Abstract
The phenotypic consequences of a given mutation can vary across individuals. This so-called “background effect” is widely observed, from mutant fitness of loss-of-function variants in model organisms to variable disease penetrance and expressivity in humans; however, the underlying genetic basis often remains unclear. Taking insights gained from recent large-scale surveys of genetic interaction and suppression analyses in yeast, we propose that the genetic network context for a given mutation may shape its propensity of exhibiting background-dependent phenotypes. We argue that further efforts in systematically mapping the genetic interaction networks beyond yeast will not only provide key insights into the functional properties of genes, but also a better understanding of the background effects and the (un)predictability of traits in a broader context.
Keywords: Genetic interaction, Background effect, Variable phenotypic expression, Conditional gene essentiality
Genetic background and the (un)predictability of traits
A particular mutation in a given gene often leads to different phenotypes in different individuals. This observation dates back almost to the beginning of modern genetics [1, 2], and yet geneticists are still challenged with understanding specific genotype-phenotype relationships [3]. In many human monogenic disorders, individuals carrying a well-defined mutation in a disease gene frequently show variable penetrance and expressivity in terms of clinical symptoms, severity and age-of-onset [4–9]. While multiple factors can contribute to this effect, the specific makeup of an individual’s genome, or the genetic background, is increasingly recognized as a primary source of variable phenotypic expression [3, 10–13]. In fact, when individual background is considered, “simple” traits, such as those primarily caused by one or a few genes, can actually be considered as genetically complex [4, 14–17].
The phenotypic consequence of a primary mutation can be influenced by genome variants termed “modifiers”, which can aggravate or mask the expected phenotypic outcome. To date, modifier genes have been identified for some of the best-studied monogenic diseases, including cystic fibrosis and sickle cell anemia [8, 16, 18]. However, systematic identification of genetic modifiers in humans is hampered by the low population frequency of rare traits and diseases, a lack of pedigree data, and other uncontrolled environmental confounders [4]. On the other hand, genetically tractable model organisms are particularly suitable for systematic approaches with precise experimental controls. Recently, large-scale reverse genetic screens revealed the extensive landscape of background-specific phenotypic expression across various model systems including yeasts [19], nematodes [20, 21], drosophila [13, 22, 23], mice [24, 25] and human cell lines [26–28]. These studies revealed loss-of-function mutations that cause a severe fitness defect in one individual, but no discernable defect in another due to background-specific genetic modifiers.
Precise phenotypic prediction for a given individual essentially requires the knowledge of all primary variants and their modifiers that collectively drive the phenotypic outcome. Here, we consider recent large-scale genetic interaction studies in yeast to discuss the molecular basis of genetic modification. We focus on gene essentiality as a conditional phenotype and discuss how the complexity of a genetic interaction network, e.g. the number of interactions observed between a primary variant and its potential modifiers, could shape the propensity of the phenotypic outcome. We argue that further understanding of the genetic interaction network across different yeast individuals and beyond may hold a key to unraveling the molecular and genetic mechanisms of background effects that lead to the apparent “unpredictability” of traits.
Genetic modifiers as part of functional networks related to primary mutations
Although it has only recently become possible to trace modifier genes in natural populations, experiments to map modifiers for a defined primary variant in a controlled experimental system have been common practice for many genetic studies. For example, genetic interactions, where the combined effect of different mutations deviates from the expected effect of each mutation on its own, have been studied systematically in the budding yeast, Saccharomyces cerevisiae. In the most comprehensive analysis so far, loss-of-function alleles for nearly all yeast genes were combined, using an automated form of yeast genetics called Synthetic Genetic Array (SGA) analysis, to generate ~18 million double mutant strains [29–31]. Genetic interactions were scored by measuring colony size as a proxy for cell fitness: a negative genetic interaction occurs when a double mutant has a more severe fitness defect than expected, while a positive genetic interaction is scored when the double mutant is more fit than expected [32]. Synthetic lethality is an extreme negative genetic interaction in which mutations in two different genes, neither of which is lethal on its own, combine to cause a lethal double mutant phenotype. In contrast, genetic suppression is an extreme positive interaction, which occurs when the phenotype caused by a mutated gene is rescued by mutation in another gene [33]. Using the colony size read-out, approximately ~500,000 negative genetic interactions and ~350,000 positive double mutant genetic interactions have been catalogued, enabling visualization of the first comprehensive genetic network for any model system [31].
