immunofluorescence, the neurons were grown on polylysine (1 mg/ml)-coated glass coverslips (no. 1.5). Polylysine was from Sigma (P2636). The age of the cultures used was 14–22 days.
Immunoblotting
Rat brains or cultured neurons were lysed in RIPA buffer (20 mM Tris–HCl, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 2.5 mM sodium pyrophosphate, 1 mM sodium orthovanadate and 1% sodium deoxycholate) containing protease inhibitors (Halt Protease Inhibitor Cocktail, Thermo Scientific). The lysed tissues or cells were centrifuged at 10,000 × g for 10 min at 4 °C. The supernatant was collected, and the amount of total proteins was estimated with a Pierce BCA Protein Assay Kit (Pierce Biotechnology). Protein samples were separated by 4–20% Bis–Tris SDS-PAGE and transferred to nitrocellulose membranes. Following incubation with blocking buffer (5% dry milk and 0.05% Tween20 in PBS), the membranes were incubated overnight at 4 °C in the blocking buffer containing the primary antibody being tested. The membranes were then washed (0.1% Tween20 in PBS) and incubated with appropriate peroxidase-conjugated secondary antibodies. The proteins were visualized using a chemiluminescence kit from Pierce.
Mitochondria were isolated using a mitochondria isolation kit for cultured cells (Thermo Scientific, #89874) following the manufacturer’s instructions. Approximately 108 cultured neurons were used for each sample. Soluble mitochondrial proteins were extracted from isolated mitochondria using 2% CHAPS in TBS.
Immunocytochemistry, Fluorescence Microscopy, Image Processing and Image Analysis
Neurons grown on glass coverslips were washed with PBS, fixed with 4% paraformaldehyde (Electron Microscopy Services) and 4% sucrose in PBS, and permeabilized with 0.2% Triton X-100 (Sigma) in PBS. After blocking with 10% bovine serum albumin in PBS, neurons were incubated with primary antibodies followed by species-appropriate secondary antibodies conjugated with Alexa Fluor-488 or Alexa Fluor-568. Coverslips were mounted on slides using ProLong Anti-fade mounting medium (Thermo Fisher).
Images were acquired with an Apochromat 63×/1.4 numerical aperture objective lens on a Zeiss LSM 880 microscope with Airyscan (Carl Zeiss). For each antibody labeling, the image acquisition settings were kept the same between different experiments. The brightness, contrast and levels of the images were adjusted in Adobe Photoshop and compiled in Adobe Illustrator. Fluorescence intensity profiles were measured from the images without any processing in Adobe. No additional digital image processing was performed. Control cells omitting the primary antibody showed no fluorescence labeling.
We used ImageJ (NIH) software to estimate the relative proportion of immunolabeled puncta for PFAS, PAICS or ATIC that contact (<50 overlapping green and red pixels) or co-localize (≥50 overlapping green and red pixels) with immunolabeled puncta for the mitochondrial markers Tom20 or CoxIV. Randomly selected neurons from two to three different cultures were used for the analysis. For each neuron, 200 immunolabeled puncta were counted (100 from soma and 100 from neurites). Counting was performed by an observer (J.W.) who was familiar with the analyzing criteria, but who was unaware of the experimental details until after analyses were complete.
Immunogold Electron Microscopy
Postembedding immunogold labeling was performed as described previously (Petralia and Wenthold 1999; Petralia et al. 2010; Yao et al. 2015). Briefly, following cryoprotection and embedding, rat brain sections were processed and embedded in Lowicryl HM-20 resin using a Leica AFS freeze-substitution instrument. After blocking, the sections were incubated with primary antibody followed by 10-nm gold-conjugated secondary antibody. Following thorough washes, the sections were stained with uranyl acetate and lead citrate. All electron micrographs were stored in their original formats. For micrographs presented for figures, their levels, brightness and contrast were minimally and evenly adjusted in Adobe Photoshop. Control sections omitting the primary antibody showed only rare gold labeling.
Presentation of Data and Statistics
All graphs were produced using KaleidaGraph (Synergy) software. Statistical comparisons were calculated using the unpaired Student’s t test. All results are expressed as mean ± SEM.
Supplementary Material
Acknowledgments
We thank Drs. James P. Lata and Stephen J. Benkovic for discussions in the initial stage of this work. We also thank Dr. Fred E. Indig for assistance in confocal Airyscan imaging. This study was supported by the Intramural Research Programs of the National Institutes of Health, National Institute on Aging, and the National Institutes of Health, National Institute on Deafness and Other Communication Disorders. The Advanced Imaging Core code is ZIC DC000081.
Abbreviations
- PFAS
Phosphoribosylformylglycinamidine synthase
- PAICS
Phosphoribosyl aminoimidazole succinocarboxamide synthetase
- ATIC
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s12017-017-8466-6) contains supplementary material, which is available to authorized users.