The global yeast network revealed that genetic interactions are more likely to occur among functionally related genes [31], a feature that is likely shared between a primary mutation and its modifiers within individuals of an outbred population. Indeed, recent large-scale analyses of genetic suppression revealed the same trend [34]. Compared to the positive genetic interactions mapped by large-scale SGA analysis, genetic suppression, which is often driven by spontaneous mutations, is not limited to loss-of-function alleles, but can also reflect gain-of-function mutations, changes in chromosome copy number, or gene dosage [34–36]. A recent analysis combined literature curation and experimental analysis to map ~2,500 unique pairwise suppression interactions for ~1,000 hypomorphic (partial loss-of-function) query alleles in yeast [34]. This sub-network of suppression interactions showed significant overlap with the global genetic interaction network mapped by SGA analysis [24]. Notably, genes pairs on the suppression network displayed a closer functional relatedness than for gene pairs on the global SGA network [27].
Intuitively, it is not surprising that modifiers and the primary mutation tend to be functionally related. Indeed, genes scarcely work as discrete units, but often operate in functional modules that coordinate with other such modules to ensure cellular processes. Thus, with a general understanding of the functional wiring diagram of cells and organisms, the prediction of modifiers should be achievable to a reasonable degree, especially when the primary mutation involves a loss-of-function allele.
Network connectivity and the modifiability of traits
Because genes tend to interact with functionally-related genes, often in the same general bioprocess [31,34], an understanding of gene function may provide insight into the genetic background effect. However, not all phenotypes are modifiable. In fact, a recent study tested the “evolvability” of all yeast essential genes in the reference S288c strain background and revealed that a subset of ~9% of yeast essential genes can rapidly acquire secondary mutations and escape essentiality [35]. In this study, a large number of meiotic offspring were generated for ~1,000 different diploid S288c strains, each carrying a heterozygous deletion of an essential gene. As essential genes are required for viability, half of the haploid offspring carrying the deletion alleles should be non-viable. However, haploid individuals derived from 88 essential gene deletion strains were able to grow after a short period of time through acquisition of chromosomal aberrations, such as aneuploidies and segmental duplications. This result enabled classification of the yeast genome into evolvable essential genes, non-evolvable essential genes, and non-essential genes, which facilitated an analysis of the shared molecular properties of these three gene sets. Interestingly, within the global yeast protein-protein interaction network, proteins encoded by non-evolvable essential genes had the highest number of interacting partners, whereas the evolvable essential genes tend to present an intermediate number of interactions, contrasting to non-essential genes, which have the least protein interactions [35]. Thus, the evolvable essential genes displayed a protein interaction degree that was intermediate, between that of the non-evolvable essential genes and the nonessential genes.
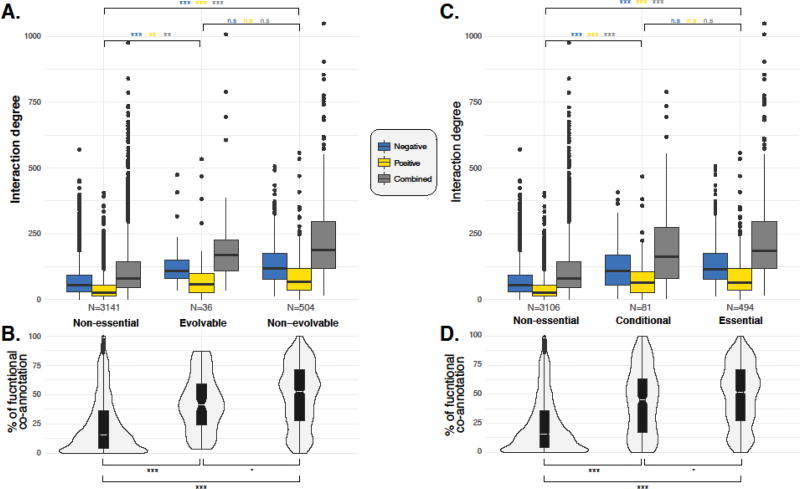
To further investigate the properties of evolvable essential genes, we analyzed the genetic interaction degrees, defined as the number of partners on the global yeast genetic interaction network [31], for all three classes of genes mentioned above. As for the protein network, there is a gradient of connectivity with non-essential genes showing relatively few connections, evolvable essential genes showing an intermediate connectivity, and non-evolvable essential genes showing the highest connectivity (Figure 1A). These trends were evident whether negative and positive genetic interactions were considered separately or as a combined dataset. We also analyzed the percentage of interacting partners that functionally co-annotated with each primary gene, and found that evolvable essential genes and their genetic partners had a significantly higher co-annotation rate than non-essential genes, and significantly lower co-annotation rate than non-evolvable essential genes (Figure 1B). These trends suggest that mutations in a gene functioning in a highly connected genetic interaction network, i.e. defined by the high number of genetic interactions, would cause a strong functional perturbation, one that is not easily suppressed by a relatively small number of modifiers. In other words, the complexity of the genetic network in which a primary mutation participates may constrain its genetic modifiability.
Figure 1. Genetic interaction degree and functional co-annotation of interacting genes.
The number of genetic interactions observed for non-essential, evolvable and non-evolvable gene categories (A) and non-essential, conditional and essential gene categories (C) are shown in boxplots. The types of interaction are color-coded and the numbers of observations are indicated at the bottom of each plot. For each gene category, the corresponding functional co-annotation rate and distribution are presented (B and D). One-tailed t-test has been performed for all pairwise comparison between assigned gene categories and percentages of functional co-annotation. The significance levels are indicated on the graph. Significance levels: n.s (p-value > 0.1), • (0.05 < p-value < 0.1), * (0.005 < p-value < 0.05), ** (0.0005 < p-value < 0.005), *** (p-value < 0.0005). Data compiled from [31, 35].
Genetic network complexity may also influence the effects of genetic background in two individuals sampled from a natural outbred population. Specifically, mutation in a gene involved in a highly complex genetic network may be less likely to exhibit different phenotypes in different individuals due to constraints imposed by the network. In contrast, mutations in less-connected genes may cause more background-specific phenotypes, as relatively few modifier loci would be required to accentuate or suppress the defect in the primary gene. To test this hypothesis, we compiled a list of 81 conditional essential genes across 6 different genetic backgrounds in S. cerevisiae (Table S1) [35]. These genes are classified as conditional essential because they are required for viability in at least one but not all of the considered backgrounds. We used this analysis to organize the yeast genome into three different groups: non-essential, conditional essential and essential gene categories. In accordance with our hypothesis, conditional essential genes showed intermediate interaction degrees and functional co-annotation rates compared to non-essential (lowest degree, lowest co-annotation rate) and essential genes (highest degree, highest co-annotation rate) (Figure 1C–D).
Overall, these analyses suggest that the background effect on phenotypic expression, at least in the case of gene essentiality in yeast, is not only influenced by the presence of background specific modifiers, but may also be constrained by the complexity of the underlying genetic network.