References
- An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320(5872):103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented neurobasal, a new serum-free medium combination. Journal of Neuroscience Research. 1993;35(5):567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- Buchanan JM, Hartman SC. Enzymic reactions in the synthesis of the purines. In: Ford FF, editor. Advances in Enzymology and Related Areas of Molecular Biology. London: Wiley; 1959. pp. 199–261. [Google Scholar]
- Endo T, Kohda D. Functions of outer membrane receptors in mitochondrial protein import. Biochimica et Biophysica Acta. 2002;1592(1):3–14. doi: 10.1016/s0167-4889(02)00259-8. [DOI] [PubMed] [Google Scholar]
- French JB, Jones SA, Deng H, Pedley AM, Kim D, Chan CY, et al. Spatial colocalization and functional link of purinosomes with mitochondria. Science. 2016;351(6274):733–737. doi: 10.1126/science.aac6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg GR, Jaenicke L. On the activation of the one-carbon unit for the biosynthesis of purine nucleotides. In: Wolstenholme GEW, O’Connor CM, editors. Ciba Foundation Symposium—Chemistry and Biology of Purines. London: Wiley; 1957. pp. 204–232. [Google Scholar]
- Hartman SC, Buchanan JM. Nucleic acids, purines, pyrimidines (nucleotide synthesis) Annual Review of Biochemistry. 1959;28:365–410. doi: 10.1146/annurev.bi.28.070159.002053. [DOI] [PubMed] [Google Scholar]
- Hoogenraad NJ, Ward LA, Ryan MT. Import and assembly of proteins into mitochondria of mammalian cells. Biochimica et Biophysica Acta. 2002;1592(1):97–105. doi: 10.1016/s0167-4889(02)00268-9. [DOI] [PubMed] [Google Scholar]
- Jaeken J, Van den Berghe G. An infantile autistic syndrome characterised by the presence of succinylpurines in body fluids. Lancet. 1984;2(8411):1058–1061. [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nature Protocols. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Marie S, Heron B, Bitoun P, Timmerman T, Van Den Berghe G, Vincent MF. AICA-ribosiduria: a novel, neurologically devastating inborn error of purine biosynthesis caused by mutation of ATIC. American Journal of Human Genetics. 2004;74(6):1276–1281. doi: 10.1086/421475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Murrain M, Guthrie PB, Kater SB. Fibroblast growth factor and glutamate: opposing roles in the generation and degeneration of hippocampal neuroarchitecture. Journal of Neuroscience. 1989;9:3728–3740. doi: 10.1523/JNEUROSCI.09-11-03728.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW. The biological significance of purine salvage. Annual Review of Biochemistry. 1971;40:811–826. doi: 10.1146/annurev.bi.40.070171.004115. [DOI] [PubMed] [Google Scholar]
- Natsumeda Y, Prajda N, Donohue JP, Glover JL, Weber G. Enzymic capacities of purine de Novo and salvage pathways for nucleotide synthesis in normal and neoplastic tissues. Cancer Research. 1984;44(6):2475–2479. [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134(1):112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley AM, Benkovic SJ. A new view into the regulation of purine metabolism: The purinosome. Trends in Biochemical Sciences. 2017;42(2):141–154. doi: 10.1016/j.tibs.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, Wang YX, Hua F, Yi Z, Zhou A, Ge L, et al. Organization of NMDA receptors at extrasynaptic locations. Neuroscience. 2010;167:68–87. doi: 10.1016/j.neuroscience.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, Wenthold RJ. Immunocytochemistry of NMDA receptors. Methods in Molecular Biology. 1999;128:73–92. doi: 10.1385/1-59259-683-5:73. [DOI] [PubMed] [Google Scholar]
- Wang X, Yang K, Xie Q, Wu Q, Mack SC, Shi Y, et al. Purine synthesis promotes maintenance of brain tumor initiating cells in glioma. Nature Neuroscience. 2017;20:661–673. doi: 10.1038/nn.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka T, Kondo M, Honda S, Iwahana H, Moritani M, Ii S, et al. Amidophosphoribosyltransferase limits the rate of cell growth-linked de novo purine biosynthesis in the presence of constant capacity of salvage purine biosynthesis. Journal of Biological Chemistry. 1997;272(28):17719–17725. doi: 10.1074/jbc.272.28.17719. [DOI] [PubMed] [Google Scholar]
- Yao PJ, Manor U, Petralia RS, Brose RD, Wu RT, Ott C, et al. Sonic hedgehog pathway activation increases mitochondrial abundance and activity in hippocampal neurons. Molecular Biology of the Cell. 2017;28(3):387–395. doi: 10.1091/mbc.E16-07-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao PJ, Petralia RS, Ott C, Wang YX, Lippincott-Schwartz J, Mattson MP. Dendrosomatic sonic hedgehog signaling in hippocampal neurons regulates axon elongation. Journal of Neuroscience. 2015;35:16126–41611. doi: 10.1523/JNEUROSCI.1360-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.