Genetic network complexity and the background effects of monogenic variants in natural yeast populations
Natural genetic variation in yeast has not only been assessed using defined isogenic mutant collections, but also by assessing variable phenotypic expression at the population level [15, 38–40]. In a recent systematic effort, a species-wide survey identified Mendelian traits across 41 crosses and 30 stress conditions [15]. In total, 1,105 cross/trait combinations were analyzed, of which 8.9% (98/1,105) were Mendelian, resulting from a single causal variant. Interestingly, the identified Mendelian cases can be divided into “stable” and “modifiable” based on the inheritance patterns across the population. Stable Mendelian traits, dominated by genes related to the osmotic stress condition, consistently displayed monogenic inheritance across all crosses tested. In contrast, modifiable Mendelian traits identified by the response to antifungal drugs displayed unexpected inheritance patterns, with increased genetic complexity ranging from a single modifier to more than 3 modifiers depending on the genetic background [15].
The primary, causal, monogenic variants in these cases have been identified: variation in ENA1, which encodes a P-type ATPase sodium pump, caused the stable phenotypic variation in the osmotic stress condition, while variation in the “transcription regulator of pleiotropic drug resistance” gene PDR1 explained the modifiable or variable response to drugs. We examined the yeast global genetic interaction data, and discovered that ENA1 has a low network complexity, with a combined interaction degree of 31. By contrast, PDR1 has a combined interaction degree of 246, indicating a medium to high network complexity. Albeit the network complexity presented here could be biased due to the fact that the genetic interactions were mapped using loss-of-function mutations in a different environmental context and strain background, these observations support our hypothesis that the phenotypic modifiability of a monogenic mutation may be shaped by the complexity of the underlying genetic network. While more systematic studies are required, the general rules concerning the genetic principles of phenotypic modifiability do seem to apply for natural genetic variants as well.
Yeast genetic network complexity and conservation of gene essentiality in other organisms
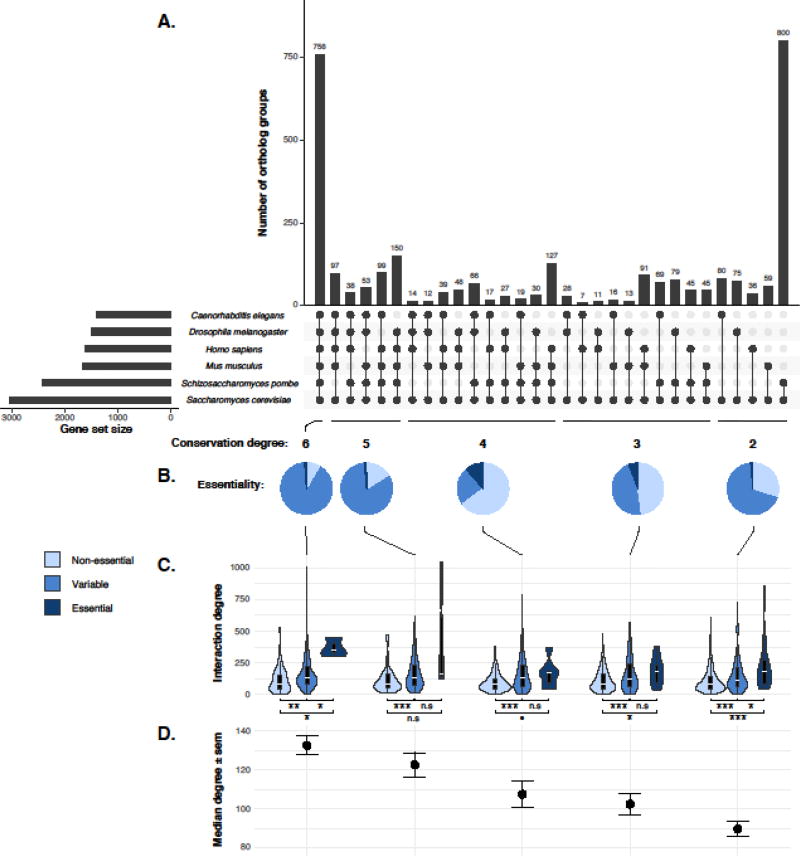
In addition to S. cerevisiae, genome-wide screens of loss-of-function mutations have been recently completed in other model organisms, including fission yeast (Schizosaccharomyces pombe) [42, 43], fly (Drosophila melanogaster) [44], worm (Caenorhabditis elegans) [45] and mouse (Mus musculus) [46], as well as several human cell lines [26–28]. These screens allowed global annotations of gene essentiality across multiple species spanning hundreds of millions of years of evolution. To further investigate how genetic interaction network properties impact gene function and evolution, we compiled a list of 3408 ortholog groups consisting of orthologous genes from at least one of the aforementioned species relative to the corresponding S. cerevisiae ortholog from the InParanoid database [47] (Figure 2A, Table S2). Each ortholog group is further categorized by the number and identity of the species that shared the same ortholog (Figure 2A). For each gene in each ortholog group, the essentiality annotation was obtained from the OGEE database for the corresponding species [48]. In addition to essential (E) and non-essential (NE), we also defined genes as conditional essential (E/NE), if multiple datasets gave different essentiality annotations for the same gene; and non-determined (ND) in the case of missing data (Table S2). Using annotations at the gene level, we further assigned an essentiality qualifier to the entire ortholog group: an ortholog group is considered as “non-essential”, if all members are consistently annotated as non-essential across different species. By contrast, if all members are consistently annotated as essential, the ortholog group is annotated as “essential”. The qualifier “variable” is then assigned for cases where different essentialities have been observed across different species or different datasets within the same species. Using these criteria, we categorized all ortholog groups into “essential”, “non-essential” and “variable” by different levels of conservation i.e. the number of species observed in each ortholog group (Figure 2B).
Figure 2. The impact of genetic network complexity on the evolution of gene essentiality in orthologs across six distantly related species.
A. Ortholog conservations across six species. A list of 3408 S. cerevisiae genes and their orthologs from human (Homo sapiens), mouse (Mus musculus), fly (Drosophila melanogaster), worm (Caenorhabditis elegans) and fission yeast (Schizosaccharomyces pombe) was compiled from the InParanoid database [47]. The gene set sizes correspond to the number of orthologs found per species and are indicated on the left side of the plot. The intersections among different species are shown by connective dots, which is ranked by the number of shared species (conservation degree). The number of orthologs within each intersection is indicated as bar plot at the top panel. B. Pie chart distribution of gene essentiality within each degree of ortholog conservation groups. Gene essentiality for each ortholog from each of the species examined was obtained from the OGEE database [48]. Genes that are consistently non-essential across different species are annotated as “Non-essential” for the entire ortholog group and is color-coded as light blue. Genes that are consistently essential are annotated as “Essential” (dark blue) and genes display variable essentialities across different species are annotated as “Variable” (blue). C. Combined genetic interaction degree observed in S. cerevisiae [31] relative to the degree of conservation and gene essentiality annotations. Interaction degree data were available for 1994/3408 yeast orthologs. For each degree of conservation, one-tailed t-test has been performed for all pairwise comparison between “Non-essential”, “Variable” and “Essential” qualifiers. The significance levels are indicated on the graph. Significance levels: n.s (p-value > 0.1), • (0.05 < p-value < 0.1), * (0.005 < p-value < 0.05), ** (0.0005 < p-value < 0.005), *** (p-value < 0.0005). D. Median interaction degree observed in each group of conservation degree. Error-bars indicate the standard error. All data can be found in Table S2.
This cross-species analysis of ortholog essentiality allowed us to further test the hypothesis that genetic network complexity may constraint the modifiability of the associated phenotype for a given gene. For example, if all members in an ortholog group are consistently essential or non-essential, this ortholog group may be less phenotypically modifiable than an ortholog group with variable essentiality across different member species. In this case, we expect the “variable” groups to display an intermediate level of network complexity compared to the “essential” and “non-essential” ones. Using the yeast genetic interaction data [31], we examined the interaction degrees (combined number of positive and negative interactions) for “essential”, “non-essential” and “variable” ortholog groups for different conservation levels (Figure 2C). Indeed, we consistently observed an intermediate level of interaction degree for the “variable” ortholog groups compared to “essential” groups, which displayed the highest interaction degrees, and “non-essential” groups, which displayed the lowest interaction degrees, across all levels of conservation (Figure 2C). Furthermore, we also observed a linear relationship between the mean interaction degree and the level of conservation: the higher the level of conservation, the higher the mean number of interaction and vice versa (Figure 2D). These observations fit well with our general model, and illustrate how data from one species, in this case the genetic interaction degrees from S. cerevisiae, can be extrapolated to predict patterns of gene evolution across a broad evolutionary scale.
Concluding remarks
At the current stage, precisely predict traits from genomes is still a goal to be reached. On the one hand, advances in sequencing technologies allow us to easily probe thousands of genomes across various populations and species; on the other hand, ever-growing datasets from systems biology studies provide deeper understanding of cellular functions. Bridging the gap between such vast functional genomic data and precise phenotypic prediction at the individual level requires a paradigm shift in our view of quantitative genetics: the genotype-to-phenotype relationship is far from linear but results from interconnected and dynamic networks. This “omnigenic” view of complex traits [49], such as most human diseases, therefore invokes a better understanding of how genes are connected within and across different genetic backgrounds, and appears to be critical for unraveling the underlying genetic architecture of traits.
Large-scale forward genetics in model systems has demonstrated the importance and prevalence of genetic interactions in basic cellular functions. In the context of gene essentiality as a measure of phenotypic variability, we showed that the phenotypic modifiability, both in regard to gene essentiality across different backgrounds in yeast and in regard to ortholog essentiality and conservation across different species, can be recapitulated by the connectivity of genes on the global yeast genetic interaction network. However, it is impractical to consider mapping the functional wiring of all genes for large numbers of individuals using current methods, even in a genetically tractable system like yeast. Moving forward, the focus should be drawn on amenable strategies to better understand network architecture and dynamics at the population level. On the experimental side, increasingly efficient editing techniques, such as those involving CRISPR-Cas9, opens the door to analysis of genetic interactions in more complex systems such human cells. Other genome-wide information such as co-expression patterns and PPIs should also be more readily integrated.
More importantly, we should take advantage of the richness of population genomics data and explore genome-wide signatures of co-evolving functional modules. Natural genetic variants comprise not only loss- or gain-of-function mutations that directly impact protein function, but also other types of mutations that may change expression patterns, localizations, protein conformations etc. Understanding the interaction networks beyond the context of loss-of-function variants and gene essentiality, for example diseases in human populations, will offer new insights into how network complexity shapes phenotypic outcome within a population.
Supplementary Material
Trends Box.
-
-
The same mutation often does not lead to the same phenotype in different individuals due to other genetic variants that are specific to each individual genome
-
-
Background modifiers can arise through gain- or loss-of-function mutations in genes involved in related cellular functions
-
-
Our ability to predict phenotypes relies on expanding our knowledge of the complex networks of genetic interactions underlying traits; however, the structure and complexity of the network itself may be variable across individuals
-
-
Deeper understanding of the conservation and variability of functional genetic networks among individuals may be key to trait prediction
Outstanding Questions Box.
-
-
Modifiers are enriched for genes that are functionally related to the primary variant; however, most functional connections among genes remain obscure. How do we integrate systems genetic information to better understand gene function?
-
-
How much do functional wiring diagrams of genetic interactions differ among individuals?
-
-
Is there a core set of genetic interactions common to all individuals?
-
-
How can other factors, such as environment, perturb genetic interaction network structure and shape phenotypic outcomes?
Acknowledgments
Functional genomics work in the Boone and Andrews labs is supported primarily by the Canadian Institutes for Health Research (CIHR) [grant numbers FDN-143264, FDN-143265], the National Institutes of Health [grant number R01HG00583], and the Ontario Ministry of Research Innovation and Science [grant number RE07-037]. J.v.L. was supported by a postdoctoral fellowship from the CIHR. C.B. holds a Canada Research Chair (Tier 1) in Proteomics, Bioinformatics and Functional Genomics. C.B. and B.A. are Senior Fellows and co-Director (C.B.) of the Canadian Institutes for Advanced Research Genetic Networks Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bateson W. Facts Limiting the Theory of Heredity. Science. 1907;26:649–660. doi: 10.1126/science.26.672.649. [DOI] [PubMed] [Google Scholar]
- 2.Altenburg E, Muller HJ. The Genetic Basis of Truncate Wing,-an Inconstant and Modifiable Character in Drosophila. Genetics. 1920;5:1–59. doi: 10.1093/genetics/5.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sackton TB, Hartl DL. Genotypic Context and Epistasis in Individuals and Populations. Cell. 2016;166:279–287. doi: 10.1016/j.cell.2016.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen R, et al. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nature biotechnology. 2016;34:531–538. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- 5.Kammenga JE. The background puzzle: how identical mutations in the same gene lead to different disease symptoms. The FEBS journal. 2017;284:3362–3373. doi: 10.1111/febs.14080. [DOI] [PubMed] [Google Scholar]
- 6.Cooper DN, et al. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Human genetics. 2013;132:1077–1130. doi: 10.1007/s00439-013-1331-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dipple KM, McCabe ER. Modifier genes convert 0022;simple0022; Mendelian disorders to complex traits. Molecular genetics and metabolism. 2000;71:43–50. doi: 10.1006/mgme.2000.3052. [DOI] [PubMed] [Google Scholar]
- 8.Steinberg MH, Sebastiani P. Genetic modifiers of sickle cell disease. American journal of hematology. 2012;87:795–803. doi: 10.1002/ajh.23232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thein SL. Genetic modifiers of sickle cell disease. Hemoglobin. 2011;35:589–606. doi: 10.3109/03630269.2011.615876. [DOI] [PubMed] [Google Scholar]
- 10.Sittig LJ, et al. Genetic Background Limits Generalizability of Genotype-Phenotype Relationships. Neuron. 2016;91:1253–1259. doi: 10.1016/j.neuron.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lachance J, et al. Genetic background and GxE interactions modulate the penetrance of a naturally occurring wing mutation in Drosophila melanogaster. G3. 2013;3:1893–1901. doi: 10.1534/g3.113.007831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandler CH, et al. How well do you know your mutation? Complex effects of genetic background on expressivity, complementation, and ordering of allelic effects. PLoS genetics. 2017;13:e1007075. doi: 10.1371/journal.pgen.1007075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chari S, Dworkin I. The conditional nature of genetic interactions: the consequences of wild-type backgrounds on mutational interactions in a genome-wide modifier screen. PLoS genetics. 2013;9:e1003661. doi: 10.1371/journal.pgen.1003661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonarakis SE, et al. Mendelian disorders and multifactorial traits: the big divide or one for all? Nature reviews. Genetics. 2010;11:380–384. doi: 10.1038/nrg2793. [DOI] [PubMed] [Google Scholar]
- 15.Hou J, et al. The Hidden Complexity of Mendelian Traits across Natural Yeast Populations. Cell reports. 2016;16:1106–1114. doi: 10.1016/j.celrep.2016.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorfman R. Modifier gene studies to identify new therapeutic targets in cystic fibrosis. Current pharmaceutical design. 2012;18:674–682. doi: 10.2174/138161212799315920. [DOI] [PubMed] [Google Scholar]
- 17.Fournier T, Schacherer J. Genetic backgrounds and hidden trait complexity in natural populations. Current opinion in genetics & development. 2017;47:48–53. doi: 10.1016/j.gde.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cutting GR. Modifier genes in Mendelian disorders: the example of cystic fibrosis. Annals of the New York Academy of Sciences. 2010;1214:57–69. doi: 10.1111/j.1749-6632.2010.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dowell RD, et al. Genotype to phenotype: a complex problem. Science. 2010;328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vu V, et al. Natural Variation in Gene Expression Modulates the Severity of Mutant Phenotypes. Cell. 2015;162:391–402. doi: 10.1016/j.cell.2015.06.037. [DOI] [PubMed] [Google Scholar]
- 21.Paaby AB, et al. Wild worm embryogenesis harbors ubiquitous polygenic modifier variation. eLife. 2015;4 doi: 10.7554/eLife.09178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chow CY, et al. Candidate genetic modifiers of retinitis pigmentosa identified by exploiting natural variation in Drosophila. Human molecular genetics. 2016;25:651–659. doi: 10.1093/hmg/ddv502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He BZ, et al. Effect of genetic variation in a Drosophila model of diabetes-associated misfolded human proinsulin. Genetics. 2014;196:557–567. doi: 10.1534/genetics.113.157800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamilton BA, Yu BD. Modifier genes and the plasticity of genetic networks in mice. PLoS genetics. 2012;8:e1002644. doi: 10.1371/journal.pgen.1002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorman A, et al. Genetic analysis of intestinal polyp development in Collaborative Cross mice carrying the Apc (Min/+) mutation. BMC genetics. 2016;17:46. doi: 10.1186/s12863-016-0349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blomen VA, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 27.Hart T, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 28.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 31.Costanzo M, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353 doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baryshnikova A, et al. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nature methods. 2010;7:1017–1024. doi: 10.1038/nmeth.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novick P, et al. Suppressors of yeast actin mutations. Genetics. 1989;121:659–674. doi: 10.1093/genetics/121.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Leeuwen J, et al. Exploring genetic suppression interactions on a global scale. Science. 2016;354 doi: 10.1126/science.aag0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu G, et al. Gene Essentiality Is a Quantitative Property Linked to Cellular Evolvability. Cell. 2015;163:1388–1399. doi: 10.1016/j.cell.2015.10.069. [DOI] [PubMed] [Google Scholar]
- 36.Patra B, et al. A genome wide dosage suppressor network reveals genomic robustness. Nucleic acids research. 2017;45:255–270. doi: 10.1093/nar/gkw1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edwards MD, et al. Interactions between chromosomal and nonchromosomal elements reveal missing heritability. Proc Natl Acad Sci U S A. 2014;111:7719–7722. doi: 10.1073/pnas.1407126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou J, et al. Comprehensive survey of condition-specific reproductive isolation reveals genetic incompatibility in yeast. Nature communications. 2015;6:7214. doi: 10.1038/ncomms8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou J, Schacherer J. Fitness Trade-Offs Lead to Suppressor Tolerance in Yeast. Molecular biology and evolution. 2017;34:110–118. doi: 10.1093/molbev/msw225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hope EA, Dunham MJ. Ploidy-regulated variation in biofilm-related phenotypes in natural isolates of Saccharomyces cerevisiae. G3. 2014;4:1773–1786. doi: 10.1534/g3.114.013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome research. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han TX, et al. Global fitness profiling of fission yeast deletion strains by barcode sequencing. Genome Biol. 2010;11:R60. doi: 10.1186/gb-2010-11-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim DU, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boutros M, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 45.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 46.Blake JA, et al. Mouse Genome Database (MGD)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res. 2017;45:D723–D729. doi: 10.1093/nar/gkw1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonnhammer EL, Ostlund G. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015;43:D234–239. doi: 10.1093/nar/gku1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen WH, et al. OGEE v2: an update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res. 2017;45:D940–D944. doi: 10.1093/nar/gkw1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyle EA, et al. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.